 To address the rising threat of multidrug-resistant (MDR) bacteria, new therapeutic strategies must be developed.
To address the rising threat of multidrug-resistant (MDR) bacteria, new therapeutic strategies must be developed.
Abstract
To address the rising threat of multidrug-resistant (MDR) bacteria, new therapeutic strategies must be developed. Efficacious drug combinations consisting of existing antibiotics and enhancer biomolecules called adjuvants offers a viable strategy. We have previously reported antibiotic hybrids consisting of tobramycin appended to different fluoroquinolones that possess potential as stand-alone antimicrobials as well as adjuvants. Herein, we report the synthesis of polybasic peptide–levofloxacin conjugates based on these tobramycin–fluoroquinolone hybrids. It was found that conjugating polybasic peptides to the fluoroquinolone levofloxacin, along with the addition of an aliphatic hydrocarbon tether, resulted in the ability of these compounds to potentiate fluoroquinolones and other antibiotics against MDR Gram-negative bacteria. The conjugates were able to potentiate ciprofloxacin, levofloxacin and moxifloxacin against MDR clinical isolates of Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae and to a lesser extent, Acinetobacter baumannii. Preliminary data revealed that the conjugates interfered with active efflux of fluoroquinolones in P. aeruginosa. In addition, synergy was observed with a wide array of other antibiotics against P. aeruginosa, including those that suffered from restricted outer membrane penetration, suggesting that in addition to blocking active efflux, the polybasic peptide–levofloxacin conjugates possessed the ability to disrupt and permeabilize the outer membrane of Gram-negative bacteria.
Introduction
Multidrug-resistant (MDR) bacteria have become increasingly widespread over the past decades to a point where one can say that we are steadily approaching a “post-antibiotic era” in which common bacterial infections can result in patient morbidity and mortality.1–4 To mitigate this looming threat, new therapeutic strategies are currently being developed. The development of efficacious drug combinations consisting of existing antibiotics and enhancer biomolecules called adjuvants is one such strategy. Adjuvants may be inhibitors of efflux pumps, inhibitors of degradation enzymes or membrane permeabilizers. Ideally, the adjuvant should be devoid of potent intrinsic antibacterial activity to reduce the risk of selecting for resistance. Overall, the function of an adjuvant is to augment activity and ideally efficacy of a partner antibiotic(s). For instance, membrane permeabilizers derived from certain cationic amphiphilic peptides5 perturb the integrity of bacterial membranes that effectively enhance intracellular passage of their partner antibiotic.
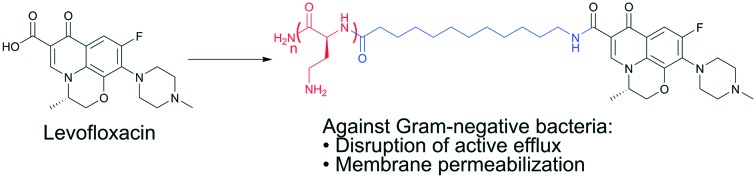
We have previously described antibiotic hybrids consisting of tobramycin appended to the fluoroquinolones ciprofloxacin or moxifloxacin via an aliphatic hydrocarbon tether.6,7 Antibiotic hybrids are constructs of two antibiotics covalently fused by molecular linkers to form heterodimeric entities that exhibit certain biological activity.8 Irrespective of their resultant activity as a stand-alone agent, we found that certain tobramycin–fluoroquinolone hybrids can serve as an adjuvant in combination with existing antibiotics (including fluoroquinolones, rifampicin and minocycline) against MDR Gram-negative bacteria. Gram-negative bacteria possess two restrictive (outer and inner) membranes that greatly limit entry of incoming molecules. Both outer and inner membranes, along with overexpressed efflux systems, comprise intrinsic resistance, a major defense weapon of Gram-negative bacteria against antibiotics.9 We demonstrated that tobramycin–fluoroquinolone hybrids were able to negate these intrinsic resistance mechanisms and as a result potentiated select partner antibiotics, which neither tobramycin nor the fluoroquinolones were able to do alone. Moreover, we provided evidence that the hybrids perturb the outer and inner membrane of P. aeruginosa.6 In addition, we demonstrated that tobramycin–fluoroquinolone hybrids dissipate the proton motive force on the inner membrane that may result in diminished energy production (and therefore may de-energize efflux systems).7 We have also reported other tobramycin based conjugates that are able to potentiate antibiotics against MDR Gram-negative bacteria.10–12 During our previous studies, a question arose whether the tobramycin moiety could be replaced by other polybasic moieties. Tobramycin consists of five protonatable primary amine groups that electrostatically interact with negatively charged lipopolysaccharide (LPS) studded in the outer membrane of Gram-negative bacteria.13 This electrostatic interaction results in the displacement of divalent cations (such as Mg2+ and Ca2+), which function to stabilize the polymeric LPS structure, leading to outer membrane destabilization and perturbation often referred to as the self-promoted uptake mechanism of aminoglycosides.14 Outer membrane imperfections induced upon tobramycin–fluoroquinolone hybrid's action then facilitate the entry of other molecules into the periplasm. Notably, this molecular interplay that results in displacement of divalent cation bridges of LPS is also observed in polybasic/cationic antimicrobial peptides.15,16 Once the tobramycin–fluoroquinolone hybrid reaches the periplasm, we hypothesize that its aliphatic linker interacts with the phospholipid bilayer effectively challenging the structural integrity of the inner membrane.
In this report we describe polybasic analogs in which the tobramycin fragment of tobramycin–fluoroquinolone hybrids is replaced by short polybasic peptides to explore whether the membrane permeabilizing activity, and thus adjuvant properties, against MDR Gram-negative bacteria can be retained. Replacing the tobramycin fragment of these molecules may be advantageous due to the inherent nephrotoxicity and ototoxicity associated with aminoglycosides,17 although the in vitro toxicity of our tobramycin–fluoroquinolone hybrids has not been assessed. We found that polybasic peptide–fluoroquinolone conjugates exhibited similar ability as tobramycin–fluoroquinolone hybrids to potentiate fluoroquinolones including ciprofloxacin (CIP), levofloxacin (LVX) and moxifloxacin (MXF) against MDR Gram-negative bacteria. Moreover, we demonstrated that the number of protonatable amine groups and the presence of an aliphatic hydrocarbon linker were critical for their ability to synergize with fluoroquinolones. Potentiation induced by the polybasic peptide–fluoroquinolone conjugates was not limited to only fluoroquinolones but also extended to other classes of antibiotics. This work provides an attempt to further develop antibiotic hybrids/conjugates as an adjuvant in combination therapy against MDR Gram-negative bacteria.
