A maize RNA splicing factor promotes normal cell differentiation and stops excessive cell proliferation in the kernel via the removal of minor introns during gene expression.
Abstract
The last eukaryotic common ancestor had two classes of introns that are still found in most eukaryotic lineages. Common U2-type and rare U12-type introns are spliced by the major and minor spliceosomes, respectively. Relatively few splicing factors have been shown to be specific to the minor spliceosome. We found that the maize (Zea mays) RNA binding motif protein 48 (RBM48) is a U12 splicing factor that functions to promote cell differentiation and repress cell proliferation. RBM48 is coselected with the U12 splicing factor, zinc finger CCCH-type, RNA binding motif, and Ser/Arg rich 2/Rough endosperm 3 (RGH3). Protein-protein interactions between RBM48, RGH3, and U2 Auxiliary Factor (U2AF) subunits suggest major and minor spliceosome factors required for intron recognition form complexes with RBM48. Human RBM48 interacts with armadillo repeat containing 7 (ARMC7). Maize RBM48 and ARMC7 have a conserved protein-protein interaction. These data predict that RBM48 is likely to function in U12 splicing throughout eukaryotes and that U12 splicing promotes endosperm cell differentiation in maize.
INTRODUCTION
Eukaryotic protein-coding genes generally are interrupted by introns, and splicing of precursor messenger RNA (pre-mRNA) is fundamental for protein expression. It has been proposed that eukaryotic introns originate from bacterial, self-splicing Group II introns (Burge et al., 1998; Roy and Irimia, 2009). However, eukaryotic introns are not self-splicing, and dynamic RNA-protein complexes, known as spliceosomes, direct accurate recognition of splice sites (Simpson and Filipowicz, 1996; Brown and Simpson, 1998; Lorković et al., 2000; Rappsilber et al., 2002; Ru et al., 2008). The last eukaryotic common ancestor had two classes of introns that were spliced by different spliceosome complexes (Irimia and Roy, 2014). The vast majority of introns, termed U2-type introns, are spliced by the major spliceosome (Lee and Rio, 2015). There are also rare, U12-type introns, which are spliced by the minor spliceosome (Will and Lührmann, 2005). U12-type introns have a higher degree of sequence conservation at the 5′ splice site and branch point sequence relative to U2-type introns (Turunen et al., 2013).
The minor spliceosome is at a low concentration in the cell, and splicing of U12-type introns is significantly slower than U2-type introns both in vitro and in vivo (Montzka and Steitz, 1988; Tarn and Steitz, 1995; Patel et al., 2002). Consequently, splicing of U12-type introns can limit the abundance of mature transcripts from minor intron-containing genes (MIGs; Patel et al., 2002; König et al., 2007; Younis et al., 2013 ; Niemelä and Frilander, 2014; Niemelä et al., 2014). In human cells, U12 splicing efficiency is responsive to a stress-activated signal transduction pathway (Younis et al., 2013). These data suggest that U12 splicing efficiency could be a deeply conserved regulatory mechanism. However, evolutionary evidence indicates the minor spliceosome is dispensable in many eukaryotes. The minor spliceosome has been lost in multiple independent lineages including model organisms such as nematodes (Caenorhabditis elegans), yeast (Saccharomyces cerevisiae), slime mold (Dictyostelium discoideum), and algae (Chlamydomonas reinhardtii; Dávila López et al., 2008). Among metazoan species with U12 splicing, the number of MIGs varies drastically, ranging from 758 in humans to only 19 in Drosophila melanogaster (Alioto, 2007). Loss of U12-type introns is the most common mechanism for reduction of MIGs within a genome (Burge et al., 1998).
Despite species variation for the presence and number of MIGs, U12 splicing appears to have roles in cell proliferation, cell differentiation, and development across divergent eukaryotes. A striking example is revealed by mutations of zinc finger CCCH-type, RNA binding motif and Ser/Arg rich 2 (ZRSR2) orthologs in human and maize (Zea mays). Somatic mutations in human ZRSR2 primarily affect U12 splicing and cause myelodysplastic syndrome, a disease that reduces differentiation of mature blood cell types (Madan et al., 2015). The rough endosperm3 (rgh3) mutation of the maize ZRSR2 ortholog has a conserved U12 splicing function and inhibits differentiation of endosperm cells (Fouquet et al., 2011; Gault et al., 2017). Other U12 splicing mutants in humans and mouse models demonstrate roles in hematopoiesis, muscle strength, as well as bone and neural development (Edery et al., 2011; He et al., 2011; Horiuchi et al., 2018). A mutation in the zebrafish (Danio rerio) RNA binding region (RNP1, RNA recognition motif [RRM]) containing 3 locus disrupts digestive organ development (Markmiller et al., 2014). Arabidopsis (Arabidopsis thaliana) knockdowns of minor spliceosome proteins show defects in leaf development and flowering time (Kim et al., 2010; Jung and Kang, 2014; Xu et al., 2016). The different pleiotropic phenotypes of U12 splicing factors have made it difficult to identify unifying developmental functions for MIGs.
Moreover, there are relatively few splicing factors that have been identified as specific to the minor spliceosome. Biochemical characterization of human small nuclear ribonucleoproteins (snRNPs) showed that most minor spliceosome proteins are shared with the major spliceosome with only seven novel proteins uniquely associated with the minor spliceosome: programmed cell death 7, RNA binding region (RNP1, RRM) containing 3, SNRNP25, SNRNP35, SNRNP48, zinc finger CCCH-type and RNA binding motif containing 1, and zinc finger matrin-type 5 (Schneider et al., 2002; Will et al., 2004). Genetic analysis of human ZRSR2, human FUS RNA binding protein, mouse survival motor neuron 1 (SMN1), and maize RGH3 indicates that these factors predominantly affect U12-type intron splicing even though there is substantial evidence for the proteins to participate in other RNA metabolic processes (Madanieh et al., 2015; Reber et al., 2016; Gault et al., 2017). Here we identify the maize RNA binding motif protein 48 (RBM48) as a minor spliceosome factor, which functions to promote cell differentiation and repress cell proliferation. RBM48 is conserved in organisms that retain the minor spliceosome, predicting that this protein will have a function in U12 splicing throughout eukaryotes. Protein-protein interactions and co-localization between RBM48, RGH3, and U2 Auxiliary Factor (U2AF) subunits suggest major and minor spliceosome factors may form complexes as part of recognizing introns.
RESULTS
Loss of RBM48 Causes a Rough Endosperm (rgh) Defective Kernel Phenotype
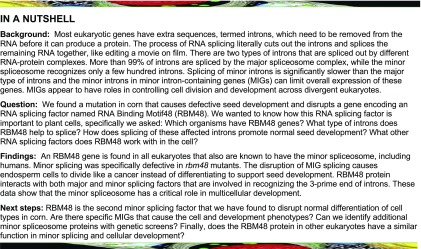
The rbm48-umu1 reference allele was identified from a collection of 144 UniformMu rgh mutants (McCarty et al., 2005; Fouquet et al., 2011). The rgh locus was mapped to the long arm of chromosome 4 with bulked segregant analysis (Liu et al., 2010) and compared with Robertson’s Mutator transposon flanking sequence tags from the rgh mutant line (Supplemental Figure 1). The rbm48-umu1 insertion in GRMZM2G163247 was the only novel insertion in this line that co-segregated with the rgh phenotype (Supplemental Figure 1). The rbm48-umu2 allele was obtained from the Maize Genetics Co-operative Stock Center (McCarty et al., 2013). Self-pollination of rbm48-umu2 heterozygotes also segregate for a rgh kernel phenotype (Supplemental Figure 1). Both alleles show similar defective kernel phenotypes and segregate at ratios consistent with a recessive mutant (Figure 1; Supplemental Table 1). The rbm48-umu1 allele transmits fully through both male and female gametes but does not give a seed phenotype when crossed to normal inbred lines (Supplemental Table 2). Crosses of the two alleles failed to complement the rgh mutant phenotype, indicating that rbm48 mutations disrupt maize seed development (Supplemental Figure 2).
Figure 1.
Mutant Alleles of rbm48.
(A) and (B) Segregating self-pollinated ears for rbm48-umu1 and rbm48-umu2 alleles. Arrowheads indicate rbm48 mutant kernels.
(C) to (L) Mature kernel phenotypes for rbm48-umu1 (C) to (G) and rbm48-umu2 (H) to (L). Abgerminal view of normal siblings (C) and (H), rbm48-umu1 (D), and rbm48-umu2 (I). Sagittal sections of normal siblings (E) and (J), rbm48-umu1 (F) and (G), and rbm48-umu2 (K) and (L). Scale bars = 0.5 cm. White arrowheads indicate shoot and root of the embryo. Orange arrowheads indicate vitreous endosperm.
(M) Schematic of the Rbm48 locus, GRMZM2G163247, and protein domain structure. Triangles indicate transposon insertions causing rbm48-umu1 and rbm48-umu2. Arrows indicate primers for RT-PCR in panel (N).
(N) RT-PCR analysis of Rbm48 and actin1 control in rbm48-umu1 and rbm48-umu2, and their normal siblings. T is a no template DNA negative control.
(O) Proportional Venn diagram showing the number of species in the NCBI Conserved Domain Database with RRM domains from RGH3 (RRM_U2AFBPL), RBM48 (RRM_RBM48), and U2AF1 (RRM_U2AF35 union with RRM_U2AF35B).
Mature rbm48 kernels have reduced endosperm size, and embryos typically fail to develop (Figures 1A to 1L). Consistent with these morphological defects, mutant kernel composition is affected with reduced oil, starch, and seed density (Supplemental Figure 2). A small fraction of rbm48 kernels have a less severe phenotype and develop a viable embryo that germinates. Mutant seedlings developed only one to two narrow leaves, stunted roots, and died around 20 d after sowing (Supplemental Figure 2). Thus, both rbm48 alleles are lethal mutations.
Reverse transcription PCR (RT-PCR) of normal Rbm48 complementary DNA (cDNA) from etiolated roots and shoots of W22, B73, and Mo17 inbred seedlings identified a common transcript isoform coding for a 219 amino acid protein (Figure 1M, Supplemental Figure 3). Alternatively spliced isoforms had premature termination codons that are predicted to be targets of nonsense mediated decay (Shaul, 2015). The predicted RBM48 protein has an N-terminal RRM specific to the RBM48 protein family and a 30 amino acid C-terminal RS-rich motif (Figure 1M; Supplemental Figure 3). This domain structure is common for SR proteins involved in pre-mRNA splicing (Graveley, 2000). We were not able to detect rbm48 transcripts in mutant seedlings for either allele, and we infer that the two alleles are likely null mutations (Figure 1N).
