Abstract
In this study, we propose a method for quantification of average hydrogen peroxide concentration within a living cell that is based on the use of genetically encoded H2O2 biosensor HyPer. The method utilizes flow cytometric measurements of HyPer fluorescence in H2O2-exposed cells to analyze the biosensor oxidation kinetics. Fitting the experimental curves with kinetic equations allows determining the rate constants of HyPer oxidation/reduction which are used further for the calculation of peroxide concentrations in the cells of interest both in the presence and absence of external H2O2. Applying this method to K562 cells, we have estimated the gradient as about 390-fold between the extracellular and intracellular level of exogenous H2O2 in cells exposed to the micromole doses of peroxide, as well as the average basal level of H2O2 in the cytosol of undisturbed cells (). The method can be extended to other H2O2-sensitive redox probes or to procedures in which, rather than adding external peroxide, intracellular production of peroxide is triggered, providing a tool to quantitate not only basal average H2O2 concentrations but also the concentration of peroxide build up in the vicinity of redox probes.
Keywords: Hydrogen peroxide, H2O2, Genetically encoded biosensors, HyPer, Kinetics, Rate constants, Gradient
Abbreviations: cpYFP, circularly permuted yellow fluorescent protein; DTT, dithiothreitol
1. Introduction
Modulation of intracellular concentration of H2O2 can cause a variety of cellular responses ranging from cell growth to cell death [1]. Although the spatio-temporal distribution of H2O2 local concentration in cells is constantly changing, being determined by the plenty of local events of peroxide generation and elimination, the cell carefully maintains its redox homeostasis and retains a dynamic balance between the number of H2O2 molecules produced and consumed per unit time. The product of this dynamic equilibrium is a macroscopic parameter, overall peroxide concentration averaged over time and intracellular space. Being able to determine the average intracellular peroxide concentration would be useful to monitor the functional state of the cells. However, only rough estimations of the average basal H2O2 level, as well as the range of its fluctuations in living cells, exist to the moment. In various publications, one can find different estimates of intracellular peroxide concentration: 1–700 nM [2], 1–10 nM [3], picomolar range [4], etc. Available evaluations are often based on the quantification of extracellular H2O2 followed by the subsequent calculation of intracellular H2O2 concentration using theoretical assessments of the extracellular-to-intracellular peroxide gradient establishing across the plasma membrane [2]. Recent elaboration of a wide range of genetically encoded redox biosensors (reviewed in Refs. [5,6]) allows going forward in quantifying the H2O2 level within a cell. We suggest to determine intracellular peroxide concentrations by using H2O2 biosensor HyPer.
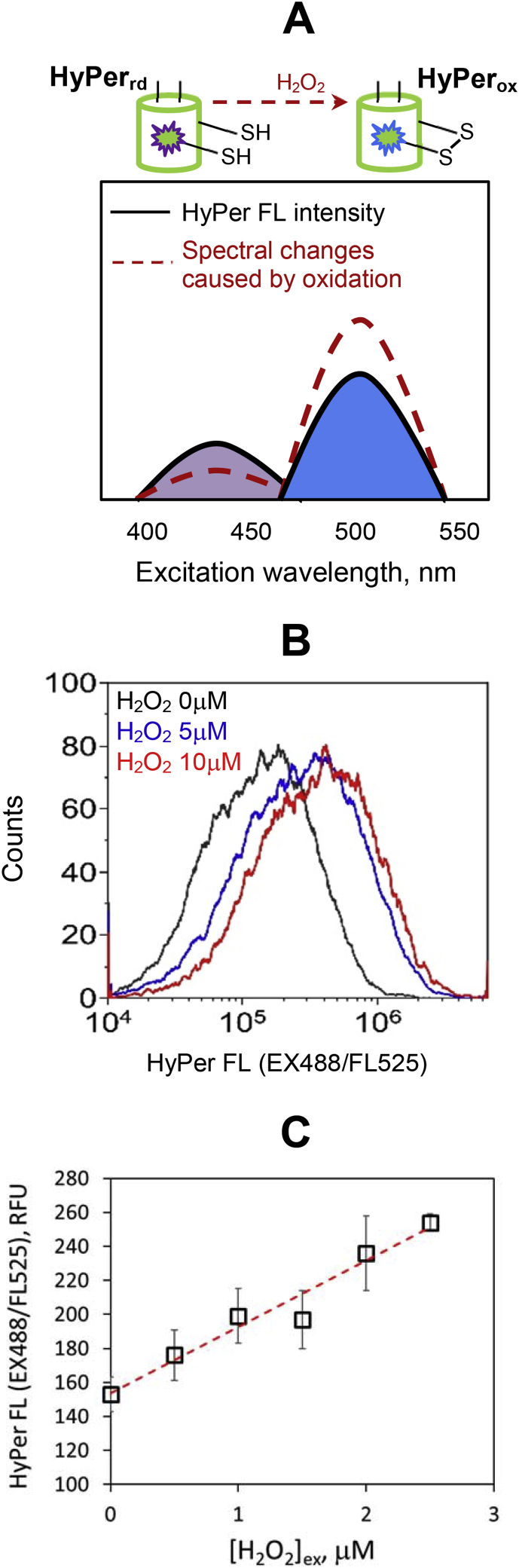
HyPer is a chimeric protein derived from the bacterial transcription factor OxyR by inserting the circularly permuted yellow fluorescent protein (cpYFP) into the regulatory domain of OxyR, which is specifically sensitive to H2O2 [7]. HyPer exhibits two excitation peaks at 420 and 500 nm in the violet and blue spectral regions, corresponding to the protonated and deprotonated form of cpYFP, respectively. Upon HyPer exposure to H2O2, the OxyR domain undergoes conformational changes that are transmitted to the cpYFP domain and result in its deprotonation. Consequently, the excitation peak at 420 nm decreases in proportion to the increase in the peak at 500 nm, reflecting the accumulation of the oxidized HyPer form [8] (see the scheme in Fig. 1 A). In this work, we use flow cytometric measurements of HyPer fluorescence signal for the analysis of HyPer oxidation kinetics in cells exposed to H2O2 and show that fitting the experimental data with kinetic equations enables quantification of intracellular hydrogen peroxide in both presence and absence of external H2O2.
