Abstract
miR-155 has recently emerged as an important promoter of antitumor immunity through its functions in T lymphocytes. However, the impact of T cell–expressed miR-155 on immune cell dynamics in solid tumors remains unclear. In the present study, we used single-cell RNA sequencing to define the CD45+ immune cell populations at different time points within B16F10 murine melanoma tumors growing in either wild-type or miR-155 T cell conditional knockout (TCKO) mice. miR-155 was required for optimal T cell activation and reinforced the T cell response at the expense of infiltrating myeloid cells. Further, myeloid cells from tumors growing in TCKO mice were defined by an increase in wound healing genes and a decreased IFN-γ–response gene signature. Finally, we found that miR-155 expression predicted a favorable outcome in human melanoma patients and was associated with a strong immune signature. Moreover, gene expression analysis of The Cancer Genome Atlas (TCGA) data revealed that miR-155 expression also correlates with an immune-enriched subtype in 29 other human solid tumors. Together, our study provides an unprecedented analysis of the cell types and gene expression signatures of immune cells within experimental melanoma tumors and elucidates the role of miR-155 in coordinating antitumor immune responses in mammalian tumors.
Keywords: Immunology, Oncology
Keywords: Adaptive immunity, Cancer, Expression profiling
MicroRNA-155 coordinates the immunological landscape within murine melanomas and broadly defines an immune-enriched phenotype in human solid tumors.
Introduction
MicroRNAs (miRs) are short regulatory RNA molecules that inhibit the translation of target messenger RNAs (mRNAs) based on sequence complementarity. Biogenesis of miRs involves transcription of miR host genes, followed by a series of posttranscriptional processing steps within the nucleus and cytoplasm that result in an approximately 22-nucleotide-long mature miR that is loaded into RNA-induced silencing complex (RISC) and capable of target repression (1). Through interacting with multiple targets, miRs participate in numerous cellular processes ranging from embryonic development and carcinogenesis to immunity. miR-155 is a classic example of a multifunctional miR that plays essential roles in hematopoietic development (2, 3), inflammatory responses (4–6), autoimmunity (7), and cancer progression (8, 9). In the context of immunity, miR-155 is upregulated in T cells, B cells, macrophages, and dendritic cells (DCs) upon cellular activation and it contributes to effector responses by regulating target genes (10). Recent studies have shown that T cell–specific expression of miR-155 is necessary for optimal antitumor immunity in various experimental models (11–14), but a detailed understanding of how T cell–expressed miR-155 impacts the tumor microenvironment (TME) is lacking. Further, the impact of miR-155 on the prognosis of human solid tumors remains to be elucidated.
The TME is composed of several different immune cell types with protumorigenic and antitumorigenic properties that play an active role in cancer development and progression. Understanding the dynamics of the TME is essential for devising effective new cancer therapeutics and predicting which patients are likely to respond to the immunotherapies (15). The advent of flow cytometry has enabled the characterization of cells infiltrating the tumor by using cell-specific antibodies, but it is unable to capture the true heterogeneity of the TME and the activation states of infiltrating immune cells. Emerging single-cell RNA sequencing (SCseq) technologies provide a powerful alternative to investigate the characteristics of the TME and study antitumor immune responses at an unprecedented resolution. Recent studies have employed SCseq technology to dissect the molecular features of the TME in both preclinical and clinical models and have unveiled the commonalities and context-dependent aspects of antitumor immunity (16, 17). Expanding the utilization of SCseq to new cancer models will be essential for obtaining a more complete understanding of how the immune system responds to developing tumors, with the eventual goal of revealing pathways that can be leveraged for therapeutic purposes.
In this study, we used SCseq to characterize the murine melanoma immune microenvironment in the presence or absence of T cell–specific miR-155 at two different time points. In support of our previous results (11), miR-155 was found to be essential for antitumor immune responses, as evidenced by faster tumor growth in T cell conditional miR-155–knockout mice (miR-155 TCKO). SCseq demonstrated a complex landscape of tumor-infiltrating immune cells including multiple populations of T cells, natural killer (NK) cells, neutrophils, DCs, and macrophages that were defined by distinct activation states in a time- and genotype-dependent manner. T cell–specific expression of miR-155 was critical not only for T cell effector responses but also for programming several myeloid cell populations in the TME. Interestingly, analysis of The Cancer Genome Atlas (TCGA) data revealed that miR-155 expression correlates with an improved clinical outcome in human melanoma and defined an immune-enriched tumor subtype. Furthermore, miR-155 expression marked an immune-enriched solid tumor subpopulation across the TCGA solid cancer cohorts and positively correlated with multiple immune signature parameters. We also found that miR-155 expression was associated with an improved clinical outcome, especially in cancers with high mutational burden such as melanoma and lung cancer. Taken together, our results describe the critical role of T cell–specific miR-155 in shaping the TME for effective antitumor immunity. These findings also delineate the characteristics and kinetics of miR-155–mediated antitumor immunity in murine melanoma for the first time to our knowledge and suggest that an antitumor immune response can be defined by miR-155 expression in human cancers.
Results
SCseq reveals the immune cell diversity within the TME in melanoma.
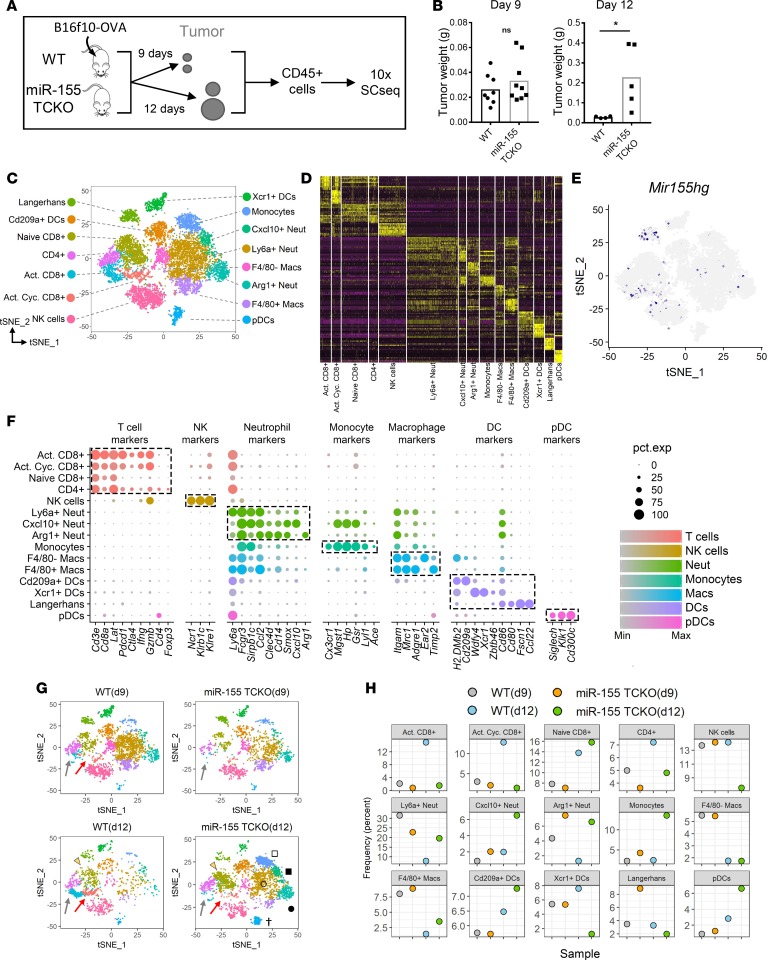
The TME is composed of a variety of immune cells that can have both pro- and antitumorigenic functions. miR-155 was previously shown to be a critical mediator of antitumor immunity in mice (11–14, 18), but the effects of T cell–specific expression of miR-155 on the dynamics of the TME are unknown. To analyze the composition of the TME and the phenotypes of tumor-infiltrating immune cells, we employed 10× Genomics SCseq technology. B16F10 murine melanoma cells expressing chicken ovalbumin (OVA) model antigen were injected subcutaneously into wild-type (WT) and miR-155 TCKO mice. Nine or 12 days after tumor injection, live CD45+ immune cells were sorted from the pooled tumors (n > 4 per time point) via flow cytometry and subjected to SCseq (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126543DS1). Consistent with our previous findings (11), we did not observe a major difference in tumor growth on day 9, whereas on day 12, miR-155 TCKO mice exhibited a higher tumor burden (Figure 1B). This suggested a lack of productive antitumor immunity in mice when T cell–specific expression of miR-155 is lost. We aggregated data from 11,054 individual cells [3,624 cells-WT(d9); 1,956 cells-miR-155 TCKO(d9); 1,759 cells-WT(d12); and 3,715 cells-miR-155 TCKO(d12)] and performed unsupervised clustering analysis based on the similarity of gene expression signatures by using the Seurat single-cell genomics R package (19). This analysis revealed 15 distinct cell clusters representative of both lymphoid and myeloid lineages (Figure 1, C and D, and Supplemental Figure 2).
Figure 1. Single-cell RNA sequencing reveals cellular dynamics within the tumor immune microenvironment in the presence and absence of T cell–specific miR-155.
