Abstract
Angiosperm mitochondrial genomes (mitogenomes) are notable for their extreme diversity in both size and structure. However, our current understanding of this diversity is limited, and the underlying mechanism contributing to this diversity remains unclear. Here, we completely assembled and compared the mitogenomes of three kiwifruit (Actinidia) species, which represent an early divergent lineage in asterids. We found conserved gene content and fewer genomic repeats, particularly large repeats (>1 kb), in the three mitogenomes. However, sequence transfers such as intracellular events are variable and dynamic, in which both ancestral shared and recently species-specific events as well as complicated transfers of two plastid-derived sequences into the nucleus through the mitogenomic bridge were detected. We identified extensive whole-genome rearrangements among kiwifruit mitogenomes and found a highly variable V region in which fragmentation and frequent mosaic loss of intergenic sequences occurred, resulting in greatly interspecific variations. One example is the fragmentation of the V region into two regions, V1 and V2, giving rise to the two mitochondrial chromosomes of Actinidia chinensis. Finally, we compared the kiwifruit mitogenomes with those of other asterids to characterize their overall mitogenomic diversity, which identified frequent gain/loss of genes/introns across lineages. In addition to repeat-mediated recombination and import-driven hypothesis of genome size expansion reported in previous studies, our results highlight a pattern of dynamic structural variation in plant mitogenomes through global genomic rearrangements and species-specific fragmentation and mosaic loss of intergenic sequences in highly variable regions on the basis of a relatively large ancestral mitogenome.
Keywords: Actinidia, asterids, genomic rearrangement, intracellular gene transfer, mitochondrial evolution, structural variation
Introduction
The evolutionary diversity of angiosperm mitochondrial genomes (mitogenomes) remains elusive despite the complete sequencing of nearly 100 mitogenomes (supplementary table S1, Supplementary Material online). Angiosperm mitogenomes contain the largest and most complex genomes of any organelle, with extensive variations in both size and structure. The known size ranges from <100 kb (e.g., ∼66-kb mitogenome of Viscum scurruloideum) (Skippington et al. 2015) to over 10 Mb (e.g., 11.3 Mb for Silene conica) (Sloan et al. 2012), even between closely related species (Alverson et al. 2010; Mower et al. 2012; Gualberto and Newton 2017). The variations in genomic components are also dynamic, including extensive and lineage-specific loss or rearrangement of mitochondrial genes (Adams et al. 2002). So far, few angiosperms have been shown to contain the full set and order of ancestral plant mitochondrial genes (Adams and Palmer 2003).
Most of the size and structural variations in angiosperm mitogenomes are related to the uptake of foreign sequences, including plastid or nuclear DNA acquired via intracellular transfer or extrinsic mitochondrial DNA (mtDNA) acquired via horizontal transfer (Mower et al. 2012). In mitochondria, the plastid-derived sequences are generally carrying loss-of-function genes (Cummings et al. 2003; Sloan and Wu 2014) or functional tRNA genes (Miyata et al. 1998). However, numbers of nucleus-derived sequences in mitogenomes are poorly conserved across species, and the direction of sequence transfer is difficult to determine because these are generally noncoding and featureless (Notsu et al. 2002; Mower et al. 2012). The horizontal gene transfer (HGT) phenomenon also plays a major role in angiosperm mitochondrial evolution (Bergthorsson et al. 2003). As an extreme case occurring in Amborella, its mitogenome captured a large amount of extrinsic DNAs from diverse eukaryotes (Rice et al. 2013). Therefore, an import-driven hypothesis of mitogenome expansion in seed plants was suggested (Goremykin et al. 2012), although alternative evidence of low mutation rate or gain of entire chromosomes underlying increases in genome size has been reported (Alverson et al. 2010; Wu et al. 2015).
Although generally assembled as a single “master circle” molecule containing all genes, angiosperm mitogenomes have multifarious other structures (Sloan 2013). Linear-mapping molecules (e.g., CMS-S line of maize) (Allen et al. 2007), two (e.g., Saccharum officinarum) (Shearman et al. 2016), or three (e.g., Cucumis sativus) circular molecules (Alverson et al. 2011) and multichromosomal genomes (e.g., Silene noctiflora and Amborella) were frequently reported (Rice et al. 2013; Wu et al. 2015). Many of these structural variations reflect differences in repetitive DNA content, leading to intragenomic recombination and structural dynamism characterized by multiple subgenomic and/or isomeric forms (Gualberto and Newton 2017). Among these variations, crossing over and reciprocal exchanges across large (>1 kb) repeats play important roles in shaping the co-occurrence of multichromosomal genomes, whereas recombination across intermediate-sized (0.1–1 kb) and smaller repeats tend to be asymmetric due to substoichiometric shifting (Arrieta-Montiel et al. 2009; Davila et al. 2011). However, some multichromosomal conformations in plants are autonomous with little or no evidence of repeat-mediated recombination between genomes, and the underlying mechanism remains unclear (Sloan 2013).
The kiwifruit (Actinidia Lindl.) genus, which belongs to the Actinidiaceae family in Ericales, is an early divergent lineage within asterids (Byng et al. 2016). This genus consists of ∼54 species (Li et al. 2007) and is mainly distributed in central and southern China (Li et al. 2007). Despite recent progress in both nuclear and chloroplast genomic sequencing of kiwifruit species (Huang et al. 2013; Yao et al. 2015; Lin et al. 2018; Pilkington et al. 2018), information on kiwifruit mitogenomes remains limited (Liu et al. 2017). Notably, a recaptured HGT event involving the rps2 gene, which was absent in all other eudicots, was documented in Actinidia, with the suggested donor being a monocot (Bergthorsson et al. 2003). Currently, only a few mitogenomes of asterids were sequenced, including one sample of Vaccinium macrocarpon from the Ericales (Fajardo et al. 2014; Park et al. 2014). Further research on both the structure and evolutionary pattern of asterid mitogenomes is needed.
To elucidate the evolutionary mechanisms underlying plant mitogenomic diversity, one important approach is to conduct comparative analysis among closely related species. Here, we sequenced and assembled the complete mitogenomes of three economically and phylogenetically important kiwifruit species, Actinidiachinensis, Actinidiaarguta, and Actinidiaeriantha (Liu et al. 2017). Compared with A. arguta and A. eriantha, we found that the mitogenome of A. chinensis had a significantly larger genome size and a two-chromosomal conformation. We investigated the genomic repeats, RNA editing sites, and gene transfer events within the three kiwifruit mitogenomes and compared their genomic structures. We discussed and concluded that the observed variations between kiwifruit mitogenomes were well characterized by an evolutionary pattern of globally genomic rearrangements and extensive and species-specific mosaic loss of intergenic sequences within a highly variable region.
Materials and Methods
Plant Materials and Sequencing
We obtained plant materials of A. arguta, A. chinensis, and A. eriantha from the National Actinidia Germplasm Repository (NAGR; Wuhan, China). For both A. arguta and A. chinensis, intact and pure mitochondria were isolated as previously reported (Tanaka et al. 2004). The cetyltrimethylammonium bromide method (Doyle 1987) was used to extract mtDNA from purified mitochondria. Two different strategies were used for sequencing, in which both a 250-bp paired-end library and 3-kb mate-pair library were prepared and sequenced on an Illumina HiSeq 2500 platform for A. arguta, whereas the PacBio RS II long reads were used for A. chinensis. For A. eriantha, genomic DNA was extracted from fresh leaves, and both Illumina paired-end short reads and PacBio long reads were obtained. The raw Illumina reads were trimmed to exclude low-quality bases and adapter sequences using Trimmomatic v0.36 (Bolger et al. 2014).
