Abstract
Circular RNAs (circRNAs) are non-coding RNAs that form covalently closed continuous loops, and act as gene regulators in physiological and disease conditions. To test our hypothesis that proatherogenic lipid lysophosphatidylcholine (LPC) induce a set of circRNAs in human aortic endothelial cell (HAEC) activation, we performed circRNA analysis by searching our RNA-Seq data from LPC-activated HAECs, and found: (1) LPC induces significant modulation of 77 newly characterized cirRNAs, among which 47 circRNAs (61%) are upregulated; (2) 34 (72%) out of 47 upregulated circRNAs are upregulated when the corresponding mRNAs are downregulated, suggesting that the majority of circRNAs are upregulated presumably via LPC-induced “abnormal splicing” when the canonical splicing for generation of corresponding mRNAs is suppressed; (3) Upregulation of 47 circRNAs is temporally associated with mRNAs-mediated LPC-upregulated cholesterol synthesis-SREBP2 pathway and LPC-downregulated TGF-β pathway; (4) Increase in upstream chromatin long-range interaction sites to circRNA related genes is associated with preferred circRNA generation over canonical splicing for mRNAs, suggesting that shifting chromatin long-range interaction sites from downstream to upstream may promote induction of a list of circRNAs in lysoPC-activated HAECs; (5) Six significantly changed circRNAs may have sponge functions for miRNAs; and (6) 74% significantly changed circRNAs contain open reading frames, suggesting that putative short proteins may interfere with the protein interaction-based signaling. Our findings have demonstrated for the first time that a new set of LPC-induced circRNAs may contribute to homeostasis in LPC-induced HAEC activation. These novel insights may lead to identifications of new therapeutic targets for treating metabolic cardiovascular diseases, inflammations, and cancers.
Keywords: circular RNAs, human aortic endothelial cell activation, proatherogenic lipid lysophosphatidylcholine, RNA-Seq, chromatin long–range interaction
Introduction
Cardiovascular diseases remain some of the most prevalent health challenges today. Predominant among these, atherosclerosis and resultant ischemic heart disease, stroke, and peripheral artery disease are estimated to cause 8.9 million deaths per year, making it the single most significant cause of death worldwide (Wang H. et al., 2016). We and others have reported that hyperlipidemia, together with other CVD risk factors, such as hyperglycemia, chronic kidney disease, obesity, and hyperhomocysteinemia (HHcy), promotes atherosclerosis development via several mechanisms. These mechanisms include endothelial cell (EC) activation and injury (Shao et al., 2014; Yin et al., 2015; Li et al., 2016a, 2018a); monocyte recruitment and differentiation (Combadière et al., 2008; Fang et al., 2014); decreased function of regulatory T cells (Tregs) (Ait-Oufella et al., 2006; Yan et al., 2008; Li et al., 2017); transdifferentiation of Tregs into antigen-presenting cell (APC) like Tregs (Xu et al., 2018); impaired vascular repairability of bone marrow-derived progenitor cells (Du et al., 2012; Barbier et al., 2015; Li et al., 2016b); increased migration and proliferation of vascular smooth muscle cells (Monroy et al., 2015; Ferrer et al., 2016), and high fat-induced adipocyte hypertrophy and metabolic healthy obesity (Virtue et al., 2017). Human aortic endothelial cell (HAEC) activation is widely understood to contribute substantively to the early development of atherosclerosis by increasing leukocyte recruitment through upregulation of adhesion molecules such as ICAM1 (Tabas et al., 2015; Vestweber, 2015; Gimbrone and García-Cardeña, 2016). We have recently reported that exposure to proatherogenic stimuli, such as the proinflammatory lysophospholipid lysophosphatidylcholine (LPC), can induce prolonged HAEC activation as part of early atherogenesis (Li et al., 2016a,c, 2018b); this phenomenon is one of a larger class of conditional danger-associated molecular patterns (DAMPs) that trigger the onset of oxidative, autoimmune, and inflammatory tissue damages (Wang X. et al., 2016; Shao et al., 2017). However, the regulatory mechanisms underpinning these physiological changes remain poorly understood; and whether and how non-coding RNAs (ncRNAs) mechanisms control HAEC activation remains poorly characterized.
For many years, proteins have represented the primary functional end product of genetic information, though the genes that encode them account for less than 2% of the genome (Anastasiadou et al., 2018). As new sequencing technology advances, as many as six types of ncRNAs have been identified, including three small ncRNA types [i.e., RNA interference (RNAi), tRNA-derived stress-induced RNA (tiRNA), and small nuclear RNA (snRNA)] and three long RNAs types [i.e., transfer RNA (tRNA), ribosomal RNA (rRNA), and long non-coding RNAs (lncRNA)] (Gomes et al., 2018). RNAi can be categorized into five subgroups including small interfering RNA (siRNA), piwi-interacting RNA (piRNA), and microRNA (miRNA), repeat associated siRNA (rasiRNA), and agotron. LncRNAs can be further classified into six subtypes: sense lncRNA, intronic lncRNA, antisense lncRNA, intergenic lncRNA, enhancer RNA (eRNA), and circular RNA (circRNA). These ncRNAs together constitute almost 60% of the transcriptional output in human cells, and they have been shown to regulate cellular processes and pathways in developmental and pathological contexts through mRNA degradation or modification, transcription regulation, translation regulation, and miRNA sponging, among other functions (Mattick and Makunin, 2006; Bartel, 2009; Cech and Steitz, 2014; Lasda and Parker, 2014). We previously reported that a group of anti-atherogenic miRNAs might suppress proatherogenic gene expression (Virtue et al., 2011, 2012; Mai et al., 2012). We also found that microRNA-155 (miR155) is among several miRs significantly upregulated in atherogenic apolipoprotein E deficient (ApoE-/-) mouse aortas. MiR155 promotes HAEC activation and mouse atherosclerosis, whereas the deficiency of miR155 in ApoE-/- background leads to high fat-induced adipocyte hypertrophy and establishes a novel metabolically healthy obese (MHO) mouse model (Virtue et al., 2017). Among 109 miRNAs that we examined in all four hyperlipidemia-related diseases (HRDs), namely atherosclerosis, non-alcoholic fatty liver disease (NAFLD), obesity and type II diabetes (T2DM), miR-155 and miR-221 are significantly modulated in these HRDs. MiR-155 is significantly upregulated in atherosclerosis and decreased in other HRDs. MiR-221 is increased in three HRDs but reduced in obesity. These findings led to our new classification of types I and II MHOs, which are regulated by miR-221 and miR-155, respectively. We also found that the proinflammatory adipokine, resistin, is significantly increased in white adipose tissues (WAT) of the MHO mice, revealing our newly proposed, miR-155-suppressed “secondary wave inflammatory state (SWIS),” characteristic of MHO transition to classical obesity (CO) (Johnson et al., 2018). Our and others’ results demonstrate that ncRNAs play significant roles in regulating the pathogenesis and progression of cardiovascular diseases and other diseases.
Two types of lncRNAs (i.e., linear and circular forms) have been found in the lncRNA family. In contrast to previous models, recent work has shown that circRNAs are relatively abundant and may be the predominant transcripts of hundreds of genes. The spliceosome machinery produces circRNAs by direct back-splicing of exons or introns within pre-mRNA or by exon skipping. As a result of a direct back-splicing event, an exon is not associated with an adjacent downstream exon, as seen in linear splicing, but with an upstream exon or intron. This back-splicing gives rise to circular RNA molecules harboring exons or introns that are out-of-order from the genomic context (Figure 1A). These circular molecules are ubiquitously expressed in human and more than 30,000 different circRNAs have been identified. They are regulated in several disease states and contribute to disease development and progression (Gomes et al., 2018). Although the importance of circRNAs in regulating EC function has been reported, the comprehensive analyses of circRNAs in EC activation, EC function, and cardiovascular diseases remain at their infancy; additionally, the characterization of circRNAs in atherosclerosis-relevant HAECs has not been reported (Maass et al., 2017; Huang et al., 2018; Shang et al., 2018).
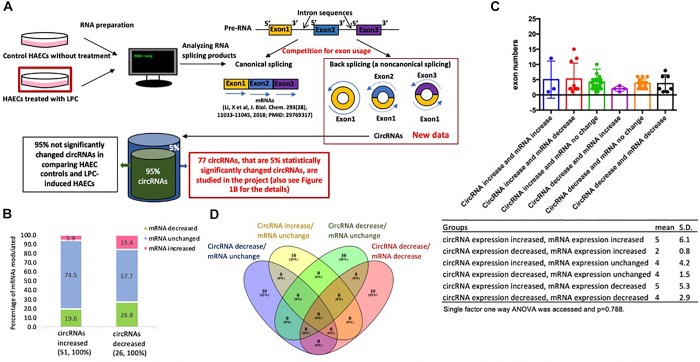
FIGURE 1.
(A) Lysophosphatidylcholine (LPC)-induced top 5% significantly changed circular RNAs (circRNAs) are studied in human aortic endothelial cells (HAECs) in comparison to that of control HAECS. This is a flow chart to outline how we analyzed the 5% top significantly changed circular RNAs from LPC-treated HAECs in comparison to that from control HAECs. (B) The majority (48 out of 51,94.1%) of circRNA expression increases while related mRNA expressions remain constant or decrease. (C) There is no significant differences in exon numbers among circRNA groups. (D) There are no overlaps among the top 10 pathways in the following four groups.
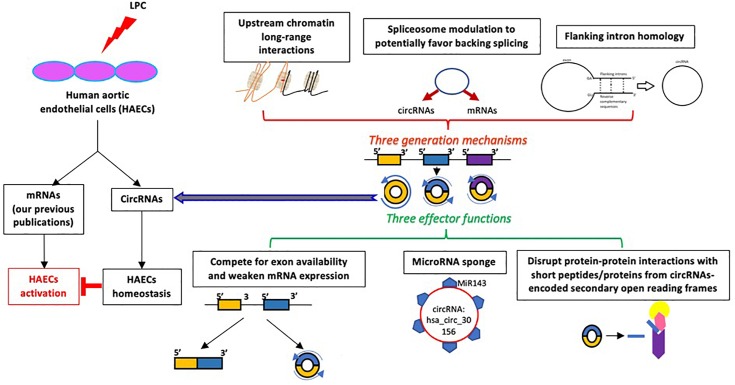
Regardless of the progress, we identified several important knowledge gaps to be filled in this study. We hypothesized that circRNAs might play an important role in mediating homeostatic status in HAECs activated by proatherogenic LPC. We analyzed our RNA-Seq raw data sets, obtained from control-, and LPC- treated HAECs (Li et al., 2018b), using two well-accepted circular RNA databases such as CircExplorer2 and CIRI to obtain a list of significantly changed circular RNAs (circRNAs) (Gao et al., 2015; Zhang et al., 2016) 4. We found that this list of 77 circRNAs may play homeostatic roles in activated HAECs via novel mechanisms including exon usage competition, miRNA sponging (Hansen et al., 2013; Memczak et al., 2013) and potential generation of short proteins for protein interaction disruption. We also found that novel mechanisms, such as (a) flanking intron homology, (b) LPC-induced modulation of spliceosome components, and (c) upstream chromatin long-range interaction, play important roles in promoting the generation of circRNAs in LPC-activated HAECs. These novel insights may lead to the identification of new therapeutic targets for treating metabolic cardiovascular diseases, inflammations, and cancers.
Materials and Methods
Generation of CircRNA Datasets
Raw RNA-Seq data was obtained from a previous study (Li et al., 2018b). Raw reads generated by the Illumina HiSeq control software were assessed using FastQC (Andrews, 2010). Samples with poor sequence quality were discarded. Sequence reads were mapped to the hg38 genome using STAR (Dobin et al., 2013). Fusion junction reads were parsed using CIRCexplorer2 and captured back-splicing junction reads were then annotated with UCSC annotation files (Zhang et al., 2016). For additional detections, sequence reads were mapped to the hg38 genome using the Burrows-Wheeler Alignment tool (Li and Durbin, 2009), circRNAs were identified using CIRI (Gao et al., 2015). Gene annotation was performed using org.Hs.eg.db (Carlson, 2017). A list of circRNAs common to both control and LPC-stimulated HAECs was generated using the VLOOKUP function in Microsoft Excel. To filter out significantly changed circRNAs, ratios of control to LPC-stimulated circRNA read numbers were calculated, and base 2 logarithms taken of these ratios. Mean and standard deviation of logarithm values were calculated using MATLAB and used to generate a 95% confidence interval (mean ± 2∗SD). Using previously obtained mRNA expression data (Li et al., 2018b), an mRNA fold change confidence interval was similarly calculated using the ten housekeeping genes: C1orf43, CHMP2A, GAPDH, EMC7, GPI, PSMB2, PSMB4, RAB7A, SNRPD3, and VPS29; and it was used to separate circRNAs by corresponding mRNA expression change.
Comparison of Flanking Intron Sequences
The generated circRNA data included flanking intron coordinates; a Python script was used to parse these coordinates and feed them into an NIH NCBI BLAST+ command line query (blastn -max_hsps 1 -outfmt “10 pident e-Value bitscore”) comparing the sequences of 3′ and 5′ flanking intron sequences using local copies of hg38 human genome primary assembly chromosome sequences1 (Camacho et al., 2009). This returned percent identity, the expected value (number of matches of equal strength expected by chance), and bitscore values (normalized score for match strength) for each pair of flanking introns. These were then exported to a text file and imported into a Microsoft Excel spreadsheet for tabulation. Bitscore values were further used for statistical analysis (described below under “Statistical Analysis”).
Matching CircRNAs to Database Entries
The genomic length of each significantly changed circRNA was calculated by subtracting its genomic start coordinate from its genomic end coordinate. A MATLAB script was then used to find circRNAs with matching length and gene locus from a local copy of the circRNAdb database and to export all results to a Microsoft Excel spreadsheet (Schliebs et al., 1996; Chen et al., 2016). Notably, genomic coordinates in circRNAdb were given with respect to the older hg37 human genomic annotation. A local copy of the hg37 genomic annotation was therefore obtained for comparison2. Then, NIH NCBI BLAST+ queries between original hg38 coordinates and CircRNAdb hg37 coordinates (blastn -max_hsps 1 -outfmt “10 pident”) were performed using a MATLAB script (Schliebs et al., 1996; Camacho et al., 2009), taking only the single best match for each pair of sequences, the percent identity values of which confirmed sequence alignment for all length matches found. CircInteractome also uses the hg37 human genome annotation but a different set of IDs; matches in CircInteractome were determined by hand due to incomplete downloadable data sets.