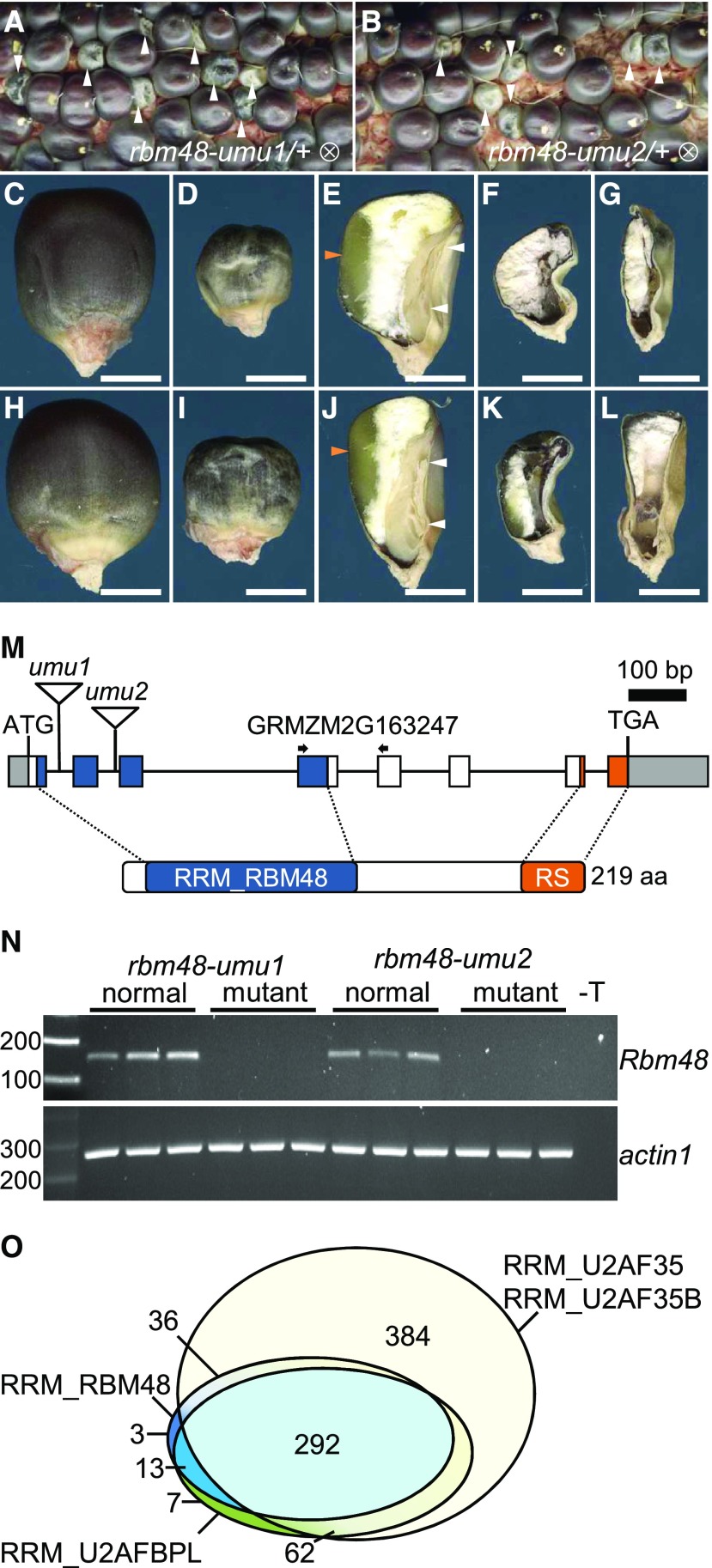
Based on the National Center for Biotechnology Information (NCBI) Conserved Domains Database (Marchler-Bauer et al., 2017), the RRM domain of RBM48 is found in 344 eukaryotic species (Figure 1O). The RRM domains of RBM48 and ZRSR2/RGH3 appear to be coselected, with 89% of species having an RBM48 domain also containing a RGH3/ZRSR2 RRM domain. By contrast, 50% of species with an RRM domain from the core U2 splicing factor, U2AF1, lack both ZRSR2 and RBM48 RRM domains. Like RGH3, the RBM48 RRM domain is not found in model organisms that have lost MIGs and the U12 splicing machinery. Phylogenetic analysis of 16 representative species shows the distribution of RBM48 orthologs across multiple eukaryotic kingdoms with RBM48 absent in clades lacking a U12 spliceosome including algae, nematodes, and slime molds (Figure 2). In Arabidopsis and Arabidopsis lyrata, a second copy of the RBM48 domain is fused to a predicted chloroplast-localized pentatricopeptide repeat protein (Supplemental Figure 3). This gene fusion is limited to the Arabidopsis genus. Interestingly, D. melanogaster has a divergent RBM48-like gene (CG34231) and a highly reduced number of MIGs. These phylogenetic data suggest a potential role for RBM48 in U12 splicing.
Figure 2.
RBM48 Is Missing in Some Eukaryotic Clades.
(A) Species tree including significant eukaryotic model organisms. Gray boxes indicate lineages that have lost U12-type introns. Drosophila melanogaster (fly) encodes a hypothetical gene with an atypical RBM48 RRM-like domain.
(B) Maximum likelihood tree of the RBM48_RRM domain. Bootstrap values ≥50 are reported in the corresponding nodes. The scale bar indicates the number of substitutions per site. Protein schematics show the RBM48 domain in blue. Protein sequences are from Zea mays (maize), Arabidopsis thaliana (Arabidopsis), Amborella trichopoda (Amborella), Ginkgo biloba (Ginkgo), Physcomitrella patens (Moss), Cyanophora paradoxa (Glaucophyta), Homo sapiens (Human), Mus musculus (Mouse), Gallus gallus (Chicken), Xenopus tropicalis (Frog), Danio rerio (Zebrafish), Rhizophagus irregularis DAOM 181602 (Fungi), and Phytophthora sojae (Oomycetes).
(C) Multiple sequence alignment of representative RBM48 domains. Asterisks indicate completely conserved sites. Ribonucleoprotein domains (RNP1, RNP2) of the predicted β1- and β3-seets in the RRM are indicated.
Aberrant Splicing of U12-Type Introns in rbm48
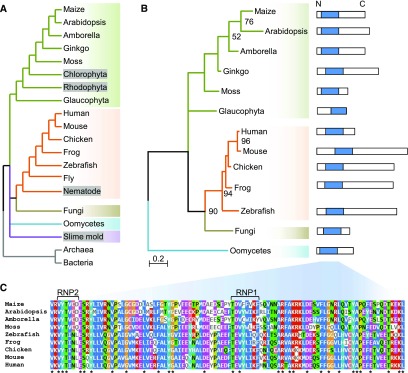
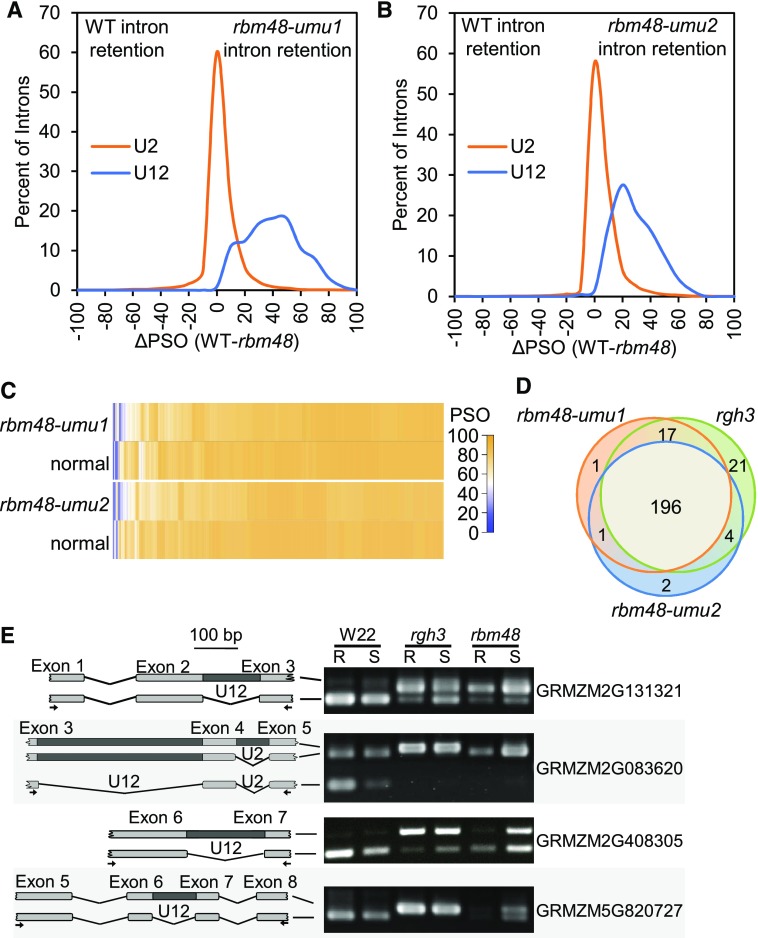
RNA splicing was assessed with messenger RNA sequencing (mRNA-seq), comparing mutant and normal sibling endosperm RNA from both rbm48 alleles. To quantify the extent of intron splicing defects, we calculated the percent spliced out (PSO) of individual introns using exon-exon junction and intron reads (Supplemental Data Set 1). PSO compares the density of exon-exon junction reads and intron reads for each intron (Figure 3A). A PSO of 100 indicates complete splicing of an intron, and a value of 0 indicates all expressed transcripts retain the intron. The difference in PSO values for individual introns between normal siblings and rbm48 mutants (ΔPSO) identifies introns that are differentially impacted. The major U2-type introns of the B73_v2 filtered gene set are largely unaffected with only 3 to 5% of introns in expressed genes having a ΔPSO > 20% (Figures 3B to 3E). By contrast, 65 and 53% of U12-type introns in rbm48-umu1 and rbm48-umu2, respectively, have a ΔPSO > 20%, suggesting that more than half of U12-type introns are retained in rbm48 mutants. Hierarchical clustering of U12-type introns based on PSO values for normal and rbm48 mutant alleles show highly overlapping effects on intron retention, except that the rbm48-umu1 RNA-seq experiment had a slightly stronger magnitude of defects (Figure 3F). Environmental variance between segregating ears or normal sibling contamination of the rbm48-umu2 mutant samples may account for differences in the magnitude of defects observed.
Figure 3.
Distribution of Intron Retention in rbm48 Mutants.
(A) Schematic of reads used to calculate PSO.
(B) and (C) Scatterplots of mutant and normal sibling PSO values for all introns with at least 10 exon-exon junction reads in rbm48-umu1 (B) and rbm48-umu2 (C) mRNA-seq experiments.
(D) and (E) Distribution of RNA splicing differences for U2-type and U12-type introns in rbm48-umu1 (D) and rbm48-umu2 (E) 16 to 18 DAP endosperm. Plots show the distribution of ΔPSO values for all introns with sufficient reads to calculate ΔPSO. Positive values have intron retention in mutant endosperm.
(F) Heat map of PSO values of all U12-type introns for both rbm48 mutant alleles and normal (N) sibling controls.
WT = the wild type.
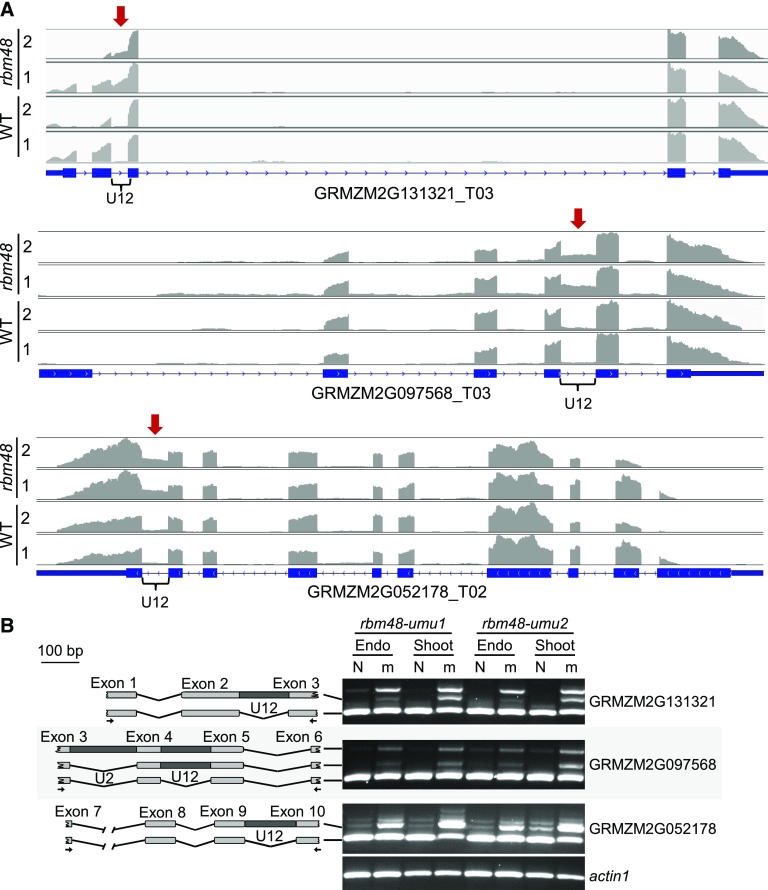
For example, the U12-type intron in GRMZM2G131321 has a ΔPSO of 65 and 50% for rbm48-umu1 and rbm48-umu2, respectively. There is a high density of U12-type intron reads in the mutant RNA-seq data (Figure 4A). The U12-type intron in GRMZM2G097568 had lower ΔPSO values, with a minimum of 23% for rbm48-umu2. No exon-exon junction reads were detected for the U12-type intron of GRMZM2G052178 in either mutant or normal samples, yet a higher density of intron reads are readily apparent in rbm48 mutants for both genes. RT-PCR assays spanning these U12-type introns show splicing defects with increased intron retention in both endosperm and shoot tissues (Figure 4B). In all cases, both rbm48 alleles appear to have similar relative ratios of splice variants amplified, indicating equivalent defects. RT-PCR assays confirmed eleven U12-type intron splicing defects (Figures 4B and 5E; Supplemental Figures 4 and 5 ).
