Abstract
Human p300 is a transcriptional co-activator and a major acetyltransferase that acetylates histones and other proteins facilitating gene transcription. The activity of p300 relies on the fine-tuned interactome that involves a dozen p300 domains and hundreds of binding partners and links p300 to a wide range of vital signaling events. Here, we report on a novel function of the ZZ-type zinc finger (ZZ) of p300 as a reader of histone H3. We show that the ZZ domain and acetyllysine recognizing bromodomain (BD) of p300 play critical roles in modulating p300 enzymatic activity and its association with chromatin. Acetyllysine binding of BD is essential for acetylation of histones H3 and H4, whereas interaction of the ZZ domain with H3 promotes selective acetylation of histone H3K27 and H3K18.
Keywords: p300, ZZ domain, histone, bromodomain, acetylation, HAT, chromatin, PTM
Introduction
P300 is critical in cell growth, differentiation, apoptosis, and DNA repair and is closely linked to many human diseases, cancer in particular1–3. It functions as a scaffolding protein that recruits numerous nuclear components, including transcription factors, co-activators and subunits of the RNA polymerase II complex to chromatin4–6. p300 possesses an intrinsic histone acetyltransferase (HAT) activity and acetylates histones, directly altering chromatin structure and promoting expression of specific genes7–9. This large ~300 kDa evolutionarily conserved enzyme is functionally related to and cooperates with paralogous CBP, and both share the same domain architecture consisting of a diverse set of protein- and DNA-binding modules. Along with the N-terminal nuclear receptor interaction domain (NRID), a transcriptional adaptor zinc-finger 1 (TAZ1), and a kinase-inducible CREB interaction region (KIX), p300 contains a combination of a bromodomain (BD), a RING finger and a plant homeodomain (PHD) finger preceding the catalytic HAT domain (Fig. 1a). The HAT domain is followed by a ZZ-type zinc finger, another TAZ domain (TAZ2), and an interferon-binding domain (IBiD). Though these domains predominantly associate with activators, basal transcription factors, and components of the multi-subunit HAT complexes, some have also been implicated in the control of the p300 catalytic activity10,11. The HAT domain itself is regulated through its autoinhibitory loop (AIL), which in a hypoacetylated form suppresses the catalytic activity but releases the inhibition upon hyper autoacetylation7,12. An additional layer of p300 regulation has been suggested based on the recent structure of the p300 region encompassing BD, RING, PHD and the HAT domain (BRPH), in which the RING finger aids in the inhibition through occluding the HAT active site in conjunction with AIL13. Owing to their crucial role in a magnitude of signaling pathways, biological activities of p300 domains have been studied extensively, however the function of its ZZ domain remains unclear. In this study, we show that the ZZ domain of p300 recognizes histone H3 tail and therefore represents a novel epigenetic reader. Our findings suggest that the histone binding function of ZZ and acetyllysine binding function of BD are essential in chromatin targeting and catalytic activities of p300. Particularly, recognition of histone H3 by the ZZ domain promotes acetylation of primarily histone H3K27 and H3K18.
Figure 1. The ZZ domain of p300 recognizes histone H3.
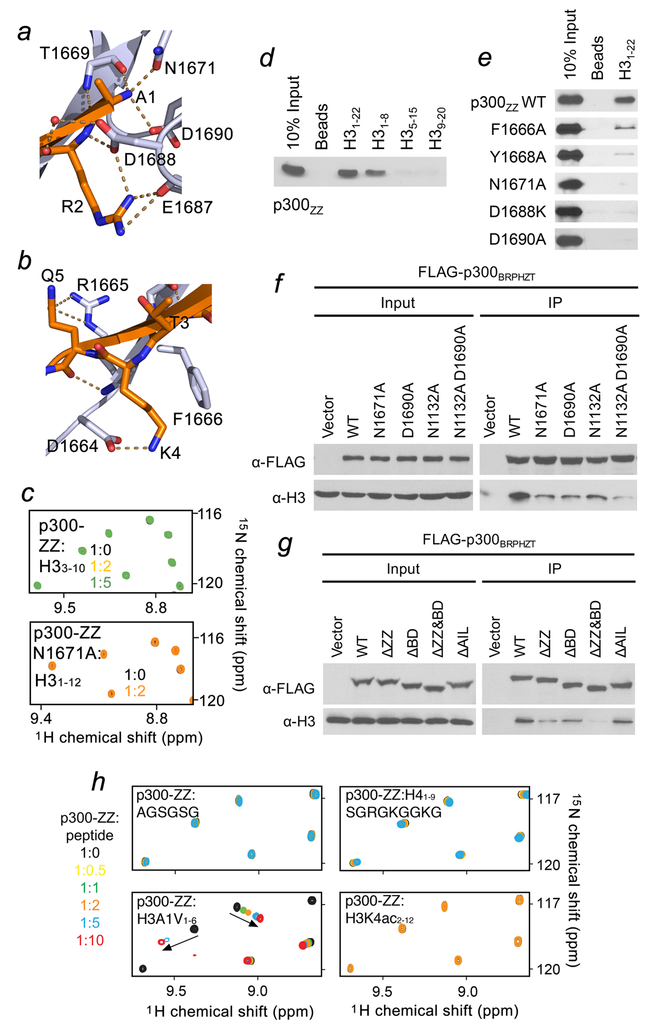
(a) Schematic representation of p300 domain architecture. (b, c) Peptide pull-down assays of the p300 ZZ domain with the indicated histone peptides. Uncropped blot images are shown in Supplementary Data Set 1. (d) Superimposed 1H,15N HSQC spectra of p300-ZZ collected upon titration with H3 peptide (residues 1–12 of H3). Spectra are color coded according to the protein-to-peptide molar ratio. (e) Binding affinities of WT p300-ZZ for the indicated histone peptides measured by tryptophan fluorescence. The experiments were carried out in triplicate for H3 and H3K4ac and in duplicate for methylated H3. (f) Representative binding curve used to determine the Kd values by fluorescence. See also Supplementary Fig. 2. (g) A ribbon diagram of the p300-ZZ domain (light blue) in complex with histone H3 tail (residues 1–6) (orange). (h) Electrostatic surface potential of p300-ZZ is colored blue (positive charge) and red (negative charge) with the bound histone H3 tail shown in stick.
Results
The ZZ domain of p300 targets histone H3.
The histone binding activity of the ZZ domain of p300 (p300-ZZ) was originally identified by peptide pulldown experiments (Fig. 1b, c). We found that GST-tagged p300-ZZ binds to the residues 1–22 of histone H3 tail but does not recognize other regions of H3 or other histones, and that common single posttranslational modifications (PTMs) on H3 do not affect this interaction (Fig. 1b, c). To characterize the binding in detail, we expressed p300-ZZ as an 15N-labeled protein and tested it in 1H,15N heteronuclear single quantum coherence (1H,15N HSQC) experiments (Fig. 1d). Gradual addition of the H31–12 peptide (residues 1–12 of H3) to the p300-ZZ NMR sample led to large resonance changes. These changes were in the intermediate exchange regime on the NMR timescale and suggested a tight binding. In agreement, an 8.8 μM binding affinity of p300-ZZ for the H31–12 peptide was measured by fluorescence assays (Fig. 1e, f). Methylation of H3K4, a common target of epigenetic readers, or acetylation of H3K4 had no effect, as almost identical patterns of resonance changes were observed in NMR titration experiments, and the comparable binding affinities were measured for the correspondingly modified peptides (Fig. 1e and Supplementary Figs. 1a and 2). Importantly, the Kd value for the p300-ZZ:H31–12 interaction was in the range of binding affinities exhibited by the majority of histone binding modules14–16, corroborating the notion that the ZZ domain of p300 is a new member of the family of epigenetic readers that select for histone H3.
