Abstract
Idiopathic pulmonary fibrosis (IPF) is a progressive disease with unremitting extracellular matrix deposition, leading to a distortion of pulmonary architecture and impaired gas exchange. Fibroblasts from IPF patients acquire an invasive phenotype that is essential for progressive fibrosis. Here, we performed RNA sequencing analysis on invasive and noninvasive fibroblasts and found that the immune checkpoint ligand CD274 (also known as PD-L1) was upregulated on invasive lung fibroblasts and was required for the invasive phenotype of lung fibroblasts, is regulated by p53 and FAK, and drives lung fibrosis in a humanized IPF model in mice. Activating CD274 in IPF fibroblasts promoted invasion in vitro and pulmonary fibrosis in vivo. CD274 knockout in IPF fibroblasts and targeting CD274 by FAK inhibition or CD274-neutralizing antibodies blunted invasion and attenuated fibrosis, suggesting that CD274 may be a novel therapeutic target in IPF.
Keywords: Pulmonology, Therapeutics
Keywords: Cell migration/adhesion, Fibrosis, Respiration
PD-L1 is upregulated in fibroblasts in a humanized idiopathic pulmonary fibrosis model, and targeting PD-L1 blunts fibroblast invasion and attenuates fibrosis.
Introduction
Tissue fibrosis is associated with severe morbidity and mortality, and idiopathic pulmonary fibrosis (IPF) is characterized by abnormal tissue remodeling and progressive scarring in the lung (1). The mechanisms leading to severe and progressive fibrosis are incompletely understood. Previous studies from our laboratory and others demonstrated that fibroblasts from IPF patients acquire an invasive phenotype that is essential for severe fibrogenesis (2–5). This phenotype is regulated by hyaluronan synthase 2, CD44, β-arrestins, as well as an α6(β1)-integrin–mediated mechanosensing mechanism (2–5).
Immune checkpoints are regulators for maintaining systemic immune homeostasis and self-tolerance (6). Among them, the PD-1 pathway is utilized by cancer cells to escape the surveillance of the immune system (7). PD-1/PD-L1 blockade with monoclonal antibodies provides significant clinical benefits for patients with various cancers (6, 8). A few studies suggest PD-1 ligands are expressed on stromal cells (9, 10). One study found increased CD4+ and CD8+ cells in lung tissues and PD-1+ lymphocytes in peripheral blood from patients with pulmonary fibrosis relative to healthy controls (11). A recent report indicated that PD-1 was upregulated in CD4+ T cells and promoted bleomycin-induced pulmonary fibrosis (12). However, to date there are no studies to our knowledge that link PD-L1 to IPF lung fibroblasts and its functions in pulmonary fibrosis.
In the present study, we found a significant increase in expression of PD-L1 (also known as CD274) in the subset of invasive human lung fibroblasts isolated from explant lung tissues. IPF fibroblasts with high CD274 expression (CD274hi) showed greater migration and invasive capacity than the CD274-negative (CD274–) fibroblasts. Interestingly, we found that the invasive capacity of CD274hi fibroblasts was regulated by p53 and focal adhesion kinase (FAK). In vivo, CD274hi fibroblasts promoted pulmonary fibrosis relative to CD274– fibroblasts in a humanized IPF model in mice. Genetic activation of CD274 significantly increased fibroblast migration and invasion, as well as lung fibrosis. Blocking FAK signaling by a small-molecular inhibitor, VS4718, inhibited fibroblast migration and invasion, as well as lung fibrosis. Moreover, targeting CD274hi fibroblasts by CRISPR knockout or anti-CD274 neutralizing antibodies significantly inhibited fibroblast migration and invasion in vitro and attenuated lung fibrosis in vivo. These data suggest that upregulation of CD274 is a profibrotic factor in lung fibroblasts and targeting PD-L1 might be a potential therapeutic approach for IPF.
Results
Invasive lung fibroblasts promoted interstitial lung fibrosis in a humanized SCID IPF model.
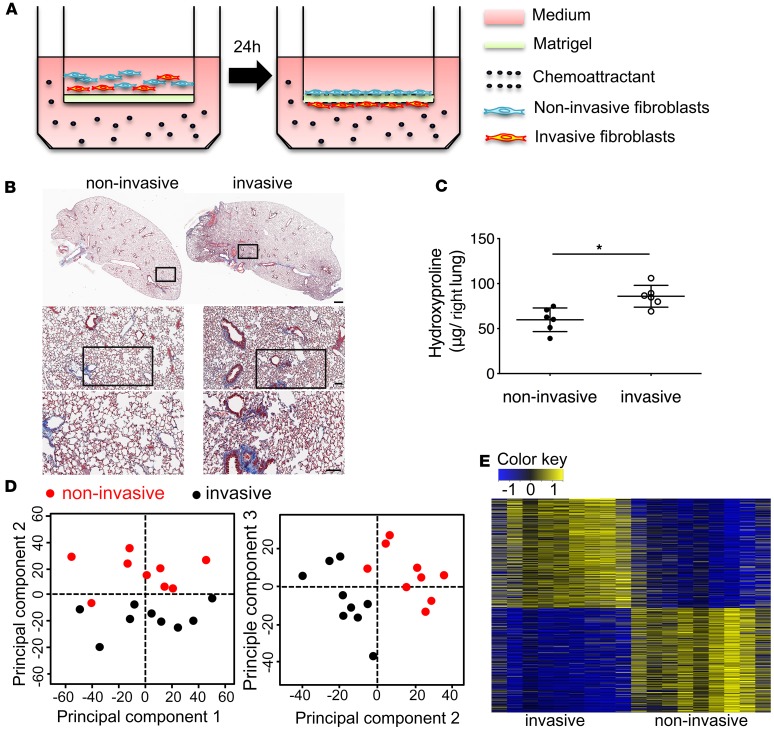
To demonstrate the fibrogenic potential of invasive fibroblasts in vivo, we isolated invasive and noninvasive IPF lung fibroblasts using the matrigel invasion assay (Figure 1A) and injected them intravenously into NOD-scid-IL2Rγc–/– (NSG) mice (humanized severe combined immunodeficient [SCID] IPF model) (13). After 50 days, mice injected with invasive IPF lung fibroblasts showed more diffuse interstitial fibrosis and increased hydroxyproline in the lung than mice injected with noninvasive IPF lung fibroblasts (Figure 1, B and C). To gain insights into mechanisms that regulate invasion, we compared invasive and noninvasive IPF lung fibroblast gene expression using RNA sequencing (RNA-seq). A total of 1,405 differentially expressed (DE) genes were identified; among them, 719 DE genes were upregulated, and 686 DE genes were downregulated (Figure 1, D and E). Among the DE genes, HAS2 and IL11 were found significantly upregulated in invasive fibroblasts (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125326DS1), which confirmed the work from our laboratory and others (2, 14, 15).
Figure 1. Invasive lung fibroblasts promoted interstitial lung fibrosis.
(A) Schematic representation of in vitro invasion assay. Lung fibroblasts were seeded in the upper part of transwells. Cells attached to the bottom of Matrigel-coated membrane after 24 hours were considered invasive fibroblasts. Cells remaining on top of the Matrigel-coated membrane were considered noninvasive fibroblasts. Invasive and noninvasive IPF lung fibroblasts (n = 9 per group) were isolated using the matrigel invasion assay. Masson’s trichrome staining of collagen on lung sections (B) and hydroxyproline content in lung tissues (C) from NSG mice 50 days after injection with invasive and noninvasive IPF lung fibroblasts on day 50 after fibroblast injection (n = 6 per group). Scale bars: 1 mm (top panel), 100 μm (middle and lower panels). (D) Principal component analysis of RNA-seq data. (E) Heatmap of all differentially expressed (DE) genes in RNA-seq data. A total of 1,405 DE genes were identified with FDR < 0.01 and |log2 FC| > 0.5; among them, 719 DE genes were upregulated, and 686 DE genes were downregulated. *P < 0.05 by Student’s t test (C).
