IncA and IncC conjugative plasmids drive antibiotic resistance dissemination among several pathogenic species of Gammaproteobacteria due to the diversity of drug resistance genes that they carry and their ability to mobilize antibiotic resistance-conferring genomic islands such as SGI1 of Salmonella enterica. While historically grouped as “IncA/C,” IncA and IncC replicons were recently confirmed to be compatible and to abolish each other’s entry into the cell in which they reside during conjugative transfer. The significance of our study is in identifying an entry exclusion system that is shared by IncA and IncC plasmids. It impedes DNA transfer to recipient cells bearing a plasmid of either incompatibility group. The entry exclusion protein of this system is unrelated to any other known entry exclusion proteins.
KEYWORDS: antibiotics, conjugation, conjugative plasmid, entry exclusion, IncA, IncA/C, IncC, resistance, T4SS, VirB6
ABSTRACT
Conjugative plasmids of incompatibility group C (IncC), formerly known as A/C2, disseminate antibiotic resistance genes globally in diverse pathogenic species of Gammaproteobacteria. Salmonella genomic island 1 (SGI1) can be mobilized by IncC plasmids and was recently shown to reshape the conjugative type IV secretion system (T4SS) encoded by these plasmids to evade entry exclusion. Entry exclusion blocks DNA translocation between cells containing identical or highly similar plasmids. Here, we report that the protein encoded by the entry exclusion gene of IncC plasmids (eexC) mediates entry exclusion in recipient cells through recognition of the IncC-encoded TraGC protein in donor cells. Phylogenetic analyses based on EexC and TraGC homologs predicted the existence of at least three different exclusion groups among IncC-related conjugative plasmids. Mating assays using Eex proteins encoded by representative IncC and IncA (former A/C1) and related untyped plasmids confirmed these predictions and showed that the IncC and IncA plasmids belong to the C exclusion group, thereby explaining their apparent incompatibility despite their compatible replicons. Representatives of the two other exclusion groups (D and E) are untyped conjugative plasmids found in Aeromonas sp. Finally, we determined through domain swapping that the carboxyl terminus of the EexC and EexE proteins controls the specificity of these exclusion groups. Together, these results unravel the role of entry exclusion in the apparent incompatibility between IncA and IncC plasmids while shedding light on the importance of the TraG subunit substitution used by SGI1 to evade entry exclusion.
IMPORTANCE IncA and IncC conjugative plasmids drive antibiotic resistance dissemination among several pathogenic species of Gammaproteobacteria due to the diversity of drug resistance genes that they carry and their ability to mobilize antibiotic resistance-conferring genomic islands such as SGI1 of Salmonella enterica. While historically grouped as “IncA/C,” IncA and IncC replicons were recently confirmed to be compatible and to abolish each other’s entry into the cell in which they reside during conjugative transfer. The significance of our study is in identifying an entry exclusion system that is shared by IncA and IncC plasmids. It impedes DNA transfer to recipient cells bearing a plasmid of either incompatibility group. The entry exclusion protein of this system is unrelated to any other known entry exclusion proteins.
INTRODUCTION
Conjugative plasmids of incompatibility group C (IncC) are large, broad-host-range plasmids found globally in diverse species of Gammaproteobacteria isolated from food products, food-producing animals, and humans (1). IncC plasmids circulate not only in several pathogenic species of Enterobacteriaceae but also in seventh pandemic African O1 El Tor isolates of Vibrio cholerae, the infectious agent of the diarrheal disease cholera, and other Vibrionaceae (2–4). IncC plasmids may carry transposons and integrons that confer resistance to clinically important families of antibiotics, including β-lactams and β-lactamase inhibitors, cephalosporins, carbapenems, aminoglycosides, tetracyclines, quinolones, trimethoprim, and sulfonamides (1, 4). IncC plasmids also participate in the emergence of drug-resistant bacteria through the mobilization of unrelated multidrug resistance-conferring genomic islands, such as MGIVchHai6 of V. cholerae and Salmonella genomic island 1 (SGI1) (5, 6).
Plasmid incompatibility and exclusion are two distinct processes preventing a bacterium that carries a plasmid from being a suitable recipient for the acquisition of an identical or similar plasmid. Incompatibility results from the inability of similar plasmids to be propagated stably in the same cell line due to interference between their respective replication or partition functions. The process of exclusion presents a barrier against DNA transfer during conjugation between two bacterial cells carrying related elements (7). Conjugation is mediated by a type IV secretion system (T4SS), a multiprotein nanomachine that spans the cell envelope to translocate proteins and DNA from a donor cell to a recipient cell (8). Two mechanisms of the process of exclusion occurring during conjugation have been described previously (9). Surface exclusion prevents close contact between cells, whereas entry exclusion impedes DNA transfer after formation of the mating pair. Entry exclusion has been studied in F-like conjugative plasmids (10) and in plasmids of the IncHI1 (11), IncI1/IncIγ (12), IncN/IncW (13, 14), and IncPα incompatibility groups (15), as well as in integrative and conjugative elements (ICEs) of the SXT/R391 family (16). Apart from IncHI1 plasmids, these conjugative elements encode an entry exclusion (Eex) protein that is required in the recipient cell but not in the donor cell to exert entry exclusion. For F-like plasmids and SXT/R391 ICEs, the Eex protein in the recipient cell interacts with a protein of the T4SS in the donor cell, namely, the VirB6-like mating-pair stabilization protein TraG (10, 16). For IncI1/IncIγ plasmids, the Eex protein interacts with TraY in the donor cell (12). This VirB6-like protein plays the same role as TraG but was given a different name due to inconsistencies in the naming of T4SS proteins encoded by different plasmid types (17). Related conjugative elements carrying orthologous and yet divergent exclusion systems can fall into different exclusion groups that do not exclude each other (12, 18, 19). Entry exclusion has been shown to be an essential feature of conjugative plasmids and is thought to be ubiquitous among conjugative elements (20). However, the mechanisms by which entry exclusion factors interact with each other and interfere with DNA transfer are not yet understood.
IncC plasmids are closely related to IncA conjugative plasmids such as pRA1 from the fish pathogen Aeromonas hydrophila (21). Upon initial characterization in the early 1970s, pRA1 was found to be compatible with plasmids of all known compatibility groups, including IncC plasmids (22, 23). However, marked entry exclusion was also observed when the IncC pR57b plasmid or pIP40a plasmid was transferred to a recipient carrying the IncA plasmid pRA1 (24). At the time, these observations led Hedges (25) to combine IncA and IncC plasmids to form the “A-C complex.” The term “IncA/C” was introduced later for reasons that remain unclear and has become widespread since then in the literature (1). Ambrose et al. (26) recently confirmed the compatibility of IncA and IncC replicons and recommended avoiding the use of the term “IncA/C” because it causes unnecessary confusion. Those authors also showed that the apparent “incompatibility” likely results from IncA and IncC plasmids exerting entry exclusion effects on one another, thereby suggesting the existence of a shared mechanism of exclusion. By analogy with F-like plasmids and the SXT/R391 ICEs, they proposed a small open reading frame (ORF) that is conserved in IncA and IncC plasmids and immediately downstream of traG as a potential candidate for an entry exclusion determinant.
Our group recently showed that SGI1 encodes three T4SS subunit proteins, TraNS, TraHS, and TraGS, that are distantly related to the corresponding TraNC, TraHC, and TraGC proteins encoded by IncC plasmids (27). The three protein subunits alone are insufficient to form a functional conjugative T4SS, and SGI1 requires a helper IncA or IncC plasmid for transfer (28). However, the substitution of TraGS of SGI1 for the VirB6-like TraGC subunits of the IncC F-type T4SS in donor cells has been shown to allow SGI1 transfer to a recipient strain carrying an IncC plasmid, thereby suggesting that entry exclusion cannot occur in this context (27). Since the Eex determinant of IncC and related plasmids was unknown, we investigated the exclusion system of IncC plasmids to identify the eex gene. Phylogenetic analyses and exclusion assays revealed that all known IncC and IncA plasmids belong to the same exclusion group and allowed us to define additional exclusion groups in more distantly related, untyped conjugative plasmids.
RESULTS
Identification of the gene mediating IncC plasmid entry exclusion in recipient cells.