Assembly and Verification of Mitogenomes
The A. arguta mitogenome was de novo assembled using SPAdes v3.10.1 (Bankevich et al. 2012). We performed multiple SPAdes runs with different k-mer values (k = 77, 101, and 127) and used QUAST (Gurevich et al. 2013) to evaluate and choose the best k-mer value of 127 for multiple assemblies. Finally, we identified only one candidate mitochondrial scaffold, which can be mapped as a circular molecule indicated by a pair of direct repeats at its both ends. We conducted Sanger sequencing to verify the connector and filled the seven remaining gaps in this scaffold (supplementary table S2, Supplementary Material online). We used the same approach to assemble the mitogenome of A. eriantha. For the A. chinensis mitogenome, the PacBio RS II reads were de novo assembled using Canu v1.4 (Koren et al. 2017), and the PacBio sequencing errors in the final assembly were fixed using Quiver (Chin et al. 2013). Two candidate mitochondrial contigs were identified by a BlastN (Camacho et al. 2009) search, and these were further verified by Sanger sequencing (supplementary table S2, Supplementary Material online).
To verify the quality and accuracy of our assemblies, the original reads were mapped back to the corresponding mitogenomes using BWA v0.7.16 (Li 2013), and the depth of read coverage was assessed via SAMtools v1.5 (Li et al. 2009). The Pilon v1.22 (Walker et al. 2014) was used to polish the final assemblies, including repair of a total of 11 mismatches and one deletion of bases in the A. eriantha mitogenome, and 25 deletions and three insertions of bases in the A. chinensis mitogenome. We further designed polymerase chain reaction (PCR) primers to confirm the regions with relatively low read coverage and randomly selected regions (supplementary table S2, Supplementary Material online).
Gene Annotations
Mitochondrial protein-coding genes were predicted using the MITOFY webserver (Alverson et al. 2010). The tRNA and rRNA genes were identified using tRNAscan-SE v1.21 (Lowe and Eddy 1997) and RNAmmer 1.2 Server (Lagesen et al. 2007), respectively. We manually revised the start/stop codons and the exon–intron boundaries of genes. ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to analyze open reading frames (ORFs) longer than 300 bp within the intergenic sequences, and BlastN (Camacho et al. 2009) was used to identify repeats with >95% identity in each mitogenome. We investigated the Guanine-Cytosine (GC) content with a shell script and visualized the circular physical map of all mitogenomes using Circos v0.69 (Krzywinski et al. 2009).
Identification of RNA Editing Sites
We predicted RNA editing sites in the three kiwifruit mitogenomes using both the PREP-Mt approach (Mower 2009) and the single nucleotide polymorphism information obtained by mapping RNA-Seq reads. We downloaded RNA-Seq data (Illumina HiSeq2000 paired-end data) from the NCBI Short Read Archive under the accessions SRX1924007, SRX1924022, and SRX1924015 for A. arguta, A. chinensis, and A. eriantha, respectively (Wang et al. 2017). These RNA-Seq reads were then aligned to the assembled mitogenomes using HISAT2 v2.1.0 (Kim et al. 2015). The resulted files were converted into the binary alignment map files using SAMtools v1.5 (Li et al. 2009). RNA variants were called from the binary alignment map files using FreeBayes v1.2.0 (Garrison and Marth 2012) and those in the protein-coding regions, which represented the RNA editing sites, were extracted according to our gene annotation information. The RNA editing level for each site was evaluated by the percentage of the edited reads compared with the all covering that site (Picardi et al. 2010). The effects of the variants (synonymous edits, nonsynonymous edits, and fatal edits) were annotated using SnpEff v4.3s (Cingolani et al. 2012).
Identification and Verification of Intracellular Gene Transfers
Plastid-derived sequences were identified by searching the mtDNA against the A. chinensis cpDNA (after removing one copy of the large inverted repeats) (Yao et al. 2015) using BlastN (Camacho et al. 2009) with an e-value of 1e-10, a minimum sequence identity of 80% and a minimum length of 100 bp as thresholds. The identified sequences were further verified using the PacBio long reads and/or Sanger sequencing of PCR amplicons (supplementary table S2, Supplementary Material online). Plastid-derived genes were annotated according to the annotation of A. chinensis cpDNA (Yao et al. 2015). Nuclear-shared sequences were identified by performing BlastN (Camacho et al. 2009) searches of mitogenomes against A. chinensis nucDNA (Pilkington et al. 2018) and filtering matches based on the thresholds of an e-value of 1e-50 and a minimum hit length of 100 bp.
Analysis of the rps2 Gene
The rps2 gene of A. arguta was used to search for homologous sequences in the NCBI nr database using BlastN. In order to construct a phylogenetic tree, the top 14 matched sequences of the rps2 gene were selected and downloaded from the GenBank (supplementary table S3, Supplementary Material online). Multiple sequence alignment was performed using the software MAFFT v7.310 (Katoh et al. 2002), and a phylogenetic tree was reconstructed using the maximum likelihood method in MEGA7 (Kumar et al. 2016) with 1,000 bootstrapping replicates.
Synteny and Phylogenetic Analysis
Locally collinear blocks (LCBs) among the three mitogenomes were analyzed using Mauve v2.0 (Darling et al. 2010). The Mauve backbone file was used for statistical analysis of homologous fragments. Dot plots were generated using NUCmer v3.1 (Kurtz et al. 2004). To investigate the phylogenetic distribution of both genes and introns during asterid evolution, we first reannotated all collected mitogenomes and the boundaries of genes and introns were corrected manually. Subsequently, the exons of 23 core protein-coding genes (atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFc, ccmFn, cob, cox1, cox2, cox3, matR, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9) were extracted from the 24 asterid mitogenomes and two species (Vitis vinifera and Nelumbo nucifera) serving as outgroups. Sequences were aligned with MAFFT v7.310 (Katoh et al. 2002). Gblocks v0.91b (Talavera and Castresana 2007) was used to eliminate poorly aligned positions. A supermatrix was constructed using SequenceMatrix v1.7.8 (Vaidya et al. 2011) to concatenate different genes, resulting in a final alignment of 23,235 positions. The Akaike information criterion was used in jModelTest v2.13 (Darriba et al. 2012) to compare models of character evolution in the supermatrix. A maximum likelihood–based phylogenetic tree of the concatenated supermatrix was constructed using RAxML v8.2.10 with 1,000 bootstraps (Stamatakis 2014).
