Abstract
Antibodies are the primary correlate of protection for most licensed vaccines; however, their mechanisms of protection may vary, ranging from physical blockade to clearance via the recruitment of innate immunity. Here, we uncover striking functional diversity in vaccine-induced antibodies driven by immunization site and associated with reduced risk of SIV infection in nonhuman primates. While equivalent levels of protection were observed following intramuscular (IM) and aerosol (AE) immunization with an otherwise identical DNA prime, Ad5 boost regimen, reduced risk of infection was associated with IgG-driven antibody-dependent monocyte-mediated phagocytosis in the IM vaccinees, but via vaccine-elicited IgA-driven neutrophil-mediated phagocytosis in AE immunized animals. Thus, while route-independent correlates indicate a critical role for phagocytic Fc-effector activity in protection from SIV, the site of immunization may drive this Fc-activity via distinct innate effector cells and antibody isotypes. Moreover, the same correlates predicted protection from SHIV infection in a second non-human primate vaccine trial using a disparate IM canarypox prime, protein boost strategy, analogous to that used in the first moderately protective human HIV vaccine trial. These data identify orthogonal functional humoral mechanisms, initiated by distinct vaccination routes and immunization strategies, pointing to multiple, potentially complementary correlates of immunity that may support the rational design of a protective vaccine against HIV.
Introduction:
Among the HIV-1 vaccine concepts evaluated in human efficacy trials, marginal protection was observed following intramuscular canarypox/protein vaccination1 (RV144 trial) in the absence of broadly neutralizing antibodies or cytotoxic T cell responses, but in association with other aspects of the humoral response2. Likewise, emerging data from multiple non-human primate (NHP) studies point to a critical role for non-neutralizing antibody activities, including antibody dependent cellular cytotoxicity (ADCC) and antibody dependent cellular phagocytosis (ADCP), in protection from infection upon viral challenge3–7. Moreover, passive antibody transfer experiments in humanized mice and NHPs have also illustrated the critical role for antibody functions, beyond neutralization, where the elimination of Fc-activity from broadly neutralizing HIV-specific antibodies results in compromised protection8,9. Furthermore, in humans, antibodies to the envelope glycoprotein (Env) V1/V2 variable loops and antibodies that were able to induce ADCC (in the absence of IgA responses), were enriched among RV144 vaccinees that resisted HIV infection2. However, questions remain regarding the specific humoral functions that track with protection against SIV, SHIV or HIV and whether they are consistent across studies.
In a previously published NHP vaccine trial, DNA priming followed by recombinant adenovirus type 5 (rAd5) vaccination resulted in significant antibody-mediated protection from SIV infection10. While protection was linked to the selective neutralization of sensitive clones within the challenge viral swarm, we sought to define whether specific antibody functional mechanisms, beyond the observed low neutralizing antibody titers, were enriched among macaques that resisted infection. Moreover, we aimed to further determine the consistency or divergence of humoral correlates of protection induced with the same vaccine regimen administered mucosally. Using a comprehensive and objective antibody Fc-profiling approach, we defined the functional antibody correlates of protection. Phagocytosis was a consistent and key correlate of protection across vaccine arms, but was associated with distinct innate immune effector cells and antibody isotypes, driven by the route of immunization. These data define potential mechanisms of protection against viral infection, and provide important insights for HIV vaccine design.
Results:
Equivalent protection from infection observed using distinct routes of vaccination
As previously described10, 80 Indian origin rhesus macaques were immunized intra-muscularly (IM) with one of four different DNA prime, rAd5 boost regimens. These four arms included an empty vector control, SIV Gag, SIV Env (IM mac239 Env), and a combination of two mosaic SIV Env constructs designed to optimize T cell epitope coverage (IM mosaic Env). Twenty additional animals were subsequently enrolled and received the SIVmac239 DNA/rAd5 regimen mucosally via aerosol (AE) administration (AE mac239 Env). As previously reported, IM vaccination with DNA/rAd5 SIVmac239 Env (IM mac239 Env) provided significant protection from infection (69% vaccine efficacy), whereas IM vaccination with mosaic DNA/rAd5 Env (IM mosaic Env) did not10. Interestingly, we report for the new study arm designed to address efficacy in the context of a different route of administration, that mucosal AE vaccination with DNA/rAd5 SIVmac239 Env (AE mac239 Env) conferred significant protection from infection (Fig. 1A: 70% vaccine efficacy). Thus, both IM and AE routes induced immune responses that provided equivalent levels of protection from infection. As previously reported, no association was observed between T cell immunity and risk of infection10. Whereas peak but not set point viral loads were reduced in both IM regimens, no reduction in virus was observed following AE SIVmac239 immunization (Supplementary Table 1).
Figure 1. Equivalent protection from SIV acquisition in SIVmac239 immunized animals despite striking Fc-profile differences induced via distinct routes of immunization. (A-D) Mucosal vaccination induces protective immunity.
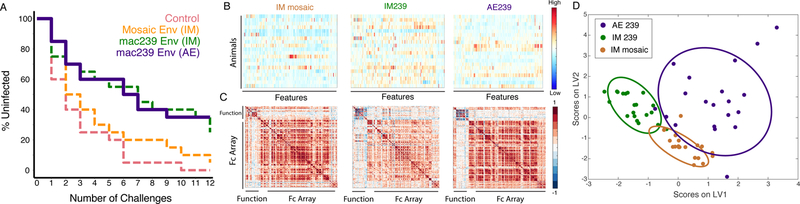
(A) Kaplan-Meier curves depicting the fraction of animals uninfected in Env-vaccinated and control arms following successive challenges (n = 80 animals). One animal in the IM mosaic, five animals in the IM239, and five animals in the AE239 arm remained uninfected after 12 challenges. Dashed lines correspond to data previously reported in Roederer et al Nature 201410, the solid line corresponds to newly reported data. (B–D) Divergent antibody responses result from modification of route of immunization and immunogen. (B) Heatmaps of humoral responses assessed for Env-vaccinated animals. Rows correspond to animals and columns correspond to the normalized (centered and scaled) Fc array and functional assay readouts (features), where high responses are indicated in red and low responses in blue. (C) Feature versus feature correlation matrices in heatmap form illustrate correlative relationships between all feature pairs within each Env-vaccinated study arm, with correlated responses indicated in red and anti-correlated responses in blue. (D) Partial Least Squares (PLS) latent variable (LV) scores biplot using features selected from an arm classification model illustrating that the groups can be separated accurately based on differences in humoral responses. Ellipses correspond to 95% confidence intervals for each group.
Distinct humoral immune profiles induced by different routes of vaccination
Given the strong associations previously reported between multiple antibody assessments and the probability of infection in the IM mosaic and IM SIVmac239 arms10, SIV-specific serum antibody responses were systematically profiled to define humoral correlates of reduced risk of infection. The ability of serum antibodies to induce effector functions was quantified by a suite of functional assays including measures of ADCC and NK cell activation, phagocytic activity mediated by neutrophils (ADNP) and monocytes (ADCP), and complement-dependent cytotoxicity3. Lastly, glycosylation and Fc receptor binding profiles of antigen-specific antibodies were defined11–13 to link these functional activities to the biophysical and biochemical profiles of the vaccine-specific humoral responses. Responses to a number of variants of SIV envelope proteins (gp140, gp120) as well as those directed at specific epitopes, including scaffolded and synthetic variable (V) loop peptides and constant (C) C1, C2, and C3 region peptides were assessed (Supplementary Table 2). No SIV Env-specific humoral immune responses were observed in animals vaccinated with empty vectors or Gag-containing vectors (Supplementary Fig. 1A), and these animals were not considered in humoral immune profiling analyses. In contrast, Env-specific humoral immune responses were observed among each of the three Env-immunized vaccine groups (IM mosaic Env, IM mac239 Env, AE mac239 Env), albeit Env-specific IgG titers were lower in the mosaic vaccine group (Supplementary Fig. 1A), as previously reported10. Beyond this difference in magnitude, the overall architecture of the humoral immune response was remarkably different among the three Env-containing vaccine regimens, as evident both in variability across individual humoral response features (Fig. 1B), as well as in the correlative relationships observed between antibody features (Fig. 1C) including functional and biophysical (Fc Array) assessments. In multivariate space, all three regimens could be robustly distinguished (Fig. 1D, Supplementary Fig. 1B–I), confirming that the route of immunization shapes the Fc-profile of the vaccine-induced immune response.