Figure 4.
U12-Type Intron Retention in rbm48 Mutants.
(A) RNA-seq read depth of GRMZM2G131321, GRMZM2G097568, and GRMZM2G052178, homologs of human SPCS2, TRAPPC2, and BYSL, respectively. Each panel is the summed read depth of four biological replicate libraries of mutant and normal sibling (WT, wild type) endosperm tissues for rbm48-umu1 and rbm48-umu2 segregating ears. Red arrow and brace symbol indicate the U12-type intron with increased read depth in rbm48 mutants.
(B) RT-PCR of normal (N) and rbm48 mutant (m) RNA from endosperm and seedling shoot tissues. Schematics show amplified products with PCR primers indicated by arrows.
Figure 5.
Statistically Defined Splicing Defects in rbm48 Mutants Overlap with rgh3.
(A) and (B) Distribution of ΔPSO values for introns with significant (<0.05 FDR) Fisher’s Exact Test statistics for rbm48-umu1 (A) and rbm48-umu2 (B).
(C) Heat maps showing PSO values for U2-type introns with significant Fisher’s Exact Test statistics.
(D) Venn diagram of U12-type introns that are significantly retained in rbm48-umu1, rbm48-umu2, and rgh3 RNA-seq experiments. Introns that were tested in all three experiments were compared.
(E) RT-PCR of root (R) and shoot (S) tissues from W22 inbred, rgh3, and rbm48-umu1 seedlings. Schematics show amplified products with PCR primers indicated by arrows. Gene names are from the maize B73_v2 annotation.
WT = the wild type.
Fisher’s exact tests of intron and exon-exon junction read counts were used to identify statistically significant splicing differences for individual introns (Supplemental Data Set 1). Approximately 63% of U12-type introns tested had a significant retention in both alleles (FDR [false discovery rate] < 0.05). U2-type introns were largely unaffected, with only 6.7% of U2-type introns having significant differences in both alleles. The median ΔPSO in these significant U2-type introns is 3%, while mis-spliced U12-type introns show a median ΔPSO of 33% (Figures 5A to 5C).
To identify U2-type introns with large magnitude splicing defects, we filtered the 7535 U2-type and 201 U12-type significant introns for consistent splicing defects by requiring the ΔPSO values to be within twofold for both alleles and requiring at least one allele to have a |ΔPSO| ≥ 25%. More than 68% of significant U12-type introns (43% of all U12-type introns tested) and only 4.6% of significant U2-type introns (0.26% of all U2-type introns tested) met these criteria. These larger magnitude effects included 73 U2-type introns that were spliced with higher efficiency in rbm48 compared with normal and 279 predicted U2-type intron retention events. Twenty of the U2-type intron retention defects were in MIGs. These data indicate rbm48 mutants confer large magnitude effects on >40% of U12-type introns with rare (∼0.2%) U2-type splicing defects.
We also compared the two rbm48 alleles with prior mRNA-seq analysis of rgh3 (Gault et al., 2017). Of the 242 U12-type introns tested in all three mutant alleles, 196 (77%) were significantly affected in all experiments (Figure 5D). Only 24 (10%) of the tested U12-type introns were significant in only one of the rbm48 alleles, which is within the expected false positive limits of the FDR correction for multiple testing. Consequently, there is no statistical difference between the U12-type introns affected in either rbm48 allele. Direct RT-PCR comparisons of mutant seedling tissues confirmed similar U12 splicing defects in rgh3 and rbm48-umu1 (Figure 5E; Supplemental Figure 5). These analyses indicate that rgh3 and rbm48 have extensive overlap in U12 splicing defects.
rbm48 Disrupts Endosperm Cellular Development
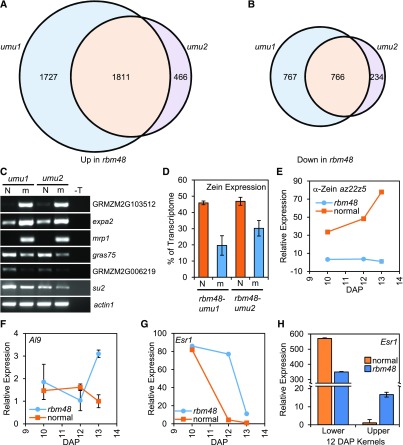
Differential gene expression analysis of the rbm48 RNA-seq data identified large-scale changes. Of 16,745 transcripts expressed, at least one transcript per million in one genotype, 5771 transcripts, were differentially expressed (FDR < 0.05, >twofold change) in either rbm48-umu1 or rbm48-umu2 (Figures 6A and 6B; Supplemental Data Set 2). There were 1811 up-regulated and 766 down-regulated transcripts in both alleles. A subset of these gene expression differences were confirmed with RT-PCR (Figure 6C). Gene Ontology (GO) term enrichment analysis identified nutrient reservoir activity as the most significantly enriched term among differentially expressed genes (Supplemental Table 3).
Figure 6.
Expression Defects in rbm48.
(A) and (B) Venn diagrams showing the overlap of up- (A) and down-regulated (B) DEGs in rbm48-umu1 and rbm48-umu2.
(C) RT-PCR of normal (N) and rbm48 mutant (m) endosperm RNA used for RNA-seq. GRMZM2G103512, expa2, and mrp1 were predicted to have increased levels in rbm48 mutants, while gras75, GRMZM2G006219, and su2 were predicted to show decreased expression in rbm48. The actin1 locus was used as a loading control. T is a no cDNA template negative control. Gene names are MaizeGDB locus identifiers.
(D) Proportion of total transcriptome represented by 39 zein transcripts based on endosperm RNA-seq. Error bars are sd of four biological replicates.
(E) to (G) RT-qPCR of total RNA extracted from whole kernels in rbm48 and normal siblings for α-Zein (E), Al9 (F), and Esr1 (G). Error bars are sd of three biological replicates.
(H) RT-qPCR of Esr1 expression in total RNA extracted from lower and upper half kernels. Error bars are sd of three biological replicates.
Nearly 80% of the nutrient reservoir activity genes affected are seed storage proteins, such as zeins. At this stage of endosperm development, zein expression accounts for 45% of the total transcriptome in the normal sibling endosperm and only 20 to 30% of the total transcriptome in rbm48 mutants (Figure 6D). We confirmed that α-zein expression was reduced in rbm48-umu1 using RT-qPCR in a developmental series of whole kernel RNA (Figure 6E). The rgh3 mutant also reduces and delays zein accumulation in the endosperm as part of an overall disruption of endosperm cell differentiation (Fouquet et al., 2011).
A focused analysis of endosperm cell-type marker genes revealed that cell-type specific genes had aberrant expression from 10 to 13 days after pollination (DAP). Cell-type markers for the aleurone (Al9), the embryo surrounding region (Esr1), and the basal endosperm transfer cell layer (Betl2, Tcrr1) had increased expression late in development (Figures 6F and 6G; Supplemental Figure 6).
Many of these whole kernel expression patterns were strikingly different from those observed from the lower half of kernels analyzed in the rgh3 mutant (Fouquet et al., 2011). For example, Esr1 is expressed at a basal level in the lower half of rgh3 mutants, while Esr1 is upregulated in whole rbm48 kernels. Extractions of the upper and lower half of 12 DAP kernels showed that this discrepancy is due to reduced expression of Esr1 in the lower half of the kernel, where Esr1 is normally expressed, and increased expression of Esr1 in the upper half of the kernel (Figure 6H). Quantitative expression analysis of the lower half of 12 DAP kernels also showed that, while Al9 is induced, Betl2 and Esr1 are reduced consistent with similar observations of rgh3 (Supplemental Figure 6). These results indicate that rbm48 endosperm cells fail to express cell-type specific markers in the correct domains of the kernel.
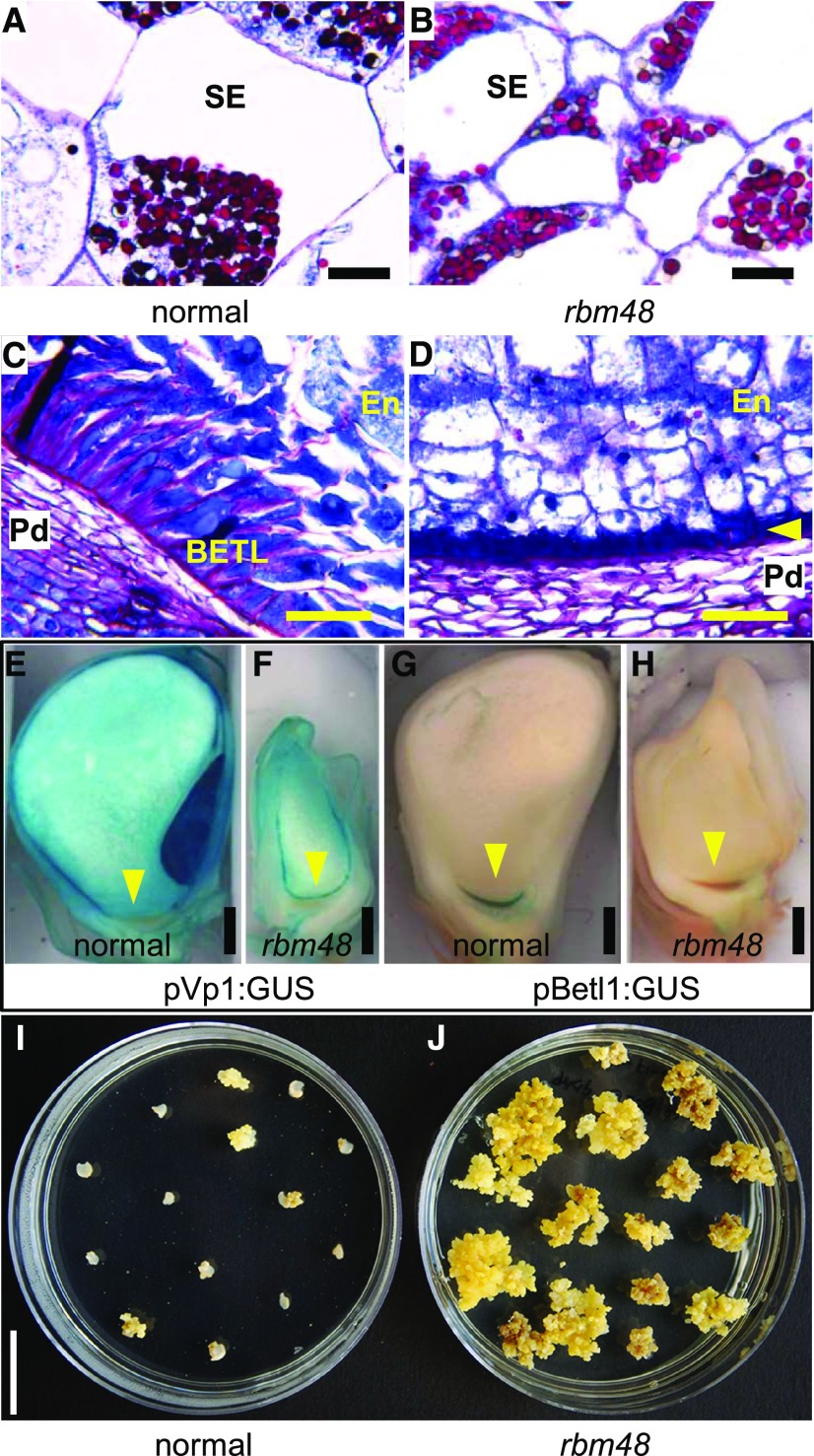
Cellular phenotypes of rbm48 endosperm are also consistent with cell differentiation defects. Transverse sections of 12 DAP kernels show that rbm48 starchy endosperm cells are smaller than normal siblings (Figures 7A and 7B). At this stage, the basal endosperm transfer layer (BETL) is differentiated in normal kernels, visible as elongated cells with intense Schiff’s staining due to secondary cell wall ingrowths (Figure 7C). The BETL in rbm48 is not developed into elongated cells (Figure 7D). Cell-type specific reporter lines indicate that the rbm48 BETL region differentiates into aleurone. A GUS reporter gene driven by the Viviparous1 promoter (pVP1:GUS) marks ABA responsive cells including the aleurone and embryo, while the Betl1 promoter (pBetl1:GUS) marks BETL cells. These reporter transgenes have aberrant expression in rbm48-umu1 mutants, with pVp1:GUS being expressed in the basal epidermal endosperm and pBetl1:GUS having low expression in the same region (Figures 7E to 7H; Supplemental Figure 7). These data support the model that epidermal endosperm cells express aleurone markers regardless of position in the endosperm.
