Abstract
Aim:
Presenting the first clinical case of Wickerhamomyces myanmarensis.
Patients & methods:
Yeast cells were isolated from blood and central venous catheter of a 5.5-year-old male subject. API 20C AUX, MALDI-TOF MS, ITS and LSU rDNA sequencing, and our qPCR assay were used for identification and the MIC values were determined by CLSI M27-A3.
Results:
ITS and LSU rDNA sequencing identified both isolates as W. myanmarensis, while API 20C AUX and MALDI-TOF MS did not identify them correctly. Our qPCR specifically distinguished W. myanmarensis from W. anomalus. Isolate obtained from blood showed a higher MIC value for fluconazole, voriconazole and posaconazole.
Conclusion:
Utilization of reliable identification tools might reveal the genuine spectrum of opportunistic yeast species.
Keywords: : antifungal susceptibility testing, blood, central venous catheter, specific multiplex qPCR, Wickerhamomyces myanmarensis
Number of patients who are at risk for candidemia continues to escalate [1]. Moreover, misuse of antifungals including fluconazole, and the emergence of rare yeast species posed a challenge for treatment of bloodstream infections [2]. For instance, nowadays Candida auris due to it being a multidrug resistant, a persistent colonizer in ICU wards, and causing a high mortality rate of 30–60% created a challenge for public health agencies [3].
Wickerhamomyces (Pichia) myanmarensis for the first time was isolated from sugar palm in 2005 [4]. Due to its high level of similarity at LSU rDNA with W. anomalus, it was thought to be either W. anomalus or its sister species [4]. However, its ability to grow at 37 and 40°C, assimilation of D-arabinose, detection by specific DNA hybridization probes, the production of hat-shaped ascospores, and presence of Q7-ubiquinone convinced investigators to consider it as a new species, Pichia myanmarensis [4]. Recent study considered P. myanmarensis as W. myanmarensis [5]. Herein, we reported W. myanmarensis as a novel opportunist yeast species that for the first time has been isolated from blood and central venous catheter samples.
Case
A 5.5-year-old boy who presented with recurrent GI tract bleeding and abnormal liver function was admitted to the pediatric surgery ward at Namazi Hospital, Shiraz, Iran on 26 October 2017. The patient lived in a rural area in the Northern part of Iran in a middle-class family (Babol, Mazandaran, Iran). The clinical history of the patient showed that he experienced esophageal and gastric fundal varices and splenectomy. Endoscopic documentation suggested diffuse erosion and convulsion in the stomach. His international normalized ratio test (INR) of 3.9 and prothrombin time (PT) of 45 along with sudden hemoglobin drop suggested portal hypertension and gastrointestinal hemorrhage. In order to compensate blood loss, the patient was supplied with several blood transfusions. Sonography examination revealed small-sized liver with mild coarse parenchymal echogenicity. One day after admission (27 October 2017), in order to control gastroesophageal reflux disorder, the patient was treated orally with omeprazole twice a day. On 1 November 2017, patient underwent laparotomy and distal splenorenal shunt and he was intravenously (RT jugular triple lumen) prescribed with prophylactic treatment of 200 mg of vancomycin once a day, 400 mg of meropenem, and 40 mg of cefazolin three-times a day. From 1 to 17 November 2017, the patient manifested several fever episodes, hence, frequent blood, urine and abdominal fluid samples were taken, which all yielded negative results. However, one blood sample taken on 17 November 2017 and one double-lumen catheter sample on 18 November yielded positive growth after 48 h of incubation in BD Bactec Bacton device (MD, USA). Streaking 100 μl of positive blood bottles on blood agar, EMB, Sabouraud dextrose agar and CHROMagar (24–48 h, 37°C) yielded yeast colonies. Presence of yeast cells in the blood sample (positive blood bottle) was confirmed by direct smear testing and germ tube testing was negative. On 2 December 2017, patient was discharged with omeprazole treatment, while he did not receive any antifungal treatments. According to the latest follow-up of the patient (18 November 2018) on 24 January 2018 due to gastrointestinal bleeding, he was referred to the same hospital and on 16 February 2018 a distal splenectomy shunt was inserted to control his hematemesis (vomiting blood). His family members mentioned that, since then, he was not re-referred to the hospital and his general health condition was satisfactory.
