Abstract
Purpose:
Despite the challenge to directly target mutant KRAS due to its high GTP affinity, some agents are under development against downstream signaling pathways, such as MEK inhibitors. However, it remains controversial whether MEK inhibitors can boost current chemotherapy in KRAS-mutant lung tumors in clinic. Considering the genomic heterogeneity among patients with lung cancer, it is valuable to test potential therapeutics in KRAS mutation–driven mouse models.
Experimental Design:
We first compared the pERK1/2 level in lung cancer samples with different KRAS substitutions and generated a new genetically engineered mouse model whose tumor was driven by KRASG12C, the most common KRAS mutation in lung cancer. Next, we evaluated the efficacy of selumetinib or its combination with chemotherapy, in KRASG12C tumors compared with KRASG12D tumors. Moreover, we generated KRASG12C/p53R270H model to explore the role of a dominant negative p53 mutation detected in patients in responsiveness to MEK inhibition.
Results:
We determined higher pERK1/2 in KRASG12C lung tumors compared with KRASG12D Using mouse models, we further identified that KRASG12C tumors are significantly more sensitive to selumetinib compared with KrasG12D tumors. MEK inhibition significantly increased chemotherapeutic efficacy and progression-free survival of KRASG12C mice. Interestingly, p53 co-mutation rendered KRASG12C lung tumors less sensitive to combination treatment with selumetinib and chemotherapy.
Conclusions:
Our data demonstrate that unique KRAS mutations and concurrent mutations in tumor-suppressor genes are important factors for lung tumor responses to MEK inhibitor. Our preclinical study supports further clinical evaluation of combined MEK inhibition and chemotherapy for lung cancer patients harboring KRASG12C and wild-type p53 status.
Introduction
KRAS is a membrane-bound small GTPase that switches between active GTP-binding and inactive GDP-binding forms, whose activity is tightly regulated by GTPase-activating proteins (GAP) and guanine nucleotide exchange-factors (GEFs) in normal cells (1). KRAS gain-of-function mutations are present in approximately 30% of all cancer types, with high prevalence in lung cancer, colon cancer, and pancreatic cancer (1). In cancer cells, these mutations keep KRAS persistently in the active state and activate downstream signaling. In lung cancer, most KRAS mutations are missense mutations with substitutions at codons 12 (91%), 13 (5%), or 61 (0.3%; ref. 2), and KRAS is mutated in approximately 30% of lung adenocarcinomas (3). In lung adenocarcinomas with EGFR mutations and ALK rearrangements, small-molecule tyrosine kinase inhibitors (TKIs) have shown superior benefits over chemotherapy (4, 5). However, there is no current therapy targeting mutant KRAS directly, and chemotherapies remain the standard of care for patients with lung cancer with KRAS mutations. Recently, the immunotherapy has become a novel treatment option for this group of patients (6,7), thanks to the development of immune checkpoint inhibitors.
Overactivation of the mitogen-activated protein kinase (MAPK) pathway is a key feature of KRAS-mutated lung cancer. Although different KRAS mutations share some common signaling, such as the KRAS–Raf–MEK–ERK cascade, some substitution-specific characteristics lead to unique therapeutic responses (8). Therefore, tumor heterogeneity remains a challenge in treating KRAS-mutant lung cancer. Most KRAS mutations in lung cancer reside in codon 12, with major populations including G12C (44%), G12D (17%), and G12V (23%) of all KRAS mutations (9). KRASG12C tumors show greater MEK-ERK dependence compared with KRASG12D and may thus be more sensitive to MEK inhibitors (8, 10). Besides the difference between substitutions, genetic alterations concomitant with KRAS, such as in TP53 and STK11, make KRAS-mutant tumors more heterogeneous (11,12). These co-occurring genomic changes could also lead to different therapeutic responses and may render KRAS-mutant tumors even more chemoresistant (11, 12). This may be one mechanism leading to inconsistent outcomes among clinical trials for KRAS-mutant lung cancers.
Multiple inhibitors are being explored to target MEK/ERK and PI3K/AKT (13), two major signaling pathways downstream of KRAS. MEK inhibitors have been widely tested in multiple cancer types, including lung cancer (14, 15), and trametinib and cobimetinib have both been approved to treat BRAF-mutant melanoma (16, 17). Although no MEK inhibitors are currently approved for lung cancer treatment, selumetinib and trametinib are two potential therapeutic MEK inhibitors. Selumetinib is an ATP-independent and orally active inhibitor for MEK1/2 (18) with demonstrated efficacy in tumor types, including neurofibroma, thyroid cancer, and lung cancer (19–22). In KRAS-mutant lung cancers, clinical trials are evaluating the efficacy of selumetinib either as a single agent or in combination with standard chemotherapy. A phase II clinical trial indicated better efficacy of selumetinib plus docetaxel in comparison with docetaxel alone (21). In contrast, a phase III clinical trial in KRAS-mutant lung cancer did not show benefit in patients treated with docetaxel combined with selumetinib compared with docetaxel alone (22). However, inconsistency with previous class II trial results may be due to multiple mechanisms, such as genomic heterogeneity within recruited patients with lung cancer with KRAS-mutant tumors. Besides, there were some evidence showing that not all KRAS-mutant tumor cells are dependent on KRAS in both lung and pancreatic cancers (23, 24), providing another mechanism for different responses to MEK inhibition. Therefore, these results do not rule out the possibility that selumetinib may be efficacious in a subset of patients with KRAS-mutant tumors. Thus, clinical progress may benefit from evaluating the efficacy of MEK inhibitors in genetically engineered mouse models (GEMM) with a clear genomic signature to identify potential responders to MEK inhibition.
To address an unmet clinical need to compare the efficacy of MEK inhibitors against different KRAS mutations, we used two KRAS-mutant GEMMs, KRASG12C and KrasG12D, to test the efficacy of selumetinib in combination with cisplatin/pemetrexed chemotherapy. Furthermore, we also evaluated the effect of p53 co-mutation on the response of KRASG12C-mutant lung cancer to selumetinib and chemotherapy combinations. Taken together, our results shed light on further clinical evaluation of MEK inhibitors in a genetically selected subset of patients with lung cancer with KRAS-mutant tumors.