Figure 7.
Endosperm Cell Differentiation Defects in rbm48.
(A) to (D) Transverse sections of 12 DAP normal sibling (A) to (C) and rbm48-umu1 (B) to (D) kernels stained with Schiff's reagent and aniline blue-black. Insoluble carbohydrates in cell walls and starch grains stain fuchsia; nucleoli, nuclei, and cytoplasm stain different intensities of blue. Scale bar = 50 µm. (A) and (B) Internal starchy endosperm cells (SE). (C) and (D) BETL region with inner endosperm (En) and maternal pedicel (Pd). Yellow arrowheads indicate small epidermal cells in the BETL region. Scale bar = 50 µm.
(E) to (H) Maize transgene reporter lines for Vp1 (pVp1:GUS) and Betl2 (pBetl2:GUS) in normal siblings (E) and (G) and rbm48-umu1 mutants (F) and (H) at 13 DAP. Blue stain indicates GUS expression. Yellow arrowhead indicates the BETL region. Scale bar = 1 mm.
(I) and (J) Tissue culture response of nine DAP endosperm from normal (I) sibling and rbm48-umu1 (J) seeds. Plates show growth response after 35 days of culture. Scale bar = 2 cm.
Mutant rgh3 endosperm retains the ability to proliferate in cell culture at late time-points in seed development (Fouquet et al., 2011). Endosperm tissue culture responses for rbm48-umu1 mutants and normal siblings were tested. At 6 DAP, normal and mutant endosperm grow equally well in the culture assay, but at 9 to 18 DAP, only rbm48 endosperms are able to grow as a callus in the culture assay (Figures 7I and 7J; Supplemental Figure 7). Thus, rbm48 mutants have prolonged endosperm cell proliferation and delayed or aberrant cell differentiation, similar to rgh3 mutants.
RBM48 Interacts with U12 and U2 Splicing Factors
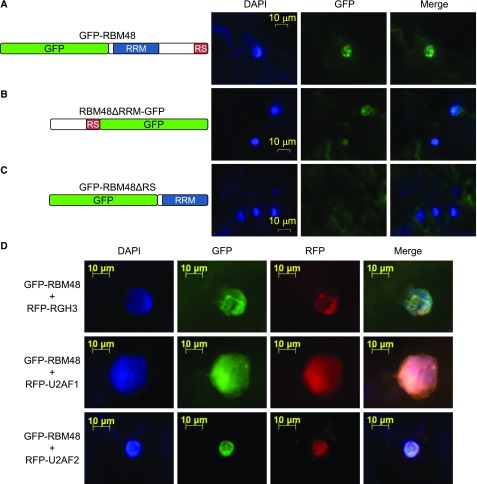
The evolutionary coselection of the RGH3 UHM and RBM48 RRM domains, as well as the U12 splicing and endosperm cell differentiation defects in rbm48 and rgh3 mutants, raises the possibility that RGH3 and RBM48 interact. We investigated the subcellular localization of RBM48 with GFP fusion proteins that were transiently expressed in Nicotiana benthamiana leaves. Full-length RBM48 is localized to the nucleus and is associated with the nuclear speckles (Figure 8A). Deletion of the RRM domain still localized the protein in nuclear speckles, albeit at a lower expression level, while deletion of the C terminus of RBM48 resulted in diffuse cytoplasmic signal (Figures 8B and 8C). These results are consistent with the localization of SR proteins in both plants and animals to sub-nuclear speckles via the RS domain (Cazalla et al., 2002; Tillemans et al., 2005, 2006; Mori et al., 2012; Saitoh et al., 2012).
Figure 8.
RBM48 Localizes to the Nucleus with U2AF and RGH3.
(A) to (C) Subcellular localization of RBM48 and RBM48 domain deletions. (A) N-terminal GFP fusion of the full length RBM48 protein. (B) C-terminal GFP fusion deleting the RBM48_RRM domain. (C) N-terminal GFP fusion deleting the C-terminal 109 amino acids including the RS domain.
(D) Transient co-expression of GFP-RBM48 with RFP-tagged RGH3, U2AF1, and U2AF2 in N. benthamiana leaves. DAPI staining of DNA marks nuclei in blue.
Co-expression of GFP-RBM48 with red fluorescent protein (RFP)-tagged RGH3 or the U2AF1 and U2AF2 subunits of U2AF resulted in extensive overlap of fluorescent signals (Figure 8D). Human U2AF2 interacts with U2AF1 and the RGH3 ortholog, ZRSR2 (Tronchère et al., 1997; Kielkopf et al., 2001). The co-localization of RBM48 with these factors suggests that RBM48 may interact in a larger complex. Human U2AF is involved in the selection of 3′ splice sites of U2-type introns, while ZRSR2 binds the 3′ splice sites of U12-type introns (Tronchère et al., 1997; Guth et al., 2001; Shen et al., 2010).
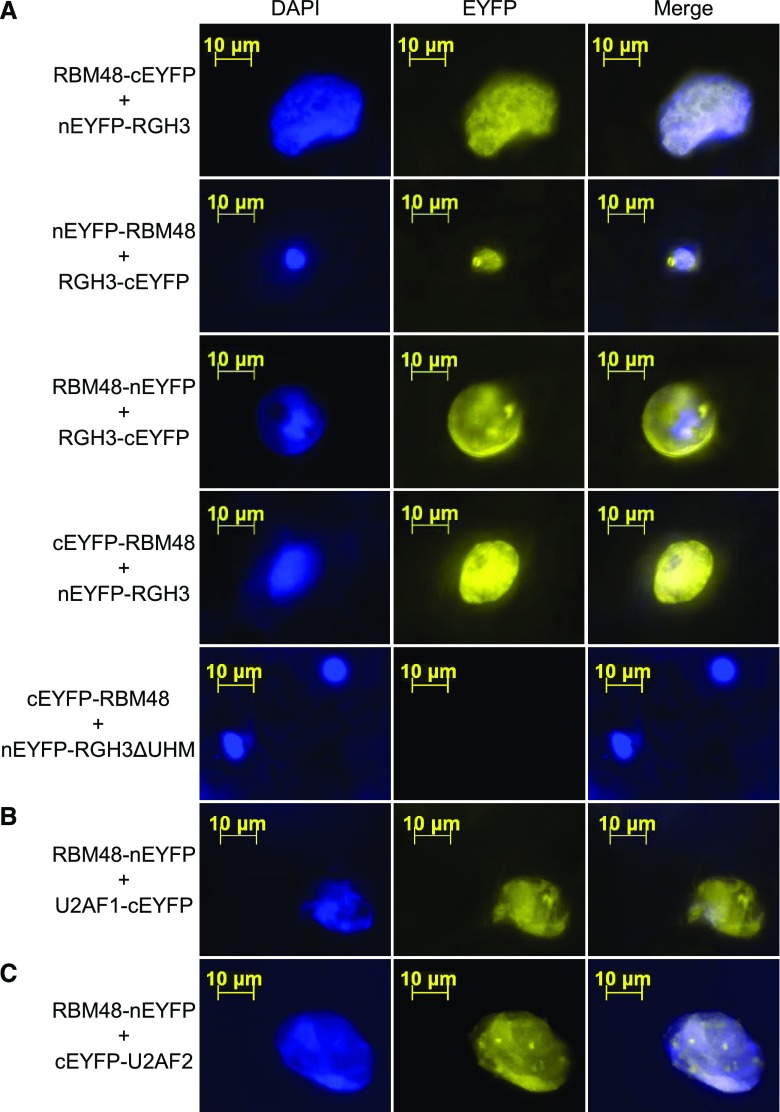
Bimolecular fluorescence complementation (BiFC) assays of RBM48 with RGH3, U2AF1, and U2AF2 support the model of a larger protein complex of these factors. BiFC tests for fluorescence complementation of split, nonfluorescent fragments of yellow fluorescent protein (YFP) that are brought in close vicinity by interacting fusion proteins (Citovsky et al., 2006; Kerppola, 2008). The N-terminal or C-terminal fragments of YFP were fused with RBM48 and transiently co-expressed in different pairwise combinations with N- or C-terminal YFP fusion constructs of RGH3, U2AF1, and U2AF2 in N. benthamiana leaves. All combinations of RBM48 with full-length RGH3, U2AF1, and U2AF2 resulted in YFP signal indicating these proteins are in close proximity in plant cells (Figure 9). An in-frame deletion of the RGH3 RRM domain (RGH3ΔUHM), also known as the U2AF homology motif (UHM), fails to complement YFP, indicating that the UHM domain is needed for RBM48 and RGH3 association (Figure 9A). When expressed with U2AF2 fusions, the RGH3ΔUHM construct results in BiFC signal, showing that the construct is capable of reporting close proximity of proteins in the nucleus (Gault et al., 2017).
Figure 9.
BiFC Interactions of RBM48 with RGH3, U2AF1, and U2AF2.
BiFC assays showing transient co-expression in N. benthamiana leaves of fusion proteins with N-terminal EYFP (nEYFP) or C-terminal EYFP (cEYFP). Yellow signal indicates reconstituted EYFP. DAPI staining of DNA marks nuclei in blue.
(A) Co-expression of four combinations of RBM48 and RGH3 N- and C-terminal fusion proteins as well as the RGH3ΔUHM in-frame domain deletion.
(B) Co-expression of RBM48 and U2AF1 fusions.
(C) Co-expression of RBM48 and U2AF2 fusions.