Microbiology
Transferring single colonies obtained from CVC and blood samples on CHROMagar (24–48 h at 37°C) yielded small pink colonies. Presence of yeast cells was confirmed by direct smear testing (Figure 1) of the positive blood bottle and the germ tube test was negative. As the first line of identification, colonies were subjected to API 20C AUX (Biomeriux, France) and the API strips were read after incubation of 72 h at 30°C. API 20C AUX identified both isolates as Wickerhamomyces anomalus.
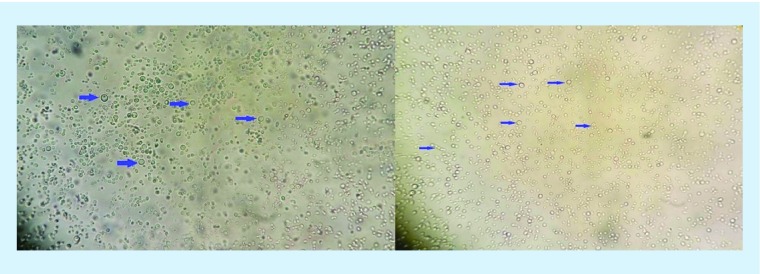
Figure 1. . Direct microscopy revealed presence of yeast cells in positive blood bottle sample.
Pictures were taken with 40× lens. Arrows clearly indicate yeast cells.
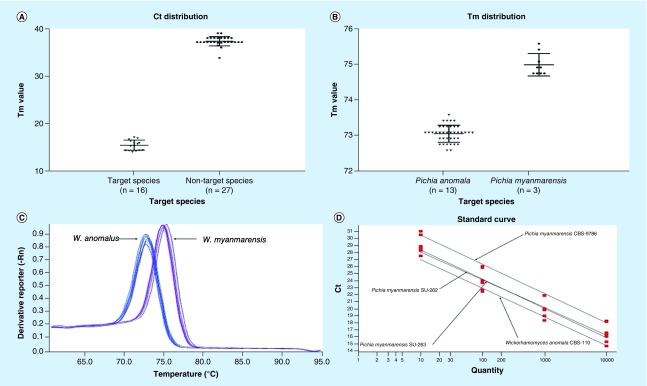
Using full extraction method [6], the proteins of clinical strains were extracted, and 1 μl of supernatants were evaluated by MALDI-TOF MS using a Bruker device (MicroFlex LT, Bruker Daltonics, Bremen, Germany). Bruker MALDI-TOF MS device failed to identify these two isolates and categorized them as not reliable identification with red scores (<1.6) matching with Kytococcus sedentarius. Repeated full extraction method and identification by MALDI-TOF MS showed the same results. Consequently, DNA sequencing of large subunit (LSU) using LROR and LR5 primers and internal transcribed sequence (ITS) domains of ribosomal DNA (rDNA) using ITS1 and ITS4 primers were performed as gold standard technique [7]. Obtained sequences were searched by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and LSU showed 100% similarity with W. myanmarensis, 99% with W. anomalus and 98% with W. edaphicus, while ITS showed 100% similarity with W. myanmarensis and 98% with W. anomalus. For further confirmation, using concatenated sequences of LSU and ITS, a phylogenetic tree was constructed (Figure 2). Subsequently, relevant sequences of the other closely related yeast species were retrieved from NCBI and aligned using MEGA v7.0. Phylogenetic trees using neighbor-joining method and 1000 bootstraps with MEGA clustered our clinical isolates of W. myanmarensis with the environmental isolate of W. myanmarensis (CBS 9786 and BCRC 23287) and both isolates (one from CVC and the other one from blood) were placed in the same clade. Obtained sequences of LSU and ITS rDNA for both strains were deposited in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) and they were designated with the following accession numbers, MH236218, MH236219, MH236220 and MH236221. Due