Materials and Methods
Mouse generation
pBabe K-Ras 12V plasmid was purchased from Addgene (#12544), and G12C mutation was introduced using a Quick-Change kit (Agilent, #200521). Kozak and StuI sites were introduced to human KRASG12C by PCR and digested with a StuI blunt cutter. Coding region of hKRASG12C with the Kozak sequence (GCCGCCACC) was introduced into pGV at the EcoRI cloning site using blunt-end cloning. Sequencing-confirmed pGV-hKRASG12C vectors were co-electroporated with plasmid-expressing FLP recombinase into mouse ES cells, and positive clones were identified by PCR. Positive ES clones were injected into mouse blastocysts for chimera generation. Chimera mice were crossed with wild-type mice to generate mice with germline mutations. The mouse gDNA was used as PCR template and the KRASG12C sequence was confirmed with sanger sequencing. The genotyping primersusedare: KC-forward: GAAGTTATCTCGACGCTGATCAG and KC-reverse: GCTGTATCGTCAAGGCACTC. A detailed strategy was previously described (25). p53R270H (mutant homologous to human p53R273H) and KrasG12D were obtained from The Jackson laboratory were bred as previously described (11,26–28). All animal experiments, including breeding and treatment studies, were performed with approval of the DFCI Animal Care and Use Committee.
Generation of NIH3T3 cell lines
KRASG12C and KRASG12D retroviral plasmids were created by point mutagenesis of the pBABE KRASWT plasmid (Addgene, plasmid #75282). Retroviruses were generated by co-transfection of pBABE plasmids together with pAmpho plasmid into 293T cells using FuGENE HD transfection reagent (Promega). Retro-viruses were transduced into NIH3T3 murine fibroblast cells, and transduced cells were selected with puromycin (1 μg/mL) for 2 weeks.
Quantitative PCR.
Total cellular RNA (1 μg), extracted by RNeasy Mini Kit (QIAGEN) was reverse-transcribed by random primers using Quantitect Reverse Transcription Kit (Quiagen, Cat#205313) according to the manufacturer’s instructions. The reverse transcription reaction (1 μL) was then subjected to PCR amplification using PowerUp SYBR Green Master Mix (Thermo Fisher). PCR signals were recorded in technical triplicates on a StepOnePlus Cycler (Applied Biosystems) and analyzed using the StepOne version 2.2 software (Applied Biosystems). Primer sets were the following:
Murine ActinB Fw: GGCTGTATTCCCCTCCATCG-Murine ActinB Rev: CCAGTTGGTAACAATGCCATGT
Human KRAS Fw: GGACTGGGGAGGGCTTTCT-Human KRAS Rev: GCCTGTTTTGTGTCTACTGTTCT
Growth assessment by IncuCyte.
Growth rate of KRASG12C and KRASG12D NIH3T3 cells was assessed as previously described (29). Briefly, cells (1 × 103) were seeded in 96-well plates in 100-μL DMEM complete medium. The following day, plates were incubated in the IncuCyte Zoom for real-time imaging, with three fields imaged per well under ×10 magnification every 2 hours for a total of 150 hours. Data were analyzed using the IncuCyte Confluence version 1.5 software, which quantified cell surface area coverage as confluence values. IncuCyte experiments were performed in triplicate. A single representative growth curve is shown for each condition.
NIH3T3 allograft and selumetinib treatment
We injected 106 NIH3T3 cells expressing KRASG12C or KRASG12D into 6-week-old female SHO mice (Crl:SHO-PrkdcscidHrhr; Charles River Laboratories). Treatments were started when tumor volume reached approximately 250 mm3. Selumetinib (AZD6244; ARRY-142886) was purchased from Selleckchem (S1008) and dissolved in 0.5% methylcellulose with 0.4% polysorbate-80. Tumor-bearing mice were administered 25 mg/kg selumetinib by oral gavage twice daily, and tumor volume was measured and calculated as length × width × width/2.
Cell proliferation assay
Cell proliferation and growth assays were performed with colorimetric MTS assays as previously described (30). Briefly, cells were seeded in 96-well plates at a density of 500 cells/well in DMEM containing 10% FBS. Once adherent, cells were treated with increasing doses of the indicated drugs (10 nmol/L–10 μmol/L) for 72 hours and then analyzed by MTS colorimetric assay. All experimental points resulted from three to six replicates, and all experiments were repeated at least three times. Data were graphically plotted using GraphPad Prism 5 for Windows (GraphPad Software). Each point (mean ± SEM) represents growth of treated cells compared with untreated cells.
Western blot
Frozen lung tumor nodules or NIH3T3 cells were homogenized in RIPA buffer (Thermo Fisher Scientific) supplemented with Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). Approximately 20 μg of protein extracts were separated by SDS-PAGE (Bio-Rad), transferred to a PVDF membrane, and blotted with primary antibodies raised against pERK (Cell Signaling Technology), ERK (Cell Signaling Technology), β-actin (Cell Signaling Technology), HA-Tag (6E2; Cell Signaling Technology, Cat), HSP90 (H114; Santa Cruz Biotechnology), phosphorylated MEK (Cell Signaling Technology), and MEK (Cell Signaling Technology). Secondary anti-mouse or anti-rabbit antibodies were purchased from Amersham. Quantification was done using ImageJ.
3D murine-derived organotypic tumor spheroids
Lung tumor nodules from GEMMs were dissected and transferred to 10-cm dishes (on ice) with RPMI-1640 medium. Nodules were cut into small pieces and digested with collagenase at 37°C for 30 minutes. Spheroid fractions (40–100 μm) were collected for further ex vivo culture as previously described (31, 32). The 40 to 100 μm spheroid fraction was centrifuged and pellets were suspended with type I rat tail collagen (Corning) before injection into the central region of a 3D cell culture CHIP (DAX-1, AIM Biotech PET LTD.). The device was incubated at 37° C for 30 minutes, and then murine-derived organotypic tumor spheroids (MDOTS) were treated with cisplatin (500 nmol/L) and/or selumetinib (500 nmol/L) in RPMI-1640 medium with 10% FBS on day 0. DMSO was used as a control. Fresh medium with drugs was changed on day 1 and 3, and live/dead staining was performed on day 6.