Results and discussion
Design and synthesis of peptide–levofloxacin conjugates
To explore the necessity of both the tobramycin fragment and the aliphatic hydrocarbon linker on the tobramycin–fluoroquinolone hybrid scaffold, we conjugated short peptides to LVX on the fluoroquinolone's carboxyl group via solid-phase peptide synthesis (SPPS) (Fig. 1). LVX provided a compatible fluoroquinolone for SPPS conditions since it only contained one available reactive carboxyl group. However, it is known that the carboxyl group of fluoroquinolones is required to inhibit their DNA/DNA gyrase and topoisomerase IV intracellular targets18 and thus we foresee that our covalent appendage of peptides will diminish antibacterial activity, a critical requirement for the function of an adjuvant to avoid selective pressure that may lead to resistance selection.19 In future studies, we will explore the adjuvant effect by attaching peptides to fluoroquinolones at a site different from the carboxyl group.
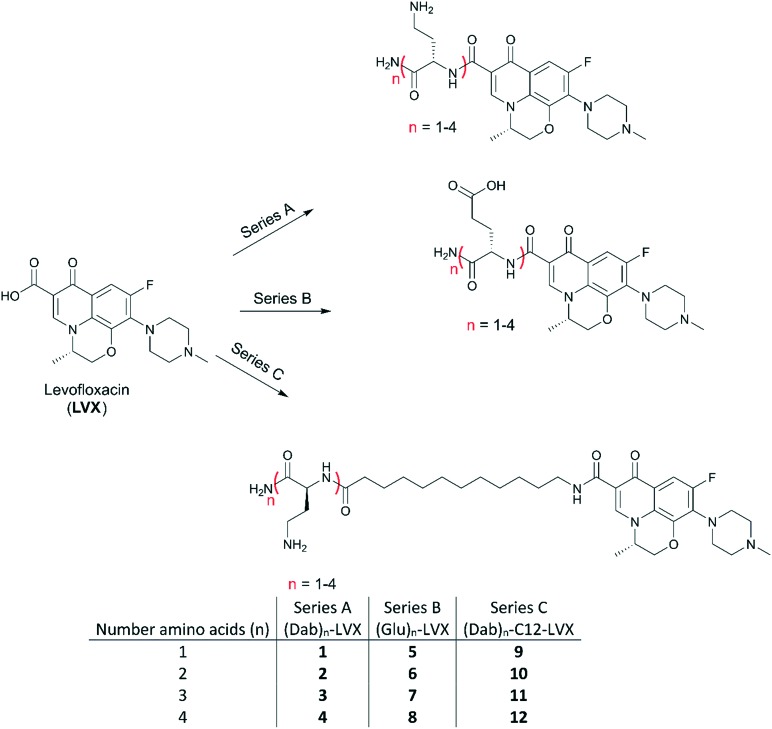
Fig. 1. Peptide–levofloxacin conjugates under study. Three cohorts were prepared that included series (A) comprised of Dabn–LVX, series (B) comprised of Glun–LVX and series (C) comprised of Dabn-C12-LVX.
Three series of peptide–levofloxacin conjugates (Fig. 1) were prepared through SPPS following a fluorenylmethyloxycarbonyl (Fmoc) strategy using 4-methylbenzhydrylamine (MBHA) Rink amide resin. It should be noted that all the peptide–levofloxacin conjugates have an amidated C-terminus, owing to the use of this resin. Each series was comprised of conjugates that only differ in the amount of amino acids, ranging from one to four amino acids long. Short polybasic peptides consisting of l-diaminobutyric acid (Dab) were appended to LVX to yield series A, where conjugates 1–4 contained increasing numbers of Dab up to four residues. Dab was selected because of its presence in polybasic peptides like the polymyxins. We purposely attached polybasic peptides that contain protonatable primary amine side-chains as this functional group appears to play a key role in enhancing outer membrane permeability.20 Notably, tobramycin contains a total of five primary amines. Through this, we explored the optimal number of protonatable amine groups necessary to result in a conjugate capable of permeabilizing the bacterial outer membrane, but also determined whether the tobramycin carbohydrate structure was necessary. To serve as a control to series A, we prepared series B by linking one to four l-glutamic acid (Glu) to LVX resulting in conjugates 5–8. We hoped to see the effect of imparting amino acids with differing side-chains, either basic or acidic functional groups. We elected Glu as it has the same number of ethylene carbon atoms in its side-chain as Dab. At physiological pH, the acidic Glu and the basic Dab would confer an overall anionic and cationic charge, respectively. In addition, we also constructed conjugates 9–12 of series C in which the polybasic peptide of series A was separated by a twelve carbons-long tether (C12) as was done in our previously described tobramycin–fluoroquinolone hybrids.6,7 Our previous studies6,7 demonstrated that a molecular linker length of C12 was optimal for the adjuvant properties.
Polybasic peptide–levofloxacin conjugates potentiated fluoroquinolones against wild-type Pseudomonas aeruginosa
We initially evaluated the activity of peptide–levofloxacin conjugates against a panel of Gram-positive and Gram-negative bacteria (Table 1), among which were wild-type strains and clinical isolates. The lowest concentration of the agent to inhibit visible bacterial growth was determined to be the minimum inhibitory concentration (MIC). All twelve conjugates displayed poor antibacterial activity as stand-alone agents, in comparison to LVX (Table 1). As we expected, the attachment of peptides at the carboxylic acid group of LVX rescinded the fluoroquinolone's inherent antibacterial activity. However, our previous experience working with tobramycin–fluoroquinolone hybrids study suggested that a free carboxylic acid moiety in the fluoroquinolone is not critical for the membrane destabilizing activity.6,7 Therefore, we explored herein the necessary structural components of the hybrid scaffold that were required to yield agents suitable to be adjuvants.