to a high degree of similarity on the rDNA gene with W. anomalus and the fact that there is no specific and rapid molecular test for identification of W. myanmarensis, a SYBR-Green I-based qPCR was developed (Table 1). Our multiplex PCR based on melting temperature of PCR products unequivocally distinguished W. myanmarensis (74.91 ± 0.31°C) from W. anomalus (73.04 ± 0.23°C) (Figure 3B & C). When subjected to our multiplex qPCR, W. edaphicus yielded high Ct values (Ct = 39) for (Figure 3A), which further confirmed that our isolates were W. myanmarensis. Specificity testing with 27 various opportunistic yeast species (Table 2) resulted in 100% specificity (Figure 3A). Yeast species that were the most common cause of yeast infections or those that were both clinically important and had a close genetic background to the target species were included in the specificity testing. Obtaining average R2 value of 0.99 indicated a high degree of reproducibility of our qPCR assay (Figure 3D). CBS-type strains of W. myanmarensis (CBS 9786; n = 1), W. edaphicus (CBS 10408; n = 1) and W. anomalus (n = 13 CBS reference strains mentioned in Table 2) were subjected to API 20C AUX, MALDI-TOF MS, ITS and LSU rDNA sequencing, and our real-time PCR assay.
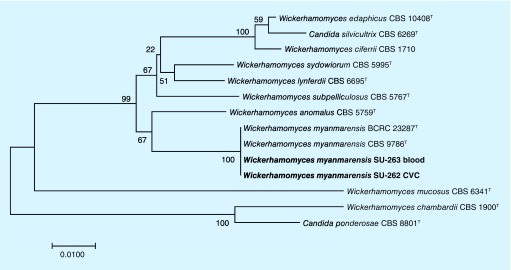
Figure 2. . Phylogenetic tree based on concatenated sequences of ITS and LSU D1/D2 domains of rDNA.
The tree was constructed using neighbor-joining method and 1000 bootstraps. Bar shows one nucleotide substitution in 100 nucleotides. Phylogenetic tree obviously placed our clinical isolates within the cluster of environmental strains of Pichia myanmarensis (CBS 9786 and BCRC 23287).
Table 1. . List of primers utilized in this study.
| Primer name | Primer sequence | Target loci | Target species | Melting temperature | PCR product size |
|---|---|---|---|---|---|
| PF-Universal | GAAATAATGTATTAGGTTCTTCCAAC | ITS | W. anomalus/W. myanmarensis | NA | NA |
| PR-Anomala | GCCGAGCCTAAAATACTTCT | ITS | W. anomalus | 73.04 ± 0.23 | 71 bps |
| PR-myanmar | ACTTTGTGTATATGTTATTGGGC | ITS | W. myanmarensis | 74.91 ± 0.31 | 93 bps |
Figure 3. . Properties of designed qPCR are depicted.
(A) Ct values for target species and nontarget species, (B) Tm distribution for W. myanmarensis and W. anomalus, (C) Individual melting curve for W. anomalus and W. myanmarensis, and (D) obtained standard curves for target species.
Table 2. . Reference strains of various opportunistic yeast species were used for specificity testing.