Live/dead staining
Live/dead staining of 3D MDOTS was performed using Nexcelom ViaStain acridine orange/propidium iodide (AO/PI) staining solution (Nexcelom, CS2–0106) as previously described (32). MDOTS were incubated with AO/PI reagents for 20 minutes in the darkness, and then images were taken with a Nikon Eclipse 80i fluorescence microscope equipped with z-stack (Prior) and a CoolSNAP CCD camera (Roper Scientific). Total area of each dye was measured to quantify live/dead cells.
GEMM treatment studies
Cisplatin and pemetrexed were purchased from Dana-Farber Cancer Institute Pharmacy. KRASG12C, KrasG12D, or KRASG12C/p53R270H mice were monitored by MRI for tumor development after intranasal induction with adeno-Cre (2.5 × 10^6 pfu for KRASG12C and KRASG12C/p53R270H; 5 × 10^6 pfu for KrasG12D mice). Tumor-bearing mice were dosed with selumetinib (25 mg/kg, twice daily), either alone or in combination with pemetrexed (50 mg/kg) and cisplatin (4 mg/kg), and monitored by MRI every 2 weeks. Cisplatin and pemetrexed were dosed intraperitoneally once a week for 6 weeks. For mice under cisplatin/pemetrexed treatment, 400 μL of saline was dosed intraperitone-ally twice every week to offset the nephrotoxicity.
Patient samples and immunohistochemistry analysis
The study included 66 lung cancer patient samples from Tongji Hospital (Shanghai, China). Clinicopathological characteristics are listed in Supplementary Table S1. Operation or biopsy samples were obtained from treatment-naïve lung cancer patients and tissue samples were genotyped for KRAS mutation subtype using Human KRAS Gene 7 Mutations fluorescence PCR diagnostic kit (Beijing Accb Biotech Ltd.). All patients provided written informed consent. Tissue collection and the following tissue studies were approved by the Ethical Committee of Tongji Hospital (Shanghai, China). Tissue samples were fixed with 4% paraformaldehyde and embedded in paraffin for sectioning. Tissue sections were stained via standard immunohistochemical protocols for phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; D13.14.4E; XP rabbit mAb #437, Cell Signaling Technology), phospho-MEK1/2 (Ser221; 166F8; rabbit mAb #2338, Cell Sig mAb #4060, Cell Signaling Technology). Results were independently scored by two pathologists using multiplicative quick systems (33). Briefly, the expression score of each marker was calculated by multiplying a score indicating percentage of positively stained cells within tumor cells counted (1 = 0%−4%; 6 = 80%−100%) by the intensity grade of staining (0 = negative; 1 = weak; 2 = moderate; 3 = strong).
Results
Human lung tumors harboring KRASG12C mutation are associated with higher ERK1/2 phosphorylation
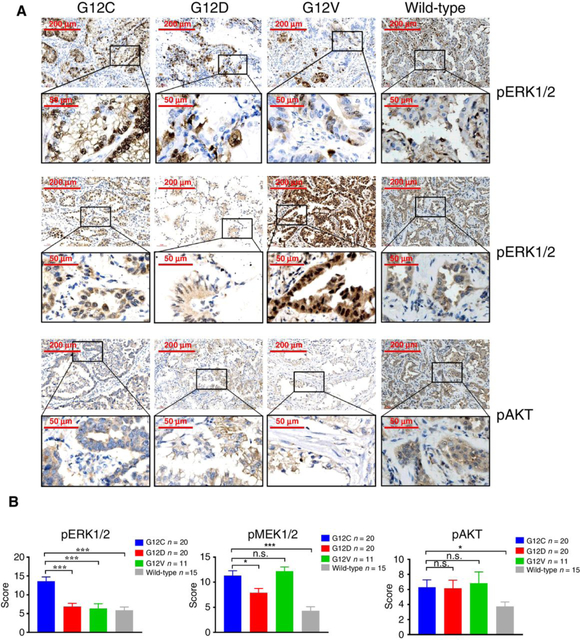
Previous studies have demonstrated that different substitutions within mutant KRAS leads to altered association with downstream MAPK and AKTsignaling pathways (8). To investigate the canonical signaling pathways mediated by different KRAS mutations, we analyzed 66 genotype-confirmed treatment-naïve human lung cancer samples (KRASG12C, n = 20; KRASG12D, n = 20; KRASG12V n = 11 and KRASwt, n = 15; Supplementary Table S1) by immunohistochemical staining for pERK1/2, pMEK1/2, and pAKT. Staining results showed that ERK1/2 phosphorylation was significantly higher in KRASG12C lung tumors compared with other KRAS subtypes (Fig. 1A and B). Both KRASG12C and KRASG12V tumors showed higher MEK phosphorylation compared with KRASG12D and KRASwt tumors (Fig. 1A and B). However, there was no significant difference in AKT phosphorylation levels across the three subtypes of KRAS-mutated tumors. These data suggest that KRASG12C tumors was preferentially associated with ERK1/2 activation compared with other mutant KRAS tumors and we, therefore, reasoned that this may result in heterogeneous therapeutic response.
Figure 1.
The ERK1/2 phosphorylation in KRAS-mutant human lung tumors. A, Representative images of immunohistochemical staining for pERK1/2, pMEK1/2, and pAKT in KRASG12C, KRASG12D, KRASG12V and KRASwt human tumor tissue samples. Scale bars, 200 or 50 μm. B, Multiplicative quick scores for quantification of pERK1/2, pMEK1/2, and pAKT staining in KRASG12C, KRASG12D, KRASG12V and KRASwt human tumor tissue samples. *, P < 0.05; ***, P < 0.001; n.s., not significant.