Vaccine-induced humoral immune responses predict protection across arms
To further define the unique biophysical humoral correlates associated with reduced risk of infection, a Cox proportional hazards model was trained and tested7, using aggressive feature filtering, along with repeated cross-validation and permutation testing. The final model accurately captured overall survival trends, including differential protection among the arms, in the absence of any knowledge of arm identities (Fig. 2A). In repeated cross-validation, predictions of risk of infection for individual animals were highly concordant with observed times to infection (representative C-index: 0.73, P < 10−11; Supplementary Fig. 2A) and distinguished the significantly higher protection observed in the IM and AE SIVmac239 study arms as compared to the IM mosaic vaccine recipients (Fig. 2B). The robustness of this result was established by the significant and substantial difference in predictive accuracy between models trained with actual challenge data compared to permuted data (Supplementary Fig. 2B). Specifically, the final model employed four features as a representative minimal set of features that were most predictive of protection (Fig. 2C, Supplementary Fig. 2C–D). Of the three positive correlates of protection, two corresponded to the ability of V1a (variable loop) and C1.TR (a constant region 1 sequence variant that was sensitive to neutralization10) peptide-specific antibodies to bind to rhesus FcγR2A.4, and one corresponded to the ability of SIVmac239 gp140-specific antibodies to bind to the complement cascade initiating C1q protein. Conversely, C1q-binding SIVsmE543 gp140-specific antibodies were predictive of increased risk of infection, pointing to both the importance of fine specificity in the humoral response (peptide versus whole protein) as well as particular Fc-binding characteristics (FcγR2 versus complement) for predicting protection. Thus models built on Fc-correlates could predict protection from infection, raising the possibility that Fc-effector function could play a critical part in protection from infection following DNA/rAd5 vaccination.
Figure 2. Fc-biophysical antibody binding profiles accurately predict protection across immunogens and routes of administration.
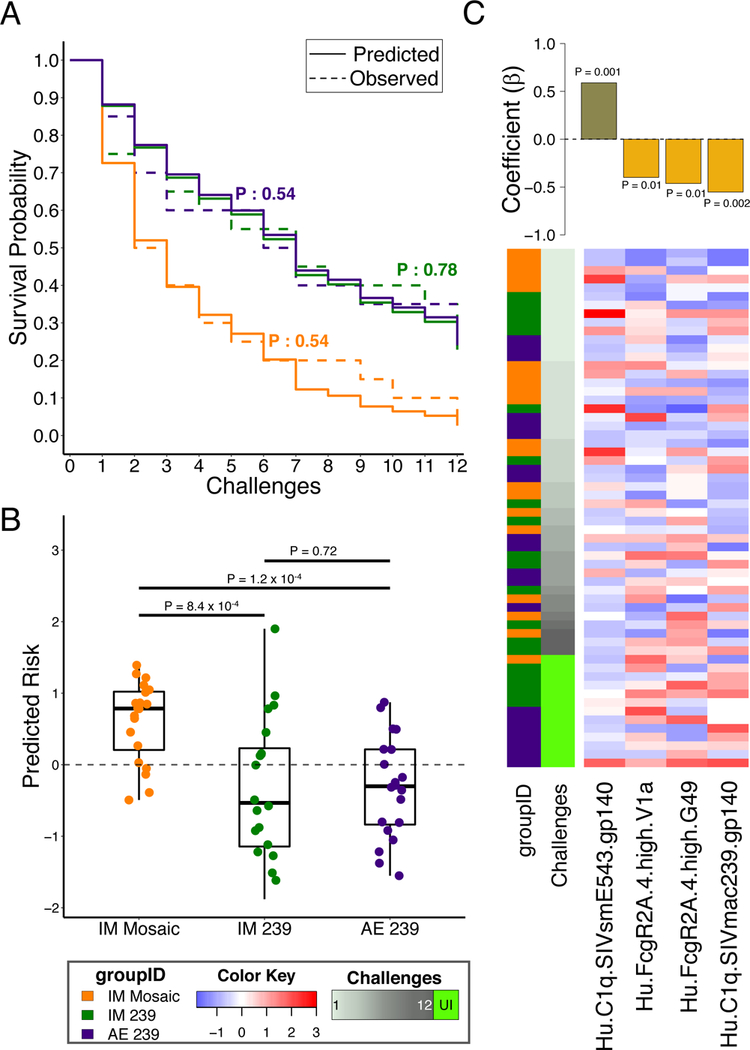
(A) The predicted infection probabilities in the final Cox Proportional Hazards (PH) model closely match observed Kaplan-Meier curves for each of the three Env-immunized study arms (n = 20 animals in each of three arms; P-values: two-sided log-rank test). (B) Animals in the IM mosaic arm have significantly higher predicted risk of infection than those in the other arms in the representative cross-validation run (n = 20 animals in each of three arms; P-values: two-sided Wilcoxon-Mann-Whitney). Boxplots depict the median and interquartile range. (C) Heatmap and feature coefficient plot of the humoral response features (columns) contributing to the final model (coefficients in bars), with one predictive of risk and three of protection (P-values: Cox PH). The animals (rows, n = 60) are ordered in ascending order of time-to-infection. Centered and scaled antibody feature values are presented in heatmap form, with high responses indicated in red and low responses in blue.
Arm-specific correlates point to distinct mechanisms of humoral immune protection against SIV
Given the differences in humoral responses between arms (Fig. 1), the specific humoral profiles, including Fc-functions, associated with protection within the two SIVmac239-immunized vaccine arms were further probed using combined feature selection and classification approaches. Distinct Fc-profiles were observed among “susceptible” animals (infected with four or fewer challenges) and more “resistant” animals (uninfected, or infected after ten or more challenges) in both the IM and AE vaccine arms, with animals with “intermediate resistance” falling between these groups (Fig. 3A,D, Supplementary Fig. 3A–D).
Figure 3. Phagocytic vaccine-specific functional antibodies predict protection from infection.
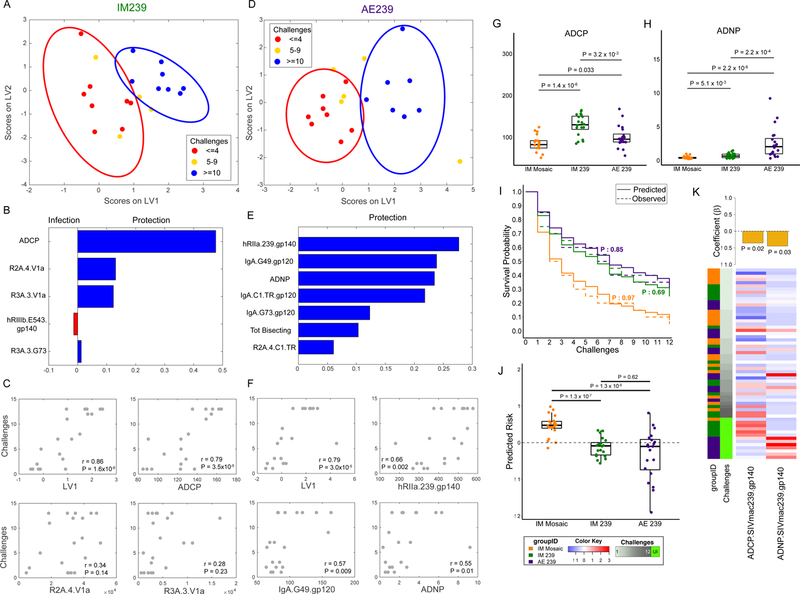
(A,D) PLS LV scores biplot using features selected from a challenge outcome classification model for IM239 (A) and AE239 (D) illustrating stratification of animals based on number of challenges survived. Ellipses correspond to 95% confidence intervals for each group. (B,E) Variable Importance in the Projection (VIP) plot for IM239 (B) and AE239 (E) models illustrating the contribution of each humoral response feature used to stratify the animals. (C,F) Scatter plots illustrating the correlation between number of challenges and I) LV1 from the PLS model and II) the three individual variables making the greatest contribution to the model (n = 20 animals in each of the two arms; two-sided P-values for Spearman’s rho). (G,H) ADCP (G) and ADNP (H) activity by arm (n = 20 animals in each of the two arms; P-values: two sided Wilcoxon-Mann-Whitney). Boxplots depict the median and interquartile range. (I) Comparison of observed KM curves with those predicted by Cox models learned from the functional antibody features (n = 20 animals in each of the two arms; two-sided log-rank test P-values). (J) Animals in the IM Mosaic arm have significantly higher predicted risk of infection than those in the other arms in the representative cross-validation run (n = 20 animals in each of the three arms; P-values: two-sided Wilcoxon-Mann-Whitney). Boxplots depict the median and interquartile range. (K) Heatmap and feature coefficient plot of the two functional features. (P-values: Cox PH). The animals (rows, n = 60) are ordered in ascending order of time-to-infection. Centered and scaled phagocytic activity values are presented in heatmap form with high responses indicated in red and low responses in blue.
