Abstract
Background
Pasteurella multocida is responsible for significant economic losses in pigs worldwide. In clinically diseased pigs, most P. multocida isolates are characterised as subspecies multocida, biovar 2 or 3 and capsular type A or D; however, there is little information regarding subspecies, biovars, and other capsular types of P. multocida isolates in Korea. Here, we provided information covering an extended time period regarding P. multocida in pigs with pneumonia in Korea using phenotypic and genotypic characterisations and data associated with the minimum inhibitory concentrations.
Results
The overall prevalence of P. multocida between 2008 and 2016 was 16.8% (240/1430), with 85% of the P. multocida isolates (204/240) coinfected with other respiratory pathogens. Of the 240 isolates, 166 were included in this study; all of these P. multocida isolates were characterised as subspecies multocida and the most prevalent phenotypes were represented by biovar 3 (68.7%; n = 114) and capsular type A (69.9%; n = 116). Additionally, three capsular type F isolates were identified, with this representing the first report of such isolates in Korea. All biovar 1 and 2 isolates were capsular types F and A, respectively. The virulence-associated gene distribution was variable; all capsular type A and D isolates harboured pmHAS and hsf-1, respectively (P < 0.001), with type F (biovar 1) significantly correlated with hsf-1 (P < 0.05) and pfhA (P < 0.01), biovar 2 highly associated with pfhA and pmHAS, and biovar 3 significantly correlated with hsf-1, pmHAS, and hgbB (P < 0.001), whereas biovar 13 was related only to hgbB (P < 0.05). The highest resistance rate was found to be to oxytetracycline (63.3%), followed by florfenicol (16.3%).
Conclusions
P. multocida subspecies multocida, biovar 3, and capsular type A was the most prevalent isolate in this study, and our findings indicated the emergence of capsular type F in Korea. Moreover, prudent use of oxytetracycline and florfenicol is required because of the identified high resistance rates. Further studies are required for continuous monitoring of the antimicrobial resistance, prevalence, and epidemiological characterisation of P. multocida, and experimental infection models are needed to define the pathogenicity of capsular type F.
Keywords: Antimicrobial resistance, Biovar, Capsular type, Pasteurella multocida, Virulence-associated gene
Background
Pasteurella multocida (P. multocida) is a commensal and opportunistic pathogen of the oral, nasopharyngeal, and upper respiratory tract [1] and the causative agent of a wide range of infections leading to high economic impact [2]. In pigs, P. multocida is associated with progressive atrophic rhinitis (PAR), and together with other respiratory pathogens, plays a significant role in porcine respiratory disease complex (PRDC) [3–6]. P. multocida prevalence has been reported as 8.0% in diseased pigs with pneumonia or PAR in China, and from 10.3 to 15.6% in pigs with pneumonia in Korea. Additionally, P. multocida constitutes 15.6% of isolated respiratory pathogens in the United States [3, 5, 7, 8].
P. multocida can be divided into three subspecies (multocida, septica, and gallicida) and 13 biovars (1–10 and 12–14) based on carbohydrate fermentation and production of the ornithine decarboxylase (ODC) enzyme [9–11]. The majority of swine isolates are subspecies multocida and mostly assigned as biovars 2 or 3 [1, 10, 12, 13]. Additionally, five capsular types based on capsular antigens (A, B, D, E, and F) have been described in P. multocida, with capsular types A, B, D, and F recovered from swine [1, 14]. Capsular types A and D are most commonly cultured from pneumonic lungs and PAR, respectively, whereas capsular types B and F are rarely isolated from pigs [3, 14–16]. In Korea, numerous studies suggest that capsular type A is more prevalent in porcine pneumonia than type D [7, 8, 15]; however, limited information is available regarding subspecies, biovars, and other capsular types of P. multocida isolates in Korea.
P. multocida reportedly possesses various virulence factors that play a significant role in pasteurellosis and survival in the host environment [3, 17, 18]. Furthermore, there is a clear correlation between certain virulence factors and capsular types or biovars [1, 3]. The functions and target genes of these factors are detailed in Table 1 and include those encoding outer membrane and porin proteins (oma87, ompH, plpB, and psl), adhesins (fimA, pfhA, ptfA, hsf-1, and hsf-2), superoxide dismutases (sodA and sodC), iron-acquisition-related factors (exbB, exbBD-tonB, fur, tbpA, hgbA, and hgbB), neuraminidases (nanB and nanH), hyaluronidase (pmHAS), and toxin (toxA). Identifying which virulence factors are prevalent is necessary to predict the pathogenic behaviour of the isolates and select potential future vaccine candidates.
Table 1.