Isogenic cell lines expressing KRASG12C indicates higher sensitivity to selumetinib in vitro and in vivo
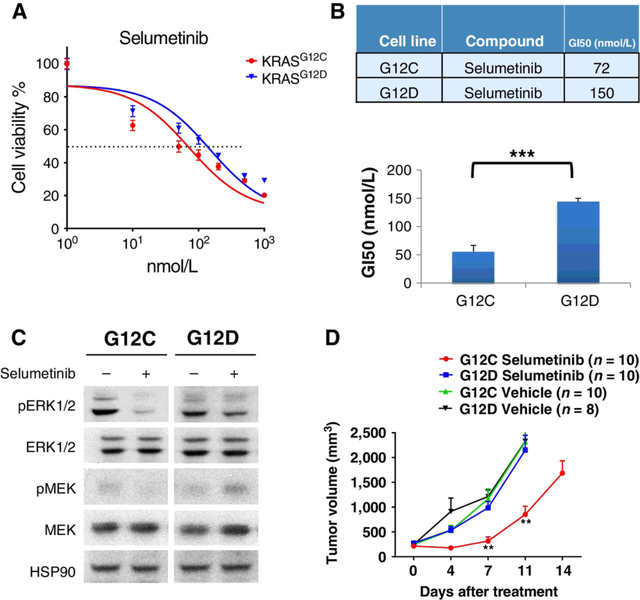
To test the efficacy of MEK inhibitors against KRAS mutations with different substitutions, we next transduced NIH3T3 mouse fibroblast cells with retrovirus expressing either human KRASG12C or KRASG12D and generated isogenic KRAS cell lines after puromycin selection. We confirmed the hKRAS expression was comparable at both the transcriptional level (Supplementary Fig. S1A) and at protein level (Supplementary Fig. S1B). The growth of both isogenic cell lines was monitored with IncuCyte live cell analysis and there was no significant difference between these two lines (Supplementary Fig. S1C). Next, we treated these isogenic cell lines with increasing doses of MEK inhibitor selumetinib for 3 days and calculated 50% growth inhibition (GI50) for each mutation. This in vitro assay demonstrated that KRASG12C cells (GI50 = 72 nmol/L) were more sensitive to selumetinib inhibition compared to KRASG12D cells (GI50 = 150 nmol/L; Fig. 2A and B). We further treated these cell lines with selumetinib at 10 nmol/L for 12 hours and found that KRASG12C is more sensitive to selumetinib treatment than KRASG12D, as shown by pERK1/2 reduction (Fig. 2C). Next, we transplanted these isogenic NIH3T3 cell lines into immunodeficient SHO mice and treated mice with 25 mg/kg selumetinib in vivo. Consistent with in vitro proliferation assays, we observed significantly increased sensitivity to MEK inhibition in KRASG12C tumors compared with KRASG12D tumors (Fig. 2D).
Figure 2.
In vitro and in vivo sensitivity of isogenic cell lines to selumetinib treatment. A, Cell viability as measured by MTS colorimetric assays of NIH3T3-KRASG12C and NIH3T3-KRASG12D cells after treatment with increasing doses of selumetinib for 3 days. B, Concentration of selumetinib that caused 50% growth inhibition (GI50) of NIH3T3-KRASG12C and NIH3T3-KRASG12D cells after treatment with increasing doses of selumetinib for 3 days. C, NIH3T3 cells transduced with either KRASG12C or KRASG12D were treated for 12 hours with selumetinib (10 nmol/L), lysed and blotted with the indicated antibodies. D, 106 NIH3T3-KRASG12C or NIH3T3-KRASG12D cells were injected into female SHO mice. Mice were treated with 25 mg/kg selumetinib when tumors reached approximately 250 mm3. Tumor volume was measured twice every week until the end of treatment. **, P < 0.01; ***, P < 0.001.
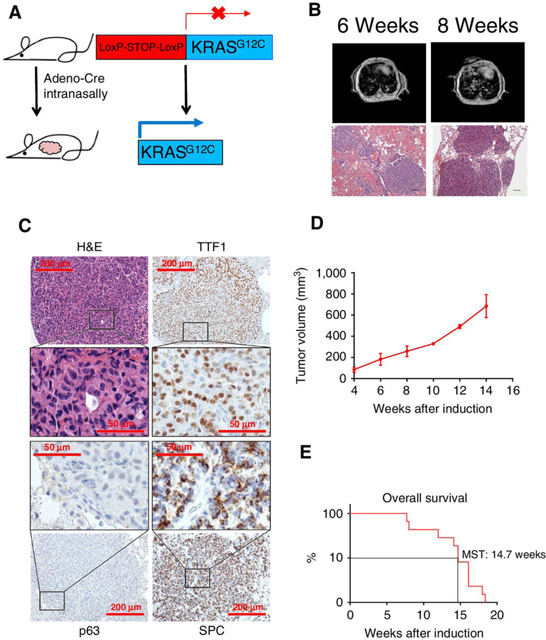
Cre-induced KRASG12C expression drives development of lung adenocarcinoma in vivo
To test the efficacy of MEK inhibition against KRASG12C in vivo, we first generated a GEMM with inducible expression of hKRASG12C. We electroporated mouse ES cells with a DNA construct containing LoxP-Stop-LoxP-hKRASG12C and then injected these positive ES cells into blastocysts to get chimeras. The chimera mice were further crossed with wild-type mice to generate germlines. We intranasally delivered adeno-Cre virus into adult KRASG12C mice to initiate hKRASG12C expression specifically in murine lung epithelial cells (Fig. 3A). MRI monitoring showed that lung tumors appeared after 6 weeks (Fig. 3B). Furthermore, IHC staining with unique adenocarcinoma and squamous cancer markers demonstrated these KRASG12C tumors were positively stained by adenocarcinoma markers TTF1 and SPC and negatively stained for squamous cell carcinoma (SCC) marker p63 (Fig. 3C). This histology was consistent with the fact that most KRAS-mutant lung tumors are adenocarcinomas. Long-term monitoring showed the growth kinetics of these tumors (Fig. 3D) and indicated that mice had a median survival time of 14.7 weeks after adeno-Cre induction (Fig. 3E).