Analysis of Long–Range Interactions
A complete list of long–range chromatin interaction sites in the human genome was obtained from the 4DGenome database3 as a tabulated text file (Teng et al., 2015). The grep command line utility was used to filter for entries detected using Hi-C methodology and involving at least one circRNA-related gene. The resulting filtered data was imported into Microsoft Excel and raw interaction distances calculated as the differences between gene start coordinates. An AWK script was used to determine whether the circRNA-related gene was downstream or upstream of its partner in each interaction pair and to add this information to the data file. The signs of distance values were then updated, with downstream entries designated as positive and upstream values designated as negative, using a Python script. These updated distance values were separated into the same six groups as their corresponding circRNA-related genes and used in pairwise two-sample Kolmogorov–Smirnov tests (described further below under “Statistical Analysis”). Distance distributions for all upregulated and all downregulated circRNAs were compared by groups overall as well as by only upregulated and downregulated circRNAs with increased, unchanged, or decreased corresponding mRNA expression, respectively.
Characterizing Open Reading Frame Sequences
Open reading frame (ORF) peptide sequences and internal ribosomal entry site (IRES) data were obtained to identify significantly changed circRNAs from circRNAdb (Chen et al., 2016). A MATLAB script was used to call remote NIH NCBI BLAST+ peptide alignments for all ORF sequences against the NCBI non-redundant protein sequences database (nr4) (Schliebs et al., 1996; Camacho et al., 2009), restricted to entries for Homo sapiens (blastp -db nr -remote -entrez_query “Homo sapiens [Organism]” -max_target_seqs 1 -outfmt “10 sacc pident e-Value”). This returned the subject accession, percent identity, and expected value of the single best database match, which was then exported with the gene name and circRNAdb ID to a Microsoft Excel spreadsheet. The obtained accessions were individually and manually searched on the NCBI protein database5 to determine if they corresponded to canonical mRNA transcripts from the same gene locus as the corresponding circRNA. A modified version of the hg38/hg37 comparison MATLAB script was also used to run NIH NCBI BLAST+ searches for the Kozak consensus sequence gccRccAUGG in genomic sequences of all significantly changed circRNAs (Kozak, 1987; Schliebs et al., 1996; Camacho et al., 2009).
Pathway Analysis
QIAGEN Ingenuity Pathway Analysis (IPA) software, which constructs predicted upstream and downstream causal networks for input datasets from a curated research literature base, was used to elucidate potential downstream pathways for sponged miRNAs (Krämer et al., 2014). Lists of miRNAs were input into IPA and ran through the miRNA target filter to generate a list of potential mRNA targets. The list of mRNA target genes was then run through an IPA core analysis. All canonical downstream pathways returned in the resulting output were exported into a Microsoft Excel spreadsheet. The top ten pathways were extracted for further qualitative consideration.
Graphical Figure Generation
For three-group Venn diagrams, lists of genes were input into an online Venn diagram generator (6Evolutionary Genomics, Ghent University, Gent, Belgium) (Draw Venn Diagram). This tool was used to produce both diagrams and lists of overlapped genes between groups. For six-group Venn diagrams, gene lists were input into the InteractiVenn online tool7 to generate diagrams (Heberle et al., 2015), while the above Ghent University tool was again used to produce lists of overlapped genes between groups (Draw Venn Diagram). Explanatory and conceptual graphics were produced using Microsoft Paint.
Statistical Analysis
Descriptive summary statistics were reported by group. Data were checked for normality assumption and, if found to be not normally distributed, subsequently transformed using various functions such as the log10 and cubic-root to find the optimal transformation for the underlying chromatin long-range interaction distance distribution data. Chromatin long-range interaction distance density functions were then estimated and plotted by group under the optimal transformation using the non-parametric kernel density approach with a normal weight function (Jones et al., 1996; Hollander and Wolfe, 1999; Silverman, 2018). Pairwise comparisons of median chromatin long-range interaction distance among the six groups were performed with multiple comparison adjustments using the Dwass, Steel, and Critchlow-Fligner method based on the Wilcoxon test for downstream and upstream data separately (Douglas and Michael, 1991; Conover, 1999; Hollander and Wolfe, 1999; Shalabh, 2011). Pairwise comparisons between groups for chromatin long-range interaction distance distribution, median distance location shift, and distance distribution scale were implemented using the Kolmogorov–Smirnov two-sample test, Hodges–Lehmann estimation method and Fligner-Policello test, and Ansari-Bradley test, respectively, again for downstream and upstream data separately (Lehmann, 1963; Douglas and Michael, 1991; Conover, 1999; Hollander and Wolfe, 1999; Shalabh, 2011). SAS version 9.4 was used to perform these analyses and generate density function plots for the chromatin long-range interaction distance data.
For rest of the data, single-factor ANOVA and non-parametric Kruskal–Wallis tests were conducted for all multi-group analyses using the Real Statistics Resource Pack add-in for Microsoft Excel8 (Zaiontz, 2013). For confidence intervals, MATLAB was used to calculate mean and standard deviations for data sets (Schliebs et al., 1996). Probabilities of ratios of upregulation to downregulation were calculated by summing binomial coefficients and dividing by the appropriate power of 2 in MATLAB (Schliebs et al., 1996), as given by the formula , where u is the total number of genes considered and n is the large number of upregulated or downregulated genes.
Results
LPC Modulates the Expression of a List of 77 Circular RNAs in HAECs; and the Majority of Which, 94.1% of Increased CircRNAs, Compete With Related mRNAs for Exon Usage
Our preliminary data processing detected 3,170 distinct circRNAs in control cells and 3,486 circRNAs in a proatherogenic lipid LPC-treated HAECs as we reported (Li et al., 2016a). To better analyze our data set, we derived a list of 1,093 circRNAs found in both the control and LPC-treated data sets before calculating read number ratio – the quotient of the read number in the LPC-treated data set and the corresponding read number in the control data set – for each circRNA in the list to serve as a measure of expression change (Figure 1A). We then took the base-2 logarithm of each readnumber ratio (log ratio) and calculated a 95 percent confidence interval (-0.3815 ± 2.3395) for log ratio, taking all the 77 circRNAs with log ratios outside this confidence interval to have significantly changed levels of expression in LPC-treated HAECs (Table 1A). Then, we separated these circRNAs into two groups of increased and decreased circRNA expression. To assess potential relationships between circRNAs and their corresponding mRNAs published, we then further divided each of these groups into three subgroups based on increased, unchanged, or decreased expression of corresponding mRNA transcripts, with all fold change levels within a 95 percent confidence interval (1.01 ± 0.1) determined by the housekeeping genes C1orf43, CHMP2A, GAPDH, EMC7, GPI, PSMB2, PSMB4, RAB7A, SNRPD3, and VPS29 considered to indicate unchanged mRNA expression (Table 1B), thereby obtaining six groups of significantly changed circRNAs classified by changes in both circRNA and corresponding mRNA expression.
Table 1A.
LPC-induced 77 circRNAs in HAECs can be classified into 6 groups based on expression change of circRNAs and corresponding (related) mRNAs.
| Gene ID | Gene | circRNAdb ID | Readnumber (LPC) | Readnumber (Control) | Ratio | Log(2) ratio | FC (mRNA) | Index (circRNA exons) | Exon count | Exon count (genomic) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| circRNA expression increased, mRNA expression increased (Shao et al., 2014) | |||||||||||
| 23094 | SIPA1L3 | hsa_circ_10577 | 9 | 2 | 4.5 | 2.17 | 1.12 | 11,12 | 2 | 24 | |
| 117583 | PARD3B | N/A | 4 | 1 | 4 | 2 | 1.19 | 4,5,6,7,8,9,10,11, 12,13,14,15 | 12 | 33 | |
| 116840 | CNTROB | hsa_circ_09218 | 4 | 1 | 4 | 2 | 1.11 | 13 | 1 | 22 | |
| circRNA expression decreased, mRNA expression increased (Li et al., 2016a) | |||||||||||
| 2530 | FUT8 | hsa_circ_02544 | 1 | 7 | 0.14 | –2.81 | 1.2 | 3 | 1 | 15 | |
| 768211 | RELL1 | hsa_circ_08846 | 2 | 15 | 0.13 | –2.91 | 1.15 | 6,5,4 | 3 | 8 | |
| 51150 | SDF4 | hsa_circ_21644 | 1 | 9 | 0.11 | –3.17 | 1.15 | 4,3 | 2 | 8 | |
| 64778 | FNDC3B | hsa_circ_15231 | 1 | 15 | 0.07 | –3.91 | 1.13 | 5,6 | 2 | 30 | |
| circRNA expression increased, mRNA expression unchanged (Huang et al., 2018) | |||||||||||
| 55852 | TEX2 | hsa_circ_31354 | 15 | 1 | 15 | 3.91 | 0.98 | 7,6,5 | 3 | 15 | |
| 5820 | PVT1 | hsa_circ_16350 | 8 | 1 | 8 | 3 | 2 | 1 | 9 | ||
| 51232 | CRIM1 | hsa_circ_16396 | 15 | 2 | 7.5 | 2.91 | 1.03 | 2,3,4 | 3 | 22 | |
| 1793 | DOCK1 | hsa_circ_00689 | 7 | 1 | 7 | 2.81 | 1.05 | 2,3,4,5,6,7,8,9,10,11, 12,13,14,15,16,17, 18,19,20,21,22,23 | 22 | 56 | |
| 4670 | HNRNPM | hsa_circ_20744 | 7 | 1 | 7 | 2.81 | 1.01 | 2,3,4,5 | 4 | 16 | |
| 22937 | SCAP | hsa_circ_19380 | 7 | 1 | 7 | 2.81 | 0.94 | 8,7,6,5,4 | 5 | 25 | |
| 9341 | VAMP3 | hsa_circ_18051 | 7 | 1 | 7 | 2.81 | 0.94 | 3,4 | 2 | 5 | |
| N/A | RP11-146B14.1 | N/A | 6 | 1 | 6 | 2.58 | #N/A | 12,13,14,15 | 4 | ||
| 9380 | GRHPR | hsa_circ_03935 | 6 | 1 | 6 | 2.58 | 1.06 | 2,3,4 | 3 | 11 | |
| 27086 | FOXP1 | hsa_circ_19469 | 6 | 1 | 6 | 2.58 | 1.01 | 12,11,10,9,8 | 5 | 32 | |
| 64750 | SMURF2 | hsa_circ_01208 | 5 | 1 | 5 | 2.32 | 1.11 | 10,9,8,7,6 | 5 | 19 | |
| 84456 | L3MBTL3 | N/A | 5 | 1 | 5 | 2.32 | 1.03 | 4,5,6,7,8,9,10,11, 12,13,14,15,16, 17,18,19,20,21 | 18 | 30 | |
| 862 | RUNX1T1 | N/A | 5 | 1 | 5 | 2.32 | 0.99 | 5,4,3,2 | 4 | 20 | |
| 151050 | KANSL1L | hsa_circ_06859 | 5 | 1 | 5 | 2.32 | 0.96 | 4,3,2 | 3 | 23 | |
| 11138 | TBC1D8 | hsa_circ_32540 | 5 | 1 | 5 | 2.32 | 0.94 | 15,14,13,12,11,10 | 6 | 25 | |
| 84328 | LZIC | hsa_circ_31394 | 5 | 1 | 5 | 2.32 | 0.94 | 4,3,2 | 3 | 10 | |
| 23331 | TTC28 | hsa_circ_22261 | 5 | 1 | 5 | 2.32 | 0.93 | 3,2 | 2 | 29 | |
| 1456 | CSNK1G3 | hsa_circ_08140 | 14 | 3 | 4.67 | 2.22 | 0.93 | 2,3,4 | 3 | 18 | |
| 8460 | TPST1 | hsa_circ_22617 | 9 | 2 | 4.5 | 2.17 | 0.98 | 2,3 | 2 | 8 | |
| #N/A | PRKY | hsa_circ_30664 | 4 | 1 | 4 | 2 | N/A | 4,5,6,7 | 4 | 8 | |
| #N/A | PCNXL2 | hsa_circ_10712 | 4 | 1 | 4 | 2 | N/A | 23,22 | 2 | ||
| 1523 | CUX1 | hsa_circ_12403 | 8 | 2 | 4 | 2 | 1.11 | 21 | 1 | 34 | |
| 9701 | PPP6R2 | hsa_circ_07544 | 4 | 1 | 4 | 2 | 1.11 | 2,3 | 2 | 29 | |
| 54919 | DNAAF5 | N/A | 4 | 1 | 4 | 2 | 1.1 | 9,10,11 | 3 | 14 | |
| 84293 | FAM213A | N/A | 4 | 1 | 4 | 2 | 1.06 | 2,3,4 | 3 | 9 | |
| 7813 | EVI5 | hsa_circ_11187 | 4 | 1 | 4 | 2 | 1.04 | 19,18,17 | 3 | 30 | |
| 81608 | FIP1L1 | hsa_circ_18069 | 4 | 1 | 4 | 2 | 1.02 | 11,12,13 | 3 | 19 | |
| 51295 | ECSIT | hsa_circ_00599 | 4 | 1 | 4 | 2 | 1 | 4,3 | 2 | 9 | |
| 152002 | XXYLT1 | hsa_circ_02143 | 4 | 1 | 4 | 2 | 1 | 3,2 | 2 | 11 | |
| 6310 | ATXN1 | hsa_circ_13397 | 4 | 1 | 4 | 2 | 0.99 | 8 | 1 | 11 | |
| 1024 | CDK8 | hsa_circ_27078 | 4 | 1 | 4 | 2 | 0.98 | 10,11,12 | 3 | 14 | |
| 6259 | RYK | hsa_circ_26048 | 4 | 1 | 4 | 2 | 0.98 | 13,12,11,10,9,8,7 | 7 | 15 | |
| 92 | ACVR2A | hsa_circ_04751 | 4 | 1 | 4 | 2 | 0.98 | 2,3,4 | 3 | 12 | |
| 22828 | SCAF8 | hsa_circ_06913 | 4 | 1 | 4 | 2 | 0.98 | 2,3,4,5,6 | 5 | 22 | |
| 29086 | BABAM1 | hsa_circ_04303 | 4 | 1 | 4 | 2 | 0.96 | 7,8 | 2 | 9 | |
| 23244 | PDS5A | hsa_circ_29001 | 4 | 1 | 4 | 2 | 0.95 | 21,20,19,18,17 | 5 | 36 | |
| 7443 | VRK1 | hsa_circ_08594 | 4 | 1 | 4 | 2 | 0.94 | 2,3,4,5,6,7,8,9,10,11 | 10 | 15 | |
| 8826 | IQGAP1 | hsa_circ_10932 | 4 | 1 | 4 | 2 | 0.93 | 6,7,8,9 | 4 | 39 | |
