Abstract
About one-third of dilated cardiomyopathy (DCM) cases are caused by mutations in sarcomere or cytoskeletal proteins. However, treating the cytoskeleton directly is not possible because drugs that bind to actin are not well tolerated. Mutations in the actin binding protein CAP2 can cause DCM and KO mice, either whole body (CAP2-KO) or cardiomyocyte-specific KOs (CAP2-CKO) develop DCM with cardiac conduction disease. RNA sequencing analysis of CAP2-KO hearts and isolated cardiomyocytes revealed overactivation of fetal genes, including serum response factor–regulated (SRF-regulated) genes such as Myl9 and Acta2 prior to the emergence of cardiac disease. To test if we could treat CAP2-KO mice, we synthesized and tested the SRF inhibitor CCG-1423-8u. CCG-1423-8u reduced expression of the SRF targets Myl9 and Acta2, as well as the biomarker of heart failure, Nppa. The median survival of CAP2-CKO mice was 98 days, while CCG-1423-8u–treated CKO mice survived for 116 days and also maintained normal cardiac function longer. These results suggest that some forms of sudden cardiac death and cardiac conduction disease are under cytoskeletal stress and that inhibiting signaling through SRF may benefit DCM by reducing cytoskeletal stress.
Keywords: Cardiology, Therapeutics
Keywords: Transcription
In mice, loss of the actin binding protein CAP2 causes dilated cardiomyopathy and serum response factor (SRF) activation, which can be targeted using an SRF inhibitor.
Introduction
Sudden cardiac death is responsible for 180,000–450,000 deaths a year in the US and is one of the leading causes of mortality (1). Nonischemic cardiomyopathy, including dilated cardiomyopathy (DCM), is responsible for 5%–20% of these deaths. About half of the cases of DCM are caused by mutations in the cytoskeletal proteins of the sarcomere (2, 3).
Heart disease can lead to remodeling of the heart, characterized by cardiomyocyte hypertrophy, fibrosis, and expression of fetal genes in cardiomyocytes or fetal reprogramming. The transcription factors that drive the fetal program include MEF2, NKX, NFAT, NKX2-5, SMAD, GATA, NRSF, and SRF (4, 5). It is not known if the fetal reprogramming protects the heart from further damage or contributes to the pathology of heart disease.
One feature of fetal reprogramming is activation of the set of genes regulated by the serum response factor (SRF). Serum activates SRF through Rho signaling. However, Rho does not activate SRF through typical kinase cascades but, instead, by promoting polymerization of actin into the filaments of the actin cytoskeleton (6). By responding to actin, SRF can be activated directly by changes in actin dynamics or activated indirectly by other causes of mechanical stress. Activation by modulating actin dynamics makes SRF unique among signaling molecules. A core component of SRF are the 2 forms of myocardin-related transcription factor (MRTF), MRTFA and MRTFB, which are both regulated by an actin monomer–binding site. Through MRTF, SRF senses the balance of monomers and filaments and serves as an actin rheostat, permitting the heart to respond to levels of cytoskeletal stress (7).
CAP2 is an actin monomer binding protein that associates with the sarcomere and promotes actin depolymerization. CAP proteins were originally isolated in yeast as a subunit of adenylyl cyclase — hence the family name, cyclase associated proteins (8, 9). In yeast, CAP assists Ras in activating cyclase by binding to the farnesyl lipid tail on Ras (10), while mammalian CAP1 can bind to the Ras-related protein Rap1, again through its lipid tail (11). All CAPs also bind to actin monomers, partner with cofilin to depolymerize actin filaments, and recharge actin-ADP with ATP (12, 13). The yeast CAPs also transduce signals from the cytoskeleton to Ras, but no role has been found for mammalian CAPs in transducing signals from the cytoskeleton (14). There are 2 CAP isoforms in mammals, CAP1 and CAP2 (15).