Molecular mechanism of the p300-ZZ association with H3.
To elucidate the molecular basis for the recognition of the H3 tail, we determined a 2.0 Å resolution crystal structure of the p300-ZZ:H3 complex using the chimeric construct containing residues 1–6 of H3 linked to the residues 1663–1713 of p300 (Fig. 1g, h). The 1H,15N HSQC spectra of the linked H3-ZZ construct and of the isolated ZZ domain in the presence of a 10-fold excess of H31–12 peptide overlaid well, confirming that the linked and unlinked ZZ:H3 complexes adopt identical structures in solution (Supplementary Fig. 3). The structure showed two H3-ZZ molecules forming the complex in which the H3 region of one molecule is bound to the ZZ domain of another molecule (Table 1 and Supplementary Fig. 4a).
Table 1.
Data collection and refinement statistics for the ZZ:H3 complex
| H3-ZZ (PDB 6DS6) | |
|---|---|
| Data collection | |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 44.0, 44.0 85.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Wavelength | 1.278 |
| Resolution (Å)a | 39.18–1.95 (1.98–1.95)a |
| Rmerge | 7.7 (29.6) |
| I/σ(I) | 71.4 (2.5) |
| Completeness (%) | 99.2 (94.5) |
| Redundancy | 22.3 (9.2) |
| Refinement | |
| Resolution (Å) | 21.32–1.95 |
| No. reflections | 11533 |
| Rwork / Rfree | 0.2070/0.2532 |
| No. atoms | |
| Protein | 465 |
| Zinc Chloride |
2 1 |
| Water | 56 |
| B factors | |
| Protein | 44.9 |
| Zinc Chloride |
53.7 65.7 |
| Water | 56.0 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.922 |
Values in parentheses refer to data in the highest resolution shell. Datasets collected from a single crystal.
The p300-ZZ fold is stabilized by two zinc-binding clusters, a couple of twisted two- and three-stranded anti-parallel β-sheets, and a short α-helix (Fig. 1g). The histone residues pair with the first β-strand (residues 1665–1669 of ZZ), creating the third anti-parallel β-stand and make extensive intermolecular contacts with the domain (Fig. 1g). Characteristic β-sheet interactions are observed between the backbone amides of Ala1, Thr3 and Gln5 of H3 and T1669, V1667 and R1665 of the ZZ domain. Ala1 occupies a highly negatively charged binding site where the NH3+ group of Ala1 is anchored through the hydrogen bonds with T1669 and N1671 and a salt bridge with D1690 (Figs. 1g, h and 2a). The guanidinium moiety of Arg2 is restrained via a salt bridge with the carboxyl group of D1688 and hydrogen bonds with the backbone carbonyl oxygen of E1687, whereas the backbone amide of Arg2 is involved in direct and water-mediated hydrogen bonding contacts with D1688. The side chain of Lys4 lays in a groove formed by the aromatic ring of F1666 and the side chain of D1664, the negatively charged carboxylic group of which forms a salt bridge with the ε-amino group of Lys4 (Figs. 1g and 2b). The formation of the salt bridge most likely accounts for the indifference of p300-ZZ towards PTMs on H3K4, which would augment cation-π and hydrophobic interactions with F1666 but diminish the electrostatic contact with D1664. The side chain of Gln5 is hydrogen bonded to the guanidinium group of R1665. This mechanism for the coordination of H3 is distinctly different and distinguishes the ZZ domain from all currently known readers, including those that bind to the N-terminus of H3 (Supplementary Fig. 4b, c).
Figure 2. Molecular basis for the recognition of H3 tail by p300-ZZ.
(a) Zoom-in view of the histone H3 Ala1 binding site. (b) Zoom-in view of the histone H3 Lys4 binding site. (c) Superimposed 1H,15N HSQC spectra of p300-ZZ (wild type and N1671A mutant) collected upon titration with indicated H3 peptides. Spectra are color coded according to the protein-to-peptide molar ratio. (d) Peptide pull-down assays for p300-ZZ using indicated histone H3 peptides. (e) Peptide pull-down assays using wild type and mutated p300-ZZ. (f, g) Western blot analysis of chromatin immunoprecipitation (ChIP) of wild type FLAG-p300BRPHZT and indicated point- and deletion-mutants in H1299 stable cells. (h) Superimposed 1H,15N HSQC spectra of p300 ZZ collected upon titration with indicated peptides. Spectra are color coded according to the protein:peptide molar ratio shown on the left. Uncropped blot images (d-g) are shown in Supplementary Data Set 1.
The critical role of the Ala1 recognition was substantiated by testing truncated variants of H3 and mutating the binding site residues in p300-ZZ (Fig. 2). Peptide pull-down and NMR experiments with various fragments of histone H3 confirmed that at least first two residues of H3 are required for the interaction (Fig. 2c, d and h). The absence of chemical shift changes in 1H,15N HSQC spectra of p300-ZZ upon titration with either the AGSGSG peptide, Ac-RTKacQTARKSTG (H3K4ac2–12) or Ac-H33–10 suggested that both the H3 sequence and the presence of the positive charge on Ala1 are necessary for the interaction (Fig. 2h). Replacement of Ala1 with a larger residue, valine, in H3A1V1–6 resulted in a reduction in the binding ~3-fold, and very small resonance perturbations in p300-ZZ upon addition of H41–9(Ser1) indicated an almost negligible binding. Substitution of the Ala1-binding site residues, N1671 or D1690 with an alanine, abolished the interaction of p300-ZZ with H3 peptides or calf thymus histones in pulldown assays (Fig. 2e and Supplementary Fig. 5) and NMR titrations (Fig. 2c). Further, mutation of D1688 and F1666 or deletion of the first five (1663–1667) residues in p300-ZZ substantially reduced the binding, pointing to the essential role of the Arg2 and Lys4 coordination and formation of the β-sheet, respectively (Fig. 2e and Supplementary Figs. 1b and 5b).
Recruitment of p300 to chromatin depends on synergistic histone binding functions of ZZ and BD.
The importance of the histone binding activity of the ZZ domain for the association of p300 with chromatin was examined by chromatin immunoprecipitation (ChIP) followed by western blot and salt fractionation assays in H1299 cells stably expressing p300 fragment containing BD-RING-PHD-HAT-ZZ-TAZ2 (BRPHZT) (Fig. 2f, g and Supplementary Fig. 5c–e). Bromodomain of p300/CBP was previously shown to target acetylated lysine residues in histone and non-histone proteins, and the BD-RING-PHD (BRP) region binds hyperacetylated nucleosomes and acetyllysine-containing peptides13,17–23. In agreement, our NMR titration experiments confirmed that BD within BRP associates with an acetylated peptide but does not recognize unmodified H31–19, which is the ligand of ZZ (Supplementary Fig. 6a). Deletion or loss-of-function mutations of either ZZ (ΔZZ, N1671A and D1690A) or BD (ΔBD and N1132A) substantially reduced the association of p300 BRPHZT with chromatin containing acetyllysine marks, and deletion or mutation of both domains had an additive effect (Fig. 2f, g and Supplementary Fig. 5c–e). These data indicate that both readers, BD and ZZ, contribute to the interaction of p300 BRPHZT with chromatin though their ligands differ: ZZ binds to the N-terminus of H3 independent of PTMs, whereas BD binds to various histone sequences acetylated at lysine residues, favoring poly-acetylated H413,17,21. The fact that the impaired histone binding activity of either ZZ or BD led to such a notable decrease in chromatin binding of p300 BRPHZT suggests that the two interactions cooperate in the recruitment or retention of p300 at chromatin. Deletion of AIL appears to have no effect on the association of BRPHZT with chromatin (Fig. 2g and Supplementary Fig. 5d).