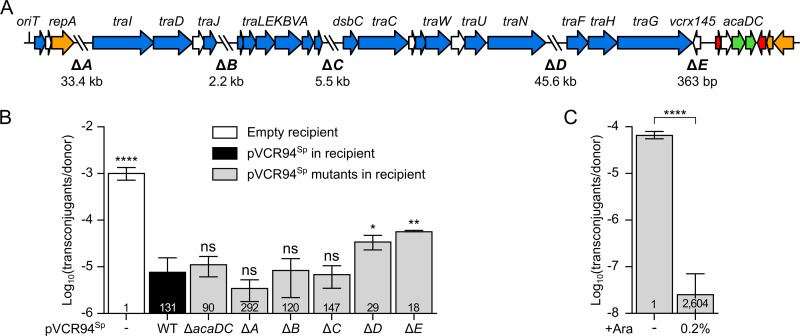
BLASTP analysis of the predicted translation products of all open reading frames of IncC plasmid pVCR94ΔX (GenBank accession number KF551948.1) (Table 1) failed to reveal any significant homology with any known exclusion proteins encoded by conjugative plasmids or ICEs. To examine whether this IncC plasmid was able to mediate entry exclusion, we used two differentially marked derivatives carrying either a kanamycin resistance gene (pVCR94Kn) or a spectinomycin resistance gene (pVCR94Sp). pVCR94Kn and pVCR94Sp were introduced into a rifampin-resistant Escherichia coli K-12 strain and a nalidixic acid-resistant E. coli K-12 strain, respectively. Since incompatibility between these two virtually identical plasmids prevented the isolation of transconjugant colonies, we monitored the transfer of ampicillin-resistant, broad-host-range mobilizable plasmid pCloDF13 (pClo), which relies entirely on the mating pore of pVCR94Kn for transfer (27). pClo was thus used as a proxy to assess entry exclusion occurring between the two strains, each bearing a different differentially marked plasmid. The frequency of transfer of pClo to a plasmid-free recipient strain was ∼130 times higher than the frequency of transfer to an isogenic recipient strain carrying pVCR94Sp (Fig. 1B). This value represents the exclusion index (EI) as defined by others (16, 20) and confirms that our pVCR94 derivatives exert entry exclusion effects on each other. In this study, we concluded that no exclusion was present when EI values were below 3.
TABLE 1.
E. coli K-12-derivative strains, plasmids, and genomic islands used in this study
| Strain. plasmid, or genomic island |
Relevant genotype or phenotypea | Source or reference(s) |
|---|---|---|
| Strains | ||
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 (Tc) | 54 |
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | 37 |
| GG56 | Nxr derivative of BW25113 (Nx) | 55, 56 |
| VB112 | Rfr derivative of MG1655 (Rf) | 57 |
| Plasmids | ||
| pAsa4c | Cmr conjugative plasmid from A. salmonicida JF2267 (Cm) | 58 |
| pVCR94 | IncC conjugative plasmid from V. cholerae O1 El Tor (Su Tm Cm Ap Tc Sm) | 59 |
| pVCR94ΔX | Sur derivative of pVCR94, lacking the large fragment containing the multidrug resistance gene cluster | 59 |
| pVCR94Sp | Spr derivative of pVCR94ΔX (pVCR94ΔX2) (Sp Su) | 29 |
| pVCR94Kn | Knr derivative of pVCR94ΔX (pVCR94ΔX3) (Kn Su) | 29 |
| pVCR94SpΔacaCD | pVCR94Sp ΔacaCD (Sp Su) | 29 |
| pVCR94SpΔA | pVCR94Sp Δ(vcrx004-vcrx028) Δ(vcrx030-vcrx059) (Sp Su) | This study |
| pVCR94SpΔB | pVCR94Sp Δ(vcrx064-vcrx069) (Sp Su) | This study |
| pVCR94SpΔC | pVCR94Sp Δvcrx076 (Sp Su) | This study |
| pVCR94SpΔD | pVCR94Sp Δ(vcrx085-vcrx141) (Sp Su) | This study |
| pVCR94SpΔE | pVCR94Sp Δvcrx145 (Sp Su) | This study |
| pVCR94SpΔtraGC | pVCR94Sp ΔtraGC (Sp Su) | 27 |
| pBAD30 | orip15A araC PBAD (Ap) | 39 |
| peexC | pBAD30::eexC (vcrx145 of pVCR94ΔX) (Ap) | This study |
| peexCrev | pBAD30::eexCrev (vcrx145 in reverse orientation) (Ap) | This study |
| ptraGS | pBAD30::traGS (traGS from SGI1) (Ap) | 27 |
| peexC2 | pBAD30::eexC2 (YR71pYR1_0178 of pYR1) (Ap) | This study |
| peexC3 | pBAD30::eexC3 (pKHM-243_0211 of pKHM-1) (Ap) | This study |
| peexC1 | pBAD30::eexC1 (pRA1_0150 of pRA1) (Ap) | This study |
| peexD | pBAD30::eexD (sequence 139329 to 139709 of pAhD4-1) (Ap) | This study |
| peexE | pBAD30::eexE (sequence 156821 to 157174 of pAsa4c) (Ap) | This study |
| ptraGE | pBAD30::traGE (sequence 153216 to 156770 of pAsa4c) (Ap) | This study |
| pCmE | pBAD30::eexCmE (Ap) | This study |
| pEmC | pBAD30::eexEmC (Ap) | This study |
| pClo | CloDF13::TnAΔEcoRV (pSU4628) (Ap) | 60 |
| pSIM6 | Thermoinducible expression of λRed recombination (orits Ap) | 61 |
| pKD3 | cat (Cm) template for one-step chromosomal gene inactivation | 37 |
| pKD4 | aph (Kn) template for one-step chromosomal gene inactivation | 37 |
| pTT01 | tetM (Tc) template for one-step chromosomal gene inactivation | 62 |
| pE-FLP | Constitutive expression of Flp recombinase (orits Ap) | 38 |
| Genomic islands | ||
| SGI1 | SGI1 inserted into the 3′ end of trmE (Ap Cm Sp Sm Su Tc) | 29 |
| SGI1Kn | ΔIn104::aph mutant of SGI1 devoid of the integron In104 (Kn) | 27 |
| SGI1KnΔtraGS | ΔIn104::aph ΔtraGS mutant of SGI1 (Kn) | 27 |
Ap, ampicillin; Cm, chloramphenicol; Kn, kanamycin; Nx, nalidixic acid; Rf, rifampin; Sp, spectinomycin; Sm, streptomycin; Su, sulfamethoxazole; Tc, tetracycline; Tm, trimethoprim; ts, thermosensitive.
FIG 1.
Mapping of IncC exclusion functions. (A) Linear schematic representation of pVCR94ΔX gene map. The positions and orientations of open reading frames (ORFs) are indicated by arrows. Colors depict the function deduced from functional analyses and BLAST comparisons as follows: orange, replication and partition; blue, conjugative transfer; red, transcriptional repression; green, transcriptional activation; white, unknown function. The positions of fragments deleted in pVCR94Sp (ΔA to ΔE) are indicated by backslashes, and lengths of deletions are indicated below the schematic representation. (B) Exclusion of pClo transferred from a donor strain bearing pVCR94Kn by a set of deletion mutants of pVCR94Sp in recipient cells. The donor strain was E. coli VB112 bearing pVCR94Kn and pClo (Apr). All recipient strains were derivatives of E. coli GG56 that were either plasmid free (−) or bearing pVCR94Sp (wild type [WT]) or its deletion mutants. (C) Expression of eexC (vcrx145) in a recipient strain is sufficient to exclude transfer of pVCR94Sp. The donor strain E. coli GG56 bearing pVCR94Sp was mated with E. coli CAG18439 carrying peexC as the recipient without or with arabinose (+Ara). Each bar represents the mean of results from three independent experiments, with error bars indicating the standard deviations. Exclusion indices (EI) are indicated at the bottom of each bar. Statistical analyses were carried out on the logarithm of the values using one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test (B) and an unpaired t test (two-tailed) (C) to compare induced transfer and noninduced transfer. Statistical significance is indicated as follows: ****, P < 0.0001; **, P < 0.01; *, P < 0.05; ns, not significant.
We next used the same approach to map the pVCR94Sp genes that mediate exclusion in recipient cells using recipient strains harboring a set of deletion mutations of pVCR94Sp. These mutants lack either the acaCD transcriptional activator genes or five regions encompassing mostly genes of unknown function (ΔA to ΔE in Fig. 1A). None of the mutations was found to affect pVCR94Sp stability as long as spectinomycin was used as the selective agent. Deletion of acaCD had no effect on exclusion (EI = 90) of pClo (Fig. 1B), suggesting that expression of entry exclusion in recipient cells is not controlled by the master activator of IncC conjugative transfer genes (29). Similar levels of exclusion were also observed using the ΔA, ΔB, and ΔC mutants (EI > 120), showing that none of those three regions contains exclusion genes. Although the frequency of transfer of pClo was not restored to the level observed in using a plasmid-free recipient strain, exclusion was reduced when the recipient carried either the ΔD (EI = 29) or ΔE (EI = 18) mutants of pVCR94Sp (Fig. 1B). This result indicates that the absence of region D or region E in the recipient facilitates the transfer of pClo, either by directly abolishing an exclusion mechanism or by destabilizing pVCR94Sp in the recipient cells, thereby weakening exclusion.