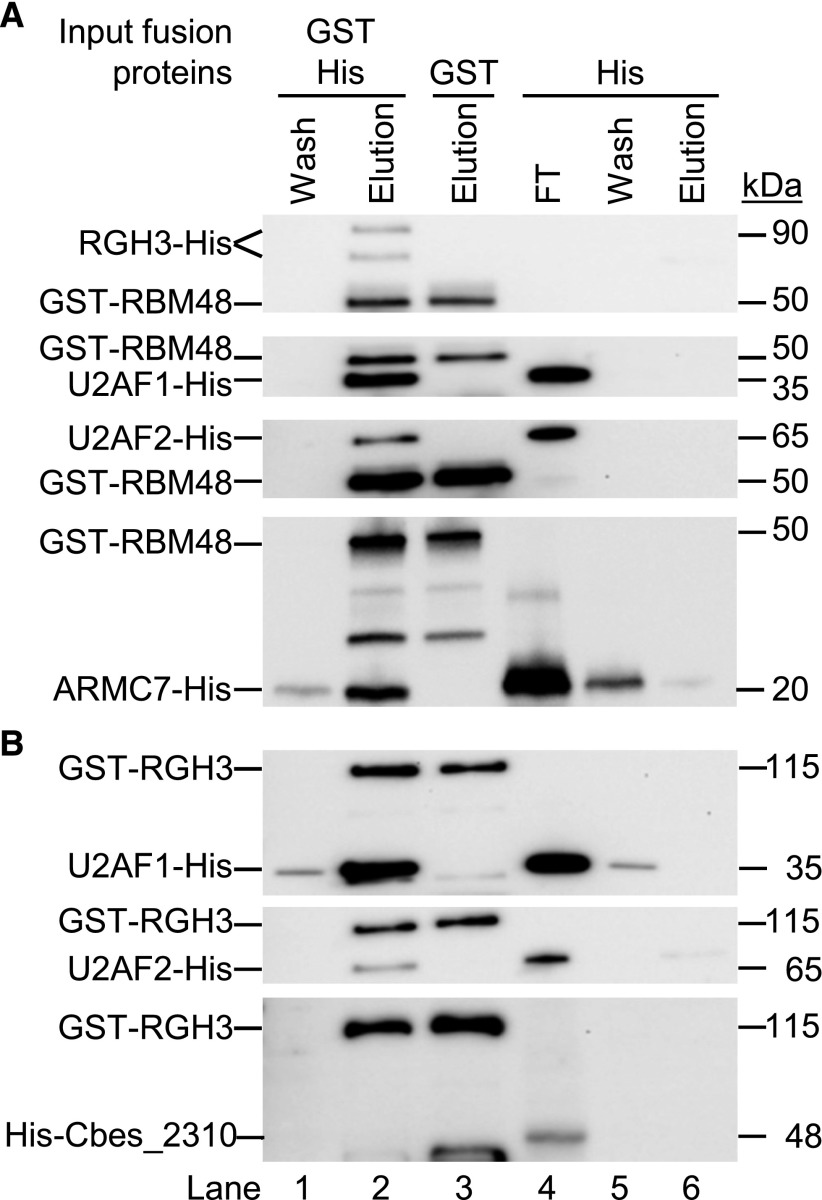
We then tested direct protein-protein interactions with in vitro pull-down assays. Recombinant glutathione S-transferase (GST)-His-RBM48 fusion protein was immobilized to glutathione magnetic beads and incubated with Escherichia coli lysate containing recombinant His-tagged RGH3, U2AF1, U2AF2, or Armadillo Repeat Containing 7 (ARMC7, GRMZM2G106137). Human RBM48 interacts with ARMC7 (Hart et al., 2015). In all cases, RBM48 can purify the His-tagged proteins from bacterial lysates (Figure 10A). Moreover, GST-His-RGH3 pulls down His-tagged U2AF1 and U2AF2, but not Cbes_2310, a protein originating from Caldicellulosiruptor bescii (Figure 10B). These results support conserved interactions in maize for RBM48 with ARMC7, and RGH3 with U2AF2. In addition, we found that RBM48 and RGH3 interact with each other as well as with U2AF1.
Figure 10.
Direct Protein-Protein Interactions with RBM48.
(A) Immunoblots of in vitro pull down assays with GST-His-tagged RBM48. GST-His-RBM48 immobilized on glutathione beads was incubated with bacterial lysate containing His-tagged RGH3, U2AF1, U2AF2, or ARMC7.
(B) Immunoblots of in vitro pull down assays with GST-His-RGH3 immobilized on glutathione beads and bacterial lysates containing His-tagged U2AF1, U2AF2, or Cbes_2310. Recombinant proteins are detected with αHis monoclonal antibodies. Lanes 1 to 2 show the wash and elution of the pull down assay with both recombinant proteins. Lane 3 is the elution of the GST-His-tagged protein alone. Lanes 4 to 6 show the level of binding of the His-tagged protein with unbound glutathione beads. FT is the flow through lysate after incubation with unbound beads.
DISCUSSION
RBM48 Is a U12 Splicing Factor
Genetic and molecular analysis of maize rbm48 revealed extensive defects in U12-type intron splicing, with more than 60% of MIGs significantly mis-spliced. Although RNA-seq analysis showed a greater magnitude of U12-type intron splicing defects in rbm48-umu1 (Figures 3 and 5), we did not find statistically significant different sets of U12-type introns affected in the two alleles of rbm48. Moreover, kernel and seedling phenotypes of the two alleles are similar at qualitative and quantitative levels (Figure 1; Supplemental Figure 2). Both alleles show a range of kernel phenotypes throughout development, and it is likely that environmental differences explain much of the difference in magnitude of U12 splicing defects observed for the two alleles in the RNA-seq experiment.
MIG splicing defects are highly overlapping in rbm48 and rgh3, with 90% of mis-spliced U12-type introns being affected in common between both mutant loci (Figure 5D). Mis-spliced U12-type introns in rgh3 show no differences in 5′ splice site, 3′ splice site, or branch point sequence motifs relative to all maize U12-type introns (Gault et al., 2017). Thus, no specific sequence motif that separates rbm48/rgh3-affected versus nonaffected U12-type introns has yet been identified. Both expression level and sequencing depth do limit the sensitivity of identifying mis-spliced introns. For example, the U12-type intron in GRMZM2G052178 is clearly mis-spliced in rbm48, but no exon-exon junction reads were detected for this splicing event (Figure 4). This intron could not be tested for statistical significance in rbm48, but it is significantly affected in rgh3 (Gault et al., 2017).
Defects in a small fraction of U2-type introns were also observed in rbm48 mutants, but these defects are predominantly of small magnitude, with >95% of significantly affected U2-type introns splicing at ±25% of normal levels. When filtered for larger magnitude effects, only 0.3% of U2-type introns have a significant splicing difference in rbm48, and U2-type introns in MIGs are ninefold enriched in this set. Altered splicing of neighboring U2-type introns in minor splicing factor mutants has been observed in multiple species (Madan et al., 2015; Horiuchi et al., 2018). These combined observations indicate that maize RBM48 is a minor splicing factor. No RNA processing role has been assigned yet to RBM48, and the human ortholog was not identified in biochemical purification of U12 spliceosomes (Schneider et al., 2002; Will et al., 2004). However, RBM48 is conserved among organisms that have a U12 spliceosome and has been coselected with ZRSR2/RGH3 throughout eukaryotic evolution. Based on this evolutionary conservation and our evidence that maize RBM48 is a U12 splicing factor, we predict that animal RBM48 proteins will also function in U12 splicing.
Significance of RBM48 Interactions with RGH3, U2AF, and ARMC7
A comparison of minor spliceosome mutants from multiple biological systems suggests that U12-type intron recognition defects have a greater impact on U2-type introns when compared with minor splicing factors required to complete the U12-dependent splicing reactions (Verma et al., 2018). For example, mutant alleles of human U4atac, which are expected to recognize U12-type introns but not complete splicing, appear to have exclusive U12 splicing defects. By contrast, a low frequency of U2-type intron splicing defects, similar to rbm48 mutants, have been observed for mutations in ZRSR2 homologs and RNPC3 (Argente et al., 2014; Madan et al., 2015; Gault et al., 2017; Horiuchi et al., 2018). RGH3/ZRSR2 and RNPC3 are required for U12-type intron recognition (Frilander and Steitz, 1999; Shen et al., 2010).
Human ZRSR2 was known to interact with U2AF2 (Tronchère et al., 1997), but we found more extensive connection between U12 splicing factors and U2AF. RBM48 interacts with RGH3, U2AF1, U2AF2, and ARMC7, while RGH3 interacts with U2AF1 and U2AF2. Both U2AF and RGH3/ZRSR2 are responsible for 3′ splice site recognition of U2-type and U12-type introns, respectively (Shen et al., 2010). Thus, the 3′ splice site recognition splicing factors of both the major and minor spliceosomes interact in maize. It is possible that RBM48 could participate in U12-type intron recognition, but a direct test of this hypothesis requires an in vitro RNA splicing assay, which has only recently been developed for the major spliceosome in plants (Albaqami and Reddy, 2018).
Our study also illustrates the importance of genetic analysis of RNA splicing factors to determine RNA processing functions. Recently, mutant models for several splicing factors including ZRSR2, ZRSR1, SMN, and FUS RNA binding protein showed a predominant in vivo function in minor intron splicing even though the proteins are thought to have broader roles based on in vitro splicing assays or protein-protein interactions (Madan et al., 2015; Doktor et al., 2017; Gault et al., 2017; Jangi et al., 2017; Horiuchi et al., 2018). RNA-seq analysis of mutant transcriptomes was able to resolve the predominant defects as impacting U12-type introns.
U12 Splicing Mutants in Plants and Animals Have Divergent Developmental Effects
In plants, genetic analysis of U12 splicing factors has defined two phenotypic syndromes. Transgenic knockdown alleles for multiple U12 splicing factors in Arabidopsis thaliana develop serrated leaves, delay senescence, and dwarf inflorescence branches, which can be complemented by exogenous application of gibberellins (Kim et al., 2010; Jung and Kang, 2014; Xu et al., 2016). Strong alleles of U12 splicing factors in either Arabidopsis thaliana or maize disrupt seed development (Figure 1; Kim et al., 2010; Fouquet et al., 2011). In maize, aberrant U12-type intron splicing disrupts both endosperm and embryo development. However, reduced U12 splicing efficiency does not cause cellular lethality, because rgh3 and rbm48 both show increased cell proliferation in endosperm tissue culture (Figure 7; Fouquet et al., 2011). These mutant phenotypes argue that efficient U12 splicing is required for cell differentiation rather than cell viability. Neither rbm48 nor rgh3 completely block U12 splicing, and it is likely that a complete block would cause cellular lethality.
Mutations disrupting U12 splicing in human cell lines reduce proliferation and are classified as core fitness genes (Blomen et al., 2015; Hart et al., 2015). Interestingly, ARMC7, an RBM48 interacting protein, was also found to be a core fitness gene in these human cell culture studies. It is somewhat surprising that homologous mutations in maize and humans have contrasting cell proliferation defects in tissue culture.
Although MIGs represent a small fraction of eukaryotic genes, plants and animals have MIGs in conserved cellular and biochemical pathways such as autophagy, secreted protein glycosylation, DNA damage repair, cell cycle, and histone methyltransferases (Gault et al., 2017). Disruption of multiple cell cycle and DNA damage repair genes would be expected to promote cell proliferation. Although rbm48 and rgh3 promote cell proliferation, ZRSR2 mutants reduce proliferation. However, ZRSR2 mutations are considered drivers toward cancer (Cazzola et al., 2013). Gault et al. (2017) suggested that divergence in MIGs or divergence in the location of U12-type introns within conserved MIGs are likely causes for the arrest of animal cell proliferation versus increased plant cell proliferation. For example, a subset of E2F cell-cycle transcription factors are MIGs in both maize and humans, but the U12-type introns are in different positions relative to the open reading frames. Retention of the human U12-type intron is predicted to knockout function through nonsense mediated decay, while retention of the U12-type intron in maize does not impact RNA stability and is unlikely to disrupt protein function (Gault et al., 2017).