This prediction of protection in the IM arm was achieved with a minimal set of select Fc-features (Fig. 3B), one of which was shared with the Cox proportional hazards model (Figure 2C). These features included four variables that were higher in the “resistant” animals and one variable that was higher in the “susceptible” animals. The protective features included one antibody effector function – the ability of gp120-specific antibodies to drive antibody dependent cellular phagocytosis (ADCP) via monocytes, as well as three peptide-specific Fc receptor-binding features including V1a-specific antibodies able to engage the rhesus macaque FcγR2A.4 (shared with the CoxPH model) and FcγR3A.3 variants, and G73 (C2 region peptide)-specific antibody binding to rhesus FcγR3A.3. Conversely, the ability of SIVsmE543-specific antibodies to bind to human FcγR3b was a negative predictor of protection, highlighting peptide-specific responses, and the balance between recognition of Env variants associated with protection (SIVmac239) and susceptibility (SIVsmE543). Although the SIVmac239 and SIVsmE543 sequences have an overall identity of 83%, sequence variability at sites of immune pressure, such as within the V1V2 loop10, may provide a rationale as to why recognition of SIVmac239 is associated with protection, and recognition of SIVsmE543 is associated with susceptibility, suggesting potentially novel sites of functional antibody vulnerability on the viral envelope. Because phagocytes typically express FcγR2 receptors, but may also express FcγR3 upon maturation, the array of biomarkers selected by the model suggest that “protective” humoral responses access both activating FcγR2 and FcγR3 receptors to mediate their “protective” functions. Moreover, while the five-feature multivariate profile provided highly significant prediction of protection (r=0.86, P=1.6×10−6), the level of ADCP alone was also highly correlated with the number of challenges required to infect animals (r=0.79, P=3.5×10−5), highlighting the mechanistic value of this functional assay in resolving protective IM-induced humoral immune profiles against SIV (Fig. 3C).
Despite use of an identical immunogen, distinct biomarkers were enriched among “protected” animals in the AE arm (Fig. 3E), including the ability of SIVmac239 gp140-specific antibodies to interact with human FcγR2a, multiple IgA specificities, the ability of antibodies to drive antibody dependent neutrophil phagocytosis (ADNP), the level of glycan bisection among antigen-specific antibodies, and C1.TR (neutralization sensitive C1 region peptide variant)-specific antibody binding to the rhesus FcγR2A.4 variant. Human neutrophils express both the FcγR3b and the FcαR, enabling them to respond to immune complexes consisting of both or either vaccine-specific IgG and IgA antibodies14. Moreover, the addition of the bisecting GlcNAc has been linked to improved FcγR3a binding and ADCC15, potentially also potentiating ADNP via enhanced FcγR3b binding, which share nearly identical extracellular domains16. Importantly, both SIVmac239 gp140-specific FcγR2a-binding antibodies and G49 (V1b peptide)-specific IgA were strongly associated with protection (Fig. 3F), arguing for a potential mechanistic collaboration between AE induced vaccine-specific IgG and IgA in driving ADNP and protection against SIV.
Thus collectively, phagocytic activity was associated with protection in both the IM and AE vaccine arms, albeit through distinct innate immune effector cells, and via distinct antibody isotype profiles.
Vaccine route directs unique functional profiles that predict protection from infection
The striking finding that protection was associated with the recruitment of distinct antibody effector functions suggested that distinct antibody effector mechanisms may contribute to reducing the risk of SIV acquisition. Univariate examination of ADCP and ADNP (Fig. 3G,H, Supplementary Fig. 3E) activity among the three Env-immunized arms highlighted the clear enhancement of ADCP induced by the SIVmac239 IM immunization compared to both the AE and IM mosaic immunization, as well as the significantly higher levels of ADNP activity induced via AE immunization as compared to the other two vaccine arms. This functional diversity highlights the importance of vaccine route in driving antibody effector profiles. Moreover, Cox modeling analysis, using just these two functional features, yielded a final model (Fig. 3K) that closely and robustly matched the survival outcomes (Fig. 3I,J, Supplementary Fig. 3F,G), and differentiated arms. Importantly, ADCP was not elevated among protected animals in the AE arm, nor was ADNP elevated in protected animals in the IM arm, highlighting the distinct phagocytic functional correlates of protection against SIV driven by the route of immunization (Supplementary Fig. 3H–J).
Vaccine route directs specific isotype selection differences governing antibody functional activity
IgA has been implicated both as a predictor of risk of HIV infection2 as well as linked to protection from infection in NHPs17. Thus to further probe the nature of the IgG and IgA biomarkers associated with protection in the AE arm, both IgG and IgA antibodies, levels of which varied significantly across arms (Fig. 4A,B), were selectively depleted from the SIVmac239 AE and IM immunized animal plasma samples and both ADCP and ADNP were tested. As expected given its high serum abundance, IgG depletion resulted in a significant reduction in ADCP and ADNP activity across both arms (Fig. 4C,D), pointing to the critical role of IgG as the dominant isotype in driving immune complex formation and antibody-effector function. Conversely, despite its lower serum concentration, IgA depletion resulted in a significant reduction in ADNP activity in the AE vaccine arm (Fig. 4D). In contrast, IgA depletion did not significantly impact ADNP in the IM arm (Fig. 4D), consistent with the lower levels of IgA induced by IM immunization. Collectively, the near complete loss of ADNP function following either IgG or IgA depletion suggests that in serum, despite differences in their induction associated with route of immunization, these isotypes can work collaboratively to drive antibody effector function(s) linked with reduced risk of SIV acquisition.
Figure 4. Dissecting mechanisms of arm-specific protection.
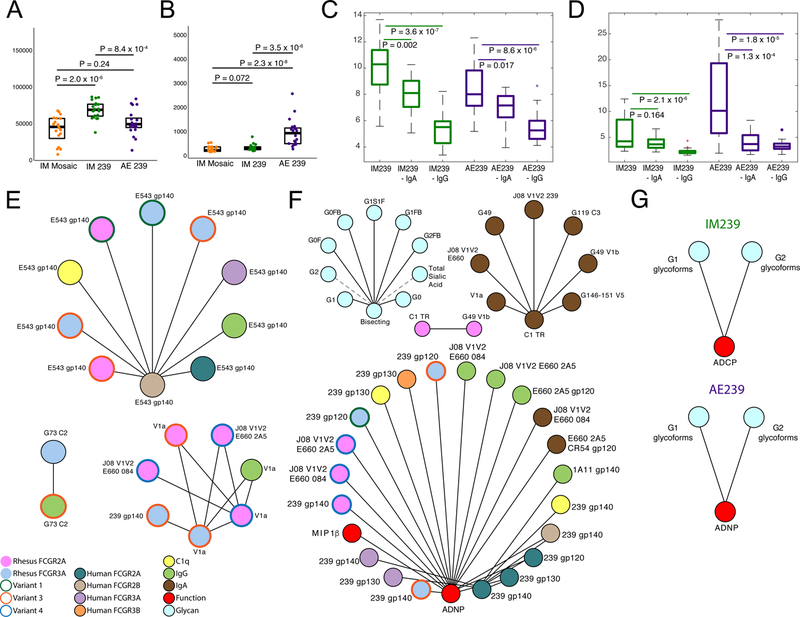
(A&B) Arm-wise comparison of IgG (A) and IgA (B) measurements (n = 20 animals in each of the two arms; P-values: two sided Wilcoxon-Mann-Whitney) indicating distinct isotype profiles associated with route of administration. (C&D) Arm-wise comparison of the impact of IgA and IgG depletion on ADCP (C) and ADNP (D) activity (n = 20 animals in each of the two arms; P-values: two-sided Wilcoxon-Mann-Whitney for pairwise comparisons). (E&F) Co-correlate networks illustrating variables that are significantly correlated with biomarkers linked to protection for the IM239 (E) and AE239 (F) study arms. (G) Arm-specific non-linear relationships between functional correlates of protection and glycans. Boxplots depict the median and interquartile range.