Primers used for the detection of capsular types and virulence-associated genes in Pasteurella multocida isolates
| Gene function | Target gene | Description | Sequence (5′ – 3′) | size (bp) | Reference |
|---|---|---|---|---|---|
| Capsule serotypes | kmt1 | Identification of all P. multocida isolates | ATCCGCTATTTACCCAGTGG GCTGTAAACGAACTCGCCAC |
460 | [19] |
| hyaD-hyaC | Serogroup A cap gene | TGCCAAAATCGCAGTGAG TTGCCATCATTGTCAGTG |
1044 | [20] | |
| bcbD | Serogroup B cap gene | CATTTATCCAAGCTCCACC GCCCGAGAGTTTCAATCC |
760 | [20] | |
| dcbF | Serogroup D cap gene | TTACAAAAGAAAGACTAGGAGCCC CATCTACCCACTCAACCATATCAG |
657 | [20] | |
| ecbJ | Serogroup E cap gene | TCCGCAGAAAATTATTGACTC GCTTGCTGCTTGATTTTGTC |
511 | [20] | |
| fcbD | Serogroup F cap gene | AATCGGAGAACGCAGAAATCAG TTCCGCCGTCAATTACTCTG |
851 | [20] | |
| Outer membrane and porin proteins | oma87 | Outer membrane protein 87 | ATGAAAAAACTTTTAATTGCGAGC TGACTTGCGCAGTTGCATAAC |
948 | [17] |
| ompH | Outer membrane protein H | CGCGTATGAAGGTTTAGGT TTTAGATTGTGCGTAGTCAAC |
438 | [17] | |
| plpB | Outer membrane protein | TTTGGTGGTGCGTATGTCTTCT AGTCACTTTAGATTGTGCGTAG |
282 | [18] | |
| psl | Porin protein | TCTGGATCCATGAAAAAACTAACTAAAGTA AAGGATCCTTAGTATGCTAACACAGGACGACG |
470 | [17] | |
| Adhesins | fimA | Fimbriae | CCATCGGATCTAAACGACCTA AGTATTAGTTCCTGCGGGTG |
866 | [18] |
| pfhA | Filamentous haemagglutinin | AGCTGATCAAGTGGTGAAC TGGTACATTGGTGAATGCTG |
275 | [21] | |
| ptfA | Fimbriae | TGTGGAATTCAGCATTTTAGTGTGTC TCATGAATTCTTATGCGCAAAATCCTGCTGG |
488 | [17] | |
| hsf-1 | Autotransporter adhesion | TTGAGTCGGCTGTAGAGTTCG ACTCTTTAGCAGTGGGGACAACCTC |
654 | [18] | |
| hsf-2 | Autotransporter adhesion | ACCGCAACCATGCTCTTAC TGACTGACATCGGCGGTAC |
433 | [18] | |
| Superoxide dismutases | sodA | Superoxide dismutases | TACCAGAATTAGGCTACGC GAAACGGGTTGCTGCCGCT |
361 | [17] |
| sodC | Superoxide dismutases | AGTTAGTAGCGGGGTTGGCA TGGTGCTGGGTGATCATCATG |
235 | [17] | |
| Iron acquisition related factor | exbB | Iron regulated and acquisition factors | TTGGCTTGTGATTGAACGC TGCAGGAATGGCGACTAAA |
283 | [18] |
| exbBD-tonB | Iron acquisition related factors | GGTGGTGATATTGATGCGGC GCATCATGCGTGCACGGTT |
1144 | [17] | |
| fur | Iron regulated and acquisition factors | GTTTACCGTGTATTAGACCA CATTACTACATTTGCCATAC |
244 | [18] | |
| tbpA | Iron acquisition related factor | TGGTTGGAAACGGTAAAGC TAACGTGTACGGAAAAGCC |
728 | [21] | |
| hgbA | Haemoglobin binding protein | TGGCGGATAGTCATCAAG CCAAAGAACCACTACCCA |
419 | [17] | |
| hgbB | Haemoglobin binding protein | TCATTGAGTACGGCTTGAC CTTACGTCAGTAACACTCG |
499 | [21] | |
| Neuraminidases | nanB | Neuraminidases | GTCCTATAAAGTGACGCCGA ACAGCAAAGGAAGACTGTCC |
554 | [17] |
| nanH | Neuraminidases | GAATATTTGGGCGGCAACA TTCTCGCCCTGTCATCACT |
360 | [17] | |
| Hyaluronidase | pmHAS | Hyaluronidase | TCAATGTTTGCGATAGTCCGTTAG TGGCGAATGATCGGTGATAGA |
430 | [18] |
| Toxin | toxA | Dermonecrotic toxin | TCTTAGATGAGCGACAAGG GAATGCCACACCTCTATAG |
846 | [21] |
Antimicrobial resistance in pathogenic bacteria from food animals and the environment has become a global public health issue. Although beta-lactams, trimethoprim combination, florfenicol, macrolides, and tetracyclines have been shown to be the best antimicrobials for treating PRDC [6], resistance to these antimicrobials has been detected previously in P. multocida in many countries [3, 22–24]. In Korea, P. multocida isolates from pigs are sensitive to most antimicrobial agents, including ampicillin, ceftiofur, tilmicosin, and enrofloxacin, other than tiamulin [7].
To the best of our knowledge, only short-term studies have been performed to characterise porcine P. multocida isolates in Korea. This long-term study was carried out to provide baseline information regarding a large collection of P. multocida isolates from clinically diseased pigs by determining the distribution and association of capsular types, biovars, extensive virulence-associated gene profiles, and antimicrobial-resistance patterns.
Results
Prevalence of P. multocida in porcine pneumonic lungs
In total, 240 P. multocida isolates (16.8%) were recovered (Table 2); P. multocida was the second most frequently isolated bacterial pathogen in this study. Most isolates (85.0%; 204/240) were detected simultaneously with other respiratory pathogens, such as porcine reproductive and respiratory syndrome virus (PRRSV; 61.3%), porcine circovirus type 2 (PCV2; 37.5%), or Streptococcus suis (S. suis; 20.0%). Mycoplasma hyorhinis (MHR), Actinobacillus pleuropneumoniae (APP), Mycoplasma hyopneumoniae (MHP), Haemophilus parasuis (HPS), Trueperella pyogenes (T. pyogenes), and swine influenza virus (SIV) were detected to a lesser extent (19.2, 14.2, 10.4, 10.0, 4.6, and 3.8%, respectively). Of the P. multocida isolates, 166 were included in this study.
Table 2.