(A) Diagram showing the method employed for tumor-infiltrating immune cell single-cell RNA sequencing (SCseq). At the experimental endpoint, cells from 4 mice per group were combined and equal numbers were processed for 10× SCseq. (B) Tumor weights at the experimental endpoints of days 9 and 12, showing a higher tumor burden in miR-155 TCKO mice on day 12. Two-tailed t test was used for statistical comparisons. *P ≤ 0.05; ns, P > 0.05. (C) T-distributed stochastic neighbor embedding (t-SNE) plots of SCseq data showing 15 distinct cell clusters (aggregate data from WT and miR-155 TCKO samples from days 9 and 12). (D) Gene expression heatmap showing the top 10 differentially expressed genes in clusters. Columns indicate clusters and rows indicate genes. The column widths are proportional to the numbers of cells in clusters. Each vertical bar within the columns represents an individual cell. (E) Expression pattern of miR-155 host gene (Mir155hg) is shown. (F) Dot charts showing the expression of selected genes in cell clusters. The size of the dots represents the frequency of cells within the cluster expressing the gene of interest, while the color intensity indicates the level of expression. Dashed boxes indicate genes that are selectively expressed within clusters. (G) SCseq t-SNE plots showing the immune landscape in WT and miR-155 TCKO animals at 2 different time points. Activated CD8+ T cell (gray arrows), activated cycling CD8+ T cell (red arrows), naive CD8+ T cell (yellow arrowhead), pDC (dagger), Ly6a+ neutrophil (unfilled circle), Arg1+ neutrophil (filled circle), monocyte (unfilled square), and F4/80– macrophage (filled square) clusters are indicated. (H) Frequency of cell clusters in WT and miR-155 TCKO tumor microenvironment at days 9 and 12.
Due to the high dimensionality of the data and overlapping expression of marker genes, identifying the biological nature of cell clusters from SCseq experiments can be a challenging task (20). Surveying the expression of known markers can aid in the description of broad immune cell populations (Supplemental Figure 2), but a more granular analysis of cell clusters requires examining multiple genes simultaneously. To assist with the identification of cell clusters, we developed an algorithm that compares the gene expression signatures of SCseq cell clusters with the publicly available Immunological Genome Project (ImmGen) database (see Methods). The ImmGen database hosts microarray and RNA sequencing (RNAseq) data from more than 200 purified immune cell subsets under various experimental conditions. This algorithm calculates an aggregate identity score for each SCseq cell cluster as a measure of molecular similarity to the ImmGen subsets (Supplemental Figure 3). With this algorithm, certain cell clusters including plasmacytoid DCs (pDCs), activated and naive CD8+ T cells, NK cells, and Langerhans cells were confidently identified (Supplemental Figures 3 and 4). For cell clusters in which the algorithm could not make a clear call such as macrophage and neutrophil populations, we resorted to differential expression analysis between clusters to identify distinguishing markers. By combining these 2 approaches, among 15 distinct clusters in our data set, we identified 3 CD8+ T cell subsets (naive, activated, and activated cycling), 1 CD4+ T cell subset, 3 neutrophil/granulocyte subsets (Ly6a+, Cxcl10+, and Arg1+), 2 macrophage subsets (F4/80– and F4/80+), 4 DC subsets (Cd209+, Xcr1+, Langerhans, and plasmacytoid), and subsets of NK cells and monocytes (Figure 1C and Supplemental Figure 5). Differential expression analysis between clusters revealed distinct gene signatures in support of unique molecular characteristics of clusters (Figure 1D). Among these clusters, miR-155 host gene (Mir155hg) expression was detected in activated T cells, NK cells, and Langerhans DCs (Figure 1E).To verify the algorithm-assisted identification of cell clusters, we examined the expression of known cellular markers in our data set. As expected, the expression of these markers corresponded with the respective identities of clusters (Figure 1F). Not surprisingly, a considerable overlap was observed in the gene expression profiles of closely related cell clusters. These results underscore the need for a multidimensional analytical approach to characterize cellular heterogeneity in the TME and suggest that lymphoid and myeloid cell subpopulations in the TME exhibit distinct molecular characteristics.
Upon confidently identifying cell clusters, we analyzed the differences between WT and miR-155 TCKO tumor-infiltrating immune cells on days 9 and 12 after tumor injection. When the data from 2 time points and genotypes were plotted side by side, time- and genotype-dependent dynamics within the TME started to emerge (Figure 1G). As early as day 9, the activated CD8+ T cell cluster was enriched in WT samples compared with miR-155 TCKO counterparts; however, on day 12, this difference was much more pronounced (Figure 1, G and H). At this later time point, the activated T cell cluster was present in miR-155 TCKO mice but was represented at a lower frequency (Figure 1G, gray arrows). A distinct population of activated CD8+ cells characterized by the expression of cell cycle genes (hence named “activated cycling CD8+ cluster”) was also enriched in WT mice on day 12. Interestingly, the activated cycling CD8+ T cell cluster was more pronounced in WT mice on day 12 compared with WT mice on day 9 (Figure 1G, red arrows). NK cells and the Xcr1+ DC subset were also enriched in WT samples on day 12, although they looked similar between WT and miR-155 TCKO samples on day 9 (Figure 1, G and H). In contrast, several myeloid populations were expanded in tumors grown in miR-155 TCKO mice by day 12. This was true for all 3 neutrophil subsets and monocytes in our data set (Figure 1, G and H). The levels of F4/80+ macrophages and pDCs were also elevated, albeit to a lower extent, in miR-155 TCKO tumors on day 12. Taken together, these data suggest that antitumor immune cells expand within the TME between days 9 and 12 of tumor growth in WT mice, whereas in miR-155 TCKO mice, the TME shifts toward a protumorigenic state in the absence of a productive T cell response.
T cell–intrinsic miR-155 is essential for antitumor immune activation.
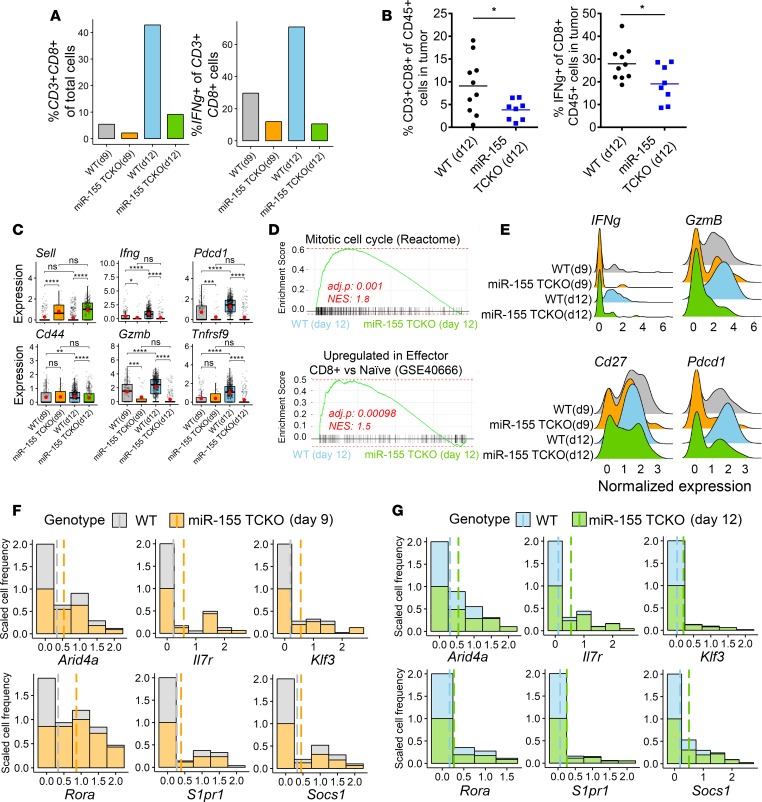
miR-155 was previously shown to be important for optimal T cell antitumor responses (6, 11, 12), but the molecular characteristics of miR-155–competent and –deficient T cell subsets in the TME have not been studied temporally. Next, we focused our analysis on infiltrating T cells to investigate the effects of miR-155 loss on T cell activation phenotypes. To complement our findings at the cluster level, we first subsetted single-cell gene expression data based on concomitant expression of CD3e and CD8a, as broad markers of cytotoxic T cells. As expected, the loss of miR-155 resulted in lower frequencies of intratumoral CD3+CD8+ T cells at both days 9 and 12 (Figure 2A). Interestingly, the overall frequency of CD3+CD8+ cells increased in miR-155 TCKO mice from day 9 to day 12, but this increase was attenuated compared with the increase observed in WT mice. When we analyzed interferon-γ (Ifng) expression within these CD3+CD8+ cells, we observed an approximately 2-fold induction in WT mice from day 9 to day 12, while T cells from miR-155 TCKO mice lacked the ability to induce IFN-γ. We validated our findings from SCseq by using multicolor flow cytometry and observed lower percentages of CD3+CD8+ T cells and a diminished production of IFN-γ by these cells in tumors grown in miR-155 TCKO versus WT mice (Figure 2B). We then proceeded to analyze the expression levels of known T cell activation markers and effector molecules within the CD3+CD8+ subset. CD44 and L-selectin (CD62L, encoded by the Sell gene) are 2 commonly used markers to distinguish activated (CD44hiCD62Llo) and naive (CD44loCD62Lhi) T cell subsets. Supporting our findings in cluster analysis, we observed higher levels of CD44 and lower levels of CD62L in WT CD3+CD8+ T cells, suggesting an activated phenotype (Figure 2C). Both at day 9 and day 12, we observed higher expression levels of Ifng and granzyme B (Gzmb) in WT CD3+CD8+ T cells compared with miR-155 TCKO counterparts, on average (Figure 2C). Both IFN-γ and Gzmb are critical mediators of T cell cytotoxicity against tumors and pathogens (21). An elevated expression of other activation marker genes such as Pdcd1 (encoding PD-1) and Tnfrsf9 (encoding 4-1BB) were observed in WT T cells, particularly by day 12 of tumor progression. These findings suggest that the intratumoral T cell compartment in WT mice is composed of more activated cells compared with miR-155 TCKO mice. In further support of this interpretation, gene set enrichment analysis (GSEA) of CD3+CD8+ intratumoral T cells from WT and miR-155 TCKO mice on day 12 revealed an enrichment for cellular proliferation and effector T cell gene expression signatures for WT samples (Figure 2D). Further, when we limit the analysis to only the activated T cell cluster (as identified in Figure 1), we observed higher expression frequency of multiple activation marker genes including Ifng, GzmB, Pdcd1, and Cd27 (Figure 2E and Supplemental Figure 6). Taken together, these findings suggest that antitumor T cell responses evolve over time and cell-intrinsic expression of miR-155 is essential for T cells to infiltrate the tumor and reach an activated state.