Since a single open reading frame, vcrx145, is present in the deletion region ΔE, further experiments were carried out to determine its function. By analogy with SXT/R391 ICEs and F-like plasmids, this small ORF downstream of traG (Fig. 1A) was recently proposed as a potential candidate for an entry exclusion determinant in IncC and IncA plasmids (26). vcrx145 was cloned under the control of the arabinose-inducible PBAD promoter and introduced into a tetracycline-resistant (Tcr) E. coli recipient strain to test whether its expression was sufficient to mediate exclusion. The frequency of transfer of pVCR94Sp to the recipient strain expressing vcrx145 was considerably lower (EI = 2,604) than the frequency of transfer to the same recipient strain in the absence of arabinose (Fig. 1C). Therefore, expression of vcrx145 in recipient cells is sufficient to mediate entry exclusion of pVCR94Sp. On the basis of these observations and evidence presented below, we renamed vcrx145 “eexC” (for “entry exclusion of IncC plasmids”).
TraG is the entry exclusion factor in donor cells.
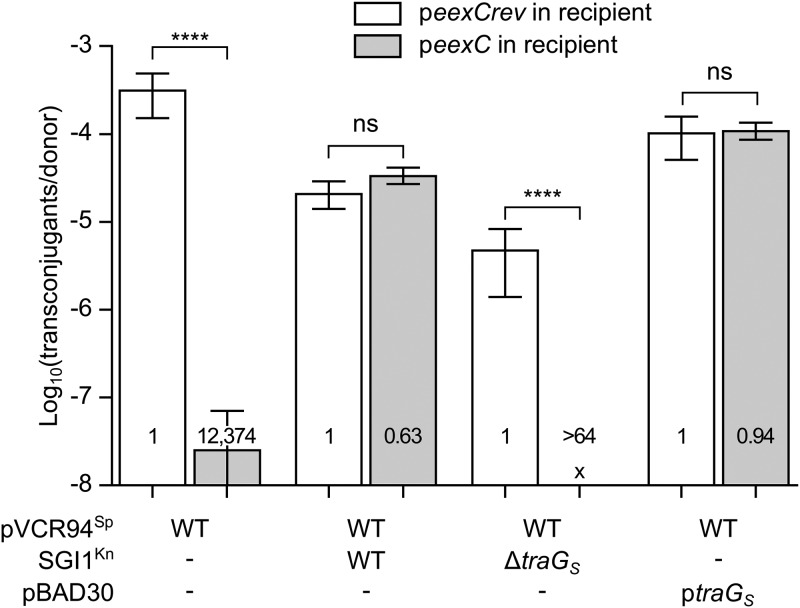
We have previously reported that SGI1 encodes three T4SS subunits, namely, TraNS, TraHS and TraGS, that are expressed under the control of the IncC-encoded AcaCD master activator (27). We found that despite weak relatedness (37% identity), the TraGS subunit encoded by SGI1 displaces the IncC-encoded TraGC in the T4SS of IncC plasmids (27). Consequently, SGI1 transfers efficiently between donor and recipient cells that both carry an IncC plasmid. This observation suggests that TraGC in the donor cells and EexC in the recipient cells are interacting partners that together mediate entry exclusion. To test this hypothesis, we monitored the transfer of pVCR94Sp from donor strains carrying or devoid of a chromosomal copy of SGI1Kn into a recipient strain expressing eexC. SGI1Kn is a kanamycin-resistant derivative of SGI1 that lacks the multidrug resistance complex integron In104 (27).
When SGI1Kn was present in the donor strain, exclusion was deficient (EI = 0.63), as pVCR94Sp transfer rates were virtually identical between the recipients expressing eexC and those not expressing eexC (Fig. 2). In contrast, when SGI1KnΔtraGS was used in the donor, transfer of pVCR94Sp was excluded by recipients expressing eexC (EI > 64). As reported previously (27), transfer of pVCR94Sp was also impaired by the coresident SGI1Kn (150-fold reduction compared to a donor lacking SGI1Kn). This impairment is likely due to the alteration of the T4SS by the SGI1-encoded TraHS subunit that displaces the pVCR94Sp-encoded TraHC subunit (64% identity between TraH subunits) (27). TraH subunits are localized in the periplasm/outer membrane and predicted to be involved in mating apparatus assembly and stabilization (30). Therefore, to confirm that TraGS alone could alleviate exclusion mediated by EexC interacting with TraGC, we used a donor strain devoid of SGI1Kn and containing ptraGS to express only traGS. When TraGS was produced in the donor, no exclusion was exerted toward the IncC plasmid (EI = 0.94) (Fig. 2). Taken together, these results strongly suggest that TraGC interacts with EexC to mediate exclusion. Considering that, like other F-type Eex/TraG pairs, EexC and TraGC are predicted to be inner membrane proteins (PSORT-B cytoplasmic membrane scores of 10 and 4.12, respectively) and that TraGC is required in the donor while EexC acts in the recipient, the exclusion mechanism is most likely entry exclusion rather than surface exclusion (10, 16, 20).
FIG 2.
TraGS of SGI1 abolishes EexC-mediated IncC entry exclusion. Mating assays were carried out using as donors GG56 bearing pVCR94Sp alone or in association with SGI1Kn or its ΔtraGS mutant or ptraGS. CAG18439 strains bearing peexC or peexCrev (negative control that contains eexC in the reverse orientation relative to the PBAD promoter) were used as recipient strains. Each bar represents the mean of results from three independent experiments, with error bars indicating standard deviations. Exclusion indices (EI) are indicated at the bottom of each bar. Statistical analyses were carried out on the logarithm of the values using one-way ANOVA with Sidak's posttest to compare each bar to its corresponding control. “x” indicates that the frequency of transfer was below the detection limit (<10−8). ****, P < 0.0001; ns, not significant.
Clustering of EexC and TraGC orthologues suggests multiple-entry exclusion groups.
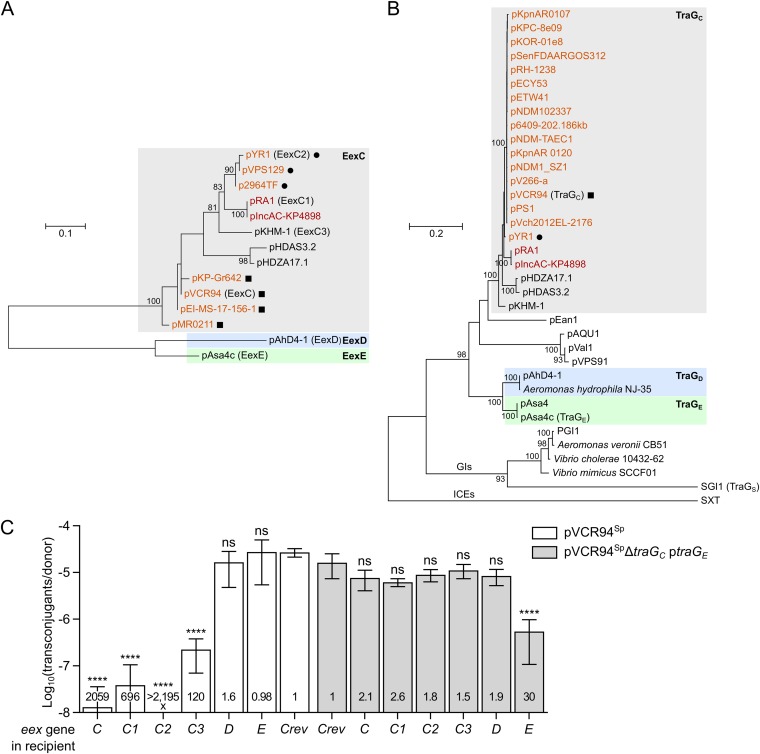
Conjugative elements of the same family can belong to different entry exclusion groups. For instance, F-like plasmids F and R100 do not exclude each other (18). Likewise, cells bearing the ICE SXT inhibit the entry of a second copy of the ICE, but not R391, and vice versa (16). To assess the diversity of entry exclusion groups and determine their specificity among genetic elements related to IncC conjugative plasmids, we searched the GenBank database for EexC and TraGc homologs. In several instances, such as pAhD4-1 of Aeromonas hydrophila, eex loci were not annotated in the nucleotide sequences. Therefore, we performed searches using tblastn (see Data Sets S1 and S2 in the supplemental material). A phylogenetic analysis of the unique representatives of EexC and TraGC homologs was then carried out. In both the TraG and Eex trees, IncA and IncC plasmids clustered together (Fig. 3A and B), which is consistent with recent data confirming that IncC and IncA plasmids exclude each other and therefore belong to the same exclusion group (26). In fact, Eex homologs encoded by all IncC and IncA plasmids seem to form a unique lineage of EexC-like proteins that also contain putative Eex proteins encoded by untyped plasmids pHDAS3.2 and pHDZA1.1 of the aphid endosymbiont “Candidatus Hamiltonella defensa” and pKMH-1 of Citrobacter freundii (Fig. 3A). This analysis also revealed that plasmids such as pVCR94, pMR0211, pKP-Gr642, and pEI-MS-17-156-1, which code for strictly identical TraGC proteins (Fig. 3B; see also Data Set S1), can encode relatively divergent EexC proteins (Fig. 3A), suggesting that the small eex genes are evolving at a higher rate than their cognate traG partners. However, eexC genes exhibit very little diversity among IncC plasmids, as more than 96% of these plasmids in our data set (254 of 263) encode the same EexC protein as pVCR94 (Data Set S2). A strictly identical EexC protein is also encoded by non-IncC elements, such as the hybrid IncC/IncX3 plasmid pSL131_IncA/C-IncX3 of Salmonella enterica subsp. enterica serotype Lomita and the IncFIB plasmid pYT3 of S. Typhimurium, or by those present on the chromosome of Providencia stuartii, S. Newport, and S. Typhimurium (Data Set S2). The 8 IncA plasmids included in our data set encode either the EexC1 variant or the EexC4 variant, which seem to be specific to IncA plasmids, as neither variant was found to be encoded by any IncC plasmids (Data Set S2). Nevertheless, we predict that all the conjugative elements cited above belong to entry exclusion group C (EexC/TraGC).