Figure 3.
Cre-induced KRASG12C expression drives development of mouse lung adenocarcinomas. A, Schematic of transgene insertion strategy to generate inducible LSL-KRASG12C mice via intranasal adeno-Cre induction. B, Representative MRI images and H&E-stained lung tissue sections of KRASG12C mice, 6 or 8 weeks after adeno-Cre inductionp; scale bars, 50 μm. C, Representative images of immunohistochemistry staining of KRASG12C tumors with H&E, adenocarcinoma markers TTF1 and SPC, or squamous cancer marker p63; scale bars, 200 or 50 μm. D, Tumor growth curve based on MRI results of KRASG12C lung tumors after adeno-Cre induction. E, Overall survival of KRASG12C mice after adeno-Cre induction.
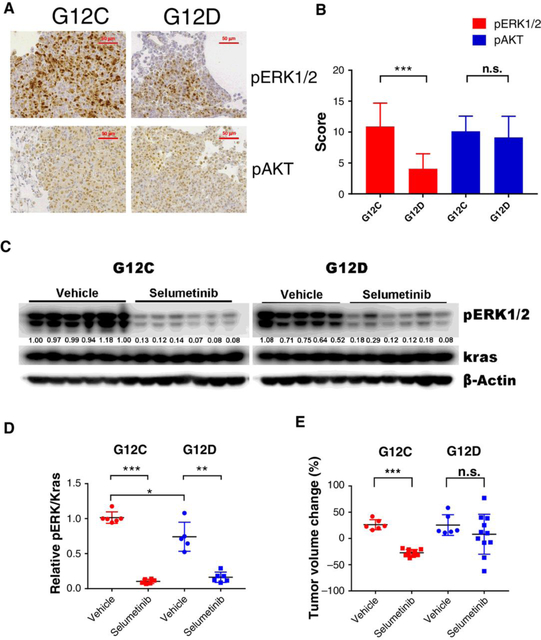
KRASG12C mouse lung tumors show higher ERK1/2 phosphorylation
To compare pERK1/2 levels between KRASG12C and KrasG12D tumors, we harvested lung tumors from KRASG12C and KrasG12D mice and stained tissues for ERK1/2 phosphorylation. ERK phosphorylation was significantly stronger in KRASG12C tumors than KrasG12D tumors (Fig. 4A and B), which was consistent with data from patients with KRAS-mutant lung cancer. Meanwhile, KRASG12C and KrasG12D tumors indicated similar AKT phosphor-ylation levels (Fig. 4A and B).
Figure 4.
ERK1/2 phosphorylation and selumetinib efficiency in KRASG12C and KrasG12D mouse lung tumors. A, Representative images of immunohistochemistry staining of pERK1/2 and pAKT in lung tissues of KRASG12C and KrasG12D mice; Scale bars, 50 μm. B, Quantification of immunohistochemistry staining scores for pERK1/2 and pAKT in KRASG12C and KrasG12D lung tissue samples. (n ≥ 8 for each group; C, Western blots of pharmacodynamic markers from tumor nodules of KRASG12C and KrasG12D mice treated with 25 mg/kg selumetinib or vehicle for 3 days (n = 5–6 for each group). D, Relative pERK1/2 level normalized to KRAS in the samples from (C) was quantified using ImageJ. E, MRI-based tumor volume changes of KRASG12C or KrasG12D mice treated with 25 mg/kg selumetinib monotherapy for a week. Each dot represents one mouse; *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
KRASG12C mouse lung tumors indicate better response to selumetinib
A three-day pharmacodynamics (PDs) study of selumetinib treatment in tumor-bearing KRASG12C and KrasG12D mice indicated that selumetinib treatment efficiently suppressed ERK1/2 phosphorylation in both KRAS-mutant mouse strains and more significant pERK reduction occurred in KRASG12C tumors (Fig. 4C and D). Western blotting also confirmed higher ERK1/2 phosphorylation in KRASG12C tumors than KrasG12D tumors, despite similar KRAS expression levels. Further, after just one week of selumetinib treatment, KRASG12C mice had significantly reduced tumor volumes compared to vehicle controls (Fig. 4E; Supplementary Fig. S2A–S2C). In contrast, selumetinib treatment had no significant effect on tumor volumes of KrasG12D mice. Thus, selumetinib demonstrated greater short-term antitumor efficacy in KRASG12C mice, whereas KrasG12D lung tumors were more resistant to selumetinib treatment.
Anti-tumor efficacy of cisplatin/pemetrexed chemotherapy plus selumetinib in KRASG12C lung tumors
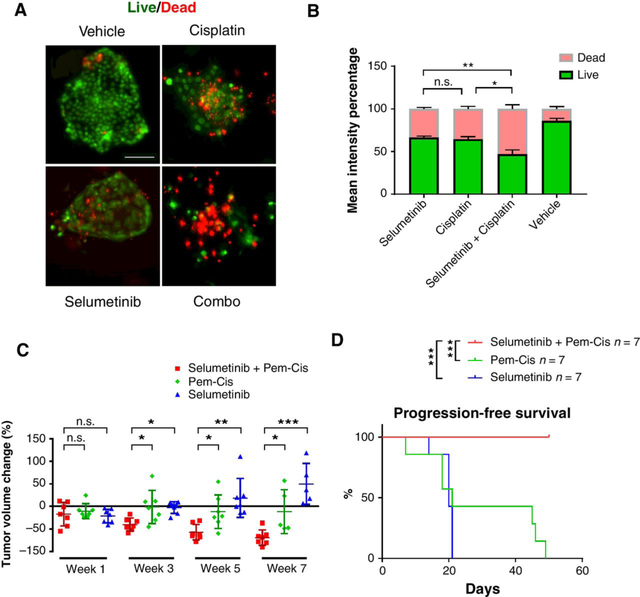
Platinum-based doublet chemotherapy, such as cisplatin plus pemetrexed, remains the best regimen for patients with KRAS-mutant lung cancer or those without known driver mutations. To test the efficacy of cisplatin/pemetrexed and its combination with selumetinib in KRASG12C and KrasG12D tumors, we cultured mouse lung tumors using 3D murine-derived organotypic tumor spheroids (MDOTS; ref. 32) and added cisplatin (500 nmol/L) and/or selumetinib (500 nmol/L). Selumetinib augmented chemotherapy-induced apoptosis in KRASG12C tumors (Fig. 5A and B).