| Species | Origin |
|---|---|
| Candida albicans | CBS 2704 |
| Candida tropicalis | CBS 2313 |
| Candida parapsilosis | CBS 11045 |
| Candida glabrata | CBS 138 |
| Pichia kudriavzevii | CBS 5147 |
| Pichia norvegensis | CBS 6564 |
| Clavispora lusitaniae | CBS 6936 |
| Debaromyces hansenii | CBS 767 |
| Candida dubliniensis | CLF 10 |
| Pichia guilliermondii | CBS 7099 |
| Kluyvermyces marxianus | STA 63 |
| Candida rugosa | CBS 613 |
| Yarrowia lipolytica | CBS 6124 |
| Pichia fermentans | CBS 187 |
| Pichia kluyveri | CBS 188 |
| Pichia membranifaciens | CBS 107 |
| Kluyveromyces marxianus | CBS 712 |
| Lodderomyces elongisporus | CBS 2605 |
| Magnusiomyces capitatus | CBS 162.8 |
| Meyerozyma caribbica | CBS 9966 |
| Cryptococcus neoformans | CBS 8710 |
| Cryptococcus gattii | CBS 7229 |
| Rhodotorula mucilaginosa | CBS 316 |
| Saccharomyces cerevisiae | CBS 1171 |
| Trichosporon asahii | CBS 2479 |
| Trichosporon inkin | CBS 5585 |
| Wickerhamomyces edaphicus | CBS 10408 |
| Wickerhamomyces myanmarensis | CBS 9786 |
| Wickerhamomyces anomalus | CBS 5759; CBS 6417; CBS 110; CBS 2870; CBS 7338; CBS 2871; CBS 260; CBS 5702; CBS 262; CBS 263; CBS 257; CBS 605; CBS 6407 |
Regarding antifungal susceptibility testing, five antifungal drugs, including posaconazole (Sigma–Aldrich, Switzerland), itraconazole (Janssen Research Foundation, Beerse, Belgium), voriconazole (Pfizer, Central Research, UK), fluconazole (Pfizer, CT, USA), and amphotericin B (AMB, Sigma–Aldrich, MO, USA) were utilized. Broth microdilution method as instructed by CLSI M27-A3 was performed [8]. The strain isolated from CVC showed a high MIC value for AMB (MIC = 2), while it was susceptible to ITC (MIC = 0.06), FLU (MIC = 0.5), PSC (MIC = 0.125) and VRC (MIC = 0.06). Surprisingly, strain isolated from blood showed only low MIC value for ITC (MIC = 0.06), while high MIC values were exhibited for FLU (MIC = 16), PSC (MIC = 0.5), VRC (MIC = 1) and AMB (MIC = 2). Unfortunately, due to financial constraints and unavailability, the main echinocandin agents, namely micafungin and anidulafungin, were not included for antifungal susceptibility testing. In addition, interlaboratory variability shown by caspofungin convinced us not to use this agent in our antifungal susceptibility testing [9].
Discussion
Advances in the medicine and therapeutic options such as surgeries and utilization of a diverse range of immunosuppressant drugs aided in increase in frequency and spectrum of isolation of yeast species in clinical settings [10]. In this study, for the first time we have recovered two isolates of W. myanmarensis from blood and CVC samples. W. anomalus (also known as Candida pelliculosa), is one of the most prominent species in this genus that can constitute up to 4.4% of total clinical isolates of non-Candida albicans Candida species [11] and even can cause fungemia outbreaks [12].
Our patient was a 5.5-year-old boy with several underlying conditions, including surgery, CVC placement, parenteral nutrition, broad-spectrum antibiotic therapy, thymic hyperplasia, mucosal erosion of GI tract and liver abnormalities. In agreement with previous studies, surgery, broad-spectrum antibiotic therapy and utilization of CVC that are associated with the development of candidemia [1]. Besides, our patient underwent splenectomy operation and spleen as an important lymphoid tissue homes for macrophages and is involved in clearance of infection [13].
As both CVC and blood samples yielded the same species, it could be transmitted through the hands of healthcare workers, but no samples were taken from the hands of healthcare workers and environmental sampling was not performed to prove this speculation. Moreover, studies have shown that W. anomalus as a close relative of W. myanmarensis is a member of human gut microbiota [14] and revealed that inflamed epithelial tissues are positively correlated with the proportion of W. anomalus in the gut of patients suffering from ulcerative colitis [14]. As a result, probably W. myanmarensis could be either among the human microbiota or transient inhabitants delivered by food components and inflamed mucosa has encouraged its colonization and outgrowth followed by invasion. Hence, we investigated the daily diet of the patient and his parents acknowledged that he did not consume dates (as the main biological niches of this species primarily was found to be sugar palm). However, he regularly ate honey, from which the causative agent has never been isolated. Additionally, patient was treated with omeprazole (proton pump inhibitor) for a long period of time, which decreases the acidity of the stomach providing a more favorable environment for growth and colonization of microorganisms in GI tract [15,16]. Consequently, utilization of omeprazole, in combination with erosions of mucosal barriers of GI tract and the possibility of taking foods contained W. myanmarensis [4,17], may have allowed the fungus to find its way from the GI tract into the bloodstream. As there is scarcity in global isolation of this yeast and there is no evidence supporting the fact that it was isolated from environmental samples in Iran, we speculate that considering these isolates as environmental contaminant is less unlikely. Despite of antibiotic therapy and lack of isolation of any other etiological agents, patient presented with persistent fever, which is one of the possible signs of infection [18].