Prevalence of respiratory pathogens and the frequency of Pasteurella multocida co-infection with other pathogens
| No. of sample | Bacteriaa | Virusb | Nonec | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PM | SS | MHR | APP | HPS | MHP | TP | PRRSV | PCV2 | SIV | |||
| Total no. (%) of samples in which pathogens were detected | 1430 | 240 (16.8) | 251 (17.6) | 199 (13.9) | 130 (9.1) | 110 (7.7) | 96 (6.7) | 47 (3.3) | 715 (50.0) | 456 (31.9) | 49 (3.4) | 323 (22.6) |
| Total no. (%) of sample co-infected with P. multocida | 240 | 240 (100) | 48 (20.0) | 46 (19.2) | 34 (14.2) | 24 (10.0) | 25 (10.4) | 11 (4.6) | 147 (61.3) | 90 (37.5) | 9 (3.8) | – |
aPM Pasteurella multocida, SS Streptococcus suis, MHR Mycoplasma hyorhinis, APP Actinobacillus pleuropneumoniae, HPS Haemophilus parasuis, MHP Mycoplasma hyopneumoniae, TP Trueperella pyogenes
bPRRSV porcine reproductive and respiratory syndrome virus, PCV2 porcine circovirus type 2, SIV swine influenza virus
cNone, none of the respiratory pathogen was detected from pneumonic lungs
Subspecies, biovar, and capsular type determination
The distribution of biovars and capsular types among the studied P. multocida isolates is shown in Table 3. All 166 isolates were identified as P. multocida subspecies multocida, which produces acid from sorbitol and glucose but not from dulcitol, lactose, and maltose. Most ODC-producing isolates belonged to biovar 3 (68.7%), followed by biovars 2 (21.1%) and 1 (1.8%). Interestingly, 14 isolates (8.4%) displayed identical carbohydrate fermentation results to biovar 3, except for ODC activity, and were thus assigned to biovar 13. All biovar 1 and 2 isolates comprised capsular type F and A, respectively (P < 0.001), whereas biovar 3 isolates comprised capsular types A and D (P < 0.001), and biovar 13 comprised capsular types A and D (P > 0.05). Capsular type A (69.9%) isolates were the most prevalent, followed by types D (28.3%) and F (1.8%), with none of the isolates in this study identified as type B or E. Importantly, this is the first report of capsular type F/biovar 1 isolation since 2014 (Table 3).
Table 3.
Distribution of biovars and capsular types among P. multocida isolates from 2008 to 2016
| Biovar | Capsular type | Total No. (%) |
No. (%) of positive isolates within the following years | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | |||
| Biovar 1 | Cap F | 3 (1.8) *** | 0 | 0 | 0 | 0 | 0 | 0 | 2 (14.3) | 1 (4.0) | 0 |
| Biovar 2 | Cap A | 35 (21.1) *** | 1 (10.0) | 0 | 8 (34.8) | 2 (18.2) | 4 (23.5) | 8 (25.8) | 3 (21.4) | 5 (20.0) | 4 (13.3) |
| Biovar 3 | Cap A | 68 (41.0) *** | 6 (60.0) | 2 (40.0) | 8 (34.8) | 2 (18.2) | 3 (17.6) | 14 (45.2) | 6 (42.9) | 15 (60.0) | 12 (40.0) |
| Biovar 3 | Cap D | 46 (27.7) *** | 3 (30.0) | 2 (40.0) | 4 (17.4) | 4 (36.4) | 7 (41.2) | 9 (29.0) | 3 (21.4) | 3 (12.0) | 11 (36.7) |
| Biovar 13 | Cap A | 13 (7.8) | 0 | 0 | 3 (13.0) | 3 (27.3) | 3 (17.6) | 0 | 0 | 1 (4.0) | 3 (10.0) |
| Biovar 13 | Cap D | 1 (0.6) | 0 | 1 (20.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 166 | 10 | 5 | 23 | 11 | 17 | 31 | 14 | 25 | 30 | |
***P < 0.001
Distribution of virulence-associated genes
Results of polymerase chain reaction (PCR) analysis of 21 virulence-associated genes showed that all P. multocida isolates harboured 14 genes (oma87, ompH, plpB, psl, fimA, hsf-2, sodA, sodC, exbB, ExbBD-tonB, fur, hgbA, nanB, and nanH), whereas tbpA was absent. Notably, the distribution of toxA (5.4%), pfhA (22.9%), hsf-1 (34.9%), pmHAS (69.9%), hgbB (78.3%), and ptfA (99.4%) varied among the 166 P. multocida isolates; this information and the distribution of virulence-associated genes according to capsular type and biovar are presented in Table 4. All capsular type A isolates harboured pmHAS (P < 0.001), and all capsular type D isolates harboured hsf-1 (P < 0.001), with most (97.9%) also harbouring hgbB (P < 0.001). Additionally, capsular type F (biovar 1) was significantly correlated with pfhA (P < 0.01) and hsf-1 (P < 0.05). Notably, toxA was present in only 5.4% (n = 9) of P. multocida isolates, which mainly belonged to biovar 3 (n = 9) and capsular type A (n = 8) (Table 4). Biovar 2 was highly associated with pfhA and pmHAS, whereas biovar 3 was significantly correlated with hsf-1, pmHAS, and hgbB (P < 0.001; Table 4). Most biovar 13 isolates harboured pmHAS and hgbB. The distribution of virulence-associated gene profiles of toxA, hgbB, and pfhA in the different biovars is shown in Table 5. All biovar 1 and 2 isolates were toxA−hgbB+pfhA+ and toxA−hgbB−pfhA+, respectively (P < 0.001), with the toxA−hgbB+pfhA− profile present in most biovar 3 (P < 0.001) and all biovar 13 (P < 0.05) isolates.
Table 4.