Humoral network analyses distinguish arm-specific protective antibody profiles
We next aimed to define the features linked to the modeled biomarkers of protection, as they may provide further mechanistic clues related to antibody-mediated restriction of SIV. Antibody features that were significantly correlated (|r| >0.7, Benjamini-Hochberg FDR < 0.01) with the biomarkers identified here were defined for the IM (Fig. 4E) and AE (Fig. 4F) vaccine arms. Specifically, humoral immune network analyses of the IM group highlighted the critical nature of antibodies that interact with FcγR2 and FcγR3, and those targeting the SIVmacE660 and 239 antigen, V1a and G73 (C2) peptides as correlates of the biomarkers associated with protection. Conversely, humoral responses targeting SIVsmE543 were associated with the correlate of risk. Similarly, network analyses of the AE group pointed to recognition of SIVmac239, E660 and V1V2 specificities via IgA or FcγR binding IgG responses. Additionally, analyses were performed to define the specific glycoforms associated with each protective function (ADCP activity in IM and ADNP in AE arms). Shared SIV gp120-specific IgG G1 and G2 glycoforms, were associated with each mode of phagocytosis (Fig. 4G, Supplementary Fig 4), suggesting that IgG antibodies induced within both arms may have shared FcγR binding characteristics whose activity was further tuned in the AE arm by the additional induction of IgA antibodies. Thus, reduced risk of infection, in both the IM and AE arms, was driven by IgA or FcγR-binding antibodies targeting specific regions of the viral envelope glycoprotein, distinct from those present in SIVsmE543.
The robust functional humoral correlates of immunity against SIV identified in this study target known sites of vulnerability2,10, but diverge functionally by route of immunization, providing a strong rationale for identifying functional correlates and co-correlates of humoral immunity beyond in vitro neutralization potency. Overall, these data show that distinct innate immune effector mechanisms, which both lead to phagocytic clearance, may contribute to restriction of SIV.
Cross-study conservation of vaccine correlates
The ultimate identification of a set of minimal correlates of protection, able to predict protection within and across studies, could provide critical insights for the evaluation and development of next generation vaccines. To define minimal correlates, features associated with protection across study arms and modeling approaches were considered. Four key humoral response characteristics were consistently associated with reduced risk of infection, capturing biologically sound, non-overlapping, but unique, variables. These four features included ADNP and ADCP as functional markers, and IgA and FcγR2A binding activities of vaccine-specific Abs as biophysical features associated with these functions. These four aspects of the humoral response were both distinct among study arms and predictive of challenge outcomes, capturing both mucosal and parenteral correlates of immunity. To validate the predictive significance of correlates defined by the DNA/rAd5 regimen in an independent vaccine and challenge setting, we assessed the four features in a cohort of animals that were primed with ALVAC and boosted with HIV recombinant gp120 proteins, analogous to the RV144 HIV vaccine trial in humans7. Important to rigorously evaluating the potential breadth of these correlates beyond SIV, in this study, animals were immunized against HIV, and challenged with a tier 2 SHIV. Remarkably, two of the four correlates, including FcγR2a binding and ADCP, showed significant (P < 0.05) univariate associations with reduced risk of infection (Fig. 5A). Notably, protection outcomes were accurately predicted in the ALVAC/protein immunized animals using a model based on the four features identified as correlates of protection in the DNA/rAd5 immunized animals (ADCP, ADNP, FcγR2A, and IgA), and trained using data only from the DNA/rAd5 immunized animals (Fig. 5B). This combination of the four DNA/rAd5-defined biomarkers defined in the SIV study exhibited better agreement with the challenge outcome (rmodel = 0.74) than any individual biomarker (rbest individual feature = 0.67), for the ALVAC/protein HIV vaccine, further supporting the robustness of the co-correlate signature across vaccine strategies. Finally, the co-correlates accurately captured overall survival trends (Fig. 5C). These data argue for conserved correlates of immunity against both SIV and SHIV, applicable across different immunization regimens and associated with different routes of administration. Protection appears to be governed by the ability of virus-specific antibodies to drive rapid phagocytic clearance, likely through the recruitment of distinct innate immune effector cells.
Figure 5. DNA/rAd5 minimal biomarkers of vaccine response also predict protection in ALVAC-protein immunized NHPs.
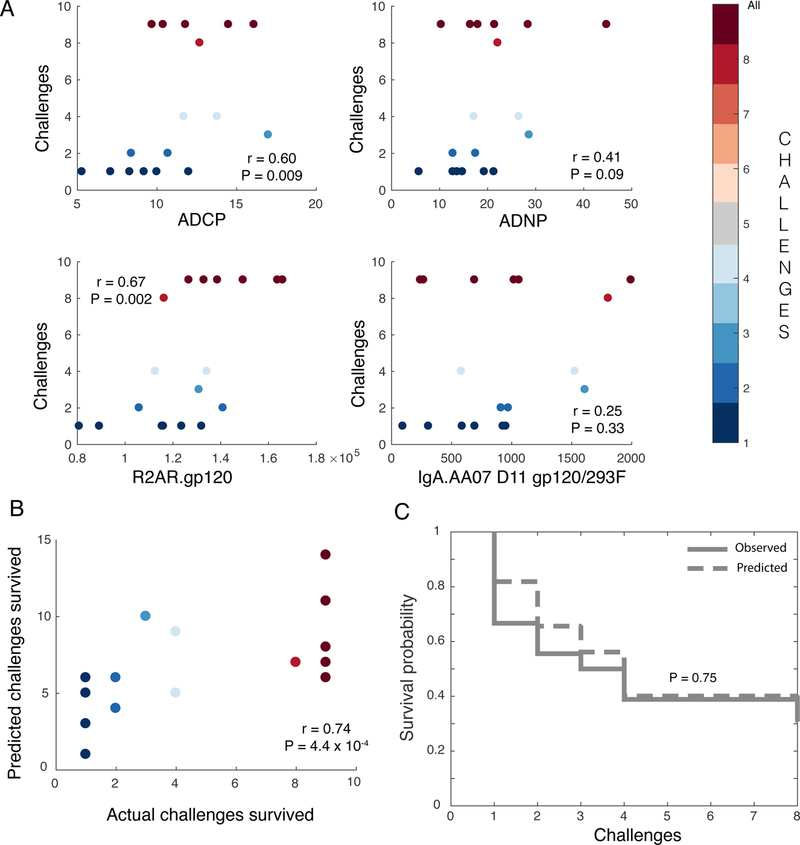
(A) Scatter plots illustrating the correlation (Spearman) of the four individual co-correlates (ADCP, ADNP, FcγR2A, and IgA) identified in the DNA/rAd5 vaccine groups with protection (i.e., number of challenges required to achieve infection) in the ALVAC/protein immunized animals (n = 18 animals; two-sided P-values for Spearman’s rho). (B) Biplot depicting the correlation between observed and predicted challenge outcomes of ALVAC/protein-immunized animals based on the challenge outcome regression model learned from the DNA/rAd5 vaccine (n = 18 animals; two-sided P-values for Spearman’s rho). (C) Survival curves depict actual and predicted infections using a Cox PH model utilizing the integrated four co-correlates (n = 18 animals; P-value: two-sided log-rank test).
Discussion
Here we aimed to define whether functional humoral correlates could explain the previously observed differences in vaccine efficacy following IM immunization with different Env antigens, while simultaneously defining whether these correlates are conserved or diverge in the setting of mucosal vaccination with the same vaccine product. Reduced risk of infection following both IM and AE vaccination were strongly associated with the induction of unique Fc-effector profiles among the vaccinated animals, driven by route of immunization. Remarkably, despite the striking differences in IM and AE vaccine induced Fc-profiles, protection was associated with phagocytosis in both vaccine arms. However, within-arm protection was associated with unique Fc-biomarker profiles, marked by elevated IgG and ADCP in the IM immunized animals versus elevated IgA and ADNP in the AE immunized animals. Importantly, each correlate did not predict protection in the converse arm, and total IgG responses had no relationship to challenge outcomes overall. Moreover, a combined correlate signature defined from both the AE and IM arm could accurately predict protection in a separate vaccine study using distinct immunogens (ALVAC/protein) and a SHIV challenge. These data point to multiple specific functional correlates of protective humoral immunity against SIV or SHIV; antibody-mediated phagocytosis associated with the deployment of different innate effector cells and induced in a vaccine route-specific manner was significantly associated with a reduced risk of infection.