Fig. 1.
Analysis of HyPer fluorescence in K562 cells exposed to extracellular H2O2. (A) Scheme demonstrating the changes in the excitation spectrum of HyPer upon oxidation. (B) Flow cytometry histograms of K562 cells measured after two-minutes exposure to different concentrations of H2O2. (C) Dependence of the mean EX488/FL525 HyPer signal intensity on the extracellular H2O2 concentration applied to cells. Abbreviations: HyPerrd reduced form of HyPer; HyPerox, oxidized form of HyPer; HyPer FL, HyPer fluorescence; EX488/FL525, HyPer green fluorescence signal measured at blue laser (488 nm) excitation; [H2O2]ex, extracellular peroxide concentration.
2. Methods
2.1. Cell cultures
For the generation of a stable cell line expressing HyPer in cell cytosol, K562 cells were transduced with lentiviral vector encoding pHyPer-cyto (Evrogen, Russia). After transduction, cells were cultivated in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin-streptomycin. Cells were subcultured twice a week at a split ratio of 1:4 for up to 15 passages after transduction. In addition to HyPer, we used SypHer [9], redox-inactive modification of cpYFP-OxyR protein, which is an optimum control molecule for HyPer [7,8], as it is characterized by identical fluorescence properties, pH sensitivity, intracellular localization, etc. K562 cells were transfected with SypHer-encoding plasmid kindly gifted by Dr. V. Belousov (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow).
2.2. Measurements of HyPer oxidation kinetics upon H2O2 exposure
Before the experiments, cells were resuspended in PBS (∼15 000 cells/ml) and incubated at standard growth conditions for 30 min to adapt to the new environment. After the bolus addition of H2O2 at concentrations ranged from 0.5 to 2.5 μM, cell samples were maintained at 37 °C and 5% of CO2. Within 10 min after the peroxide addition, at different time points, cell sample aliquots were analyzed with CytoFLEX flow cytometer (Beckman Coulter). HyPer fluorescence in gated HyPer-positive cells was examined by monitoring fluorescence signal at 525 nm registered at 488 nm excitation (hereafter denoted as EX488/FL525 signal), which reflects the accumulation of oxidized form of sensor [10] (Fig. 1 A). For the quantification of the oxidized HyPer fraction in cells (described in Results section), the EX488/FL525 fluorescence signal was measured also in the cells treated with high doses of DTT (10 mM, 10 min) and H2O2 (0.5 mM, 5 min) to achieve total HyPer reduction and oxidation, respectively. Cell treatments were performed at standard growth conditions (37 °C and 5% of CO2) in cell culture plates, or non-conical tubes to prevent cell deoxygenation. SypHer fluorescence was analyzed according to the same protocol as used for measuring HyPer signal.
To monitor the extracellular H2O2 concentration in the course of kinetic measurements, Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen) was used in accordance with the procedure described in Ref. [11]. At H2O2 concentration of 5 μM and cell density of 15 000 cells/ml, the decrease in H2O2 concentration in the extracellular medium did not exceed several percent within 10 min after the peroxide addition to cells and therefore was neglected in the analysis of HyPer oxidation.
2.3. Analysis of HyPer oxidation kinetics
Generally, under the steady-state conditions of cell exposure to both exogenous and endogenous H2O2, kinetics of HyPer oxidation can be described by the following equations [12]:
| (1) |
| (2) |
| (3) |
| (4) |
In these equations, refers to the fraction of HyPer in the reduced form observed in a cell at time t. is the steady state level of the reduced HyPer fraction established as a result of concurrent processes of HyPer oxidation and reduction. and are apparent first-order rate constants for the oxidation and reduction of HyPer (corresponding to and in Ref. [12]), respectively. is determined by , the intracellular peroxide concentration averaged over the cites of HyPer localization (in this study it is a cell cytosol), and the second-order rate constant for the reaction between HyPer and hydrogen peroxide, [13]. , generally, consists in the sum of the basal H2O2 level () and additional intracellular H2O2 concentration arising from the cell exposure to peroxide (). Finally, is the basal fraction of HyPer in the reduced form accessible for oxidation at time t = 0.