Distribution of virulence-associated (VA) genes according to capsular type and biovar in 166 P. multocida isolates
| VA genesa | No. (%) of VA genes within the following capsular types | No. (%) of VA genes within the following biovars | Total No. (% of 166) | |||||
|---|---|---|---|---|---|---|---|---|
| Type A (69.9%, n = 116) | Type D (28.3%, n = 47) | Type F (1.8%, n = 3) | Biovar 1 (1.8%, n = 3) | Biovar 2 (21.1%, n = 35) | Biovar 3 (68.7%, n = 114) | Biovar 13 (8.4%, n = 14) | ||
| toxA | 8 (6.9) | 1 (2.1) | 0 | 0 | 0 | 9 (7.9) * | 0 | 9 (5.4) |
| pfhA | 35 (30.2)*** | 0 | 3 (100) ** | 3 (100) ** | 35 (100) *** | 0 | 0 | 38 (22.9) |
| hsf-1 | 8 (6.9) *** | 47 (100) *** | 3 (100) * | 3 (100)* | 0 | 54 (47.4) *** | 1 (7.1) * | 58 (34.9) |
| pmHAS | 116 (100) *** | 0 | 0 | 0 | 35 (100) *** | 68 (59.6) *** | 13 (92.9) | 116 (69.9) |
| hgbB | 81 (69.8) *** | 46 (97.9) *** | 3 (100) | 3 (100) | 0 | 113 (99.1) *** | 14 (100) * | 130 (78.3) |
| ptfA | 116 (100) | 46 (97.9) | 3 (100) | 3 (100) | 35 (100) | 113 (99.1) | 14 (100) | 165 (99.4) |
*P < 0.05, **P < 0.01, ***P < 0.001
aAll isolates contained the following genes: oma87, psl, ompH, sodA, sodC, ExbBD-tonB, hgbA, nanB, nanH, hsf-2, plpB, fur, fimA, exbB. However, tbpA was not found in any of the isolates
Table 5.
Distribution of the toxA, hgbB, and pfhA gene profiles according to biovars
| Gene profile of toxA/hgbB/pfhA | No. (%) of gene profiles within the following biovars | Total (n = 166) | |||
|---|---|---|---|---|---|
| Biovar 1 (n = 3) | Biovar 2 (n = 35) | Biovar 3 (n = 114) | Biovar 13 (n = 14) | ||
| toxA − hgbB + pfhA + | 3 (100)*** | 0 | 0 | 0 | 3 |
| toxA − hgbB − pfhA + | 0 | 35 (100)*** | 0 | 0 | 35 |
| toxA + hgbB + pfhA − | 0 | 0 | 9 (7.9)* | 0 | 9 |
| toxA − hgbB + pfhA − | 0 | 0 | 104 (91.2)*** | 14 (100)* | 118 |
| toxA − hgbB − pfhA − | 0 | 0 | 1 (0.9) | 0 | 1 |
*P < 0.05, *** P < 0.001
Antimicrobial susceptibility
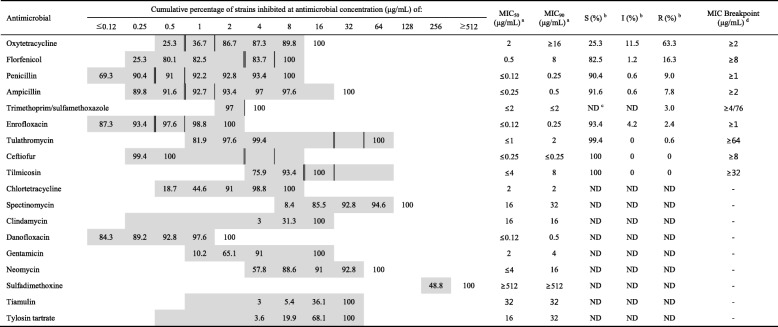
The antimicrobial-resistance patterns, cumulative minimum inhibitory concentrations (MICs), MIC50, and MIC90 of P. multocida isolates from diseased pigs are shown in Table 6. Of the 18 antimicrobials tested, isolates exhibited the highest level of resistance to oxytetracycline (63.3%), followed by florfenicol (16.3%), penicillin (9.0%), ampicillin (7.8%), trimethoprim-sulfamethoxazole (3.0%), enrofloxacin (2.4%), and tulathromycin (0.6%), whereas all isolates were susceptible to ceftiofur and tilmicosin. The MIC90 values of antimicrobials for which breakpoints had not been determined according to Clinical and Laboratory Standards Institute (CLSI) criteria were as follows: chlortetracycline (2 μg/mL), spectinomycin (32 μg/mL), clindamycin (16 μg/mL), danofloxacin (0.5 μg/mL), gentamicin (4 μg/mL), neomycin (16 μg/mL), sulfadimethoxine (≥512 μg/mL), tiamulin (32 μg/mL), and tylosin (32 μg/mL).
Table 6.
Antimicrobial susceptibility and cumulative percentage of P. multocida isolates (n = 166) for 18 antimicrobials
The grey zone represents the tested concentration range of each antimicrobial provided in the BOPO6F plate
Susceptibility and resistance breakpoints are indicated by double vertical (sensitive) and single vertical (resistant) lines according to the guidelines of each reference
aMIC50 and MIC90, concentrations at which the growth of 50 and 90%, respectively, of the isolates is inhibited
bS susceptible, I intermediate, R resistant
cND not determined
dMIC breakpoints applied were those recommended by the Clinical and Laboratory Standards Institute (CLSI); trimethoprim/sulfamethoxazole interpretation was based on a previous study [25]
Discussion
Our findings showed that P. multocida isolates were prevalent (16.8%) in pig farms and the second most frequently isolated bacterial pathogen from diseased pigs, following S. suis (17.6%). This was consistent with previous studies in Korea that reported the prevalence of P. multocida to be between 10 and 15.6% [7, 8]. The infections in this study comprised of a mix of P. multocida (85.0%) with other respiratory pathogens, particularly PRRSV (61.3%; P = 0.0001). Therefore, veterinary practitioners and surveillance stakeholders should consider coinfection with various pathogens that might exist in a given herd for PRDC control.