Upregulated PD-1 ligands on invasive fibroblasts and IPF fibroblasts.
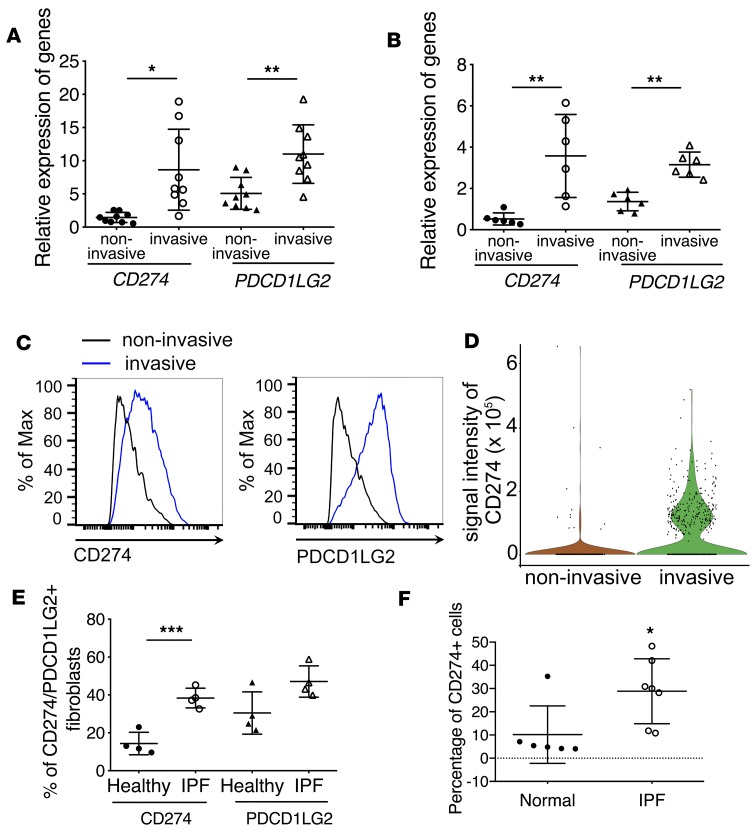
Surprisingly, by RNA-seq data analysis we found that mRNAs for both checkpoint PD-1 ligands, CD274 (PD-L1) and PDCD1LG2 (PD-L2, CD273), were significantly upregulated in invasive fibroblasts (Figure 2A, Supplemental Table 1, and Supplemental Table 2). Expression of RGMB, a binding partner for PD-L2 (16), was also upregulated in IPF invasive lung fibroblasts (Supplemental Table 2). The expression of other stimulatory or inhibitory checkpoint molecules was either not detected or not altered in IPF invasive lung fibroblasts (Supplemental Table 2). The expression of PDCD1 (PD-1), the receptor for both CD274 and PDCD1LG2, was not detected in IPF lung fibroblasts (Supplemental Table 2). Upregulation of CD274 and PDCD1LG2 was confirmed by qRT-PCR (Figure 2B). Also, we validated the upregulated-RNA data using flow cytometric analysis (Figure 2C) and single-cell Western blot (Figure 2D).
Figure 2. Upregulation of PD-1 ligands in invasive fibroblasts.
(A and B) Upregulation of immune checkpoint CD274 and PDCD1LG2 in invasive lung fibroblasts. RNA-seq (n = 9 per group) (A) and qRT-PCR analysis (n = 6 per group) (B) of CD274 and PDCD1LG2 expression in invasive and noninvasive IPF lung fibroblasts. (C) Cell surface expression of CD274 and PDCD1LG2 in invasive and noninvasive IPF lung fibroblasts. (D) Single-cell Western blot analysis of CD274 expression in invasive and noninvasive lung fibroblasts. (E) Cell surface expression of CD274 and PDCD1LG2 in primary IPF fibroblasts and healthy controls by flow cytometry. (F) Flow cytometry analysis of lung single-cell homogenate for CD274 expression in CD31–CD45–EPCAM– cells from IPF (n = 7) or healthy (n = 6) samples. Throughout, data are the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA (A, B, and E) or Student’s t test (F).
Our previous reports revealed that severe lung fibrosis required an invasive phenotype and fibroblasts from IPF lung showed higher invasive capacity (2). Next, we wanted to determine if CD274 and PDCD1LG2 are upregulated on IPF fibroblasts. We performed flow cytometric and Western blot analyses on IPF fibroblasts and healthy controls, and found that the percentage of CD274+ and PDCD1LG2+ fibroblasts (Figure 2E) and total protein expression (Supplemental Figure 1A) of CD274 was higher on the IPF lung fibroblasts than that of healthy controls. Furthermore, flow cytometric analysis on IPF and normal lung homogenates confirmed the higher percentage of CD274+ cells in CD31–CD45–EPCAM– mesenchymal cells in IPF homogenates than normal control (Figure 2F and Supplemental Figure 1B). Moreover, immunofluorescence showed that CD274 expression was colocalized with a small portion of PDGFRβ+ (lung fibroblast marker) and endomucin+ (endothelial cell marker) cells, but not obviously with α-smooth muscle actin–positive (α-SMA+) cells (myofibroblast marker). CD274 expression was also found adjacent to CD8+ T cells (Supplemental Figure 1C).
The expression level of CD274 was closely related with fibroblast invasion.
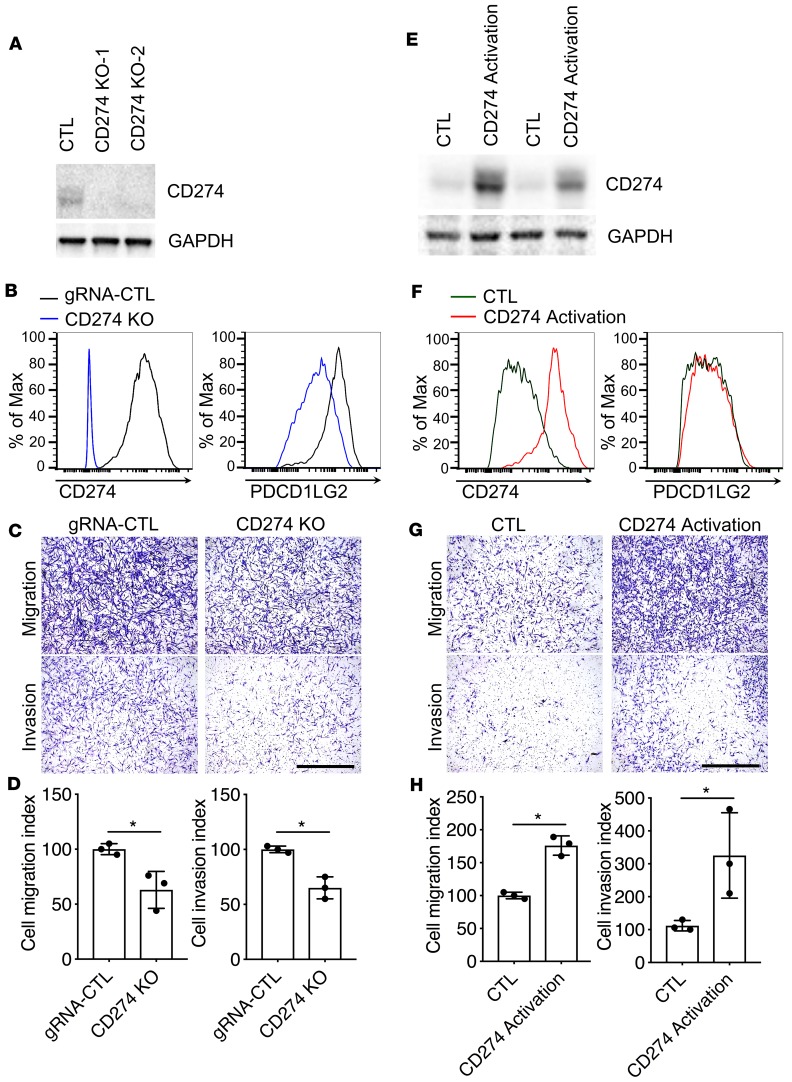
CD274 was upregulated in invasive fibroblasts, which suggested that CD274 might be necessary for the invasion of lung fibroblasts. Using CRISPR technology, we generated CD274-knockout (CD274-KO) and -overexpression stable lines in IPF lung fibroblasts. The knockout and overexpression efficiency was confirmed by qRT-PCR (Supplemental Figure 2, A and B), Western blot (Figure 3, A and E), and flow cytometric analysis (Figure 3, B and F). Functionally, CD274 deletion blunted cell migration and invasion (Figure 3, C and D), whereas CD274 activation promoted migration and invasion (Figure 3, G and H) in IPF lung fibroblasts. These gain- and loss-of-function analyses confirmed the functions of CD274 on fibroblast invasion.