Substantial evidence points to the importance of CAP2 in cardiac function. CAP2 is located on chromosome 6 at 6p22, near a locus that confers a risk for sudden cardiac death and conduction disease (16–19). Deletion of 6p22 is associated with cardiac abnormalities (20). A mutation in CAP2 is likely the cause of childhood DCM in 2 related patients in a large pedigree that was analyzed by exome sequencing. Both affected family members were the only ones in the pedigree having the same homozygous CAP2 mutation, while 12 heterozygotes had no evidence of DCM. Therefore, the CAP2 mutations segregate as an autosomal recessive DCM gene (21), though some mutations may be dominant or semidominant.
The DCM caused by CAP2 loss has been modeled in mice. CAP2-KO mice are characterized by DCM with conduction disease. Males are more severely affected than females. CAP2-KO mice are also short and develop eye abnormalities (22, 23). The various phenotypes in CAP2-KO mice are recessive. Cardiomyocytes are the cells that are primarily affected in DCM since cardiomyocyte-specific CAP2 KOs (CAP2-CKO) are especially prone to sudden cardiac death (23).
Here, we show that both CAP2-KO mice and CAP2-CKO mice have elevated levels of SRF gene activation in their hearts. Furthermore, treatment of mice with an SRF inhibitor improves their cardiac function and extends their lifespan. This study suggests that SRF activation may be an indication of cytoskeletal stress and that reducing levels of SRF activation can prevent fetal reprogramming and benefit some forms of DCM.
Results
Gene expression profiling in hearts from CAP2-KO mice.
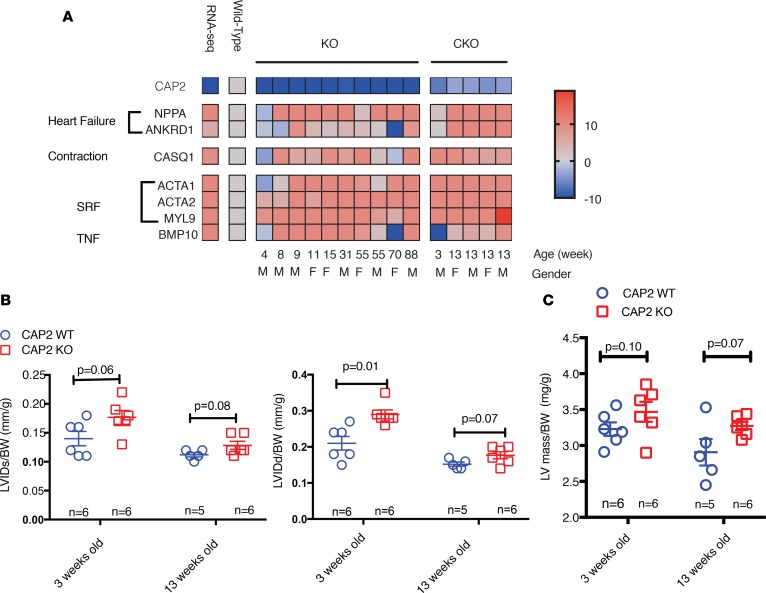
To identify the gene expression differences caused by loss of CAP2, we isolated hearts and carried out RNA sequencing (RNA-seq) analysis in both CAP2-KO and -CKO mice. The RNA-seq identified 788 and 2,274 differentially expressed genes (>1.2-fold change), with 50.3% and 41.2% upregulated in the CAP2-KO and -CKO mice, respectively. Figure 1A shows the top 50 regulated genes in the CAP2-KO mice and CAP2-CKO mice. Volcano plots of the changes in gene expression of CAP2-KO or -CKO mice versus WT mice are shown in Supplemental Figure 1A (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124629DS1). Comparing the 2 gene lists by Venn diagram gives 325 genes found in both 2 lists, with 172 genes upregulated and 153 downregulated (Figure 1C). The top upregulated gene was Natriuretic peptide A (Nppa), a common marker of heart failure and hypertrophy. Other upregulated genes were heart failure and hypertrophy related genes such as α-actin-1 (Acta1), ankyrin repeat domain 1 (Ankrd1), and myosin light chain kinase 2 (Mylk2), as well as several genes implicated in arrhythmia such as calsequestrin 1 (Casq1), potassium voltage-gated channel subfamily A member 5 (Kcna5), and potassium channel subfamily K member 2 (Kcnk2). CAP2 was downregulated more than 20-fold in the mutants.