Results
The A. chinensis Mitogenome Is Comparatively Large and Consists of Two Mapped Chromosomal Genomes
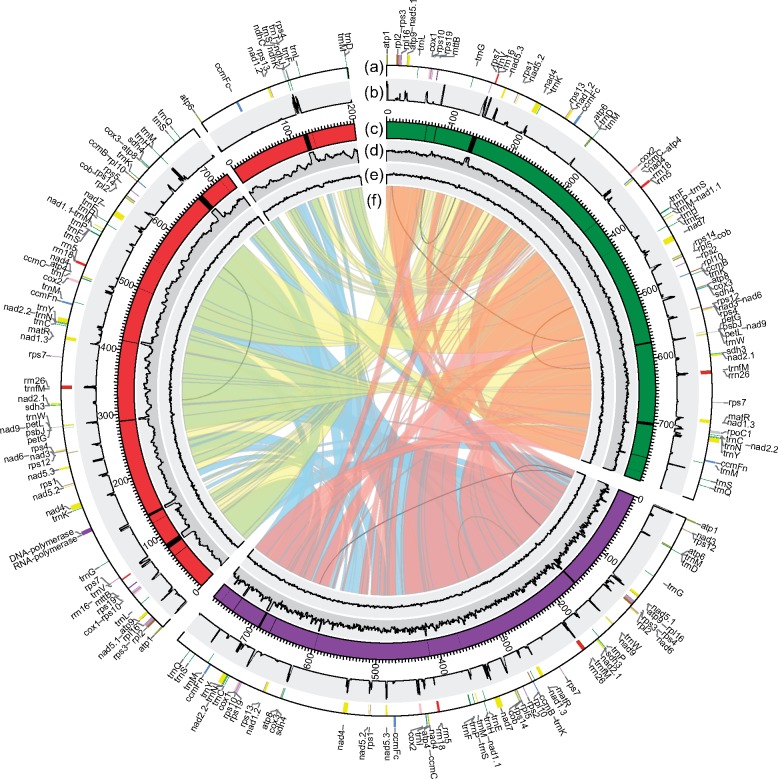
We sequenced the kiwifruit mitogenomes by three slightly different strategies and evaluated the mean depth of reads on each mitogenome respectively (supplementary table S4, Supplementary Material online). The de novo genome assembly resulted in a single circular-mapping chromosome for each of A. arguta (792 kb) and A. eriantha (773 kb), whereas two distinct circular chromosomal genomes (724 and 201 kb) were assembled for A. chinensis (fig. 1 and table 1). The total genome size of A. chinensis (925 kb) was significantly larger than those of A. arguta and A. eriantha. Moreover, a slightly lower GC content was observed in A. chinensis (∼45.87%; table 1), whereas both A. arguta and A. eriantha mitogenomes exhibited similar GC contents (46.22% vs. 46.16%).
Fig. 1.
—Circular diagram showing the information on kiwifruit mitogenomes. The Actinidia chinensis (red), Actinidia eriantha (purple) and Actinidia arguta (green) mitogenomes are circular but are shown here as linear starting from the atp1 gene (from the atp6 gene in the small chromosome of Actinidia chinensis). (a) Gene blocks shown on the outside and inside the circle were transcribed clockwise and counter clockwise, respectively. Genes from the same complex are similarly colored. (b) RNA-Seq depth ratio* supporting the annotated protein-coding genes. (c) Plastid-derived fragments characterized by the black blocks inlaid in the karyotypes. (d) DNA-Seq depth ratio* supporting the assemblies and identified plastid-derived regions. (e) The GC content in 1,000-bp windows. (f) The colored bands in the center show links between syntenic blocks among the three mitogenomes. The inner repeats in each mitogenome (>300 bp) are represented by black lines. *The depth ratio = average depth in 100-bp windows/average total depth.
Table 1.
Summary of Features in Kiwifruit Mitogenomes
| Feature | Actinidia arguta | Actinidia eriantha | Actinidia chinensis |
|---|---|---|---|
| Genome size (bp) | 792,319 | 772,626 | chr1: 724,257; chr2: 200,567 |
| Number of chromosomes | 1 | 1 | 2 |
| GC content (%) | 46.22 | 46.16 | chr1: 45.9%; chr2: 45.80% |
| Length of coding region (%) | 33,192 (4.19%) | 33,192 (4.30%) | 33,192 (3.59%) |
| Length of tRNA genes (%) | 1,800 (0.23%) | 1,657 (0.21%) | 2,261 (0.24%) |
| Length of rRNA genes (%) | 5,718 (0.72%) | 5,713 (0.74%) | 5,702 (0.62%) |
| Length of cis-spliced introns (%) | 27,857 (3.52%) | 27,516 (3.56%) | 27,857 (3.01%) |
| Length of intergenic sequences (%) | 725,720 (91.59%) | 706,516 (91.44%) | 857,780 (92.75%) |
| Length of repeats (%) | 29,833 (3.77%) | 25,247 (3.27%) | 32,855 (3.55%) |
| Longest repeat (bp) | 636 | 483 | 337 |
| Length of plastid-derived sequences (%) | 18,628 (2.35%) | 8,937 (1.16%) | chr1: 29,657; chr2: 7,826 |
| Length of nuclear-shared sequences (%) | 323,621 (40.84%) | 315,920 (40.89%) | 388,517 (42.01%) |
| Number of rRNA genes (native) | 3 | 3 | 3 |
| Number of rRNA genes (plastid derived) | 1 | 0 | 1 |
| Number of tRNA genes (native) | 22 | 21 | 23 |
| Number of tRNA genes (plastid derived) | 2 | 1 | 7 |
| Number of protein genes (native) | 39 | 39 | 39 |
| Number of protein genes (plastid derived) | 5 | 3 | 12 |
| Number of gene acquired by HGT | 1 | 1 | 1 |
| Total genes | 74 | 69 | 89 |
Note.—chr1, large chromosome of A. chinensis mitogenome; chr2, small chromosome of A. chinensis mitogenome.
To ensure the correct assembly of the two chromosomes in A. chinensis, we first verified the circularization of each chromosome using PCR amplification (supplementary table S2, Supplementary Material online). Moreover, we identified repetitive sequences (see details in the next section) and found no recombining repeats presented between them. This result was further supported by mapping of the PacBio long reads on both chromosomes, which showed rare read interactions across the two chromosomes. We then investigated the sequencing depth on each chromosome using both PacBio- and MiSeq-obtained reads (not used for assembly), resulting in similar sequencing depths for them (supplementary table S4, Supplementary Material online). This was consistent with the presence of the mitochondrial genes on both chromosomes (fig. 1a). Finally, we assembled another version of the A. chinensis mitogenome using previously published resequencing data (Liu et al. 2017). Eight mitochondrial candidate contigs were filtered from the assembly results, and synteny analysis of two assemblies supported the two-chromosomal conformations in A. chinensis (supplementary fig. S1, Supplementary Material online). These collectively provided strong evidence for the presence of a relatively large genome and two mitochondrial chromosomal genomes in A. chinensis.
Conservation of Gene Content and Lack of Large Genomic Repeats
We annotated functional regions of protein-coding genes, rRNA, and tRNA genes, accounting for 7.25–8.56% of the three mitogenomes, whereas more than 90% of the sequences belonged to intergenic regions (table 1). The kiwifruit mitochondrial gene contents revealed the evolutionary constant of native genes, in which a set of 39 protein-coding genes, 3 rRNA genes, and 21–23 tRNA genes were shared (fig. 1a and supplementary table S5, Supplementary Material online). All protein-coding genes and rRNA genes were single copies and highly conserved, and all tRNA genes produced the same 15 amino acids despite the tRNA-Lys and tRNA-Leu gene copies varying across the three species (supplementary table S5, Supplementary Material online). We further identified 23 group II introns (18 cis- and 5 trans-spliced introns) within 10 intron-containing genes (supplementary table S5, Supplementary Material online), while no group I introns were detected, including the cox1 gene group I intron that was widely distributed across angiosperms (Sanchez-Puerta et al. 2008). Interestingly, both chromosomes of the A. chinensis mitogenome had protein-coding capability, in which three intact protein-coding genes (atp6, ccmFc, and rps13) and three tRNA genes (tRNA-Asp, tRNA-Leu, and tRNA-Met) were annotated on the small chromosome, whereas the nad1 gene was separated by the two chromosomes (fig. 1a).