| circRNA expression decreased, mRNA expression unchanged (Li et al., 2016b) | |||||||||||
| 23362 | PSD3 | hsa_circ_02851 | 1 | 7 | 0.14 | –2.81 | 1.05 | 8,7,6,5 | 4 | 29 | |
| 55833 | UBAP2 | hsa_circ_30156 | 1 | 7 | 0.14 | –2.81 | 0.94 | 12,11,10,9 | 4 | 30 | |
| 10443 | N4BP2L2 | hsa_circ_12240 | 1 | 7 | 0.14 | –2.81 | 0.93 | 6,5,4,3 | 4 | 22 | |
| 6904 | TBCD | hsa_circ_00754 | 2 | 15 | 0.13 | –2.91 | 1.05 | 14,15,16,17,18,19 | 6 | 61 | |
| 5747 | PTK2 | hsa_circ_23485 | 2 | 15 | 0.13 | –2.91 | 1.03 | 5,4,3 | 3 | 44 | |
| 23603 | CORO1C | hsa_circ_01619 | 1 | 9 | 0.11 | –3.17 | 1.06 | 8,7 | 2 | 20 | |
| 5917 | RARS | hsa_circ_21209 | 1 | 9 | 0.11 | –3.17 | 1.01 | 2,3,4,5 | 4 | 15 | |
| 11174 | ADAMTS6 | hsa_circ_28538 | 3 | 29 | 0.1 | –3.27 | 1.05 | 7,6,5,4,3,2 | 6 | 33 | |
| 22955 | SCMH1 | hsa_circ_12377 | 1 | 10 | 0.1 | –3.32 | 1.09 | 10,9 | 2 | 22 | |
| 51306 | FAM13B | hsa_circ_11413 | 1 | 10 | 0.1 | –3.32 | 1.01 | 10,9,8 | 3 | 30 | |
| 2050 | EPHB4 | hsa_circ_08510 | 1 | 10 | 0.1 | –3.32 | 1 | 12,11 | 2 | 17 | |
| 444 | ASPH | hsa_circ_10828 | 1 | 11 | 0.09 | –3.46 | 1 | 3,2 | 2 | 33 | |
| 57488 | ESYT2 | hsa_circ_25060 | 1 | 11 | 0.09 | –3.46 | 0.95 | 13,12,11,10,9 | 5 | 26 | |
| 10905 | MAN1A2 | hsa_circ_02643 | 1 | 15 | 0.07 | –3.91 | 1.06 | 2,3,4,5 | 4 | 14 | |
| 50807 | ASAP1 | hsa_circ_09642 | 1 | 15 | 0.07 | –3.91 | 0.93 | 14,13,12,11,10,9 | 6 | 37 | |
| circRNA expression increased, mRNA expression decreased (Yan et al., 2008) | |||||||||||
| 4649 | MYO9A | N/A | 8 | 1 | 8 | 3 | 0.75 | 39,38,37,36,35,34, 33,32,31,30,29,28 | 12 | 48 | |
| 10564 | ARFGEF2 | N/A | 5 | 1 | 5 | 2.32 | 0.91 | 5,6,7 | 3 | 39 | |
| 51444 | RNF138 | hsa_circ_01057 | 5 | 1 | 5 | 2.32 | 0.9 | 3,4 | 2 | 8 | |
| 152006 | RNF38 | hsa_circ_32756 | 5 | 1 | 5 | 2.32 | 0.79 | 4,3 | 2 | 19 | |
| 23047 | PDS5B | hsa_circ_01546 | 4 | 1 | 4 | 2 | 0.91 | 2,3 | 2 | 38 | |
| 55719 | SLF2 | N/A | 4 | 1 | 4 | 2 | 0.9 | 5,6 | 2 | 21 | |
| 55247 | NEIL3 | hsa_circ_00262 | 4 | 1 | 4 | 2 | 0.84 | 8,9 | 2 | 10 | |
| 56886 | UGGT1 | N/A | 4 | 1 | 4 | 2 | 0.78 | 2,3,4,5,6,7,8,9,10, 11,12,13,14,15,16 | 15 | 44 | |
| 23499 | MACF1 | N/A | 4 | 1 | 4 | 2 | 0.68 | 42,43,44,45,46,47, 48,49,50,51,52 | 11 | 111 | |
| 11123 | RCAN3 | hsa_circ_22201 | 4 | 1 | 4 | 2 | 0.67 | 2 | 1 | 7 | |
| circRNA expression decreased, mRNA expression decreased (Combadière et al., 2008) | |||||||||||
| 659 | BMPR2 | hsa_circ_06837 | 1 | 7 | 0.14 | –2.81 | 0.84 | 2,3 | 2 | 13 | |
| 23253 | ANKRD12 | hsa_circ_31497 | 1 | 7 | 0.14 | –2.81 | 0.78 | 2,3,4,5,6,7 | 6 | 18 | |
| 10725 | NFAT5 | hsa_circ_16704 | 1 | 8 | 0.13 | –3 | 0.64 | 15 | 1 | 19 | |
| 26057 | ANKRD17 | hsa_circ_01636 | 1 | 9 | 0.11 | –3.17 | 0.91 | 29 | 1 | 37 | |
| 55125 | CEP192 | hsa_circ_26363 | 1 | 9 | 0.11 | –3.17 | 0.85 | 2,3,4,5,6,7,8,9 | 8 | 46 | |
| 121512 | FGD4 | hsa_circ_21030 | 1 | 9 | 0.11 | –3.17 | 0.85 | 5,6,7,8,9,10 | 6 | 27 | |
| 22836 | RHOBTB3 | hsa_circ_19124 | 1 | 17 | 0.06 | –4.09 | 0.91 | 6,7 | 2 | 14 | |
Table 1B.
LysoPC does not significantly modulate the expressions of housekeeping mRNAs in human aortic endothelial cells (HAECs), suggesting that the RNA-Seq experiments in HAECs treated with lysoPC were well performed.
| Housekeeping genes | Fold change |
| C1orf43 | 0.97 |
| CHMP2A | 1 |
| GADPH | 1 |
| EMC7 | 0.98 |
| GPI | 1 |
| PSMB2 | 0.98 |
| PSMB4 | 1.02 |
| RAB7A | 0.98 |
| SNRPD3 | 1 |
| VPS29 | 1.04 |
We also matched each circRNA, if possible, with its corresponding ID in the circRNA database CircRNAdb (Chen et al., 2016), to provide for more consistent identification, and for later analysis of the database’s open reading frame data. Of note, among 51 circRNAs increased, 48 out of 51 related mRNAs (94.1%) are either unchanged (38 out of 51, 74.5%) or decreased (10 out of 51, 19.5%), while only three increased both mRNA and circRNA expression (5.9%, Figure 1B). Similarly, among 26 circRNAs decreased, 19 out of 26 related mRNAs (73.1%) are either unchanged (15 out of 26, 57.7%) or increased (4 out of 26, 15.4%), while only seven decreased both mRNA and circRNA expression (26.9%, Figure 1B). The results suggest that a dominant inversely correlative relationship between circRNA expression and mRNA expression levels potentially indicates exon usage competition during splicing for either circRNA expression or mRNA expression.
We also compared the average number of exons per circRNA transcript of each group using single-factor ANOVA to assess any potential impact of circRNA length on expression patterns, but no significant differences were found (p = 0.788) (Figure 1C). Considering the slightly right-skewed distributions of exon numbers, we also performed a Kruskal–Wallis test of the same data, which also failed to indicate significance (p = 0.46309). These results suggest that transcript length has little to no role in modulating circRNA expression following LPC treatment in HAECs. Taken together, our results suggest that the proatherogenic lipid LPC induces a list of circRNAs in HAECs; and that a dominant inversely correlative relationship between circRNA and mRNA expression levels potentially indicates exon usage competition during splicing.
CircRNA-Related Genes Do Not Share Any Top Pathways With LPC-Induced mRNA Pathways; and Genes Associated With Significantly Changed CircRNAs Exhibit Little Relationship With Potential Effects of Inflammation-Related Cytokine Stimulation
To determine the signaling pathways associated with circRNA-related genes, we performed the IPA. As shown in Table 1C, we found the following: first, six top pathways related to the unchanged genes which related circRNAs were increased, including pyridoxal 5′-phosphate salvage pathway, Fcγ receptor-mediated phagocytosis in macrophage and monocyte, transforming growth factor-β (TGF-β) signaling, salvage pathways of pyrimidine ribonucleotides, epithelial adherens junction signaling, and Wnt/β-catenin signaling; second, ten top pathways related to the unchanged mRNAs which related circRNAs were decreased, including ephrin B signaling, focal adhesion kinase (FAK) signaling, axonal guidance signaling, ephrin receptor signaling, integrin signaling, interleukin-15 (IL-15) production, tRNA charging, semaphoring signaling in neurons, ephrin A signaling, and regulation of cellular mechanics by calpain protease; third, ten top pathways related to the unchanged RNAs which related circRNAs were increased, including pyridoxal 5′-phosphate salvage pathway, Fcγ receptor-mediated phagocytosis, TGF-β signaling, salvage pathways of pyrimidine ribonucleotides, epithelial adherens junction signaling, Wnt/β-catenin signaling, actin cytoskeleton signaling, cell cycle control of chromosomal replication, unfolded protein response, remodeling of epithelial adherens junctions; and fourth, ten top pathways related to the decreased mRNAs which related circRNAs were decreased, including role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis, neuroinflammation signaling pathway, cardiomyocyte differentiation via bone morphogenetic protein (BMP) receptors, April mediated signaling, B cell activation factor signaling, Wnt/Ca++ pathway, netrin signaling, BMP signaling pathway, regulation of IL-2 expression in activated and anergic T lymphocytes and factors promotion of cardiogenesis in vertebrates. The Vann Diagram analysis found that no signaling pathways are overlapped among the four groups (Figure 1D). The results suggest that, although we cannot determine directly circRNAs functional pathways in this LPC-induced HAEC activation, LPC-induced mRNAs-related HAEC activation pathways, that we reported (Li et al., 2018b), have no overlaps with any signaling pathways for circRNAs-related mRNAs.
Table 1C.
There are no overlaps among the top 10 pathways in the following four groups #, suggesting that the functions of circRNAs-related mRNAs do not participate in LPC-induced human aortic endothelial cell activation.
| Number | LPC upregulated mRNAs (promoting endothelial activation, PMID) | CircRNA increased and mRNA unchanged genes (inflammation, PMID) |
|---|---|---|
| 1 | Superpathway of cholesterol biosynthesis (HAEC activation, 29371247) | Pyridoxal 5′-phosphate salvage pathway (neurological pathologies, 22201923) |
| 2 | Cholesterol biosynthesis I | Fcγ Receptor-mediated phagocytosis in macrophages and monocytes (immune responses, 28117787) |
| 3 | Cholesterol biosynthesis II (via 24,25-dihydrolanosterol) | TGF-β signaling (anti-inflammation, 22438968) |
| 4 | Cholesterol biosynthesis III (via Desmosterol) | Salvage pathways of pyrimidine ribonucleotides (repair and inflammatory signal, 28053879) |
| 5 | Granulocyte adhesion and diapedesis | Epithelial adherens junction signaling (epithelial cell migration, 30010460) |
| 6 | Hepatic fibrosis/hepatic stellate cell activation | Wnt/β-catenin signaling (immune response, 25136145) |
| 7 | Role of macrophages, fibroblasts, and endothelial Cells in rheumatoid arthritis | Actin cytoskeleton signaling (immune response, 30274167) |
| 8 | Role of IL-17F in allergic inflammatory airway diseases | Cell cycle control of chromosomal replication (immune response, 29908233) |
| 9 | Agranulocyte adhesion and diapedesis | Unfolded protein response (inflammatory response, 30278941) |
| 10 | p38 MAPK signaling | Remodeling of epithelial adherens junctions (epithelial cell migration, 30010460) |
| Number | CircRNA decreased and mRNA unchanged genes | CircRNA decreased and mRNA decreased genes |
| 1 | Ephrin B signaling | Role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis |
| 2 | FAK signaling | Neuroinflammation signaling pathway |
| 3 | Axonal guidance signaling | Cardiomyocyte differentiation via BMP receptors |
| 4 | Ephrin receptor signaling | April mediated signaling |
| 5 | Integrin signaling | B Cell activating factor signaling |
| 6 | IL-15 production | Wnt/Ca+ pathway |
| 7 | tRNA charging | Netrin signaling |
| 8 | Semaphorin signaling in neurons | BMP signaling pathway |
| 9 | Ephrin A signaling | Regulation of IL-2 expression in activated and anergic T lymphocytes |
| 10 | Regulation of cellular mechanics by calpain protease | Factors promoting cardiogenesis in vertebrates |
CircRNAs may play roles in weakening the roles of related mRNAs via their roles in exon usage competition. #Of note, there were no top ten pathways identified by the Ingenuity Pathway Analysis in three other groups in Table 1A.
We further hypothesized that LPC-induced circRNA expression could be positively associated with changes in gene expression under other pro-inflammatory stimuli. Thus, to compare the effects of LPC stimulation with that of prototypical inflammation-related responses, we obtained mRNA expression change data for cells stimulated with the pro-inflammatory cytokines tumor necrosis factor alpha (TNF-α), interferon gamma (IFN-γ), and interleukin 6 (IL-6) and the anti-inflammatory cytokines TGF-β, IL-4, and IL-10 from the database ArrayExpress (Braun et al., 2013; Teles et al., 2013; Bald et al., 2014; Jin et al., 2014; Kharma et al., 2014; Montoya et al., 2014; Polak et al., 2014, 2017; Duncan et al., 2015; Kolesnikov et al., 2015; Tan et al., 2015). We found all circRNA-related genes significantly changed after stimulation in each data set (not shown) and results indicate that cytokines with similar effects did not tend to regulate circRNA-related genes in similar directions, nor did pro-inflammatory and anti-inflammatory cytokines tend to regulate circRNA-related genes in opposing directions (not shown). This in turn suggests a limited or absent canonical role for circRNA-related genes in inflammatory response and fails to indicate general trends for pro- or anti-inflammatory cytokine modulation of circRNA-related genes. However, it neither supports nor disproves the roles of inflammatory signaling in regulating circRNA levels, as circRNA levels are largely independent of the mRNA expression levels used in this analysis and, furthermore, circRNAs may fulfill different physiological functions such as homeostasis than their canonically spliced mRNA counterparts do such as promotion of HAEC activation induced by LPC.