In the limit , Eq. (1) becomes:
| (5) |
Thus, at long incubation times, reduced HyPer fraction settles in a steady-state, which by taking into account Eq. (2) and Eq. (3) is expressed as a non-linear function of :
| (6) |
It is important to note that in the absence of external peroxide, Eq. (6) can be used for determining the basal H2O2 concentration. For this purpose, the following substitutions are made: and .
When the experimental set up entails the addition of extracellular peroxide, the dependence of on the extracellular peroxide concentration, , is made explicit in Eq. (7), in which is the ratio between the extracellular and the intracellular exogenous H2O2 concentrations. Combination of Eq. (3) and Eq. (7) allows expressing as a function of (Eq. (8)). If is independent on the extracellular peroxide concentration, then this function is linear, but, generally, this is not the case because may depend on as well.
| (7) |
| (8) |
2.4. Statistical analysis
Experimental data are presented as the mean values of at least three independent experiments with standard deviations.
3. Results
To test the applicability of HyPer for quantitative measurements, we exposed HyPer-expressing K562 cells suspended in PBS to steady-state micromolar doses of extracellular H2O2. Near steady-state conditions were attained by bolus addition of peroxide to highly diluted cell suspensions (∼15 000 cells/mL). At such cell densities, the drop of extracellular H2O2 concentration due to the peroxide scavenging by cells was negligible during the first 10 min after H2O2 addition (see Methods section). To start with, we analyzed HyPer fluorescence after 2-min incubation of cells with H2O2 and measured EX488/FL525 signal, which corresponds to the oxidized form of HyPer (Fig. 1 B). At H2O2 concentrations exceeding 10 μM, sensor became totally oxidized and signal intensity saturated. However, up to the 2.5 μM of H2O2, the signal increased linearly with H2O2 concentration (Fig. 1 C), demonstrating a sensitive response under the experimental conditions used.
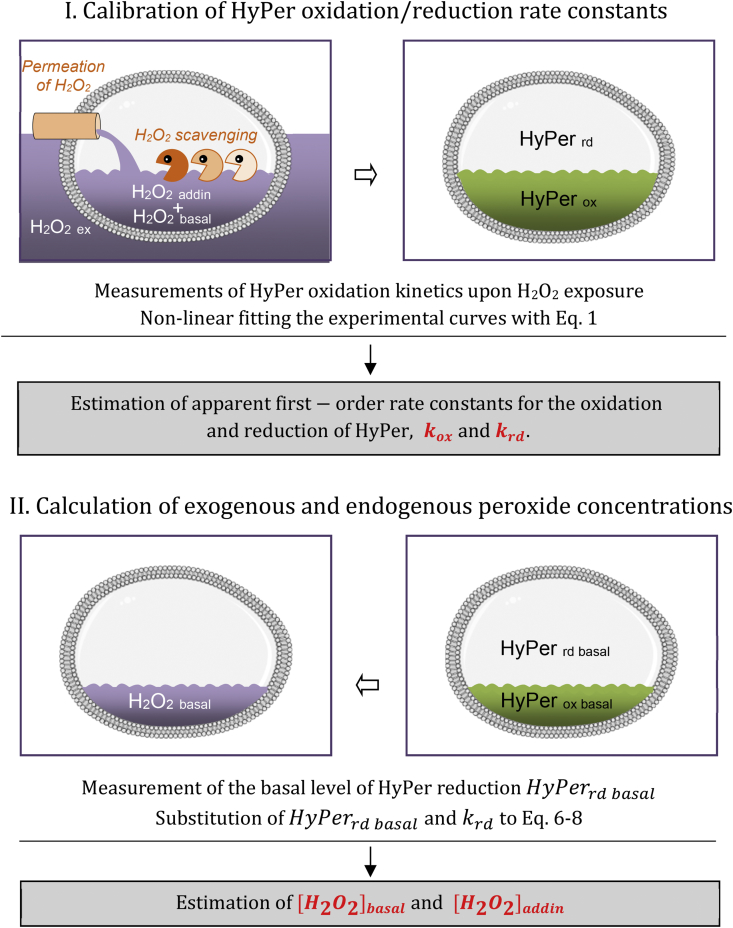
Next, to measure the intracellular levels of peroxide in both disturbed or undisturbed cells, we calibrated the HyPer fluorescence signal using exogenous peroxide. The experimental design is shown in Fig. 2. At first, the drop in the reduced HyPer fraction in cells is measured at different time points after the addition of H2O2 to cell suspensions. Oxidation kinetics is determined by the rate of sensor oxidation with intracellular peroxide and also by the capacity of cells to reduce the sensor. When the rates of HyPer reduction and oxidation balance each other, reduced HyPer fraction reaches the steady state level. Accordingly, non-linear fitting the experimental curves with kinetic equations (Eq. (1) in Methods section) enables simultaneous deriving two parameters, namely apparent first-order rate constants for the oxidation and reduction of HyPer, and , respectively. This procedure is performed for different concentrations of extracellular peroxide. After that, using the estimated rate constants and measured values of the steady state levels of HyPer reduction in exposed and non-exposed cells, both basal and additional intracellular H2O2 concentration, and , are determined (Eqs. (6)–(8)). In fact, the main idea of the proposed method is to use the extracellular peroxide for calibrating not the HyPer signal itself, but the rate constants of sensor oxidation/reduction, which are used further for estimation of intracellular H2O2 levels in both disturbed and undisturbed cells.