We also identified several large ORFs (>300 bp) in the three kiwifruit mitogenomes (supplementary table S6, Supplementary Material online). Among these, two ORFs, named Ac_RNA_pol (1,743 bp) and Ac_DNA_pol_B (2,814 bp), were unique to the A. chinensis mitogenome. These have two conserved domains, namely, pfam00940 (encoding a DNA-dependent RNA polymerase) and pfam03175 (a type B DNA polymerase), and no kiwifruit nuclear genomic reads can cover the two ORFs. However, we found their full-length homologous sequences in the mitogenomes of Clematoclethra lasioclada (a species in the sister genus of Actinidia within the Actinidiaceae family) and V.macrocarpon (a species commonly belonging to the Ericales) and in some other higher plants such as Populus tremula, Hesperelaea palmeri, and Olea europaea (supplementary table S7, Supplementary Material online). The phylogenetic analysis of these sequences (supplementary fig. S2, Supplementary Material online) suggested that both ORFs were present in the ancestral mitogenome before the divergence of Actinidia from other Ericales, and its species-specific loss occurred in both A. arguta and A. eriantha. No transcripts of the two ORFs were detected in our RNA-Seq data (fig. 1b), suggesting that they are pseudogenes or are only transcribed under special conditions.
Remarkably, the three mitogenomes have fewer genomic repeats (∼3.27–3.77% of the three mitogenomes), particularly the lack of large repeats (>1 kb) (supplementary table S8, Supplementary Material online). The largest repeats identified in the A. arguta, A. eriantha, and A. chinensis mitogenomes are only 636, 483, and 337 bp in size, respectively, and the majority of the repeats (77–80%) are only 50–99 bp in length, accounting for 1.93–2.27% of the genomes (supplementary table S8, Supplementary Material online). Although the A. chinensis mitogenome has slightly more repeats (based on total length) compared with the other two species, these repeats could not have contributed to the increase in mitogenome size of A. chinensis (supplementary table S8, Supplementary Material online).
Abundance and Efficiency of RNA Editing
About 80% of the RNA editing sites predicated by the PREP-Mt approach and the RNA-Seq data are the same (supplementary tables S9–S11, Supplementary Material online). Moreover, a total of 528 sites were shared among the three kiwifruit species on the basis of the RNA-Seq information, whereas the rest were unique to one or two of them, suggesting that species specificity of RNA editing exists even between the mitogenomes of closely related species. Notably, all editing sites showed a cytidine to uridine (C-to-U) transition, and no “reverse” U-to-C editing was detected (supplementary table S11, Supplementary Material online).
Approximately 90% of the editing sites had an editing frequency >50% (supplementary fig. S3a, Supplementary Material online), and 90–93% of the editing sites occurred at the first two codon positions (supplementary fig. S3b, Supplementary Material online). The most frequent amino acid changes modified by RNA editing were serine-to-leucine (S-to-L), proline-to-leucine (P-to-L), and serine-to-phenylalanine (S-to-F) (supplementary fig. S3c, Supplementary Material online). Moreover, the ribosomal protein genes tended to be less edited than the other genes (supplementary fig. S3d, Supplementary Material online). We detected five fatal RNA editing sites in each of the three kiwifruit mitogenomes, including CGA-to-UGA and CAG-to-UAG transitions in the atp9, ccmFc, rpl16, and rps10 genes and a CAA-to-UAA transition in the atp6 gene. These transitions caused premature stop codons and regulated gene expression.
Integration of Foreign DNA Fragments through Diversified Sequence Transfers
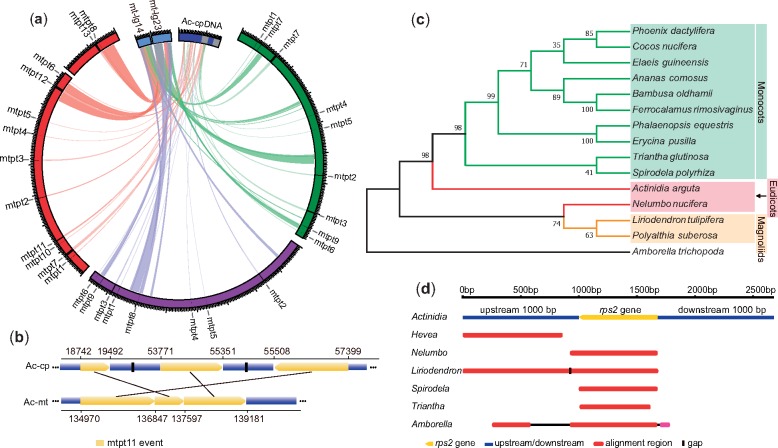
We observed multiple plastid-derived and nuclear-shared sequence transfer events in the three kiwifruit mitogenomes (table 1). The plastid-derived sequences comprised inverted repeat and long single copy regions in the A. chinensis plastid genome (cpDNA) (Yao et al. 2015) and covered 5.7–24.0% of the cpDNA (fig. 2a). Comparatively, the A. chinensis mitogenome had the largest insertions of plastid-derived sequences (∼37 kb, 78% located in the large chromosome) (table 1) as well as the maximum number of plastid-derived genes (12 protein-coding genes, 7 tRNA genes, and 1 rRNA gene) (supplementary table S12, Supplementary Material online). According the coordinates of each insertion into the respective mitochondrial and plastid genomes, we identified a total of 13 plastid-derived sequence (also called mtpt for mitochondrial plastid DNA) transfer events, of which six transfer events (named mtpt1 to mtpt6 in supplementary table S12, Supplementary Material online) were shared among the three species (fig. 2a). Interestingly, the difference in the insertion size of the mtpt3 event was the result of a 1.2-kb deletion in both the A. chinensis and A. eriantha mitogenomes (supplementary table S12, Supplementary Material online). Given that A. arguta is an earlier divergent lineage within the Actinidia genus (Liu et al. 2017), this deletion possibly occurred after the divergence of A. arguta from the common ancestor of A. chinensis and A. eriantha. Three transfer events (mtpt7, mtpt8, and mtpt9) were detected in only two of the three mitogenomes, and four events (from mtpt10 to mtpt13) were unique to A. chinensis (supplementary table S12, Supplementary Material online). Notably, the mtpt11 was derived from multiple-step integrations of plastid-derived sequences (fig. 2b and supplementary table S12, Supplementary Material online).
Fig. 2.
—Gene transfer events in kiwifruit mitogenomes. (a) IGT events in kiwifruit mitogenomes. The mtDNA of Actinidia chinensis (red), Actinidia eriantha (purple), and Actinidia arguta (green). The cpDNA of A. chinensis (Ac-cpDNA; blue) and related nuclear fragments (mt-lg23, mt-lg14; light blue) are depicted with a circular diagram. (b) The mtpt11 event revealed a complex intracellular gene transfer event between the A. chinensis mitogenome and cpDNA. (c) The phylogenetic tree of the Actinidia rps2 gene and homologous genes from 14 species. (d) The top six BLAST results using the Actinidia rps2 gene and its upstream/downstream 1,000-bp flanking sequences against the NCBI nr database.