FIG 3.
Multiple-entry exclusion groups among IncC-related conjugative plasmids. (A and B) Maximum likelihood phylogenetic analysis of Eex (A) and TraG (B) homologs. Trees with the highest log likelihoods (−987.74 and −16,264.82 for Eex and TraG, respectively) are shown. Bootstrap supports are indicated as percentages at the branching points only when >80%. Branch length represents the number of substitutions per site over 114 and 1,204 amino acid positions for Eex and TraG proteins, respectively. Only one representative per cluster of identical proteins is shown in each tree (Data Sets S1 and S2). IncC and IncA plasmids are shown in orange and red, respectively. Circles and squares indicate plasmids coding for identical TraG proteins. GIs indicates a lineage of TraG proteins encoded by SGI1-like genomic islands. TraG of the ICE SXT (TraG/EexS exclusion system) was used as the outgroup. (C) Experimental confirmation of exclusion groups. Mating assays were performed using as donor strains GG56 bearing either pVCR94Sp or its ΔtraGC mutant with ptraGE. Recipient strains were CAG18439 strains expressing the different putative eex genes or carrying peexCrev used as the nonexclusion control. Each bar represents the mean of results from three independent experiments, with error bars indicating the standard deviations. Exclusion indexes (EI) are indicated at the bottom of each bar. One-way ANOVAs with Dunnett's multiple-comparison test were carried out on the logarithm of the values to compare each bar to the nonexclusion control for each donor. ****, P < 0.0001; ns, not significant.
Other untyped plasmids, including pAsa4-like or pAhD4-1-like plasmids found in Aeromonas species, encode more distantly related Eex and TraG proteins that form distinct lineages (Fig. 3A and B). However, the strong divergence of these pairs of proteins suggests that two distinct entry exclusion groups exist within this lineage: pAsa4c-like plasmids likely form exclusion group E (EexE/TraGE) whereas pAhD4-1-like plasmids would form exclusion group D (EexD/TraGD).
Confirmation of exclusion groups by mating assays.
Since three different pairs of Eex/TraG exclusion partners were identified, we hypothesized that EexC proteins would exclude pVCR94Sp transfer and not pAsa4c transfer and that EexE would exclude pAsa4c transfer and not pVCR94Sp transfer. We also hypothesized that EexD should exclude neither. To confirm the existence of three distinct exclusion groups, we cloned five additional eex genes originating from IncA pRA1 (eexC1), IncC pYR1 (eexC2), and untyped plasmids pKHM-1 (eexC3), pAsa4c (eexE) and pAhD4-1 (eexD), under the control of the PBAD promoter and then assessed whether their expression in a recipient strain excluded the transfer of pVCR94Sp or pAsa4c. Expression of any of the four eexC alleles (including the IncA-derivated eexC1) strongly inhibited or even abolished transfer of pVCR94Sp (EI > 120), confirming that IncC and IncA plasmids belong to exclusion group C (Fig. 3C). In contrast, EexE and EexD were unable to exclude pVCR94Sp, as shown by the low EIs that were not statistically different from the control, confirming that neither protein belongs to exclusion group C.
Direct assessment of pAsa4c exclusion was challenging using E. coli as a host. Despite multiple attempts, intraspecific E. coli transfer of pAsa4c remained near the detection threshold (10−8 transconjugant/donor), even using different donor and recipient strains and temperatures closer to the optimal growth temperature of its original host, the fish pathogen Aeromonas salmonicida subsp. salmonicida. This observation suggests that physiological differences limit the transfer of pAsa4c in E. coli. Therefore, we used instead a donor strain bearing pVCR94SpΔtraGC complemented with traGE of pAsa4c expressed from ptraGE and then monitored the transfer into the recipient strains expressing the different eex genes. Using this traG substitution, we observed that transfer of pVCR94Sp and its ΔtraGC mutant complemented with traGE occurred at frequencies that were indistinguishable (Fig. 3C, compare Crev bars). Moreover, these assays revealed that exclusion of the complemented mutant occurred only when eexE was expressed in the recipient (EI = 30) (Fig. 3C). None of the four eexC variants or eexD was able to exclude pVCR94SpΔtraGC complemented with traGE (EI ranging between 1.5 and 2.6). This result confirms that EexC and TraGC of the IncA and IncC plasmids form an entry exclusion group distinct from the EexE/TraGE system of untyped pAsa4c. Furthermore, since neither TraGC nor TraGE showed an interaction with EexD from pAhD4-1 (EI = 1.6 or 1.9, respectively), this plasmid likely belongs to a third distinct entry exclusion group.
The variable C terminus of EexC defines the specificity of entry exclusion.
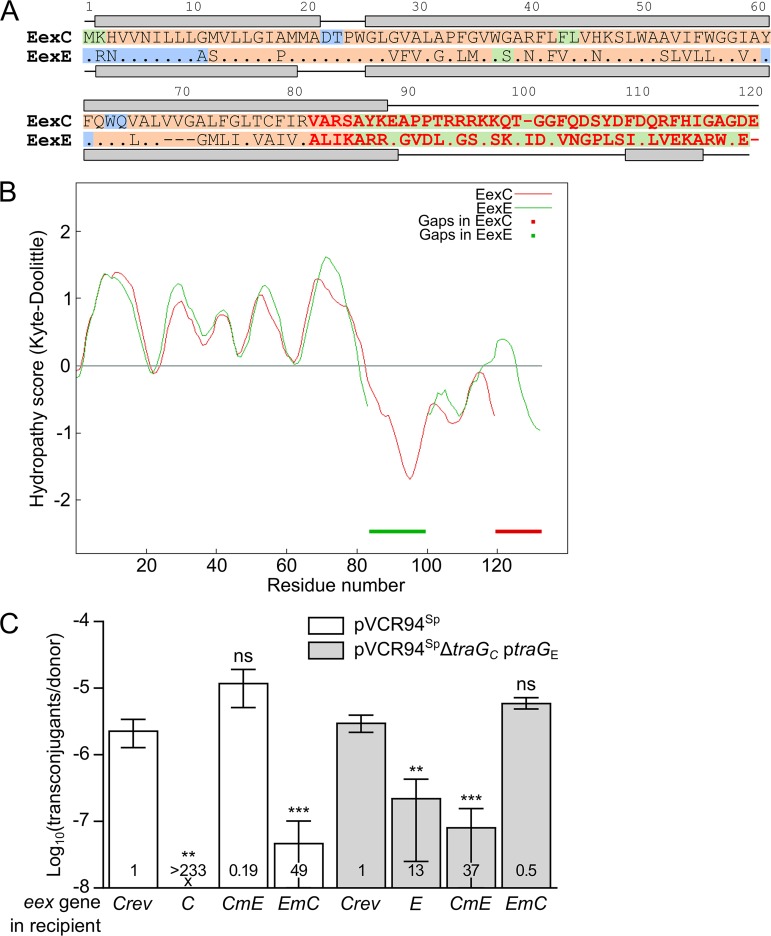
BLASTP alignment of EexC and EexE revealed that the two proteins share only 52% identity over 83% coverage. However, the 62 amino acid (aa) residues of the N terminus of both proteins share 69% identity. The predicted secondary structures of both N termini are strikingly similar and consist of an α-helix folding that is predicted to be located in the inner membrane (PSORTb cytoplasmic membrane scores of 10.0 and 9.82 for EexC and EexE, respectively) (Fig. 4A). Comparison of the hydropathy profiles of the two proteins also shows strikingly similar features, particularly over the 80 N-terminal amino acid residues (Fig. 4B). These observations suggest that specificity of entry exclusion is defined by the highly variable C terminus of both Eex proteins. To test this hypothesis, we swapped the C termini of EexC at position 82 and EexE at position 79 (Fig. 4A) and then assessed whether the expression of the two resulting chimeric genes, EmC and CmE, in a recipient strain would exclude pVCR94Sp or the corresponding ΔtraGC mutant complemented with traGE. We observed that CmE, which contains the C terminus of EexE, did not exclude pVCR94Sp (EI = 0.19) whereas it excluded pVCR94SpΔtraGC complemented with traGE (EI = 37) (Fig. 4C). Conversely, EmC, which contains the C terminus of EexC, excluded pVCR94Sp (EI = 49) but not pVCR94SpΔtraGC complemented with traGE (EI = 0.5). Furthermore, exclusion activities mediated by the C terminus of the chimeras toward the cognate TraG were comparable to those seen with the native proteins (Fig. 4C; compare C to EmC and E to CmE). Together, these results confirm that the C termini of EexE and EexC define entry exclusion specificity.