68.8% of the CircRNAs Feature Significant Flanking Intron Homology
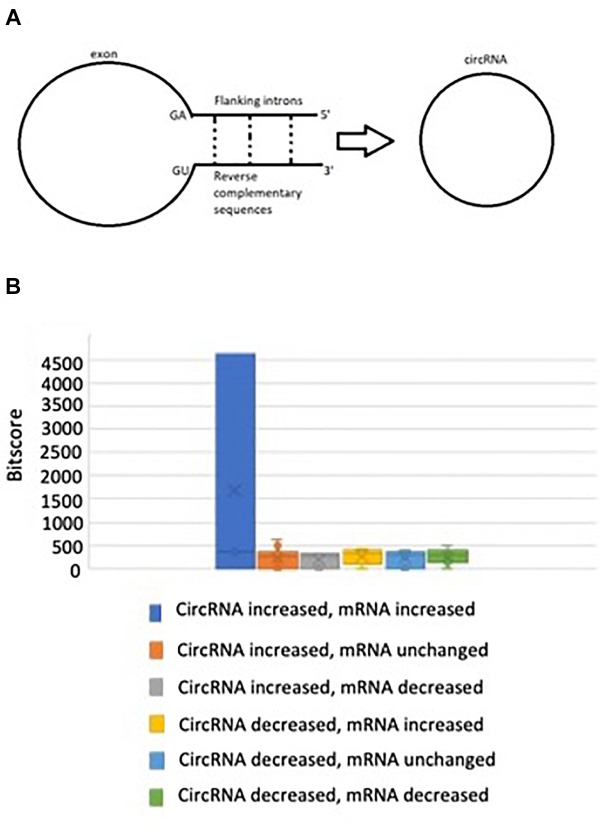
To identify the potential mechanism underlying the generation of circRNAs, previous reports found that many circRNA transcripts feature reverse complementary flanking intron sequences, which enable self-pairing that brings 3′ and 5′ exonic sequences into close proximity, an arrangement that favors head-to-tail alternative splicing and circRNA biogenesis (Figure 2A; Albury et al., 2015). To confirm this pattern in our dataset, we ran NIH NCBI BLAST sequence alignments using NIH NCBI BLAST+ command line tools (blastn -max_hsps 1 -outfmt “10 pident e-Value bitscore”) comparing the 3′ and 5′ flanking intron sequences for each circRNA, as previously described (Albury et al., 2015). We found that 53 out of 77 circRNAs (68.8%) possessed flanking intron sequences with significant reverse complements as determined by expected value (number of matches expected given random sequences) <1∗10-20 (Table 2). This number is lower than that of human 88% circRNA with the Alu repeats in flanking regions (Albury et al., 2015), and that in C. elegans circRNAs (96.3%); and high than that of Zebrafish circRNAs (2.5%) (Ivanov et al., 2015). To test potential association of reverse complementarity of flanking introns with different expression patterns of circRNA and mRNA, we performed a single-factor ANOVA using the bitscores (normalized values reporting strength of alignment between sequences) of circRNAs from each of the six groups sorted previously, assigning a bitscore of zero (indicating no matches better than those resulting from chance) for all circRNAs whose flanking introns did not demonstrate reverse complementarity. This initial analysis indicated significant differences between groups (p = 0.0011), but further inspection revealed the presence of an outstanding data point (bitscore = 4617) in the “circRNA increased, mRNA increased” group, which combined with the small size (n = 3) of the group greatly weakened the generalizability of the test (Figure 2B). We thus performed a non-parametric Kruskal–Wallis test on the same data set, but this failed to reach significance (p = 0.74053). We also performed another single-factor ANOVA with the outstanding point removed, the results of which marginally failed to indicate any significant difference in bitscore between groups (p = 0.07877). Thus, we cannot conclude that there is an association between flanking intron homology and circRNA expression patterns, although that our statistical analysis approaches significance suggests that the area may warrant further study, especially with a larger sample. Our results indeed demonstrate that most circRNAs feature significant flanking intron homology, suggesting that this mechanism may be underlying selection for the expressions of circRNAs versus mRNAs.
FIGURE 2.
(A) Pre-splicing RNA transcripts with reverse complementary matches in flanking intron sequences tend to favor circRNA biogenesis, (B) There is no significant difference in bitscore between groups when non-parametric testing is performed.
Table 2.
53 out of 77 (68.8%) circRNAs have significant homologies between flanking introns, which may facilitate chromatin looping and circRNA generation.
| Gene | circRNAdb ID | % identity | e-Value | Bitscore |
|---|---|---|---|---|
| circRNA expression increased, mRNA expression increased | ||||
| PARD3B | N/A | 98.44 | 0.00 | 4617 |
| SIPA1L3 | hsa_circ_10577 | 90.24 | 0.00 | 374 |
| circRNA expression decreased, mRNA expression increased | ||||
| FUT8 | hsa_circ_02544 | 93.17 | 0.00 | 429 |
| RELL1 | hsa_circ_08846 | 88.03 | 0.00 | 335 |
| FNDC3B | hsa_circ_15231 | 84.59 | 0.00 | 311 |
| circRNA expression increased, mRNA expression unchanged | ||||
| PCNXL2 | hsa_circ_10712 | 98.65 | 0.00 | 525 |
| RUNX1T1 | hsa_circ_02162 | 89.74 | 0.00 | 427 |
| DOCK1 | hsa_circ_00689 | 89.61 | 0.00 | 387 |
| TTC28 | hsa_circ_22261 | 89.6 | 0.00 | 375 |
| ATXN1 | hsa_circ_13397 | 89.45 | 0.00 | 300 |
| XXYLT1 | hsa_circ_02143 | 89.26 | 0.00 | 521 |
| ECSIT | hsa_circ_00599 | 88.63 | 0.00 | 361 |
| CUX1 | hsa_circ_12403 | 87.25 | 0.00 | 333 |
| EVI5 | hsa_circ_11187 | 86.53 | 0.00 | 363 |
| SMURF2 | hsa_circ_01208 | 86.23 | 0.00 | 324 |
| IQGAP1 | hsa_circ_10932 | 86.13 | 0.00 | 291 |
| TEX2 | hsa_circ_31354 | 85.26 | 0.00 | 316 |
| TBC1D8 | hsa_circ_32540 | 84.04 | 0.00 | 263 |
| TPST1 | hsa_circ_22617 | 83.83 | 0.00 | 560 |
| LZIC | hsa_circ_31394 | 83.55 | 0.00 | 278 |
| SCAF8 | hsa_circ_06913 | 83.05 | 0.00 | 261 |
| KANSL1L | hsa_circ_06859 | 82.95 | 0.00 | 191 |
| VRK1 | hsa_circ_08594 | 82.95 | 0.00 | 115 |
| DNAAF5 | N/A | 82.87 | 0.00 | 254 |
| PDS5A | hsa_circ_29001 | 82.68 | 0.00 | 257 |
| PPP6R2 | hsa_circ_07544 | 82.34 | 0.00 | 499 |
| ACVR2A | hsa_circ_04751 | 81.88 | 0.00 | 255 |
| VAMP3 | hsa_circ_18051 | 81.76 | 0.00 | 250 |
| PRKY | hsa_circ_30664 | 81.66 | 0.00 | 230 |
| CSNK1G3 | hsa_circ_08140 | 80.89 | 0.00 | 226 |
| PVT1 | hsa_circ_16350 | 76.23 | 0.00 | 619 |
| circRNA expression decreased, mRNA expression unchanged | ||||
| FAM13B | hsa_circ_11413 | 89.9 | 0.00 | 379 |
| SCMH1 | hsa_circ_12377 | 89.74 | 0.00 | 387 |
| N4BP2L2 | hsa_circ_12240 | 88.45 | 0.00 | 363 |
| ADAMTS6 | hsa_circ_28538 | 88.41 | 0.00 | 353 |
| PSD3 | hsa_circ_02851 | 87.66 | 0.00 | 351 |
| UBAP2 | hsa_circ_30156 | 87.63 | 0.00 | 326 |
| PTK2 | hsa_circ_23485 | 85.49 | 0.00 | 396 |
| ASAP1 | hsa_circ_09642 | 84.95 | 0.00 | 294 |
| MAN1A2 | hsa_circ_02643 | 84.03 | 0.00 | 272 |
| EPHB4 | hsa_circ_08510 | 82.08 | 0.00 | 257 |
| circRNA expression increased, mRNA expression decreased | ||||
| UGGT1 | N/A | 87.2 | 0.00 | 326 |
| RNF38 | hsa_circ_32756 | 86.81 | 0.00 | 318 |
| RNF138 | hsa_circ_01057 | 86.3 | 0.00 | 315 |
| SLF2 | N/A | 85.76 | 0.00 | 315 |
| RCAN3 | hsa_circ_22201 | 83.84 | 0.00 | 278 |
| PDS5B | hsa_circ_01546 | 82.06 | 0.00 | 254 |
| circRNA expression decreased, mRNA expression decreased | ||||
| RHOBTB3 | hsa_circ_19124 | 90.64 | 0.00 | 392 |
| ANKRD12 | hsa_circ_31497 | 85.86 | 0.00 | 313 |
| ANKRD17 | hsa_circ_01636 | 85.37 | 0.00 | 296 |
| CEP192 | hsa_circ_26363 | 84.76 | 0.00 | 307 |
| BMPR2 | hsa_circ_06837 | 81.99 | 0.00 | 503 |
| FGD4 | hsa_circ_21030 | 78.65 | 0.00 | 115 |
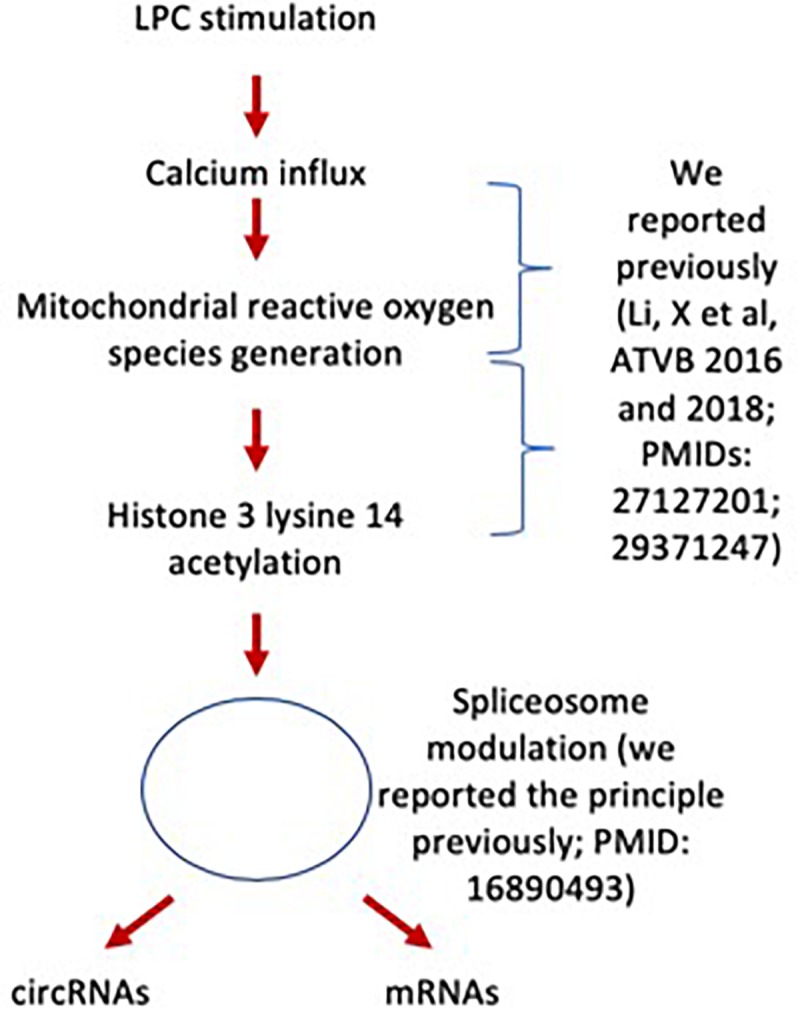
LPC Stimulation Disrupts the Expression Levels of 17 Spliceosome Components, Potentially Promoting Back Splicing for CircRNA Formation Over Canonical Splicing for mRNAs
We and others reported that downregulation of certain spliceosome components promotes circRNA formation over canonical mRNA splicing (Shen et al., 2017; Wilusz, 2018); and that proinflammatory cytokine TNF-a, inflammation (Liang et al., 2017), autoimmune disease (Xiong et al., 2006b), and tumors modulate alternative splicing (Ng et al., 2004) and spliceosome (Yan et al., 2004). To determine whether this potential mechanism plays any roles in shaping LPC-modulated circRNA expression patterns, we compared the list of significantly changed mRNAs from RNA-Seq (determined via housekeeping gene confidence interval, as previously described) with lists of known spliceosome proteins (Deckert et al., 2006; Yang et al., 2006; Behzadnia et al., 2007; Bessonov et al., 2008; Table 3). Six spliceosome components were significantly downregulated after LPC stimulation, while another 11 were significantly upregulated. Given that 51 (66.2%) circRNAs were significantly upregulated while only 26 (33.8%) were significantly downregulated (p = 0.0029, where p represents the probability of a ratio equal to or more extreme than the experimental value, assuming circRNA upregulation and downregulation equally likely) and that among upregulated circRNAs, 10 corresponding mRNAs experienced significant downregulation while only 3 experienced significant upregulation (p = 0.0461, where p represents the probability of a ratio equal to or more extreme than the experimental value, assuming mRNA upregulation and downregulation equally likely) (Figure 1A and Table 3), this disruption may have been sufficient to shift the splicing patterns from canonical mRNA splicing toward back splicing for circRNA formation (Figure 3), as reported previously (Wilusz, 2018). While the data currently available is insufficient to establish a causal link between LPC-induced spliceosome disruption and a shift from canonical splicing to circRNA formation, it does demonstrate a strong association suitable for more intensive study. Taken together, our results suggest that LPC stimulation disrupts the expression levels of 17 spliceosome components, potentially promoting back splicing for circRNA formation over canonical splicing for mRNAs.
Table 3.
Seventeen spliceosome genes out of approximately 80 proteins have fold changes outside a housekeeping gene-determined 95 percent confidence interval in RNA-Seq data.
| Gene | FC |
|---|---|
| FUS | 0.68 |
| PCBP2 | 0.68 |
| RBM7 | 0.77 |
| THOC1 | 0.82 |
| LSM8 | 0.83 |
| PCBP1 | 0.87 |
| RBM10 | 1.11 |
| EIF4A3 | 1.12 |
| CCDC130 | 1.12 |
| RBM42 | 1.13 |
| DGCR14 | 1.13 |
| MFAP1 | 1.18 |
| PUF60 | 1.18 |
| THOC3 | 1.20 |
| LSM6 | 1.21 |
| RNF113A | 1.24 |
| FAM50B | 1.25 |
There are 6 protein downregulation and 11 proteins upregulation.