We characterised 166 P. multocida isolates by determining their subspecies, biovar, capsular type, virulence-associated genes, and MIC. To the best of our knowledge, this is the first report of biovar prevalence in Korea. All isolates belonged to subspecies multocida, and the most prevalent type was biovar 3 (68.7%), which is consistent with the results of previous studies of P. multocida recovered from pigs [1, 10, 13]. P. multocida biovar 1 is frequently isolated from poultry, but not pigs [1]. We found that the prevalence of biovar 13 was 8.4%, which is slightly higher than that in other countries, such as Australia (2.0%) and Hungary (4.8%) [10, 11]. In agreement with numerous previous studies, the dominant P. multocida capsular types recovered from pneumonic pig lungs were capsular types A (69.9%) and D (28.3%) [1, 15, 16, 26]. Additionally, capsular type B is the etiological cause of septicaemic pasteurellosis, whereas type F is rarely reported in pigs [1, 14]. Interestingly, capsular type F has been isolated in Korea post 2014, although at relatively low proportions (n = 3; 1.8%), the prevalence of which is consistent with that reported in other European studies [0.3% (Germany), 1.0% (UK), and 2.4% (Spain)] [1, 2, 16]. A recent Chinese experimental study indicated that pig-origin capsular type F isolates are associated with porcine pneumonia and exhibit high pathogenicity in pigs [27]. Additionally, we found that P. multocida capsular type F was the only relevant respiratory pathogen isolated from three growing pigs with moderate-to-severe suppurative bronchopneumonia with fibrous/fibrinous pleuritis. This represents the first report identifying capsular type F isolates in Korea; therefore, the pathogenic significance of type F in pigs needs to be elucidated.
Virulence genotyping is a useful typing method for molecular characterisation of bacterial pathogens and has been previously applied to P. multocida [1, 3]. Although oma87, ompH, plpB, psl, fimA, hsf-2, sodA, sodC, exbB, ExbBD-tonB, fur, hgbA, nanB, nanH, and ptfA were uniformly distributed among the isolates tested, none possessed tbpA, which agreed with the results of previous pig studies [1–3, 17]. The wide distribution of these genes indicates their importance for the survival of P. multocida within the host environment. Additionally, the virulence factors involved in cross-protection might constitute potential vaccine candidates, regardless of capsular type [3]. However, previous studies demonstrated that several non-uniformly distributed virulence-associated genes exhibit significant relatedness with specific capsular types [1, 3, 8, 17]. As shown in Table 4, all capsular type A and D isolates harboured pmHAS and hsf-1, respectively, and most type D isolates harboured hgbB (P < 0.001). In this study, capsular type F displayed virulence-associated gene profiles similar to those of capsular type D (hsf-1+hgbB+), except for pfhA. Previous studies reported toxA as clearly associated with type D [1, 3, 7, 17]; however, we found that only one of the 47 type D (2.1%) isolates and 6.9% of type A isolates harboured toxA. These results, however, are not significant because most of the isolates were from pneumonic lesions and not from turbinates with PAR. Similar to a previous report, distinct associations were observed between the virulence-associated gene profiles of toxA, hgbB, and pfhA and biovars, except for biovar 13 [1]. All biovar 1, 2, and 13 isolates exhibited toxA−hgbB+pfhA+ (P < 0.001), toxA−hgbB−pfhA+ (P < 0.001), and toxA−hgbB+pfhA− (P < 0.05) profiles, respectively, and most biovar 3 isolates displayed a toxA−hgbB+pfhA− profile (P < 0.001). Additionally, toxA was found only in biovar 3 isolates (toxA+hgbB+pfhA−; P < 0.05).
Swine diseases have become co-infected with immunosuppressive diseases, leading to antimicrobial treatment failure and frequent resistance occurrence. Treatment against P. multocida infections commonly includes broad-spectrum antimicrobials [3]. In this study, beta-lactams (penicillin, ampicillin, and ceftiofur), macrolides (tulathromycin and tilmicosin), and fluoroquinolone (enrofloxacin) were found to be more effective than oxytetracycline and florfenicol. Therefore, these agents are recommended as empirical antimicrobials for the treatment of P. multocida infection. Tetracycline resistance has previously been reported in P. multocida isolates worldwide [3, 6, 25, 28, 29]. Its prevalence in the present study was found to be 63.3%, which is similar to the prevalence in China (58.0%) and North America (53.4%) [3, 28] but higher than that in Australia (28.0%) and European countries (20.4%). Previous studies recommended the use of florfenicol for the treatment of infections caused by P. multocida, because florfenicol-resistance rates are very low (0–2%) in China, North America, Australia, and Europe [6, 25]; however, the present study showed a relatively higher resistance (16.3%). According to the Korea Animal Health Products Association, tetracyclines and florfenicol are the most commonly used antibiotics in Korean pig husbandry [30], with their frequent use reflected in the resistance rates in the present study. Based on the occurrence of high rates of tetracycline and florfenicol resistance, these antimicrobial agents should be used carefully and accompanied by susceptibility tests. Additionally, continuous surveillance of antimicrobial resistance in respiratory pathogens, including P. multocida, is required due to the increasing use of therapeutic antimicrobials and emergence of new resistant strains.