Figure 3. The expression level of CD274 was closely associated with fibroblast invasion.
(A–D) CD274 in control (CTL) and CD274-KO IPF lung fibroblasts was assessed by Western blot analysis along with GAPDH (A), and cell surface expression along with PDCD1LG2 (B), and the fibroblasts evaluated for migration and invasion (C and D). (E–H) CD274 in CTL and CD274-activated IPF lung fibroblasts was assessed by Western blot analysis along with GAPDH (E), cell surface expression along with PDCD1LG2 (F), and the fibroblasts evaluated for migration and invasion (G and H). GAPDH served as loading control. See complete unedited blots in the supplemental material. Representative images of migration and invasion are shown. Scale bars: 1 mm. Data are the mean ± SEM (n = 3 per group). *P < 0.05 by Student’s t test.
Reciprocal negative regulatory loop between PD-1 ligands and p53 in IPF lung fibroblasts.
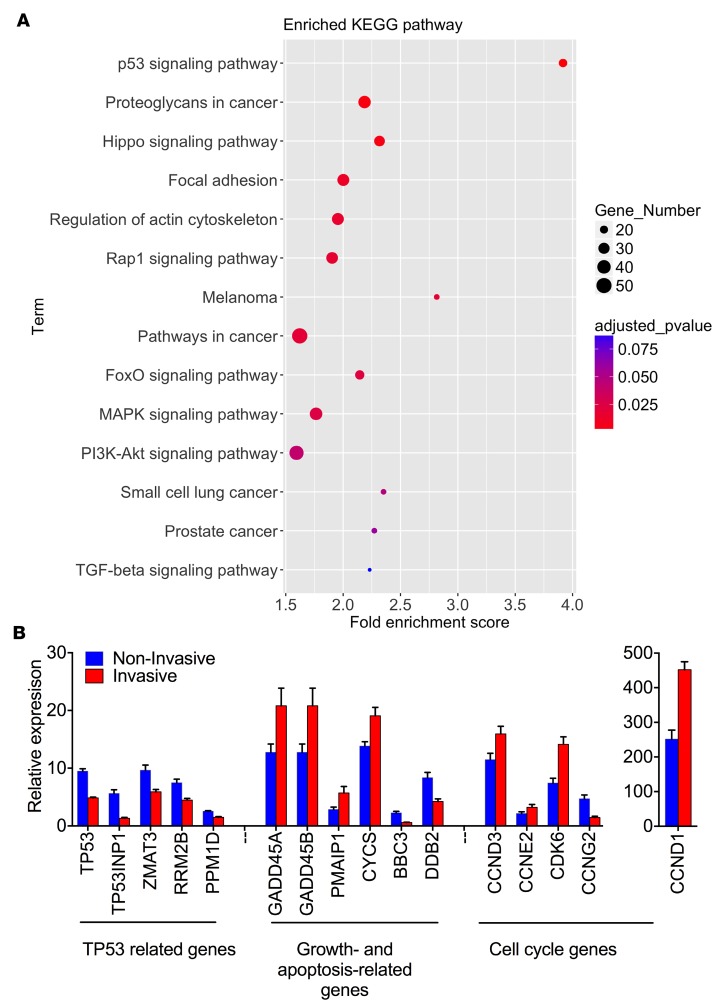
To uncover potential signaling pathways that regulate fibroblast invasion, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. KEGG analysis for DE genes between invasive and noninvasive IPF lung fibroblasts by RNA-seq revealed that the p53 signaling pathway, focal adhesion, regulation of actin cytoskeleton, MAPK, and cancer signaling pathways were significantly correlated with the lung fibroblast invasive phenotype (Figure 4A).
Figure 4. Analyses of RNA-seq data.
(A) KEGG pathway enrichment analysis of 1,405 DE genes. (B) Relative gene expression of p53 signaling pathways in RNA-seq (n = 9 per group) analysis. The P value for each gene between invasive and noninvasive was less than 0.05 by Student’s t test.
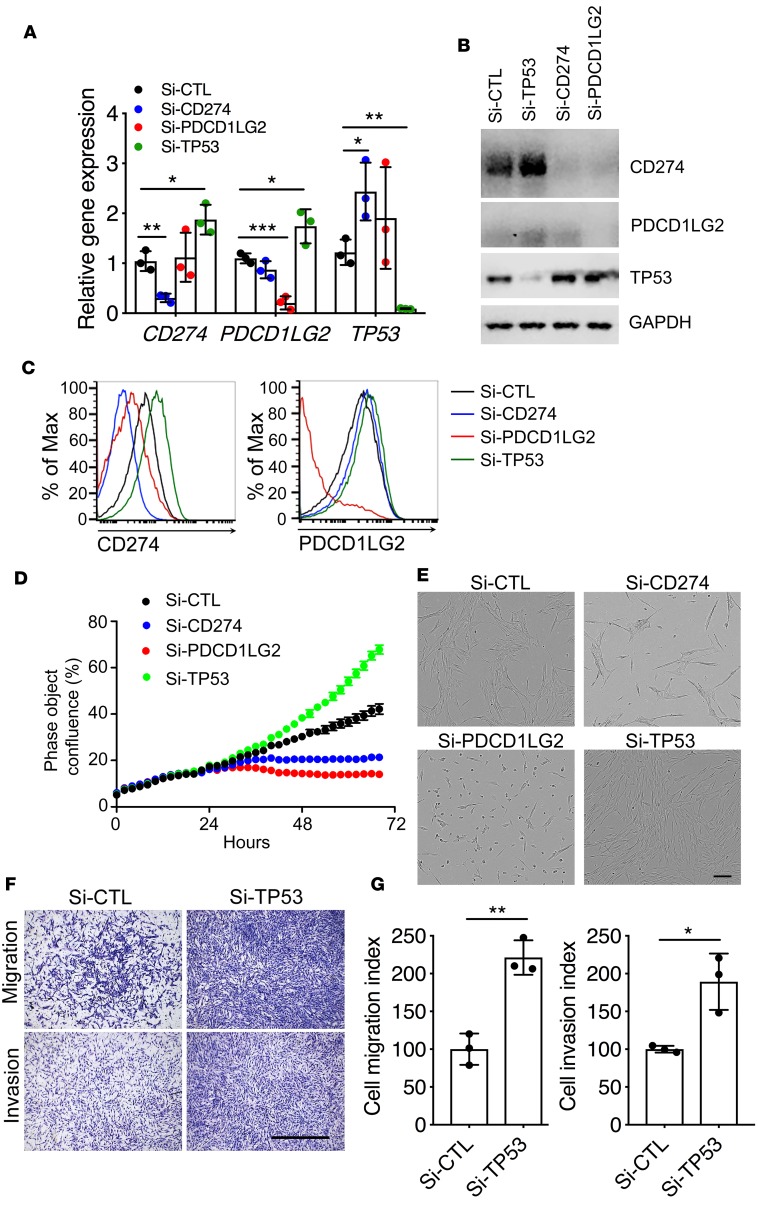
Tumor-suppressor p53 (encoded by the TP53 gene) modulates the tumor immune response by regulating CD274 (PD-L1) expression (17). Interestingly, analysis of RNA-seq data also revealed significant differential expression of TP53, growth- and apoptosis-related genes, and cell cycle genes (Figure 4B). We wondered if the upregulation of CD274 was related to the p53 signaling pathway and we performed small interfering RNA (siRNA) knockdown assays. Knockdown of TP53 in lung fibroblasts from IPF patients upregulated mRNA levels, total protein, and cell surface expression of CD274 and PDCD1LG2 as determined by qRT-PCR (Figure 5A), Western blot (Figure 5B), and flow cytometric analysis (Figure 5C). On the other hand, knockdown of CD274 or PDCD1LG2 in lung fibroblasts upregulated TP53 gene expression (Figure 5, A–C), suggesting a reciprocal negative regulatory loop between PD-1 ligands and p53. Functionally, knockdown of TP53 significantly promoted fibroblast growth and adhesion, while knockdown of CD274 or PDCD1LG2 inhibited fibroblast growth and adhesion (Figure 5, D and E). TP53 controls cell invasion in cancer cells (18) and knockdown of TP53 also enhanced the migration and invasive capacities of IPF lung fibroblast (Figure 5, F and G).