P300-ZZ and HAT cooperate in binding to H3 tail.
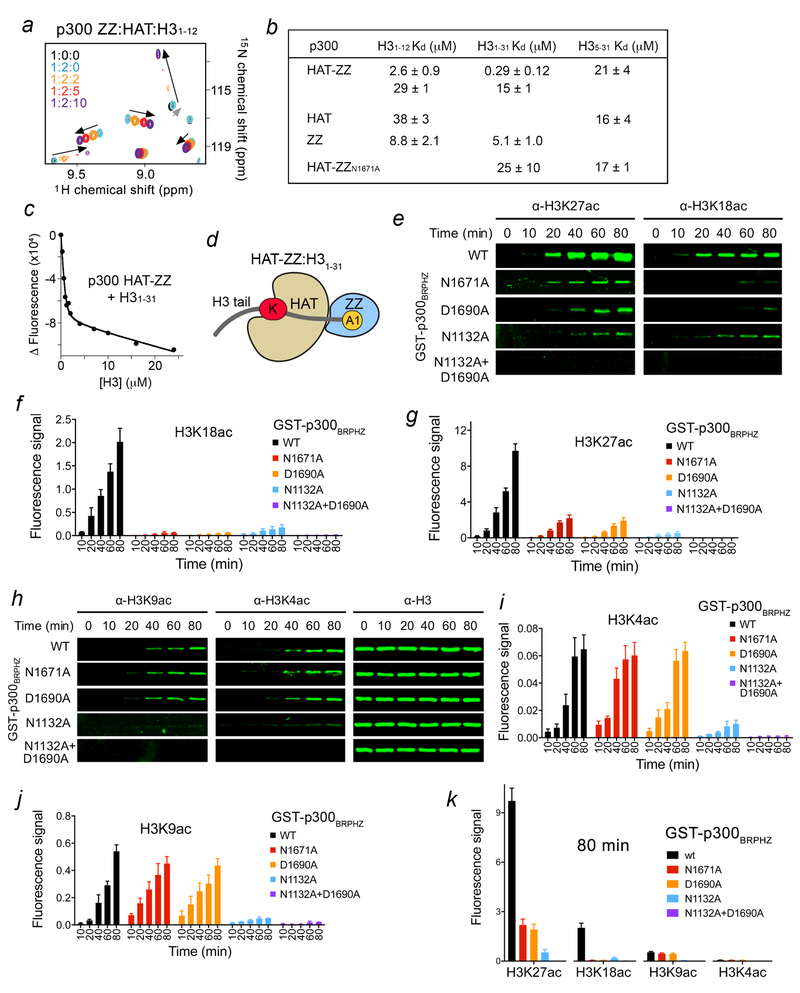
Much like the BRP region preceding the HAT domain, the following ZZ domain is in close proximity to the HAT domain with only ~two residues connecting the domains. We found that the isolated ZZ and HAT domains interact only weakly, as the HAT domain caused very small changes in ZZ, however subsequent titration of H31–12 peptide resulted in large resonance perturbations, suggesting that the histone binding function of ZZ is preserved in the presence or absence of HAT (Fig. 3a). Analysis of the fluorescence-derived binding curves for the interaction of the natively linked HAT-ZZ construct with H31–12 peptide required a two-site binding model, which implies that both HAT and ZZ are engaged with the peptide. The fitting yielded two Kd values of 2.6 μM and 29 μM, which are comparable to Kds measured for the isolated ZZ and HAT domains (8.8 μM and 38 μM, respectively) (Fig. 3b, c). The longer H3 peptide (residues 1–31, H31–31), however, was bound by HAT-ZZ substantially tighter, revealing a cooperative binding and an in cis mechanism (Fig. 3b–d and Supplementary Fig. 2b). In support, the cooperativity was no longer observed for binding of the HAT-ZZN1671A construct that harbors loss-of-function ZZ to the H31–31 peptide (Fig. 3b). The cooperative binding was also eliminated in WT HAT-ZZ when the histone H35–31 peptide, in which the ZZ-targeted sequence is deleted, was used as ligand. The binding affinities of WT HAT-ZZ and HAT-ZZN1671A for the H35–31 peptide (Kds=21 μM and 17 μM, respectively) were essentially the same as the binding affinity of the isolated HAT domain for H35–31 (Kd=16 μM).
Figure 3. ZZ binding to H3 facilitates HAT activity on the distal lysine residues in H3 tail.
(a) Superimposed 1H,15N HSQC spectra of p300 ZZ collected upon titration with the unlabeled HAT domain (1:2 molar ratio) first and then with H3 peptide. Spectra are color coded according to the protein:ligands molar ratio. (b) Binding affinities of the HAT domain, the ZZ domain and HAT-ZZ of p300 for the indicated histone peptides as measured by tryptophan fluorescence. Kds were calculated from triplicate measurements with the exception of H31–31/ZZ (duplicate). (c) Representative binding curve used to determine the Kd values by fluorescence. See also Supplementary Fig. 2. (d) Cartoon representation of HAT-ZZ in complex with H31–31. (e) in vitro HAT assays using WT and mutated p300BRPHZ and the reconstituted nucleosome. (f, g) Quantification of the HAT activity on H3K18 and H3K27 based on the fluorescence signal in (e) from three biological replicates. A common standard sample is used for normalization in each replicate. (h) H3K9ac, H3K4ac and H3 western blot analysis of in vitro HAT assays using WT and mutated p300BRPHZ and the reconstituted nucleosome. (i, j) Quantification of the HAT activity on H3K4 and H3K9 based on the fluorescence signal from three biological replicates as in (h). A common standard sample is used for normalization in each replicate. (k) Comparison of the fluorescence signal for the indicated histone acetylation marks at 80 min of in vitro HAT reaction. Error bars in (f, g, i-k) represent s.e.m. from 3 independent HAT assays using different batches of enzymes (n = 3 biological replicates). Uncropped blot images (e, h) are shown in Supplementary Data Set 1.
Interaction of p300-ZZ with H3 directs acetylation of H3K27 and H3K18 in vitro.