Figure 1. Genes and pathways altered in whole body CAP2-KO mice and CAP2-CKO mice.
For KO mice and its controls, RNA was isolated from female WT mice (n = 3) and mutant CAP2 KO hearts (n = 3), ages ~86 weeks old. For CKO mice, RNA was isolated from male (n = 2) and female (n = 3) rescued mice and male (n = 1) and female (n = 3) CAP2-CKO mice hearts, ages ~13 weeks old. The samples were subjected to RNA-seq analysis, and genes were ranked by differential expression P values as determined by the limma-voom software. Pathway analysis by Ingenuity Pathway Analysis (IPA) software was performed on the list of all genes that were differentially expressed genes with FDR < 0.2 and fold change at least 1.2 in both KO and CKO experiments. (A) Top 50 altered genes in hearts from CAP2 KO mice (n = 3) compared with WT (n = 3) (left) and CAP2-CKO mice (n = 4) compared with age-matched rescued mice (n = 5) (right). (B) The RNA-seq analysis pipeline used in this study. (C) The overlap genes with altered expression levels in both CAP2 KO and CKO mice. (D) Regulators analysis shows serum responsive factor (SRF), TGFβ, and TNF signaling were significantly activated; red means predicted activation by IPA software.
Next, we filtered the gene list with the pipeline shown in Figure 1B and performed pathway analysis with Ingenuity Pathway Analysis (IPA; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) on the list of genes shown upregulated in both KO and CKO mice. The top activated canonical pathways were actin related, such as RhoA signaling and actin cytoskeleton signaling, and several pathways related to heart disease, such as cardiac hypertrophy signaling and calcium signaling. The top upstream regulators were SRF and TGFβ. Among these, SRF was the top transcription regulator by IPA Z-score calculation (Figure 1D). IPA identified the SRF genes overexpressed in the hearts from CAP2-KO mice (Supplemental Figure 1B).
SRF activation precedes heart failure in CAP2-KO mice.
To confirm expression levels of genes identified by RNA-seq and trace the progression of changes as mice age, we used quantitative PCR (qPCR) to quantify the expression of genes representative of the most altered pathways. Myl9 is regulated by SRF, Bmp10 is regulated by TGFβ, Nppa and Ankrd1 are markers of heart failure, and Casq1 is a marker of arrhythmia and calcium signaling. Figure 2A shows expression of these marker genes in hearts from CAP2-KO and -CKO mice at different ages. Each data point shows the fold-change in mutant mice compared with age-matched WT mice. For genes overexpressed more than 10-fold in the mutants, we assigned a 10-fold change for better data visualization. These data confirm the expression changes that were identified by RNA-seq analysis. In comparing expression over time, we found that SRF genes Myl9, Acta1, and Acta2 were activated up to 10-fold at 3 weeks of age and remained elevated until 90 weeks of age. Heart failure and arrhythmia indicators Nppa, Ankrd1, and Casq1 were elevated, but at 8 weeks of age, they were delayed compared with SRF genes. The TNF gene Bmp10 was activated sporadically.
Figure 2. SRF pathway activation is an early step in CAP2 mutant hearts and precedes dilated cardiomyopathy.
RNA was isolated from mouse hearts at the indicated ages, and individual gene expression levels were determined by qPCR. Normalization was done by using GAPDH expression as a standard. (A) qPCR analysis of representative genes of each pathway and associated phenotypes were selected and tested in the hearts of 10 pairs of WT and CAP2-KO and 5 pairs of WT and CAP2-CKO age-matched mice. Heatmap shows the fold-change in gene expression levels with a cut-off of 10-fold; CAP2-KO or -CKO mice compared with age-matched control mice. The age and sex of each pair of mice are labeled in the figure. Three technical triplicates were used in each sample. (B and C) Left ventricle (LV) function parameters were measured from M-mode recordings in the short-axis view. (B) LV diameter during diastole (LVIDd) and systole (LVIDs) (n = 5–6) and (C) LV mass, corrected for body weight, was increased in CAP2-KO mice at 3 weeks and 13 weeks of age, compared with WT mice (n = 5–6). Bars represent mean values ± SEM. P value between groups was based on Student’s t test.