About 40% of each mitogenome have homologous sequences in the nuclear genome (nucDNA) (table 1). The shared nuclear-mitochondrial sequences on two nuclear chromosomes, LG14 and LG23, were significantly longer than the others (supplementary fig. S4, Supplementary Material online), of which two large fragments (47 and 68 kb in size) were highly homologous to sequences located in the two mitochondrial chromosomes of A. chinensis, but the corresponding sequences in both the A. arguta and A. eriantha mitogenomes were truncated and then dispersed across the mitogenome (fig. 2a and supplementary table S13, Supplementary Material online). Although no characteristic features (genes or transposable elements) presented to determine the direction of sequence transfers, the lack of rearrangements in both large fragments (named mt-lg14 and mt-lg23) in the A. chinensis mitogenome suggested their transfers from the A. chinensis mitogenome to the nucDNA. Furthermore, two plastid-derived sequences (mtpt12 and mtpt13) unique to the A. chinensis mitogenome may also have been retransferred into the nucDNA through a “hitchhiking” effect because mtpt13 was adjacent to the edge of mt-lg14, and mtpt12 was located within mt-lg23 (fig. 2a).
We investigated the classical rps2 gene HGT event in Actinidia (Bergthorsson et al. 2003) on the basis of the complete mitogenomes assembled. We found that all rps2 genes and the large flanking sequences in the three kiwifruit mitogenomes are highly homologous. Notably, these rps2 genes contain the same internal stop codon without transcription, which indicates that the rps2 gene in Actinidia is a pseudogene. Further analysis based on resequencing data (Liu et al. 2017) showed that this gene is present in all Actinidia species but is absent in closely related species C.lasioclada (Actinidiaceae), suggesting that an Actinidia-specific HGT event occurred in their common ancestor. To trace the HGT event of the rps2 gene in Actinidia, we searched for the A. arguta rps2 gene against the NCBI nr database, resulting in 14 corresponding rps2 gene sequences that were used for phylogenetic reconstruction (supplementary table S3, Supplementary Material online). This provided strong support for the Actinidia rps2 gene as a sister group to all monocots (fig. 2c). We also extracted the plus/minus 1-kb flanking sequences of the rps2 gene from the A. arguta mitogenome (total length of 2,678 bp) to further search for homologous sequences. The top six hits showed high homology between the Actinidia rps2 gene (and the upstream 1,000-bp sequence) and the two fragments in the Liriodendron tulipifera mitogenome from magnoliids (fig. 2d).
Abundant Rearrangements and Local Mosaic Losses of Intergenic Sequences
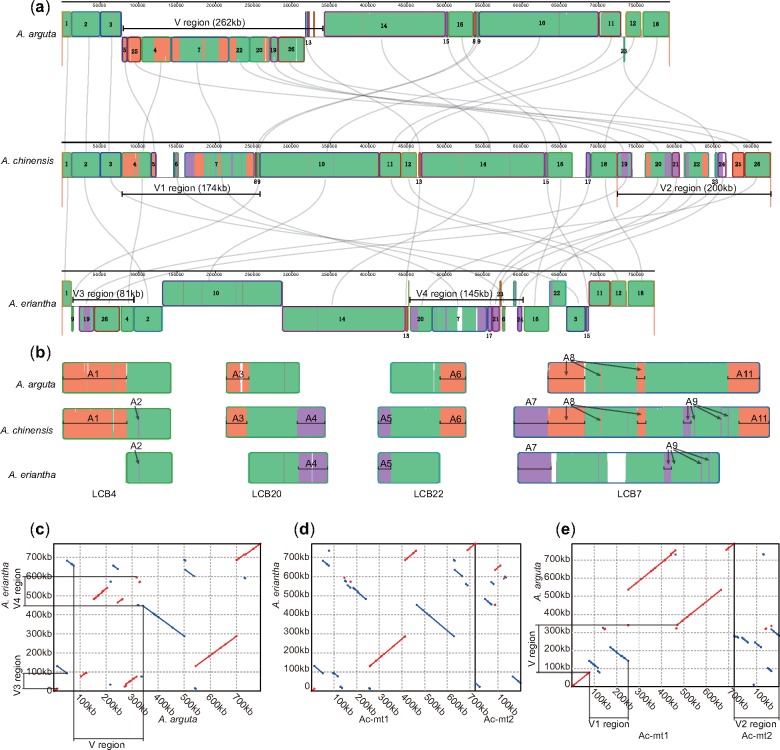
We examined the structural variations among kiwifruit mitogenomes using Mauve analysis (Darling et al. 2010). By setting the A. chinensis mitogenome as a reference, a total of 26 large LCBs (>5 kb) were identified, and numerous rearrangements and indels were detected (fig. 3a). The interstices between LCBs represented the unique fragments in each mitogenome. Comparatively, A. chinensis carried more unique fragments, which may have partially contributed to the increase in genome size (fig. 3a). We extracted four chimeric LCBs (LCB4, LCB7, LCB20, and LCB22) for further analysis (fig. 3b). We set markers to indicate those regions shared between only two of three kiwifruit species. In LCB4, region A1 was absent in A. eriantha, and region A2 was not detected in A. arguta, but both regions were present in A. chinensis. Similar patterns were observed in both LCB20 and LCB22 blocks (fig. 3b). LCB7 revealed more complicated structural variations due to scattered simultaneously deleted regions (e.g., regions A8 and A9; fig. 3b). Overall, the A. chinensis mitogenome carried characterized regions from both A. arguta and A. eriantha. This was further supported by mapping Illumina paired-end reads of A. arguta and A. eriantha to the A. chinensis mitogenome, which indicated that the identified interstices and mosaic LCBs were not the result of artificial assembly (supplementary fig. S5, Supplementary Material online).
Fig. 3.
—The structural comparison of kiwifruit mitogenomes. (a) Genome alignment of the three mitogenomes. Conserved LCBs among the three species are shown in green. Regions conserved only among subsets of the three mitogenomes are color coded. Specifically, the LCBs conserved between Actinidia arguta and Actinidia chinensis are orange; the LCBs conserved between Actinidia eriantha and Actinidia chinensis are purple; and the LCBs conserved between Actinidia arguta and Actinidia eriantha are red. (b) The details show the evolution of four chimeric LCBs. The markers inlaid in the LCBs indicate homologous fragments between two of the three kiwifruit species. (c–e) Synteny analysis of pairs of the three mitogenomes.
Statistical analysis of homologous fragments at the whole-genome level among the three mitogenomes was performed for further verification (supplementary fig. S6, Supplementary Material online). Obviously, fewer unique sequences were detected in the A. arguta (31 kb) and A. eriantha (29 kb) mitogenomes than in the A. chinensis mitogenome (86 kb; supplementary fig. S6, Supplementary Material online). Interestingly, although A. arguta and A. eriantha had similar genome sizes (792 and 773 kb), only 661 kb of these sequences were homologous sequences. More homologous sequences were shared between A. arguta and A. chinensis (756 kb) and between A. eriantha and A. chinensis (739 kb). If the unique sequences of A. arguta and A. eriantha were regarded as independent evolution after the divergence of the three species, then the A. chinensis mitogenome used to contain almost the entire mitogenomes of A. arguta and A. eriantha.