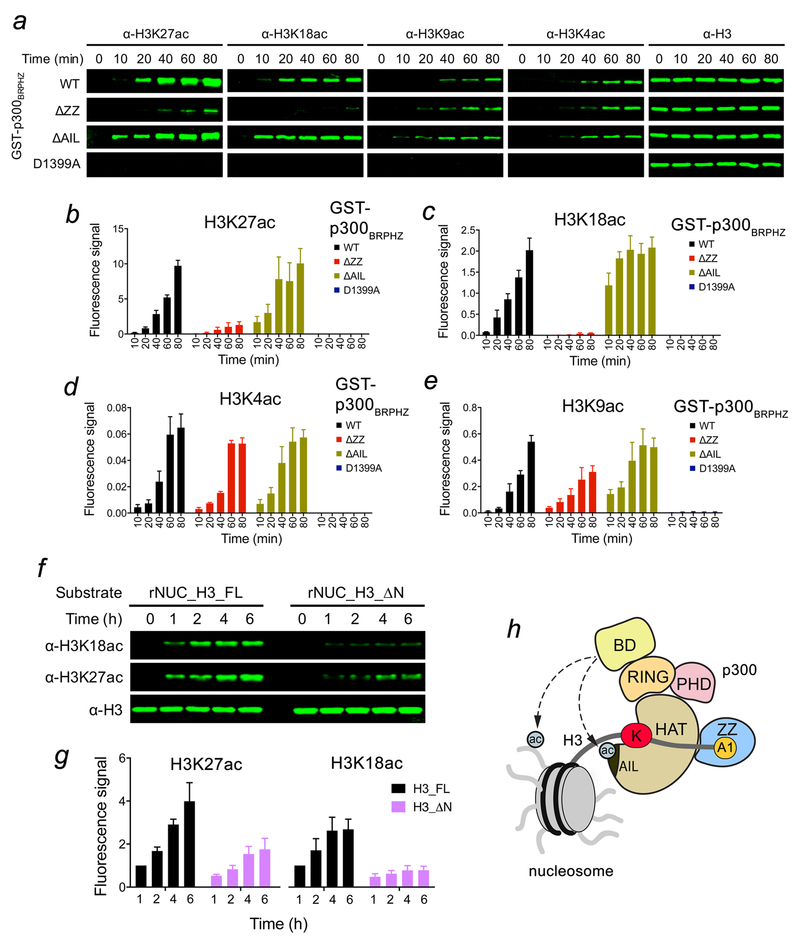
The BRP region has been shown to associate with the HAT domain with the binding affinity of 1.6 μM13. We confirmed the direct and robust interaction through monitoring rapid disappearance of resonances of the 15N-labeled BD-RING-PHD region induced by the unlabeled HAT domain in 1H,15N HSQC experiments (Supplementary Fig. 6). However, this interaction was weakened when the linked HAT-ZZ was titrated in, indicating a conformational mobility within the HAT-ZZ construct that may alter the priming of BRP (Supplementary Fig. 6b). The dynamic organization of the adjacent to the HAT domain modules may facilitate substrate and acyl-CoA association, enlarge the substrate pool of p300, and mediate its HAT activity. To determine whether the ZZ:H3 interaction affects the catalytic activity of p300, we carried out in vitro HAT assays using the p300-BRPHZ fragment and reconstituted unmodified nucleosomes. We found that WT p300-BRPHZ acetylates H3 on H3K27 and H3K18 robustly, but very little acetylation was detected on H3K9 and H3K4 (Fig. 3e–k and Supplementary Fig. 7a). Mutation of the ZZ residues critical for binding to H3, N1671 and D1690, to an alanine in p300-BRPHZ substantially reduced the HAT activity on H3K27 and H3K18 but did not affect acetylation on H3K4 and H3K9. Likewise, the deletion of ZZ led to a considerable reduction of acetylation on H3K27 and H3K18 however had no effect on acetylation of H3K4 and H3K9 (Fig. 4a–e). Furthermore, the HAT activity of p300-BRPHZ on the nucleosome, in which first two residues of H3 (Ala1-Arg2, that are required for the binding of ZZ) were deleted, was severely compromised (Fig. 4f, g). In contrast, mutation of N1132 that impairs acetyllysine binding of BD led to a substantial decrease in acetylation of all lysine residues tested, H3K4, K3K9, H3K18 and H3K27 (Fig. 3e–k). These results suggest that the acetyllysine binding function of BD is necessary for the overall catalytic activity of p300-BRPHZ on the nucleosome, whereas the ability of p300-BRPHZ to acetylate primarily H3K27 and H3K18 is due to binding of the ZZ domain to H3.
Figure 4. p300 ZZ-H3 interaction promotes acetylation of H3K27 and H3K18.
(a) In vitro HAT assays using p300BRPHZ, wild type and counterparts harboring either ZZ deletion, AIL deletion or loss-of-catalytic activity HAT mutation (D1399A), and the reconstituted nucleosome. (b-e) Quantification of the HAT activity based on the fluorescence signal from three biological replicates. A common standard sample is used for normalization in each replicate. (f) In vitro HAT assays using WT p300BRPHZ and the reconstituted nucleosome carrying either intact H3.1 (rNUC_H3_FL) or N-terminally truncated H3.1 (rNUC_H3_ΔN, Ala1 and Arg2 of H3.1 are deleted). (g) Quantification of the HAT activity on H3K18 and H3K27 based on the fluorescence signal in (f) from three biological replicates. The 1 hr sample of rNUC_H3_FL is used for normalization. (h) A model for the regulation of the p300 HAT activity by the BD and ZZ. Potential intermolecular interactions of BD with acetylated histones and acetylated AIL are indicated by dashed lines. Error bars in (b-e, g) represent s.e.m. from 3 independent HAT assays using different batches of enzymes (n = 3 biological replicates). Uncropped blot images (a, f) are shown in Supplementary Data Set 1.
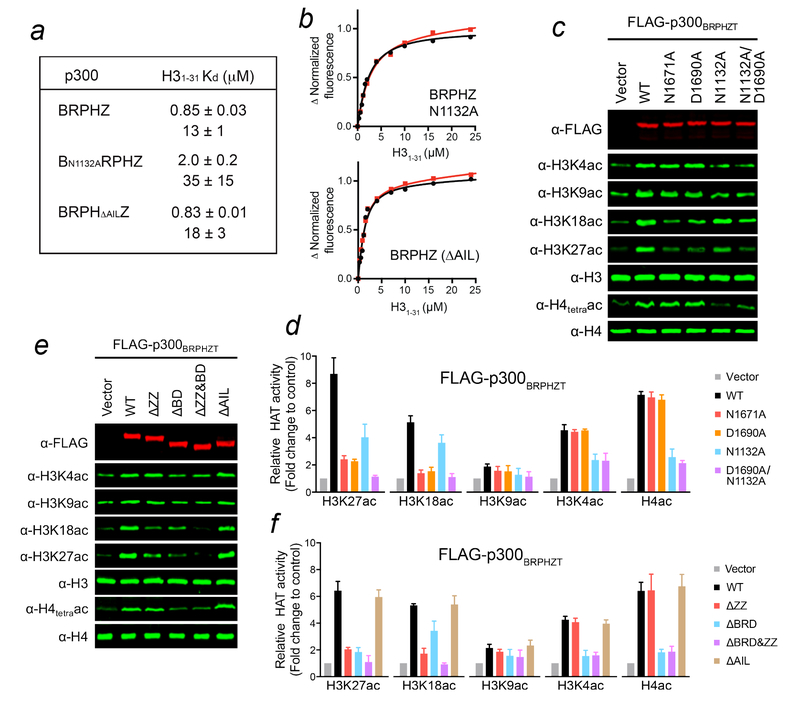
p300 BD was previously proposed to bind acetylated substrates to facilitate acetylation at other sites13,19. Although we used unmodified nucleosomes in the HAT assays, the initial acetylation of histones by p300-BRPHZ can provide a feed-forward mechanism, supporting this idea. Potentially, AIL of the HAT domain can also be a ligand for BD (Fig. 4h), as this loop is autoacetylated in endogenous and recombinant p30012, and BD of CBP interacts with the AIL peptide acetylated at K159619. Although binding affinities of WT p300-BRPHZ, p300-BN1132ARPHZ and p300-BRPHΔAILZ for H31–31 suggest that abrogating acetyllysine binding of BD or deleting AIL has little effect on the association of p300-BRPHZ with this peptide (Fig. 5a, b), comparison of acetylation levels produced by WT p300-BRPHZ and p300-BRPHΔAILZ in 10 min reveals that the deletion of AIL stimulates the HAT activity of p300-BRPHZ (Fig. 4a–e). These data are in agreement with the previous reports on inhibition of the p300 catalytic activity by hypoacetylated AIL12. It was proposed that the hypoacetylated AIL might contact a negatively charged patch on the HAT domain surface blocking the active site, whereas acetylation of AIL releases this loop freeing the active site7,12. At least 10 lysine residues in AIL can be acetylated12, however it remains unclear as to how many of these lysines should be acetylated to regulate equilibrium between an inhibitory (less acetylated, more positively charged) state of AIL and an activation (more acetylated, less positively charged) state of AIL. Further studies are required to establish the effect of poly-autoacetylation of AIL on this equilibrium and explore whether BD is capable of targeting it, which would shift the equilibrium towards the AIL activation state facilitating the catalytic activity (Fig. 4h).