While there are a number of known and likely additional yet-to-be-characterized differences in human and macaque antibodies18,19, FcγRs20,21, and effector cells expression profiles22, the systems show evidence of converging evolution, and studies in both species have suggested relationships between antibody effector functions and protection. For example, previous correlates analyses following Ad26 vaccination in NHP pointed to a role for ADCP in protection3,4. Here, ADCP was associated with reduced risk of infection in both DNA/rAd5 IM and ALVAC/protein immunized animals, suggesting that ADCP may represent a common correlate of protection following IM vaccination. Furthermore, vaccine-specific IgG3 antibody levels, which were associated with a reduced risk of infection in the moderately protective RV144 vaccine23, also correlated with robust ADCP24, further suggesting that ADCP may also track with enhanced protection in humans. Given the abundance of phagocytic cells within mucosal tissues25, as well as their recruitment among the first infiltrators upon tissue assault26,27, ADCP represents a promising mechanism for antibody-mediated control and clearance, of both cell-free and cell-associated virus.
Surprisingly, while ADCP was linked to protection following IM vaccination, protection following mucosal vaccination was associated with a remarkably different set of biomarkers, including elevated levels of IgA and ADNP. Decades of research have highlighted the opportunities to skew immunity via immunization within different immune compartments including muscle, the skin, and within different mucosal compartments (mouth, nose, vagina, etc)28. For example, nasal immunization has been associated with the induction of both IgA and IgG responses in human cervicovaginal secretions29–31, potentially providing a unique opportunity to harness particular innate immune effector cell functions localized at sites of vulnerability to infection. Moreover, given the abundance of neutrophils, both in the blood and within the female reproductive tract, as well as their ability to be rapidly migrate to sites of injury and infection, the combination of IgA and IgG may uniquely leverage neutrophil activity and deploy a broad array of anti-pathogen activities including phagocytosis, degranulation, and the formation of DNA NETs involved in trapping and preventing infectious spread32. However, given both the immunoprotective and immunopathological roles of neutrophils, balanced IgG:IgA responses may be key to leveraging the speed and anti-pathogen activity of these innate effectors.
Immune correlates of risk in the RV144 trial suggested that among vaccine recipients, IgA levels were associated with enhanced risk of infection2, suggesting the possibility that they may compete for antigen and block IgG activity33. However, network analyses in case:control samples from the RV144 vaccine trial34 suggested that subjects that generated elevated IgA levels had lower IgG levels, and specifically generated lower IgG3 antibody responses, which were associated with reduced risk of infection23 and polyfunctional activity24. Moreover, previous passive transfer studies in NHP have demonstrated the protective activity of IgA antibodies35. Moreover, consistent with the possibility that elevated IgA might be a surrogate, rather than mechanistically involved in reducing protection, IgA levels were not associated with risk of infection in ALVAC/protein immunized monkeys7. Here we observed that IgA can work collaboratively with IgG to drive both ADNP and ADCP. Moreover, IgA-mediated immunity in the AE vaccine arm was linked to viral targets within the V1V2 loop, highlighting the importance of targeting key sites of vulnerability via functional antibody isotypes. Thus, the induction of IgA antibodies that synergize with IgG may provide an opportunity to direct additional innate immune effector function(s) and enhance protection from infection.
Overall, this comprehensive immune correlates analysis strongly argues for a role for functional antibodies in reducing the risk of SIV and SHIV acquisition. Moreover, while several antibody effector functions including ADCC and complement activation were also interrogated in this study, only phagocytic functions mediated by monocytes or neutrophils, and associated with IgA or IgG, were associated with reduced risk of infection across vaccine regimens and challenge viruses. These results demonstrate that protection against SIV challenge may be achievable via the induction of antibodies that promote phagocytosis via distinct innate Fc-effector cell types, and whose isotypes are naturally controllable via the route of immunization. Further, such effector activities are potentially required for distal eradication of the virus upon viral/infected-cell escape beyond the mucosal compartment36. Importantly, we demonstrate that multiple mechanisms may protect against viral infection, suggesting that vaccines eliciting a combination of antibody types and a combination of phagocytic activities may be more potent. Thus, efforts able to direct the humoral immune response to sites of vulnerability while simultaneously leveraging the potent antiviral activity of antibody Fc-effector functions may contribute to the rational design of a protective vaccine against HIV.
Online Methods
Study Cohort
DNA/rAd5 study:
80 Indian origin rhesus macaques were immunized intra-muscularly (IM) with a DNA prime, rAd5 boost with four varying immunogens10. Twenty additional animals were subsequently enrolled and received one of the same vaccine regimens but delivered mucosally via aerosol (AE) administration. The control group received empty vectors, whereas the IM and AE arms received SIVmac239 Env sequence inserts (83% sequence identify with challenge strain), and the “mosaic” Env group received two SIV Env mosaic immunogens37 (78% and 87% sequence identity to the challenge virus) designed to optimize coverage of T cell epitopes present among diverse virus strains, via the IM route. The fifth group received only SIV Gag immunization. All animals received three injections of DNA at four week intervals, followed by rAd5 vaccination at week 30, and were randomized across groups for TRIM5α alleles, gender, age, and weight. Animals were challenged each week beginning at week 53 with an intrarectal administration of smE660 at a dose that infected 30% of control animals per exposure38 for either a period of 12 weeks or until they exhibited detectable infection as defined by plasma viremia. Virology assessments were performed as previously described via real-time PCR and single genome amplification10. No differences were observed in FcR genotype across groups or with protection. Antibody assessments were performed at the peak (two weeks) post-boost time point.
ALVAC/protein study:
26 Indian origin male and female rhesus monkeys (Macaca mulatta) were vaccinated intramuscularly (IM) with ALVAC (vCP1521; Sanofi Pasteur) and recombinant Env gp120 protein(s) in GLA-SE (IDRI-EM107)7 Animals were then challenged intrarectally with SHIV-1157(QNE)Y173H weekly for a period of eight weeks, or until they exhibited detectable viremia7. Antibody assays for this study were reported previously7.
All animals were handled in accordance with the standards of the American Association for the Accreditation of Laboratory Animal Care (AAALAC) and work met NIH standards as set forth in the Guidelines for Care and Use of Laboratory Animals.
Effector Function Assays
The functional activity of SIV-specific antibodies was determined in a number of cell-based assays3,39,40 (Supplementary Table s2). Functional assays were performed in triplicate, with the exception of the NK activation assay, which was performed in duplicate. Replicates of assays performed using primary cells or primary plasma as a source of effectors or complement were performed across multiple donors. Averages across replicates are reported.
ADCP.
Antibody dependent cellular phagocytosis (ADCP) was determined using antigen-coated fluorescent beads, as described41,42. Briefly, fluorescent, streptavidin-conjugated microspheres were coated with chemically biotinylated SIVmac239 gp120 (Immune Technology) for 2 hrs at 37°C and washed with 0.1% BSA in PBS. The antigen-coupled beads were incubated with diluted plasma for 2 hrs at 37°C, washed with PBS to remove unbound antibodies, and then incubated with 2.5 x 104 THP-1 cells overnight at 37°C. Cells were then fixed with 4% paraformaldehyde solution and analyzed on a flow cytometer. A phagocytic score was derived as an integrated MFI by multiplication of the fraction of the percent and mean fluorescence intensity of bead-positive THP-1 cells.
ADCC.
Antibody dependent cellular cytotoxicity (ADCC) was tested using a rapid fluorescent ADCC assay, which assesses the ability of antibodies to drive primary NK cells to lyse gp120-pulsed target cells43. Briefly, CEM.NKr target cells were pulsed with SIVmac239 gp120 protein (Immune Technology) at 60 μg/mL and then labeled with the membrane dye PKH26 and the intracellular dye CFSE. NK cells were enriched from healthy donor whole blood by negative selection using RosetteSep (Stem Cell Technologies). Purified IgG (100 μg/mL) was mixed with the labeled CEM.NKr cells and then NK cells were added at an E:T ratio of 10:1 and incubated for 4 hrs at 37°C. Cells were then fixed, and analyzed by flow cytometry. gp120-specific lysis was measured as the percentage of PKH26+ CFSE– CEM.NKr target cells after incubation.