FIGURE 3.
LPC modulates the expressions of spliceosome components and may facilitate the generation of circRNAs.
Shifting Chromatin Long–Range Interaction Sites From Downstream to Upstream Is Associated With Induction of a List of Circular RNAs in LysoPC-Activated Human Aortic Endothelial Cells; and in Contrary, Shifting Chromatin Long–Range Interaction Sites From Upstream to Downstream Is Associated With Induction of a List of mRNAs
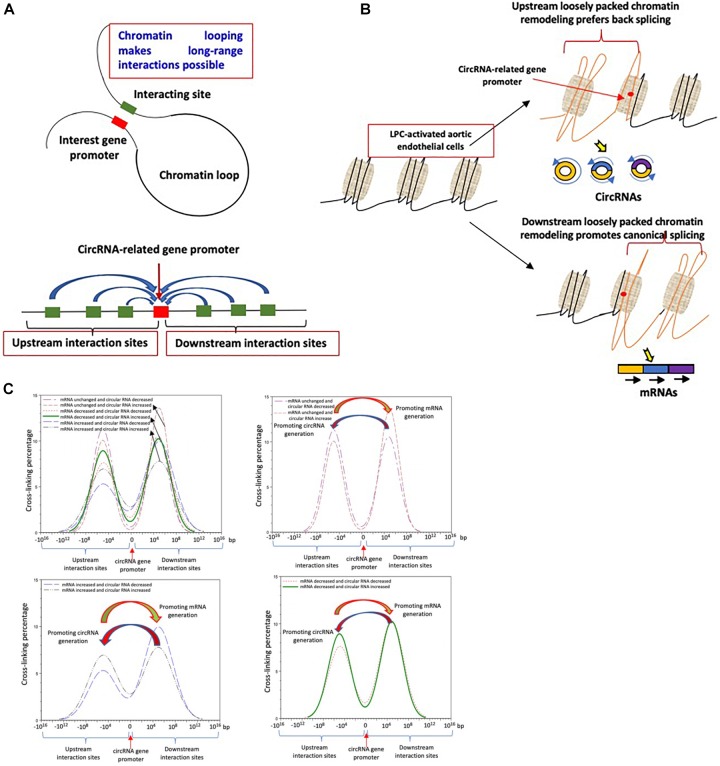
Rapid development of technologies in capturing chromosome conformations such as chromosome conformation capture sequencing (3C-Seq) (Wahl et al., 2009), chromosome conformation capture-on-chip (4C-Seq) (Dekker et al., 2002; Splinter et al., 2011), and chromosome conformation capture carbon copy sequencing (5C-Seq) (Stadhouders et al., 2013) allows determination of the relative frequency of direct physical contact between a pair of linearly separated chromatin segments with 3C-Seq, and genome–wide interactions involving a single anchor region, and interactions involving multiple genomic regions with 4C-Seq and 5C-Seq, respectively (Figure 4A). Differences in chromatin long-range interaction patterns between genes have previously been hypothesized to influence alternative splicing (Dostie et al., 2006) and the transcription of inflammatory genes such as cytokine (Acuña and Kornblihtt, 2014) and cytokine receptor (Park et al., 2016) and cardiovascular disease causative genes (Deligianni and Spilianakis, 2012). We hypothesize that chromatin long–range interactions differentially regulate the gene promoters to differentiate the mRNA increase from circRNA increase versus decrease, derived from the same gene, under LPC stimulation in HAECs. To test this hypothesis with respect to circRNA biogenesis, we obtained long-range interaction data for all genes related to significantly changed circRNAs from the well-accepted 4DGenome database with a huge collection of 4,433,071 experimentally derived chromatin long–range interactions (Teng et al., 2015). We then calculated the distances between interacting genes with respect to the circRNA-related gene, with distances designated as positive if the circRNA-related gene was localized downstream of its partner and as negative if the circRNA-related gene was localized upstream of its partner. The two-sample Kolmogorov–Smirnov test of the chromatin long-range interaction distance between genes corresponding to downregulated and upregulated circRNAs indicated a significant difference between the two distance distributions (p < 0.001). First, the majority of long-range interaction sites with circRNA-related genes were focused on 106 kb upstream and downstream of the circRNA-related gene, suggesting a common feature of the long-range chromatin interaction sites with LPC-induced circRNA-related genes; Second, in the three groups with circRNA related genes such as (a) upregulation; (b) downregulation; or (c) no changes induced by LPC, the group with LPC-induced upregulation of mRNAs had the lowest peak, the group with LPC-induced downregulation of mRNAs had the middle peak, the group with LPC-induced no changed expression of mRNAs had the highest peak, suggesting that high concentration of chromatin long-range interaction sites prevents HAECs from responding to LPC stimulation in changing gene expressions (Figure 4B,C); Third, in both groups of mRNA upregulation and mRNA downregulation induced by LPC, circRNA increase was associated with increased upstream chromatin long-range interaction sites, suggesting that an increase in upstream chromatin long-range interaction sites is associated with circRNA generation presumably by favoring back-splicing in generating circRNAs over canonical splicing in generating mRNAs (Figure 4C). In contrast, a decrease in upstream chromatin long-range interaction sites is associated with mRNA generation presumably by favoring canonical splicing in generating mRNAs over back-splicing in generating circRNAs (Figure 4C). Altogether, these results suggest that different long-range interaction mechanisms are likely to govern circRNA upregulation and downregulation. Indeed, upregulated circRNAs tended to interact at somewhat longer distances than downregulated circRNAs did (Figure 4B,C). Since the 4DGenome database contains the experimental data derived from human non-aortic ECs (Teng et al., 2015), the future work will be needed to use circular chromosome conformation capture sequencing (4C-Seq) to examine HAECs stimulated with LPC to map the specific upstream interaction sites for a favorable selection of circRNA over mRNAs. Therefore, our results may suggest that certain ideal long-range interaction distances exist that are more likely to promote circularization; and that favorable distances tend to be greater in magnitude than unfavorable ones.
FIGURE 4.
(A) Chromatin long range interaction may regulate the expressions of circRNAs-related genes via forming chromatin loops. (B) Loosely packed chromatin regions either upstream or downstream from circRNAs related gene promoters may differentially prefer back splicing to generate circRNAs to canonical splicing to generate mRNAs. (C) Shifting chromatin long-range interaction sites from downstream to upstream may promotes a list of circular RNAs; and in contrast, shifting chromatin long-range interaction sites from upstream to downstream promotes induction of a list of mRNAs. The data were collected from the experimental data deposited in the 4DGenome database (https://4dgenome.research.chop.edu) containing the data from human umbilical vein endothelial cells (HUVECs).
Six Significantly Changed CircRNAs Sponge miRNAs, Which May Be Associated With Inhibition of Type 2 Diabetes Mellitus, Inhibition of Migration, and Inhibition of Vascular Dysfunction and Immune Responses in Endothelial Cells
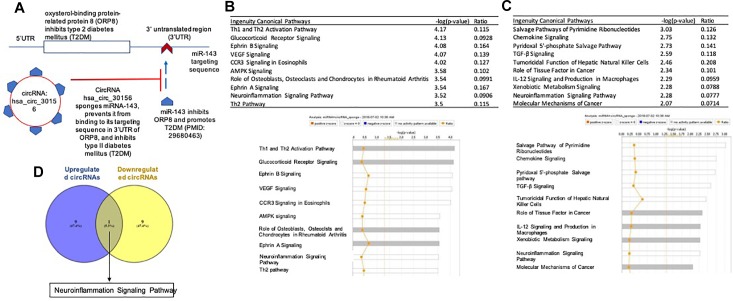
A number of circRNAs can sponge miRNAs, preventing miRNA-mediated degradation of mRNA transcripts and disruption of canonical signaling and effector pathways (Hansen et al., 2013; Memczak et al., 2013). Thus, by regulating miRNA activity, significantly changed circRNAs can act through canonical pathways, potentially playing a significant role in HAEC activation. We examined the sponging potential of all significantly changed circRNAs using the CircInteractome database (Montefiori et al., 2018), recording two miRNAs with four or more predicted binding sites in a single circRNA transcript, a threshold above which meaningful sponging activity is likely to occur Memczak et al. (2013). Another four significantly changed circRNAs are experimentally shown to sponge miRNAs (Dudekula et al., 2016; Chen et al., 2017; Yan et al., 2017; Wang et al., 2018), for six total circRNAs with miRNA sponging activity including miR125, miR143, miR1272, miR153, miR515-5p, and miR196a-5p (Table 4). In Figure 5A, to demonstrate the proof of principle, we showed how circRNA has_circ_30156 could inhibit type 2 diabetes mellitus (T2DM) by sponging miR143 and preventing miR143 from binding to oxysterol-binding protein-related protein 8 (ORP8) 3′untranslated region (3′UTR).
Table 4.
Six circRNAs demonstrate miRNA sponging activities.
| Gene | circRNAdb ID | miRNA sponged | Readnumber (LPC) | Readnumber (control) | Ratio | Function of microRNA | PMID |
|---|---|---|---|---|---|---|---|
| PVT1 | hsa_circ_16350 | miR-125 | 8 | 1 | 8 | Promote regeneration/repair | 29461585 |
| UBAP2 | hsa_circ_30156 | miR-143 | 1 | 7 | 0.14 | Inhibit autophage/promote inflammation | 29562274 |
| VAMP3 | hsa_circ_18051 | miR-1272 | 7 | 1 | 7 | T cell differentiation | 26941729 |
| VRK1 | hsa_circ_08594 | miR-153 | 4 | 1 | 4 | Inhibit migration/invasion | 29113423 |
| FGD4 | hsa_circ_21030 | miR-515-5p | 1 | 9 | 0.11 | Decrease in atopic eczema | 17622355 |
| DOCK1 | hsa_circ_00689 | miR-196a-5p | 7 | 1 | 7 | Delay lymph node metastasis | 29434944 |
FIGURE 5.
(A) CircRNA has the potential role to inhibit type 2 diabetes mellitus (T2DM)by sponging miRl43 and preventing miRl43 from binding to oxysterol-binding protein-related protein 8 (ORP8) ˆuntranslated region (3′UTR). (B) Upregulated circRNAs sponge miRNAs involved in both pro- and anti-inflammatory pathways. (C) Downregulated circRNAs sponge miRNAs are involved with both pro-inflammatory and anti-inflammatory pathways. (D) One pathway is overlapped in the sponged microRNAs between upregulated and downregulated circRNAs.
To elucidate potential downstream pathways for these miRNAs that might influence the LPC-induced inflammatory response, we separately performed IPA for miRNAs sponged by upregulated and downregulated circRNAs, respectively. Notably, the top two pathways for miRNAs sponged by upregulated circRNAs were the pro-inflammatory type 1 T helper cell (Th1), and Th2 activation pathway, and the anti-inflammatory glucocorticoid receptor signaling pathway (Figure 5B). Similarly, miRNAs sponged by downregulated circRNAs were prominently implicated in the anti-inflammatory transforming growth factor-β (TGF-β) signaling pathway and the pro-inflammatory interleukin-12 (IL-12) signaling and production in macrophages pathway (Figure 5C). Owing to these numerous conflicting results, the pro- and anti-inflammatory effects of the predicted downstream pathways likely cancel in large part, resulting in little overall pro- or anti-inflammatory effects (Figure 5D). Specifically, microRNA-125a plays crucial roles both in development and in the adult tissues, including it: (1) contributes to the control of phase transitions in development and/or cell differentiation; (2) regulates the expression of several target proteins that are involved in cell proliferation, apoptosis, and migration; (3) interferes with the expression of the hepatitis B virus surface antigen in liver cells, thus counteracting viral replication (Zhang et al., 2017). miR-125b plays a significant role in immune response and apoptosis (Potenza and Russo, 2013). The novel long intergenic non-coding RNA UCC promotes colorectal cancer progression by sponging miR-143 (Ribeiro and Sousa, 2014). The function of miR1272 remains unreported. miR153 is tumor suppressive (Huang et al., 2017), and miR153 targeting of KCNQ4 contributes to vascular dysfunction in hypertension (Zhao et al., 2013). miR-515-5p controls cancer cell migration through MARK4 regulation (Carr et al., 2016). Higher expression of miR-196a was both associated with poor overall survival and disease-free survival and recurrence-free survival in different kinds of cancers (Pardo et al., 2016). Taken together, sponging of most of six miRs in ECs may be associated with inhibition of migration, inhibition of vascular dysfunction and immune responses.
74% Significantly Changed CircRNAs Contain Open Reading Frames, the Majority of Which Encode Fragments of Known Proteins
Despite their classification as ncRNAs, numerous circRNAs can be translated into peptides, some of which are novel forms with uncharacterized yet potentially relevant function (Cai et al., 2016; Legnini et al., 2017; Pamudurti et al., 2017). Moreover, the circRNAdb database used to assign circRNA IDs provides ORF peptide sequence data for all circRNAs with ORFs (Chen et al., 2016). To assess the potential of significantly changed circRNAs to encode small proteins/peptides, we performed NIH NCBI BLAST peptide sequence alignments against the NCBI non-redundant protein sequences database (nr), restricted to entries for Homo sapiens, for all significantly changed circRNAs with ORF data in circRNAdb using the NIH NCBI BLAST+ command line utility (blastp -db nr -remote -entrez_query “Homo sapiens [Organism]” -max_target_seqs 1 -outfmt “10 sacc pident e-Value”) (Table 5A). Out of 77 significantly changed circRNAs, 57 (74.0%) contained ORFs; of these, 48 ORF peptide sequences matched substantially (with an expected value < 1∗10-4) to fragments of known proteins. Notably, 47 of the 48 ORFs with substantial matches mapped to a protein encoded by mRNA corresponding to the same gene as the circRNA, the sole exception originating from a circRNA corresponding to the non-coding gene PVT1. A majority of significantly changed circRNAs, therefore, have protein-coding potential and could produce fragments of known proteins. Such protein fragments remain both uncharacterized and experimentally undetected, but could theoretically serve physiological functions, including ones different from those of the full canonical protein (Figure 6). Furthermore, using predicted IRES data from circRNAdb, we found that all 57 circRNAs with ORFs contain at least one predicted IRES, and 32 of them (56.1%) contain at least one predicted IRES with an R-value (a metric indicating suitability of a sequence as an IRES) greater than a previously determined cutoff value of 1.54 and a predicted pseudoknot structure, which together indicate a greater probability of ribosomal entry and subsequent translation (Yang et al., 2017). We previously reported that this type of IRES drive secondary open reading frames is translated into short proteins (Wu et al., 2009); and that short peptides can disrupt the protein–protein interactions (Xiong et al., 2006a). The Kozak consensus sequence is a sequence which occurs on eukaryotic mRNA and has the consensus (gcc) gccRccAUGG, which plays a major role in the initiation of the translation process (Yang et al., 2005). However, NIH NCBI BLAST searches for Kozak (1996) sequences insignificantly changed circRNAs did not yield any matches.