Table 1. Antibacterial activity of peptide–levofloxacin conjugates against a panel of wild-type and multidrug-resistant clinical isolates of Gram-positive and Gram-negative bacteria.
| Organism | Minimum inhibitory concentration (MIC) (μg ml–1) |
||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | LVX | |
| S. aureus ATCC 29213 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 128 | 64 | >128 | ≤0.25 |
| MRSA ATCC 33592 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | >128 | ≤0.25 |
| MSSE CANWARD-2008 81388 | >128 | >128 | >128 | 128 | >128 | >128 | >128 | >128 | 64 | 64 | 64 | >128 | ≤0.25 |
| MRSE CAN-ICU 61589 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | 128 | 128 | >128 | >128 |
| S. pneumoniae ATCC 49619 | >128 | >128 | >128 | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 1 |
| E. faecalis ATCC 29212 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 2 |
| E. coli ATCC 25922 | 128 | 128 | 128 | >128 | >128 | 128 | >128 | 128 | >128 | 64 | 32 | >128 | ≤0.25 |
| E. cloacae 117029 | >128 | >128 | 128 | 128 | >128 | >128 | 64 | >128 | 128 | 128 | 128 | 128 | 0.125 |
| P. aeruginosa ATCC 27853 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 | >128 | 1 |
| S. maltophilia CAN-ICU 62584 | >128 | >128 | >128 | >128 | >128 | 16 | >128 | >128 | >128 | >128 | >128 | >128 | 4 |
| A. baumannii CAN-ICU 63169 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 1 |
| K. pneumoniae 116381 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 |
We then evaluated the ability of the twelve conjugates to potentiate the fluoroquinolones CIP, LVX and MXF against wild-type P. aeruginosa PAO1 (Table 2). We decided to screen for fluoroquinolone potentiation as our tobramycin–ciprofloxacin7 and tobramycin–moxifloxacin6 hybrids synergized with this antibiotic class. The peptide–levofloxacin conjugates were screened at a fixed concentration of 8 μg mL–1 in combination with either CIP, LVX or MXF. A ≥4-fold decrease in MIC of fluoroquinolone would indicate latent ability of the conjugates to act as adjuvants, which will then be confirmed by checkerboard assay. Notably, 8 μg mL–1 was chosen as the working concentration as our previous studies with peptide-based adjuvants suggest that any concentration higher than this may be too toxic for therapeutic use.5 Of the twelve peptide–levofloxacin conjugates, compounds 10, 11 and 12 were found to decrease the MIC of all three fluoroquinolones by ≥4-fold. We inferred from this initial screening that the aliphatic linker C12 was necessary to synergize with fluoroquinolones. All three conjugates that potentiated fluoroquinolones belonged to series C while none from series A were able to do so. Note that conjugates from series A (1–4) and series C (9–12) only differ in the presence of a C12 linker. In addition, there appeared to be a minimum amount of protonatable amine side-chains of the peptide fragment and thus positive charges to result to fluoroquinolone potentiation (compound 9 which only contained one Dab is not effective). As expected, compounds 5–8 that contained Glu and thus anionic character did not synergize with fluoroquinolones. This confirmed that basic protonatable amines were required for peptide–levofloxacin conjugates to potentiate fluoroquinolones. Therefore, we identified three polybasic peptide–levofloxacin conjugates as potential adjuvants for fluoroquinolones.
Table 2. Synergy evaluation with 8 μg mL–1 peptide–levofloxacin conjugates 1–12 in combination with fluoroquinolones (FQ) against wild-type P. aeruginosa PAO1.
| Conjugate | MIC (μg ml–1) of FQs in combination with 8 μg ml–1 conjugates |
||
| Ciprofloxacin | Levofloxacin | Moxifloxacin | |
| 1 | 0.125 | 0.500 | 1 |
| 2 | 0.125 | 0.250 | 1 |
| 3 | 0.125 | 0.250 | 1 |
| 4 | 0.063 | 0.250 | 1 |
| 5 | 0.125 | 0.500 | 1 |
| 6 | 0.125 | 0.500 | 1 |
| 7 | 0.125 | 0.250 | 1 |
| 8 | 0.063 | 0.250 | 1 |
| 9 | 0.063 | 0.250 | 1 |
| 10 | 0.031 | 0.063 | 0.250 |
| 11 | 0.031 | 0.125 | 0.250 |
| 12 | 0.031 | 0.125 | 0.250 |
| Antibiotic alone: | 0.125 | 0.500 | 1 |
Next, we confirmed our observed synergism against wild-type P. aeruginosa PAO1 for combinations of the three polybasic peptide–levofloxacin conjugates and the three clinically-used fluoroquinolones via checkerboard assay. The fractional inhibitory concentration (FIC) index for each combination was determined to assess interactions between the two components. FIC for the conjugates (adjuvants) were calculated by dividing the MIC of the conjugate in combination with the fluoroquinolone by the MIC of the conjugate alone. Similar calculations were performed to obtain the FIC of the fluoroquinolones (antibiotics). FIC index was then calculated by adding the FIC indices of the conjugate and the fluoroquinolone. FIC index values of ≤0.5, 0.5 < x ≤ 4, and >4 were interpreted as synergistic, additive and antagonistic interactions, respectively. Our results indicated that all combinations of polybasic peptide–levofloxacin conjugates 10, 11 and 12 with fluoroquinolones CIP, LVX and MXF were synergistic against PAO1 strain (Table 3). To compare the adjuvant potency between the three conjugates, we examined the fold potentiation that resulted upon addition of a fixed concentration of the conjugate (Table 3). The antibiotic fold potentiation was calculated by dividing the MIC of the fluoroquinolone alone by the absolute MIC of the fluoroquinolone in the presence of a fixed concentration of the conjugate. It appeared that 8 μg mL–1 of the three conjugates potentiated CIP, LVX and MXF to the same degree (4–8 fold) against P. aeruginosa PAO1 strain (Table 3). Nonetheless, we validated synergism between polybasic peptide–levofloxacin conjugates 10, 11 and 12 and fluoroquinolones against wild-type P. aeruginosa.
Table 3. Synergy evaluation for combinations of polybasic peptide–levofloxacin conjugates 10, 11, and 12 with fluoroquinolones (FQ) ciprofloxacin (CIP), levofloxacin (LVX), and moxifloxacin (MXF) against wild-type P. aeruginosa PAO1.
| Conjugate | Antibiotic | MICFQ [MICcombo] (μg ml–1) | MICconjugate [MICcombo] (μg ml–1) | FIC index | Interpretation | Absolute MICFQ a (μg ml–1) | Potentiation b |
| 10 | CIP | 0.125 [0.031] | 128 [8] | 0.313 | Synergy | 0.031 | 4-fold |
| LVX | 0.5 [0.031] | 128 [16] | 0.188 | Synergy | 0.063 | 8-fold | |
| MXF | 1 [0.125] | 128 [16] | 0.250 | Synergy | 0.250 | 4-fold | |
| 11 | CIP | 0.125 [0.031] | 128 [32] | 0.500 | Synergy | 0.031 | 4-fold |
| LVX | 0.5 [0.125] | 128 [2] | 0.266 | Synergy | 0.125 | 4-fold | |
| MXF | 1 [0.250] | 128 [8] | 0.313 | Synergy | 0.250 | 4-fold | |
| 12 | CIP | 0.125 [0.031] | 64 [16] | 0.500 | Synergy | 0.031 | 4-fold |
| LVX | 0.5 [0.063] | 64 [16] | 0.266 | Synergy | 0.125 | 4-fold | |
| MXF | 1 [0.250] | 64 [8] | 0.375 | Synergy | 0.250 | 4-fold |
aMIC of FQ in presence of 8 μg ml–1 conjugate.
bDegree of FQ potentiation in the presence of 8 μg ml–1 conjugate.