Divergence of U12 Splicing Phenotypes Likely Result from Fractionation of MIGs
U12-type introns have longer and more conserved 5′ splice site and branch point consensus sequences that are recognized through cooperative binding of the U11 and U12 snRNPs (Frilander and Steitz, 1999). Consequently, divergence in MIGs between eukaryotic species predominantly is due to U12-type intron loss or mutation to U2-type introns (Burge et al., 1998; Lin et al., 2010). A large fraction of MIGs have been maintained in a conserved genetic architecture between plant and animal species, suggesting that U12 splicing efficiency could have similar effects on the cell (Gault et al., 2017).
Minor splicing mutants in animals have been shown to cause defects in cell differentiation. A subset of human myelodysplasia patients have mutations in ZRSR2 that result in aberrant U12 splicing and reduced terminal differentiation of myeloid blood cell types (Madan et al., 2015). Mutation of the mouse ZRSR1 paralog of ZRSR2 leads to blood and sperm differentiation defects (Horiuchi et al., 2018). Patients with Roifman syndrome have mutations in the human U4atac snRNA along with defects in both myeloid and lymphoid blood cell differentiation (Heremans and National Institute for Health Research BioResource et al., 2018). These examples suggest a common requirement for efficient splicing of U12-type introns to promote the differentiation of a subset of eukaryotic cell types. However, minor splicing mutations cause a range of human diseases and diverse phenotypes in animal models (Pessa et al., 2010; Markmiller et al., 2014; Verma et al., 2018; Horiuchi et al., 2018). The similarity of endosperm defects in rbm48 and rgh3 suggest a mutant ideotype for the developmental pathways sensitive to U12-type intron splicing efficiency. Maize genetic screens for delayed endosperm cell differentiation and prolonged cell proliferation could discover both U12 splicing mutants and specific MIGs needed to promote cell differentiation.
METHODS
Genetic Stocks
The rbm48-umu1 allele was isolated as a visual rough endosperm (rgh) mutant from the UniformMu transposon-tagging population (McCarty et al., 2005). The rbm48-umu2 allele was identified from the UniformMu reverse genetics resource as Mu insertion mu1056623 in stock UFMu-07152 (Settles et al., 2007; McCarty et al., 2013). Both rbm48-umu1 and rbm48-umu2 were backcrossed five times into W22, B73, Mo17, and A636 for phenotypic analysis. An rbm48-umu1 B73 x W22 F2 mapping population was generated by self-pollinating the F1 hybrid for the B73 introgression. The pBetl1:GUS and pVp1:GUS reporter transgenics have been described (Hueros et al., 1999; Cao et al., 2007). The rgh3 mutant stock was maintained as a Pr1 rgh3/pr1 Rgh3 stock in the W22 inbred background (Fouquet et al., 2011). All plant materials were grown at the University of Florida Plant Science Research and Education Unit in Citra, Florida or greenhouses located at the Horticultural Sciences Department in Gainesville, Florida.
Molecular Cloning of rbm48
The rbm48-umu1 allele was mapped with bulked segregant analysis using a pool of 93 homozygous rgh kernels from a B73 x W22 F2 mapping population. DNA was extracted from the pooled kernels and genotyped with 144 single nucleotide polymorphism (SNP) markers using the Sequenom MassARRAY platform at the Iowa State University Genomic Technologies Facility as described in Liu et al. (2010). Relative enrichment for the W22 genotype was calculated from non-segregating pooled samples.
Mu flanking sequence tags (MuFSTs) were generated from a 12 × 12 grid of 144 UniformMu rgh mutants including rbm48-umu1. Transposon-flanking sequences were amplified from pooled DNA using MuTAIL-PCR (Settles et al., 2004). The PCR products were sequenced with a Roche Genome Sequencer FLX System using custom A primers to add pool-specific barcodes and sequence from the Mu terminal inverted repeat (TIR). Resulting sequences were filtered for intact TIR sequences and mapped to the B73_v2 genome with blastn. Identical insertion sites sequenced from a single row and a single column pool were assigned to individual rgh mutant isolates. The ten unique Mu flanking sequence tags in rbm48-umu1 were co-aligned to the bulked segregant analysis map position to identify rbm48-umu1 as the likely causative mutation. Co-segregation of rbm48-umu1 with the rgh kernel phenotype was tested by extracting DNA from segregating kernels and amplifying the wild-type or Mu-insertion alleles using gene-specific primers and the TIR5 primer as described in Settles et al. (2007))).
Kernel Phenotypes
Segregating ears and mature kernels were imaged on a flatbed scanner. Both rbm48 alleles were analyzed for kernel composition phenotypes by a single-kernel grain analyzer with a microbalance and near infrared spectrometer (Spielbauer et al., 2009; Gustin et al., 2013). At least 30 normal and 30 rgh kernels were sampled from three segregating ears of each allele. Four technical replicate kernel weight and near infrared spectra were collected from each kernel. Average single-kernel composition predictions for each biological replicate were used as the kernel composition phenotypes shown in Supplemental Figure 2.
RT-PCR
Tissues were dissected, frozen in liquid nitrogen, and ground to a powder. RNA was extracted from 100 mg of tissue and mixed with 200 μL of RNA extraction buffer (50 mM Tris-HCl, pH 8, 150 mM LiCl, 5 mM EDTA, 1% SDS in DEPC treated water). The slurry was extracted using phenol:chloroform and Trizol (Invitrogen). RNA was precipitated from the aqueous fraction using isopropanol and washed with 70% ethanol. RNA pellets were re-suspended in nuclease free water (Sigma) treated with Purelink DNase (Invitrogen). RNA was further purified using an RNeasy MinElute Cleanup Kit (Qiagen). M-MLV reverse transcriptase (Promega) used 1 μg total RNA as a template to synthesize cDNA.
RT-PCR was performed by modifying the methods from Bai and DeMason (2006). The PCR conditions were 3 min at 94°C, followed by 24 to 28 cycles of 30 s at 94°C, 30 s at the appropriate annealing temperature for the primers and 1 min at 72°C, followed by a final extension of 5 min at 72°C. RT-qPCR used a StepOnePlus real-time PCR machine (Applied Biosystems) with 1X SYBR Green PCR Master Mix (Applied Biosystems) as described in Fouquet et al. (2011) and . The normalized expression level of each gene represents the average of three technical replicates of three biologically distinct kernel pools sampled from different segregating ears. Normalization was relative to actin1 using the comparative cycle threshold (ΔΔCt) method (Livak and Schmittgen, 2001). Primer sequences are listed in Supplemental Data Set 3.
Phylogenetic Analysis
Coselection was detected by parsing the species names from the “specific protein” lists for the RRM_RBM48, RRM_U2AFBPL, RRM_U2AF35, and RRM_U2AF35B domains in the NCBI Conserved Domains Database (Marchler-Bauer et al., 2017). A representative set of RBM48 proteins was aligned with MUSCLE (version 3.8.31) using default parameters, muscle -in seqs.fa -out seqs.afa, with subsequent manual edits (Edgar, 2004). The multiple sequence alignment (Supplemental Data Set 4) of the RBM48_RRM domain was input to RAxML (version 8.2.3) (Stamatakis, 2014) to construct a maximum likelihood phylogeny with the LG4X amino acid replacement matrix model and eight tree searches to test for tree stability (Le et al., 2012). Eight tree searches were completed to further test for tree stability. The specific RAxML command was: raxmlHPC-PTHREADS-SSE3 -m PROTGAMMALG4X -s seqs.domain.afa -n RBM48.tre -p $RANDOM -N 8 -T $PBS_NP. Node stability was tested with 1000 bootstrap replicates using the RAxML command: raxmlHPC-PTHREADS-SSE3 -m PROTGAMMALG4X -s seqs.domain.afa -n RBM48_bootstrap.tre -p $RANDOM -b $RANDOM -# 1000 -T $PBS_NP. The phylogeny was rooted by the oomycetes RBM48 protein and displayed with Dendroscope (version 3.2.10; Huson and Scornavacca, 2012).
RNA-Seq Analysis
Endosperm tissue from 16 to 18 DAP normal and rbm48 kernels in the W22 genetic background was dissected and frozen in liquid nitrogen. Total mRNA was extracted from four biological replicates of paired rbm48 mutant and normal sibling pools. Each replicate was a segregating ear from different plants. Non-strand-specific TruSeq (Illumina) cDNA libraries were prepared from 1 µg total RNA input with a 200 bp median insert length. All libraries were quantified using a Qubit, pooled; and 100 bp paired-end reads were sequenced on two lanes of the HiSeq 2000 platform.
Raw RNA-seq data were screened to remove adapter sequences using Cutadapt v1.1 (Martin, 2011) with the following parameters: error rate = 0.1, times = 1, overlap = 5, and minimum length = 0. Adapter trimmed sequences were quality trimmed with Trimmomatic v0.22 (Bolger et al., 2014) using parameters (HEADCROP:0, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:40) to truncate reads for base quality <15 within 4 base windows and kept only reads ≥40 bases after trimming.
Reads were uniquely aligned to the B73 RefGen_v2 maize (Zea mays) genome assembly with GSNAP (Version 2013-07-20) using the following parameters: orientation = FR, batch = 5, suboptimal levels = 0, novel splicing = 1, local-splice dist = 8000, local-splice penalty = 0, distant-splice penalty = 4, quality protocol = sanger, npaths = 1, quiet-if-excessive–max-mismatches = 0.02, no fails-format = sam, sam-multiple-primaries–pairmax-rna = 8000, pair expect = 200, pair dev = 150, nthreads = 4. The use-splicing argument was implemented to guide GSNAP by the ZmB73_5b Filtered Gene Set annotations but was permitted to discover novel splicing events. The argument use-snps was called with an IIT map file containing W22/B73 (Gault et al., 2017) was used to allow GSNAP to perform SNP-tolerant alignments ensuring proper mapping of the W22 RNA-Seq reads to the B73 reference genome.
Read counts/gene were determined with the HTSeq-Count utility in the HTSeq package (Anders et al., 2015; Ver 0.8.0). Non-redundant introns and genomic coordinates were identified from the ZmB73_5b annotation as described, except that U12-type introns were filtered to a non-redundant set of 372 introns (Gault et al., 2017). PSO metrics for each intron were calculated from exon-exon junction spanning reads and intron reads for each unique 5′ and 3′ splice site (Katz et al., 2010; Gault et al., 2017). Fisher’s exact test was calculated for each intron from the summed exon-exon junction reads and intron reads across the four normal and four mutant libraries for each rbm48 allele. Test statistics were correct to a false discovery rate ≤0.05 using the Benjamini-Hochberg method.
Differentially expressed transcripts were detected with the DESeq2 (Love et al., 2014) Bioconductor package. Transcripts were considered expressed if transcript per million was ≥1 in at least one genotype. Differential expression criteria were an adjusted p-value ≤ 0.05 and ≥twofold change. GO term enrichment analysis used agriGO v2.0 with default parameters and the expressed transcripts as a customized reference (Du et al., 2010).
Histology
Developing kernels from three 12 DAP ears segregating for rbm48-umu1 were harvested and fixed overnight at 4°C in FAA (3.7% formaldehyde, 5% glacial acetic acid, and 50% ethanol). Kernels were embedded in JB-4 plastic embedding media (Electron Microscopy Sciences). Five normal and five mutant kernels were sectioned at 4 μm thickness as described in Bai et al. (2016). Sections were stained in Schiff’s reagent, counter-stained with 1% aniline blue-black in 7% acetic acid, washed, dried, and mounted. Imaging was completed with a Zeiss Axiophot light microscope and an Amscope digital camera.