Figure 2. T cell–intrinsic expression of miR-155 is necessary for optimal antitumor T cell activation.
(A) Proportions of cells expressing T cell and activation markers in the SCseq data set (4 mice pooled per group). (B) Flow cytometric analysis of the B16F10-OVA tumor-infiltrating immune cells on day 12 showing elevated levels of CD8+ T cells in tumors of WT mice, and higher levels of IFN-γ production by these cells. Two-tailed t test was used for statistical comparisons. *P ≤ 0.05; ns, P > 0.05. (C) Expression levels of T cell activation markers and effector genes within the CD3+CD8+ cells are shown. Sell, Pdcd1, and Tnfrsf9 encode CD62L, PD-1, and 4-1BB respectively. In these plots, each dot represents a single cell. Normalized expression values were used, and random noise was added to show the distribution of data points. The box plots show interquartile range and the median value (bold horizontal bar). Average expression value per sample is indicated by the red points. Wilcoxon’s test was used for statistical comparisons. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001; ns, P > 0.05. (D) Gene set enrichment analysis (GSEA) of CD3+CD8+ cells in SCseq data on day 12. Normalized enrichment score (NES) and adjusted P value are shown. WT CD8+ T cells were enriched for cell cycle genes and genes upregulated in effector CD8+ T cells (gene sets were derived from MSigDB) (33). (E) Analysis of activation markers within activated CD8+ T cell cluster (as defined in Figure 1) showing a more robust activation phenotype in WT T cells. (F) Analysis of the miR-155 target gene expression in activated CD8+ T cell clusters on day 9. The x axis of the stacked histograms indicates the normalized expression values of target genes and the y axis indicates the scaled frequency of cells. To account for differences in cell numbers per sample, the frequency of cells with no expression of the indicated genes was scaled to 1. Dashed lines indicate the average expression values in each group. (G) Analysis of miR-155 targets in activated CD8+ T cell clusters on day 12. Data were scaled similarly to panel F.
Lastly, we wanted to examine the expression levels of miR-155 targets at single-cell resolution. While the analysis of miR targets via commonly used methods such as qPCR, bulk RNAseq, and Western blotting can provide an overall measure from a mixed cell population, delineating the effects of miRs in individual cells and studying the heterogeneity of cellular responses to miRs are not possible with these methods. For instance, upon experimental manipulation of a miR, the target gene expression may be altered uniformly across the population to a certain extent, or it could be altered in a small subset of cells to a much greater level resulting in the same overall observed changes. In this context, SCseq is a novel tool capable of examining the expression of miR targets at the individual cell level. Since miR-155 expression is induced in T cells upon activation and floxed miR-155 in our model is deleted only in T cells, we investigated the expression levels of miR-155 targets only in activated T cell clusters within our data set. We found that these genes were not ubiquitously expressed and had variable expression across the data set. To address these issues and to show side-by-side results from WT and miR-155 samples with different numbers of T cells, we scaled the frequency of cells to the same relative abundance (the frequency of cells not expressing a gene of interest was normalized to 1, and the relative frequencies of cells expressing the gene was calculated based on this normalized value). As expected, the relative number of cells expressing the target genes was higher in miR-155 TCKO mice at both time points (Figure 2, F and G). Interestingly, the difference between WT and miR-155 TCKO T cells was consistent across the spectrum of target gene expression, suggesting a broad derepression of miR-155 targets as opposed to high levels of upregulation in a narrow subset of T cells upon deletion of miR-155. These results indicate that the expression of miR-155 target genes is similarly affected across single T cell clones, supporting the idea that miR-155 expression uniformly modifies target genes within this T cell population in our experimental model.
T cell miR-155 controls the phenotype of tumor-infiltrating myeloid cells.
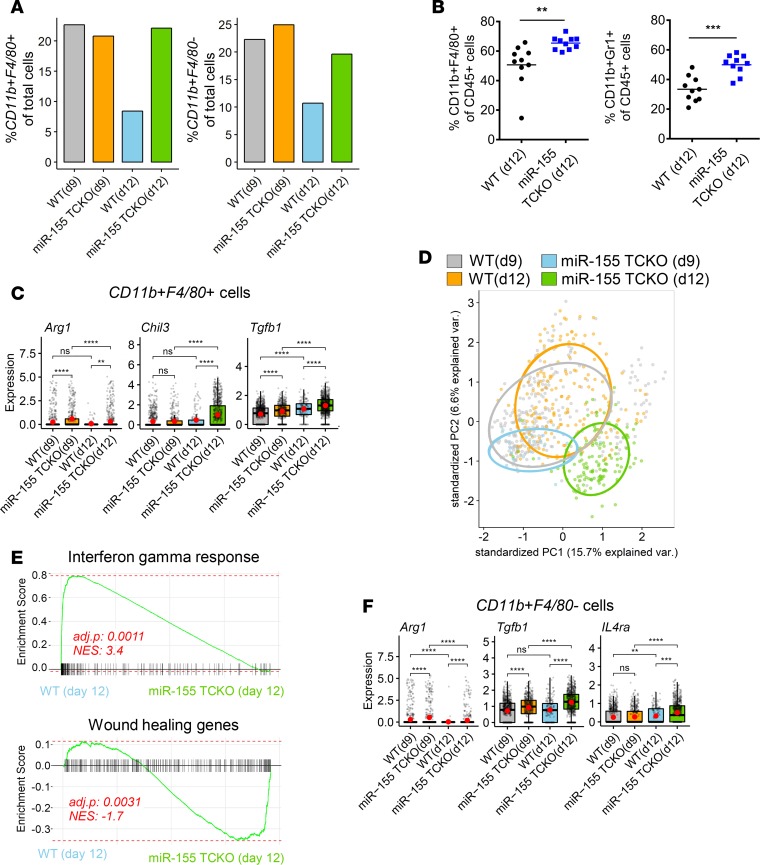
Upon activation, T cells produce several molecules that can interact with myeloid cells and regulate their function (22). Next, we proceeded to analyze the myeloid cells in tumors grown in WT and miR-155 TCKO mice. We first examined the frequency of cells expressing Itgam (encoding CD11b) and Adgre1 (encoding F4/80) markers as a measure of macrophages in the TME. We did not observe obvious differences at day 9 between WT and miR-155 TCKO mice, but on day 12, miR-155 TCKO mice had more than 2-fold higher frequency of macrophages compared with WT mice (Figure 3A). Interestingly, the levels of intratumoral macrophages in WT mice decreased from day 9 to day 12. When we limit our analysis to CD11b+F4/80– cells as a surrogate for a general myeloid cell phenotype, we also observed a higher proportion in miR-155 TCKO mice on day 12 (Figure 3A). Flow cytometric analysis of tumor-infiltrating immune cells validated the observations of SCseq and showed a higher frequency of CD11b+F4/80+ macrophages and CD11b+Gr1+ myeloid-derived suppressor cells (MDSCs) in miR-155 TCKO mice on day 12 (Figure 3B). CD11b+F4/80+ macrophages in tumors grown in miR-155 TCKO mice expressed higher levels of arginase 1 (Arg1), TGF-β (TGFb1), and chitinase-like 3 (Chil3, also known as Ym1), which are commonly used markers to define the protumorigenic M2 state (Figure 3C). Changes in the expression levels of Arg1 and Chil3 were validated by qPCR (Supplemental Figure 7) (23). Principal component analysis (PCA) of F4/80+ macrophage clusters (as defined in Figure 1) using consistently expressed immune-associated genes defined a distinct molecular phenotype for macrophages in miR-155 TCKO mice on day 12, suggesting the myeloid compartment of the TME undergoes phenotypic evolution over time (Figure 3D). When the global gene expression signatures of CD11b+F4/80+ macrophages were examined, IFN-γ–response genes were found to be enriched in WT mice, whereas miR-155 TCKO macrophages were enriched for a wound-healing phenotype at both days 9 and 12 (Figure 3E and Supplemental Figure 8A). Interestingly, when CD11b+F4/80+ cells from WT mice were compared between the 2 time points, we observed a significant enrichment of IFN-γ–response genes on day 12 (Supplemental Figure 8B), supporting the interpretation that antitumor immune response is enhanced by day 12. Additionally, the enrichment of an IFN-γ–response signature was evident in several other cell types in the WT tumor microenvironment by day 12 including neutrophils and various DC populations, suggesting IFN-γ secreted by T cells interacts with multiple cell types in the TME (Supplemental Figure 9). Complementing our observations with CD11b+F4/80+ macrophages, we also found Arg1 and TGFb1 to be elevated in CD11b+F4/80– cells within the TME of miR-155 TCKO mice (Figure 3F). Furthermore, IL4Ra, a marker of MDSCs in both mice and humans (24) was expressed at a higher frequency in CD11b+F4/80– cells of these mice (Figure 3F), suggesting a greater portion of CD11b+F4/80– cells develop into an MDSC-like phenotype when miR-155 expression is lost in T cells. These findings demonstrate how tumor-infiltrating immune cells in WT mice function together to mount a potent antitumor immune response by day 12 of tumor growth, and show that the loss of miR-155 in T cells reprograms the TME toward a protumorigenic state by affecting multiple myeloid cell subsets.