Fluoroquinolone efflux was affected by polybasic peptide–levofloxacin conjugates
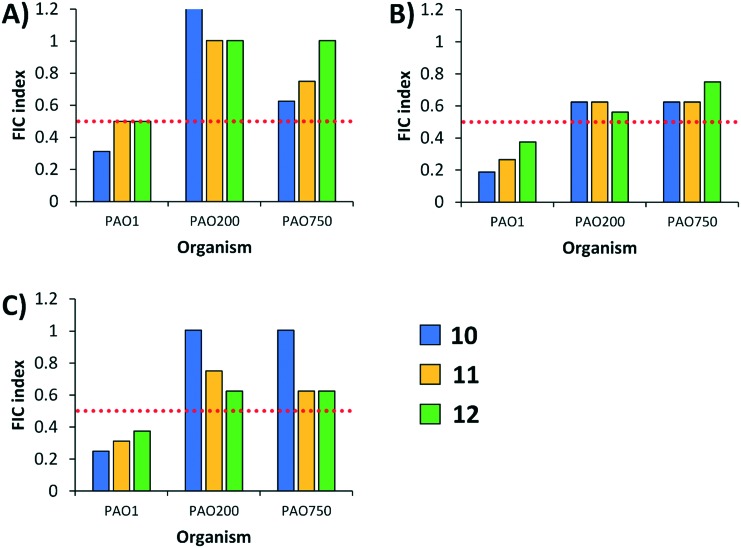
Drug efflux contributes greatly to fluoroquinolone resistance in Gram-negative bacteria.18,21–23 On that note, we wondered whether our conjugates potentiate fluoroquinolones by interfering with active efflux. Since the polybasic peptide–levofloxacin conjugates contain a fluoroquinolone fragment that is widely recognized as a substrate for efflux pumps, we hypothesized that the polybasic conjugates may ‘clog’ the transient channel of efflux pumps. Another hypothesis is that the conjugates may have the ability to dissipate proton motive force thereby de-energizing efflux systems. The latter was found true for our previously reported tobramycin–fluoroquinolone hybrids.7 To test this hypothesis, we evaluated combinations of the conjugates with the fluoroquinolones CIP, LVX and MXF against efflux pump-knockout strains isogenic to wild-type P. aeruginosa PAO1. Two efflux-deficient strains were used that included strain PAO200, which lacked the MexAB-OprM tripartite efflux system, and strain PAO750, which lacked five clinically-relevant efflux pumps and the outer membrane protein H (OpmH). If the observed potentiation was partly due to efflux pump inhibition, we expect reduction if not annulment of synergism between the combinations against the efflux-deficient strains. Indeed, synergy was lost against both efflux-deficient strains (PAO200 and PAO750) for all combinations of conjugates and fluoroquinolones (Fig. 2). While we do not dispute the idea that our conjugates possess the ability to permeabilize the outer membrane and thereby enhance intracellular accumulation of antibiotics, our findings herein suggest that conjugates 10, 11 and 12 may also disrupt drug efflux. Future work will be undertaken to decipher specific molecular interplay on how these conjugates block efflux systems, yet our previous findings from tobramycin–fluoroquinolone hybrids strongly suggest that this may be due to dissipation of proton motive force.
Fig. 2. FIC indices of polybasic peptide–levofloxacin conjugates 10, 11 and 12 in combination with either ciprofloxacin (A), levofloxacin (B), moxifloxacin (C) against wild-type (PAO1) and efflux-deficient (PAO200 and PAO750) P. aeruginosa strains. PAO200 lack the MexAB-OprM efflux pump, while PAO750 lack five clinically relevant pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexJK and MexXY) and outer membrane protein OpmH. Red dotted line denotes an FIC index value of 0.5. See ESI† for MIC and FIC values of combinations. PA = P. aeruginosa.
Synergy was retained against multidrug-resistant clinical isolates of P. aeruginosa
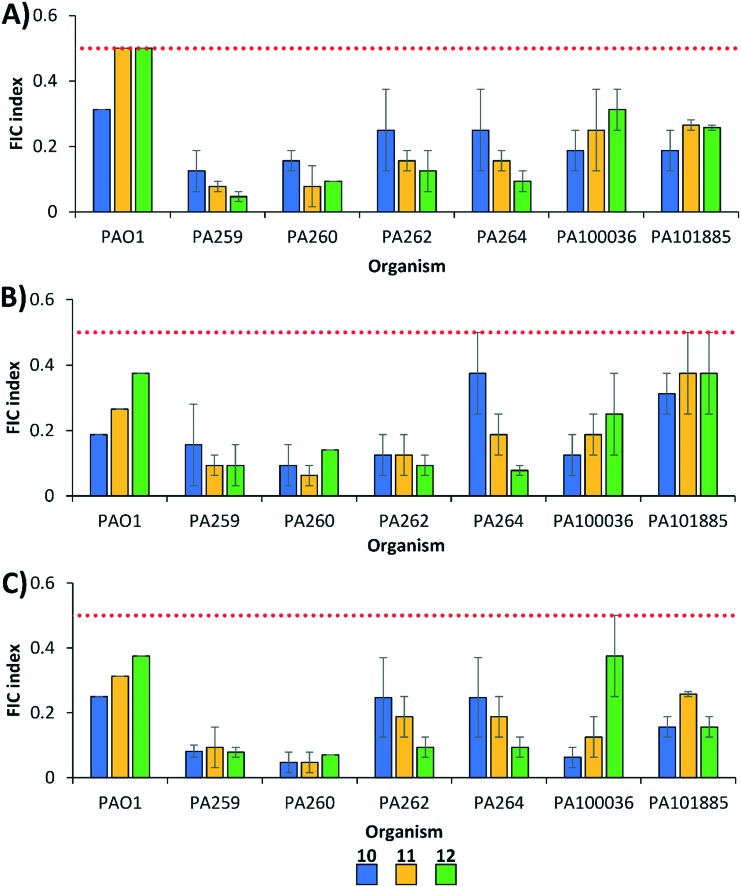
We further evaluated the therapeutic potential of conjugates 10, 11 and 12 as adjuvants in combination with fluoroquinolones against MDR fluoroquinolone-resistant clinical isolates of P. aeruginosa via checkerboard assay (Fig. 3). All combinations were found to be synergistic against tested MDR P. aeruginosa. No conjugate clearly out-performed the others, with compound 10 displaying the lowest FIC indices against three of the strains tested (PAO1, PA100036, PA101885), while compound 12 had the lowest FIC index against MDR P. aeruginosa PA259, PA262, and PA264. Compound 11 appeared to be marginally more synergistic against PA260. It appeared that above the minimum threshold of basic protonatable amine side-chain groups (>2), additional amine groups on the peptide fragment of the conjugate do not further enhance their potency for fluoroquinolone potentiation against MDR P. aeruginosa.