Dot plots further illustrated the structural variations in the kiwifruit mitogenomes. The identity of syntenic regions was extremely high, with an average identity of 99.3%, but substantial rearrangements existed among the three mitogenomes (fig. 3c–e). Clearly, the mitogenomes between A. eriantha and A. arguta underwent at least 11 large-scale rearrangements (fig. 3c), whereas 13 and 9 rearrangements occurred between the mitogenomes of A. eriantha and A. chinensis (fig. 3d) and between those of A. arguta and A. chinensis (fig. 3e). There were more similar structures between A. chinensis and A. arguta than the other comparisons. Except for one large fragment rearrangement, the structural differences between A. arguta and A. chinensis were mainly related to a 262-kb variable region (called the V region) in the A. arguta mitogenome (fig. 3a and e). The V region in A. arguta corresponded to the V1 (a 174-kb region in the large chromosome) and V2 regions (almost the entire small chromosome, ∼200 kb) in A. chinensis, and the V3 (81-kb) and V4 (145-kb) regions in A. eriantha (fig. 3c and e). The V1 and V2 regions of A. chinensis contained almost all the chimeric LCBs, except for LCB17 and fragments in its left interstice, which might have been transposed from the V1 or V2 region (fig. 3a). The V region therefore explained most of the main structural and size differences among the three kiwifruit mitogenomes.
Comparison of Actinidia Mitogenomes with Those of Other Asterids
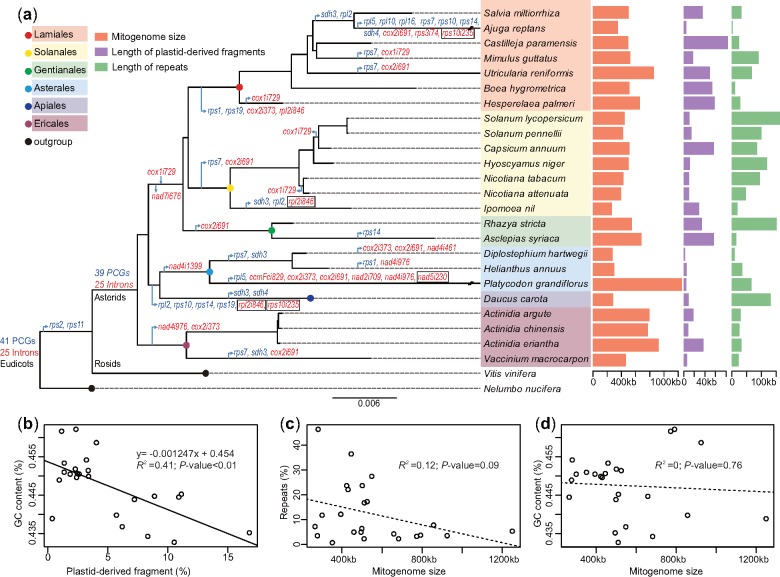
We investigated mitochondrial evolution in asterids by comparing the three Actinidia mitogenomes with 21 others in this clade. The mitogenomes of asterid species exhibited extensive variations in genome size (265–1,249 kb), GC content (43.27–46.22%), plastid-derived fragments (0.41–16.93%), and repeats (0.75–46.40%; fig. 4a and supplementary table S14, Supplementary Material online). Comparatively, the kiwifruit mitogenomes had a larger size and higher GC content but fewer repeats (supplementary table S14, Supplementary Material online). Theoretically, the GC content of the mitogenome is affected by both plastid- and nucleus-derived fragments. Notably, we found a distinct correlation between GC content and the percentage of plastid-derived fragments (fig. 4b). However, no correlation between the percentage of repeats and mitogenome size was observed (fig. 4c), and no relationship was detected between GC content and mitogenome size (fig. 4d).
Fig. 4.
—The comparative analysis of asterid mitogenomes. (a) The phylogenetic distribution of genes and introns and genomic features of asterid mitogenomes. The phylogenetic tree of asterids was constructed based on the concatenated coding regions of 24 core protein-coding genes (PCGs) with Vitis vinifera and Nelumbo nucifera as outgroups. Genes and introns are presented in red and blue, respectively. The arrow represents the loss or gain of a gene or intron from the corresponding clade. The framed introns are those in which their absence accompanied the loss of the corresponding gene. (b–d) The linear regression analysis between GC content and length of plastid-derived fragment, between mitogenome size and length of repeats, and between GC content and mitogenome size, respectively.
Phylogenetic reconstruction indicated that lineage-specific gains, losses, or HGTs of mitochondrial genes and introns frequently occurred during asterid evolution, leading to only 28 protein-coding genes and 15 group II introns being shared among the 24 species investigated (fig. 4a, supplementary fig. S7 and supplementary table S15, Supplementary Material online). Among these, Ajuga reptans has the smallest gene set due to the loss of seven genes and three introns (fig. 4a, supplementary fig. S7, Supplementary Material online). The cis-splicing nad1i728 intron, which was inferred to be present in the ancestral mitogenome of asterids, possessed an intron-encoded matR gene (Bonen 2008). In the majority of asterids (18/24), this intron had been converted into a trans-splicing intron, and the matR gene was also retained (supplementary fig. S7, Supplementary Material online). The group I cox1i729 intron was evidently absent from most asterid species, including the kiwifruit species investigated here. The presence of this intron in nine evolved asterid species therefore suggests potential HGT events (supplementary fig. S7, Supplementary Material online). The synteny analysis of the cox1i729 introns from the nine species showed high homology (supplementary fig. S8, Supplementary Material online), which indicates that the HGT of cox1i729 intron possibly occurred in their common ancestor, followed by loss of this intron in some species, such as Nicotiana, Capsicum, Solanum, Hesperelaea, and Mimulus (fig. 4a). This scenario differs from the independent captures of the cox1i729 intron proposed in Park et al. (2014).
Discussion
Gene Content and Genomic Repeats Do Not Contribute to the Size and Structural Variations between Kiwifruit Mitogenomes
We completed the de novo assembly of three kiwifruit mitogenomes and found extensive size and structural variations (table 1 and fig. 3). Among the observed size variations, the mitogenome sizes of A. arguta and A. eriantha were similar to those of two remotely related species, Vi.vinifera (773 kb) (Goremykin et al. 2009) and Populus tremula (783 kb) (Kersten et al. 2016), but the genome size of A. chinensis was almost twice that of the closely related species V.macrocarpon (459 kb) (Fajardo et al. 2014) (supplementary table S1, Supplementary Material online). Overall, the sizes of the three Actinidia mitogenomes were >90% (84/92) of those of sequenced flowering plants (supplementary table S1, Supplementary Material online). At the whole-genome level, the native gene constituents of kiwifruit mitogenomes are highly conserved, and the majority of the mitogenomic variations are presented in the intergenic regions, which is similar to that of the Vi.vinifera mitogenome (Goremykin et al. 2009).
Generally, large repeats can be involved in frequent homologous recombination (Arrieta-Montiel et al. 2009; Davila et al. 2011), which generates dynamic structural variations and extreme mitogenome sizes (Alverson et al. 2010). In the present study, the observation of a low number of genomic repeats (∼3.27–3.77% in the three mitogenomes), particularly the lack of large repeats (>1 kb) (supplementary table S8, Supplementary Material online) indicates that these may not be the main contributor to the observed size and structural variations.
Identification of RNA Editing Sites Based on RNA-Seq Data
Mitochondrial RNA editing in higher plants is related to potentially molecular functions and physiological processes (Yang et al. 2017; He et al. 2018). Initially, RNA editing sites were identified by comparing cDNA sequences with their corresponding templates (Takenaka and Brennicke 2007). Subsequently, a strategy for investigating RNA editing using RNA-Seq short reads was proposed (Picardi et al. 2010). Here, we similarly developed a simple variant-calling pipeline by mapping RNA-Seq reads onto the three kiwifruit mitogenomes to investigate their RNA editing sites. The sites identified by our approach was comparative to those predicated by the PREP-Mt method (Mower 2009), with a great improvement for examining species-specific RNA editing sites between closely related species. Overall, the RNA editing sites in Actinidia were slightly more than those inferred in other plants such as Vi.vinifera (401) (Picardi et al. 2010) and Citrullus lanatus (463) (Alverson et al. 2010), but the most frequent amino acid changes modified by RNA editing (supplementary fig. S3c, Supplementary Material online) were consistent with those found in both Arabidopsis thaliana (Giege and Brennicke 1999) and Vi.vinifera (Picardi et al. 2010).