Figure 5. ZZ and BD of p300 differentially modulate p300 activity.
(a) Binding affinities of p300-BRPHZ for H31–31 peptide measured by tryptophan fluorescence (duplicate measurements). (b) Binding curves used to determine the Kd values by fluorescence. (c) Western blot analysis of histone acetylation levels in whole cell extract of H1299 cells stably expressing wild type FLAG-p300BRPHZT or the indicated mutants. (d) Quantification of the indicated histone acetylation levels from three biological replicates as in (c). Total H3 or H4 was used for normalization. (e) Western blot analysis of histone acetylation levels in whole cell extract of H1299 cells stably expressing the indicated FLAG-p300BRPHZT fragments. (f) Quantification of the indicated histone acetylation levels from three biological replicates as in (e). Total H3 or H4 was used for normalization. Error bars in (d, f) represent s.e.m. from 3 independent batches of stable cells (n = 3 biological replicates). Uncropped blot images (c, e) are shown in Supplementary Data Set 1.
H3K18 and H3K27 acetylation requires H3-binding activity of p300-ZZ in vivo.
To determine whether the H3-binding by the ZZ domain is required for p300-mediated histone acetylation in cells, we examined acetylation marks in H1299 cells stably expressing Flag-tagged WT or mutated p300-BRPHZT. As expected, ectopic expression of WT p300-BRPHZT increased the global levels of acetylation at all sites tested (Fig. 5c–f). In support of our model, p300-BRPHZT mutations that abrogate histone binding of the ZZ domain, N1671A and D1690A, or deletion of ZZ greatly attenuated the increase of H3K18 and H3K27 acetylation without affecting H3K4, H3K9 and H4 acetylation (Fig. 5c–f). Similar changes were observed in 293T cells transiently expressing full-length p300 or the ZZ-mutated counterparts (Supplementary Fig. 7b). Mutation of N1132 that disrupts acetyllysine binding of BD or deletion of BD led to a reduction of acetylation at all sites, however acetylation of H4 was most notably impeded (Fig. 5c–f). These findings indicate that while acetylation of histones, H4 in particular, by p300 requires the acetyllysine binding activity of p300-BD, specific acetylation of the H3K18 and H3K27 sites24,25 requires the H3-binding activity of p300-ZZ.
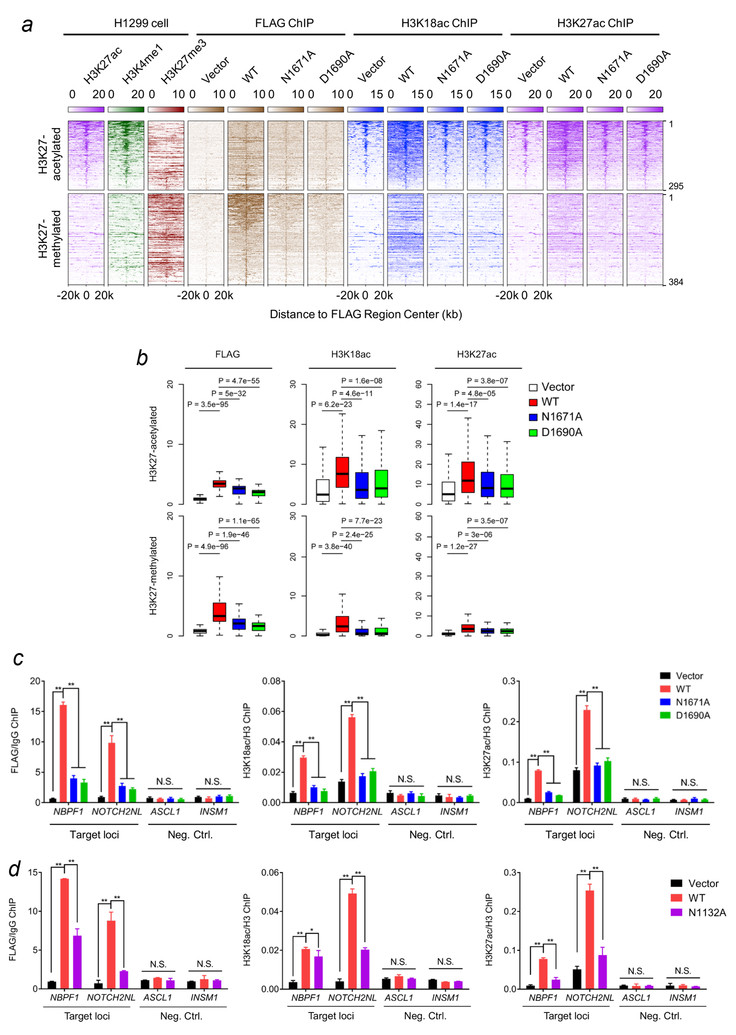
We next examined the role of the ZZ:H3 interaction in chromatin binding and acetylation function of p300-BRPHZT in H1299 cells by ChIP assays followed by high-throughput sequencing (ChIP-seq) (Fig. 6a). p300/CBP has been previously reported to associate with both H3K27 hyperacetylated active enhancers and poised regulatory regions enriched in H3K27me326–28. ChIP-seq using the M2 Flag antibody in the cells expressing Flag-tagged WT p300-BRPHZT identified 679 p300-BRPHZT binding sites in both regions. An evident increase in H3K18ac and H3K27ac occupancies, especially in the regions flanking H3K27 pre-acetylated sites, was also observed in these cells (Fig. 6a, b and Supplementary Table 1). In contrast to the WT protein, the two ZZ domain mutants, N1671A and D1690A, were incapable of gaining the chromatin binding, and no increase in H3K18ac and H3K27ac occupancies was observed in the cells expressing these mutants (Fig. 6a, b). Furthermore, the loss-of-function mutations of the ZZ domain considerably decreased binding of p300-BRPHZT to individual genes and led to a notable reduction in the H3K18 and H3K27 acetylation levels on these genes (Fig. 6c and Supplementary Fig. 7c). Similarly, the N1132A mutation that abrogates acetyllysine binding activity of BD resulted in a decrease in p300-BRPHZT occupancy and H3K18ac and H3K27ac levels at the target gene regions (Fig. 6d). Together, these results underscore the critical roles of the ZZ:H3 and BD:acetyllysine interactions in p300 function in maintaining histone H3K18 and H3K27 acetylation in cells, corroborating the in vitro findings described above.