For GUS reporter expression analysis, hybrids of pBetl1:GUS or pVp1:GUS crossed by rbm48-umu1/+ heterozygous plants were self-pollinated (Hueros et al., 1999; Cao et al., 2007). Developing kernels were sampled from segregating ears, sectioned in the sagittal plane, and stained for GUS activity as previously described in Costa et al. (2003), Gutiérrez-Marcos et al. (2006), and Bai and DeMason (2008). At least 20 kernels for three to four segregating ears were observed, and 6 to 9 kernels were imaged to ensure that GUS reporter images were representative of the staining patterns. Images were captured with a Wild Heerbrugg dissecting microscope and an Amscope digital camera.
Endosperm Tissue Culture
Endosperm callus cultures were initiated as described (Shannon, 1994; Fouquet et al., 2011). Briefly, ears segregating for rbm48-umu1 from the A636 introgression were harvested from 6-18 DAP. Ears were surface sterilized in 70% ethanol and 20% bleach for 10 min and washed with sterile water. Individual endosperm tissues were dissected and placed on Murashige and Skoog media (pH 5.7) and supplemented with 3% Suc, 4 ppm thiamine, 0.2% Asn, and 2% phytagel. Growth of 400 individual endosperm tissues per genotype and harvest date was assayed after incubating in the dark for 35 d at 29°C. Cultures were imaged with a Nikon digital camera.
Subcellular Localization
RGH3, RGH3ΔUHM, U2AF1, and U2AF2 fusion protein constructs have been described previously in Fouquet et al. (2011) and Gault et al. (2017). RBM48 fusion proteins were constructed with a similar strategy. Briefly, the full-length RBM48 ORF was amplified from B73 seedling cDNA. Primer pairs to enable Gateway cloning of N- or C-terminal fusions were used to amplify full-length (RBM48_FL), N-terminal deletion of the RRM domain (RBM48ΔRRM), or C-terminal deletion (RBM48ΔRS); Supplemental Data Set 3 shows primer sequences. PCR products were cloned into pDONR221 and recombined into Gateway destination vectors containing GFP, RFP, or split enhanced YFP (EYFP) following the manufacturer’s protocol (Invitrogen; Karimi et al., 2002, 2007). For BiFC constructs, recombined expression cassettes, pSAT5-DEST or pSAT4-DEST, were digested with I-CeuI or I-SceI and ligated into pPZP-RCS2-bar binary vector. Binary vectors were transformed in Agrobacterium tumefaciens strains ABi and GV3101 (Wise et al., 2006).
Nicotiana benthamiana plants were grown for 5 to 8 weeks in a growth chamber under ∼50 μM/m2/s photosynthetically active radiation with a 16/8 h and 26/22°C day/night photoperiod for transient expression assays. For each construct combination, Agrobacterium was infiltrated into stomata of the abaxial surface of leaves on three different plants using a needleless syringe. Leaves were imaged 24 to 48 h after infiltration. For co-expression analysis, Agrobacterium strains with individual constructs were mixed in a 1:1 ratio prior to infiltration. Leaf sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) prior to visualization of fusion protein expression (Kapila, 1997). Representative images of subcellular localization were obtained using a Carl Zeiss Axio Imager .Z2 confocal microscope (Cevik and Kazan, 2013).
Protein Pull-Down Assays
Coding sequences for maize U2AF1 (GRMZM2G177229) and U2AF2 (GRMZM2G093256) were amplified with primers containing Nco-I and Not-I restriction enzyme sites, cloned into a pCR4-TOPO vector (Invitrogen), then subcloned into pET-28b with standard restriction-ligation protocols. Escherichia coli codon-optimized coding sequences of maize RBM48, RGH3, and ARMC7 were synthesized and cloned into pET-28b using NcoI and XhoI restriction sites (Genscript, Inc.). Caldicellulosiruptor bescii Cbes_2310 (WP_015908637.1), a predicted periplasmic sugar binding protein, was cloned into pET-28b using NheI and XhoI restriction sites (Dam et al., 2011). A stop codon was included in the inserted Cbes_2310 sequence to prevent translation of the C-terminal His-tag in the pET-28b vector. RBM48 and RGH3 were also cloned in pET-42a using NcoI and XhoI restriction sites to produce N-terminal tandem GST-His-tag fusion proteins (GenScript, Inc.). A stop codon was included in the inserted RBM48 and RGH3 sequences to prevent translation of the C-terminal His-tag in the pET-42a vector.
Recombinant expression conditions for all tagged versions of RGH3, U2AF1, U2AF2, and ARMC7 were initiated with a 1:50 dilution of overnight cultures in fresh lysogeny broth media supplemented with 50 µg/mL kanamycin. Cultures were grown at 37°C with shaking at 225 rpm until OD600 = 0.45. Expression was induced by adding Isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.5 mM. Cultures were induced for 3 h at room temperature with shaking at 225 rpm. Bacterial cells were harvested by centrifugation at 2500 × g for 15 min at 4°C. U2AF1, U2AF2, and ARMC7 cell pellets were stored at −80°C. RGH3 induction was improved by storing cell pellets overnight at 4°C with supernatant media. The supernatant was removed the following morning and the pellet stored at −80°C.
RBM48 recombinant proteins were expressed by inoculating a 1:20 dilution of an overnight-grown culture into MagicMedia E. coli Expression Medium (Invitrogen), supplemented with 50 µg/mL kanamycin. Cells were grown at 37°C with shaking at 225 rpm until OD600 = 0.6. The culture was then transferred to 18°C and grown for an additional 36 h at 225 rpm to allow for auto-induction of recombinant protein expression. After incubation at 18°C, the culture was centrifuged at 2500 × g for 15 min at 4°C, and the pellet was stored at −80°C.
Cell pellets were resuspended in 2 mL of lysis buffer (50 mM Tris, 150 mM NaCl, 1X Halt Protease Inhibitor Cocktail from Thermo Fisher Scientific, 1 mg/mL lysozyme, 10 µg/mL DNase I, 1% (v/v) Triton X-100) and mixed with a rotisserie tube-rotator at room temperature for 10 min. The lysate was sonicated at 20 kHz for 30 s on/off five times, and subsequently centrifuged at 10,000 × g for 10 min at 4°C.
GST protein pull-down assays were based on Pierce Glutathione Magnetic Agarose Beads instructions (Thermo Fisher Scientific). Briefly, 400 μL bacterial lysate containing induced fusion protein was mixed with 100 μL equilibration buffer and incubated with 25 μL settled bead volume in a 1.5 mL microcentrifuge tube for 1 h at 4°C on a rotisserie tube-rotator. Beads were collected in a magnetic stand, the supernatant was discarded, and beads were washed twice with equilibration buffer. In the third wash, half of the slurry volume was transferred to a fresh microcentrifuge tube, and the GST fusion protein was eluted with 125 μL elution buffer to assay input protein for the pull-down.
The remaining protein-bound beads were separated on a magnetic stand, incubated with a mix of 200 μL of induced bacterial lysate containing His-tagged protein mixed with 50 μL of equilibration buffer. The lysate was incubated with the bound GST fusion protein for 1 h at 4°C on a rotisserie tube-rotator. The beads were collected and supernatant discarded. Beads were washed three times using 250 μL of equilibration buffer. Bound proteins were eluted with 125 μL elution buffer.
GST-His-RBM48, GST-His-RGH3, RGH3-His, U2AF1-His, U2AF2-His, ARMC7-His, and His-Cbes_2310 were detected by immunoblot analysis using a 1:1000 dilution of monoclonal His-tag antibody (Cell Signaling, cat# 2365S). Protein fragments of RGH3-His were excised from the gels, subjected to trypsin digestion, and sequenced according to the procedure described in Shevchenko et al. (1996), at the Biomolecular and Proteomics Mass Spectrometry Facility at the University of California, San Diego. Each recombinant protein was produced at least three times from independent induction cultures. Pull-down assays were replicated for each combination of proteins at least three times, using independent batches of recombinant proteins.
Accession Numbers
Sequence data from this article are available at the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA). Maize transposon flanking sequence tags for identification of rbm48-umu1 are in accession: SRX006230. Endosperm RNA-seq data are in accession: GSE118505.
Supplemental Data
Supplemental Figure 1. Molecular cloning of rgh*-00F-061-05 to the rbm48 locus (supports Figure 1).
Supplemental Figure 2. Phenotypes of rbm48 alleles (supports Figure 1).
Supplemental Figure 3. Characterization of the Rbm48 locus (supports Figures 1 and 2).
Supplemental Figure 4. RNA-seq read depth for experimentally validated genes (supports Figure 5).
Supplemental Figure 5. Overlap of RNA splicing defects in rbm48 and rgh3 mutant seedlings (supports Figure 5).
Supplemental Figure 6. DEGs identified in rbm48 mutant endosperm (supports Figure 6).
Supplemental Figure 7. Endosperm cell differentiation defects in rbm48 at additional developmental stages (supports Figure 7).
Supplemental Table 1. Segregation of rgh kernel phenotypes in self-pollinations of rbm48 heterozygotes (supports Figure 1).
Supplemental Table 2.Transmission of the rbm48-umu1 allele in reciprocal crosses with normal inbred plants (supports Figure 1).
Supplemental Table 3. Enriched GO terms from differential gene expression analysis (supports Figure 6).
Supplemental Data Set 1. Read counts and PSO statistics (supports Figures 3 to 5).
Supplemental Data Set 2. Expressed transcripts (≥1 TPM in one genotype) with DeSeq2 adjusted p-values and DeSeq2 fold change ratios (supports Figure 6).
Supplemental Data Set 3. Primers used in this study (supports Methods section).
Supplemental Data Set 4. Aligned sequences used for Figure 2.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank John Baier, Jennifer Moses, J. Paige Gronevelt, Elizabeth Jankulovski, and Laurel Levine for technical assistance. This work was supported by National Science Foundation (grants MCB-1412218 to S.L., W.B.B., and A.M.S.; IOS-1547787 to W.B.B.; and IOS-1623478 to A.M.S.), the University of Florida HHMI Science for Life undergraduate research program, and the Vasil-Monsanto Endowment.
AUTHOR CONTRIBUTIONS
A.M.S., S.L., W.B.B., F.B., and J.C. wrote the manuscript. A.M.S., F.B.,G.S., and C.-W.T. completed plant genetics and mutant mapping. F.B. characterized maize kernel, seedling, and cellular phenotypes. A.M.S., S.L., and G.F. completed protein sequence evolutionary analysis. W.B.B., R.D., F.B., and A.M.S. designed and completed RNA-seq experiments. F.B., D.N.S., J.M., and A.E.S. completed RT-PCR experiments. A.M.S. and S.L. designed and supervised protein-protein interaction experiments. D.N.S. and F.M. cloned fluorescent protein fusion constructs. D.N.S. completed co-localization and BiFC experiments. J.C., C.J.B., and F.M. developed recombinant expression constructs and methodology. J.C. completed protein pull-down experiments.