ADNP.
Antibody-dependent neutrophil phagocytosis (ADNP) was determined using an adaptation of a flow cytometry-based phagocytic assay described previously41,42. Briefly, fluorescent, streptavidin-conjugated microspheres were coated with chemically biotinylated SIVmac239 gp120 (Immune Technology) for 2 hrs at 37°C and washed with 0.1% BSA in PBS. The antigen-coupled beads were incubated with a 1:100 dilution of plasma for 2 hrs at 37°C and washed with PBS to remove unbound antibodies. WBCs were isolated from healthy donor blood anti-coagulated with acid citrate dextrose by ACK lysis of red blood cells. 5×104 WBCs were added to the opsonized beads and incubated for 1 hr at 37°C and then stained with anti-CD66b-Pacific Blue (Biolegend). Cells were then fixed with 4% paraformaldehyde solution and analyzed on a flow cytometer. A phagocytic score was derived as an integrated MFI by multiplication of the fraction of neutrophils that phagocytosed one or more opsonized beads by the MFI of this population.
NKA.
An assay to determine NK cell activation (NKA) state based on the expression of surface CD107a and intracellular production of IFNγ and MIP1β was performed as previously described3. NK cells were isolated from whole blood from healthy donors using negative selection with RosetteSep (STEMCELL Technologies), added to a SIVmac239 gp120-absorbed plate with purified IgG, anti-CD107a-phycoerythrin (PE)-Cy5 (BD), brefeldin A (Sigma), and GolgiStop (BD), and incubated for 5 hrs at 37°C. After the incubation, cells were stained for surface markers using anti-CD16-allophycocyanin (APC)-Cy7 (BD), anti-CD56-PE-Cy7 (BD), and anti-CD3-Alexa Fluor 700 (BD) and then stained intracellularly with anti-IFNγ-APC (BD) and anti-MIP1β-PE (BD) after treatment with Perm A and B solutions (Invitrogen). Cells were then fixed in 4% paraformaldehyde and analyzed by flow cytometry. NK cells were defined as CD3-negative and CD16-positive and/or CD56-positive, and the percent of NK cells positive for each marker was determined.
ADCD.
The ability of donor antibodies to induce complement component C3b deposition on SIVmac239 gp120-coated target cells was assessed by flow cytometry as previously described3. Briefly, CEM-NKr target cells were pulsed with gp120SF162 (60 μg/ml), then incubated with purified antibody at a concentration of 100 μg/mL and freshly harvested HIV negative donor plasma diluted 1:10 with veronal buffer 0.1% gelatin as a source of complement for 20 min at 37˚C. Following a wash with 15 mM EDTA in PBS, cells were fixed and complement deposition was detected by staining with anti-C3b-FITC (Cedarlane). The proportion of C3b-positive cells was determined based on negative controls in which heat inactivated donor plasma was used.
Fc Array
The quantity and qualitative features of HIV-specific antibodies were evaluated using a custom multiplex array in which SIV-specific antibodies were characterized according to their titer (anti-IgG, anti-IgA), and ability to interact with human and/or rhesus innate immune antibody FcγR (FcγRIIa, IIIa, IIIb and their major allotypic variants20,44), and human C1q, an initiator of the complement cascade, essentially as previously described13. Previous studies show that these low affinity human and rhesus FcγR molecules generally exhibit consistent binding preferences for rhesus IgG types20. Biotinylated FcR (FcγR and C1q) were biotinylated either chemically at a molar ratio of 5 mols biotin per mol of protein using EZ-Link Sulfo-NHS-SS-Biotin (Pierce) or site-specifically via BirA-mediated enzymatic modification (Avidity). Immediately prior to use, each biotinylated detection reagent was mixed with a 1/4th molar ratio of Streptavadin-PE (Prozyme), and diluted to a final concentration of 1.0 µg/mL in Assay Buffer (PBS + 0.1% BSA + 0.05% Tween). The ability of antigen-specific antibodies to bind to these detection reagents was assessed across a diverse panel of HIV antigens coupled to uniquely fluorescently-coded magnetic beads (Luminex Corp.) (Supplementary Table 2) as follows.
A working mixture of coupled microspheres was added to dilute plasma in black, clear bottom 384-well plates (Greiner Bio One). Following incubation for two hrs at RT on an XYZ plate shaker (IKA), plates were washed and antigen-specific antibody was detected using the tetrameric PE-conjugated detection reagents described above, at 1.0 µg/mL, or R-phycoerthrin (PE)-conjugated anti-Ig secondary reagents, at 0.65 µg/ml, with 50 µL/well. After a one hr incubation at room temperature on a shaker, beads were washed five times with 65 µL of sheath fluid (Luminex 40–75680), and microspheres were resuspended in 40 µL of sheath fluid. A Bio-Plex array reader (FlexMap 3D, Luminex Corp) detected the microspheres and binding of each PE-functionalized detection reagent was measured to calculate a Median Fluorescence Intensity (MFI). These features were named according to the following convention: “detection reagent.antigen”. Fc Array measurements were pre-processed for quality, eliminating measurements that had a per-arm median MFI values less than 500 across the three vaccine arms. After this filtering step, a total of 109 features remained, which were considered in the classification and survival analyses.
Antibody Depletions
IgG and IgA were depleted using isotype specific CaptureSelect Affinity Matrix resin (ThermoFisher). Specifically, 100 µL of 1:10 diluted plasma was added to 50 µL of resuspended and washed resin (based on 8 mg/mL binding capacity), and incubated for 1 hour with end-to-end mixing, at room temperature. The depleted fraction (flow through) was collected by centrifugation for 2 minutes at 1500 rpm to eliminate the target isotype. Depletion was confirmed by luminex analysis and the concentration of sample was assessed by ELISA to establish concentrations of flow through for functional experiments in comparison to undepleted (control) plasma.
Glycan Analysis
A capillary electrophoresis-based technique for analyzing plasma-derived polyclonal IgG-glycosylation was used to determine the relative abundance of glycan structures decorating gp120-specific and total plasma IgG, as previously described12. Briefly, glycans were released from IgG with the N-linked glycosidase PNGase F, were fluorescently labeled by reductive amination with 8-aminopyrene-1,3,6-trisulfonic acid, separated from unreacted dye, and analyzed on an Agilent 3130XL ABI DNA sequencer. Peak identities were determined by exoglycosidase reactions and use of glycan standards, and the prevalence of each glycan species was determined by peak area integration with a custom script. The prevalence of 19 individual glycoforms and 6 summary glycosylation state assessments were determined (Supplementary Table 2). Summary glycosylation characteristics, such as level of galactosylation (G) and sialylation (S), and presence of absence of fucose (F) or bisecting GlcNAc (B) were compiled by summing individual glycoforms.
Data Analysis and Visualization
Basic data analysis and visualization were performed using GraphPad Prism along with in-house scripts developed either for the R statistical computing environment45 (supported by standard R packages caret, ggplot2, and gplots) or the Matlab statistical computing environment (supported by the Statistics and Optimization toolboxes). Network visualizations were performed using Cytoscape46. Raw data are available in Supplementary Information.
Classification of vaccine arms
Classification models were trained to distinguish animals between the three vaccine arms based on measured functional, biophysical and biochemical humoral immune responses. Models were built using an approach similar to one described previously34 – a combination of the least absolute shrinkage and selection operator (LASSO)47 for feature selection and then classification using the LASSO-selected features (Supplementary Code File 1). The robustness of this modeling approach was evaluated using repeated five-fold cross validation. For each five-fold cross validation run, primates were randomly divided into five subsets. This was done such that for each fold, four subsets served as the training set, and the fifth one served as the test set, with each subset serving as the test set once. For each such fold, LASSO-based feature selection was performed using only the four subsets designated as the training set for that fold, with the coefficient for the LASSO penalty term determined via an additional internal five-fold cross validation using only the fold-specific training dataset. A fold-specific support vector machine (SVM)48 classifier (linear kernel with default Matlab parameters) was trained using the LASSO-selected features and training data for that fold. (Thus each fold may have a different LASSO penalty parameter, different features and different coefficients.) This fold-specific classifier was then used to predict labels for the primates in the test set for that fold. This process was performed for each of the five folds, and repeated 100 times for different five-fold splits. Visualizations of latent variables from a partial least square discriminant analysis (PLSDA)34,49 model trained on the LASSO-selected features are reported.