FIG 4.
Localization of the specificity domain of Eex proteins. (A) MUSCLE alignment of EexC and EexE. EexC was used as the reference, and identical amino acids in EexE are represented by a dot. Predicted secondary structures are shown above and below the sequence of each protein, with lines and gray boxes representing coils and helices, respectively. Background colors indicate amino acid residues predicted to be cytoplasmic (green) or periplasmic (blue) or present within a transmembrane helix (orange). Variable C termini of EexC and EexE shown in red were swapped to construct the chimeric proteins CmE and EmC. (B) Aligned Kyte-Doolittle hydropathy plots of EexC and EexE. Thick lines indicate alignment gaps in EexC and EexE. (C) Specificity of Eex chimeras toward TraGC and TraGE. Mating assays were performed using as the donor strains GG56 bearing either pVCR94Sp or its ΔtraGC mutant with ptraGE. All recipient strains were derivatives of CAG18439 expressing eexC or eexE or either of the two chimeras eexCmE and eexEmC. CAG18439 bearing peexCrev was used as the nonexclusion control. Each bar represents the mean of results from three independent experiments, with error bars indicating the standard deviations. Exclusion indexes (EI) are indicated at the bottom of each bar. One-way ANOVAs with Dunnett's multiple-comparison test were carried out on the logarithm of the values to compare each bar to the nonexclusion control for each donor. ***, P < 0.001; **, P < 0.01; ns, not significant.
DISCUSSION
Entry exclusion is a feature conserved among conjugative plasmids of different incompatibility groups and has also been reported in one family of ICEs to date (20). Entry exclusion has been proposed to confer immunity to lethal zygosis, i.e., death of recipient cells resulting from exposure to an excess of donor cells that could increase cell permeability by inducing extensive damage in the recipient membrane. It also likely increases donor fitness by limiting excessive energy consumption due to transfer to another donor. Moreover, entry exclusion improves plasmid stability by impeding entry of highly similar, incompatible replicons that can lead to plasmid displacement and loss. IncC and IncA plasmids were long suspected to exclude each other’s entry, but the genetic determinants responsible for exclusion were unknown (22–25). Confusion between exclusion and incompatibility has plagued the nomenclature and typing of these plasmids since the late 1980s (26, 31). Nevertheless, Ambrose et al. (26) recently confirmed that IncA and IncC replicons are compatible and that the apparent incompatibility between these two plasmid groups most likely resulted from entry exclusion. Here, we identified the genes involved in entry exclusion of IncA and IncC plasmids and showed that the product of eexC in recipient cells recognizes the cognate TraGC protein in donor cells, exerting entry exclusion toward both IncC and IncA plasmids. Therefore, the C entry exclusion group spans at least two different plasmid incompatibility groups, a discrepancy that led to the grouping of IncA and IncC plasmids into the incorrect and now obsolete “IncA/C” group, alias the “A/C” group (1, 26). Moreover, at least one IncFIB plasmid, pYT3, also seems to belong to the C exclusion group (see Data Sets S1 and S2 in the supplemental material).
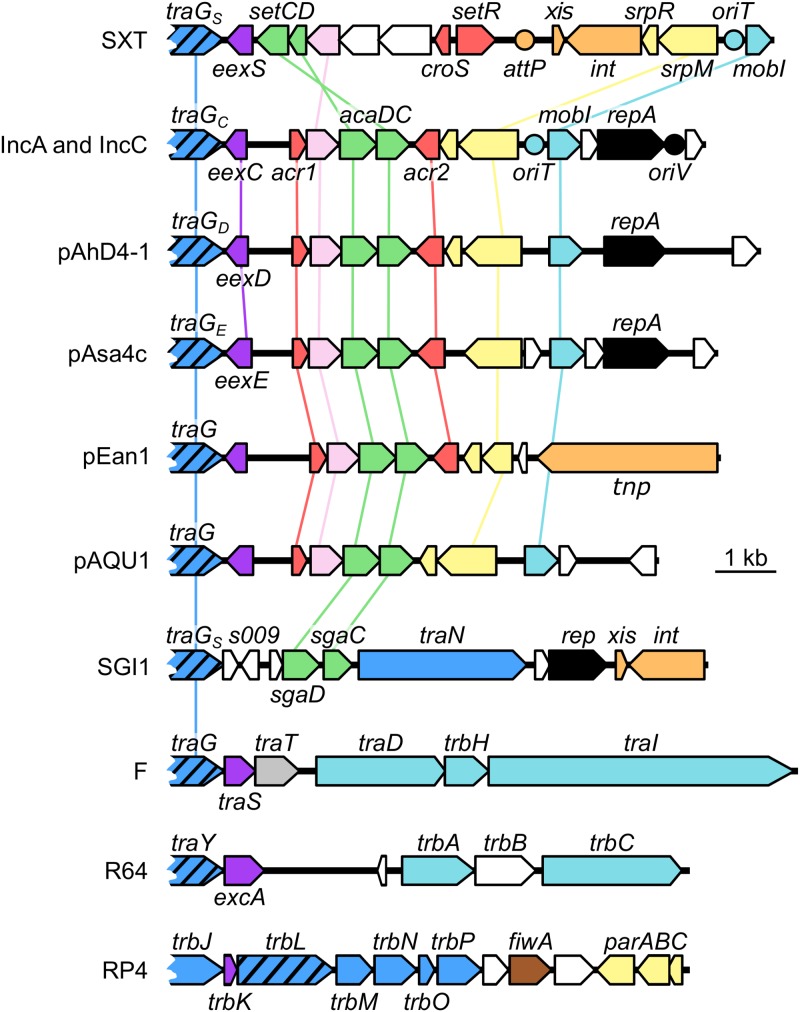
The identification of EexC revealed the existence of homologous exclusion proteins, such as EexE and EexD, that are encoded by untyped conjugative plasmids that are related either closely (e.g., pAsa4c) or more distantly (e.g., pAhD4-1) to IncA and IncC plasmids. These Eex proteins define additional exclusion groups that do not exclude IncC and IncA plasmids. Furthermore, several genomic islands and plasmids such as pEan1 or pVPS91 were found to code for closely related TraG homologs and yet encode predicted Eex proteins related distantly (<40% identity) to EexC, EexD, or EexE (Fig. 3A and B; see also Fig. 5). Consistent with this observation, EexC has no detectable sequence similarity to EexS of SXT, TraS of the F-like plasmids F and R100, or ExcA of the IncI1/IncIγ plasmids R64 and R621a (Fig. 5). The lack of homology between Eex proteins from one entry exclusion system and those from another suggests that eex genes evolve faster than most tra genes, including virB6 homologs such as traG (20). According to Sakuma et al. (12), the association of VirB6-like proteins within a high-order multimeric structure that requires interactions with several other transfer proteins in the T4SS imposes a higher level of conservation than Eex proteins, which likely interact only with their cognate TraG partner. While SGI1 TraGS is a functional substitute for IncC TraGC despite being only 37% identical, no entry exclusion gene has been identified in SGI1 to date. Nevertheless, we predict that SGI1 s009 codes for an Eex protein on the basis of its location relative to traGS and of the partial identity of its translation product with F TraS (38% identity and 69% similarity of aa 45 to 57 of S009 to aa 50 to 62 of TraS) (Fig. 5).
FIG 5.
Comparison of the genetic contexts of entry exclusion genes in diverse mobile genetic elements. A schematic representation of demonstrated or putative exclusion loci of the ICE SXT, IncC plasmid pVCR94; untyped plasmids pAhD4-1, pAsa4c, pEan1 and pAQU1; genomic island SGI1; IncFI plasmid F; IncI1 plasmid R64; and IncPα plasmid RP4 is shown. Arrows of identical colors represent genes predicted to have similar functions and are color coded as follows: dark blue, T4SS subunit; hatched blue, VirB6-like subunit; light blue, DNA processing; violet, entry exclusion; gray, surface exclusion; green, transcriptional activator; pink, lytic transglycosylase; red, transcriptional repressor; yellow, partition; orange, site-specific recombination and transposition; black, replication; white, unknown function. Circles indicate the position of origins of transfer (oriT), of the origin of replication (oriV), of pVCR94 based on identity with pRA1, and of the attP site used for chromosomal integration of SXT by site-specific recombination.