This study was conducted to determine the phenotypic and genotypic characteristics of swine P. multocida isolates in Korea. However, the collected samples cannot be representative of current P. multocida isolates in Korean swine farms, given that the number of isolates submitted annually varies, and the isolates used in this study originated from diagnostic samples with unknown antimicrobial-treatment history. However, a large-scale study for the characterisation of clinical lung samples of P. multocida isolates would sufficiently broaden the understanding of P. multocida as a respiratory pathogen.
Conclusions
This represents a comprehensive report of P. multocida isolates in pigs in Korea. Our findings provide scientific information for further research, including development of vaccine candidates and guidelines for antimicrobial use in veterinary medicine. Moreover, the low discriminatory power of phenotypic characterisation limits the scope of adequate epidemiological information; therefore, different genotyping techniques using pulsed-field gel electrophoresis or multilocus sequence typing might be required to further elucidate the epidemiology of P. multocida and its genetic relatedness.
Methods
Bacterial isolation and identification
In total, 1430 lung samples were collected from pigs (suckling pigs, 9%; weaning pigs, 49%; growing-finishing pigs, 23%; and unknown, 19%) with pneumonic gross lesions from 514 farms nationwide between 2008 and 2016. All lung samples were submitted to the Animal and Plant Quarantine Agency for differential diagnosis of porcine respiratory diseases, including APP, HPS, S. suis, T. pyogenes, MHP, MHR, PRRSV, PCV2, and SIV. Following gross and histopathologic examination, samples were cultured on 5% sheep blood agar, chocolate agar (Asan Pharm. Co., Ltd., Seoul, Korea), and MacConkey agar (Becton Dickinson, Sparks, MD, USA) and then incubated aerobically at 37 °C for 48 h. Suspected mucoid and non-haemolytic colonies were subjected to Gram staining and biochemical identification using the VITEK II system (BioMérieux, Marcy l’Etoile, France). Identification was further confirmed by species-specific PCR assay for amplification of kmt1 (Table 1) [19]. All P. multocida isolates were stored at − 80 °C until use to determine the subspecies, biovar, and capsular type. Previously reported methods were used to differentiate between P. multocida and other pathogens [3, 31, 32].
Subspecies and biovar determination
The confirmed P. multocida isolates were classified into three subspecies (multocida, septica, and gallicida) based on sorbitol and dulcitol fermentation [9]. Additionally, isolates were assigned to one of the established biovars based on their ability to ferment carbohydrates (sorbitol, dulcitol, maltose, xylose, glucose, trehalose, lactose, and arabinose) and produce the ODC enzyme [10].
PCR assay for capsular typing and virulence-associated gene detection
P. multocida isolates were inoculated into brain-heart infusion broth (Becton Dickinson) and cultured for 18 h. Genomic DNA was extracted using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to manufacturer instructions. The capsular types of the isolates were determined by multiplex PCR using the capsule-specific primers shown in Table 1 [20]. PCR analysis of 21 virulence-associated genes, including oma87, ompH, plpB, psl, fimA, pfhA, ptfA, hsf-1, hsf-2, sodA, sodC, exbB, exbBD-tonB, fur, tbpA, hgbA, hgbB, nanB, nanH, pmHAS, and toxA (Table 1) [3, 17, 18, 21], was conducted as previously described. PCR amplification was performed using a Mastercycler ep Gradient S (Eppendorf, Hamburg, Germany), and amplified products were analysed with a capillary electrophoresis system (QIAxcel Advanced System; Qiagen). All tests were performed in duplicate in parallel with the relevant positive and negative controls.
Antimicrobial-susceptibility testing
The MIC of all isolates (n = 166) was determined using the standard broth microdilution method with the Sensititre system (TREK Diagnostic System; Thermo Fisher Scientific, Cleveland, OH, USA) and commercially prepared 96-well antimicrobial testing plates containing 18 different agents (BOPO6F; TREK Diagnostic Systems). The following antimicrobials were tested: penicillin, ampicillin, ceftiofur, florfenicol, gentamicin, neomycin, chlortetracycline, oxytetracycline, clindamycin, enrofloxacin, danofloxacin, trimethoprim/sulfamethoxazole, sulfadimethoxine, spectinomycin, tulathromycin, tylosin tartrate, tilmicosin, and tiamulin. Escherichia coli ATCC 25922 was tested for quality control purposes. As shown in Table 6, the MICs were interpreted according to CLSI guidelines for oxytetracycline, florfenicol, penicillin, ampicillin, enrofloxacin, tulathromycin, ceftiofur, and tilmicosin or those of a previous study describing analysis of trimethoprim/sulfamethoxazole, for which CLSI breakpoints were not available [25, 33]. The overall MIC50 and MIC90 values (i.e., the lowest concentrations at which growth was inhibited by 50 and 90%, respectively) for each antimicrobial were determined for all isolates.
Statistical analysis
Statistical testing was performed using GraphPad Prism (v5.01; GraphPad Software, San Diego, CA, USA) and SPSS (v22.0; IBM Corp., Armonk, NY, USA). Pearson’s chi-squared and Fisher’s exact tests were used to assess associations among capsular types, biovars, and virulence-associated genes. A P < 0.05 was considered statistically significant.
Acknowledgements
The authors would like to thank Yu-Ran Lee and Bun Seung Jo for their technical assistance.
Funding
This work was supported by a grant from the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs of the Republic of Korea. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- APP
Actinobacillus pleuropneumoniae
- CLSI
Clinical and Laboratory Standards Institute
- HPS
Haemophilus parasuis
- MHP
Mycoplasma hyopneumoniae
- MHR
Mycoplasma hyorhinis
- MIC
Minimum inhibitory concentration
- ODC
Ornithine decarboxylase
- P. multocida
Pasteurella multocida
- PAR
Progressive atrophic rhinitis
- PCR
Polymerase chain reaction
- PCV2
Porcine circovirus type 2
- PRDC
Porcine respiratory disease complex
- PRRSV
Porcine reproductive and respiratory syndrome virus
- S. suis
Streptococcus suis
- SIV
Swine influenza virus
- T. pyogenes
Trueperella pyogenes
Authors’ contributions
JK and SIO were involved in study design, performed sample collection, conducted laboratory work, and drafted the manuscript. JWK helped design the study and interpret the data. BS and WIK were involved in designing and analysing the data. HYK interpreted the data and wrote the manuscript. All authors reviewed the article and approved it.