Fig. 3. FIC indices of polybasic peptide–levofloxacin conjugates 10, 11 and 12 in combination with (A) ciprofloxacin, (B) levofloxacin, and (C) moxifloxacin against wild-type and MDR P. aeruginosa. All strains with the exception of PAO1 are fluoroquinolone-resistant. Red dotted line denotes an FIC index value of 0.5. See ESI† for MIC and FIC values of combinations. PA = P. aeruginosa.
Synergy was retained against Enterobacteriaceae but was reduced against A. baumannii
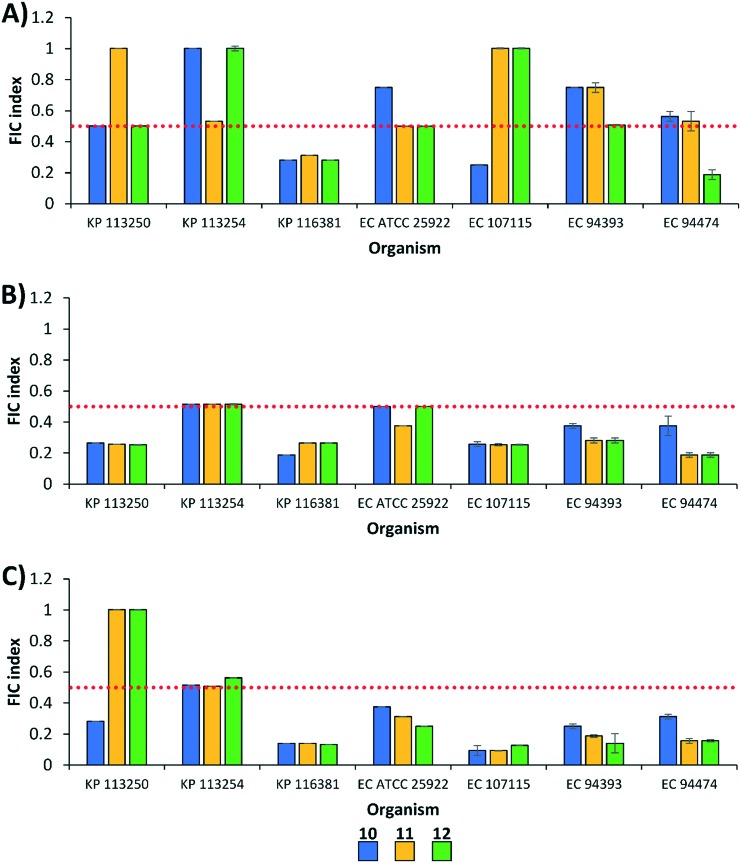
We then determined whether polybasic peptide–levofloxacin conjugates 10–12 were able to potentiate fluoroquinolones against other Gram-negative bacteria, aside from P. aeruginosa. First, we evaluated the combinations against wild-type and MDR strains of Enterobacteriaceae (Fig. 4), including Klebsiella pneumoniae (3) and Escherichia coli (4). The degree of fluoroquinolone potentiation seemed to differ against Enterobacteriaceae, with combinations of CIP being additive in most strains (Fig. 4). For instance, at least one combination of conjugates and CIP was additive against the four tested E. coli strains (Fig. 4A). On the other hand, all combinations of LVX (Fig. 4B) and MXF (Fig. 4C) were synergistic against E. coli. Fluoroquinolone potentiation appeared to be strain specific against K. pneumoniae (Fig. 4). For instance, all conjugates potentiated the three fluoroquinolones against K. pneumoniae 116381 while the opposite was observed against K. pneumoniae 113254. Similar to what we observed against P. aeruginosa, conjugates 10, 11 and 12 seemed to have similar adjuvant potency against Enterobacteriaceae.
Fig. 4. FIC indices of polybasic peptide–levofloxacin conjugates 10, 11 and 12 in combination with (A) ciprofloxacin, (B) levofloxacin, and (C) moxifloxacin against wild-type and MDR Enterobacteriaceae. Red dotted line denotes an FIC index value of 0.5. See ESI† for MIC and FIC values of combinations. KP = K. pneumoniae; EC = E. coli.
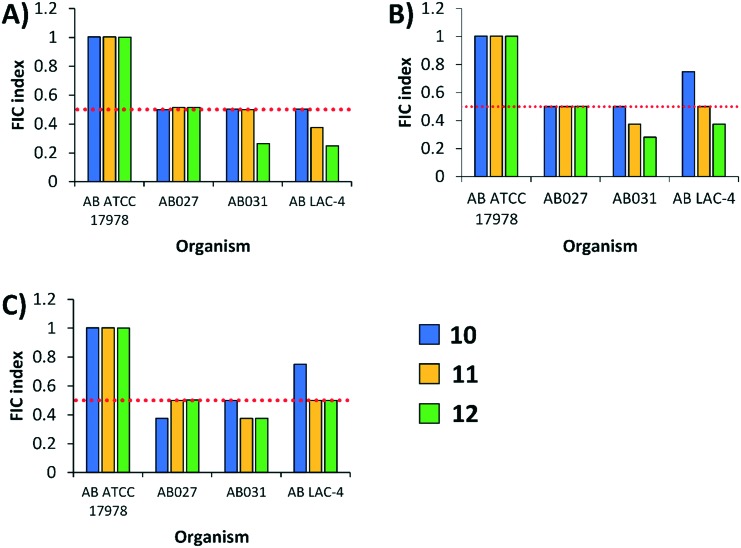
Combinations of conjugates and fluoroquinolones were then evaluated against Acinetobacter baumannii (4), including one wild-type and three MDR clinical isolates. Surprisingly, fluoroquinolone potentiation by the conjugates was reduced against A. baumannii (Fig. 5). For instance, none of the combinations were synergistic against wild-type A. baumannii ATCC 17978. Conjugate 10 synergized with all three fluoroquinolones against MDR A. baumannii AB027 (FIC indices ranged from 0.37 to 0.5), while conjugates 11 and 12 potentiated all three fluoroquinolones against MDR A. baumannii AB031 (FIC indices ranged from 0.265 to 0.5) and LAC-4 (FIC indices ranged from 0.25 to 0.5). Most combinations of conjugates and fluoroquinolones yielded FIC indices of 0.5 against A. baumannii which is considered to be borderline synergistic. Overall, the ability of polybasic peptide–levofloxacin conjugates 10, 11 and 12 to potentiate fluoroquinolones was conserved against other Gram-negative bacteria.