Figure 5.
Antitumor efficacy of cisplatin/pemetrexed chemotherapy alone or in combination with selumetinib in KRASG12C mouse lung tumors. A, Representative images of live/dead cell staining of 3D murine-derived organotypic tumor spheroid (MDOTS) cultures of KRASG12C lung tumors treated with vehicle control or selumetinib (500 nmol/L) and/or cisplatin (500 nmol/L) for 6 days; scale bars, 100 μm. B, Quantification of live/dead cell staining with each treatment. (n = 5 for each group; C, Volume change of KRASG12C tumors 1 to 7 weeks after indicated treatment. Each dot represents one mouse. D, Progression-free survival of KRASG12C mice treated with cisplatin/pemetrexed alone, selumetinib alone or their combination. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
Next, we treated KRASG12C mice with cisplatin/pemetrexed chemotherapy alone or selumetinib alone or in combination and evaluated the antitumor efficacy by MRI every 2 weeks from the first week (Supplementary Fig. S3A). MRI quantification of tumor volumes showed that addition of selumetinib significantly increased the efficacy of cisplatin/pemetrexed chemotherapy (Fig. 5C; Supplementary Fig. S3B and S3C). Mice treated with cisplatin/pemetrexed plus selumetinib showed sustained responses for at least 7 weeks. However, cisplatin/pemetrexed treatment alone had little efficacy, and mice began dying after week 7 (Fig. 5C). Treatment with selumetinib alone showed some efficacy after 1 week; however, the tumors relapsed afterwards with the following treatment (Fig. 5C). Overall, mice treated with selumetinib in addition to cisplatin/pemetrexed chemotherapy had significantly prolonged progression-free survival compared with treatment with either selumetinib or cisplatin/pemetrexed alone (Fig. 5D). These in vivo experiments together with 3D MDOTS culture data demonstrated that MEK inhibition could increase the antitumor efficacy of chemotherapy in KRASG12C lung cancers.
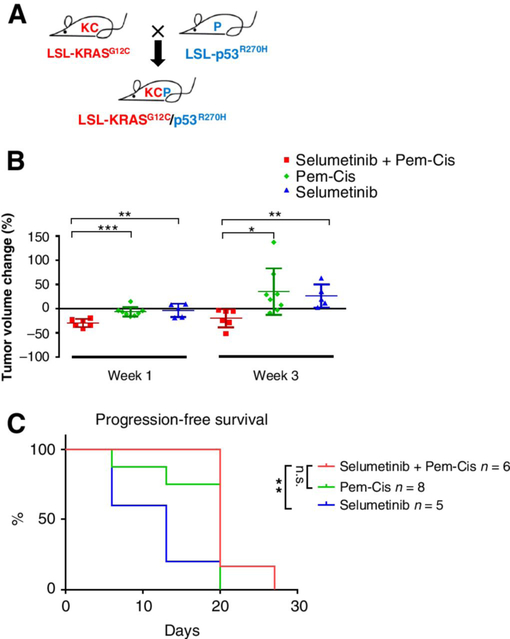
Resistance to cisplatin/pemetrexed plus selumetinib combination by p53 co-mutation in KRASG12C tumors
To evaluate the effect of co-occurring mutations in KRASG12C in response to selumetinib, we crossed KRASG12C mice with a p53R270H strain to generate KRASG12C/p53R270H (KCP) mice (Fig. 6A). KCP mice had a shorter median overall survival time of 9 weeks compared with KRASG12C mice (Supplementary Fig. S4A). KCP tumors also showed similarly strong pERK1/2 levels as KRASG12C tumors (Supplementary Fig. S4B and S4C). Treatment of tumor-bearing KCP mice with cisplatin/pemetrexed plus selumetinib showed that KCP lung tumors were less sensitive to combination treatment (Fig. 6B; Supplementary Fig. S5) compared with KRASG12C tumors with wild-type p53 as tumors began to relapse after 3 weeks. Besides, the selumetinib alone also showed less efficacy in KCP mice, compared with in the KRASG12C model. Moreover, cisplatin/pemetrexed plus selumetinib treatment in KCP mice showed no significant benefit for progression-free survival compared with cisplatin/pemetrexed alone (Fig. 6C). These data show that co-occurring genomic events, especially commonly mutated tumor suppressors, can dramatically alter the KRASG12C tumor responses. This emphasizes the need to subdivide KRASG12C patients based on the status of concurrent tumor suppressors, such as p53 and STK11, to select potential responders to MEK inhibition.
Figure 6.
Effect of p53 co-mutation in KRASG12C mouse lung tumors on efficacy of cisplatin/pemetrexed treatment alone or in combination with selumetinib. A, Schematic of KRASG12C/p53R270H breeding strategy. B, KCP tumor volume change 1 to 3 weeks after treatment with cisplatin/pemetrexed alone (n = 8), selumetinib alone (n = 5) or in their combination (n = 6). Each dot represents one mouse. C, Progression-free survival of KCP mice treated with cisplatin/pemetrexed alone, selumetinib alone or their combination.