Heart function was evaluated by echocardiography at 3 weeks and 13 weeks (n = 6 in each group). Loss of CAP2 caused mild left ventricular (LV) chamber dilation, with an increase in end-diastolic LV internal diameter (LVIDd) and end-systolic LV internal diameter (LVIDs) at 3 weeks and 13 weeks (Figure 2B), corrected for body-weight. At 13 weeks, the ratio of LV mass/body weight was increased in CAP2-KO (Figure 2C). We note that most parameters measured in CAP2-KO indicated a trend toward significance, and we did not observe a difference in ejection fraction (EF) or fractional shortening (Supplemental Figure 2). In comparing the transcription data with the functional cardiac data, we note that, though all genes are eventually activated, only SRF genes were activated at all ages tested, suggesting that SRF signaling activation precedes heart failure.
SRF signaling is activated through MRTFB translocation in CAP2-KO mice.
To determine if SRF is activated by loss of CAP2 in other tissues where CAP2 is highly expressed, we examined expression of Acta2 and Myl9 in brain and muscle but found that there were no changes in mRNA levels in these tissues (Figure 3A). Therefore, even though CAP2 is expressed in brain and skeletal muscle, as well as in the heart, SRF pathway activation is exclusive to the heart. SRF can drive 2 mutually exclusive gene regulation programs by associating with different transcriptional cofactors, Elk or MRTF (also known as Mkl1/2). To address which cofactor is most likely regulating the affected genes, we compared the expression of the Elk-dependent gene cfos with MRTF-dependent genes Acta2 and Myl9. Levels of cfos did not change (Figure 3B). These data suggest that only MRTF/SRF signaling was activated in the hearts of CAP2-KO mice.
Figure 3. The SRF pathway is specifically activated in hearts from CAP2-KO mice and associated with MRTFB translocation.
(A) SRF target genes are activated in hearts but not brains or skeletal muscle from CAP2-KO mice. Expression levels of SRF targets Myl9 and Acta2 were determined in CAP2-KO mice (n = 3); data is presented as mean ± SEM. Three technical triplicates were used in each sample. (B) Relative expression level of Myl9 and cfos in hearts from CAP2-KO mice (n = 5) and age-matched WT mice (n = 5). SRF regulation through 2 pathways. Note the difference in scales from A. Data is presented as mean ± SEM. P value compared with WT mice was based on the Student’s t test. Three technical triplicates were used in each sample. (C) Representative Western blots and quantification showing MRTFB nuclear accumulation in CAP2-KO mice hearts (n = 2). Data is presented as the mean of the values; P value was based on Student’s t test. Western blot of SRF and MRTFA and MRTFB in nuclear and cytoplasmic extracts from mice hearts (n = 2). Right panel shows quantification of the Western blot using ImageJ software. Three technical replicates were used in each sample.
To address if activation was cell autonomous or specific to the CAP2 isoform, we investigated whether CAP loss could activate SRF in other cells (Supplemental Figure 3, A–C). We performed microarray experiments in HeLa cells and found that CAP1 knockdown caused several SRF-regulated genes, including Acta2, to be upregulated. We did not observe Acta2 activation when CAP2 alone was knocked down, but when CAP2 was knocked down in the CAP1 knockdowns, we observed additional activation of SRF markers. Thus, loss of CAP activation of SRF is cell autonomous, though — in some cells and tissues — CAP1 is rate limiting.
SRF is activated by MRTF, which has 2 isoforms, MRTFA and MRTFB. To address these factors, we isolated the protein from the nuclei in the hearts from CAP2-KO mice, and we performed Western blots for MRTFA, MRTFB, and SRF. Compared with age-matched CAP2 WT mice, MRTFB accumulated in the nuclei in the CAP2-KO mice, while there was no difference with MRTFA. SRF was also upregulated in the CAP2-KO mice (Figure 3C). This suggests that CAP2 loss primarily activates MRTFB.
MRTF/SRF signaling is activated only in the cardiomyocytes of CAP2-KO mice.