Fig. 5. FIC indices of polybasic peptide–levofloxacin conjugates 10, 11 and 12 in combination with (A) ciprofloxacin, (B) levofloxacin, and (C) moxifloxacin against wild-type and MDR A. baumannii. Red dotted line denotes an FIC index value of 0.5. See ESI† for MIC and FIC values of combinations. AB = A. baumannii.
Polybasic peptide–levofloxacin conjugates did not lyse red blood cells
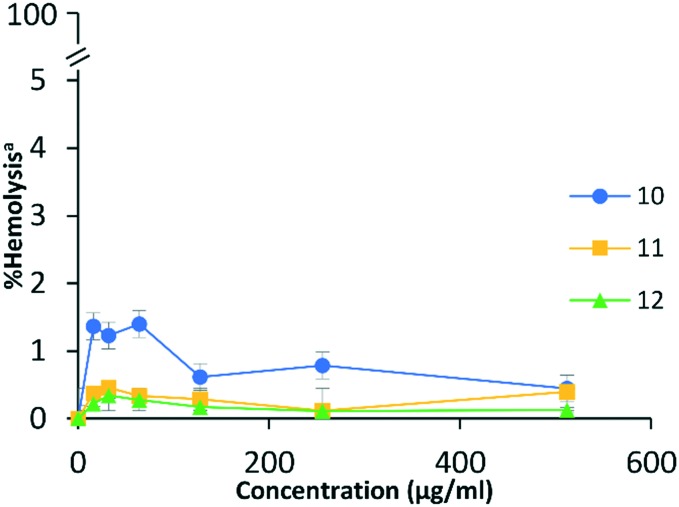
Certainly, there is a need to address toxicity of compounds intended for therapeutic use. An initial attempt to evaluate toxicity was performed by assessing the ability of the polybasic peptide–levofloxacin conjugates to lyse eukaryotic red blood cells. This is important as amphiphilic compounds such as our conjugates, especially those that permeabilize membranes, have the tendency to indistinguishably lyse bacterial and eukaryotic membranes.24 In the assay, 0.1% Triton X-100 was used as a positive control and conjugate concentration was tested up to 512 μg mL–1. All three polybasic peptide–levofloxacin conjugates were found to be non-hemolytic even at the highest concentration tested (Fig. 6). It appeared that conjugate 10 was slightly more hemolytic than conjugates 11 and 12. However, <1% hemolysis was observed for all three conjugates at 512 μg mL–1 (Fig. 6).
Fig. 6. Concentration dependent hemolysis induced by polybasic peptide–levofloxacin conjugates against red blood cells. a% hemolysis relative to control (0.1% Triton X-100).
Polybasic peptide–levofloxacin conjugates potentiated multiple classes of antibiotics against P. aeruginosa
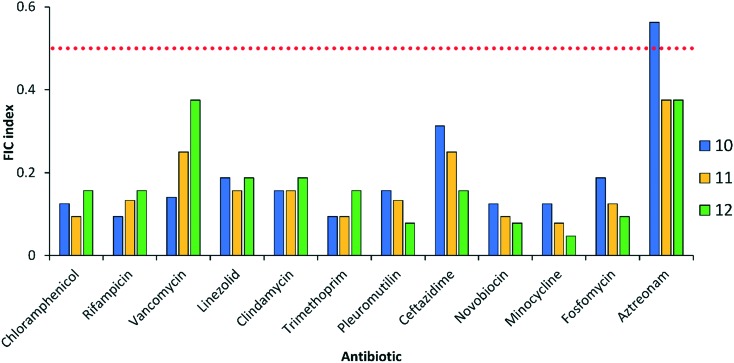
In addition to the three fluoroquinolones evaluated above, we explored whether conjugates 10, 11 and 12 could potentiate other classes of antibiotics against wild-type P. aeruginosa PAO1. Our panel consisted of twelve clinically-used antibiotics (Fig. 7). Conjugates 11 and 12 potentiated all twelve antibiotics that included those that are significantly affected by efflux (chloramphenicol, linezolid, clindamycin, trimethoprim, pleuromutilin, ceftazidime, minocycline, fosfomycin and aztreonam) and those that are perceived to have limited outer membrane penetration (rifampicin, vancomycin and novobiocin). Similarly, conjugate 10 potentiated almost all antibiotics with the exception of aztreonam (Fig. 7). This suggested that in addition to blocking active efflux, the polybasic peptide–levofloxacin conjugates possessed the ability to disrupt and permeabilize the outer membrane of Gram-negative bacteria. Our previously reported tobramycin–fluoroquinolone hybrids also potentiated the same set of antibiotics. This suggests that the mechanism of adjuvant action found in tobramycin–fluoroquinolone hybrids was retained in polybasic peptide–levofloxacin conjugates. Collectively, our results suggest that the pentabasic tobramycin portion in the tobramycin–fluoroquinolone hybrid is not necessary to potentiate antibiotics but rather the protonatable amines and aliphatic hydrocarbon linker are important. Nevertheless, the overall potency of the previously tobramycin–fluoroquinolone hybrid adjuvants is superior when compared to the compounds described in this study.
Fig. 7. FIC indices of polybasic peptide–levofloxacin conjugates 10, 11 and 12 in combination with twelve clinically used antibiotics wild-type P. aeruginosa PAO1. Red dotted line denotes an FIC index value of 0.5. See ESI† for MIC and FIC values of combinations.