Figure 5. Regulation of cell growth and invasion by p53.
Gene expression (n = 3 per group) (A) and Western blot analysis (B) of CD274, PDCD1LG2, TP53, and GAPDH in IPF lung fibroblasts treated with Si-CTL, Si-CD274, Si-PDCD1LG2, or Si-TP53. See complete unedited blots in the supplemental material. Cell surface expression (C) of CD274 and PDCD1LG2 in IPF lung fibroblasts treated with Si-CTL, Si-CD274, Si-PDCD1LG2, or Si-TP53 after 68 hours. (D) Representative cell growth curve of lung fibroblasts treated with Si-CTL, Si-CD274, or Si-PDCD1LG2. (E) Representative images of lung fibroblasts treated with Si-CTL, Si-CD274, or Si-PDCD1LG2 after 68 hours. Scale bar: 150 μm. (F and G) In vitro migration and invasion assay. Equal numbers of cells were seeded in the upper part of transwells. (F) Representative images of migrated and invasive Si-CTL or Si-TP53 lung fibroblasts. Scale bar: 1 mm. (G) Cell migration or invasion index was calculated as the number of cells attached to the bottom of control or Matrigel-coated membranes after 24 hours, normalized to respective Si-CTL lung fibroblasts (n = 3 per group). Throughout, data are the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.01 by 1-way ANOVA (A) or Student’s t test (G).
FAK1 regulated the invasion and migration of lung fibroblasts.
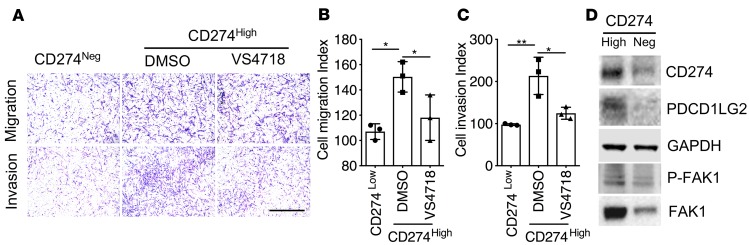
The focal adhesion pathway was highly enriched in invasive fibroblasts according to RNA-seq data (Figure 4A), which suggested PD-1 ligands might be regulated by these pathways. To confirm our hypothesis, we harvested CD274hi and CD274– fibroblasts by fluorescence-activated cell sorting from IPF explant tissue (Supplemental Figure 3) and found that increased expression of CD274 on the cell surface is associated with increased fibroblast cell adhesion (Supplemental Figure 4, A and B), migration, and invasion (Figure 6, A–C). FAK, a nonreceptor tyrosine kinase, plays an essential role in multiple biological functions, including cell survival, proliferation, migration, adhesion, and invasion (19). FAK signaling also has been implicated in pathologic fibrosis in several tissues (20, 21). We found that phosphorylated FAK1 and total FAK1 expression was also increased in IPF lung fibroblasts (Supplemental Figure 1A), as well as in the CD274hi lung fibroblasts (Figure 6D and Supplemental Figure 4C). VS4718, a small-molecule inhibitor of FAK, significantly blocked cell migration and invasion of lung fibroblasts (Figure 6, A–C).
Figure 6. CD274 regulates lung fibroblast invasion via FAK1 signaling.
(A–C) Equal numbers of cells were seeded in the upper part of transwells and cell migration and invasion assays were performed (n = 3 per group). (A) Representative images of migrating and invasive CD274– and CD274hi IPF fibroblasts treated with VS4718 or vehicle (DMSO). Scale bar: 1 mm. (B and C) Cell migration or invasion index was calculated as the number of cells attached to the bottom of control or Matrigel-coated membranes after 24 hours, normalized to respective CD274– lung fibroblasts. (D) Western blot analyses of CD274, PDCD1LG2, p-FAK1, and FAK1. GAPDH served as loading control. See complete unedited blots in the supplemental material. Neg, negative. Scale bars: 1 mm. Throughout, data are the mean ± SEM. *P < 0.05; **P < 0.01 by 1-way ANOVA (B and C).
CD274 was required for lung fibroblast invasion and lung fibrosis.
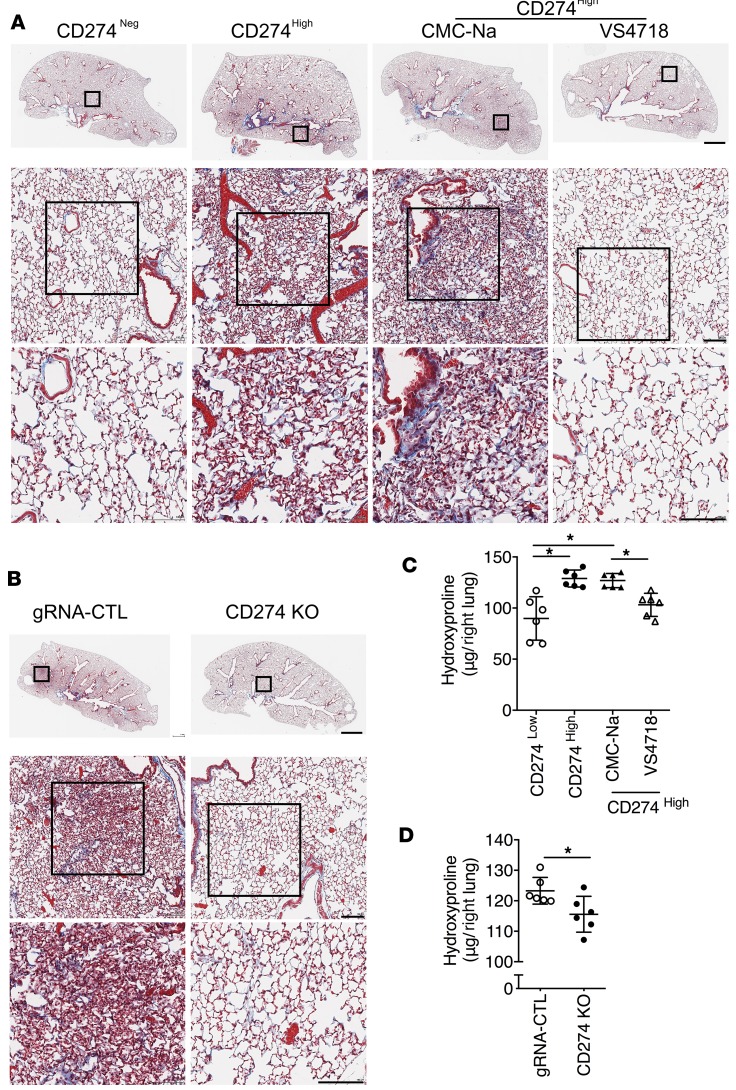
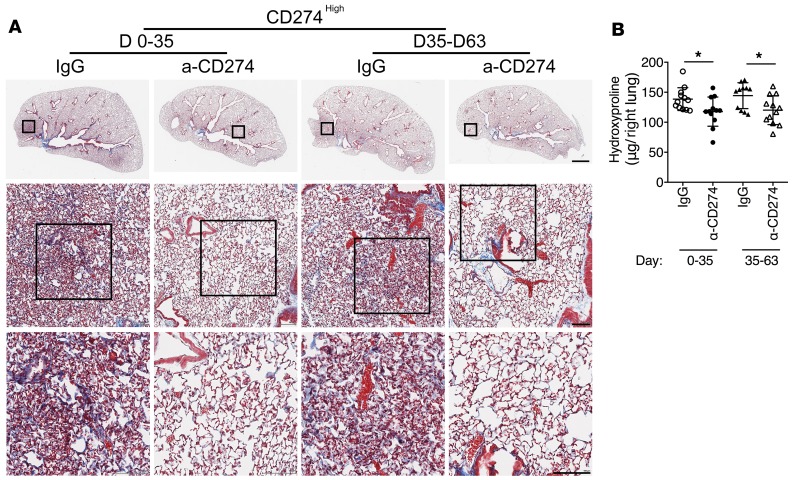
We then injected CD274hi or CD274– lung fibroblasts into NSG mice to investigate the role of CD274 in lung fibrosis in vivo. Mice receiving CD274hi fibroblasts developed significantly more lung fibrosis than the mice receiving CD274– fibroblasts (Figure 7, A and C). We further found that there was less diffuse interstitial fibrosis and a decrease in hydroxyproline in the lungs of the NSG mice injected with CD274-KO lung fibroblasts compared with mice receiving control guide RNA (gRNA) lung fibroblasts (Figure 7, B and D). Moreover, VS4718 treatment prevented the development of fibrosis in the mice receiving CD274hi lung fibroblasts, compared with mice treated with vehicle (carboxymethyl cellulose sodium [CMC-Na]) (Figure 7, A and C). Furthermore, blocking CD274 with an anti-CD274 neutralizing antibody attenuated the development of fibrosis at both early (days 0–35) and late stages (days 35–63) of fibrogenesis (Figure 8, A and B), which was confirmed by the expression level of fibrosis-related genes.
Figure 7. CD274 is required for lung fibrosis in a humanized SCID mouse model.
Masson’s trichrome staining of collagen in lung sections (A and B) and hydroxyproline content in lung tissues (C and D) from NSG mice injected with CD274– and CD274hi IPF fibroblasts treated with VS4718, vehicle control CMC-Na, or from NSG mice that received gRNA control (gRNA-CTL) or CD274-KO lung fibroblasts (n = 6 per group). Neg, negative. Scale bars (A and B): 1 mm (top panel), 100 μm (middle and lower panels). Throughout, data are the mean ± SEM. *P < 0.05 by 1-way ANOVA (C) or Student’s t test (D).
Figure 8. Blocking CD274 attenuates lung fibrosis.
Masson’s trichrome staining of collagen in lung sections (A) and hydroxyproline content in lung tissues (B) from NSG mice injected with CD274hi IPF fibroblasts treated with anti-CD274 (α-CD274) antibody (n = 12 per group) or isotype control IgG (n = 12 for days 0–35 IgG, n = 11 for days 35–63 IgG) on day 63 after fibroblast injection. Scale bars: 1 mm (top panel), 100 μm (middle and lower panels). Data are the mean ± SEM. *P < 0.05 by 2-way ANOVA (B).