Discussion
Although previous studies indicated a potential benefit for MEK inhibition in KRAS-mutant lung cancer (21, 34), the recent phase III SELECT-1 clinical trial did not show better progression-free survival in KRAS-mutant lung cancer patients treated with selumetinib plus docetaxel compared with docetaxel alone (22). However, it remains an important question whether a subset of KRAS-mutant lung cancer patients could benefit from MEK inhibitor treatment. Using our newly generated KRASG12C lung cancer mouse model, we compared responses to MEK inhibitor selumetinib between KRASG12C and KrasG12D lung tumors. Our data reveal that, compared with KrasG12D, KRASG12C tumors are more sensitive to selumetinib either alone or in combination with cisplatin/pemetrexed. Importantly, a combination of cisplatin/pemetrexed and selumetinib significantly improved survival of KRASG12C mice. Furthermore, we identified that concomitant p53 mutation renders KRASG12C lung tumors less sensitive to selumetinib and cisplatin/pemetrexed combination treatment, which may be one of the potential reasons underlying negative results in the SELECT-1 clinical trial. Our current study also indicates that KRASG12C tumors have higher ERK1/2 phosphorylation than KRASG12D or KRASG12V tumors and demonstrates the benefit of adding selumetinib to cisplatin/pemetrexed chemotherapy in KRASG12C mouse models of lung cancer, thus shedding light on future screening for responders to MEK inhibition in patients with KRAS-mutant lung cancer.
Though rarely found in other cancer types, G12C substitution is the most common KRAS mutation in NSCLC (9). A previous study indicated that patients with lung cancer with KRASG12C or KRASG12V mutations in the BATTLE trial had worse progression-free survival than those with other KRAS mutations or wild-type KRAS under enrolled treatments (8). Therefore, it is of great significance to boost the responses of current chemotherapies in this subset of patients with lung cancer. Our previous study demonstrated the benefit of adding MEK inhibitor selumetinib to docetaxel chemotherapy in KrasG12D lung cancer (11). Our current results show that KRASG12C tumors have both higher ERK1/2 phosphorylation and better response to selumetinib treatment.
It is of note that the two GEMMs used in this research, KRASG12C and KrasG12D, were generated with different strategies. In the newly generated KRASG12C mice, two wild-type mouse Kras alleles remain intact, and a single copy of human KRAS with a G12C mutation was inserted at the COL1A locus. In KrasG12D mice, one allele of wild-type mouse Kras was endogenously replaced with KrasG12D, and the other allele remains wild-type (28). Thus, KRASG12C and KrasG12D mouse tumors were driven by human KRASG12C and mouse KrasG12D, respectively. Mouse and human KRAS have very high homo-logy, with only five out of 188 different amino acids, four of which reside in the C-terminal membrane-binding motif. To further address this concern, we generated NIH3T3 cells expressing human KRASG12C or human KRASG12D and used these isogenic cell lines in allograft studies to compare their responses with selumetinib. Isogenic cell data indicated better selumetinib response in KRASG12C tumors than KRASG12D tumors, which was consistent with efficacy data from GEMMs.
Another concern may be that there is an additional wild-type Kras allele in KRASG12C mice, which could lead to higher KRAS expression. However, we found similar KRAS expression in tumor nodules from the two mouse models, indicating that different downstream signaling mostly resulted from different KRAS mutations. In addition, patient sample analysis also indicated greater ERK phosphorylation in KRASG12C tumors than in KRASG12D or KRASG12V tumors, which was consistent with mouse data. Taken together, KRASG12C and KrasG12D GEMMs were good in vivo models to compare responses to KRAS therapeutics.
Docetaxel was used in our previous selumetinib study (11) and the SELECT-1 trial. Pemetrexed-based regimens have been clinically used to treat lung adenocarcinoma as the taxane-based chemotherapy drugs such as docetaxel indicated higher potential for neurotoxicity (35). Therefore, we used cisplatin/pemetrexed chemotherapy in our current GEMM treatment study. Actually, all these chemotherapeutic drugs are used commonly in clinics to treat patients with KRAS-mutant lung cancer, although the drugs target tumor cells via different mechanisms. Docetaxel has microtubule depolymerizing capacity (36), and cisplatin, an alkylating-like agent, can crosslink DNA and interfere with DNA replication and mitosis (37). Pemetrexed can inhibit enzymes in thymidylate synthesis, thus suppressing DNA replication and repair (38). As MAPK signaling may play different roles in these processes, MEK inhibitors may also cooperate with these chemotherapeutic drugs via different mechanisms, leading KRAS-mutant tumors to respond differently to MEK inhibition. The molecular mechanism underlying the synergy between MEK inhibitors and different chemotherapeutic drugs needs further investigation. In addition, a previous report indicated that MEK inhibition could overcome cisplatin resistance in squamous cell carcinoma (39). Furthermore, another study identified cisplatin treatment could increase ERK phosphorylation and contribute to chemoresistance in melanoma (40). Whether cisplatin is better synergized with selumetinib than docetaxel remains an open question to be addressed both pre-clinically and clinically.
Genomic co-alteration in tumor suppressors may decrease response of KRASG12C tumors to selumetinib, as indicated by our treatment results in KRASG12C/p53R270H mice. There is hope that KRASG12C combined with wild-type p53 status could be used as a marker to screen potential responders, although this possibility requires further clinical evaluation. It is also helpful for retrospective studies to analyze the correlation between p53/STK11 status and progression-free survival data for previous clinical trials, including SELECT-1. Our data also indicate a positive correlation between ERK1/2 phosphorylation and MEK inhibitor efficacy, and it will be meaningful to further evaluate this correlation in KRAS-mutant lung cancer therapy.
Supplementary Material
Translational Relevance.
Although a few clinical trials have used MEK inhibitors to treat KRAS-mutant lung cancers, it remains unclear whether MEK inhibition could clinically benefit current therapeutics in at least a subset of patients with lung cancer with KRAS-mutant tumors. This study identified higher ERK1/2 phosphorylation in KRASG12C-mutant lung tumors compared with KRASG12D or KRASG12V tumors. Using both in vitro isogenic cell lines and in vivo GEMMstudies, we further demonstrated that KRASG12C tumors are more sensitive than KRASG12D tumors to the MEK inhibitor selumetinib either alone or in combination with cisplatin/pemetrexed chemotherapy. Moreover, p53 co-mutation rendered KRASG12C tumors insensitive toMEKinhibition. Our current research provides evidence that different activating point mutations of KRAS display differential sensitivity to MEK inhibition and that patients with lung cancer with KRASG12C/p53wt tumors may therapeutically benefit from MEK inhibition plus chemotherapy, which needs to be further evaluated in the clinic.