Fig. 2.
Scheme describing the method for quantification of exogenous and endogenous H2O2 concentrations in HyPer-expressing cells. Abbreviations: HyPerox and HyPerrd, oxidized and reduced HyPer fractions; kox and krd, apparent first-order rate constants of HyPer oxidation and reduction; [H2O2]basal basal endogenous hydrogen peroxide concentration, [H2O2]addin, additional intracellular hydrogen peroxide concentration arising from the cell exposure to external peroxide.
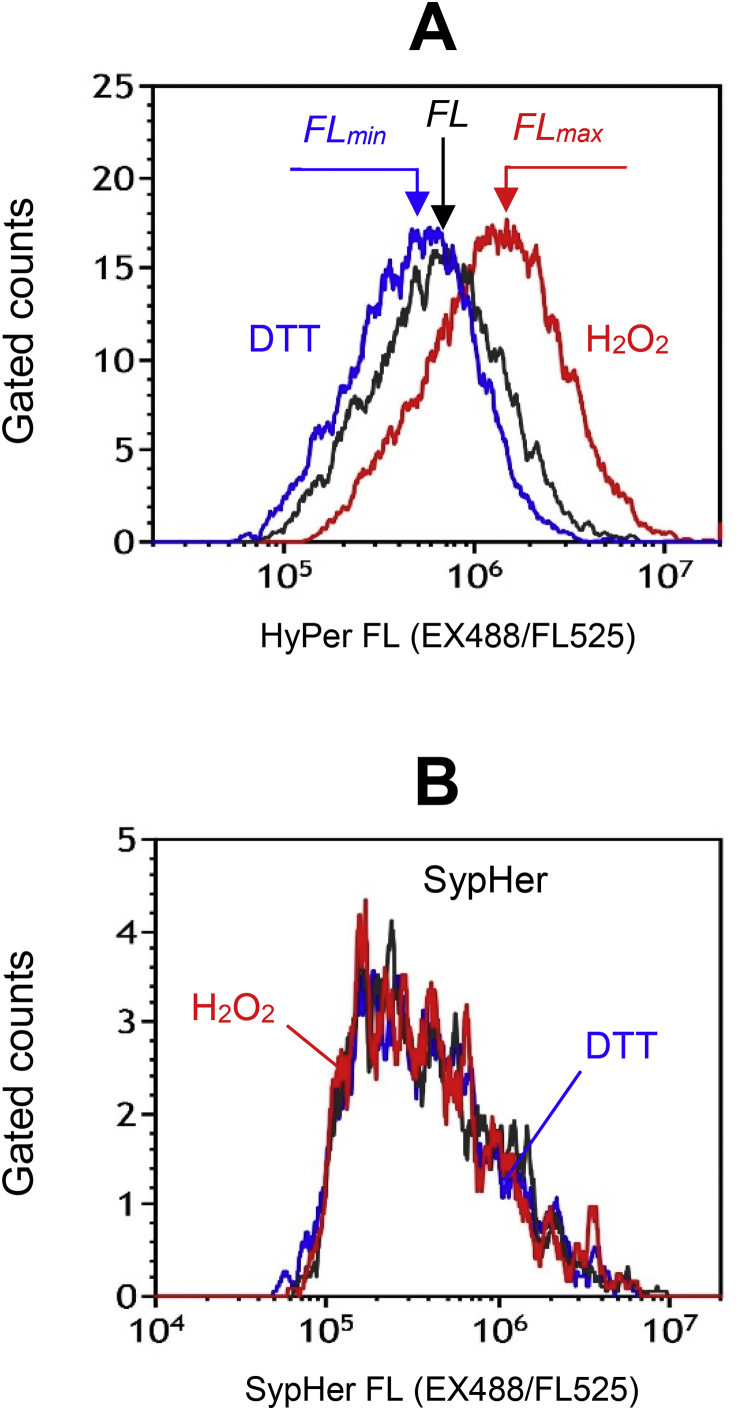
For approbation of this method, we performed experiments on K562 cells suspended in PBS. The reduced HyPer fraction in cells, , was derived from the fluorescence measurements (Fig. 3 A):
| (9) |
where FL is a mean EX488/FL525 signal measured in the cells of interest, whereas FLmin and FLmax are mean EX488/FL525 signals measured in cells incubated with high doses of DTT (to achieve total HyPer reduction) and H2O2 (to achieve total HyPer oxidation), respectively. To ensure that the changes in HyPer fluorescence after the DTT or H2O2 exposure are caused by the reduction/oxidation of HyPer redox-active cysteine residues, in parallel experiments we examined the response of SypHer-expressing cells to the same treatments. SypHer [9] is a redox-inactive modification of HyPer that differs from the parental protein by single mutation (Cys-199 was replaced by Ser-199) and thus has identical spectral characteristics [8]. Due to this fact, SypHer is a best control to HyPer, as it is characterized by similar pH sensitivity, intracellular localization, etc. Contrary to HyPer-expressing K562 cells, SypHer-expressing K562 cells did not respond to high dosage DTT/H2O2 treatments (Fig. 3 B), as well as to micromole H2O2 concentrations, giving confidence on the reliability of the quantification method used.