Multiple Intracellular Sequence Transfer Events Diversifying Kiwifruit Mitogenomes
The plastid-derived sequences occupy 3–6% of the known angiosperm mitogenomes (Mower et al. 2012). This was also observed among the closely related kiwifruit species (table 1), in which plastid-derived fragments were frequently integrated into the mitogenomes (supplementary table S12, Supplementary Material online). The observed sequence transfer events further reflected an ongoing process in relation to continuous DNA integrations, in which ancestrally shared events and recently species-specific ones simultaneously occurred in kiwifruit (supplementary table S12, Supplementary Material online). Considering both plastid and nuclear DNA transfers in the plant mitogenome, five possible directions have already been reported in other studies (supplementary fig. S9, Supplementary Material online) (Nakazono and Hirai 2008; Smith 2014). Interestingly, we hereby present evidence that the mitochondrial sequences with integrated plastid-derived fragments were transferred into the nucleus of A. chinensis (Pilkington et al. 2018) via route A (fig. 2a and supplementary fig. S9, Supplementary Material online), which provides insights into the complex intracellular transfers between plastid and nuclear genomes through a mitochondrial bridge that occurred during plant mitochondrial evolution.
The previous investigation on the rps2 gene-related HGT event (Bergthorsson et al. 2003) suggested that the possible donor of the rps2 gene in kiwifruit mitogenomes was derived from monocots. Our phylogenetic analysis also supported the Actinidia rps2 gene as a sister group to all monocots (fig. 2c). Moreover, our analysis based on the flanking sequences of the rps2 gene provided additional clues of the homology between the kiwifruit rps2 gene and those of the Liriodendron lineage, suggesting a potentially complex origin of the kiwifruit rps2 gene. Collectively, the sequence transfers, particularly the intracellular events, have substantially diversified kiwifruit mitogenomes to some degree, which is indicated in the comparison of both A. chinensis and A. eriantha mitogenomes, where about 28 kb of the total 152 kb difference between them are plastid-derived fragments.
Global Genomic Rearrangements and Local Mosaic Loss of Intergenic Sequences Are the Main Contributors of the Observed Structural Variations
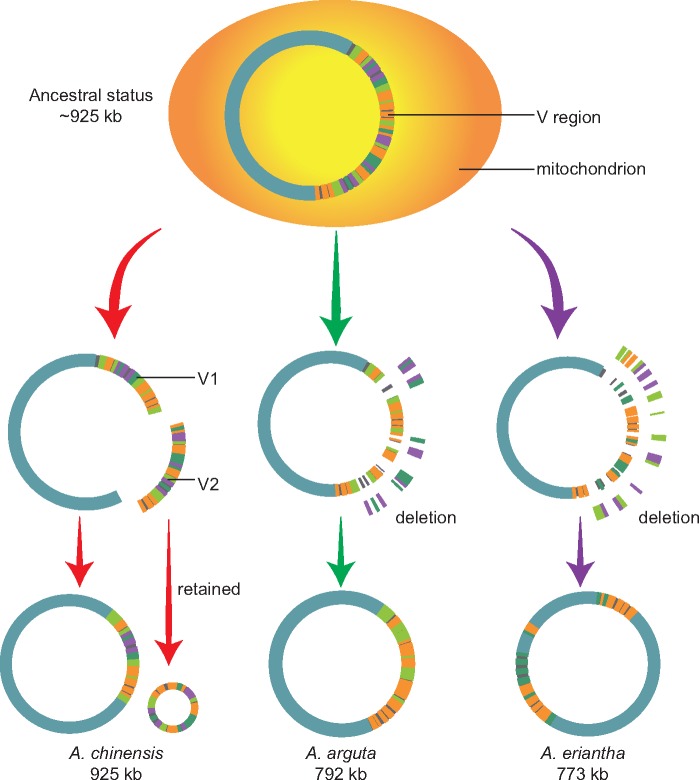
The relatively large genome size and the presence of two-chromosomal genomes in the A. chinensis mitogenome were surprising. Comparatively, the A. chinensis mitogenome contained almost the full combination of mitogenome components of A. arguta and A. eriantha, whereas it lacked the sequences unique to both A. arguta and A. eriantha (<5 kb, excluding the region shared by the three species) (supplementary fig. S6, Supplementary Material online). Besides the observed genomic rearrangements at the whole-genome level, we identified a highly variable genomic region (fig. 3e) that contributes to the main structural variations detected among the three kiwifruit mitogenomes. We additionally assembled seven draft mitogenomes from previous resequencing data (supplementary table S16, Supplementary Material online). We found that the size of these draft genomes was ∼820 kb, and all of these were smaller than that of A. chinensis (925 kb). When the corresponding V region in each genome was excluded, the variations in mitogenome size were greatly reduced (standard deviation = 13 kb; supplementary table S16, Supplementary Material online). Moreover, all seven species showed less sequence sharing with A. arguta or A. eriantha compared with that with A. chinensis (supplementary table S16, Supplementary Material online). Together, these results suggest that the mitogenome of A. chinensis is more closely related to the ancestral state of the Actinidia mitogenome. This could be further evidenced by the two identified ORFs (Ac_RNA_pol and Ac_DNA_pol_B) that are present in A. chinensis and its sister lineages of C.lasioclada and V.macrocarpon in the Ericales and other higher plants (supplementary table S7 and supplementary fig. S2, Supplementary Material online).
Under this hypothesis, a dynamic process of evolution and diversification of kiwifruit mitogenomes and the origin of two-chromosomal genomes in A. chinensis could be inferred (fig. 5). First, the ancestral mitogenome of Actinidia was regarded to consist of a single circular chromosome with an approximate size of 925 kb like A. chinensis and a large V region. Subsequently, during the evolution and divergence of Actinidia species, the V region underwent several mosaic deletions of intergenic sequences accompanied by few insertions, and the whole genome underwent a couple of rearrangements, except for that of A. chinensis, the fragments of which were largely retained because these included several crucial genes (fig. 5). In this process, the ancestral mitogenome lost ∼130–150 kb of sequences, leading to generation of A. eriantha and A. arguta mitogenomes with a few species-specific insertions. (supplementary fig. S6, Supplementary Material online). Remarkably, during the evolution of A. chinensis, the small chromosome was isolated from the V region in the ancestral mitogenome, which resulted in coexistence of two chromosomes (fig. 5). The reality of this evolutionary process may have been more complicated than our inference, but repeat-driven recombination contributed less to the production of two chromosomes in A. chinensis due to the lack of large recombining repeats in its mitogenome (supplementary table S8, Supplementary Material online). Our results thus revealed that the abundant mosaic loss of intergenic sequences in a highly variable region and the rearrangements at the whole-genome level served as the main driving force for the evolution and diversification of kiwifruit mitogenomes.
Fig. 5.
—Putative dynamic evolution and diversification of three kiwifruit mitogenomes.