Ethics approval and consent to participate
Non-experimental study has been performed on animals. Diagnostic investigation and additional characterisation have been conducted on samples that are submitted by veterinarians and pig owners. The specimens used in this research are field samples originating from natural respiratory symptoms and sent to our laboratory for official diagnosis.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jongho Kim, Email: whdgh2339@naver.com.
Jong Wan Kim, Email: biotics@korea.kr.
Sang-Ik Oh, Email: ohsangik@korea.kr.
ByungJae So, Email: bjso@korea.kr.
Won-Il Kim, Email: kwi0621@jbnu.ac.kr.
Ha-Young Kim, Phone: +82-54-912-0491, Email: kimhy@korea.kr, Email: jnm4jesus@naver.com.
References
- 1.García N, Fernández-Garayzábal JF, Goyache J, Domínguez L, Vela AI. Associations between biovar and virulence factor genes in Pasteurella multocida isolates from pigs in Spain. Vet Rec. 2011;169:362. doi: 10.1136/vr.d4869. [DOI] [PubMed] [Google Scholar]
- 2.Bethe A, Wieler LH, Selbitz HJ, Ewers C. Genetic diversity of porcine Pasteurella multocida strains from the respiratory tract of healthy and diseased swine. Vet Microbiol. 2009;139:97–105. doi: 10.1016/j.vetmic.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 3.Tang X, Zhao Z, Hu J, Wu B, Cai X, He Q, Chen H. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J Clin Microbiol. 2009;47:951–958. doi: 10.1128/JCM.02029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fablet C, Marois C, Kuntz-Simon G, Rose N, Dorenlor V, Eono F, Eveno E, Jolly J, Le Devendec L, Tocqueville V, Quéguiner S, Gorin S, Kobisch M, Madec F. Longitudinal study of respiratory infection patterns of breeding sows in five farrow-to-finish herds. Vet Microbiol. 2011;147:329–339. doi: 10.1016/j.vetmic.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opriessnig T, Giménez-Lirola LG, Halbur PG. Polymicrobial respiratory disease in pigs. Anim Health Res Rev. 2011;12:133–148. doi: 10.1017/S1466252311000120. [DOI] [PubMed] [Google Scholar]
- 6.Dayao DAE, Gibson JS, Blackall PJ, Turni C. Antimicrobial resistance in bacteria associated with porcine respiratory disease in Australia. Vet Microbiol. 2014;171:232–235. doi: 10.1016/j.vetmic.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Lee KE, Jeoung HY, Lee JY, Lee MH, Choi HW, Chang KS, Oh YH, An DJ. Phenotypic characterisation and random amplified polymorphic DNA (RAPD) analysis of Pasteurella multocida isolated from Korean pigs. J Vet Med Sci. 2012;74:567–573. doi: 10.1292/jvms.11-0418. [DOI] [PubMed] [Google Scholar]
- 8.Lee KE, Choi HW, Jo HY, Kim HH, Yang DK. Pasteurella multocida isolation from pigs with respiratory disease in Korea. Korean J Vet Res. 2016;56:37–40. doi: 10.14405/kjvr.2016.56.1.37. [DOI] [Google Scholar]
- 9.Mutters R, Ihm P, Pohl S, Frederiksen W, Mannheim W. Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis, and Pasteurella langaa. Int J Syst Bacteriol. 1985;35:309–322. doi: 10.1099/00207713-35-3-309. [DOI] [Google Scholar]
- 10.Blackall PJ, Pahoff JL, Bowles R. Phenotypic characterisation of Pasteurella multocida isolates from Australian pigs. Vet Microbiol. 1997;57:355–360. doi: 10.1016/S0378-1135(97)00111-9. [DOI] [PubMed] [Google Scholar]
- 11.Varga Z, Sellyei B, Magyar T. Phenotypic and genotypic characterisation of Pasteurella multocida strains isolated from pigs in Hungary. Acta Vet Hung. 2007;55:425–434. doi: 10.1556/AVet.55.2007.4.2. [DOI] [PubMed] [Google Scholar]
- 12.Blackall PJ, Fegan N, Pahoff JL, Storie GJ, McIntosh GB, Cameron RD, O’Boyle D, Frost AJ, Bara MR, Marr G, Holder J. The molecular epidemiology of four outbreaks of porcine pasteurellosis. Vet Microbiol. 2000;72:111–120. doi: 10.1016/S0378-1135(99)00192-3. [DOI] [PubMed] [Google Scholar]
- 13.Jamaludin R, Blackall PJ, Hansen MF, Humphrey S, Styles M. Phenotypic and genotypic characterisation of Pasteurella multocida isolated from pigs at slaughter in New Zealand. N Z Vet J. 2005;53:203–207. doi: 10.1080/00480169.2005.36505. [DOI] [PubMed] [Google Scholar]
- 14.Cardoso-Toset F, Gómez-Laguna J, Callejo M, Vela AI, Carrasco L, Fernández-Garayzábal JF, Maldonado A, Luque I. Septicaemic pasteurellosis in free-range pigs associated with an unusual biovar 13 of Pasteurella multocida. Vet Microbiol. 2013;167:690–694. doi: 10.1016/j.vetmic.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Choi C, Kim B, Cho WS, Kim J, Kwon D, Cheon DS, Chae C. Capsular serotype, toxA gene, and antimicrobial susceptibility profiles of Pasteurella multocida isolated from pigs with pneumonia in Korea. Vet Rec. 2001;149:210–212. doi: 10.1136/vr.149.7.210. [DOI] [PubMed] [Google Scholar]