Discussion
The mechanisms that control IPF are not fully understood and new therapeutic targets are still needed. This study further supports the concept that invasive fibroblasts drive progressive lung fibrogenesis, suggesting a previously unrecognized nonimmune regulatory role of PD-L1 in invasive fibroblasts in the setting of lung fibrosis. PD-L1 is overexpressed in many human cancers and promotes T cell tolerance and escape from host immunity. Although PD-L1 has been shown to be expressed on fibroblasts (9, 10, 22, 23), to the best of our knowledge there are no data that connect its expression on fibroblasts to fibrogenesis. Our study demonstrates that PD-L1 mediates fibroblast adhesion and invasion independently of immune regulation, since the in vivo studies were performed in immune-deficient mice. This, of course, does not preclude an immune-mediated mechanisms in patients with IPF. This study also provides circumstantial evidence that IPF and lung cancer share a number of similarities genetically or epigenetically. Targeting the immune checkpoint components has been a treatment breakthrough in a number of cancers, albeit not without complications including pneumonitis (24). The role of the immune checkpoint in stromal regulation of tumor growth and metastasis is an area of active investigation and the hypothesis has been developed that part of the efficacy of immune checkpoint inhibition may be due to effects on the tumor microenvironment (7).
Recently, human mesenchymal stem cells were reported to attenuate lung fibrosis through the PD-1/PD-L1 pathway in bleomycin-induced pulmonary fibrosis in humanized mice (11), although the mechanisms differ. During the preparation of this manuscript, another publication suggested that PD-1+CD4+ T cells were able to induce fibroblasts to produce collagen and blocking PD-1 in these T cells reduced bleomycin-induced pulmonary fibrosis (12). Neutralizing antibodies have been widely used to target CD274 in cancer therapy (25–27). As CD274 is activated in IPF lung fibroblasts and contributes to the progress of pulmonary fibrosis, targeting CD274 at both early stages and late stages has the potential to significantly reduce IPF fibroblast invasion and not only abrogate lung fibrosis development but potentially reverse fibrosis, as we saw in the humanized SCID IPF model.
Immunodeficient mice have become increasingly important as small preclinical animal models for the study of human diseases, as these mice can also be engrafted with human tissues such as islet cells, liver, skin, and most solid and hematologic cancers (28). In this study, we used a humanized SCID mouse model of IPF, which is an established mouse model, to test the fibrogenic potential of human lung fibroblasts. This humanized IPF model was first used to study the in vivo role of CC chemokine receptor 7 (CCR7) in IPF (29); this model allows for cell trafficking during different stages of fibrosis development and progression, and offers unique insights into different fibroblast populations (13). Since the establishment of this model, several more studies have used it to study the heterogeneity of IPF fibroblasts and to explore antifibrotic agents with human specificity (13, 29–32).
The tumor suppressor protein p53 induces cell cycle arrest, apoptosis, senescence, and innate immunity. Induction of p53 is associated with lung injury and development of pulmonary fibrosis (33–35). Type II alveolar epithelial cell (AEC II) apoptosis is associated with acute lung injury and the development of pulmonary fibrosis. Active induction of p53 and apoptosis are found in AEC II from IPF patients (35). Lung injury due to exposure to DNA-damaging agents like bleomycin rapidly induces p53 expression in AEC II (34). Induction of p53 leads to apoptosis in AECs and subsequent activation and overgrowth of activated fibroblasts (36). p53 has been actively studied as a major barrier against cancer development in the past decade and has been found to regulate several key stages of metastatic progression, such as cell cycle checkpoint controls, apoptosis, cell migration, and invasion (37). Moreover, as a major component of the tumor stroma, fibroblasts in the metastatic progression of cancer are also regulated by p53 and suppression of p53 in normal fibroblasts promotes acquisition of a cancer-associated fibroblast phenotype (38). Inactivation of p53 in fibroblasts augments the expression of several proteins, including TSPAN12 (39) and SDF-1/CXCL12 (40), which might enhance tumor proliferation, migration, and invasion. In addition to the functions in regulating cell apoptosis, invasion, and DNA repair, the role of p53 in regulating metabolic pathways has also recently been identified (41). Several reports suggest a mechanism of p53 activation during cellular senescence that may be a significant factor to block cell growth by inducing the expression of p21 (42, 43). Changes in p53 expression in mouse fibroblasts can modify motility and extracellular matrix organization (44). Different TP53-mutant proteins showed different effects on glycolysis and mitochondrial metabolism in experimental models of human cancer and normal lung fibroblasts (45).