Table 5A.
All but one circRNA open reading frames with significant BLAST matches encode peptides from the same gene as their corresponding mRNAs.
| Gene | circRNAdb ID | Accession | % ID | E-value | Same translation as mRNA? |
|---|---|---|---|---|---|
| Near-perfect overlap with known sequences | |||||
| VAMP3 | hsa_circ_18051 | XP_016858330 | 100 | 0.00 | Yes |
| DOCK1 | hsa_circ_00689 | Q14185 | 100 | 0.00 | Yes |
| HNRNPM | hsa_circ_20744 | BAG57075 | 100 | 0.00 | Yes |
| FOXP1 | hsa_circ_19469 | BAC11065 | 100 | 0.00 | Yes |
| TBC1D8 | hsa_circ_32540 | BAA76517 | 100 | 0.00 | Yes |
| KANSL1L | hsa_circ_06859 | XP_011509011 | 100 | 0.00 | Yes |
| RUNX1T1 | hsa_circ_02162 | AAB34820 | 100 | 0.00 | Yes |
| CSNK1G3 | hsa_circ_08140 | XP_016864559 | 100 | 0.00 | Yes |
| TPST1 | hsa_circ_22617 | XP_011514936 | 100 | 0.00 | Yes |
| RCAN3 | hsa_circ_22201 | NP_001238914 | 100 | 0.00 | Yes |
| NEIL3 | hsa_circ_00262 | Q8TAT5 | 100 | 0.00 | Yes |
| PPP6R2 | hsa_circ_07544 | EAW73534 | 100 | 0.00 | Yes |
| VRK1 | hsa_circ_08594 | 2LAV_A | 100 | 0.00 | Yes |
| BABAM1 | hsa_circ_04303 | BAG61451 | 100 | 0.00 | Yes |
| RYK | hsa_circ_26048 | AAH21700 | 100 | 0.00 | Yes |
| ATXN1 | hsa_circ_13397 | NP_000323 | 100 | 0.00 | Yes |
| ACVR2A | hsa_circ_04751 | AAH67417 | 100 | 0.00 | Yes |
| PRKY | hsa_circ_30664 | O43930 | 100 | 0.00 | Yes |
| UBAP2 | hsa_circ_30156 | BAG64527 | 100 | 0.00 | Yes |
| RELL1 | hsa_circ_08846 | XP_016864079 | 100 | 0.00 | Yes |
| NFAT5 | hsa_circ_16704 | XP_016878362 | 100 | 0.00 | Yes |
| CEP192 | hsa_circ_26363 | XP_016881293 | 100 | 0.00 | Yes |
| SDF4 | hsa_circ_21644 | XP_011539858 | 100 | 0.00 | Yes |
| CORO1C | hsa_circ_01619 | BAH14436 | 100 | 0.00 | Yes |
| ADAMTS6 | hsa_circ_28538 | XP_011541429 | 100 | 0.00 | Yes |
| ASAP1 | hsa_circ_09642 | CAD97831 | 100 | 0.00 | Yes |
| MAN1A2 | hsa_circ_02643 | XP_016855604 | 100 | 0.00 | Yes |
| ANKRD17 | hsa_circ_01636 | XP_016863500 | 99.796 | 0.00 | Yes |
| ANKRD12 | hsa_circ_31497 | BAB15014 | 99.683 | 0.00 | Yes |
| CRIM1 | hsa_circ_16396 | XP_016859748 | 99.448 | 0.00 | Yes |
| PSD3 | hsa_circ_02851 | XP_016868754 | 99.333 | 0.00 | Yes |
| PDS5B | hsa_circ_01546 | AAH70274 | 99.048 | 0.00 | Yes |
| FNDC3B | hsa_circ_15231 | EAW78473 | 98.864 | 0.00 | Yes |
| FUT8 | hsa_circ_02544 | BAA92858 | 98.551 | 0.00 | Yes |
| PCNXL2 | hsa_circ_10712 | BAG51507 | 98.387 | 0.00 | Yes |
| RHOBTB3 | hsa_circ_19124 | EAW96050 | 98.148 | 0.00 | Yes |
| PTK2 | hsa_circ_23485 | NP_001339677 | 98.052 | 0.00 | Yes |
| SCAP | hsa_circ_19380 | EAW64826 | 97.5 | 0.00 | Yes |
| EPHB4 | hsa_circ_08510 | 4AW5_A | 97.5 | 0.00 | Yes |
| IQGAP1 | hsa_circ_10932 | AAH05906 | 97.458 | 0.00 | Yes |
| XXYLT1 | hsa_circ_02143 | XP_005269343 | 97.403 | 0.00 | Yes |
| SMURF2 | hsa_circ_01208 | EAW94194 | 97.03 | 0.00 | Yes |
| CNTROB | hsa_circ_09218 | XP_016879630 | 97.015 | 0.00 | Yes |
| LZIC | hsa_circ_31394 | NP_001303904 | 96.97 | 0.00 | Yes |
| TTC28 | hsa_circ_22261 | XP_011528324 | 96.947 | 0.00 | Yes |
| PVT1 | hsa_circ_16350 | EAW92103 | 95.918 | 0.00 | No (non-coding gene) |
| FAM13B | hsa_circ_11413 | XP_016865040 | 87.786 | 0.00 | Yes |
| N4BP2L2 | hsa_circ_12240 | EAX08513 | 78.261 | 0.00 | Yes (fragment of same gene) |
| Partial overlap with known sequences | |||||
| BMPR2 | hsa_circ_06837 | NP_057675 | 56 | 3.10 | |
| ESYT2 | hsa_circ_25060 | EAW95833 | 48 | 3.20 | |
| GRHPR | hsa_circ_03935 | EAW60274 | 39.286 | 3.00 | |
| No overlap with known sequences | |||||
| RNF38 | hsa_circ_32756 | ||||
| SIPA1L3 | hsa_circ_10577 | ||||
| SCAF8 | hsa_circ_06913 | ||||
| CDK8 | hsa_circ_27078 | ||||
| ECSIT | hsa_circ_00599 | ||||
| FGD4 | hsa_circ_21030 | ||||
FIGURE 6.
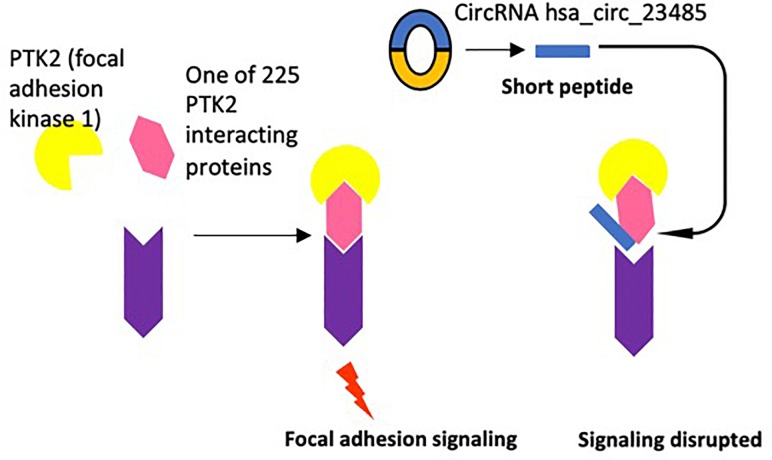
One example of potential disruption of protein–protein interactions by circRNA encoded short peptides is demonstrated.
We previously reported that short peptides with high homology to the interaction domain sequences of protein interaction partners could interfere the protein interaction (Yang et al., 2005). To demonstrate the proof of principle, in Figure 6, as suggested by the experimental data in the NIH-NCBI-Gene database9, focal adhesion kinase 1 (PTK2) has as many as 225 interaction proteins (Table 5B and Figure 6), thus, the circRNA hsa_circ_23485 encoded short peptide(s) could disrupt the protein interaction between PTK2 and interaction proteins. Taken together, 74% significantly changed circRNAs contain open reading frames, the majority of which encode fragments of known proteins, which may interfere with protein interaction-based signaling pathways. Of note, in addition to perform experiments in the future to verify the circRNAs, we also notice that various novel computational methods for finding ncRNAs versus coding RNAs (Abbas et al., 2016) and identifying small peptides (Pauli et al., 2015) have been developed, so that we will collaborate in the future with bioinformatics experts to verify the generation of those potential short peptides in HAECs.
Table 5B.
75 of 77 circRNA-related genes have numerous interactions with other proteins, suggesting that functional interactions between the circRNAs-related proteins and their interaction proteins may be disrupted by circRNA-encoded short peptides.
| Gene name | circRNAdb ID | Interactions |
|---|---|---|
| TEX2 | hsa_circ_31354 | 7 |
| MYO9A | N/A | 27 |
| PVT1 | hsa_circ_16350 | 2 |
| CRIM1 | hsa_circ_16396 | 3 |
| VAMP3 | hsa_circ_18051 | 74 |
| DOCK1 | hsa_circ_00689 | 37 |
| SCAP | hsa_circ_19380 | 29 |
| HNRNPM | hsa_circ_20744 | 341 |
| GRHPR | hsa_circ_03935 | 34 |
| FOXP1 | hsa_circ_19469 | 47 |
| RP11-146B14.1 | N/A | N/A |
| RNF38 | hsa_circ_32756 | 17 |
| SMURF2 | hsa_circ_01208 | 201 |
| ARFGEF2 | N/A | 53 |
| RNF138 | hsa_circ_01057 | 45 |
| TTC28 | hsa_circ_22261 | 25 |
| LZIC | hsa_circ_31394 | 13 |
| L3MBTL3 | N/A | 38 |
| TBC1D8 | hsa_circ_32540 | 6 |
| KANSL1L | hsa_circ_06859 | 0 |
| RUNX1T1 | hsa_circ_02162 | 96 |
| CSNK1G3 | hsa_circ_08140 | 38 |
| SIPA1L3 | hsa_circ_10577 | 39 |
| TPST1 | hsa_circ_22617 | 13 |
| MACF1 | N/A | 85 |
| RCAN3 | hsa_circ_22201 | 14 |
| UGGT1 | N/A | 65 |
| PARD3B | N/A | 7 |
| NEIL3 | hsa_circ_00262 | 19 |
| CNTROB | hsa_circ_09218 | 123 |
| SLF2 | N/A | 9 |
| PDS5B | hsa_circ_01546 | 54 |
| CUX1 | hsa_circ_12403 | 69 |
| DNAAF5 | N/A | 66 |
| IQGAP1 | hsa_circ_10932 | 294 |
| PPP6R2 | hsa_circ_07544 | 69 |
| VRK1 | hsa_circ_08594 | 34 |
| PDS5A | hsa_circ_29001 | 76 |
| BABAM1 | hsa_circ_04303 | 67 |
| FAM213A | N/A | 23 |
| EVI5 | hsa_circ_11187 | 18 |
| SCAF8 | hsa_circ_06913 | 22 |
| RYK | hsa_circ_26048 | 93 |
| FIP1L1 | hsa_circ_18069 | 81 |
| CDK8 | hsa_circ_27078 | 120 |
| ATXN1 | hsa_circ_13397 | 416 |
| ACVR2A | hsa_circ_04751 | 48 |
| ECSIT | hsa_circ_00599 | 117 |
| XXYLT1 | hsa_circ_02143 | 10 |
| PRKY | hsa_circ_30664 | 8 |
| PCNXL2 | hsa_circ_10712 | 3 |
| ANKRD12 | hsa_circ_31497 | 17 |
| BMPR2 | hsa_circ_06837 | 99 |
| N4BP2L2 | hsa_circ_12240 | 29 |
| UBAP2 | hsa_circ_30156 | 34 |
| PSD3 | hsa_circ_02851 | 8 |
| FUT8 | hsa_circ_02544 | 30 |
| RELL1 | hsa_circ_08846 | 19 |
| TBCD | hsa_circ_00754 | 64 |
| PTK2 | hsa_circ_23485 | 225 |
| NFAT5 | hsa_circ_16704 | 10 |
| FGD4 | hsa_circ_21030 | 5 |
| CEP192 | hsa_circ_26363 | 78 |
| SDF4 | hsa_circ_21644 | 92 |
| ANKRD17 | hsa_circ_01636 | 40 |
| CORO1C | hsa_circ_01619 | 109 |
| RARS | hsa_circ_21209 | 146 |
| ADAMTS6 | hsa_circ_28538 | 2 |
| SCMH1 | hsa_circ_12377 | 41 |
| FAM13B | hsa_circ_11413 | 6 |
| EPHB4 | hsa_circ_08510 | 30 |
| ESYT2 | hsa_circ_25060 | 34 |
| ASPH | hsa_circ_10828 | 44 |
| FNDC3B | hsa_circ_15231 | 12 |
| ASAP1 | hsa_circ_09642 | 44 |
| MAN1A2 | hsa_circ_02643 | 17 |
| RHOBTB3 | hsa_circ_19124 | 35 |
Discussion
We previously published experimental data outlining the role of LysoPC (LPC) on HAEC (Li et al., 2018c). In this previous publication we demonstrated that LysoPC stimulation leads to: (1) the upregulation of inflammatory adhesion molecules in HAEC; (2) upregulation of cytokine transcript expression; and (3) promotion of prolonged HAEC activation and expression of innate immune co-stimulation molecule expression. We believe that this RNA-Seq (RNA sequencing) data analyses provide strong experimental evidence for the role of LysoPC stimulation of proinflammatory mRNAs. Given that alternative splicing events lead to the generation of circRNAs from mRNA precursors in other cell types (Wilusz, 2018), there is strong justification to explore the role of LysoPC-induced circRNAs.
Although the importance of circRNAs in regulating EC function has been reported, the comprehensive analyses of circRNAs in EC activation, EC function, and cardiovascular diseases remain at their infancy; and the characterization of circRNAs in atherosclerosis-relevant HAECs has not been reported (Maass et al., 2017; Huang et al., 2018; Shang et al., 2018). To fill in this important knowledge gap, we profiled the circRNAs in HAECs stimulated with we proposed “conditional danger associated molecular pattern” LPC (Wang X. et al., 2016), and compared the circRNAs significantly modulated in HAECs by LPC with the mRNAs modulated in the same sets of RNA-Seq samples (Li et al., 2018b).