W. myanmarensis did not form germ tube and, like W. anomalus, yielded colonies with pink coloration on CHROMagar. Application of API 20C AUX identified both isolates as W. anomalus. Repeated experimentation of MALDI-TOF MS even using full extraction method resulted in failed identification, which is due to the lack of reference spectrum of this species in the MALDI-TOF MS library. However, DNA sequencing of ITS and LSU D1/D2 rDNA and the subsequent constructed phylogenetic tree, definitively clustered our clinical strains with the environmentally obtained isolates of W. myanmarensis and differentiated them from W. anomalus and W. edaphicus. In addition, as MALDI-TOF MS failed in identification of this species, biochemical and phenotypic assays wrongly identified both isolates as W. anomalus and due to close genetic background between W. myanmarensis and W. anomalus, we developed a rapid multiplex qPCR that reliably and specifically can distinguish these two species in less than 4 h (including DNA extraction step). Although, the application of our multiplex PCR can be restricted to rare cases, it could be a rapid and reliable alternative to time-consuming and inaccurate phenotypic and biochemical assays, where they have identified an isolate as W. anomalus. There is only one study that using DNA hybridization probe could distinguish W. myanmarensis from W. anomalus [4].
Antifungal susceptibility testing of isolate of W. myanmarensis obtained from blood showed high MIC values for fluconazole, voriconazole, posaconazole and AMB, while it was susceptible to itraconazole. However, isolate obtained from CVC showed high MIC value only for AMB. The observed difference in antifungal susceptibility patterns of the two isolates might be due to the fact that they belonged to two distinctive clones. Although, the patient was not treated with antifungal, the follow-up of blood samples remained negative and this infection was manifested as a self-limiting and transient one. We suppose that this phenomenon could be linked to a combination of multiple factors, including active immune system, low virulence attributes of W. myanmarensis as a not fully established and adapted opportunistic yeast species, and low sensitivity of culture to capture low quantity of fungal cells. There are studies that showed that blood samples of 50% of patients suffering from candidemia contain 1 CFU/ml and the rest of the 50% harbor less than or equal to 1 CFU/ml [19]. As a result, we estimated that the active immune system has thwarted the multiplication of the low virulent W. myanmarensis strains and the low sensitivity of culture showed false-negative results. Secondary to insufficient sensitivity of the culture as the gold standard method, unlucky timing of blood withdrawal is another factor playing role in transient candidemia cases [20].
Unfortunately, we could not trace the source of infection whether if the etiologic agent was acquired from the hands of healthcare workers or it was acquired through ingestion of specific fermented foods. However, as shown in other studies, conducting environmental screening does not always guarantee solving the source of infection [21] and in some cases finding source of infection requires huge efforts and is quite time consuming [22]. Moreover, due to the lack of authority for offering intervention and the research-based nature of the study, the patient was left untreated with antifungal drugs. As a result, we were deprived from observing the efficacy of appropriate antifungal for clearance of infection.
Conclusion
As the number of yeast species causing infection in human is on the rise and the majority of the rare and emerging yeast species are less susceptible to routinely prescribed antifungal drugs, this is highly important to create species-specific breakpoints to acquire a clear idea about their susceptibility patterns. Unfortunately, clinical breakpoints and epidemiological cut-off values are just restricted to a handful number of Candida species. Moreover, MALDI-TOF MS should accommodate the spectra of emerging and rare yeast species to identify them correctly. Advances in nucleic acid-based technologies such as real-time PCR and utilization of appropriate pan fungal, pan yeasts and pan filamentous fungi will aid in timely prescription of appropriate antifungal drugs, and hence, reducing mortality rate associated with fungal infections. Utilization of metagenomics technologies could revolutionize our understanding about the human microbiome, which is followed by ease of tacking of the sources of newly emerged infectious yeast species.