Several reports suggest a role for p53 in the regulation of fibroblast metabolism. Furthermore, p53 is reported to regulate PD-L1 via regulation of miR-34a and miR-200 microRNA families (17, 46, 47), which indicates that p53 might function in the lung fibroblasts to regulate the expression of CD274. In the present study, we confirmed a negative regulatory loop between CD274 and p53 in the lung fibroblasts and the underlying mechanism are still actively being pursued.
FAK is a nonreceptor tyrosine kinase involved in various biological functions, including cell survival, proliferation, migration, and adhesion. Lung epithelial cell FAK signaling regulates the fate determination of lung epithelial cells and inhibits lung injury and fibrosis (48, 49). FAK is also an essential factor for TGF-β to induce myofibroblast differentiation (50). Blocking FAK effectively inhibited the growth of lung fibroblasts and attenuated the expression of α-SMA and type I collagen in vitro and bleomycin-induced lung fibrosis in vivo (20). Moreover, FAK-related non-kinase (FRNK) plays a key role in limiting the development of lung fibrosis (51, 52). FRNK expression is significantly reduced in IPF lung fibroblasts, while activation of FAK is increased in IPF lung fibroblasts (51, 52). Loss of FRNK function results in increased fibroblast migration and myofibroblast differentiation. Exogenous FRNK expression abrogates the increased cell migration and blocks the activation of FAK and Rho GTPase in IPF lung fibroblasts, which suggests that FRNK plays a role in promoting cell migration through FAK and Rho GTPase in fibrotic IPF lung fibroblasts (51). In the present study, FAK signaling was significantly upregulated in IPF fibroblasts and invasive fibroblasts and blocking FAK signaling reduced the increase of the invasion of CD274hi fibroblasts, which are consistent with previous studies.
We believe our data are the first to identify PD-L1 as a regulator of mesenchymal cell invasion in human disease and may have implications for cancer progression. Furthermore, these data are the first to our knowledge to suggest that targeting CD274-expressing fibroblasts in IPF could be a promising approach to inactivating invasive fibroblasts and attenuating and potentially reversing established pulmonary fibrosis.
Methods
Study approval.
All human lung experiments were approved by the Cedars-Sinai Medical Center Institutional Review Board (IRB) and were in accordance with the guidelines outlined by the IRB. Informed consent was obtained from each subject (IRB: Pro00032727). All animal experiments were approved by the Institutional Animal Care and Use Committee at Cedars-Sinai Medical Center (protocol IACUC005136). All mice were housed in a pathogen-free facility at Cedars-Sinai Medical Center and had access to autoclaved water and pelleted mouse diet ad libitum.
Human lung fibroblast culture.
Human lung fibroblasts were isolated from surgical lung biopsies or lung transplant explants obtained from patients with IPF (Supplemental Table 2). The diagnosis of IPF was arrived at by standard accepted American Thoracic Society recommendations (ATS/ERS, 2000). The tissues were minced, digested, and cultured in DMEM supplemented with 15% FBS and Antibiotic-Antimycotic (Thermo Fisher Scientific).
Humanized SCID mouse model of IPF.
Female NOD-scid-IL2Rγc–/– (NSG) mice (6 to 8 weeks old) were purchased from the Jackson Laboratory). The NSG mice received single-cell preparations of invasive, noninvasive, CD274hi and CD274lo IPF lung normal fibroblasts (0.5 × 106 cells) via tail vein injection. Lung fibrosis was assessed on day 50 or day 63 after fibroblast transfer. For FAK inhibitor studies, mice were treated with 0.5% CMS-Na (control group) or 50 mg/kg VS4718 (Chemietek) every other day from day 35. For the anti-CD274 studies, mice were injected with IgG (InVivoMAb mouse IgG2b isotype control, clone MCP-11) or anti-CD274 (InVivoMab anti–human PD-L1, clone 29E.2A3) (both Bio X Cell) twice per week, 100 μg/mouse from days 0 to 35 or from days 35 to 63. Mice were sacrificed on day 63 after the human lung fibroblast transfer. The left lobe was used for histology and right lobes were used for hydroxyproline assay.
Western blotting.
Cell samples were lysed in RIPA buffer (Thermo Fisher Scientific). The proteins were resolved by SDS-PAGE using gradient gels (4%–20%) and electroblotted onto PVDF membranes (Thermo Fisher Scientific). The membranes were probed with a rabbit monoclonal anti-CD274 antibody (Cell Signaling Technology, clone E1L3N, 1:1,000 dilution), anti-PDCD1LG2 antibody (Cell Signaling Technology, clone E1L3N, 1:1,000 dilution), anti-p53 antibody (Cell Signaling Technology, clone 48818, 1:1,000 dilution), anti–phospho-FAK (Tyr397) antibody (Thermo Fisher Scientific, clone 31H5L17, 1:1,000 dilution), polyclonal anti-FAK antibody (Cell Signaling Technology, 1:1,000 dilution), and then probed with the appropriate second antibody. GAPDH (Cell Signaling Technology, clone 14C10, 1:1,000 dilution) was used as a loading control.
Single-cell Western blot.
Single-cell Western blot for CD274 was performed according to the manufacturer’s instructions. Briefly, single-cell suspensions of noninvasive and invasive IPF lung fibroblasts were loaded into single-cell chips (Standard scWest Kit, K600-1, Proteinsimple) and the chips were run with the Milo system. After washing with buffer, the chips were incubated with anti-CD274 antibody (Cell Signaling Technology, clone E1L3N, 1:40 dilution) and anti–β-tubulin antibody (GenScript, clone 2G7D4, 1:40 dilution) for 2 hours and then with secondary antibody for 1 hour at room temperature. The chip was read and the data were analyzed with the Scout system. The expression of CD274 was normalized to that of β-tubulin.
Gene knockdown assay.
The primary cells were used from 4 to 6 generations for invasion assays and siRNA interference assays. Si-CTL (1022076), Si-CD274 (SI03021158), Si-PDCD1LG2 (SI00681149), and Si-TP53 (SI02655170) were all from Qiagen. siRNA was transfected in cells using Lipofectamine RNAiMAX Transfection Reagent following the manufacturer’s protocol (Thermo Fisher Scientific).
CD274-KO and activation with CRISPR.
The fibroblasts were also immortalized with expression of telomerase reverse transcriptase (TERT) protein (hTERT Cell Immortalization Kit; CILV02, ALSTEM). The immortalized cells were used to generate CD274-KO and -activated cell lines. For CD274-KO, we first generated a Cas9-expressing cell line (Invitrogen LentiArray Cas9 Lentivirus, A32069, Thermo Fisher Scientific), and then sgRNA expression clones targeting CD274 (HCP208443-SG01-3-10, GeneCopoeia) and scrambled sgRNA control plasmid (CCPCTR01-SG01-10, GeneCopoeia) were used to generate CD274-KO and control cells. For CD274 activation, Pdcd-1L1 Lentiviral Activation Particles (sc-401140-LAC, Santa Cruz Biotechnology) were used.
Cell invasion and migration assay.