Figure 6. ZZ is required for binding of p300 to chromatin and its acetylation activity on H3K18 and H3K27 in cells.
(a) Heatmap of normalized H3K27ac (purple), H3K4me1 (green), H3K27me3 (scarlet), FLAG (brown) and H3K18ac (blue) ChIP-seq signals centered on FLAG binding sites in a ±20kb window in H1299 control cells and H1299 cells stably expressing wild type FLAG-p300BRPHZT or the indicated ZZ mutants. The color keys represent signal density. The peaks are divided into two groups (H3K27 acetylated or H3K27 methylated) based on the pre-existing H3K27 modifications at the peak center. (b) Box plots compare FLAG, H3K18ac and H3K27ac occupancies in different samples at FLAG binding peaks. All FLAG peaks are divided into two groups as in (a). The center line of box represents the median and box limits indicate the 25th and 75th percentiles. Two-tailed paired Student’s t-test was used for statistical analyses. (c) qPCR analysis of the FLAG-p300BRPHZT, H3K18ac and H3K27ac ChIP at target loci (locations indicated by black line in Supplementary Fig. 7c) and two negative control loci in H1299 cells stably expressing wild type FLAG-p300BRPHZT or the indicated mutants. IgG or H3 ChIP was used for normalization. (d) qPCR analysis of the FLAG-p300BRPHZT, H3K18ac and H3K27ac ChIP at target loci and two negative control loci in H1299 cells stably expressing wild type FLAG-p300BRPHZT or the N1132A BD mutant. IgG or H3 ChIP was used for normalization. Error bars in (c, d) represent s.e.m. from 3 individual experiments (n = 3 experimental repeats).
Discussion
In conclusion, we have identified a novel function for the ZZ domain of p300. It recognizes histone H3 tail through a unique and previously uncharacterized mechanism. The H3 binding activity of ZZ and the acetyllysine binding activity of BD are both essential for the association of p300 with chromatin. We show that the acetyllysine binding of BD is necessary for p300 to catalyze acetylation of virtually all lysine residues in H3 and H4 tested, whereas the ability of p300 to acetylate primarily H3K27 and H3K18 is due to the binding of the ZZ domain to the N-terminus of H3. A model of the p300-BRPHZ:H3 complex generated using the simulated annealing method and the crystal structures of p300 BRPH13 and H3-ZZ revealed a ~38 Å distance between the H3A1-binding site of ZZ and the catalytic site in the HAT domain (Supplementary Fig. 7d). This distance is too long for Lys9 or Lys14 of H3 to occupy the active site of the HAT domain and thus be acetylated in cis when the N-terminus of H3 is locked through the interaction with ZZ, however other lysine residues (K18, K23, K27) in the H3 tail can reach the active site. Furthermore, the substrate-binding groove of the HAT domain is highly negatively charged and would favor the positively charged H3 tail. Collectively, these results suggest that binding of p300-ZZ to the N-terminus of H3 provides selectivity of the HAT domain toward the distal lysine sites in H3, such as H3K18 and H3K27. Intriguingly, we found that homologous CBP-ZZ binds to H3 weakly (Supplementary Fig. 8), and it will be interesting in future studies to differentiate the biological roles of p300 and CBP that are due to different activities of their ZZ domains.
ONLINE METHODS
Materials
The coding DNA sequences (CDS) encoding human full-length p300 protein and the p300 BD-RING-PHD-HAT-ZZ-TAZ2 region (BRPHZT, amino acids 1035–1830) were cloned into pENTR3C vector and subsequently cloned into p3FLAG and pCDH-FLAG destination vectors using Gateway techniques (Invitrogen), respectively. The CDS encoding the human p300 BD-RING-PHD-HAT-ZZ region (BRPHZ, amino acids 1035–1720) and ZZ domain (amino acids 1650–1720) were cloned into the pGEX-6P-1 vector (GE Healthcare). Point mutations and deletions were generated using a site-directed mutagenesis kit (Stratagene) and verified by Sanger sequencing. Histone peptides bearing different modifications were synthesized at CPC, LLC. Anti-histone antibodies including anti-H3 (Ab1791), anti-H3K4ac (ab176799), anti-H3K9ac (Ab32129), anti-H3K27ac (Ab4729) and anti-H4 (Ab7311) antibodies were obtained from Abcam. Anti-H3K9ac (61251) and anti-H3K18ac (39755) antibodies were from Active Motif. Anti-acetyl-Histone H4 antibody (06–598) was from Millipore. Anti-GST (sc-459) antibody was from Santa Cruz. Anti-FLAG (M2, F1804) antibody was from Sigma. Fluorescent secondary antibodies (926–32211 and 926–68020) were from LI-COR. Mononucleosomes reconstituted from recombinant histones (16–0009) were from EpiCypher.
Protein expression and purification for pull-down assays
p300-BRPHZ and p300-ZZ cloned in the pGEX-6P-1 vector were expressed in BL21 Rosetta 2 cells. Protein production was induced with 0.2 mM IPTG and cultured overnight at 16 °C in Luria broth (LB) medium supplemented with 0.05 mM ZnCl2. The GST-tagged proteins were purified on glutathione Sepharose 4B beads (Amersham) in binding buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40, 1 mM PMSF plus protease inhibitors (Roche)) and eluted by 100 mM Tris pH 8.0 containing 15 mg/mL Reduced Glutathione (Sigma). All proteins harboring mutations or deletions were expressed and purified as WT proteins.
Peptide pull-down assays
Peptide pull-down assays were performed as described previously29. In brief, 1 μg of biotinylated histone peptides with different modifications were incubated with 1–2 μg of GST-fused p300 ZZ domain in binding buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.1% NP-40, 1 mM PMSF) for overnight with rotation at 4 °C. Streptavidin beads (Amersham) were added to the mixture, and the mixture was incubated for 1 h with rotation at 4 °C. The beads were then washed three times and the bound proteins were analyzed using SDS–PAGE and Western blotting.
Calf thymus histone pull-down assay
Full-length histone pull-down assays were performed as described previously30. Briefly, 2 μg GST-tagged protein were incubated with 10 μg of calf thymus total histones (Worthington) in binding buffer (50 mM Tris-HCl pH 7.5, 1M NaCl, 1% NP-40) at 4 °C for overnight with rotation. Glutathione Sepharose 4B beads (Amersham) were added to the solution and incubate for 1 h. The beads were then washed six times and bound histones were detected using SDS–PAGE and Western blotting.
Salt fractionation
Salt fractionation of nuclei was performed as previously described31. In short, cells were swollen with hypotonic buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10mM KCl) and lysed by gentle disruption to isolate nuclei. The nuclei were incubated with wash buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 0.2mM EDTA, 25% glycerol) containing 75 mM NaCl for 30 min at 4 °C and then pelleted and the supernatant fraction was collected. The nuclei were then similarly washed by wash buffer containing 150mM, 300mM and 600mM NaCl and the supernatant fraction of each step was collected. After the final wash step, the pellet was resuspended and sonicated before collection.