Figure 3. T cell–specific expression of miR-155 regulates the myeloid populations within the tumor microenvironment.
(A) The proportions of cells expressing Itgam and Adgre1, which encode myeloid/macrophage markers CD11b and F4/80, respectively. Graphs show an enrichment of macrophages (CD11b+F4/80+) and other myeloid-lineage cells (CD11b+F4/80–) in miR-155 TCKO mice on day 12. (B) Flow cytometric analysis of the tumor microenvironment shows a higher frequency of CD11b+F4/80+ macrophages and CD11b+Ly6G+ myeloid-derived suppressor cells (MDSCs) in B16F10 tumors growing in miR-155 TCKO hosts. (C) Analysis of protumorigenic M2 macrophage marker genes in SCseq data. The box plots show the interquartile range and the median value (bold horizontal bar). Average expression value per sample is indicated by the red points. (D) Principal component analysis (PCA) of the F4/80+ macrophage cluster (as defined in Figure 1) using 142 consistently expressed immune-associated genes. Genes were selected based on “mouse immune process” GO annotation and by filtering out genes with average expression counts less than 1 per cell. F4/80+ macrophages in miR-155 TCKO mice on day 12 have a unique gene expression profile as evidenced by spatial separation in the PCA plot. (E) Analysis of gene set enrichment in CD11b+F4/80+ macrophages on day 12. Normalized enrichment score (NES) and adjusted P value are shown. WT macrophages were enriched for IFN-γ–response pathway genes, whereas macrophages from miR-155 TCKO mice upregulated genes related to wound healing pathways (gene sets derived from MSigDB) (33). (F) Expression levels of MDSC-associated genes within CD11b+F4/80– cells of the SCseq dataset, suggesting an MDSC-like phenotype for F4/80– myeloid cells in miR-155 TCKO mice. Graphs were prepared similarly to those in panel C. **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001; ns, P > 0.05 by Wilcoxon’s test.
miR-155 expression defines an immune-enriched subtype of human melanoma.
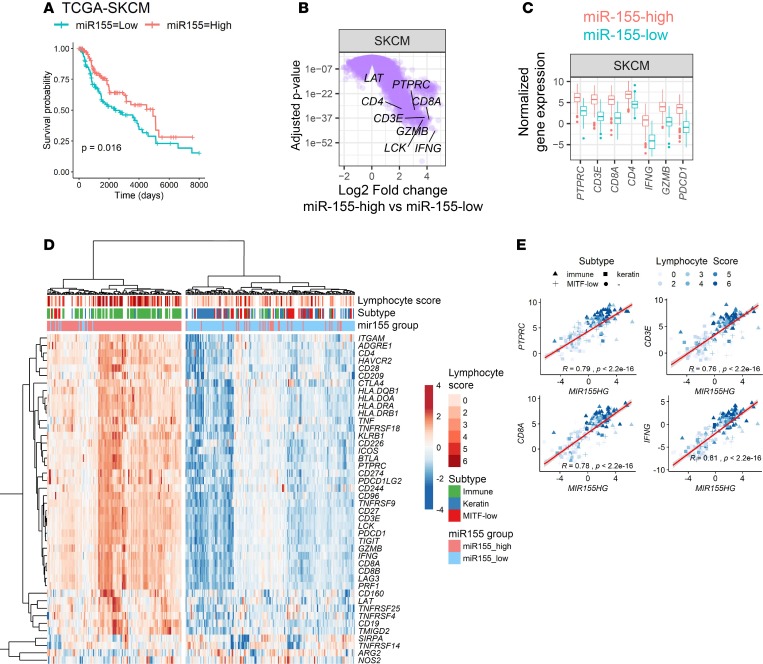
In addition to their roles in immunity, miRs are known to be important determinants of tumorigenesis (25). Specifically, miR-155 was shown to have both protumorigenic (8, 26) and antitumorigenic properties (27, 28) depending on the tumor context in a tumor cell–intrinsic manner. Because miR-155 expressed in tumor cells or in infiltrating immune cells can exert different functions, roles of miR-155 in determining tumor prognosis in human cancers remain to be elucidated. Following on our findings in murine B16 melanoma model, we first focused on miR sequencing (miRseq) data from human skin cutaneous melanoma (SKCM) in TCGA database to assess whether miR-155 expression correlated with survival outcome and clinical parameters. Expression levels of mature miR-155 across the SKCM patient group were categorized at the top and bottom thirds and Kaplan-Meier survival curves were plotted from miR-155–high and miR-155–low groups, with 148 patients each. Our analysis showed that higher expression of miR-155 significantly marks a subpopulation of patients with a favorable clinical outcome in SKCM (Figure 4A). We then turned to RNAseq data from these patients to investigate the differential gene expression between miR-155–high and miR-155–low groups. Several hundred genes were found to be differentially regulated in miR-155–high melanoma patients (1,784 genes were upregulated, and 206 genes were downregulated >2-fold, with adjusted P value of < 0.05) (Figure 4B). The PTPRC gene encoding pan-leukocyte marker CD45 was found to be among significantly upregulated genes, suggesting an immune-enriched subtype in the miR-155–high cohort (Figure 4, B and C). Interestingly, several genes associated with T cells including T cell receptor complex signal transducers (LCK and LAT), coreceptors (CD4 and CD8A), and effector molecules (IFNG and GZMB) were also upregulated, suggesting the presence of activated T cells in the TME (Figure 4B). The majority of these genes exhibited more than 4-fold upregulation in miR-155–high tumors (Figure 4, B and C).
Figure 4. miR-155 expression defines an immune-enriched phenotype in human cutaneous melanoma.
(A) Kaplan-Meier survival curves showing an improved clinical outcome in miR-155–high subpopulation of skin cutaneous melanoma (SKCM) patients (red line). For this analysis, miRseq data from The Cancer Genome Atlas (TCGA) was categorized at the top and bottom thirds, resulting in 148 patients per group. P value of the log-rank test is shown in the graph. (B) Volcano plots showing differentially expressed genes in miR-155–high SKCM subpopulation. This plot was generated by analyzing RNAseq data from the same patients categorized in panel A. T cell–associated genes are indicated, showing a significant upregulation in the miR-155–high cohort. A linear fit model was used and the P values were corrected using the Benjamini-Hochberg method. (C) Comparison of the T cell–associated gene expression between miR-155–high and miR-155–low SKCM subpopulations. (D) Heatmap showing the correlation between miR-155 expression and SKCM clinical subtype and lymphocyte infiltration scores. Lymphocyte score and tumor subtype for SKCM were defined in a study from TCGA consortium (29). Several immune-related genes showed higher levels of expression in the miR-155–high SKCM subset. Hierarchical clustering was performed by using Euclidean distances and the ward.D2 algorithm. (E) Scatter plots showing the positive correlation between miR-155 host gene (MIR155HG) and immune-associated gene expression in TCGA-SKCM. Color gradient indicates the lymphocyte infiltration score of tumors, and the symbol shape indicates the molecular subtype of tumors (▲ immune, ■ keratin, + MITF-low, ● uncategorized). Student’s t test was used for the statistical analysis of the slope of regression line.
A landmark study published by TCGA consortium in 2015 described multiple molecular and clinical characteristics of SKCM (29). In this study, the authors classified melanoma samples into 3 groups as immune-rich, keratin-rich, and microphthalmia-associated transcription factor–low (MITF-low) subclasses depending on the expression of immune- and tumor-specific markers (29). We next wanted to investigate the clinical classifications and immune-associated gene expression profiles of miR-155–high and miR-155–low tumor subgroups in SKCM. Interestingly, the miR-155–high subpopulation of SKCM tumors expressed higher levels of several immune-related genes including general immune cell markers, costimulatory and coinhibitory receptors, and effector molecules (Figure 4D). Of note, mediators of a protumorigenic phenotype such as NOS2 and ARG1 (30) were expressed at slightly higher levels in the miR-155–low group. Strikingly, the miR-155–high subpopulation of patients corresponded to an immune-rich melanoma subtype, as described in TCGA study (Figure 4D, green column annotations) (29). These findings were further supported by higher lymphocyte infiltration scores in miR-155–high tumors (Figure 4D, dark red column annotations), as described in TCGA study (29). Interestingly, when we examined the relationship between the expression levels of miR-155 and various immune genes, we noted a strong and nearly linear positive correlation, suggesting the miR-155 expression continuum overlaps with the immune signature in the TME (Figure 4E and Supplemental Figure 10).
miR-155–high human melanoma tumors are highly infiltrated by lymphocytes.