For cell invasion, human lung fibroblasts (2.5 × 104 to 10.0 × 104 cells per well for 24-well plates and 1.0 × 106 cells per well for 6-well plates) in 10% FBS were loaded into the top chamber of a BioCoat Matrigel Invasion Chamber (BD Biosciences). PDGF-BB (10 ng/ml; Peprotech) was used as a chemoattractant in the bottom chamber. After 24 hours at 37°C with 5% CO2, invasive cells had passed through the matrigel layer and clung to the bottom of the insert membrane. Cells remaining on the upper chamber were defined as noninvasive fibroblasts. For RNA-seq, the invasive and noninvasive cells were harvested separately by trypsin digestion. For quantification of the invasion index, the filters were fixed and stained with the Protocol Hema 3 stain set (Thermo Fisher Scientific). Noninvasive cells were removed from the upper side of the filter by gentle scrubbing with a cotton swab. The number of fibroblasts that invaded through the basement membrane was counted in 5 randomly chosen fields per filter from triplicate filters per sample at ×400 magnification. The cell invasion index was calculated as the number of cells able to invade through Matrigel during a 24-hour period normalized to controls (mean ± SEM). For cell migration assay, the Corning BioCoat Control Insert was used. All other settings were the same as for the cell invasion assay.
RNA-seq and data analysis.
RNA was sent to the UCLA Clinical Microarray Core for Library preparation and RNA-seq. Samples were sequenced on an Illumina HiSeq 3000 instrument using 50-bp single reads.
Tophat2 (2.0.7) (53) was applied to align sequencing reads to the reference human genome (GRCh38). The relative gene expression RPKM (reads per kb of transcript, per million mapped reads) and read counts were estimated using SAMMate (2.7.4) (54) and Ensembl database (Homo Sapiens.GRCh38.77). Protein-coding genes with at least 2 RPKM on average in either condition were used to perform the differential gene expression analysis using edgeR (55). The P values of multiple tests were adjusted using the Benjamini-Hochberg method (56) and the significance level was set at FDR < 0.01 and |log2 FC| > 0.5. KEGG pathway enrichment analysis of DE genes was performed using DAVID (57). The raw RNA-seq analyses data reported in this study have been deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE118933; the secure token for review is uraxogkwbbwtpuf).
Cell adhesion assay.
Cells were allowed to attach to collagen IV–coated 48-well plates (Cell Biolabs) for 1 hour at 100,000 cells/well in serum-free medium. Adherent cells were stained with 0.5% crystal violet and quantified at OD 560 nm after extraction.
RNA isolation and qRT-PCR analysis.
RNA was isolated using the Absolutely RNA Microprep Kit (Agilent Technologies) following the manufacturer’s recommendations. MultiScribe Reverse Transcriptase (Thermo Fisher Scientific) was used for cDNA synthesis. Gene expression was measured relative to the endogenous reference gene GAPDH using the comparative ΔCT method and the TaqMan Gene Expression Assay (Thermo Fisher Scientific). The ID of each Taqman assay was as follows: CD274 (Hs00204257_m1), PDCD1LG2 (Hs00228839_m1), TP53 (Hs01034249_m1), and GAPDH (Hs03929097_g1).
Flow cytometry.
Cells were resuspended in Hank’s balanced saline solution supplemented with 2% FBS, 10 mM HEPES, 0.1 mM EDTA, 100 IU/ml penicillin, and 100 μg/ml streptomycin (HBSS+ buffer). Labeled primary antibodies anti-CD274-PE (Biolegend, clone 29E.2A3, 1:300 dilution) and anti-PDCD1LG2–APC (Biolegend, clone 24F.10C12, 1:300 dilution) were added to the cells. Dead cells were discriminated by 7-amino-actinomycin D (7-AAD) staining. Flow cytometry was performed using an LSRFortessa cell analyzer and FACSAria III sorter (BD Immunocytometry Systems) and analyzed using FlowJo 10.2 software (Tree Star).
Hydroxyproline assay.
Collagen content in mouse lungs was measured using a conventional hydroxyproline method. Lung tissues were vacuum-dried and hydrolyzed with 6N hydrochloride acid at 120°C overnight. Hydroxyproline content is expressed as μg per right lung. The ability of the assay to completely hydrolyze and recover hydroxyproline from collagen was confirmed using samples containing known amounts of purified collagen.
Histological analysis.
After anesthesia-induced euthanasia, the left lobe from each mouse was dissected, fully inflated with 10% formalin solution, and placed in fresh formalin for 24 hours. Standard histological techniques were used to paraffin-embed each lobe, and 5-μm sections were stained with hematoxylin and eosin and Masson’s trichrome for histological analysis. For immunohistochemical staining, the protocol for fluorescent multiplex immunohistochemistry (mIHC) with Tyramide Signal Amplification (Cell Signaling Technology) was used. The following antibodies were used: anti-CD274 (Cell Signaling Technology, clone E1L3N, 1:200 dilution), anti-endomucin (Thermo Fisher Scientific, PA5-21395), anti–α-SMA (Sigma, clone 1A4, 1:4,000 dilution), and anti-CD8α (Cell Signaling Technology, clone C8/144B, 1:200 dilution). The processed sections were mounted in Vectashield (Vector Labs) containing DAPI and photographed with a confocal microscope (Carl Zeiss Microscopy).
Statistics.
Data are expressed as the mean ± SEM. All experiments were repeated 2 or more times. Student’s 2-tailed t test was used for comparing differences between 2 groups. One-way or 2-way ANOVA followed by Tukey-Kramer test was used for multiple comparisons. Significance was set at P < 0.05. GraphPad Prism software 7.0 was used for statistical analysis.
Author contributions
All authors participated in the design, execution, and interpretation of the study. PWN, DJ, JL, and YG conceived and designed the study. YG performed the majority of experiments and analyzed data. JL, XL, DMH, VK, ALC, TX, NL, GH, and AK performed experiments and analyzed data. ND, YW, ZL, JT, and CMH contributed to the analyses and interpretation of the study. YG, XL, DJ, and PWN wrote the manuscript. PWN and DJ approved the final, submitted manuscript.
Supplementary Material
Acknowledgments
The work was supported by NIH grants R01 AI052201, R01 HL060539 (to PWN), P01 HL108793 (to PWN and DJ), R01 HL122068 (to DJ), and R01 HL123899 (to CMH).
Version 1. 02/14/2019
In-Press Preview
Version 2. 03/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(6):e125326. https://doi.org/10.1172/jci.insight.125326.
Contributor Information
Yan Geng, Email: gengyan@jiangnan.edu.cn.
Xue Liu, Email: Xue.Liu@CSHS.org.
Jiurong Liang, Email: Carol.Liang@cshs.org.
David M. Habiel, Email: david.habiel@gmail.com.
Ana Lucia Coelho, Email: AnaLucia.Coelho@cshs.org.
Nan Deng, Email: Nan.Deng@cshs.org.
Ting Xie, Email: ting.xie@cshs.org.
Yizhou Wang, Email: yizhou.wang@cshs.org.
Ningshan Liu, Email: Ningshan.Liu@cshs.org.
Guanling Huang, Email: Guanling.Huang@cshs.org.
Adrianne Kurkciyan, Email: adrianne226@gmail.com.
Zhenqiu Liu, Email: liuzx0123@gmail.com.
Jie Tang, Email: jie.tang@cshs.org.
Cory M. Hogaboam, Email: Cory.Hogaboam@cshs.org.
Dianhua Jiang, Email: Dianhua.Jiang@cshs.org.
Paul W. Noble, Email: paul.noble@cshs.org.