The significance of the predictive performance of the models was assessed with two complementary control approaches. As described above, the 100 repetitions of five-fold classification generated a distribution of model classification accuracies. Corresponding model accuracy distributions were generated for two types of control models. The first control model followed a permutation testing50 approach, by randomly shuffling the data with respect to the arm labels prior to feature selection, within a cross validation framework. This generates permutation-specific control models. The features themselves were not shuffled, preserving the correlation structure of the data. The second control used, within a cross-validation framework, randomly selected size-matched subsets of features (the size-matching was fold-specific). Here too the correlation structure of the data was preserved. For each control model approach, these processes were repeated 100 times to generate a distribution of model accuracies observed in the context of permuted data and randomly selected size-matched feature sets.
The entire procedure was repeated across all 5 cross validation folds. After running through all 5 folds, we compared the predicted arm label for each primate (since each primate was in the test set in exactly one of the 5 cross validation folds) to the true arm labels, and obtained a true classification accuracy (equivalent to average classification accuracy across folds as the folds are of equal size). We computed the P value as the tail probability of the true classification accuracy in the distribution of control model classification accuracies. We reported median P values across 20 independent cross validation replicates.
LASSO-regularized multinomial logistic regression was also performed to identify humoral properties able to classify animals into their respective vaccine arm. These classifier models were trained via the R package ‘glmnet’51 with default options (Supplementary Code File 2). To enable visual inspection of a single set of model coefficients, a final model was trained using the penalty parameter (lambda) that achieved the lowest cross-validation classification error. To visualize classification confusion and log odds predictions of arm, a single run of ten-fold cross-validation was performed; the predicted class log odds were obtained from the ratio of the predicted probability for the true arm to the maximum of the predicted probabilities for the other arms. Predictive performance was assessed in terms of balanced accuracy (mean true positive rate) over 100 repetitions of ten-fold cross-validation, with training/testing splits determined randomly while ensuring that each testing set included an equal number of animals from each vaccine arm. Robustness was established through permutation testing50, randomly shuffling the arm labels and performing cross-validation following the exact same process as with the original data (i.e. training, feature selection, and classification). This process was repeated 100 times, randomly shuffling the arm labels for each repetition. Random selection models were trained by randomly selecting feature sets size-matched to the number of features in the final model and were evaluated using the same cross-validation framework as with the original data. A random size-matched subset of features was selected for each repetition of cross-validation. Robustness was quantified in terms of effect size (Cliff’s ∆) analysis of the cross-validation vs. permutation testing balanced accuracy distributions, as well as the tail probability of the average balanced accuracy from repeated cross-validation with actual data vs. the distribution from permutation testing.
Survival analysis
An updated version of a previously described multivariate survival analysis approach7 was used to train Cox proportional hazards (CoxPH) regression models52 to predict the risk of infection at each challenge point using Fc Array features (Supplementary Code File 2). Key steps are as follows:
Feature preanalysis.
To aid the modeling process in dealing with redundancy, features were clustered according to their profiles across all animals (ignoring infection status) using hierarchical agglomerative clustering53, as implemented by the R function ‘hclust’ from the ‘stats’ package54 with the ‘ward.D2’ method. The tree was cut so as to yield between 5 to 15 clusters, and a set of 9 clusters was selected as maximizing the distance between the nearest cluster centroids.
Cross-validation.
The animals were split into 10 subsets, or folds, such that each fold contained an equal number of animals from each of the vaccine arms. For each fold, the animals from the remaining folds were used as the training set, and the animals in the fold as the test set.
Feature filtering.
Within the cross-validation, the features in each cluster were ranked by their correlations with time-to-infection for the training set animals, as evaluated by polyserial correlation coefficient55, which measures correlation between discrete and continuous variables. The top ranked feature from each cluster was shortlisted for consideration in modeling. Pair-wise correlations among the shortlisted features were minimized by selecting features in decreasing order of polyserial coefficient magnitude and eliminating subsequent features that were highly correlated (Pearson correlation coefficient > 0.7) to the selected features.
Model training.
CoxPH models were trained using the R package ‘survival’56, with greedy backward feature elimination57 employed to select features during training by iteratively eliminating features as long as the training set likelihood was no more than 25% worse than an initial model trained using all of the shortlisted features.
Risk prediction.
Since the CoxPH model is semiparametric, with the baseline hazard function unspecified, the absolute risk of an individual animal cannot be estimated. Hence, risk of infection for each animal was predicted relative to the mean of all animals, which provides a constant baseline to compare risk across all animals. Predictive performance was assessed using the Concordance index (C-index) metric58 over 100 repetitions of ten-fold cross-validation.
Representative run and final model.
A representative run of ten-fold cross-validation was performed using the most frequent features, defined as those appearing in at least 50% of the models obtained over the repeated cross-validation. A final model was trained using all the animals and the same set of high-frequency features to predict arm-wise risk (aggregate KM curves) using the “mean” animal of each arm. To do so, the probability of infection at each challenge point for the three vaccine arms was estimated by making predictions on ‘mean’ animals, whose features represent mean of the features over the respective arms’ animals. The ‘survfit.coxph’ function in R package ‘survival’56 was used to estimate these probabilities. The predicted and observed probabilities for each group were compared using the log-rank statistic. A high P value of the log-rank statistic favors the null hypothesis that the two curves are similar.
Robustness.
Permutation testing was performed by randomly shuffling the challenge labels and then following the exact same cross-validation approach as was used for the actual labels (i.e. feature filtering, model training, and risk prediction). The process was repeated 100 times, randomly shuffling the challenge labels for each repetition. Random selection models were trained by randomly selecting feature sets size-matched to the number of features in the final model (see below) and were evaluated using the same cross-validation framework as with the actual data. A random size-matched subset of features was selected for each repetition of cross-validation. Robustness was quantified in terms of effect size (Cliff’s ∆) analysis of the cross-validation vs. permutation testing C-index distributions, as well as the tail probability of the average C-index from repeated cross-validation vs. the distribution of C-indices from permutation testing.
Co-correlates.
Fc features correlated to each feature in the final model (Pearson correlation coefficient > 0.75) were considered as possible substitutes. For each such possible substitute, a ten-fold cross-validation was performed to assess performance of a variant of the final model using the substituted instead of the corresponding final feature.
Analysis of functional data.
Risk predictions were made with the two functional measurements that showed distinct arm-specific responses (ADCP, and ADNP), the feature filtering and greedy backward elimination strategies were not needed, so standard bivariate CoxPH models were directly trained using the “survival”56 R package. The predictive performance and robustness of the models was again evaluated using the 100-repeated ten-fold cross-validation and permutation testing approaches described above.
Arm-specific classification models of challenge outcomes
Arm-specific models of protection were also built using a LASSO/SVM approach similar to that described above for arm classification (Supplementary Code File 1). A separate model was built for each of the two SIVmac239-immunized arms – IM239 and AE239. The target variable was the number of challenges. The model classified animals into three discrete bins based on the number of challenges survived (survived ≤ 4 challenges, survived 5–9 challenges or survived ≥10 challenges). The performance of each arm-specific model was measured in a five-fold cross-validation framework as described above. The significance of the models was assessed by comparing the real models to control models, based on either permuted data or randomly selected features, as described above (in the arm classification section). P values were computed using permutation tests, as described in the arm classification section. Visualizations of latent variables from a PLSDA model trained on the LASSO-selected features are reported.
Cross-study co-correlate validation
To investigate the ability of the four key humoral correlates identified in the IM and AE arms (ADCP, ADNP, FcγR2A, and IgA) to predict protection in an alternative regimen and challenge study – samples from a cohort of animals that were primed with ALVAC and boosted with HIV recombinant gp120 proteins, analogous to the RV144 HIV vaccine trial in humans7, were Fc-profiled. Univariate Spearman correlations were first computed for each of the 4 correlates and the number of challenges. Next, a PLS regression model was built using only the 4 co-correlates – ADCP, ADNP, FcγR2A, and IgA. The model was trained using data corresponding to only the DNA/rAd5 primates. This model was then used to predict the number of challenges required to infect animals in the second -ALVAC/protein study. This strategy provides the most stringent evaluation of the model as it is trained using one cohort, and tested using a completely different cohort. The performance of the model was evaluated in terms of the Spearman correlation between the actual number of challenges survived and the predicted number of challenges survived for the test set i.e., the primates in the ALVAC/protein study. Finally, a Cox proportional hazards model was built using the four co-correlates only. The survival curve predicted by this model was compared to the actual survival curve for the ALVAC/protein study primates.