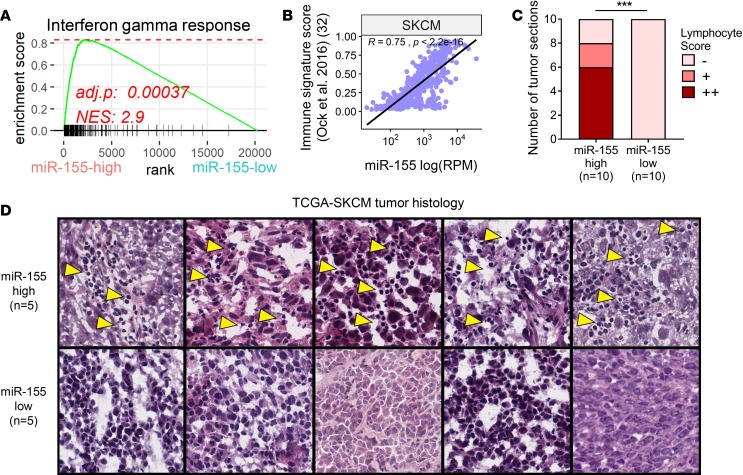
Upon observing a positive correlation between miR-155 and immune gene expression, we next performed GSEA in the miR-155–high versus miR-155–low patient cohorts to characterize active signaling networks. In agreement with higher IFNG transcript levels, we observed an enrichment of IFN-γ–response gene signature in miR-155–high tumors (Figure 5A). Furthermore, miR-155–high melanoma subset was significantly enriched for other inflammatory pathways including tumor necrosis factor α (TNFa), IFN-α (IFNa), and interleukin 2 (IL2) signaling (Supplemental Figure 11). In contrast, the miR-155–low melanoma subset was characterized by an enrichment in proliferative pathways, indicative of a higher representation of transcripts from tumoral origins in RNAseq data (Supplemental Figure 12). Of note, the miR-155–low subset also exhibited an enrichment of genes regulated by MYC, an oncogene that was shown to negatively regulate antitumor immune responses (Supplemental Figure 12) (31).
Figure 5. miR-155 expression is correlated with parameters associated with immunotherapy response.
(A) GSEA plot showing enrichment of IFN-γ–response gene signature in miR-155–high TCGA melanoma patients. Normalized enrichment score (NES) and adjusted P value are shown. (B) miR-155 expression is strongly correlated with an immune signature (IS) score (Ock et al.; ref. 32) that predicts immunotherapy response. (C) Histological assessment of 20 TCGA melanoma tumor samples with the highest and lowest miR-155 expression reveals a brisk lymphocyte infiltration in the miR-155-high subset. A (–) denotes tumor sections with no appreciable immune cell presence. A (+) indicates sections in which a non-brisk and localized lymphocyte infiltrate was observed. A (++) indicates sections with brisk and widespread lymphocyte infiltration were evident. Statistical comparison was performed by using χ2 test. ***P ≤ 0.001. (D) Representative H&E–stained tumor sections used for the quantification of lymphocyte infiltration in panel C. Five samples with the highest and 5 samples with the lowest miR-155 expression are shown. TCGA patient identifiers for these images are as follows (in descending order of miR-155 expression): TCGA-D9-A149-06A, TCGA-D3-A2JH-06A, TCGA-D3-A1QB-06A, TCGA-D3-A51F-06A, TCGA-D9-A6E9-06A, TCGA-EE-A3J8-06A, TCGA-YD-A89C-06A, TCGA-FS-A4F0-06A, TCGA-GN-A262-06A, and TCGA-WE-A8ZM-06A. Yellow arrowheads indicate lymphocyte infiltrates in the tumor. Images in D are from TCGA’s web interface (http://cancer.digitalslidearchive.net/).
Understanding which cancer patients will respond to immunotherapy is an important clinical challenge. A recent study examined gene expression profiles of TCGA tumors and defined an “immune signature (IS) score” that predicts responses to various forms of immunotherapy (32). In this study, IS score was found to positively correlate with an IFN-γ–response gene signature and tumoral lymphocyte infiltration. Since miR-155 expression also marks an immune-rich subtype in human melanoma, we next wanted to determine the association between IS score and miR-155 expression levels. Our analysis revealed that miR-155 expression is strikingly correlated with IS score in human melanoma (Figure 5B). Importantly, when the 10 highest and 10 lowest miR-155–expressing TCGA melanoma samples were examined histologically, a brisk lymphocyte infiltration was evident in the miR-155–high subset (Figure 5, C and D). Taken together, these findings further support the role of miR-155 in defining an immune-rich subtype in human melanoma and suggest that intratumoral expression of miR-155 may have important clinical implications.
miR-155 expression positively correlates with immune enrichment in a variety of distinct human solid tumors.
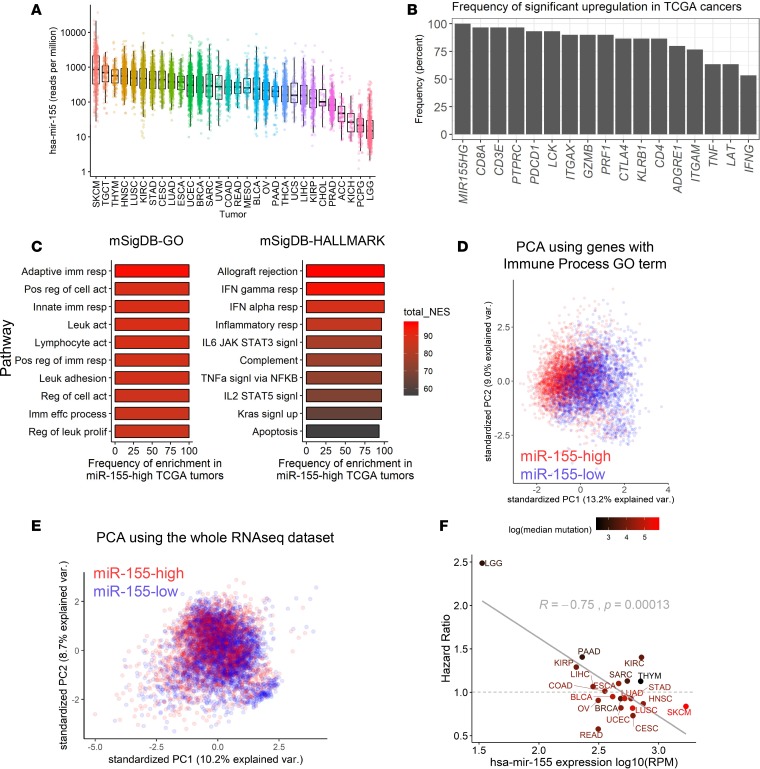
TCGA database currently hosts data from 29 other solid tumor types originating in different tissues (https://cancergenome.nih.gov/). We first surveyed miR-155 expression in TCGA miRseq data sets and observed variable levels of expression across different tumor cohorts, with SKCM having the highest median miR-155 levels (Figure 6A). We next wanted to investigate the impact of miR-155 on global gene expression and clinical outcome in different TCGA cohorts. To this end, we categorized mature miR-155 expression levels in each cancer type by dividing the patient cohort to the top and bottom thirds, as previously done for SKCM, and performed differential expression analysis by using publicly available RNAseq data. To examine the commonalities in differential gene expression among TCGA data sets, we calculated the frequency at which a gene is significantly upregulated in the miR-155–high subsets of TCGA cohorts. In this analysis, if a gene is found to be differentially expressed in the miR-155–high subsets of all TCGA cancers analyzed, the frequency of differential expression would be 100%, whereas if a gene is significantly upregulated only in half of the TCGA cancers, the frequency would be 50%. By using this approach, we summarized the frequency of differential upregulation for select immune genes in miR-155–high patient subsets (Figure 6B). As expected, expression of miR-155 host gene (MIR155HG) was elevated in the patient subsets where the mature form of miR-155 was higher (Figure 6B). Interestingly, the PTPRC (CD45) gene was found to be upregulated in 96.6% (29 of 30) of the miR-155–high TCGA tumor cohorts. Furthermore, genes encoding T cell signal transducers and coreceptors such as CD3E, LCK, and CD8A were all upregulated more than 90% of the time within miR-155–high cancers across TCGA data set (Figure 6B and Supplemental Figure 12). Transcript levels of T cell effector genes such as IFNG, GZMB, PRF1, and TNF were also elevated in at least 50% of these tumors. Of note, multiple other genes encoding canonical markers of immune cell subsets such as KLRB1 (NK1-1; NK cells), ITGAM (CD11B; macrophages and granulocytes), ITGAX (CD11C; DCs), and ADGRE1 (F4/80; macrophages) were also upregulated in miR-155–high cancer subsets (Figure 6B). To examine functional pathways in these miR-155–high tumor subsets, we performed GSEA by using gene ontology (GO) and HALLMARK gene sets from the Molecular Signature Database (MSigDB) (33). High expression of miR-155 correlated with a strong enrichment of adaptive and innate immune pathways in all solid tumors analyzed regardless of their origin (Figure 6C and Supplemental Figure 13). These observations demonstrate that miR-155 expression defines a multicellular immune-enriched phenotype in a wide variety of human solid tumors.
Figure 6. miR-155 positively correlates with immune infiltration in human solid tumors.