Summary points.
Isolation of first case of candidemia caused by Wickerhamomyces myanmarensis from both central venous catheter and blood of a 5.5-year-old boy.
Our patient was an immunocompetent with severe background conditions.
Misidentification of this yeast by using API 20C AUX, as a widely used biochemical assay in routine laboratories.
Lack of identification using MALDI-TOF MS, which is due to the lack of reference spectra of this yeast in the reference library.
Concatenated sequences of ITS and LSU rDNA followed by construction of phylogenetic tree placed our isolate in the same cluster as the environmental strains of Wickerhamomyces myanmarensis.
Development of a specific melt curve-based multiplex PCR assay that could unequivocally distinguish Wickerhamomyces myanmarensis from its close relative Wickerhamomyces anomalus.
Antifungal susceptibility testing showed higher MIC values for blood isolate.
As this yeast for the first time was isolated from sugar palm and the patient was suffering from ulceration in the gut, this infection could be transferred through ingesting some dietary products contaminated with this yeast.
Wickerhamomyces anomalus, the close relative of Wickerhamomyces myanmarensis, was found in the microbiome of a human, hence, this yeast might live within the human gut and have somehow found its way to the bloodstream.
As this infection was transiently observed, we speculate that there was lack of adaptability to the host environment and low virulence attribute of this yeast, along with immunocompetent status of the host has eradicated this yeast, hence, this infection was manifested as a self-limiting one.
Footnotes
Financial & competing interests disclosure
This work was supported by the Major National R&D Projects of the National Health Department (2018ZX10101003), European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 642095, National Natural Science Foundation of China (31770161 and 81720108026), Shanghai Science and Technology Committee (grant numbers 17DZ2270900, 19YF1448000 and 2017ZZ01024). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Cristabol L, Luis OZ, Mindy S. What is new in the clinical and diagnostic management of invasive candidiasis in critically ill patients. Intensive Care Med. 2014;40(6):808–819. doi: 10.1007/s00134-014-3281-0. [DOI] [PubMed] [Google Scholar]
- 2.Frederic L, Shawn L, Elizabeth LB, Thierry C. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018;73:4–13. doi: 10.1093/jac/dkx444. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Epidemiolgical shift and emergenece of non-Candida albicans candida species.
- 3.Bidaud AL, Anuradha C, Eric D. Candida auris: an emerging drug resistant yeast- a mini-review. J. Mycol. Med. 2018;28(3):568–573. doi: 10.1016/j.mycmed.2018.06.007. [DOI] [PubMed] [Google Scholar]; •• Candida auris as the most serious emerging fungal species.
- 4.Nagatsuka Y, Kawasaki H, Seki T. Pichia myanmarensis sp. nov., a novel cation-tolerant yeast isolated from palm sugar in Myanmar. Int. J. Syst. Evol. Microbiol. 2005;55(3):1379–1382. doi: 10.1099/ijs.0.63558-0. [DOI] [PubMed] [Google Scholar]; •• First isolation of Wickerhamomyces myanmarensis from palm sugar.
- 5.James SA, Barriga EJ, Barahona PP, et al. Wickerhamomyces arborarius f.a., sp. nov., an ascomycetous yeast species found in arboreal habitats on three different continents. Int. J. Syst. Evol. Microbiol. 2014;64(3):1057–1061. doi: 10.1099/ijs.0.059162-0. [DOI] [PubMed] [Google Scholar]; • The most updated taxonomic name for Pichia myanmarensis (Wickerhamomyces myanmarensis).