Protein Expression and Purification for structural studies
The human p300 ZZ domain (aa 1663–1713) and H3-linked ZZ domain (H3 aa 1–6, ZZ aa 1663–1713) were cloned into a pCIOX vector with N-terminal His-tag and Ulp1 cleavage site. The p300 HAT domain (aa 1287–1663, Y1467F), HAT-ZZ domain (aa 1287–1713, Y1467F) and BD-RING-PHD region (aa 1047–1278) were cloned into a pDEST-17 vector with N-terminal His-tag and PreScission cleavage site. The p300 ZZ1668 domain (aa 1668–1713) and CBP ZZ (aa 1705–1750) were cloned into a pCOOL vector with N-terminal GST tag and Thrombin cleavage site. All proteins were expressed in BL21 (DE3) RIL cells. Protein production was induced with 0.2 mM IPTG and incubated overnight at 18 °C in Luria broth (LB) or minimal media (M9) supplemented with 15NH4Cl and 0.05 mM ZnCl2. The His-tag proteins were purified on Ni-NTA beads (Qiagen) in 20 mM Tris-HCl (pH 7.5) buffer, supplemented with 300 mM NaCl and 10 mM ßME. The His-tag was cleaved overnight at 4 °C with PreScission or Ulp1 protease. The GST-tagged proteins were purified on glutathione Sepharose 4B beads (GE Healthcare) in 20 mM Tris-HCl (pH 7.0) buffer, supplemented with 100 mM NaCl and 5 mM DTT. The GST tag was cleaved overnight at 4 °C with Thrombin protease. The GST-tagged p300-BRPHZ and mutants were purified as above and cleaved overnight at 4 °C with PreScission. Unlabeled proteins were further purified by size exclusion chromatography and concentrated in Millipore concentrators. All mutants were generated by site-directed mutagenesis using the Stratagene QuickChange mutagenesis protocol, grown and purified as WT proteins.
NMR Experiments
NMR experiments were carried out at 298K on Varian INOVA 600 and 900 MHz spectrometers. NMR samples contained 0.1 mM uniformly 15N-labeled WT or mutated p300-ZZ, p300-HAT-ZZ, p300-BD-RING-PHD and CBP-ZZ proteins in 20 mM Tris-HCl (pH 7.0) buffer supplemented with 100 mM NaCl, 5 mM DTT and 8% D2O. Binding was characterized by monitoring chemical shift changes in the proteins induced by histone peptides (synthesized by SynPeptide).
X-Ray crystallography
The p300 H3-linked ZZ domain (aa 1663–1713) was crystallized. For crystallization, the purified H3-ZZ (15 mg/mL) was buffer exchanged into 5 mM HEPES (pH 7.4), 50 mM NaCl and 0.5 mM TCEP prior to crystallization. Good quality diffracting crystals were obtained at 4 °C in 0.1 M HEPES and 70% MPD (pH 7.5). Data collection was performed at the ALS 4.2.2 beamline, Berkeley. HKL3000 was used for indexing, scaling, and data reduction. Solution was found by Single-wavelength Anomalous Dispersion method with Zn anomalous signal. Manual model building was performed using Coot32 and the structure was refined using Phoenix Refine33. The final structure was verified by MOLProbity34 showing 94.1% and 5.9% of residues in the most favored regions and additionally allowed regions, respectively. The X-ray diffraction and structure refinement statistics are summarized in Table 1.
Fluorescence spectroscopy
Spectra were recorded at 25 °C on a Fluoromax-3 spectrofluorometer (HORIBA) as described previously35 with the following modifications. The samples containing 1.0 μM p300 ZZ, HAT, HAT-ZZ and HAT-ZZN1671A with Y1467F mutation, BRPHZ, BN1132ARPHZ or BRPHΔAILZ and progressively increasing concentrations of the histone peptide were excited at 295 nm. All experiments were performed in buffer containing 15 mM HEPES (pH 7.2), 150 mM NaCl and 0.5 mM TCEP. Emission spectra were recorded over a range of wavelengths between 330 and 360 nm with a 0.5 nm step size and a 1 s integration time and averaged over 3 scans. The Kd values were determined using a nonlinear least-squares analysis in Kaleidagraph and the equation:
where [L] is the concentration of the histone peptide, [P] is the protein concentration, ΔI is the observed change of signal intensity, and ΔImax is the difference in signal intensity of the free and bound states. Binding curves of HAT-ZZ, BRPHZ, BN1132ARPHZ or BRPHΔAILZ were fit by a nonlinear least-squares analysis in Kaleidagraph using a two-site binding model:
The Kd values were averaged over three or two separate experiments as stated in corresponding Figure legends, with error calculated as the standard deviation between the runs.
Modeling of the H3-bound BD-RING-PHD-HAT-ZZ region of p300
The model of the complex of p300 BRDHZ (aa 1047–1713) with H3 tail (1–31) was generated using the crystal structure of BRDH (PDB: 4BHW)13 and the H3-ZZ complex and the simulated annealing method in Xplor-NIH36. The lysine portion of the bi-substrate Lys-CoA was modified into the side chain of Lys27 of H3, and then the H3-ZZ complex was modeled onto the crystal structure with a flexible linker (residues 1661–1664). The model of the complex was treated as an initial structure and refined by simulated annealing. During the refinement, the backbone atoms of BRDH region (aa 1047–1660) were fixed, whereas the backbone atoms of the H3(aa 1–4)-ZZ(aa 1665–1713) complex were grouped and allowed to move as a rigid body. Distances between the Lys-CoA and residues W1436 and S1396 in the crystal structure were used as additional restraints to maintain the active site conformation. At all time, residues 1661–1664 of p300 and residues 5–31 of H3 were set to be fully flexible. 20 models were calculated and 6 structures with the lowest energy were selected for further analysis.
In vitro histone acetyltransferase (HAT) assays
For HAT assays on recombinant nucleosomes containing full-length histone H3.1, purified wild type or mutated p300BRPHZ (300 nM) was incubated with recombinant mononucleosome (100 nM) in HAT reaction buffer (50 mM Tris pH 8.0, 0.1 mM EDTA, 10% glycerol, 1 mM PMSF and 1mM DTT) in a total volume of 50 μL. After pre-warming at 37 °C for 5 minutes, reactions were initiated with the addition of acetyl-CoA (Sigma) to a final concentration of 0.1 mM and incubated for 10~80 minutes at 37 °C. For assays comparing the HAT activities on recombinant nucleosomes containing full-length histone H3.1 and N-terminally truncated H3.1, wild type p300BRPHZ fragment (50 nM) and monobucleosome (500 nM) were incubated under the same condition for 1 to 6 hours. Reactions were quenched by flash-freezing in liquid nitrogen and then analyzed by SDS-PAGE and Western blot analysis. Western blot results were quantified by LI-COR Odyssey and normalized to a common standard sample.
Cell culture, transfection and virus transduction
HEK293T cell (ATCC) was maintained in DMEM (Cellgro) supplemented with 10% fetal bovine serum (Sigma). Human lung cancer cell line H1299 (ATCC) was maintained in RPMI (Cellgro) supplemented with 10% fetal bovine serum (Sigma). Both cell lines were mycoplasma-negative and were tested for authentication by short tandem repeat (STR) profiling performed by the MDACC CCSG-funded Characterized Cell Line Core, NCI CA016672. Transient transfection was performed using X-tremeGENE DNA transfection reagent (Sigma). 48 h after transfection, cells were collected for Western blot analysis. Lentiviral transduction was performed as described previously37. Briefly, 293T cells were co-transfected with pMD2.G, pPAX2 (Addgene) and pCDH cDNA constructs using X-tremeGENE DNA transfection reagent (Roche). For infections, H1299 cell was incubated with viral supernatants in the presence of 8 mg/ml polybrene; after 48 hr, the infected cells were selected with blasticidin (10 mg/ml) for 4–6 days before experiments.