(A) Mature miR-155 transcript reads per million in the log scale were plotted across TCGA cohorts. (B) Summary of differential upregulation of immune-associated genes in miR-155–high tumor subsets. The frequency at which the genes were found to be upregulated across TCGA tumors is plotted. (C) Summary of top GSEA results from miR-155–high subsets of TCGA tumors. Color scale indicates the cumulative normalized enrichment score (NES) across 30 TCGA cohorts. (D) PCA of immune-associated gene expression using miR-155–high versus miR-155–low subsets of TCGA tumors. Data from 30 different TCGA cohorts were aggregated, resulting in 6,414 patients (3,201 belonging to miR-155–high subset and 3,213 belonging to miR-155–low subset). A total of 2,038 genes with human “immune process” GO annotation were used for this analysis. Separation of red and blue points indicates that miR-155–high tumors have a shared immune-related gene expression profile that is distinct from miR-155–low tumors across TCGA data sets. (E) PCA of TCGA tumor samples by using all shared genes in RNAseq data sets (15,151 genes analyzed). (F) Scatter plot showing an inverse correlation between the univariate Cox proportional hazard ratio (HR) of miR-155 and the average levels of miR-155 expression in TCGA tumors. Dashed line denotes HR of 1, which indicates no effect on survival. Red color gradient indicates the median genomic mutational burden in these tumors. High mutational burden was found to be loosely associated with a lower HR (i.e., improved clinical outcome). BLCA, bladder urothelial carcinoma; LGG, brain lower grade glioma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; READ, rectal adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma.
To analyze the immune landscape from a multidimensional point of view, we performed PCA with 2,038 genes annotated with the Immune Process GO term by using RNAseq data from TCGA solid tumors. Even though the tumor RNAseq data in our analysis originated from several different tissue types, high miR-155 expression designated tumors with a common immune signature distinctly from miR-155–low counterparts, as evidenced by the spatial separation of data points in the PCA plot (Figure 6D). The same trend was observed when PCA was performed with 548 immune-associated genes downloaded from the ImmPort database (Supplemental Figure 14, A and B) (34). Not surprisingly, when all the genes in the RNAseq data set were used in PCA, high miR-155 expression did not define a distinct tumor population (Figure 6E), suggesting that miR-155 expression correlates with a consistent immune gene signature rather than describing a difference in global gene expression patterns in human solid tumors. Surprisingly, the level of miR-155 expression was found to correlate positively with the IS score (reported by Ock et al., ref. 32) across TCGA solid tumors, mirroring our findings in melanoma (Supplemental Figure 15), further supporting the role of miR-155 in defining a common immune involvement in human cancer.
Lastly, we wanted to investigate the relationship between miR-155 expression in the TME and the survival outcome in human cancers. A univariate Cox proportional hazards model was used to investigate the correlation between miR-155 expression and patient survival data for each TCGA solid tumor cohort. For this analysis, we excluded TCGA tumor types (i) that consisted of fewer than 100 patients or (ii) tumors in which a consistent, time-dependent decline in patient survival was lacking (see Figure 6F and Supplemental Figure 16 for the tumor cohorts included in the analysis). Results of the survival analysis revealed that, although miR-155 expression had a variable relationship with survival depending on the tumor context, it was inversely correlated with the hazard ratio (HR) from the Cox survival model (Figure 6F and Supplemental Table 1). In these analyses, an HR larger than 1 indicates a poorer survival (i.e., increased hazard), while an HR smaller than 1 indicates an improved survival (i.e., decreased hazard or protection). SKCM had an HR value of 0.84, corresponding to a 16% decrease in clinical hazard in the miR-155–high patient subset (Figure 6F). Interestingly, when the mutational burden of these TCGA tumors was analyzed, we observed that cancers in which miR-155 correlated with a more favorable clinical outcome (HR < 1) tended to have a higher mutational load (Figure 6F, red-colored data points). Among the TCGA cohorts, SKCM exhibited the highest mutational burden, followed by lung squamous cell carcinoma (LUSC), where, in both cases, miR-155 expression correlated with an improved prognosis. In summary, miR-155 expression defines an immune-enriched subtype of human cancers and may have an impact on the tumor prognosis, although the ultimate survival outcome is determined by the contribution of other tumor-specific factors such as mutational burden and the composition of the TME.
Discussion
In this study, we described the molecular characteristics of antitumor immune responses mediated by T cell–specific miR-155 at a cellular resolution via SCseq technology. Our analysis at 2 different time points revealed distinct activation states of tumor-infiltrating immune cells and delineated the dynamic interactions of these cells within the TME. Supporting the previous literature, loss of miR-155 in T cells was detrimental to antitumor T cell responses (11, 12). Importantly, T cell–specific expression of miR-155 was also critical for shaping the intratumoral myeloid immune compartment both in terms of cellular composition and activation states. Although miR-155’s role in antitumor immunity was evident early during tumor development (day 9), its importance was more pronounced at a later time point (day 12), suggesting a continued evolution of antitumor immune responses. Analysis of TCGA data from human melanoma revealed that miR-155 expression correlates with an immune-enriched subtype and is associated with an improved clinical outcome (29). Furthermore, analysis of TCGA data revealed that miR-155 expression marked an immune-enriched phenotype in 29 other solid tumor types. Although miR-155 expression did not always correlate with an improved clinical outcome in TCGA tumors, the overall expression level of miR-155 was found to have an inverse relationship with the clinical HR in a univariate survival analysis. These findings corroborate the roles of miR-155 in promoting antitumor immunity and suggest that miR-155 can be a marker of immune enrichment in human cancers.
miR-155 is extensively studied in the contexts of inflammation and carcinogenesis. While miR-155 expression is induced in immune cells upon stimulation (4, 7), cancer cells of various origins frequently overexpress miR-155 (35). Tumor-intrinsic miR-155 was shown to have both tumor-promoting (3, 8, 26) and tumor-suppressing functions (9, 27). Although miR-155 largely contributes to inflammatory responses in a variety of immune cell types (4, 7, 13, 18, 36, 37), it can also promote the function of regulatory T cells, which are important negative regulators of the immune response (38). Therefore, miR-155 expression within the TME can have variable effects on cancer prognosis depending on which cells express miR-155. Our findings here, however, suggest that higher expression of miR-155 in the tumors correlate with an improved survival in human cancer patients. Against the backdrop of renewed interest in miR therapeutics, characterization of miR-155–expressing cells within the TME is critical to reveal which populations are dominant and can be targeted for therapeutic purposes.
Previous studies reported that miR-155 is induced in T cells upon activation (7, 12, 36). In support of these observations, SCseq data in this study revealed significant levels of Mir155hg expression in activated CD8+ T cell clusters, which constitute greater than 30% of the tumor-infiltrating immune cells in WT hosts by the 12th day of tumor growth. Deletion of miR-155 specifically in T cells resulted in lower levels of activated T cells and higher levels of myeloid cells in the TME, indicating an inverse relationship between tumor-infiltrating T cells and myeloid cell subsets. Interestingly, when the gene expression landscape of human tumors is examined, both T and myeloid cell–specific genes were frequently coexpressed within the miR-155–high tumor subsets, suggesting a broad infiltration of multiple immune cell types. This may be explained by induction and activity of miR-155 in multiple cell types in the TME. Indeed, miR-155 can be expressed in a variety of immune cells other than activated T cells including macrophages, DCs, NK cells, and B cells (4, 37, 39). Interestingly, miR-155 was shown to convert immunosuppressive tumor-associated DCs to an immunostimulatory phenotype in an ovarian cancer model, suggesting the miR-155 axis can be functional in other immune cells in the TME (40). Albeit in smaller proportions, we detected Mir155hg expression in NK cells and subsets of DCs and macrophages in support of these previous findings. However, due to low sequencing coverage of SCseq, other cells expressing Mir155hg at low levels may be present in our data set but remain under the detection limit. Similarly, we were unable detect appreciable levels of Ly6G expression in myeloid populations, a marker commonly used in conjunction with CD11b to define MDSCs, although B16 tumors were shown to be rich in MDSCs (41). Immunomodulatory markers commonly observed in MDSCs were expressed in a considerable portion of CD11b+F4/80– myeloid cells in our data set, which is consistent with the presence of MDSCs in the murine melanoma TME. These findings suggest that a deeper sequencing coverage may be needed to detect genes with low transcript counts in SCseq experiments and underscore the importance of analyzing multiple parameters to define the identity and activation state of cells in the TME.
SCseq technology is increasingly utilized in tumor immunity studies and provides an unprecedented insight into the dynamics of tumor-infiltrating immune cells. By identifying several distinct myeloid and lymphoid cell clusters, our study demonstrates the distinct activation states of immune cells in the murine melanoma TME. Most of the closely related clusters identified by SCseq through multidimensional analysis of gene expression would have otherwise been characterized as the same cell type via routine immunophenotyping methods such as flow cytometry. This emphasizes the importance of high-throughput analyses for the characterization of the cellular heterogeneity within the TME. SCseq in this study also enabled us to assess the expression levels of miR-155 targets in individual cells and describe the genes that are consistently derepressed in activated T cells upon loss of miR-155 at different time points. Therefore, our data set can serve as a resource to survey known and novel miR-155 targets in the context of an evolving antitumor immune response. In addition to describing an inept T cell phenotype, our study revealed increased levels of pDCs, and multiple subsets of neutrophils and macrophages in miR-155 TCKO mice. These cell types were suggested to promote tumor immunoevasion in various experimental settings (42, 43) and will be further investigated in the context of miR-155–dependent antitumor immunity.