Fig. 3.
Quantitation of the reduced HyPer fraction in cells using total reduction/oxidation of the sensor. (A, B) Flow cytometry histograms of HyPer-expressing (A) and SypHer-expressing (B) K562 cells: untreated and exposed to high doses of DTT (10 mM, 10min) or H2O2 (0.5 mM, 5min).
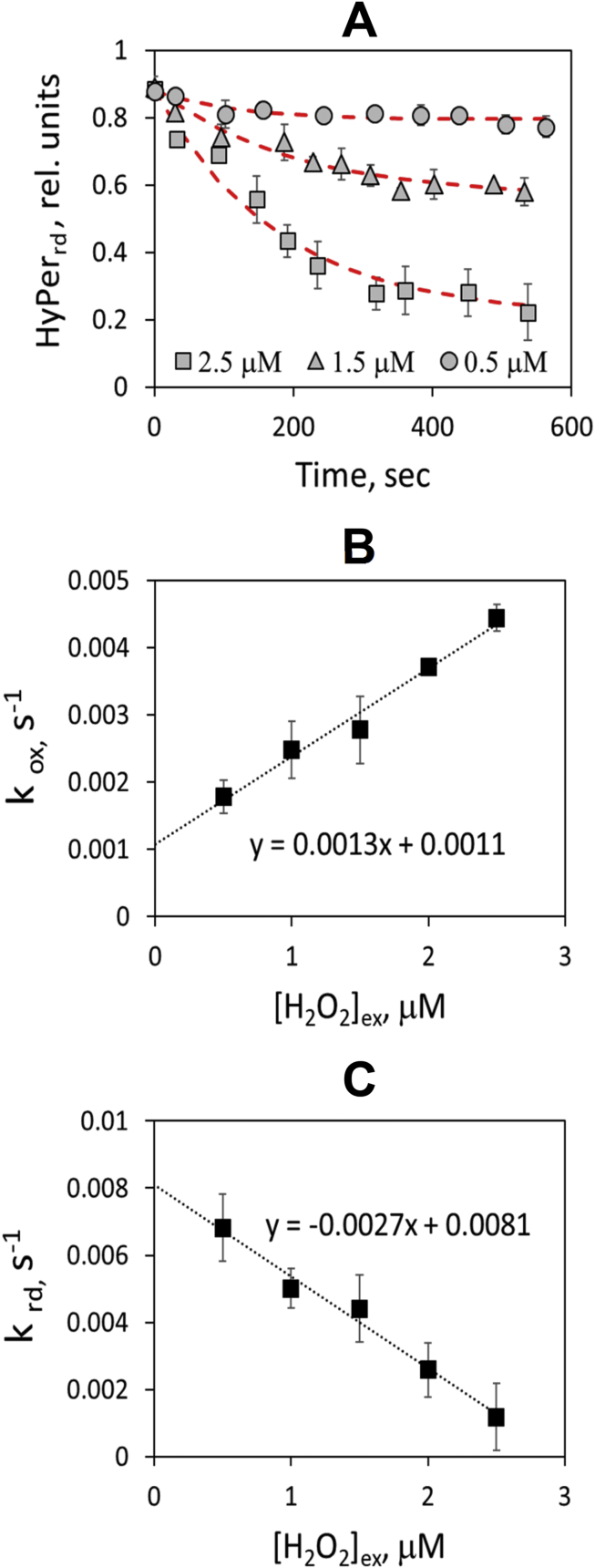
To quantify intracellular H2O2 concentrations, following the procedure shown in Fig. 2, was monitored after the addition of micromolar concentrations of peroxide to cell suspensions. Exposure of cells to H2O2 resulted in the gradual accumulation of oxidized HyPer and concomitant decrease of (Fig. 4 A), and after 10 min, the reduced HyPer fraction reached a pseudo-steady state level when the rates of HyPer reduction and oxidation were balanced. It is important to note that the kinetics of HyPer oxidation (Fig. 4 A) does not reflect the dynamics of H2O2 penetration to cells. According to recent estimations, the H2O2 concentration profile reaches a steady state in a cell within a time scale on the order of 1 ms after H2O2 addition [14]. For the description of HyPer oxidation dynamics, we applied non-linear fittings of Eq. (1) (see Methods section) to the experimental data and obtained apparent first-order rate constants for the oxidation and reduction of HyPer, and , respectively (see Table 1). Interestingly, decreased whereas increased linearly with increasing extracellular peroxide concentration (Fig. 4 B, C). In accordance with Eq. (8), as long as is proportional to , the gradient between the extracellular and intracellular concentration of exogenous peroxide () can be considered as independent on . Due to this circumstance, linear approximation of plot with Eq. (8) allowed simultaneous estimation of two parameters: the gradient and the average basal concentration of peroxide in cells . As a result, we obtained the following estimates: of and .