References
- 1.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest. 2012;122(8):2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, et al. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med. 2011;208(7):1459–1471. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovgren AK, et al. β-arrestin deficiency protects against pulmonary fibrosis in mice and prevents fibroblast invasion of extracellular matrix. Sci Transl Med. 2011;3(74):74ra23. doi: 10.1126/scitranslmed.3001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahluwalia N, et al. Fibrogenic lung injury induces non-cell-autonomous fibroblast invasion. Am J Respir Cell Mol Biol. 2016;54(6):831–842. doi: 10.1165/rcmb.2015-0040OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, et al. Mechanosensing by the α6-integrin confers an invasive fibroblast phenotype and mediates lung fibrosis. Nat Commun. 2016;7:12564. doi: 10.1038/ncomms12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci USA. 2002;99(19):12293–12297. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dezutter-Dambuyant C, et al. A novel regulation of PD-1 ligands on mesenchymal stromal cells through MMP-mediated proteolytic cleavage. Oncoimmunology. 2016;5(3):e1091146. doi: 10.1080/2162402X.2015.1091146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beswick EJ, et al. TLR4 activation enhances the PD-L1-mediated tolerogenic capacity of colonic CD90+ stromal cells. J Immunol. 2014;193(5):2218–2229. doi: 10.4049/jimmunol.1203441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni K, et al. PD-1/PD-L1 pathway mediates the alleviation of pulmonary fibrosis by human mesenchymal stem cells in humanized mice. Am J Respir Cell Mol Biol. 2018;58(6):684–695. doi: 10.1165/rcmb.2017-0326OC. [DOI] [PubMed] [Google Scholar]
- 12.Celada LJ, et al. PD-1 up-regulation on CD4+ T cells promotes pulmonary fibrosis through STAT3-mediated IL-17A and TGF-β1 production. Sci Transl Med. 2018;10(460):eaar8356. doi: 10.1126/scitranslmed.aar8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trujillo G, et al. TLR9 differentiates rapidly from slowly progressing forms of idiopathic pulmonary fibrosis. Sci Transl Med. 2010;2(57):57ra82. doi: 10.1126/scitranslmed.3001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q, et al. IL-11 receptor alpha in the pathogenesis of IL-13-induced inflammation and remodeling. J Immunol. 2005;174(4):2305–2313. doi: 10.4049/jimmunol.174.4.2305. [DOI] [PubMed] [Google Scholar]
- 15.Schafer S, et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552(7683):110–115. doi: 10.1038/nature24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, et al. RGMb is a novel binding partner for PD-L2 and its engagement with PD-L2 promotes respiratory tolerance. J Exp Med. 2014;211(5):943–959. doi: 10.1084/jem.20130790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez MA, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst. 2016;108(1):djv303. doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SP, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11(6):694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 19.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14(9):598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinoshita K, et al. Antifibrotic effects of focal adhesion kinase inhibitor in bleomycin-induced pulmonary fibrosis in mice. Am J Respir Cell Mol Biol. 2013;49(4):536–543. doi: 10.1165/rcmb.2012-0277OC. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22(8):851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinchuk, et al. PD-1 ligand expression by human colonic myofibroblasts/fibroblasts regulates CD4+ T-cell activity. Gastroenterology. 2008;135(4):1228–1237.e1. doi: 10.1053/j.gastro.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Kilani RT, Pakyari M, Leung G, Nabai L, Ghahary A. Increased expression of PD-L1 and PD-L2 in dermal fibroblasts from alopecia areata mice. J Cell Physiol. 2018;233(3):2590–2601. doi: 10.1002/jcp.26134. [DOI] [PubMed] [Google Scholar]
- 24.Friedman CF, Proverbs-Singh TA, Postow MA. Treatment of the immune-related adverse effects of immune checkpoint inhibitors: A review. JAMA Oncol. 2016;2(10):1346–1353. doi: 10.1001/jamaoncol.2016.1051. [DOI] [PubMed] [Google Scholar]
- 25.McDermott DF, et al. Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase Ia study. J Clin Oncol. 2016;34(8):833–842. doi: 10.1200/JCO.2015.63.7421. [DOI] [PubMed] [Google Scholar]
- 26.Massard C, et al. Safety and efficacy of durvalumab (MEDI4736), an anti-programmed cell death ligand-1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J Clin Oncol. 2016;34(26):3119–3125. doi: 10.1200/JCO.2016.67.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh NC, et al. Humanized mouse models of clinical disease. Annu Rev Pathol. 2017;12:187–215. doi: 10.1146/annurev-pathol-052016-100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce EM, et al. Therapeutic targeting of CC ligand 21 or CC chemokine receptor 7 abrogates pulmonary fibrosis induced by the adoptive transfer of human pulmonary fibroblasts to immunodeficient mice. Am J Pathol. 2007;170(4):1152–1164. doi: 10.2353/ajpath.2007.060649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tashiro J, et al. Exploring animal models that resemble idiopathic pulmonary fibrosis. Front Med (Lausanne) 2017;4:118. doi: 10.3389/fmed.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B Moore B, Lawson WE, Oury TD, Sisson TH, Raghavendran K, Hogaboam CM. Animal models of fibrotic lung disease. Am J Respir Cell Mol Biol. 2013;49(2):167–179. doi: 10.1165/rcmb.2013-0094TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espindola MS, et al. Targeting of TAM receptors ameliorates fibrotic mechanisms in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2018;197(11):1443–1456. doi: 10.1164/rccm.201707-1519OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shetty SK, et al. Regulation of airway and alveolar epithelial cell apoptosis by p53-induced plasminogen activator inhibitor-1 during cigarette smoke exposure injury. Am J Respir Cell Mol Biol. 2012;47(4):474–483. doi: 10.1165/rcmb.2011-0390OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhandary YP, et al. Regulation of alveolar epithelial cell apoptosis and pulmonary fibrosis by coordinate expression of components of the fibrinolytic system. Am J Physiol Lung Cell Mol Physiol. 2012;302(5):L463–L473. doi: 10.1152/ajplung.00099.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shetty SK, et al. p53 and miR-34a feedback promotes lung epithelial injury and pulmonary fibrosis. Am J Pathol. 2017;187(5):1016–1034. doi: 10.1016/j.ajpath.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhandary YP, et al. Regulation of lung injury and fibrosis by p53-mediated changes in urokinase and plasminogen activator inhibitor-1. Am J Pathol. 2013;183(1):131–143. doi: 10.1016/j.ajpath.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192(2):209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Procopio MG, et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol. 2015;17(9):1193–1204. doi: 10.1038/ncb3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otomo R, et al. TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc Natl Acad Sci U S A. 2014;111(52):18691–18696. doi: 10.1073/pnas.1412062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M. p53 attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res. 2006;66(22):10671–10676. doi: 10.1158/0008-5472.CAN-06-2323. [DOI] [PubMed] [Google Scholar]
- 41.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9(10):691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 42.Atadja P, Wong H, Garkavtsev I, Veillette C, Riabowol K. Increased activity of p53 in senescing fibroblasts. Proc Natl Acad Sci USA. 1995;92(18):8348–8352. doi: 10.1073/pnas.92.18.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macleod KF, et al. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9(8):935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 44.Alexandrova A, Ivanov A, Chumakov P, Kopnin B, Vasiliev J. Changes in p53 expression in mouse fibroblasts can modify motility and extracellular matrix organization. Oncogene. 2000;19(50):5826–5830. doi: 10.1038/sj.onc.1203944. [DOI] [PubMed] [Google Scholar]
- 45.Eriksson M, et al. Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism. Mol Cell Biol. 2017;37(24):e00328-17. doi: 10.1128/MCB.00328-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208(5):875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding Q, et al. Focal adhesion kinase signaling determines the fate of lung epithelial cells in response to TGF-β. Am J Physiol Lung Cell Mol Physiol. 2017;312(6):L926–L935. doi: 10.1152/ajplung.00121.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheaton AK, Agarwal M, Jia S, Kim KK. Lung epithelial cell focal adhesion kinase signaling inhibits lung injury and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2017;312(5):L722–L730. doi: 10.1152/ajplung.00478.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S, et al. FAK is required for TGFbeta-induced JNK phosphorylation in fibroblasts: implications for acquisition of a matrix-remodeling phenotype. Mol Biol Cell. 2007;18(6):2169–2178. doi: 10.1091/mbc.e06-12-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding Q, et al. FAK-related nonkinase is a multifunctional negative regulator of pulmonary fibrosis. Am J Pathol. 2013;182(5):1572–1584. doi: 10.1016/j.ajpath.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai GQ, et al. Downregulation of FAK-related non-kinase mediates the migratory phenotype of human fibrotic lung fibroblasts. Exp Cell Res. 2010;316(9):1600–1609. doi: 10.1016/j.yexcr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med. 2011;6(1):2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.