References
- Albaqami M., Reddy A.S.N. (2018). Development of an in vitro pre-mRNA splicing assay using plant nuclear extract. Plant Methods 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto T.S. (2007). U12DB: A database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 35: D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argente J., Flores R., Gutiérrez-Arumí A., Verma B., Martos-Moreno G.A., Cuscó I., Oghabian A., Chowen J.A., Frilander M.J., Pérez-Jurado L.A. (2014). Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol. Med. 6: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., DeMason D.A. (2006). Hormone interactions and regulation of Unifoliata, PsPK2, PsPIN1 and LE gene expression in pea (Pisum sativum) shoot tips. Plant Cell Physiol. 47: 935–948. [DOI] [PubMed] [Google Scholar]
- Bai F., DeMason D.A. (2008). Hormone interactions and regulation of PsPK2:GUS compared with DR5:GUS and PID:GUS in Arabidopsis thaliana. Am. J. Bot. 95: 133–145. [DOI] [PubMed] [Google Scholar]
- Bai F., Daliberti M., Bagadion A., Xu M., Li Y., Baier J., Tseung C.W., Evans M.M., Settles A.M. (2016). Parent-of-origin-effect rough endosperm mutants in maize. Genetics 204: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V.A., et al. (2015). Gene essentiality and synthetic lethality in haploid human cells. Science 350: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.W.S., Simpson C.G. (1998). Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 77–95. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Padgett R.A., Sharp P.A. (1998). Evolutionary fates and origins of U12-type introns. Mol. Cell 2: 773–785. [DOI] [PubMed] [Google Scholar]
- Cao X., Costa L.M., Biderre-Petit C., Kbhaya B., Dey N., Perez P., McCarty D.R., Gutierrez-Marcos J.F., Becraft P.W. (2007). Abscisic acid and stress signals induce Viviparous1 expression in seed and vegetative tissues of maize. Plant Physiol. 143: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D., Zhu J., Manche L., Huber E., Krainer A.R., Cáceres J.F. (2002). Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 22: 6871–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzola M., Della Porta M.G., Malcovati L. (2013). The genetic basis of myelodysplasia and its clinical relevance. Blood 122: 4021–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik V., Kazan K. (2013). Agroinfiltration of Nicotiana benthamiana leaves for co-localization of regulatory proteins involved in jasmonate signaling. Methods Mol. Biol. 1011: 199–208. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Costa L.M., Gutierrez-Marcos J.F., Brutnell T.P., Greenland A.J., Dickinson H.G. (2003). The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130: 5009–5017. [DOI] [PubMed] [Google Scholar]
- Dam P., et al. (2011). Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 39: 3240–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila López M., Rosenblad M.A., Samuelsson T. (2008). Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res. 36: 3001–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktor T.K., Hua Y., Andersen H.S., Brøner S., Liu Y.H., Wieckowska A., Dembic M., Bruun G.H., Krainer A.R., Andresen B.S. (2017). RNA-sequencing of a mouse-model of spinal muscular atrophy reveals tissue-wide changes in splicing of U12-dependent introns. Nucleic Acids Res. 45: 395–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P., et al. (2011). Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332: 240–243. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet R., Martin F., Fajardo D.S., Gault C.M., Gómez E., Tseung C.W., Policht T., Hueros G., Settles A.M. (2011). Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development. Plant Cell 23: 4280–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M.J., Steitz J.A. (1999). Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 13: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault C.M., Martin F., Mei W., Bai F., Black J.B., Barbazuk W.B., Settles A.M. (2017). Aberrant splicing in maize rough endosperm3 reveals a conserved role for U12 splicing in eukaryotic multicellular development. Proc. Natl. Acad. Sci. USA 114: E2195–E2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin J.L., Jackson S., Williams C., Patel A., Armstrong P., Peter G.F., Settles A.M. (2013). Analysis of maize (Zea mays) kernel density and volume using microcomputed tomography and single-kernel near-infrared spectroscopy. J. Agric. Food Chem. 61: 10872–10880. [DOI] [PubMed] [Google Scholar]
- Guth S., Tange T.O., Kellenberger E., Valcárcel J. (2001). Dual function for U2AF35 in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 21: 7673–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Evans M.M. (2006). Maternal gametophytic baseless1 is required for development of the central cell and early endosperm patterning in maize (Zea mays). Genetics 174: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T., et al. (2015). High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163: 1515–1526. [DOI] [PubMed] [Google Scholar]
- He H., et al. (2011). Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332: 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans J., et al. ; National Institute for Health Research BioResource (2018). Abnormal differentiation of B cells and megakaryocytes in patients with Roifman syndrome. J. Allergy Clin. Immunol. 142: 630–646. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., et al. (2018). Impaired spermatogenesis, muscle, and erythrocyte function in U12 intron splicing-defective Zrsr1 mutant mice. Cell Reports 23: 143–155. [DOI] [PubMed] [Google Scholar]
- Hueros G., Gomez E., Cheikh N., Edwards J., Weldon M., Salamini F., Thompson R.D. (1999). Identification of a promoter sequence from the BETL1 gene cluster able to confer transfer-cell-specific expression in transgenic maize. Plant Physiol. 121: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. (2012). Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Irimia M., Roy S.W. (2014). Origin of spliceosomal introns and alternative splicing. Cold Spring Harb. Perspect. Biol. 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M., et al. (2017). SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc. Natl. Acad. Sci. USA 114: E2347–E2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.J., Kang H. (2014). The Arabidopsis U11/U12-65K is an indispensible component of minor spliceosome and plays a crucial role in U12 intron splicing and plant development. Plant J. 78: 799–810. [DOI] [PubMed] [Google Scholar]
- Kapila J., de Rycke R., Van Montagu M., Angenon G. (1997). An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122: 101–108. [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37: 465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf C.L., Rodionova N.A., Green M.R., Burley S.K. (2001). A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Jung H.J., Kwak K.J., Kim M.K., Oh S.H., Han Y.S., Kang H. (2010). The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell 22: 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H., Matter N., Bader R., Thiele W., Müller F. (2007). Splicing segregation: The minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 131: 718–729. [DOI] [PubMed] [Google Scholar]
- Le S.Q., Dang C.C., Gascuel O. (2012). Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol. Biol. Evol. 29: 2921–2936. [DOI] [PubMed] [Google Scholar]
- Lee Y., Rio D.C. (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.F., Mount S.M., Jarmołowski A., Makałowski W. (2010). Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol. Biol. 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen H.D., Makarevitch I., Shirmer R., Emrich S.J., Dietrich C.R., Barbazuk W.B., Springer N.M., Schnable P.S. (2010). High-throughput genetic mapping of mutants via quantitative single nucleotide polymorphism typing. Genetics 184: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Wieczorek Kirk D.A., Lambermon M.H., Filipowicz W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5: 160–167. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., et al. (2015). Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 6: 6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madanieh R., Mathew S., Shah P., Vatti S.K., Madanieh A., Kosmas C.E., Vittorio T.J. (2015). Cardiac magnetic resonance imaging might complement two-dimensional echocardiography in the detection of a reversible nonischemic cardiomyopathy. Clin. Med. Insights Case Rep. 8: 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 (D1): D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S., et al. (2014). Minor class splicing shapes the zebrafish transcriptome during development. Proc. Natl. Acad. Sci. USA 111: 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- McCarty D.R., Suzuki M., Hunter C., Collins J., Avigne W.T., Koch K.E. (2013). Genetic and molecular analyses of UniformMu transposon insertion lines. Methods Mol. Biol. 1057: 157–166. [DOI] [PubMed] [Google Scholar]
- Montzka K.A., Steitz J.A. (1988). Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. USA 85: 8885–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Yoshimura K., Nosaka R., Sakuyama H., Koike Y., Tanabe N., Maruta T., Tamoi M., Shigeoka S. (2012). Subcellular and subnuclear distribution of high-light responsive serine/arginine-rich proteins, atSR45a and atSR30, in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 76: 2075–2081. [DOI] [PubMed] [Google Scholar]
- Niemelä E.H., Frilander M.J. (2014). Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol. 11: 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä E.H., Oghabian A., Staals R.H., Greco D., Pruijn G.J., Frilander M.J. (2014). Global analysis of the nuclear processing of transcripts with unspliced U12-type introns by the exosome. Nucleic Acids Res. 42: 7358–7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.A., McCarthy M., Steitz J.A. (2002). The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 21: 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessa H.K., Greco D., Kvist J., Wahlström G., Heino T.I., Auvinen P., Frilander M.J. (2010). Gene expression profiling of U12-type spliceosome mutant Drosophila reveals widespread changes in metabolic pathways. PLoS One 5: e13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A.I., Mann M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber S., Stettler J., Filosa G., Colombo M., Jutzi D., Lenzken S.C., Schweingruber C., Bruggmann R., Bachi A., Barabino S.M., Mühlemann O., Ruepp M.D. (2016). Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 35: 1504–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.W., Irimia M. (2009). Splicing in the eukaryotic ancestor: Form, function and dysfunction. Trends Ecol. Evol. (Amst.) 24: 447–455. [DOI] [PubMed] [Google Scholar]
- Ru Y., Wang B.B., Brendel V. (2008). Spliceosomal proteins in plants. Curr. Top. Microbiol. Immunol. 326: 1–15. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Sakamoto C., Hagiwara M., Agredano-Moreno L.T., Jiménez-García L.F., Nakao M. (2012). The distribution of phosphorylated SR proteins and alternative splicing are regulated by RANBP2. Mol. Biol. Cell 23: 1115–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Will C.L., Makarova O.V., Makarov E.M., Lührmann R. (2002). Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol. 22: 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles A.M., et al. (2007). Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles A.M., Latshaw S., McCarty D.R. (2004). Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res. 32: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon J.C. (1994). Establishment and culture of maize endosperm. In The Maize Handbook, Freeling M., Walbot V., eds (New York: Springer-Verlag; ), pp. 719–722. [Google Scholar]
- Shaul O. (2015). Unique aspects of plant nonsense-mediated mRNA decay. Trends Plant Sci. 20: 767–779. [DOI] [PubMed] [Google Scholar]
- Shen H., Zheng X., Luecke S., Green M.R. (2010). The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 24: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Filipowicz W. (1996). Splicing of precursors to mRNA in higher plants: Mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32: 1–41. [DOI] [PubMed] [Google Scholar]
- Spielbauer G., Armstrong P., Baier J.W., Allen W.B., Richardson K., Shen B., Settles A.M. (2009). High-throughput near-infrared reflectance spectroscopy for predicting quantitative and qualitative composition phenotypes of individual maize kernels. Cereal Chem. 86: 556–564. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W.Y., Steitz J.A. (1995). Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc. Natl. Acad. Sci. USA 92: 2504–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillemans V., Dispa L., Remacle C., Collinge M., Motte P. (2005). Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J. 41: 567–582. [DOI] [PubMed] [Google Scholar]
- Tillemans V., Leponce I., Rausin G., Dispa L., Motte P. (2006). Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 18: 3218–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchère H., Wang J., Fu X.D. (1997). A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 388: 397–400. [DOI] [PubMed] [Google Scholar]
- Turunen J.J., Niemelä E.H., Verma B., Frilander M.J. (2013). The significant other: Splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 4: 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma B., Akinyi M.V., Norppa A.J., Frilander M.J. (2018). Minor spliceosome and disease. Semin. Cell Dev. Biol. . 79: 103–112. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. (2005). Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 386: 713–724. [DOI] [PubMed] [Google Scholar]
- Will C.L., Schneider C., Hossbach M., Urlaub H., Rauhut R., Elbashir S., Tuschl T., Lührmann R. (2004). The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A.A., Liu Z., Binns A.N. (2006). Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol. Biol. 343: 43–53. [DOI] [PubMed] [Google Scholar]
- Xu T., Kim B.M., Kwak K.J., Jung H.J., Kang H. (2016). The Arabidopsis homolog of human minor spliceosomal protein U11-48K plays a crucial role in U12 intron splicing and plant development. J. Exp. Bot. 67: 3397–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I., Dittmar K., Wang W., Foley S.W., Berg M.G., Hu K.Y., Wei Z., Wan L., Dreyfuss G. (2013). Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife 2: e00780. [DOI] [PMC free article] [PubMed] [Google Scholar]