Our findings suggest that miR-155 expression may be a general marker of immune infiltration in human cancer. However, since miR-155 can be expressed by both tumor cells and immune cells in the TME, elucidating the cellular origins of the miR-155 transcript in bulk RNAseq is challenging. Deconvolution approaches that aim to distinguish gene signatures of cancer and immune cells in RNAseq data can potentially be employed to address this issue and reveal detailed immunobiological characteristics of miR-155–high and –low tumors (44, 45). Our data showing a strong positive correlation between the expression levels of miR-155 and various immune-associated genes suggest that the miR-155 transcript in bulk tumor RNAseq originates from the immune component. This is further supported by the results of PCA that revealed a consistent immune gene signature within miR-155–high tumor subsets, and by the results of GSEA, which showed a significant enrichment of several inflammatory pathways upon high expression of miR-155. Moreover, miR-155–high tumor subsets were characterized by a more predominant immune cell infiltration at the histological level in SKCM. Observing similar trends in both molecular and histological analyses of miR-155–high tumor subsets is intriguing because, while gene expression profiling requires extraction of RNA from a considerably sized tumor tissue, a histological preparation only samples a few micrometers of the tumor. Despite the caveat of subsampling, histological assessment of immune infiltration is a common clinical practice in human melanoma where studies have revealed a positive correlation between intratumoral lymphocyte levels and the clinical prognosis (46). Our findings suggest that the enhanced immune signature marked by higher expression of miR-155 did not always correlate with an improved clinical outcome across TCGA data sets, suggesting that tumor-specific features can play an important role in shaping the antitumor immune response and tumor prognosis. Two tumor types from TCGA data set in which miR-155 expression was associated with a better clinical outcome were SKCM and LUSC, which are characterized by the highest mutational burden among TCGA cohorts. High genomic instability and mutational burden are thought to generate neoantigens and contribute to tumor immunogenicity (47). Supporting this notion and findings of this study, previous multivariate analyses of TCGA data revealed that tumor mutational burden correlated with immune infiltration overall, but the clinical outcome was determined by the contribution of other factors including the nature of tumor antigens, and the characteristics of the immune pathways involved (48, 49).
The central tenet of cancer immunotherapy is not only enhancing immune cell infiltration of tumors but also reprogramming the intratumoral immune landscape for effective tumor killing. By marking an immune-enriched subtype in human cancer, miR-155 expression may identify patients who are likely to benefit from immunotherapies that reprogram the TME for tumor elimination. Indeed, miR-155 expression within TCGA tumors exhibited a remarkable correlation with a previously calculated immune score that strongly predicts clinical responses to immunotherapy (32). In support of miR-155’s role in immunotherapeutic responses, our findings in preclinical cancer models revealed that, although the effects of miR-155 loss were largely rescued by immune checkpoint blockade therapy, the optimal responses to immunotherapy required a functional miR-155 axis (11). Therefore, by characterizing the dynamics of miR-155–mediated antitumor immunity and by identifying correlations between miR-155 expression and clinical parameters in human solid cancers, our study opens a new avenue for the pursuit of miR-155–targeting therapeutics and immunotherapy.
Methods
Mice
miR-155–floxed mice were described previously (5). To achieve T cell–specific knockout of miR-155, floxed animals were crossed with CD4-Cre mice on the same genetic background. Due to the CD4/CD8 double-positive phase in thymic T cell development, Cre-mediated excision of floxed miR-155 occurs in both helper and cytotoxic T cell subsets (50). As miR-155 WT controls, mice containing a floxed miR-155 cassette, but lacking the Cre recombinase were used. Five to 10 mice per group were used in the experiments, which were repeated at least 2 times.
Cell culture and tumor experiments
B16F10 murine melanoma cells (ATCC, CRL-7475) or B16F10-OVA cells expressing chicken OVA model antigen were used in the tumor experiments (11). Cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin-streptomycin, and L-glutamine. B16F10-OVA cells were provided by Mingnan Chen (University of Utah). To derive B16F10-OVA cells, parental B16F10 cells were transfected with an OVA-expressing plasmid and selected in media containing 0.1 mg/ml G418. Cells were passaged at least once prior to injection and were 80%–90% confluent on the day of injection. Adherent cells were trypsinized and washed 3 times in PBS and adjusted to 1 × 106 cells per 100 μl injection volume in PBS. Cells were injected into shaved flanks of mice under isoflurane anesthesia. Tumor growth was monitored throughout the experimental duration of 9–12 days. At the experimental endpoints of days 9 and 12 following tumor injection, tumors were collected for flow cytometric analysis and SCseq, respectively.
SCseq and analysis
Sample preparation and sequencing.
Tumor tissues were dissociated using scissors and forceps and incubated in 5 ml Accumax (Innovative Cell Technologies) with constant agitation at room temperature for 30 minutes. Tumors from at least 4 mice were pooled together per group. After this enzymatic digestion, cells were stained with DAPI and APC-conjugated CD45 for 15 minutes on ice in PBS containing 2 mM EDTA and 0.5% BSA. Live CD45+ cells were sorted via BD FACSAria cell sorter and washed once in PBS containing 0.04% BSA. Samples were then processed for SCseq via a 10× platform according to the manufacturer’s instructions (10× Genomics). Paired-end RNAseq (125 cycles) was performed via an Agilent HiSeq next-generation sequencer. Sequencing reads were processed by using 10× Genomics CellRanger pipeline and further analyzed with the Seurat R package. The effect of mitochondrial gene representation and the variance of unique molecular identifier (UMI) counts were regressed out from the data set prior to analysis. Gene expression signatures defining cell clusters were analyzed after aggregating 4 samples (WT and miR-155 TCKO, days 9 and 12). The raw data from SCseq experiments in this manuscript can be found in the NCBI’s Gene Expression Omnibus database (GEO GSE121478).
Identification of cell clusters
Cells in our data set were clustered by using the FindClusters function of the Seurat analysis package, which identifies clusters via a shared nearest neighbor (SNN) modularity optimization–based algorithm. This function identified 14 distinct clusters spanning the lymphoid and myeloid cell lineages. We noted that CD4+ T cells were clustered with a larger cell cluster composed mainly of CD8+ T cells. To increase the granularity of our analysis, we manually separated this cluster and named it accordingly, resulting in the 15 clusters analyzed throughout the manuscript. The biological identities of cell clusters were annotated with the help of an immune-cell scoring algorithm (developed in house and available at http://labs.path.utah.edu/oconnell/resources.htm) and by surveying known immune cell markers in the SCseq data set. The immune-scoring algorithm compares the gene signatures of the cell clusters in this study with the publicly available microarray data hosted in the Immunological Genome Project Database (ImmGen). By using differentially expressed gene signatures from Seurat, the immune-scoring algorithm performs the following steps: (a) for each ImmGen cell population, and for each gene found in ImmGen microarrays, it calculates the ratio of normalized microarray signal to the average signal value of the gene from the whole ImmGen data; (b) applies natural log transformation to the ratio, resulting in positive numbers for upregulated genes and negative numbers for downregulated genes in ImmGen data sets; (c) multiplies ImmGen log-ratio values with the log-ratio of matching genes that are differentially expressed in each cell cluster in the SCseq dataset; and (d) sums up scores from all the genes to yield an aggregate identity score for each ImmGen cell type for a given SCseq cluster. In this approach, genes that are differentially upregulated or downregulated in both ImmGen and SCseq data sets contribute to the immune identity score more heavily (a positive number is obtained when 2 log-ratio values with the same sign are multiplied). In contrast, if a gene is inversely regulated in ImmGen and SCseq clusters, the immune identity score is reduced. Through this method, the correlation between the gene expression signatures of SCseq cell clusters in our study and ImmGen data subsets assists in determining the cluster identities. In cases where this algorithm is unable to make a clear call (as in myeloid cell subsets), we surveyed the expression of known genes in the data set and performed differential expression analyses between closely related cell clusters. This approach allowed us to further differentiate subsets of neutrophils, macrophages, and T cell clusters. Upon naming the clusters, the Seurat R package was used to create plots for the expression of selected genes. GSEA analysis was performed by using fgsea R package (51), after ranking genes using a signal-to-noise metric (52).
Flow cytometry
For the staining of tumor-infiltrating immune cells, tumor tissue was mechanically disrupted and incubated for 30 minutes at room temperature in 5 ml Accumax (Innovative Cell Technologies). Following incubation, cells were filtered through a 0.45-μm nylon filter to obtain single-cell suspensions. Cells were stained in Hank’s balanced salt solution (HBSS) supplemented with 0.5% BSA and 2 mM EDTA by using the following fluorophore-conjugated antibodies (purchased from Biolegend or eBioscience/ThermoFisher Scientific): anti-CD3e (clone 145-2C11) (Pacific Blue), anti-CD8a (clone 53-6.7) (APC), anti-CD45 (clone 30-F11) (PE-Cy7), anti-Gr1 (clone RB6-8C5) (PE), anti-CD11b (clone M1/70) (PerCp-Cy5.5), and anti-F4/80 (clone BM8) (PE) at 1:500 to 1:1,000 dilutions. After staining cell surface antigens on ice for 15 minutes, cells were washed and analyzed using a BD LSRFortessa flow cytometer. Data analysis was done using FlowJo (Tree Star) and GraphPad Prism software.
Analysis of TCGA data
Categorization of miR-155 expression and survival analysis.
Publicly available TCGA data were downloaded using TCGABiolinks and RTCGA R packages (53, 54). Reads-per-million miR counts from miRseq experiments were used for the categorization of miR-155 expression levels in cancer patients. Glioblastoma tumors (TCGA-GBM) were excluded from analysis due to the lack of miRseq. In the remaining solid tumor cohorts, data only from primary tumors were considered for all tumor types except cutaneous melanoma (TCGA-SKCM). For SKCM, both metastatic and primary tumor samples were analyzed because the majority of the samples were derived from metastatic tumors. In rare cases where both primary and metastatic tumor data are available from the same patient, only the primary tumor data were included in the analyses. For each type of tumor, hsa-mir-155 expression was categorized at the top- and bottom-third quantiles (67th and 33rd percentile), and patient subpopulations were designated as miR-155–high and miR-155–low based on these cutoffs. Survival analysis was performed by using survival and survminer R packages. Survival outcome in miR-155–high versus miR-155–low patient subpopulations was examined by using Kaplan-Meier, log-likelihood statistics, and the univariate Cox proportional hazards model. For this analysis, tumor cohorts with at least 100 patients that exhibit a gradual time-dependent decline in survival were included.