- 6.Marklein G, Michaele J, Klanke U, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 2009;47(9):2912–2917. doi: 10.1128/JCM.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamin S, Lévesque CA, Seifert KA, et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia - Mol. Phylogeny Evol. Fungi. 2015;35(1):242–263. doi: 10.3767/003158515X689135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. CLSI; Wayne, PA, USA: 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; Approved Standard – 3rd Edition: CLSI Document M27-S3. [Google Scholar]
- 9.Espinel-Ingroff A, Maiken Calvin A, Michael P, et al. Interlaboratory variability of caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob. Agents Chemother. 2013;57(12):5836–5842. doi: 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassetti M, Merelli M, Righi E, et al. Epidemiology, species distribution, antifungal susceptibility, and outcome of Candidemia across five sites in Italy and Spain. J. Clin. Microbiol. 2013;51(12):4167–4172. doi: 10.1128/JCM.01998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francesca B, Anna MT, Falconi DF, et al. Genotyping variation and antifungal susceptibility of Candida pelliculosa clinical isolates. J. Med. Microbiol. 2005;54(3):279–285. doi: 10.1099/jmm.0.45850-0. [DOI] [PubMed] [Google Scholar]; •• Clinical importance of W. anomalus (Candida pelliculosa) and its frequency of isolation.
- 12.Hsiao-Chuan L, Hsiang-Yu L, Bai-Hung S, et al. Reporting and outbreak of Candida pelliculosa fungemia in a neonatal intensive care unit. J. Microbiol. Immunol. Infect. 2013;46(6):456–462. doi: 10.1016/j.jmii.2012.07.013. [DOI] [PubMed] [Google Scholar]; •• Wickerhamomyces anomalus as a represntative of Wickerhamomyces genus was associated with outbreaks.
- 13.Prabhu DS. Overwhelming post splenectomy infection syndrome – review study. Int. J. Surg. 2014;12(12):1314–1316. doi: 10.1016/j.ijsu.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Xinyun Q, Jingjing M, Chunhua J, et al. Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis. Oncotarget. 2017;8(64):107577–107588. doi: 10.18632/oncotarget.22534. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Presence of W. anomalus in the GI tract and its positive association with ulcerative colitis.
- 15.Tauseef A, David NR, William T. Long-term safety concerns with proton pump inhibitors. Am. J. Med. 2009;122(10):896–903. doi: 10.1016/j.amjmed.2009.04.014. [DOI] [PubMed] [Google Scholar]; •• Long-term utilization of proton pump inhibitors and concerns with development of gastrointestinal infections.
- 16.Roberto BC, Pia C, Paola R, et al. Therapy with gastric acidity inhibitors increases the risk of acute gastroenteritis and community-acquired pneumonia in children. J. Pediatr. Gastroenterol. Nutr. 2006;43(4):545. doi: 10.1097/01.mpg.0000239994.20270.a8. [DOI] [PubMed] [Google Scholar]
- 17.Jonathan B, Peera H, Romney H, Theodoros K. First report of ventriculoperitoneal shunt infection due to Cyberlindnera fabianii . Case Rep. Infect. Dis. 2015;2015:630816. doi: 10.1155/2015/630816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Candida Infection of the Bloodstream – Candidemia. Am. Thorac. Soc. 2013;185:4. [Google Scholar]
- 19.Christopher DP, Gregory PS, Wiley AS, Barth R, Jhon RP, Barbara DA. Quantitation of Candida CFU in initial positive blood cultures. J. Clin. Microbiol. 2011;49(8):2879–2883. doi: 10.1128/JCM.00609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford H. New molecular method for detection of candidemia, but don't forget the blood cultures. Clin. Infect. Dis. 2018;66(11):1687–1688. doi: 10.1093/cid/cix1101. [DOI] [PubMed] [Google Scholar]
- 21.Noura AS, Suhail A, Seema K, Leena J, Mohammad A, Ziauddin K. Cyberlindnera fabianii fungemia outbreak in preterm neonates in Kuwait and letrature review. Mycoses. 2019;62(1):51–61. doi: 10.1111/myc.12846. [DOI] [PubMed] [Google Scholar]
- 22.David WE, Anna ES, Hilary M, et al. A Candida auris outbreak and its control in an intensive care setting. N. Engl. J. Med. 2018;379(14):1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]