Entry exclusion systems of IncC-like, F-like, and IncI1/IncIγ plasmids as well as SXT/R391 ICEs exhibit striking functional similarities. In all these systems, exclusion involves the VirB6-like protein TraG (or TraY for IncI1/IncIγ plasmids) in the donor and Eex in the recipient (12, 16, 18). F and SXT TraG proteins have been shown to be polytopic inner membrane proteins, with large hydrophilic C-terminal domains that are predicted to localize in the cytoplasm (10, 16). Like TraY (12), TraGC is predicted to be an inner membrane protein. Though phylogenetically unrelated, F TraS, R64 ExcA, and SXT EexS have all been shown or predicted to be inner membrane proteins (10, 12, 16). Like their counterparts, EexE, EexD, and EexC are predicted to be bound to the inner membrane. The segments of TraGS and EexS of SXT that mediate entry exclusion specificity were shown to be cytoplasmic, thereby suggesting a model of entry exclusion in which a cytoplasmic segment of TraG would be translocated into the recipient cell cytoplasm (32). Indeed, exclusion specificity was shown to be controlled by the C-terminal parts of SXT EexS and of closely related R391 EexR and only three amino acid residues (aa 606 to 608) located in the cytoplasmic, central region of their cognate TraG (16, 32). Precise mapping of the amino acid residues that control exclusion specificity was facilitated by the low divergence between TraGS and TraGR (98% identity) (16). The TraS exclusion proteins of the F-like plasmids F and R100 share only 17% identity and lack regions of higher dissimilarity to aid in predicting the features that are recognized by TraG (10, 18). Moreover, attempts to construct stable TraS chimeras failed, preventing the identification of the region of TraS responsible for exclusion specificity. Audette et al. (10) were, however, able to locate the segment of TraG that interacts with TraS within a discrete segment (aa 610 to 673) on the basis of its lower conservation (55.7% identity versus 93% overall identity) and the use of TraG chimera proteins. For IncC-like plasmids, TraGC and TraGE share only 76% overall identity, with 89% identity over the predicted inner membrane segment (aa 1 to 456) and only 68% identity over the predicted cytoplasmic segment (aa 457 to 752). Poor conservation of TraG cytoplasmic segments could hamper the mapping of the amino acid residues interacting with Eex, especially if different segments of TraGC and TraGE are recognized by their cognate EexC and EexE proteins. Such a feature has been reported for the entry exclusion system of IncI1/IncIγ plasmids. Indeed, the internal variable segment of R64 TraY has been shown to recognize the R64 ExcA whereas the C-terminal variable segment of R621a TraY recognizes R621a ExcA (12). Here, we mapped the region that controls exclusion specificity to the C-terminal third of EexC and EexE. Swapping of shorter fragments could help identify the exact amino acid residues responsible for entry exclusion specificity. However, the strong divergence between the C-terminal regions of EexC and EexE (Fig. 4A) suggests that these amino acid residues might not be equivalent or even contiguous in both proteins, thereby precluding precise identification by fragment swapping. Alternative methods such as cross-linking or immunoprecipitation could be attempted, although such approaches have not been successful in attempts to decipher the interactions between TraS and TraG of F and R100 (10). Comparisons of structural data and three-dimensional (3D) models of EexC and EexE, which are not currently available, would help shed new light on the specific interactions between Eex proteins and their cognate TraG partners and provide a better understanding of the mechanism of entry exclusion, which remains to be established.
We found an unexpected lack of diversity of eexC variants in IncC plasmids, considering the diversity of geographical locations and bacterial species from which the plasmids of our data set originated. IncC plasmids have been typed in two main groups, type 1 and type 2, based on differences within the resistance gene clusters or loss of backbone regions adjacent to them (1). Hybrid plasmids that share features of both types have also been described, as well as subtypes 1a and 1b, which differ by the presence or absence of a 14.5-kb diverged segment (31). We found no clear correlation between IncC plasmid type and the variant of the eexC gene that they carry. In fact, plasmids of all IncC types and subtypes most frequently carry the variant eexC (Data Set S2). Among 280 IncC plasmids, only 12 distinct EexC variants were found. In striking contrast, SXT/R391 ICEs encode a much more diverse set of Eex proteins, with two exclusion groups, S and R (16, 33). Among 31 ICEs of the S group, 18 distinct EexS variants have been described to date. Among those ICEs, 32% encode the same EexS2 protein (19, 33). A total of 19 distinct EexR variants have also been described among 31 ICEs of the R group, and the EexR1 protein variant is encoded by a small majority of only 19% of these ICEs. One hypothesis that could explain the different distribution patterns of eexC and eexS/R variants proposes a relatively clonal expansion of drug resistance-associated IncC plasmids in pathogenic species. In contrast, acquisition of SXT/R391 ICEs seems to have occurred multiple times in recent history from diverse reservoirs, especially in the seventh-pandemic V. cholerae lineage (34).
In our setup using pVCR94 derivatives, we observed that overexpression of different eexC alleles in recipients yielded different levels of exclusion effects on traGC, ranging from complete reduction (eexC2) to only 120-fold reduction (eexC3) (Fig. 3C). This suggests that not all pairs of eexC and traGC alleles confer the same levels of exclusion. A similar observation was reported for the ICE R391, which was strongly excluded by a recipient expressing the variant EexR1 and weakly excluded when variant EexR or variant EexR3 was used (19). Assessment of the exclusion strength of the 12 distinct eexC variants identified here against each of the traGC variants identified in IncC plasmids could provide valuable information on the interactions between these two proteins and the amino acid residues involved in recognition and the specificity of exclusion. pVCR94SpΔtraGC, which can be complemented by traG genes as distant as traGE or traGS, could be an interesting platform for such a study to prevent inconsistency due to the variable segments present in different IncC plasmids.
In experiments reported by Ambrose et al. (26), assays of mating between strains bearing the IncA plasmid pRA1 (eexC1/traGC1) and the IncC plasmid pRMH760 (eexC/traGC) did not yield any transconjugants, thereby suggesting complete exclusion. In contrast, Datta and Hedges (24) reported that pRA1 in a recipient excluded transfer of pIP40a (eexC/traGC) by only about 10-fold. Clearly, different plasmids carrying identical eex/traG pairs do not exert similar levels of exclusion. This observation suggests that other IncC-encoded factors may influence exclusion. This hypothesis is supported by results from our exclusion assay performed using the ΔD mutant of pVCR94Sp. We found that pClo mobilization was more efficient toward a recipient bearing pVCR94SpΔD or pVCR94SpΔE (ΔeexC) but that neither deletion was sufficient to restore the observed rate of transfer toward an empty recipient (Fig. 1B), thereby suggesting that the eexC/traGC system is not the only exclusion system encoded by IncC plasmids. One or several of the 57 genes that contained the missing ΔD segment seem to reduce the transfer of pClo when present in the recipient strain. This reduction could have been the result of surface exclusion, like that mediated by TraT of plasmid F (35). However, since IncC plasmids do not seem to encode any TraT homolog, further investigations of the phenotype resulting from the ΔD mutation will be necessary to decipher its mechanism and how this observation relates to data published by others (24, 26).
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are described in Table 1. Strains were routinely grown in lysogeny broth (LB-Miller; EMD) at 37°C in an orbital shaker/incubator and were preserved at −80°C in LB broth containing 15% (vol/vol) glycerol. The following antibiotics and concentrations were used: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; kanamycin (Kn), 50 μg/ml; nalidixic acid (Nx), 40 μg/ml; spectinomycin (Sp), 50 μg/ml; tetracycline (Tc), 12 μg/ml; rifampin (Rf), 50 μg/ml.
Mating assays.
Conjugation assays were performed by mixing 100 μl of donor cells and 100 μl of recipient cells (typically ∼2 × 109 cells/ml each) that were grown overnight in LB broth at 37°C with suitable antibiotics to ensure retention of plasmids and/or SGI1 derivatives. Cells were pelleted by centrifugation for 3 min at 1,200 × g, washed once in 200 μl of LB broth, and resuspended in 10 μl of LB broth. Mating mixtures were then deposited as drops on LB agar plates and incubated at 37°C for 2 h. The cells were recovered from the plates in 800 μl of LB broth, subjected to vortex mixing, and diluted via serial 10-fold dilutions before plating on LB agar containing suitable antibiotics was performed. Donor, recipient, and transconjugant colonies were selected using antibiotics as described in corresponding figure legends. To induce the expression of traG and eex genes, as well as eex chimeric genes, in complementation or expression assays, mating was carried out on LB agar plates with 0.2% arabinose. Frequencies of transfer were expressed as the number of transconjugant CFU per donor CFU from data obtained from three independent mating experiments. Exclusion indexes (EI) were calculated as the ratio of an element’s frequency of transfer to a plasmid-free recipient to its frequency of transfer to the tested recipient, as defined elsewhere (16, 20).
Molecular biology methods.