Conclusion
Polybasic peptide–levofloxacin conjugates were synthesized based on our previously reported tobramycin–fluoroquinolone hybrids to determine whether the tobramycin fragment is required for membrane permeabilization. We found that >2 protonatable primary amine groups and the aliphatic hydrocarbon C12 tether were necessary for the conjugates to potentiate fluoroquinolones. We demonstrated the ability of the conjugates to potentiate CIP, LVX and MFX, as well as other classes of antibiotics, against wild-type and MDR Gram-negative bacteria. The conjugates were found to interfere with active efflux of fluoroquinolones. Furthermore, the polybasic peptide-conjugates were not hemolytic and therefore did not display non-specific membranolytic activity. Overall, it appeared that the tobramycin scaffold was not necessary for the ability of tobramycin–fluoroquinolone hybrids to potentiate antibiotics although our previously reported hybrids were far superior as both adjuvants and stand-alone antimicrobials. The findings in this study provide data that will be of use towards the development of antibiotic hybrids/conjugates as potential adjuvants to rescue existing antibiotics from the clutches of antimicrobial resistance.
Experimental
Reagents, solvents and antibiotics were purchased from commercial sources and were used without further purification. All peptide–levofloxacin conjugates were synthesized using solid-phase peptide synthesis on MBHA Rink amide resin following an Fmoc protocol as previously reported.25 Dab amino side-chains were protected by tert-butyloxycarbonyl (Boc) and Glu side-chains were protected by OtBu. Fmoc-Dab(Boc)-OH, Fmoc-12-aminododeconic acid or Fmoc-Glu(OtBu)-OH was pre-mixed with the coupling reagent N,N,N′,N′-tetramethyl-O-(benzotriazol-1-yl)uronium tetrafluoroborate (TBTU) and N-methylmorpholine, followed by reaction with the resin in the peptide vessel for 45 minutes. The Fmoc protecting groups were deprotected using 4 : 1 DMF : piperidine solution for 15 min with gentle agitation using N2 gas. A sample of resin was subjected to Kaiser test (2% chloranil in DMF) to qualitatively detect the presence of free amine, ensuring reaction completion. After the desired number of amino acids (and aliphatic hydrocarbon linker, if required) were added, LVX was pre-mixed with the peptide coupling reagents benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP) and hydroxybenzotriazole (HOBt), as well as the weak base N-methylmorpholine. This pre-mixed solution was then added to the resin and reacted by gentle agitation using N2 gas for 120 minutes. Cleavage of the peptide–levofloxacin conjugates from resin, as well as removal of Boc/OtBu protecting groups, was achieved by adding a 95 : 5 trifluoroacetic acid (TFA) : H2O solution followed by gentle stirring for 30 minutes. Crude products were purified using reverse-phase chromatography using C18 silica gel and a gradient eluent mixture of 0% to 50% methanol in water spiked with 0.1% TFA. Purity of the conjugates were measured using high-performance liquid chromatography (HPLC) on a Breeze HPLC Waters with 2998 PDA detector (1.2 nm resolution) coupled to Phenomenex Synergi Polar (50 × 2.0 mm) 4 μm reverse-phase column and were determined to be >95%. All purified compounds were characterized using 1-dimensional (1H and 13C) and 2-dimensional nuclear magnetic resonance (NMR) experiments as well as mass spectrometry. All NMR measurements were recorded on AMX-500 (500 MHz) Bruker instrument (Germany). High-resolution matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) experiments were done on a Bruker Ultraflextreme mass spectrometer (Germany) coupled to a time-of-flight mass analyzer. Measurements for MALDI-MS are reported to three decimal places.
Antimicrobial susceptibility assay
Bacterial isolates used in this study were obtained from the American Type Culture Collection (ATCC) [reference strains], the Canadian National Intensive Care Unit (CAN-ICU) surveillance study26 and the Canadian Ward (CANWARD) surveillance study.27 Clinical isolates belonging to the CAN-ICU and CANWARD surveillance studies were recovered from patients suffering presumed infectious diseases entering or admitted in a participating medical center across Canada during the time of study. The in vitro antibacterial activities of the conjugates were assessed the using microbroth dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.28 Bacterial cultures were grown overnight and diluted in saline to achieve a 0.5 McFarland turbidity, followed by 1 : 50 dilution in Mueller-Hinton broth (MHB) for inoculation to a final concentration of approximately 5 × 105 colony forming units per mL. Experiments were performed on 96-well plates. The tested agents were 2-fold serially diluted in MHB and incubated with equal volumes of bacterial inoculum at 37 °C for 18 h. The MIC was determined as the lowest concentration to inhibit visible bacterial growth in the form of visible turbidity, which was confirmed using an EMax Plus microplate reader (Molecular Devices, USA) at a wavelength of 590 nm. MHB with or without bacterial cells were used as positive or negative controls, respectively.