RNAseq differential expression analysis.
Raw count data from miR-155–high and miR-155–low patient subpopulations were downloaded by using the TCGABiolinks R package. For differential expression analysis, the limma R package was used, which tests differential gene expression between samples by using a linear model (55). RNAseq data were normalized via voom transformation as part of this package for each tumor type separately after removing genes expressed at low levels. Differentially expressed genes were visualized in volcano and bar plots.
GSEA
Voom-transformed RNAseq expression values from miR-155–high and miR-155–low patient subpopulations were ranked by using a signal-to-noise metric (52). Ranked gene lists were then analyzed for gene set enrichment by using the fgsea R package (51). Gene sets used in these analyses were derived from the Molecular Signature Database (MSigDB) (33) and previously published manuscripts that investigated immune signatures in various tumor contexts.
PCA
PCA was used to investigate the relationship between miR-155–high and miR-155–low patient subpopulations of TCGA data set. After limiting the analysis to the shared genes between tumor types, either the whole expression data set or only the immune-related genes were used for PCA. For these analyses, lists containing approximately 1,000 immune-specific genes were downloaded from the ImmPort database (34) or from the GO browser.
Tumor histology and the assessment of immune infiltration
miRseq data from TCGA database were analyzed and the 10 highest and 10 lowest miR-155–expressing tumors were selected for histological examination in each solid tumor cohort. H&E–stained frozen tumor sections were assessed for immune infiltration by using TCGA data repository web interface (https://portal.gdc.cancer.gov/repository). For this analysis, biospecimen IDs were randomized and provided to a board-certified anatomic pathologist (AHG), who was blinded to the miR-155 expression levels. Tumor sections devoid of immune cells were scored with (–). Tumors were scored (+) if a non-brisk localized lymphocyte infiltrate was observed. A (++) score was given if a brisk and widespread lymphocyte infiltration was present across the tumor tissue.
Statistics
Assessments of tumor growth and flow cytometry data were performed using 2-tailed Student’s t tests. Wilcoxon’s test was used for analyzing gene expression in select SCseq clusters. Reported P values were corrected for multiple comparisons by the Holm-Sidak method. P values less than or equal to 0.05 were considered statistically significant throughout (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). P values from the log-rank test are reported in SKCM survival graphs. In TCGA differential expression analyses, Benjamini-Hochberg–corrected P values calculated by limma R package are plotted.
Study approval
All experimental procedures and handling of mice were performed in accordance with the University of Utah Institutional Animal Care and Use Committee (IACUC) regulations and approved protocols. TCGA data analyzed in this study are publicly available via the National Cancer Institute Genomic Data Commons (GDC) and do not require regulated approval.
Author contributions
TBH and HAE coordinated and performed the experiments, analyzed the data, and prepared the figures. HAE wrote the manuscript and developed the immune cell scoring algorithm. HAE and AHG scored lymphocyte infiltration in TCGA melanoma samples. MAW, JLR, and WZS advised on experiments. RMO designed the research, advised on experiments, and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R01-AG047956 and R01-AI123106. TBH was supported by the NIH grant F30CA189731. We thank the University of Utah Flow Cytometry, High-Throughput Genomics, and Bioinformatics Core Facilities. We are grateful to Chris Conley and Chris Stubben at the Huntsman Cancer Institute Bioinformatics Core for invaluable discussions and their assistance in implementing computational analysis pipelines used in this manuscript. We also acknowledge Mingnan Chen (University of Utah) for providing B16F10-OVA cells. We are thankful to the authors of the Seurat Single Cell Genomics analysis package that facilitated the analysis of our single-cell sequencing data (Rahul Satija Lab, New York University).
Version 1. 02/05/2019
In-Press Preview
Version 2. 03/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(6):e126543. https://doi.org/10.1172/jci.insight.126543.
Contributor Information
H. Atakan Ekiz, Email: atakan.ekiz@hci.utah.edu.
Thomas B. Huffaker, Email: thomas.huffaker@path.utah.edu.
Allie H. Grossmann, Email: Allie.Grossmann@hsc.utah.edu.
W. Zac Stephens, Email: zac.stephens@path.utah.edu.
Matthew A. Williams, Email: Matthew.Williams@path.utah.edu.
June L. Round, Email: june.round@path.utah.edu.
References
- 1.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.Georgantas RW, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104(8):2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205(3):585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu R, et al. miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity. 2014;41(4):605–619. doi: 10.1016/j.immuni.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gracias DT, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14(6):593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu R, et al. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. 2013;190(12):5972–5980. doi: 10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang S, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70(8):3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 9.Li CL, et al. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27(6):1960–1966. doi: 10.3892/or.2012.1719. [DOI] [PubMed] [Google Scholar]
- 10.Hsin JP, Lu Y, Loeb GB, Leslie CS, Rudensky AY. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat Immunol. 2018;19(10):1137–1145. doi: 10.1038/s41590-018-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huffaker TB, et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J Biol Chem. 2017;292(45):18530–18541. doi: 10.1074/jbc.M117.808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huffaker TB, et al. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2(6):1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Y, et al. miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic γc cytokines. Proc Natl Acad Sci USA. 2015;112(2):476–481. doi: 10.1073/pnas.1422916112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudda JC, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38(4):742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zonari E, et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122(2):243–252. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 19.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews TS, Hemberg M. Identifying cell populations with scRNASeq. Mol Aspects Med. 2018;59:114–122. doi: 10.1016/j.mam.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Halle S, Halle O, Förster R. Mechanisms and dynamics of T cell-mediated cytotoxicity in vivo. Trends Immunol. 2017;38(6):432–443. doi: 10.1016/j.it.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 22.DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010;29(2):309–316. doi: 10.1007/s10555-010-9223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sica A, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18(5):349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Mandruzzato S, et al. IL4Ralpha+ myeloid-derived suppressor cell expansion in cancer patients. J Immunol. 2009;182(10):6562–6568. doi: 10.4049/jimmunol.0803831. [DOI] [PubMed] [Google Scholar]
- 25.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76(13):3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong W, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33(6):679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levati L, et al. MicroRNA-155 targets the SKI gene in human melanoma cell lines. Pigment Cell Melanoma Res. 2011;24(3):538–550. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 28.Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587(9):1434–1439. doi: 10.1016/j.febslet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grohmann U, Bronte V. Control of immune response by amino acid metabolism. Immunol Rev. 2010;236:243–264. doi: 10.1111/j.1600-065X.2010.00915.x. [DOI] [PubMed] [Google Scholar]
- 31.Casey SC, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352(6282):227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ock CY, et al. Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat Commun. 2017;8(1):1050. doi: 10.1038/s41467-017-01018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A. A description of the Molecular Signatures Database (MSigDB) web site. Methods Mol Biol. 2014;1150:153–160. doi: 10.1007/978-1-4939-0512-6_9. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya S, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 36.Dickey LL, Worne CL, Glover JL, Lane TE, O’Connell RM. MicroRNA-155 enhances T cell trafficking and antiviral effector function in a model of coronavirus-induced neurologic disease. J Neuroinflammation. 2016;13(1):240. doi: 10.1186/s12974-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532(1):1–12. doi: 10.1016/j.gene.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander M, et al. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cubillos-Ruiz JR, et al. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72(7):1683–1693. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha P, Parker KH, Horn L, Ostrand-Rosenberg S. Tumor-induced myeloid-derived suppressor cell function is independent of IFN-γ and IL-4Rα. Eur J Immunol. 2012;42(8):2052–2059. doi: 10.1002/eji.201142230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mishalian I, Bayuh R, Levy L, Zolotarov L, Michaeli J, Fridlender ZG. Tumor-associated neutrophils (TAN) develop pro-tumorigenic properties during tumor progression. Cancer Immunol Immunother. 2013;62(11):1745–1756. doi: 10.1007/s00262-013-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Mercier I, et al. Tumor promotion by intratumoral plasmacytoid dendritic cells is reversed by TLR7 ligand treatment. Cancer Res. 2013;73(15):4629–4640. doi: 10.1158/0008-5472.CAN-12-3058. [DOI] [PubMed] [Google Scholar]
- 44.Li B, Liu JS, Liu XS. Revisit linear regression-based deconvolution methods for tumor gene expression data. Genome Biol. 2017;18(1):127. doi: 10.1186/s13059-017-1256-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67(7):1031–1040. doi: 10.1007/s00262-018-2150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schatton T, Scolyer RA, Thompson JF, Mihm MC. Tumor-infiltrating lymphocytes and their significance in melanoma prognosis. Methods Mol Biol. 2014;1102:287–324. doi: 10.1007/978-1-62703-727-3_16. [DOI] [PubMed] [Google Scholar]
- 47.Lyu GY, Yeh YH, Yeh YC, Wang YC. Mutation load estimation model as a predictor of the response to cancer immunotherapy. NPJ Genom Med. 2018;3:12. doi: 10.1038/s41525-018-0051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorsson V, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/S1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 51. doi: 10.1101/060012. Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. Published June 20, 2016. [DOI]
- 52.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colaprico A, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva TC, et al. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Res. 2016;5:1542. doi: 10.12688/f1000research.8923.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.