Fig. 4.
Kinetics of HyPer oxidation in K562 cells exposed to H2O2. (A) Reduced HyPer fraction drop with time after H2O2 addition to cell suspension. HyPerrd was estimated using Eq. (9). Data fitting with Eq. (1) is shown in red. (B, C) Dependence of the apparent first-order rate constants of HyPer oxidation (B) and reduction (C) on extracellular H2O2 concentration. Both dependencies were derived from fittings of kinetic curves (C). Abbreviations: HyPerrd, reduced HyPer fraction calculated with Eq. (9); kox and krd, apparent first-order rate constants of HyPer oxidation and reduction, respectively; [H2O2]ex, extracellular peroxide concentration.
Table 1.
Endogenous and exogenous H2O2 levels, as well as rate constants of HyPer oxidation/reduction, derived from the analysis of HyPer oxidation kinetics in H2O2-exposed K562 cells.
| Parameters | Extracellular hydrogen peroxide concentration |
||||
|---|---|---|---|---|---|
| 0.5 μM | 1 μM | 1.5 μM | 2 μM | 2.5 μM | |
| (N = 3) | (6.8 ± 1) × 10−3 | (5.0 ± 0.6) × 10−3 | (4.4 ± 1) × 10−3 | (2.6 ± 0.8) × 10−3 | (1.2 ± 1) × 10−3 |
| (N = 3) | (1.8 ± 0.2) × 10−3 | (2.5 ± 0.4) × 10−3 | (2.8 ± 0.5) × 10−3 | (3.7 ± 0.1) × 10−3 | (4.4 ± 0.2) × 10−3 |
| 380 | 360 | 440 | 380 | 370 | |
| 1.3 | 2.8 | 3.3 | 5.2 | 6.7 | |
| 390 ± 40 (N = 5) | |||||
| (8.1 ± 0.5) × 10−3 (N = 3) | |||||
| 2.2 ± 0.4 (N = 15) | |||||
Abbreviations: kox and krd, apparent first-order rate constants of HyPer oxidation and reduction; krd basal, apparent first-order rate constant of HyPer reduction in undisturbed cells; , ratio between the extracellular and intracellular exogenous peroxide concentration; [H2O2]basal, basal hydrogen peroxide concentration; [H2O2]ex, external hydrogen peroxide concentration; [H2O2]addin, additional intracellular H2O2 concentration arising from the cell exposure to external peroxide.
Since these estimates were obtained in the presence of external low micromolar H2O2 concentration, it is important to evaluate whether they are physiological relevant or whether they reflect a situation where endogenous peroxiredoxins and peroxidases are overwhelmed by the added peroxide. If this last scenario is the case, estimated gradients would be less steep and the calculated would be much higher than the true average basal level. To clarify this issue, we applied an alternative procedure that determines based on the basal level of HyPer oxidation. Eq. (6) links the pseudo-steady state level of HyPer reduction and equilibrated concentration of intracellular H2O2 for both disturbed and undisturbed cells. To calculate using Eq. (6), the value of basal HyPer reduction measured in absence of exogenous peroxide as well as the rate constant value for undisturbed cells are needed. For this purpose, the dependence of (Fig. 4 C) was extrapolated to the situation of , yielding the value denoted in Table 1 as . After the substitution of measured at t = 0 and into Eq. (6), we estimated the average concentration of endogenous H2O2 in K562 cells as . This value is similar to the value calculated above, indicating that our estimates are physiologically relevant.
In general, if the plot of on is not linear, meaning that the gradient depends on peroxide concentration, an alternative approach to the linear fit to Eq. (8) has to be applied to estimate the gradient. Within this approach, the basal peroxide concentration calculated from Eq. (6) and the second-order rate constant for the reaction between HyPer and hydrogen peroxide [13] can be imputed in Eq. (8) to calculate the gradient and the intracellular level of exogenous peroxide, , for each peroxide concentration applied (see Table 1). Using this approach, we estimated the mean gradient value, averaged over all experiments, as . What is important, the values of and , which we obtained using Eqs. (6), (8), were similar, within experimental uncertainty, to those determined from the linear plot of Fig. 4 B before, giving confidence on the rigor of the methodology applied.