Checkerboard assay
The assay was performed on 96-well plates as previously described.29 The antibiotic was 2-fold serially diluted along the x axis, while the adjuvant was 2-fold serially diluted along the y axis to create a matrix in which each well contain a combination of both agents at different concentrations. Bacterial cultures grown overnight were diluted in saline to 0.5 McFarland turbidity, followed by 1 : 50 dilution in MHB and inoculation on each well to a final concentration of approximately 5 × 105 colony forming units per mL. Wells consisting of only MHB with or without bacterial cells were used as positive or negative controls, respectively. Plates were incubated for 18 h at 37 °C and inspected for visible turbidity, which was confirmed using EMax Plus microplate reader (Molecular Devices, USA) at a wavelength of 590 nm. Fractional inhibitory concentration (FIC) of antibiotic was calculated by dividing the MIC of antibiotic in the presence of conjugates by the MIC of antibiotic alone. Likewise, the FIC of conjugates was calculated via dividing the MIC of conjugates in the presence of antibiotic by the MIC of conjugates alone. The FIC index was obtained by the summation of both FIC values. The FIC indices were interpreted as synergistic, indifferent or antagonistic for values of ≤0.5, 0.5 < x ≤ 4 or >4, respectively.30
Hemolytic assay
The ability of polybasic peptide–levofloxacin conjugates 10, 11 and 12 to lyse eukaryotic red blood cells was quantified by the amount of hemoglobin released upon incubation, following published protocol.31 5 ml of fresh pig blood was drawn from the antecubital vein of healthy pigs, collected in an EDTA blood collection tube and centrifuged at 1000×g for 5 min at 4 °C. Then it was washed with phosphate-buffered saline (PBS) three times and re-suspended in the same buffer, consecutively, to prepare the working erythrocyte solution. The conjugates were then 2-fold serially diluted in PBS on 96-well plates and mixed with equal volumes of working erythrocyte solution. Post 1 h incubation at 37 °C, intact cells on the 96-well plates were pelleted by centrifugation at 1000×g for 5 min at 4 °C. The supernatant was then transferred to a new 96-well plate. The released hemoglobin in each well was then measured via EMax Plus microplate reader (Molecular Devices, USA) at 570 nm wavelength. Red blood cells in PBS with or without 0.1% Triton X-100 were used as negative or positive controls, respectively. All procedures performed in studies involving animals were in accordance with the ethical standards of the University of Manitoba Animal Care and Use policy and all experiments were approved by the Animal Care Committee.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by the Natural Sciences and Engineering Council of Canada (NSERC-DG 49467) and the University of Manitoba. We thank Dr. Richard Hodges (University of Manitoba) for generously providing fresh porcine erythrocytes, and Dr. A. Kumar (University of Manitoba) for generously providing the efflux-deficient P. aeruginosa PAO200 and PAO750 strains.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9md00051h
References
- Gulland A. BMJ [Br. Med. J.] 2014;348:g3062. doi: 10.1136/bmj.g3062. [DOI] [PubMed] [Google Scholar]
- Brown E. D., Wright G. D. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- World Health Organisation (WHO) Bull. W. H. O. 2014;51:1–256. [Google Scholar]
- Theuretzbacher U. Curr. Opin. Microbiol. 2017;39:106–112. doi: 10.1016/j.mib.2017.10.028. [DOI] [PubMed] [Google Scholar]
- Domalaon R., Sanchak Y., Cherono Koskei L., Lyu Y., Zhanel G. G., Arthur G., Schweizer F. Antimicrob. Agents Chemother. 2018;62:e02374. doi: 10.1128/AAC.02374-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorityala B. K., Guchhait G., Goswami S., Fernando D. M., Kumar A., Zhanel G. G., Schweizer F. J. Med. Chem. 2016;59:8441–8455. doi: 10.1021/acs.jmedchem.6b00867. [DOI] [PubMed] [Google Scholar]
- Gorityala B. K., Guchhait G., Fernando D. M., Deo S., McKenna S. A., Zhanel G. G., Kumar A., Schweizer F. Angew. Chem., Int. Ed. 2016;55:555–559. doi: 10.1002/anie.201508330. [DOI] [PubMed] [Google Scholar]
- Domalaon R., Idowu T., Zhanel G. G., Schweizer F. Clin. Microbiol. Rev. 2018;31:e00077. doi: 10.1128/CMR.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurskaya H. I., López C. A., Gnanakaran S. ACS Infect. Dis. 2016;1:512–522. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Domalaon R., Lyu Y., Zhanel G., Schweizer F. J. Clin. Med. 2018;7:158. doi: 10.3390/jcm7070158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Goswami S., Gorityala B. K., Domalaon R., Lyu Y., Kumar A., Zhanel G. G., Schweizer F. J. Med. Chem. 2017;60:3913–3932. doi: 10.1021/acs.jmedchem.7b00156. [DOI] [PubMed] [Google Scholar]
- Lyu Y., Yang X., Goswami S., Gorityala B. K., Idowu T., Domalaon R., Zhanel G. G., Shan A., Schweizer F. J. Med. Chem. 2017;60:3684–3702. doi: 10.1021/acs.jmedchem.6b01742. [DOI] [PubMed] [Google Scholar]
- Yadav R., Bulitta J. B., Schneider E. K., Shin B. S., Velkov T., Nation R. L., Landersdorfer C. B. Antimicrob. Agents Chemother. 2017;61:e00722. doi: 10.1128/AAC.00722-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Bell A. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7:713–720. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- Epand R. M., Walker C., Epand R. F., Magarvey N. A. Biochim. Biophys. Acta, Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Zhang L., Dhillon P., Yan H., Farmer S., Hancock R. E. W. Antimicrob. Agents Chemother. 2000;44:3317–3321. doi: 10.1128/aac.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosso M. Y., Li Y., Garneau-Tsodikova S. Med. Chem. Commun. 2014;5:1075–1091. doi: 10.1039/C4MD00163J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Hiasa H., Kerns R., Malik M., Mustaev A., Zhao X. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieren M., Tigges M. Curr. Opin. Pharmacol. 2012;12:551–555. doi: 10.1016/j.coph.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Richter M. F., Drown B. S., Riley A. P., Garcia A., Shirai T., Svec R. L., Hergenrother P. J. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A. Clin. Infect. Dis. 2005;41:S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- Willers C., Wentzel J. F., du Plessis L. H., Gouws C., Hamman J. H. Expert Opin. Ther. Targets. 2017;21:23–36. doi: 10.1080/14728222.2017.1265105. [DOI] [PubMed] [Google Scholar]
- Redgrave L. S., Sutton S. B., Webber M. A., Piddock L. J. V. Trends Microbiol. 2014;22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Hollmann A., Martínez M., Noguera M. E., Augusto M. T., Disalvo A., Santos N. C., Semorile L., Maffía P. C. Colloids Surf., B. 2016;141:528–536. doi: 10.1016/j.colsurfb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Domalaon R., Brizuela M., Eisner B., Findlay B., Zhanel G. G., Schweizer F. Amino Acids. 2018:1–11. doi: 10.1007/s00726-018-2673-9. [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., DeCorby M., Laing N., Weshnoweski B., Vashisht R., Tailor F., Nichol K. A., Wierzbowski A., Baudry P. J., Karlowsky J. A., Lagacé-Wiens P., Walkty A., McCracken M., Mulvey M. R., Johnson J., T. C. A. R. A. Canadian Antimicrobial Resistance Alliance (CARA), Hoban D. J. Antimicrob. Agents Chemother. 2008;52:1430–1437. doi: 10.1128/AAC.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban D. J., Zhanel G. G. J. Antimicrob. Chemother. 2013;68(Suppl 1):i3–i5. doi: 10.1093/jac/dkt021. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing, Clin. Lab. Stand Institute, Wayne, PA, 26th edn, 2016. [Google Scholar]
- Domalaon R., Berry L., Tays Q., Zhanel G. G., Schweizer F. Bioorg. Chem. 2018;80:639–648. doi: 10.1016/j.bioorg.2018.07.018. [DOI] [PubMed] [Google Scholar]
- Meletiadis J., Pournaras S., Roilides E., Walsh T. J. Antimicrob. Agents Chemother. 2010;54:602–609. doi: 10.1128/AAC.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R., Findlay B., Ogunsina M., Arthur G., Schweizer F. Peptides. 2016;84:58–67. doi: 10.1016/j.peptides.2016.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.