- 16.Davies RL, MacCorquodale R, Baillie S, Caffrey B. Characterisation and comparison of Pasteurella multocida strains associated with porcine pneumonia and atrophic rhinitis. J Med Microbiol. 2003;52:59–67. doi: 10.1099/jmm.0.05019-0. [DOI] [PubMed] [Google Scholar]
- 17.Ewers C, Lübke-Becker A, Bethe A, Kießling S, Filter M, Wieler LH. Virulence genotype of Pasteurella multocida strains isolated from different hosts with various disease status. Vet Microbiol. 2006;114:304–317. doi: 10.1016/j.vetmic.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Aski HS, Tabatabaei M. Occurrence of virulence-associated genes in Pasteurella multocida isolates obtained from different hosts. Microb Pathog. 2016;96:52–57. doi: 10.1016/j.micpath.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Townsend KM, Frost AJ, Lee CW, Papadimitriou JM, Dawkins HJ. Development of PCR assays for species and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J Clin Microbiol. 2001;39:924–929. doi: 10.1128/JCM.39.3.924-929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atashpaz S, Shayegh J, Hejazi MS. Rapid virulence typing of Pasteurella multocida by multiplex PCR. Res Vet Sci. 2009;87:355–357. doi: 10.1016/j.rvsc.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Vera-Lizarazo YA, Rodríguez-Ferri EF, Martin de la Fuente AJ, Gutiérrez-Martín CB. Evaluation of changes in antimicrobial susceptibility patterns of Pasteurella multocida subsp. multocida isolates from pigs in Spain in 1987-1988 and 2003-2004. Am J Vet Res. 2006;67:663–668. doi: 10.2460/ajvr.67.4.663. [DOI] [PubMed] [Google Scholar]
- 23.Nedbalcová K, Kučerová Z. Antimicrobial susceptibility of Pasteurella multocida and Haemophilus parasuis isolates associated with porcine pneumonia. Acta Vet. 2013;82:3–7. doi: 10.2754/avb201382010003. [DOI] [Google Scholar]
- 24.De Jong A, Thomas V, Simjee S, Moyaert H, El Garch F, Maher K, Morrissey I, Butty P, Klein U, Marion H, Rigaut D, Vallé M. Antimicrobial susceptibility monitoring of respiratory tract pathogens isolated from diseased cattle and pigs across Europe: the VetPath study. Vet Microbiol. 2014;172:202–215. doi: 10.1016/j.vetmic.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 25.El Garch F, de Jong A, Simjee S, Moyaert H, Klein U, Ludwig C, Marion H, Haag-Diergarten S, Richard-Mazet A, Thomas V, Siegwart E. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Vet Microbiol. 2016;194:11–22. doi: 10.1016/j.vetmic.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Rúbies X, Casal J, Pijoan C. Plasmid and restriction endonuclease patterns in Pasteurella multocida isolated from a swine pyramid. Vet Microbiol. 2002;84:69–78. doi: 10.1016/S0378-1135(01)00440-0. [DOI] [PubMed] [Google Scholar]
- 27.Peng Z, Liang W, Wang Y, Liu W, Zhang H, Yu T, Zhang A, Chen H, Wu B. Experimental pathogenicity and complete genome characterisation of a pig origin Pasteurella multocida serogroup F isolate HN07. Vet Microbiol. 2017;198:23–33. doi: 10.1016/j.vetmic.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 28.Portis E, Lindeman C, Johansen L, Stoltman G. Antimicrobial susceptibility of porcine Pasteurella multocida, Streptococcus suis, and Actinobacillus pleuropneumoniae from the United States and Canada, 2001 to 2010. J Swine Health Prod. 2013;21:30–41. [Google Scholar]
- 29.Sweeney MT, Lindeman C, Johansen L, Mullins L, Murray R, Senn MK, Bade D, Machin C, Kotarski SF, Tiwari R, Watts JL. Antimicrobial susceptibility of Actinobacillus pleuropneumoniae, Pasteurella multocida, Streptococcus suis, and Bordetella bronchiseptica isolated from pigs in the United States and Canada, 2011 to 2015. J Swine Health Prod. 2017;25:106–120. [Google Scholar]
- 30.Anonymous . Animal and plant quarantine agency report (Korea) Gimcheon: Animal and Plant Quarantine Agency (APQA); 2016. Establishment of antimicrobial resistance surveillance system for livestock. [Google Scholar]
- 31.Jung JY, Jeong JH, Kim HY, Oh SI, Ryu DY, Yoon HL, So B, Yoon SS. A case of submandibular pyogranuloma caused by Trueperella pyogenes in the slaughtered dairy cows. Korean J Vet Serv. 2016;39:65–68. doi: 10.7853/kjvs.2016.39.1.65. [DOI] [Google Scholar]
- 32.Kim SH, Park JY, Jung JY, Kim HY, Park YR, Lee KK, Lyoo YS, Yeo SG, Park CK. Detection and genetic characterization of porcine circovirus 3 from aborted fetuses and pigs with respiratory disease in Korea. J Vet Sci. 2018;19:721–724. doi: 10.4142/jvs.2018.19.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CLSI, editor. Performance Standards for Antimirobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 3rd ed. CLSI supplement VET01S. Wayne: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated or analysed during the current study are available from the corresponding author on reasonable request.