In the current study, we made the following findings: first, LPC modulates the expression of a list of 77 circRNAs in HAECs; and the majority of which, 94.1% of increased circRNAs, compete with related mRNAs for exon usage; second, circRNAs-related genes do not share any top pathways with LPC-induced mRNA pathways, and genes associated with significantly changed circRNAs exhibit little relationship with potential effects of inflammation-related cytokine stimulation; third, 68.8% of the circRNAs feature significant flanking intron homology; fourth, LPC stimulation disrupts the expression levels of 17 spliceosome components, potentially promoting back splicing for circRNA formation over canonical splicing for mRNAs; fifth, shifting chromatin long-range interaction sites from downstream to upstream promotes induction of a list of circRNAs in lysoPC-activated HAECs; and in contrary, shifting chromatin long-range interaction sites from upstream to downstream promotes induction of a list of mRNAs; sixth, six significantly changed circRNAs sponge miRNAs, which may be associated with inhibition of type 2 diabetes mellitus, inhibition of migration, inhibition of vascular dysfunction and immune responses in ECs; and seventh, 74% significantly changed circRNAs contain open reading frames, the majority of which encode fragments of known proteins, suggesting that short proteins generated from the secondary open reading frames potentially driven by internal ribosome entry sites may interfere the protein-protein interaction-based signaling.
As reported previously, LPC stimulation causes significant changes in mRNA expression levels for many genes (Li et al., 2018b). IPA of these changed genes implicates several common upstream regulatory signaling pathways, the most significant of which are the sterol regulatory element binding transcription factor 2 (SREBF2), SREBF chaperone (SCAP), and SREBF1 pathways (not shown), which jointly serve as key lipid metabolism regulators and contribute to pro-inflammatory signaling (Shao and Espenshade, 2012). This establishes a novel association between upregulated pro-inflammatory lipid metabolism pathways and significant expression changes for 77 circRNAs. Our data are insufficient to conclude any roles of lipid metabolism pathways in circRNA expression modulation. However, given the strength of the association between the significantly changed upstream pathways and the significantly changed circRNAs, more direct future investigation of the effects of lipid metabolism pathway disruption on circRNA levels will be appropriate and could confirm or extend this observation.
It is highly unlikely that the extreme ratio of upregulated to downregulated circRNAs or that of upregulated to downregulated corresponding mRNAs among upregulated circRNAs under LPC stimulation would occur by chance, as noted earlier. For this reason, spliceosome depletion, which is experimentally demonstrated to cause such responses in circRNA and mRNA expression (Liang et al., 2017; Wilusz, 2018), continues to be a compelling area for future study despite the inconclusive results presented in this study. For instance, knockdown of downregulated or overexpression of upregulated spliceosome components identified after LPC stimulation, potentially combined with more robust circRNA detection methods such as qPCR verification of back-spliced sequences (Jeck and Sharpless, 2014; Szabo and Salzman, 2016), could clarify the role of each component in LPC-induced disruption of circRNA expression patterns.
Additionally, the dramatic expression changes of many circRNAs under LPC stimulation suggests that they play currently undetermined, possibly significant roles in HAEC activation and the proatherogenic response. Indeed, the circRNA circular antisense non-coding RNA in the INK4 locus (circANRIL) performs anti-atherogenic, anti-proliferative functions in vitro by inhibiting ribosomal biogenesis through assembly factor binding (Holdt et al., 2016), although this circRNA was not detected in our data set. Nevertheless, it is possible that some significantly changed circRNAs fulfill regulatory roles in atherogenesis through similar mechanisms. As circRNAs are better characterized and prediction of RNA-protein binding becomes more sophisticated, it may be possible to detect protein-binding functions, including those related to atherogenesis or atheroprotection, in our significantly changed circRNAs.
Likewise, many of our significantly changed circRNAs could encode proteins. Although the ORFs detected match the loci of canonically spliced mRNAs, many of the peptides encoded are likely to be fragments of canonical proteins and therefore may serve different functions. Knock-in experiments of these novel predicted peptides could uncover new insights about their potential roles in LPC-induced atherogenesis. Furthermore, as our current findings do not include protein expression data, additional assay experiments under LPC stimulation, such as Western blots and high-performance liquid chromatography-linked mass spectrometer analyses specific to these novel predicted peptides, would provide additional insight on their expression patterns. Given the established coding potential of circRNAs (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017, such microproteins remain an underexplored pathway for circRNA regulatory roles in atherogenesis.
Furthermore, the field of circRNAs is still in its infancy. Software detection methods may give widely differing results, and not all circRNAs predicted from RNA-Seq data can be verified experimentally through methods such as RNase R-catalyzed degradation (Hansen et al., 2016). Simply repeating an LPC-induced HAEC activation experiment, treating with RNase R to remove spurious linear transcripts, and using additional circRNA algorithms to corroborate findings could provide more accurate data on circRNA expression change; using qPCR confirmation for particularly meaningful or significant detections would likewise further improve data reliability. Additionally, recent studies have outlined a number of novel experimental methods for verifying the expression and function (Jeck and Sharpless, 2014; Petkovic and Muller, 2015; Barrett and Salzman, 2016; Werfel et al., 2016; Liu et al., 2017; Zeng et al., 2017; Gomes et al., 2018; Li et al., 2018b). Overall, however, changes in circRNA expression levels following LPC stimulation follow unusual and poorly understood patterns, and further, more intensive profiling under otherwise identical conditions could reveal regulatory pathways not apparent in this study, which uses relatively simple detection methods. To this end, future studies are required using clinical arterial samples from atherosclerotic patients and healthy controls through the collaboration with vascular surgeons using the Temple University Institutional Review Board approved protocol.
Taken together, based on our findings, we propose a novel working model (Figure 7): first, modulation of 77 significantly changed circRNAs in LPC-activated HAECs is temporally associated with LPC-upregulated cholesterol synthesis-SREBP2 pathway and LPC-downregulated TGF-β pathway that we reported recently (Li et al., 2018b); second, the generation of circRNAs in LPC-activated HAECs may be facilitated by three novel molecular mechanisms including: (i) high flanking intron homologies, (ii) LPC-induced modulation of spliceosome components, and (iii) increased upstream chromatin long–range interactions; and third, circRNAs in LPC-activated HAECs may play homeostatic functions by acting on a few novel aspects, including: S (a) competition with related mRNAs for exon usage, (b) miRNA sponge, and (c) secondary open reading frame-generated short peptide disruption of protein-protein interaction. We acknowledge that circRNAs profiling data have brought a huge amount of new findings for detailed experimental verifications in the future. Nevertheless, these novel insights may lead to identification of new therapeutic targets for treating metabolic cardiovascular diseases, inflammations, and cancers.
FIGURE 7.
A new working model: Proatherogenic lipid, LPC, induces the upregulation of a group of circular RNAs in human aortic endothelial cells, which may be generated by three mechanisms and play homeostatic functions via three effector mechanisms.
Author Contributions
AL and YS carried out the data gathering, data analysis, and prepared the tables and figures. CD, YL, DY, YZ, XL, CJ, CY, WY, KM, XJ, JS, TR, WH, and HW aided with the analysis of the data. XY supervised the experimental design, data analysis, and manuscript writing. SP aided with the analysis of the data and drafting of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by NIH grant to XM and HW.
References
- Abbas Q., Raza S. M., Biyabani A. A., Jaffar M. A. (2016). A review of computational methods for finding non-coding RNA genes. Genes 7:E113. 10.3390/genes7120113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuña L. I. G., Kornblihtt A. R. (2014). Long range chromatin organization. Transcription 5:e28726. 10.4161/trns.28726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Oufella H., Salomon B. L., Potteaux S., Robertson A.-K. L., Gourdy P., Zoll J., et al. (2006). Natural regulatory T cells control the development of atherosclerosis in mice. Nat. Med. 12 178–180. 10.1038/nm1343 [DOI] [PubMed] [Google Scholar]
- Albury T. M., Pandey V., Gitto S. B., Dominguez L., Spinel L. P., Talarchek J., et al. (2015). Constitutively active Akt1 cooperates with KRas(G12D) to accelerate in vivo pancreatic tumor onset and progression. Neoplasia 17 175–182. 10.1016/j.neo.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadou E., Jacob L. S., Slack F. J. (2018). Non-coding RNA networks in cancer. Nat. Rev. Cancer 18 5–18. 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC. A Quality Control Tool for High Throughput Sequence Data. Babraham: Babraham Institute. [Google Scholar]
- Bald T., Quast T., Landsberg J., Rogava M., Glodde N., Lopez-Ramos D., et al. (2014). Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature 507 109–113. 10.1038/nature13111 [DOI] [PubMed] [Google Scholar]
- Barbier E., Tapocik J. D., Juergens N., Pitcairn C., Borich A., Schank J. R., et al. (2015). DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J. Neurosci. 35 6153–6164. 10.1523/JNEUROSCI.4571-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S. P., Salzman J. (2016). Circular RNAs: analysis, expression and potential functions. Development 143 1838–1847. 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadnia N., Golas M. M., Hartmuth K., Sander B., Kastner B., Deckert J., et al. (2007). Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26 1737–1748. 10.1038/sj.emboj.7601631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008). Isolation of an active step I spliceosome and composition of its RNP core. Nature 452 846–850. 10.1038/nature06842 [DOI] [PubMed] [Google Scholar]
- Braun D. A., Fribourg M., Sealfon S. C. (2013). Cytokine response is determined by duration of receptor and signal transducers and activators of transcription 3 (STAT3) activation. J. Biol. Chem. 288 2986–2993. 10.1074/jbc.M112.386573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Liu X., Lu N., Xiao M., Li Z. (2016). Prognostic value of MicroRNA-196a in Asian cancer patients: a meta-analysis. Clin. Lab. 62 2257–2265. 10.7754/Clin.Lab.2016.160425 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. (2017). Bioconductor org.Hs.eg.db. R package version 3.6.0. [Google Scholar]
- Carr G., Barrese V., Stott J. B., Povstyan O. V., Jepps T. A., Figueiredo H. B., et al. (2016). MicroRNA-153 targeting of KCNQ4 contributes to vascular dysfunction in hypertension. Cardiovasc. Res. 112 581–589. 10.1093/cvr/cvw177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Steitz J. A. (2014). The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157 77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., et al. (2017). Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 388 208–219. 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Chen X., Han P., Zhou T., Guo X., Song X., Li Y. (2016). circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 6:34985. 10.1038/srep34985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combadière C., Potteaux S., Rodero M., Simon T., Pezard A., Esposito B., et al. (2008). Combined inhibition of CCL2, CX3CR1, and CCR5 abrogates Ly6Chi and Ly6Clo monocytosis and almost abolishes atherosclerosis in hypercholesterolemic mice. Circulation 117 1649–1657. 10.1161/CIRCULATIONAHA.107.745091 [DOI] [PubMed] [Google Scholar]
- Conover W. (ed.). (1999). “Statistics of the Kolmogorov-Smirnov type,” in Practical Nonparametric Statistics, (New York, NY: John Wiley & Sons; ), 428–473. [Google Scholar]
- Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., et al. (2006). Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26 5528–5543. 10.1128/MCB.00582-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. (2002). Capturing chromosome conformation. Science 295 1306–1311. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Deligianni C., Spilianakis C. G. (2012). Long-range genomic interactions epigenetically regulate the expression of a cytokine receptor. EMBO Rep. 13 819–826. 10.1038/embor.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Richmond T. A., Arnaout R. A., Selzer R. R., Lee W. L., Honan T. A., et al. (2006). Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16 1299–1309. 10.1101/gr.5571506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C. E., Michael F. A. (1991). On distribution-free multiple comparisons in the one-way analysis of variance. Commun. Stat. Theory Methods 20 127–139. 10.1080/03610929108830487 [DOI] [Google Scholar]
- Du F., Zhou J., Ren Gong X. H., Pansuria M., Virtue A., Li X., et al. (2012). Endothelial progenitor cells in atherosclerosis. Front. Biosci. 17 2327–2349. 10.2741/4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudekula D. B., Panda A. C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. (2016). CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13 34–42. 10.1080/15476286.2015.1128065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C. J. A., Mohamad S. M. B., Young D. F., Skelton A. J., Leahy T. R., Munday D. C., et al. (2015). Human IFNAR2 deficiency: lessons for antiviral immunity. Sci. Transl. Med. 7:307ra154. 10.1126/scitranslmed.aac4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Zhang D., Cheng Z., Yan C., Jiang X., Kruger W. D., et al. (2014). Hyperhomocysteinemia potentiates hyperglycemia-induced inflammatory monocyte differentiation and atherosclerosis. Diabetes 63 4275–4290. 10.2337/db14-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer L. M., Monroy A. M., Lopez-Pastrana J., Nanayakkara G., Cueto R., Li Y.-F., et al. (2016). Caspase-1 plays a critical role in accelerating chronic kidney disease-promoted neointimal hyperplasia in the carotid artery. J. Cardiovasc. Transl. Res. 9 135–144. 10.1007/s12265-016-9683-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang J., Zhao F. (2015). CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 16:4. 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimbrone M. A., García-Cardeña G. (2016). Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 118 620–636. 10.1161/CIRCRESAHA.115.306301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C. P. C., Salgado-Somoza A., Creemers E. E., Dieterich C., Lustrek M., Devaux Y., et al. (2018). Circular RNAs in the cardiovascular system. Noncoding RNA Res. 3 1–11. 10.1016/j.ncrna.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Venø M. T., Damgaard C. K., Kjems J. (2016). Comparison of circular RNA prediction tools. Nucleic Acids Res. 44:e58. 10.1093/nar/gkv1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H., Meirelles G. V., da Silva F. R., Telles G. P., Minghim R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16 169. 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt L. M., Stahringer A., Sass K., Pichler G., Kulak N. A., Wilfert W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M., Wolfe D. A. (1999). Nonparametric Statistical Methods. Accra: Akrong Publications Limited. [Google Scholar]