Chromatin immunoprecipitation (ChIP) and ChIP-seq
ChIP analysis was performed essentially as described previously37,38. In brief, cells were cross-linked with 1% formaldehyde for 10 min and stopped with 125 mM glycine. The isolated nuclei were resuspended in nuclei lysis buffer (50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS) and sonicated using a Bioruptor Sonicator (Diagenode). The samples were immunoprecipitated with 2–4 μg of the appropriate antibodies overnight at 4 °C. Protein A/G beads (Millipore) were added and incubated for 1 hour, and the immunoprecipitates were washed twice, each with low-salt (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS), high-salt (20 mM Tris pH 8.0, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS) and LiCl buffer (20 mM Tris pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% NP-40 and 1% SDS). Eluted DNA was reverse-crosslinked, purified using PCR purification kit (Qiagen), and analyzed by quantitative real-time PCR on the ABI 7500-FAST System using the Power SYBR Green PCR Master Mix (Applied Biosystems).
For ChIP-seq, the ChIP experiments were carried out essentially the same as described above. Purified DNA was sequenced using the Illumina Solexa Hiseq 3000. The raw reads were mapped to human reference genome NCBI 37 (hg19) or the Drosophila melanogaster genome (dm3) by bowtie v1.1.0, allowing up to 1 mismatch. Only uniquely mapped reads were retained for peak calling. But before that, we use spike-in normalization for sample size correction as previously described39. For simplicity, the reads were downsampled to keep same spike-in reads count in different samples. Then the ChIP-seq peaks were called by MACS v1.4.2 with a cutoff of p ≤1e−8, and clonal reads were automatically removed by MACS. The ChIP-seq reads density was determined by deepTools v2.3.4, and then the average binding profile and heatmap were visualized using R v3.2.3 (Supplementary Tables 1–4).
Protein-ChIP
Protein-ChIP assays for detection of in situ p300-chromatin interactions was performed as described previously40. Briefly, cells were cross-linked with 1% formaldehyde for 10 min and stopped with 125 mM glycine. The isolated nuclei were resuspended in nuclei lysis buffer and sonicated. Anti-FLAG M2-conjugated agarose beads (Sigma) were incubated with lysates overnight at 4 °C. The beads were then washed twice, each with low-salt, high-salt and LiCl buffer, and the bound proteins were analyzed by SDS-PAGE and Western blot.
Statistical analyses
Experimental data are presented as mean ± s.e.m. unless stated otherwise. Statistical significance was calculated by two-tailed unpaired t-test on two experimental conditions with P < 0.05 considered statistically significant unless stated otherwise. Statistical significance levels are denoted as follows: *P < 0.05; **P < 0.01. No statistical methods were used to predetermine sample size.
Supplementary Material
Acknowledgements
We thank Jay Nix at beam line 4.2.2 of the ALS in Berkeley for help with X-ray crystallographic data collection. This work was supported by grants from NIH GM106416, GM125195 and GM100907 to T.G.K., CA204020 to X.S., and HG007538 and CA193466 to W.L., and from Cancer Prevention and Research Institute of Texas (RP160237 and RP160739), Welch Foundation (G1719) and Leukemia & Lymphoma Society Career Development Program Scholarship to X.S.
Footnotes
Methods
Methods and any associated references are available at XX.
Accession codes: Coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 6DS6.
Reporting Summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability. The atomic coordinates and structure factors of p300 ZZ-H3 have been deposited in the Protein Data Bank under the accession code 6DS6. The ChIP-seq data is submitted to Gene Expression Omnibus under the accession number GSE109591. Other data are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
References
- 1.Wang F, Marshall CB & Ikura M Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cellular and molecular life sciences : CMLS 70, 3989–4008, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasko LM et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer NG, Ozdag H & Caldas C p300/CBP and cancer. Oncogene 23, 4225–4231, (2004). [DOI] [PubMed] [Google Scholar]
- 4.Dancy BM & Cole PA Protein lysine acetylation by p300/CBP. Chemical reviews 115, 2419–2452, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman RH & Smolik S CBP/p300 in cell growth, transformation, and development. Genes Dev 14, 1553–1577, (2000). [PubMed] [Google Scholar]
- 6.Bedford DC, Kasper LH, Fukuyama T & Brindle PK Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics 5, 9–15, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451, 846–850, (2008). [DOI] [PubMed] [Google Scholar]
- 8.Ogryzko VV, Schiltz RL, Russanova V, Howard BH & Nakatani Y The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959, (1996). [DOI] [PubMed] [Google Scholar]
- 9.Wang Z et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraus WL, Manning ET & Kadonaga JT Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Molecular and cellular biology 19, 8123–8135, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomita A et al. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene 19, 444–451, (2000). [DOI] [PubMed] [Google Scholar]
- 12.Thompson PR et al. Regulation of the p300 HAT domain via a novel activation loop. Nature structural & molecular biology 11, 308–315, (2004). [DOI] [PubMed] [Google Scholar]
- 13.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E & Panne D Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nature structural & molecular biology 20, 1040–1046, (2013). [DOI] [PubMed] [Google Scholar]
- 14.Andrews FH, Strahl BD & Kutateladze TG Insights into newly discovered marks and readers of epigenetic information. Nature chemical biology 12, 662–668, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musselman CA, Lalonde ME, Cote J & Kutateladze TG Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology 19, 1218–1227, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taverna SD, Li H, Ruthenburg AJ, Allis CD & Patel DJ How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology 14, 1025–1040, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippakopoulos P et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ragvin A et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. Journal of molecular biology 337, 773–788, (2004). [DOI] [PubMed] [Google Scholar]
- 19.Park S et al. Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation. Proceedings of the National Academy of Sciences of the United States of America 114, E5335–E5342, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Martinez-Yamout MA, Dyson HJ & Wright PE The CH2 domain of CBP/p300 is a novel zinc finger. FEBS letters 587, 2506–2511, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plotnikov AN et al. Structural insights into acetylated-histone H4 recognition by the bromodomain-PHD finger module of human transcriptional coactivator CBP. Structure 22, 353–360, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mujtaba S et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Molecular cell 13, 251–263, (2004). [DOI] [PubMed] [Google Scholar]
- 23.Das C et al. Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation. Proceedings of the National Academy of Sciences of the United States of America 111, E1072–E1081, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Q et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. The EMBO journal 30, 249–262, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 154, 297–310, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada-Iglesias A et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buecker C & Wysocka J Enhancers as information integration hubs in development: lessons from genomics. Trends Genet 28, 276–284, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zentner GE, Tesar PJ & Scacheri PC Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21, 1273–1283, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein BJ et al. PHF20 Readers Link Methylation of Histone H3K4 and p53 with H4K16 Acetylation. Cell reports 17, 1158–1170, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen H et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teves SS et al. A dynamic mode of mitotic bookmarking by transcription factors. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography 66, 213–221, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography 66, 12–21, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatchalian J et al. Dido3 PHD modulates cell differentiation and division. Cell reports 4, 148–158, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwieters CD, Kuszewski JJ, Tjandra N & Clore GM The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160, 65–73, (2003). [DOI] [PubMed] [Google Scholar]
- 37.Li Y et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159, 558–571, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan L et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan B et al. An Alternative Approach to ChIP-Seq Normalization Enables Detection of Genome-Wide Changes in Histone H3 Lysine 27 Trimethylation upon EZH2 Inhibition. PloS one 11, e0166438, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricke RM & Bielinsky AK Easy detection of chromatin binding proteins by the Histone Association Assay. Biol Proced Online 7, 60–69, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.