The Evolution of Asterid Mitogenomes
Although a comparative analysis of eight mitogenomes in asterids was previously performed (Park et al. 2014), our understanding of the overall features of asterid mitochondrial evolution is poor. In the present study, our combined analysis of 24 asterid mitogenomes showed significant differences in genome sizes, repeats, and plastid-derived fragments (fig. 4a). Much of the variation in the mitogenome size can be attributed to the different amounts of noncoding DNA. The import-driven hypothesis has been suggested to explain mitogenomic size expansion in seed plants (Goremykin et al. 2012). In our study, we found a clear correlation between the GC content of mitogenomes and the percentage of plastid-derived fragments in asterid plants (fig. 4b). To our knowledge, the GC content of either chloroplast genome or nuclear genome is about 37% (supplementary table S17, Supplementary Material online), whereas that for most of the mitogenomes is ∼45% (supplementary table S1, Supplementary Material online). If the import-driven hypothesis is validated in asterids, we would expect decreased GC content in the expanded mitogenomes. However, the linear relationship between the GC content and genome size was not observed among the asterid mitogenomes (fig. 4d).
Furthermore, our study showed that the kiwifruit mitogenomes were larger than 90% of the size of other sequenced plant mitogenomes and possessed the highest GC content in asterid mitogenomes (supplementary tables S1 and S13, Supplementary Material online). Unless a specific import mechanism of foreign fragments occurred (e.g., selective uptake of high-GC content DNA fragments), the import-driven hypothesis may not be a main driving force on the mitogenome size of kiwifruit plants. Our conclusion of the extensive rearrangements and the mosaic loss of noncoding sequences from a large ancestral mitogenome was slightly similar to the intraspecific mitogenomic variations in Silene noctiflora, wherein mitogenomes evolved via gains or losses of entire chromosomes (Alverson et al. 2010; Wu et al. 2015). In addition, the described picture of the variation and evolution of mitochondrial genes and introns in asterids is also consistent with this pattern of species- or lineage-specific gains/losses in large ancestral mitogenomes (fig. 4a). Therefore, under this hypothesis of mitogenomic evolution and diversification mechanism, the kiwifruit mitogenomes may represent the ancestral state of the asterid mitogenome because they contain the largest genomes and the most comprehensive set of genes among asterid species.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Ying Wang from the Wuhan Benagen Tech Solutions Company Limited for bioinformatics support. We also thank the Associate Editor, two anonymous reviewers, and Prof. J.S. Heslop-Harrison for help of polishing the final version of this article. This work was supported by the National Natural Science Foundation of China (31870198 and 31770374) and the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2018376).
Data deposition: The raw data have been deposited at NCBI Sequence Read Archive under the accessions SRR7458400–SRR7458405. The mitogenomes have been deposited at GenBank database under accessions MH559343, MH645952, MH645953, and MH645954. All the source codes used for the analysis are provided at https://github.com/wangshuaibin1015/kiwifruit_mitogenomes.
Literature Cited
- Adams KL, Palmer JD.. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 29(3):380–395. [DOI] [PubMed] [Google Scholar]
- Adams KL, Qiu YL, Stoutemyer M, Palmer JD.. 2002. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 99(15):9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JO, et al. 2007. Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177(2):1173–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD.. 2011. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of Cucumber. Plant Cell 23(7):2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, et al. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27(6):1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA.. 2009. Diversity of the Arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics 183(4):1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Adams KL, Thomason B, Palmer JD.. 2003. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424(6945):197–201. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. 2008. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8(1):26–34. [DOI] [PubMed] [Google Scholar]
- Byng JW, et al. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20. [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 10(6):563. [DOI] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MP, Nugent JM, Olmstead RG, Palmer JD.. 2003. Phylogenetic analysis reveals five independent transfers of the chloroplast gene rbcL to the mitochondrial genome in angiosperms. Curr Genet. 43(2):131–138. doi: 10.1007/s00294-003-0378-3 [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila JI, et al. 2011. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Fajardo D, et al. 2014. The American cranberry mitochondrial genome reveals the presence of selenocysteine (tRNA-Sec and SECIS) insertion machinery in land plants. Gene 536(2):336–343. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. Quant Biol. 1207:3907. [Google Scholar]
- Giege P, Brennicke A.. 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci U S A. 96(26):15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Lockhart PJ, Viola R, Velasco R.. 2012. The mitochondrial genome of Malus domestica and the import-driven hypothesis of mitochondrial genome expansion in seed plants. Plant J. 71(4):615–626. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Salamini F, Velasco R, Viola R.. 2009. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 26(1):99–110. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ.. 2017. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol. 68(1):225–252. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, et al. 2018. Two pivotal RNA editing sites in the mitochondrial atp1 mRNA are required for ATP synthase to produce sufficient ATP for cotton fiber cell elongation. New Phytol. 218(1):167–182. [DOI] [PubMed] [Google Scholar]
- Huang S, et al. 2013. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun. 4:2640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma Ki MT.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B, et al. 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLoS One 11(1):e0147209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint 1303:3997. [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JQ, Li XW, Soejarto DD.. 2007. Actinidiaceae. Beijing (China: ): Science Press. [Google Scholar]
- Lin M, et al. 2018. The complete chloroplast genome sequence of Actinidia arguta using the PacBio RS II platform. PLoS One 13(5):e0197393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2017. Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 215(2):877–890. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Nakazono M, Hirai A.. 1998. Transcription of plastid derived tRNA genes in rice mitochondria. Curr Genet. 34(3):216–220. [DOI] [PubMed] [Google Scholar]
- Mower JP. 2009. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 37(Web Server):W253–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Sloan DB, Alverson AJ.. 2012. Plant mitochondrial genome diversity: the genomics revolution In: Wendel JF, Greilhuber J, Dolezel J, Leitch IJ, editors. Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. New York: Springer; p. 123–144. [Google Scholar]
- Nakazono M, Hirai A.. 2008. Frequent DNA transfer among mitochondrial, plastid and nuclear genomes of rice during evolution. Berlin (Germany: ): Springer. [Google Scholar]
- Notsu Y, et al. 2002. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet. Genomics. 268(4):434–445. [DOI] [PubMed] [Google Scholar]
- Park S, et al. 2014. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics. 15(1):405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, et al. 2010. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 38(14):4755–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkington SM, et al. 2018. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 19:257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DW, et al. 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the Angiosperm Amborella. Science 342(6165):1468–1473. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD.. 2008. Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Mol Biol Evol. 25(8):1762–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman JR, et al. 2016. The two chromosomes of the mitochondrial genome of a sugarcane cultivar: assembly and recombination analysis using long PacBio reads. Sci Rep. 6:31533.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD.. 2015. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci U S A. 112(27):e3515–e3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB. 2013. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 200(4):978–985. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Wu Z.. 2014. History of plastid DNA insertions reveals weak deletion and at mutation biases in angiosperm mitochondrial genomes. Genome Biol Evol. 6(12):3210–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10(1):e1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2014. Mitochondrion-to-plastid DNA transfer: it happens. New Phytol. 202(3):736–738. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Brennicke A.. 2007. RNA editing in plant mitochondria: assays and biochemical approaches. Methods Enzymol. 424:439–458. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tanaka N, et al. 2004. Proteomics of the rice cell: systematic identification of the protein populations in subcellular compartments. Mol Genet. Genomics. 271(5):566–576. [DOI] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R.. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2):171–180. [DOI] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2017. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front Plant Sci. 8:413.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Cuthbert JM, Taylor DR, Sloan DB.. 2015. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc Natl Acad Sci U S A. 112(33):10185–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. 2017. The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 175(4):1690–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, et al. 2015. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLoS One 10(6):e0129347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.