Plasmid DNA was extracted using an EZ-10 spin column plasmid DNA miniprep kit (Bio Basic) according to the manufacturer’s instructions. All enzymes used in this study were purchased from New England Biolabs. PCR assays were performed with the primers described in Table 2. PCR conditions were as follows: (i) 3 min at 94°C for initial denaturation; (ii) 30 cycles of 10 s at 98°C for denaturation, 30 s at the appropriate annealing temperature, and 30 s/kb at 72°C for elongation; and (iii) 5 min at 72°C for the final elongation step. When necessary, PCR products were purified using an EZ-10 spin column PCR products purification kit (Bio Basic) according to the manufacturer’s instructions. E. coli was transformed by electroporation as described by Dower et al. (36) in a Bio-Rad GenePulser Xcell apparatus set at 25 μF, 200 V, and 1.8 kV using 1-mm-gap electroporation cuvettes.
TABLE 2.
DNA sequences of the primers used in this study
| Primer name | Nucleotide sequence (5′ to 3′)a |
|---|---|
| vcrx076del.f | CTTATTTGCTCAAAAAGGGCACTTCCACAGCCCATGCGGAATAGGAACTTCAAGAT |
| vcrx076del.r | TCTGAACCGGGCTTTGAGAGGAATGAACCTTAGTTCAGGAACTTCAGAGCGCTT |
| 94del145.for | ATGCCGTGGTAAACTGGAGGTAAAGAGTAAGGGGGTTGATgtgtaggctggagctgcttc |
| 94del145.rev | ACCAACCAAATAATAAGGGGGCCAGCAGGCCCCCTTATTACATATGAATATCCTCCTTA |
| 94vcrx145.for | NNNNNNGAATTCAAGGAGGAATAATAAATGAAACATGTGGTCAATATTCTT |
| 94vcrx145.rev | NNNNNNGAATTCTTATTCGTCTCCAGCTCCAA |
| pAsa4cTrxA.f | GTACGAATTCAAGGAGGAATAATAATTGAGGACCTGCCTGTAT |
| pAsa4cTrxA.r | AGGTGAATTCTCATTCACCCCAACGAGCT |
| ptrxAmut.f | ATGAGGAATGTGGTTAAC |
| ptrxAmut.r | TTATTATTCCTCCTTGAATTCG |
| pAsa4cTraG.f | NNNNNNGAATTCAAGGAGGAATAATAAATGGGTACGTTCACAATCTA |
| pAsa4cTraG.r | NNNNNNGAATTCTTAATGCTTCGAATCTAAGCCT |
| eexC183.f | TGGTTGCCATTGTTCGTGCCGCGCGGTCAGCATATAAAGAG |
| eexC279.r | CTCTTTATATGCTGACCGCGCGGCACGAACAATGGCAACCA |
| eexC280.f | CTAACGTGCTTTATCCGCGTTCTCATAAAGGCGAGAAGAGAG |
| eexC182.r | CTCTCTTCTCGCCTTTATGAGAACGCGGATAAAGCACGTTAG |
| ptrxA2.f | GTCAGAATTCAAGAGGGAGAGCGTGATGAG |
| toxdel-F | GGTCTGCAATTGGGATCATTACTTCATAAAACCTTTTCTCGTGTAGGCTGGAGCTGCTTC |
| toxdel-R | CAACATGAAAGAGCTTCGGGTCCAAAGCAAAGGAGATGCCATGGTCCATATGAATATCCT |
| Del1-Kan-F | CTGCACGAACACCAGATCTGATATGTGGGAACGCTGCACGAAGATCCCCTCACGCTGCCG |
| Del1-Kan-R | GTGCCTCTAACGCCCCCGAGCAAAAGCCTCAGCCCCAGATCAGAAGAACTCGTCAAGAAG |
| Alt-del2-F | TGCATCTGTGCGACATCCCATTGAATGTCTACCAGGTGATAAGATCCCCTCACGCTGCCG |
| new-Alt-del2-R | CCGAATGTTTTTCCGAAGCCGCAACGCTAACTTCATTGGCTCAGAAGAACTCGTCAAGAA |
| new-del4-F | ACTCTGCTTTTCCCTGATCAAGAAGGTAAGGGAAAGCAACGTCGTGACTGGGAAAACCCT |
| new-del4-R | CTTCTGTCGCTTCTGCGGCCATCTTCATCTGCTTCTCGACACTCCAGCATATTCAGCGAC |
EcoRI restriction sites are underlined.
Plasmid and strain construction.
Strains, plasmids, and genomic islands used in this study are listed in Table 1. Primers are listed in Table 2.
Deletions ΔA to ΔE in pVCR94Sp were constructed using the one-step chromosomal gene inactivation technique with pSIM6 (37). ΔC and ΔE deletions were obtained using primer pair vcrx076del.f/vcrx076del.r and primer pair 94del145.for/94del145.rev, respectively, and pKD4 or pKD3 as the templates. ΔA, ΔB, and ΔD deletions were constructed in two steps. First, the vcrx028 toxin gene was deleted using primer pair toxdel-F/toxdel-R and pKD4 as the template. The FLP recombination target (FRT)-flanked kanamycin resistance cassette was then removed by Flp-catalyzed excision using pE-FLP (38). Then, deletions ΔA, ΔB, and ΔD were obtained using primer pair Del1-Kan-F/Del1-Kan-R, primer pair Alt-del2-F/new-Alt-del2-R, and primer pair new-del4-F/new-del4-R, respectively, and pKD4 (deletions ΔA and ΔB) or pTT01 (deletion ΔD) as the template. All deletions were verified by PCR and antibiotic resistance profiling.
Plasmids peexC and peexCrev were constructed by PCR amplification of eexC using EcoRI-containing primer pair 94vcrx145.for/94vcrx145.rev and pVCR94Sp as the template followed by EcoRI digestion of the amplified fragment and then cloning into EcoRI-digested pBAD30. Forward (peexC) and reverse (peexCrev) orientations of eexC relative to the PBAD promoter were verified by AfeI/AflII digestion. Plasmids peexE and ptraGE were constructed by PCR amplification of eexE and traGE using pAsa4c as the template and EcoRI-containing primer pair pAsa4cTrxA.f/pAsa4cTrxA.r and primer pair pAsa4cTraG.f/pAsa4cTraG.r, respectively. The amplified fragments were then digested by EcoRI and cloned into EcoRI-digested pBAD30. peexE was subjected to site-directed mutagenesis to remove a 78-bp untranslated region with primer pair ptrxAmut.f/ptrxAmut.r using Q5 site-directed mutagenesis (New England BioLabs) according to the manufacturer’s instructions.
eexD, eexC1, eexC2 and eexC3 were obtained by DNA synthesis (Bio Basic) of the respective open reading frames from pAhD4-1 (CP013966.1), pRA1 (FJ705807.1), pYR1 (CP000602.1), and pKHM-1 (AP014939.1) preceded by a Shine-Dalgarno sequence (5′-AAGGAGGAATAATAA-3′) and flanked by EcoRI and SalI sites. DNA fragments provided in pUC57-Amp were digested by EcoRI and SalI and were then cloned into EcoRI/SalI-digested pBAD30 (39).
Chimeric genes CmE and EmC were constructed using the gene splicing by overlap extension technique (40). CmE fragments were PCR amplified with primer pair 94vcrx145.for/eexC182.r and primer pair eexC280.f/pAsa4cTrxA.r using pVCR94Sp and pAsa4c as the templates, respectively. EmC fragments were PCR amplified with primer pair ptrxA2.f/eexC279.r and primer pair eexC183.f/94vcrx145.rev using pAsa4c and pVCR94Sp as the templates, respectively. After extension and PCR amplification, the DNA fragments containing the chimeric genes were digested with EcoRI and cloned into EcoRI-digested pBAD30.
All constructions were verified by sequencing performed by the Plateforme de Séquençage et de Génotypage du Centre de Recherche du CHUL (Québec, QC, Canada).
Phylogenetic analyses.
Primary sequences of Eex and TraG homologs were obtained by using the NCBI’s tblastn algorithm against the nr/nt database restricted to Gammaproteobacteria (taxid: 1236). Only primary sequences sharing more than 40% identity with EexC and 50% identity with TraGC over 80% minimum coverage were included in subsequent analyses. Since the SGI1 TraGS and SXT TraG proteins do not fit this criterion (37% identity with TraGC over 99% coverage [27, 41]), both were included manually in the TraG data set as an outgroup. Eex and TraG primary sequences were first clustered with CD-HIT (42) (sequence identity cutoff of 1) prior to alignment. Exhaustive data on the composition of these clusters are available in Data Set S1 in the supplemental material.
Evolutionary analyses of Eex or TraG proteins were performed within MEGA7 (43), and data were inferred by using the maximum likelihood (PhyML) (44) method based on the JTT matrix-based (Eex) and Whelan and Goldman (WAG) + Freq. (TraG) models (45, 46). Phylogenetic analyses were computed using amino acid alignments generated by MUSCLE (47). Poorly aligned regions were removed using trimAl v1.3 software and the automated heuristic approach (48) prior to phylogenetic analyses. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with the superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 3.0102]) for the TraG phylogenetic tree.