4. Discussion
Presented analysis shows that HyPer fluorescence signal can be used for the quantitation of both exogenous and endogenous H2O2 in living cells. Calibration of HyPer signal can be performed by using extracellularly added H2O2, taking into account the gradient between the extracellular and intracellular levels of the oxidant that establishes due to the effective scavenging of peroxide within a cell [[15], [16], [17]]. Previously, it has been shown that to quantify the concentration gradient, either theoretical [18], or experimental [15,17] assessments may be used. For instance, the gradient can be experimentally estimated by measuring the kinetics of consumption of H2O2 in intact cells and activities of H2O2-removing enzymes in disrupted cells [15,17]. However, enzyme activities measured in disrupted cells may not reflect fluxes in vivo, and therefore, in this study, we used an alternative approach based on the acquisition of HyPer oxidation kinetics in intact living cells, followed by calculations with only one predefined parameter being needed, the second-order rate constant for the reaction between HyPer and H2O2. The equations describing the kinetics of protein oxidation in conditions of steady state exposure to peroxide were derived in Ref. [12] and then were employed in Ref. [19] for the characterization of the peroxide-sensing transcriptional regulators and estimation of H2O2 gradients in fission yeast exposed to external peroxide. We applied these equations to describe the kinetics of HyPer oxidation after the exposure of K562 cells to the bolus micromole doses of H2O2. In general, the near steady-state approximation can be applied in cases of short-term exposures of diluted cell suspensions to H2O2, because the typical first-order rate constant of H2O2 consumption by cultivated cells is about 10 −12 s−1 cell−1 L [20] and the drop of extracellular H2O2 concentration due to the peroxide scavenging by cells can be neglected.
According to our calculations, the basal peroxide level averaged over the sites of HyPer localization in the cytosol of K562 cells is about several nanomoles. Given that numerous experiments on the visualization of HyPer fluorescence [8], demonstrated a uniform spatial distribution of the biosensor in the cytosol, our estimates can be considered as the average overall H2O2 concentration in this cellular compartment. Follow-up studies will probably result in the determination of the average level of H2O2 in other cellular compartments and/or cell lines. In addition, we prove that the rise in H2O2 that occurs in the cytosol after the steady-state exposure of cells to the micromolar concentrations of extracellular peroxide is quite comparable to the average basal level of H2O2. We show that, under our experimental conditions, scavenging of H2O2 by cellular antioxidants generates a gradient between the extracellular and intracellular level of exogenous H2O2 of about 390-fold, independently on H2O2 concentrations applied, that seems reasonable in the case of the weak perturbations of the cell redox environment employed in the study.
At the same time, we found that, even in case of these weak oxidative perturbations, the rate constant of HyPer reduction , which is determined by the interaction of oxidized sensor with thiol-reducing species, such as glutaredoxins and/or thioredoxins, is clearly influenced by the extracellularly added H2O2. In fact, the character of this dependence indicates that in H2O2-exposed cells, HyPer reduction cannot be considered as a reaction of pseudo-first order, and its rate is dependent on the level of reduced HyPer-interacting enzymes, which may be depleted after H2O2 exposure. For instance, recent theoretical considerations have shown that depletion of the reduced form of glutaredoxin can affect HyPer oxidation after a bolus exposure of HeLa cells to 20 μM of H2O2 [21]. In addition, we suggest that the level of reduced thioredoxins can also be lowered in the H2O2-exposed cells due to the massive reduction of oxidized peroxiredoxins [22]. Calibration of with the use of external peroxide enables quantification of the basal H2O2 levels (Eq. (6)).
Given the simplicity of the approach elaborated in our study, inaccuracies in the H2O2 quantification can occur due to the potential saturation of HyPer signal under the conditions of heavy oxidation, possible uncertainty in the signal calibration with the use of high doses of DTT and H2O2, as well as the simplicity of the kinetic model used. Having all this in mind, we suggest to consider the obtained estimates of the H2O2 gradient and the average basal H2O2 level in the cytosol of K562 cells as an approximate values, which nonetheless significantly restrict the range of possible assessments.
In conclusion, we proposed a method for absolute quantification of H2O2 within the cell that is based on the use of genetically encoded H2O2 biosensor HyPer. By applying this method to K562 cells, we have estimated the gradient between the extracellular and intracellular level of exogenous H2O2 in cells exposed to the micromolar doses of oxidant (), as well as the average level of H2O2 in the cytosol of undisturbed cells (). The method can be extended to other H2O2-sensitive redox probes or to procedures in which, rather than adding external peroxide, intracellular production of peroxide is triggered, providing a tool to quantitate not only basal H2O2 concentration, but also the concentration of peroxide build up in the vicinity of redox probes.
Disclosures
The authors have no conflict of interest.
Acknowledgements
The authors thank Dr. Vsevolod Belousov for cooperation and Julia Ivanova for her help in plasmid cloning. FA acknowledges support from Fundação para a Ciência e a Tecnologia (Portugal) project UID/MULTI/00612/2019. OL acknowledges support from Russian Foundation for Basic Research (Grant # 19-04-00994).