Correlation networks
Correlation networks were constructed as previously described34. Briefly, Spearman correlations between all pairs of features were computed and the significance of the correlations was assessed using the false discovery rate (post Benjamini-Hochberg multiple testing correction). Significant correlations were defined as those with |r| > 0.7 and FDR <0.01. These significant correlations were visualized as a network.
Non-linear glycan-function relationships
First, using the maximal information coefficient (MIC)59, relationships between glycan levels and ADCP for the IM239 arm, and ADNP for the AE239 arm were quantified. MIC is a maximal information-based nonparametric exploration statistic that can capture a wide range of relationships including non-linear ones. Next, an alternate metric, the randomized dependence coefficient (RDC)60 was used to validate the identified associations. RDC is a measure of nonlinear dependence between random variables of arbitrary dimension, based on correlations of random non-linear copula projections. Significant associations were defined as those with a P < 0.05 based on a permutation test. Consistent results were obtained using both metrics.
Supplementary Material
Acknowledgements
These studies were supported by the Bill and Melinda Gates Foundation (OPP1032817 and OPP1114729) and the NIH (R37 AI080289, R01 AI102291, P01 AI120756, R01 AI131975, and R01 AI102660). We would like to thank Dr. William Evan Johnson, Associate Professor of Medicine and Biostatistics, Boston University, for his help with statistical review.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Code and data availability
Code, detailed documentation, and relevant datasets are available in supplementary information.
References:
- 1.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361, 2209–2220 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366, 1275–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch DH, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349, 320–324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155, 531–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alpert MD, et al. ADCC develops over time during persistent infection with live-attenuated SIV and is associated with complete protection against SIV(mac)251 challenge. PLoS Pathog 8, e1002890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouts TR, et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc Natl Acad Sci U S A 112, E992–999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley T, et al. Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat Commun 8, 15711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449, 101–104 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Bournazos S, et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roederer M, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505, 502–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown EP, et al. Microscale purification of antigen-specific antibodies. J Immunol Methods 425, 27–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahan AE, et al. A method for high-throughput, sensitive analysis of IgG Fc and Fab glycosylation by capillary electrophoresis. J Immunol Methods 417, 34–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown EP, et al. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J Immunol Methods 443, 33–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futosi K, Fodor S & Mocsai A Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int Immunopharmacol 17, 638–650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umana P, Jean-Mairet J, Moudry R, Amstutz H & Bailey JE Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol 17, 176–180 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Bruhns P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Watkins JD, et al. Anti-HIV IgA isotypes: differential virion capture and inhibition of transcytosis are linked to prevention of mucosal R5 SHIV transmission. AIDS 27, F13–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boesch AW, et al. Biophysical and Functional Characterization of Rhesus Macaque IgG Subclasses. Front Immunol 7, 589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobsen FW, et al. Molecular and functional characterization of cynomolgus monkey IgG subclasses. J Immunol 186, 341–349 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Chan YN, et al. IgG Binding Characteristics of Rhesus Macaque FcgammaR. J Immunol 197, 2936–2947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trist HM, et al. Polymorphisms and interspecies differences of the activating and inhibitory FcgammaRII of Macaca nemestrina influence the binding of human IgG subclasses. J Immunol 192, 792–803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warncke M, et al. Different adaptations of IgG effector function in human and nonhuman primates and implications for therapeutic antibody treatment. J Immunol 188, 4405–4411 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Yates NL, et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med 6, 228ra239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung AW, et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci Transl Med 6, 228ra238 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Sips M, et al. Fc receptor-mediated phagocytosis in tissues as a potent mechanism for preventive and therapeutic HIV vaccine strategies. Mucosal immunology 9, 1584–1595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen GY & Nunez G Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol 10, 826–837 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi C & Pamer EG Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11, 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolton DL, Song K, Tomaras GD, Rao S & Roederer M Unique cellular and humoral immunogenicity profiles generated by aerosol, intranasal, or parenteral vaccination in rhesus macaques. Vaccine 35, 639–646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson EL, Wassen L, Holmgren J, Jertborn M & Rudin A Nasal and vaginal vaccinations have differential effects on antibody responses in vaginal and cervical secretions in humans. Infect Immun 69, 7481–7486 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlowski PA, Cu-Uvin S, Neutra MR & Flanigan TP Comparison of the oral, rectal, and vaginal immunization routes for induction of antibodies in rectal and genital tract secretions of women. Infect Immun 65, 1387–1394 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardelli-Haefliger D, et al. Specific antibody levels at the cervix during the menstrual cycle of women vaccinated with human papillomavirus 16 virus-like particles. J Natl Cancer Inst 95, 1128–1137 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Kolaczkowska E & Kubes P Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Tomaras GD, et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc Natl Acad Sci U S A 110, 9019–9024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung AW, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell 163, 988–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sholukh AM, et al. Defense-in-depth by mucosally administered anti-HIV dimeric IgA2 and systemic IgG1 mAbs: complete protection of rhesus monkeys from mucosal SHIV challenge. Vaccine 33, 2086–2095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, et al. Antibody-mediated protection against SHIV challenge includes systemic clearance of distal virus. Science 353, 1045–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer W, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 13, 100–106 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Letvin NL, et al. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Sci Transl Med 3, 81ra36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackerman ME, et al. Polyfunctional HIV-Specific Antibody Responses Are Associated with Spontaneous HIV Control. PLoS Pathog 12, e1005315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaccari M, et al. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22, 762–770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackerman ME, et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. Journal of immunological methods 366, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAndrew EG, et al. Determining the phagocytic activity of clinical antibody samples. J Vis Exp, e3588 (2011). [DOI] [PMC free article] [PubMed]
- 43.Gomez-Roman VR, et al. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J Immunol Methods 308, 53–67 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Boesch AW, et al. Highly parallel characterization of IgG Fc binding interactions. MAbs 6, 915–927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R: A language and environment for statistical computing. R Foundation for Statistical Computing (2013).
- 46.Smoot ME, Ono K, Ruscheinski J, Wang PL & Ideker T Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tibshirani R Regression shrinkage and selection via the Lasso. J Roy Stat Soc B Met 58, 267–288 (1996). [Google Scholar]
- 48.Cortes C & Vapnik V Support-Vector Networks. Mach Learn 20, 273–297 (1995). [Google Scholar]
- 49.Lau KS, et al. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Science signaling 4, ra16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ojala M & Garriga GC Permutation Tests for Studying Classifier Performance. J. Mach. Learn. Res 11, 1833–1863 (2010). [Google Scholar]
- 51.Friedman J, Hastie T & Tibshirani R Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 52.Cox DR Regression Models and Life-Tables. Journal of the Royal Statistical Society. Series B (Methodological) 34, 187–220 (1972). [Google Scholar]
- 53.Hastie T, Tibshirani R & Friedman JH The elements of statistical learning : data mining, inference, and prediction, (Springer, New York, 2009). [Google Scholar]
- 54.R: A language and environment for statistical computing. R Foundation for Statistical Computing (2018).
- 55.Drasgow F Polychoric and Polyserial Correlations. in Encyclopedia of Statistical Sciences (eds. Kotz S, Read CB, Balakrishnan N, Vidakovic B & Johnson NL) (John Wiley and Sons, Inc, 2006). [Google Scholar]
- 56.Therneau TM & Grambsch PM Modeling survival data : extending the Cox model, (Springer, New York, 2000). [Google Scholar]
- 57.Guyon I, Andr, #233 & Elisseeff. An introduction to variable and feature selection. J. Mach. Learn. Res 3, 1157–1182 (2003). [Google Scholar]
- 58.Harrell FE Jr., Lee KL & Mark DB Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in medicine 15, 361–387 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Reshef DN, et al. Detecting novel associations in large data sets. Science 334, 1518–1524 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez-Paz D, Hennig P & Scholkopf B The Randomized Dependence Coefficient. in 26th International Conference on Neural Information Processing Systems, Vol. 1 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