MRTF/SRF signaling is upregulated when fibroblasts are activated into myofibroblasts during heart remodeling, a process thought to be driven primarily by MRTFA. We addressed if fibroblasts are affected and found that the SRF genes Acta2 and Myl9 were not upregulated in MEF cells (Supplemental Figure 4A). We also evaluated these genes in isolated cardiomyocytes from CAP2-KO mice (Figure 4A). Myl9, Acta1, and Acta2 were all upregulated, while cfos was not changed (Figure 4B) in the isolated cardiomyocytes. We observed activation of SRF genes even in 3-week-old mice, while Nppa was not activated until 13 weeks of age (Figure 4C). Since the fibrosis pathway is activated in the RNA-seq data in both CAP2-KO and -CKO mice, we also evaluated fibrosis by Sirius Red staining, but as we previously noted, we did not observe significant fibrosis in the hearts from CAP2-KO mice (Supplemental Figure 4B) (23). This suggests that cardiomyocytes are the primary cells for SRF activation in the heart.
Figure 4. SRF genes are activated in cardiomyocytes of CAP2-KO mice.
Cardiomyocytes were isolated by the Langendorff method from CAP2-KO mice and age-matched WT mice at 3 weeks (n = 3) and 13 weeks of age (n = 4), respectively. (A) Schematic of this experiment. (B) Relative expression of SRF pathway representative genes Myl9, Acta1, Acta2, and cfos in isolated cardiomyocytes. Data is presented as mean ± SEM; P values compared with WT mice based on Student’s t test. Three technical triplicates were used in each sample. (C) Nppa and CAP2 expression in isolated cardiomyocytes. Data is presented as mean ± SEM. P values compared with WT mice, based on Student’s t test. Three technical triplicates were used in each sample.
CCG-1423-8u inhibits SRF signaling through MRTF translocation in vitro and in vivo.
Whole body CAP2-KO males rarely survive to adulthood, and the few males that survive develop DCM slowly, while females do not reliably develop DCM (23). However, CAP2-CKO mice develop a robust DCM, so we used CAP2-CKO mice to test the functional importance of SRF in CAP2 DCM. qPCR analysis confirmed deletion of CAP2 in Myh6Cre-cap2loxp/cap2loxp compared with those of control cap2loxp/cap2loxp mice (Supplemental Figure 5, A and B). CAP2-CKO mice died from heart failure during a narrow time window of 8–15 weeks (23). We found that mice sacrificed near this time window or subsequent necropsy of animals that died suddenly displayed robust heart dilation and pleural effusion. Histological analysis of CAP2-CKO hearts with H&E staining revealed marked cardiac enlargement, including both ventricular and atrial hypertrophy. MRTF/SRF gene expression was also elevated in the hearts from CAP2-CKO mice, as we observed in CAP2-KO mice (Supplemental Figure 5C).
To evaluate the contribution of overactivated MRTF/SRF signaling to the phenotype of the CAP2-CKO mice, we treated mice with the SRF inhibitor CCG-1423-8u, a more potent derivative of CCG-1423 (Figure 5A) (24, 25). CCG-1423 compounds inhibit SRF transcription and prevent MRTF translocation (Figure 5B and Supplemental Figure 6). Since CCG-1423-8u has not been used in vivo, we first performed a preliminary experiment to evaluate its effect on SRF markers. We treated 3 CAP2 WT mice and 3 CAP2-KO mice for 14 days by i.p. injection at a dose of 0.15 mg/kg/d, sacrificed the mice the day after the last injection, and then measured SRF target gene mRNA expression levels. Acta2 and Myl9 activation were attenuated after treatment, both in CAP2 WT and KO mice, while cfos was unaffected. Nppa was also downregulated in the treated CAP2-KO mice (Figure 5C). These data indicate that CCG-1423-8u inhibited SRF signaling, in vivo, without affecting Elk signaling.
Figure 5. SRF inhibition reduces severity of cardiac pathology in CAP2-KO mice.