In silico typing of plasmids.
Replicon types of plasmids listed in Data Sets S1 and S2 were determined using PlasmidFinder 2.0 (49) and the Enterobacteriaceae database (95% identity, 60% coverage).
Protein secondary structure prediction.
The secondary structures of EexE and EexC were predicted using PSIPRED protein sequence analysis workbench v3.3 (50). Subcellular localization was predicted using PSORT-B v3.0 (51). Transmembrane protein topology was predicted using MEMSAT3 (52). Comparisons of the aligned Kyte-Doolittle hydropathy plots of EexC and EexC were generated using AlignMe with the Fast method (53).
Supplementary Material
ACKNOWLEDGMENTS
We thank Steve Charette from Université Laval for the kind gift of pAsa4c. We are grateful to Nicolas Carraro and Kevin Neil for technical assistance and Alain Lavigueur for critical reading of the manuscript.
This work was supported by a Discovery Grant (2016-04365) from the Natural Sciences and Engineering Council of Canada (NSERC) and a Project Grant (PJT-153071) from the Canadian Institutes of Health Research (CIHR) to V.B. K.T.H. was supported by a postdoctoral fellowship (SPE20170336797) from the Fondation pour la Recherche Médicale (FRM, France).
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00731-18.
REFERENCES
- 1.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Weill F-X, Domman D, Njamkepo E, Tarr C, Rauzier J, Fawal N, Keddy KH, Salje H, Moore S, Mukhopadhyay AK, Bercion R, Luquero FJ, Ngandjio A, Dosso M, Monakhova E, Garin B, Bouchier C, Pazzani C, Mutreja A, Grunow R, Sidikou F, Bonte L, Breurec S, Damian M, Njanpop-Lafourcade B-M, Sapriel G, Page A-L, Hamze M, Henkens M, Chowdhury G, Mengel M, Koeck J-L, Fournier J-M, Dougan G, Grimont PAD, Parkhill J, Holt KE, Piarroux R, Ramamurthy T, Quilici M-L, Thomson NR. 2017. Genomic history of the seventh pandemic of cholera in Africa. Science 358:785–789. doi: 10.1126/science.aad5901. [DOI] [PubMed] [Google Scholar]
- 3.Miller RA, Harbottle H. 2018. Antimicrobial drug resistance in fish pathogens. Microbiol Spectr 6. doi: 10.1128/microbiolspec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 5.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 6.Carraro N, Rivard N, Ceccarelli D, Colwell RR, Burrus V. 2016. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. mBio 7:e00509-16. doi: 10.1128/mBio.00509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP. 1969. Extrachromosomal inheritance in bacteria. Bacteriol Rev 33:210–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandran Darbari V, Waksman G. 2015. Structural biology of bacterial type IV secretion systems. Annu Rev Biochem 84:603–629. doi: 10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- 9.Arutyunov D, Frost LS. 2013. F conjugation: back to the beginning. Plasmid 70:18–32. doi: 10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. 2007. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153:442–451. doi: 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- 11.Gunton JE, Ussher JER, Rooker MM, Wetsch NM, Alonso G, Taylor DE. 2008. Entry exclusion in the IncHI1 plasmid R27 is mediated by EexA and EexB. Plasmid 59:86–101. doi: 10.1016/j.plasmid.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Sakuma T, Tazumi S, Furuya N, Komano T. 2013. ExcA proteins of IncI1 plasmid R64 and IncIγ plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid 69:138–145. doi: 10.1016/j.plasmid.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Pohlman RF, Genetti HD, Winans SC. 1994. Entry exclusion of the IncN plasmid pKM101 is mediated by a single hydrophilic protein containing a lipid attachment motif. Plasmid 31:158–165. doi: 10.1006/plas.1994.1017. [DOI] [PubMed] [Google Scholar]
- 14.Bolland S, Llosa M, Avila P, de la Cruz F. 1990. General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J Bacteriol 172:5795–5802. doi: 10.1128/jb.172.10.5795-5802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haase J, Kalkum M, Lanka E. 1996. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncP alpha plasmid RP4. J Bacteriol 178:6720–6729. doi: 10.1128/jb.178.23.6720-6729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrero J, Waldor MK. 2005. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell 8:963–970. doi: 10.1016/j.devcel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Christie PJ. 4 August 2016, posting date. The mosaic type IV secretion systems. EcoSal Plus 2016. doi: 10.1128/ecosalplus.ESP-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anthony KG, Klimke WA, Manchak J, Frost LS. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J Bacteriol 181:5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrero J, Waldor MK. 2007. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol 189:3302–3305. doi: 10.1128/JB.01902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcillán-Barcia MP, de la Cruz F. 2008. Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60:1–18. doi: 10.1016/j.plasmid.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 191:4750–4757. doi: 10.1128/JB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta N, Hedges RW. 1973. R factors of compatibility group A. J Gen Microbiol 74:335–337. doi: 10.1099/00221287-74-2-335. [DOI] [PubMed] [Google Scholar]
- 23.Hedges RW, Datta N. 1971. fi− R factors giving chloramphenicol resistance. Nature 234:220. doi: 10.1038/234220b0.4943088 [DOI] [Google Scholar]
- 24.Datta N, Hedges RW. 1972. R factors identified in Paris, some conferring gentamicin resistance, constitute a new compatibility group. Ann Inst Pasteur (Paris) 123:849–852. [PubMed] [Google Scholar]
- 25.Hedges RW. 1974. R factors from Providence. J Gen Microbiol 81:171–181. doi: 10.1099/00221287-81-1-171. [DOI] [PubMed] [Google Scholar]
- 26.Ambrose SJ, Harmer CJ, Hall RM. 2018. Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 96–97:7–12. doi: 10.1016/j.plasmid.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Carraro N, Durand R, Rivard N, Anquetil C, Barrette C, Humbert M, Burrus V. 2017. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet 13:e1006705. doi: 10.1371/journal.pgen.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douard G, Praud K, Cloeckaert A, Doublet B. 2010. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One 5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. 2014. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 31.Ambrose SJ, Harmer CJ, Hall RM. 2018. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 99:40–55. doi: 10.1016/j.plasmid.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Marrero J, Waldor MK. 2007. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol 189:6469–6473. doi: 10.1128/JB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bioteau A, Durand R, Burrus V. 2018. Redefinition and unification of the SXT/R391 family of integrative and conjugative elements. Appl Environ Microbiol 84:e00485-18. doi: 10.1128/AEM.00485-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, Colombo MM, Colwell RR, Balloux F. 2014. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5:e01356-14. doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Achtman M, Kennedy N, Skurray R. 1977. Cell-cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci U S A 74:5104–5108. doi: 10.1073/pnas.74.11.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St-Pierre F, Cui L, Priest DG, Endy D, Dodd IB, Shearwin KE. 2013. One-step cloning and chromosomal integration of DNA. ACS Synth Biol 2:537–541. doi: 10.1021/sb400021j. [DOI] [PubMed] [Google Scholar]
- 39.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 41.Wozniak RAF, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 46.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinforma Oxf Engl 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 51.Gardy JL, Spencer C, Wang K, Ester M, Tusnády GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman F. 2003. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res 31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DT. 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinforma Oxf Engl 23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 53.Stamm M, Staritzbichler R, Khafizov K, Forrest LR. 2014. AlignMe—a membrane protein sequence alignment web server. Nucleic Acids Res 42:W246–W251. doi: 10.1093/nar/gku291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete genome sequence of Escherichia coli BW25113. Genome Announc 2:e01038-14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carraro N, Sauvé M, Matteau D, Lauzon G, Rodrigue S, Burrus V. 2014. Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol 5:44. doi: 10.3389/fmicb.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ceccarelli D, Daccord A, René M, Burrus V. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190:5328–5338. doi: 10.1128/JB.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka KH, Vincent AT, Trudel MV, Paquet VE, Frenette M, Charette SJ. 2016. The mosaic architecture of Aeromonas salmonicida subsp. salmonicida pAsa4 plasmid and its consequences on antibiotic resistance. PeerJ 4:e2595. doi: 10.7717/peerj.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carraro N, Sauvé M, Matteau D, Lauzon G, Rodrigue S, Burrus V. 6 February 2014. Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol. doi: 10.3389/fmicb.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cabezón E, Sastre JI, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254:400–406. [DOI] [PubMed] [Google Scholar]
- 61.Datta S, Costantino N, Court DL. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 62.Matteau D, Pepin M-E, Baby V, Gauthier S, Arango Giraldo M, Knight TF, Rodrigue S. 17 March 2017. Development of oriC-based plasmids for Mesoplasma florum. Appl Environ Microbiol 10.1128/AEM.03374-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.