Contributor Information
Olga Lyublinskaya, Email: o.lyublinskaya@mail.ru.
Fernando Antunes, Email: fantunes@fc.ul.pt.
References
- 1.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 2.Stone J.R., Yang S. Hydrogen peroxide: a signaling messenger. Antioxidants Redox Signal. 2006;8:243–270. doi: 10.1016/j.redox.2014.08.00116677071. [DOI] [PubMed] [Google Scholar]
- 3.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ezeriņa D., Morgan B., Dick T.P. Imaging dynamic redox processes with genetically encoded probes. J. Mol. Cell. Cardiol. 2014;73:43–49. doi: 10.1016/j.yjmcc.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 5.Kostyuk A.I., Panova A.S., Bilan D.S., Belousov V.V. Redox biosensors in a context of multiparameter imaging. Free Radic. Biol. Med. 2018;128:23–39. doi: 10.1016/j.freeradbiomed.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Schwarzlander M., Dick T.P., Meyer A.J., Morgan B. Dissecting redox biology using fluorescent protein sensors. Antioxidants Redox Signal. 2016;24:680–712. doi: 10.1089/ars.2015.6266. [DOI] [PubMed] [Google Scholar]
- 7.Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V., Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3(4):281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 8.Bilan D.S., Belousov V.V. HyPer family probes: state of the art. Antioxidants Redox Signal. 2016;24(13):731–751. doi: 10.1089/ars.2015.6586. [DOI] [PubMed] [Google Scholar]
- 9.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyublinskaya O.G., Antonov S.A., Gorokhovtsev S.G., Pugovkina N.A., Kornienko J.S., Ivanova J.S., Shatrova A.N., Aksenov N.D., Zenin V.V., Nikolsky N.N. Flow cytometric HyPer-based assay for hydrogen peroxide. Free Radic. Biol. Med. 2018;128:40–49. doi: 10.1016/j.freeradbiomed.2018.05.091. [DOI] [PubMed] [Google Scholar]
- 11.Lyublinskaya O.G., Ivanova J.S., Pugovkina N.A., Kozhukharova I.V., Kovaleva Z.V., Shatrova A.N., Aksenov N.D., Zenin V.V., Kaulin Y.A., Gamaley I.A., Nikolsky N.N. Redox environment in stem and differentiated cells: a quantitative approach. Redox Biol. 2017;12:758–769. doi: 10.1016/j.redox.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brito P.M., Antunes F. Estimation of kinetic parameters related to biochemical interactions between hydrogen peroxide and signal transduction proteins. Front. Chem. 2014;2:82. doi: 10.3389/fchem.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilan D.S., Pase L., Joosen L., Gorokhovatsky A.Y., Ermakova Y.G., Gadella T.W., Grabher C., Schultz C., Lukyanov S., Belousov V.V. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 2013;8(3):535–542. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 14.Lim J.B., Langford T.F., Huang B.K., Deen W.M., Sikes H.D. A reaction-diffusion model of cytosolic hydrogen peroxide. Free Radic. Biol. Med. 2016;90:85–90. doi: 10.1016/j.freeradbiomed.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Antunes F., Cadenas E. Estimation of H2O2 gradients across biomembranes. FEBS Lett. 2000;475:121–126. doi: 10.1016/j.redox.2014.08.00110858501. [DOI] [PubMed] [Google Scholar]
- 16.Marinho H.S., Cyrne L., Cadenas E., Antunes F. The cellular steady-state of H2O2: latency concepts and gradients. Methods Enzymol. 2013;527:3–19. doi: 10.1016/B978-0-12-405882-8.00001-5. [DOI] [PubMed] [Google Scholar]
- 17.Huang B.K., Sikes H.D. Quantifying intracellular hydrogen peroxide perturbations in terms of concentration. Redox Biol. 2014;2:955–962. doi: 10.1016/j.redox.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adimora N.J., Jones D.P., Kemp M.L. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxidants Redox Signal. 2010;13(6):731–743. doi: 10.1089/ars.2009.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domènech A., Ayté J., Antunes F., Hidalgo E. Using in vivo oxidation status of one- and two-component redox relays to determine H2O2 levels linked to signaling and toxicity. BMC Biol. 2018;16(1):61. doi: 10.1186/s12915-018-0523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner B.A., Witmer J.R., van’t Erve T.J., Buettner G.R. An assay for the rate of removal of extracellular hydrogen peroxide by cells. Redox Biol. 2013;1(1):210–217. doi: 10.1016/j.redox.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang B.K., Ali S., Stein K.T., Sikes H.D. Interpreting heterogeneity in response of cells expressing a fluorescent hydrogen peroxide biosensor. Biophys. J. 2015;109(10):2148–2158. doi: 10.1016/j.bpj.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Netto L.E., Antunes F. The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol. Cell. 2016;39(1):65–71. doi: 10.14348/molcells.2016.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]