- Huang F. T., Chen W. Y., Gu Z. Q., Zhuang Y. Y., Li C. Q., Wang L. Y., et al. (2017). The novel long intergenic noncoding RNA UCC promotes colorectal cancer progression by sponging miR-143. Cell Death Dis. 8:e2778. 10.1038/cddis.2017.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chen Y., Xiao J., Huang Z., He L., Xu D., et al. (2018). Identification of differentially expressed circular RNAs during TGF-ss1-induced endothelial-to-mesenchymal transition in rat coronary artery endothelial cells. Anatol. J. Cardiol. 19 192–197. 10.14744/AnatolJCardiol.2018.95142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A., Memczak S., Wyler E., Torti F., Porath H. T., Orejuela M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10 170–177. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- Jeck W. R., Sharpless N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. H., Choi D., Chun Y.-J., Noh M. (2014). Keratinocyte-derived IL-24 plays a role in the positive feedback regulation of epidermal inflammation in response to environmental and endogenous toxic stressors. Toxicol. Appl. Pharmacol. 280 199–206. 10.1016/j.taap.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Johnson C., Drummer D. I. V., Virtue A., Gao T., Wu S., Hernandez M., et al. (2018). Increased expression of resistin in MicroRNA-155-deficient white adipose tissues may be a possible driver of metabolically healthy obesity transition to classical obesity. Front. Physiol. 9:1297. 10.3389/fphys.2018.01297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. C., Marron J. S., Sheather S. J. (1996). A brief survey of bandwidth selection for density estimation. J. Am. Stat. Assoc. 91 401–407. 10.1080/01621459.1996.10476701 [DOI] [Google Scholar]
- Kharma B., Baba T., Matsumura N., Kang H. S., Hamanishi J., Murakami R., et al. (2014). STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 74 6519–6530. 10.1158/0008-5472.CAN-14-0847 [DOI] [PubMed] [Google Scholar]
- Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., Williams E., et al. (2015). ArrayExpress update—simplifying data submissions. Nucleic Acids Res. 43 D1113–D1116. 10.1093/nar/gku1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987). An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15 8125–8148. 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1996). Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome 7 563–574. 10.1007/s003359900171 [DOI] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Tugendreich S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30 523–530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E., Parker R. (2014). Circular RNAs: diversity of form and function. RNA 20 1829–1842. 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66 22–37.e9. 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann E. (1963). Nonparametric confidence intervals for a shift parameter. Ann. Math. Stat. 34 1507–1512. 10.1214/aoms/1177703882 17133379 [DOI] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Fang P., Yang W. Y., Wang H., Yang X. (2017). IL-35, as a newly proposed homeostasis-associated molecular pattern, plays three major functions including anti-inflammatory initiator, effector, and blocker in cardiovascular diseases. Cytokine 10.1016/j.cyto.2017.06.003 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Shao Y., Sha X., Fang P., Kuo Y. M., Andrews A. J., et al. (2018a). IL-35 (Interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of H3K14 (histone 3 lysine 14). Arteriosc. Thromb. Vasc. Biol. 38 599–609. 10.1161/ATVBAHA.117.310626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang L., Fang P., Sun Y., Jiang X., Wang H., et al. (2018b). Lysophospholipids induce innate immune transdifferentiation of endothelial cells, resulting in prolonged endothelial activation. J. Biol. Chem. 293 11033–11045. 10.1074/jbc.RA118.002752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-F., Huang X., Li X., Gong R., Yin Y., Nelson J., et al. (2016a). Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Front. Biosci. 21 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-F., Li R.-S., Samuel S. B., Cueto R., Li X.-Y., Wang H., et al. (2016b). Lysophospholipids and their G protein-coupled receptors in atherosclerosis. Front. Biosci. 21 70–88. 10.2741/4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Fang P., Li Y., Kuo Y.-M., Andrews A. J., Nanayakkara G., et al. (2016c). Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activationhighlights. Arteriosc. Thromb. Vasc. Biol. 36 1090–1100. 10.1161/ATVBAHA.115.306964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Tatomer D. C., Luo Z., Wu H., Yang L., Chen L.-L., et al. (2017). The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell 68 940–954.e3. 10.1016/j.molcel.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang J., Khanabdali R., Kalionis B., Tai X., Xia S. (2017). Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 14 1715–1721. 10.1080/15476286.2017.1367886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P. G., Glazar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., et al. (2017). A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 95 1179–1189. 10.1007/s00109-017-1582-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J., Virtue A., Maley E., Tran T., Yin Y., Meng S., et al. (2012). MicroRNAs and other mechanisms regulate interleukin-17 cytokines and receptors. Front. Biosci. 4 1478–1495. 10.2741/e474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S., Makunin I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15 R17–R29. 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Monroy M. A., Fang J., Li S., Ferrer L., Birkenbach M. P., Lee I. J., et al. (2015). Chronic kidney disease alters vascular smooth muscle cell phenotype. Front. Biosci. 20 784–795. 10.2741/4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori L. E., Sobreira D. R., Sakabe N. J., Aneas I., Joslin A. C., Hansen G. T., et al. (2018). A promoter interaction map for cardiovascular disease genetics. eLife 7:e35788. 10.7554/eLife.35788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D., Inkeles M. S., Liu P. T., Realegeno S., Teles R. M. B., Vaidya P., et al. (2014). IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci. Transl. Med. 6:250ra114. 10.1126/scitranslmed.3009546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B., Yang F., Huston D. P., Yan Y., Yang Y., Xiong Z., et al. (2004). Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J. Allergy Clin. Immunol. 114 1463–1470. 10.1016/j.jaci.2004.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti N. R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., et al. (2017). Translation of CircRNAs. Mol. Cell 66 9–21.e7. 10.1016/j.molcel.2017.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo O. E., Castellano L., Munro C. E., Hu Y., Mauri F., Krell J., et al. (2016). miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep. 17 570–584. 10.15252/embr.201540970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Choi Y., Song M. J., Park K., Lee J. J., Kim H. P. (2016). Dynamic long-range chromatin interaction controls expression of IL-21 in CD4(+ T cells. J. Immunol. 196 4378–4389. 10.4049/jimmunol.1500636 [DOI] [PubMed] [Google Scholar]
- Pauli A., Valen E., Schier A. F. (2015). Identifying (non-)coding RNAs and small peptides: challenges and opportunities. Bioessays 37 103–112. 10.1002/bies.201400103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic S., Muller S. (2015). RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 43 2454–2465. 10.1093/nar/gkv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M. E., Thirdborough S. M., Ung C. Y., Elliott T., Healy E., Freeman T. C., et al. (2014). Distinct molecular signature of human skin Langerhans cells denotes critical differences in cutaneous dendritic cell immune regulation. J. Investig. Dermatol. 134 695–703. 10.1038/jid.2013.375 [DOI] [PubMed] [Google Scholar]
- Polak M. E., Ung C. Y., Masapust J., Freeman T. C., Ardern-Jones M. R. (2017). Petri net computational modelling of Langerhans cell interferon regulatory factor network predicts their role in T cell activation. Sci. Rep. 7:668. 10.1038/s41598-017-00651-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza N., Russo A. (2013). Biogenesis, evolution and functional targets of microRNA-125a. Mol. Genet. Genomics 288 381–389. 10.1007/s00438-013-0757-5 [DOI] [PubMed] [Google Scholar]
- Ribeiro J., Sousa H. (2014). MicroRNAs as biomarkers of cervical cancer development: a literature review on miR-125b and miR-34a. Mol. Biol. Rep. 41 1525–1531. 10.1007/s11033-013-2998-0 [DOI] [PubMed] [Google Scholar]
- Schliebs R., Rossner S., Bigl V. (1996). Immunolesion by 192IgG-saporin of rat basal forebrain cholinergic system: a useful tool to produce cortical cholinergic dysfunction. Prog. Brain Res. 1996 253–264. 10.1016/S0079-6123(08)62109-3 [DOI] [PubMed] [Google Scholar]
- Shalabh (2011). Nonparametric Statistical Inference, 5th Edn New York, NY: JSTOR. [Google Scholar]
- Shang F. F., Luo S., Liang X., Xia Y. (2018). Alterations of circular RNAs in hyperglycemic human endothelial cells. Biochem. Biophys. Res. Commun. 499 551–555. 10.1016/j.bbrc.2018.03.187 [DOI] [PubMed] [Google Scholar]
- Shao W., Espenshade P. J. (2012). Expanding roles for SREBP in metabolism. Cell Metab. 16 414–419. 10.1016/j.cmet.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Cheng Z., Li X., Chernaya V., Wang H., Yang X.-F. (2014). Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction-a novel mechanism for maintaining vascular function. J. Hematol. Oncol. 7:80. 10.1186/s13045-014-0080-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Nanayakkara G., Cheng J., Cueto R., Yang W. Y., Park J.-Y., et al. (2017). Lysophospholipids and their receptors serve as conditional DAMPs and DAMP receptors in tissue oxidative and inflammatory injury. Antioxid. Redox Signal. 10.1089/ars.2017.7069 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Guo X., Wang W. (2017). Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 591 213–220. 10.1002/1873-3468.12500 [DOI] [PubMed] [Google Scholar]
- Silverman B. W. (2018). Density Estimation for Statistics and Data Analysis. Abingdon: Routledge; 10.1201/9781315140919 [DOI] [Google Scholar]
- Splinter E., de Wit E., Nora E. P., Klous P., van de Werken H. J., Zhu Y., et al. (2011). The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25 1371–1383. 10.1101/gad.633311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R., Kolovos P., Brouwer R., Zuin J., van den Heuvel A., Kockx C., et al. (2013). Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat. Protoc. 8 509–524. 10.1038/nprot.2013.018 [DOI] [PubMed] [Google Scholar]
- Szabo L., Salzman J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17 679–692. 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., García-Cardeña G., Owens G. K. (2015). Recent insights into the cellular biology of atherosclerosis. J. Cell Biol. 209 13–22. 10.1083/jcb.201412052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Xin X., Coffey F. J., Wiest D. L., Dong L. Q., Testa J. R. (2015). Appl1 and Appl2 are expendable for mouse development but are essential for HGF-induced akt activation and migration in mouse embryonic fibroblasts. J. Cell Physiol. 231 1142–1150. 10.1002/jcp.25211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles R. M. B., Graeber T. G., Krutzik S. R., Montoya D., Schenk M., Lee D. J., et al. (2013). Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 339 1448–1453. 10.1126/science.1233665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L., He B., Wang J., Tan K. (2015). 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics 31 2560–2564. 10.1093/bioinformatics/btv158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D. (2015). How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 15 692–704. 10.1038/nri3908 [DOI] [PubMed] [Google Scholar]
- Virtue A., Johnson C., Lopez-Pastrana J., Shao Y., Fu H., Li X., et al. (2017). MicroRNA-155 deficiency leads to decreased atherosclerosis, increased white adipose tissue obesity, and non-alcoholic fatty liver disease: a novel mouse model of obesity paradox. J. Biol. Chem. 292 1267–1287. 10.1074/jbc.M116.739839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue A., Mai J., Yin Y., Meng S., Tran T., Jiang X., et al. (2011). Structural evidence of anti-atherogenic microRNAs. Front. Biosci. 16 3133–3145. 10.2741/3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue A., Wang H., Yang X. F. (2012). MicroRNAs and toll-like receptor/interleukin-1 receptor signaling. J. Hematol. Oncol. 5:66. 10.1186/1756-8722-5-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. C., Will C. L., Lührmann R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136 701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wang H., Naghavi M., Allen C., Barber R. M., Bhutta Z. A., Carter A., et al. (2016). Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388 1459–1544. 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li Y.-F., Nanayakkara G., Shao Y., Liang B., Cole L., et al. (2016). Lysophospholipid receptors, as novel conditional danger receptors and homeostatic receptors modulate inflammation—novel paradigm and therapeutic potential. J. Cardiovasc. Transl. Res. 9 343–359. 10.1007/s12265-016-9700-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wei Y., Yan Y., Wang H., Yang J., Zheng Z., et al. (2018). CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol. Rep. 39 951–966. 10.3892/or.2017.6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werfel S., Nothjunge S., Schwarzmayr T., Strom T. M., Meitinger T., Engelhardt S. (2016). Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 98 103–107. 10.1016/j.yjmcc.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Wilusz J. E. (2018). A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA 9:e1478. 10.1002/wrna.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.-Y., Hsieh C.-C., Hong J.-J., Chen C.-Y., Tsai Y.-S. (2009). IRSS: a web-based tool for automatic layout and analysis of IRES secondary structure prediction and searching system in silico. BMC Bioinformatics 10:160. 10.1186/1471-2105-10-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Liu E., Yan Y., Silver R. T., Yang F., Chen I. H., et al. (2006a). An unconventional antigen translated by a novel internal ribosome entry site elicits antitumor humoral immune reactions. J. Immunol. 177 4907–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Shaibani A., Li Y. P., Yan Y., Zhang S., Yang Y., et al. (2006b). Alternative splicing factor ASF/SF2 is down regulated in inflamed muscle. J. Clin. Pathol. 59 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Yang W. Y., Nanayakkara G. K., Shao Y., Yang F., Hu W., et al. (2018). gaTa3, hDac6, and Bcl6 regulate FOXP3+ Treg Plasticity and Determine Treg conversion into either novel antigen-Presenting cell-like Treg or Th1-Treg. Front. Immunol. 9:45. 10.3389/fimmu.2018.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Xu H., Zhang J., Xu L., Zhang Y., Zhang L., et al. (2017). Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget 8 95704–95718. 10.18632/oncotarget.21183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Phan L., Yang F., Talpaz M., Yang Y., Xiong Z., et al. (2004). A novel mechanism of alternative promoter and splicing regulates the epitope generation of tumor antigen CML66-L. J. Immunol. 172 651–660. 10.4049/jimmunol.172.1.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Xiong Z., Song J., Yin Y., Cowan A., Wang H., et al. (2008). Expression Of TCTP antisense in CD4+ CD25 high regulatory T cells weakens the cell survival and aggravates vascular inflammation. Am. Heart. Assoc. 203 401–408. 10.1016/j.atherosclerosis.2008.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Chen I. H., Xiong Z., Yan Y., Wang H., Yang X. F. (2006). Model of stimulation-responsive splicing and strategies in identification of immunogenic isoforms of tumor antigens and autoantigens. Clin. Immunol. 121 121–133. 10.1016/j.clim.2006.06.007 [DOI] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang F., Xiong Z., Yan Y., Wang X., Nishino M., et al. (2005). An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 24 4778–4788. 10.1038/sj.onc.1208666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Li X., Sha X., Xi H., Li Y.-F., Shao Y., et al. (2015). Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arteriosc. Thromb. Vasc. Biol. 35 804–816. 10.1161/ATVBAHA.115.305282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiontz C. (2013). The Data Analysis for This Paper was Generated Using the Real Statistics Resource Pack Software (Release 5.4). Available at: www.real-statistics.com [Google Scholar]
- Zeng X., Lin W., Guo M., Zou Q. (2017). A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 13:e1005420. 10.1371/journal.pcbi.1005420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang G., Ding C., Liu P., Wang R., Ding W., et al. (2017). Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 8 61687–61697. 10.18632/oncotarget.18671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-O., Dong R., Zhang Y., Zhang J.-L., Luo Z., Zhang J., et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26 1277–1287. 10.1101/gr.202895.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Deng Y., Liu Y., Chen X., Yang G., Mu Y., et al. (2013). MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol. Biol. Rep. 40 2789–2798. 10.1007/s11033-012-2278-4 [DOI] [PubMed] [Google Scholar]