Acknowledgments
We would like to acknowledge the help of Mei Zheng (Brigham and Women’s Hospital) in immunohistochemistry studies. This work was supported by the National Cancer Institute R01 CA195740, CA205150, CA166480, CA140594, P01 CA154303, U01 CA213333 (to K.-K. Wong). This work was also funded by the National Natural Science Foundation of China (81401882, 81570053, and 81600043) and Key Medical Research of Shanghai (034119868 and 09411951600; to X.Y.). G.S. Herter-Sprie acknowledges funding from the Deutsche Forschungsgemeinschaft (HE 6897/1–1) and the Claudia Adams Barr Program for Innovative Cancer Research.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
R.W. Jenkins holds a patent owned by Dana-Farber, Microfluidic Cell Culture of Patient-derived tumor cell Spheroids. D.A. Barbie is a consultant/advisory board member for N of One. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell 2017;170:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood K, Hensing T, Malik R, Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol 2016;2:805–12. [DOI] [PubMed] [Google Scholar]
- 3.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki JComprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao BC, Lin CC, Yang JC. Second and third-generation epidermal growth factor receptor tyrosine kinase inhibitors in advanced nonsmall cell lung cancer. Curr Opin Oncol 2015;27:94–101. [DOI] [PubMed] [Google Scholar]
- 5.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013;13:685–700. [DOI] [PubMed] [Google Scholar]
- 6.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
- 7.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med 2017;377:1919–29. [DOI] [PubMed] [Google Scholar]
- 8.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 2014;13: 828–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janne PA, Smith I, McWalter G, Mann H, Dougherty B, Walker J, et al. Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non-small-cell lung cancer. Br J Cancer 2015;113:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012;483:613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov 2014;13:928–42. [DOI] [PubMed] [Google Scholar]
- 14.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 2015;15: 577–92. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol 2014;11:385–400. [DOI] [PubMed] [Google Scholar]
- 16.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014;371:1867–76. [DOI] [PubMed] [Google Scholar]
- 17.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014;371:1877–88. [DOI] [PubMed] [Google Scholar]
- 18.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer 2004;4:937–47. [DOI] [PubMed] [Google Scholar]
- 19.Ho AL, Grewal RK, Leboeuf R, Sherman EJ, Pfister DG, Deandreis D, et al. Selumetinib-enhanced radioiodine uptake in advanced thyroid cancer. N Engl J Med 2013;368:623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N Engl J Med 2016;375:2550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013;14:38–47. [DOI] [PubMed] [Google Scholar]
- 22.Janne PA, van den Heuvel MM, Barlesi F, Cobo M, Mazieres J, Crino L, et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: the SELECT-1 randomized clinical trial. JAMA 2017;317: 1844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009;15:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen PY, Muzumdar MD, Dorans KJ, Robbins R, Bhutkar A, Del Rosario A, et al. Adaptive and reversible resistance to kras inhibition in pancreatic cancer cells. Cancer Res 2018;78:985–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akbay EA, Moslehi J, Christensen CL, Saha S, Tchaicha JH, Ramkissoon SH, et al. D-2-hydroxyglutarate produced by mutant IDH2 causes cardiomyopathy and neurodegeneration in mice. Genes Dev 2014;28:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adeegbe D, Liu Y, Lizotte PH, Kamihara Y, Aref AR, Almonte C, et al. Synergistic immunostimulatory effects and therapeutic benefit of combined histone deacetylase and bromodomain inhibition in non-small cell lung cancer. Cancer Discov 2017;7:852–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119:847–60. [DOI] [PubMed] [Google Scholar]
- 28.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 2018;172:857–68 e15. [DOI] [PubMed] [Google Scholar]
- 30.Ambrogio C, Gomez-Lopez G, Falcone M, Vidal A, Nadal E, Crosetto N, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med 2016;22:270–7. [DOI] [PubMed] [Google Scholar]
- 31.Aref AR, Huang RY, Yu W, Chua KN, Sun W, Tu TY, et al. Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integr Biol 2013;5:381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov 2018;8:196–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Yu X, Xie J, Zhan M, Yu Z, Xie L, et al. ANGPTL2/LILRB2 signaling promotes the propagation of lung cancer cells. Oncotarget 2015;6: 21004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blumenschein GR Jr., Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann Oncol 2015;26:894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edelman MJ, Le Chevalier T, Soria JC. Maintenance therapy and advanced non-small-cell lung cancer: a skeptic’s view. J Thorac Oncol 2012;7:1331–6. [DOI] [PubMed] [Google Scholar]
- 36.Kavallaris M Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer 2010;10:194–204. [DOI] [PubMed] [Google Scholar]
- 37.Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol 2012;9:144–55. [DOI] [PubMed] [Google Scholar]
- 38.Wilson PM, Danenberg PV, Johnston PG, Lenz HJ, Ladner RD. Standing the test of time: targeting thymidylate biosynthesis in cancer therapy. Nat Rev Clin Oncol 2014;11:282–98. [DOI] [PubMed] [Google Scholar]
- 39.Kong LR, Chua KN, Sim WJ, Ng HC, Bi C, Ho J, et al. MEK inhibition overcomes cisplatin resistance conferred by SOS/MAPK pathway activation in squamous cell carcinoma. Mol Cancer Ther 2015;14:1750–60. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Melton DW. Cisplatin regulates the MAPK kinase pathway to induce increased expression of DNA repair gene ERCC1 and increase melanoma chemoresistance. Oncogene 2012;31:2412–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.