(A) Structure of the SRF inhibitor CCG-1423-8u. (B) The inhibitory activity of CCG-1423-8u was measured using a luciferase reporter for SRF. Three technical triplicates were used in each sample. (C) Expression of SRF target gene expression detected by qPCR in the hearts of whole body CAP2-KO (n = 6) mice or WT mice (n = 6) treated for 14 days with CCG-1423-8u (n = 3) or DMSO (n = 3), at 6 weeks of age. Data is presented as mean ± SEM. Three technical triplicates were used in each sample. (D) H&E staining of the CAP2-CKO or control mouse hearts treated with CCG-1423-8u or DMSO. Scale bar: 500 μm. (E) Heart weight to body weight and heart weight to tibia length of the treated control or CAP2-CKO mice (n = 6–11). Data is presented as mean ± SEM. P values compared with WT mice, based on the Student’s t test. (F) Detail of stained hearts. Scale bar: 50μm. (G) Myocyte cross-sectional area was measured in different groups (n = 3) using ImageJ. For each heart, the mean cross-sectional area was determined from 120 myocytes in 3 different visual fields. Data is presented as mean ± SEM. P value compared with WT mice was determined using the 2-tailed paired t test.
CCG-1423-8u restores cardiac structure and function.
After finding that CCG-1423-8u reduced the SRF marker genes in vivo, we next tested if the drug could affect the course of DCM in the CAP2-CKO mice. A total of 95% of CAP2-CKO mice died from heart failure between 12 and 15 weeks of age. For comparison, we also treated Myh6-Cre-cap2+/cap2+ mice as a control for the toxicity of CCG-1423-8u and, additionally, cap2loxp/cap2loxp mice as a control for any cardiac toxicity of the Myh6-Cre mice (26, 27). We assigned 10-week-old CAP2-CKO mice to 2 different treatment groups (DMSO, n = 10; MRTF/SRF inhibitor CCG-1423-8u, n = 10), and examined cardiac structure and function at 13 weeks of age, after 3 weeks of treatment.
The CAP2-CKO mice developed severe cardiomyopathy around 13 weeks of age, just before they died. As shown by H&E staining, hearts from CAP2-CKO mice had significantly larger hearts (Figure 5D), with a 78.2% increase in the heart weight/body weight (HW/BW) ratio and a 77.5% increase in the ratio of heart weight/tibia length (HW/TL) compared with control mice (Figure 5E). In contrast, CCG-1423-8u treatment reduced the size of the CAP2-CKO hearts (Figure 5D), with a 35.4% decrease in the HW/BW ratio and 31.6% decrease in the HW/TL ratio (Figure 5E). Morphometric analysis of the cross-sectional area of cardiomyocytes from histological sections showed that KO of CAP2 caused a 2.12-fold increase in the myocardial cell area. This hypertrophy was decreased by 21.3% via CCG-1423-8u treatment (Figure 5, F and G). These findings demonstrate that treating with CCG-1423-8u decreased progression of DCM and pathologic myocyte remodeling in CAP2-CKO mice.
We also measured cardiac dimensions and contraction by echocardiography on anesthetized mice after 3 weeks of treatment with CCG-1423-8u. M-mode transthoracic echocardiography showed increased LV end-diastolic cavity dimension (LVEDD) and LV end-systolic cavity dimension (LVESD) in CAP2-CKO mice, compared with control mice. However, the CCG-1423-8u–treated CAP2-CKO mice had significantly smaller LVEDD and LVESD (Figure 6A). The EF and fractional shortening was significantly reduced in the CAP2-CKO mice, compared with cap2loxp/cap2loxp and Myh6Cre- cap2+/cap2+, but it was restored in the CAP2-CKO mice treated with CCG-1423-8u (Figure 6B).
Figure 6. SRF inhibition delays DCM in CAP2-CKO mice.
CAP2-CKO mice and cap2loxp/cap2loxp controls were treated with CCG-1423-8u for 3 weeks starting 6–7 weeks of age, and heart function was measured by echocardiography. (A) Echocardiogram of CAP2-CKO mice and controls treated with CCG-1423-8u or DMSO, respectively. (B) Ejection fraction was significantly reduced in treated CAP2-CKO mice (n = 12) compared with age-matched controls (n = 9), while CCG-1423-8u treatment delayed the reduction in contraction. Data is presented as mean ± SEM. P value compared with WT mice was determined using the Student’s t test.
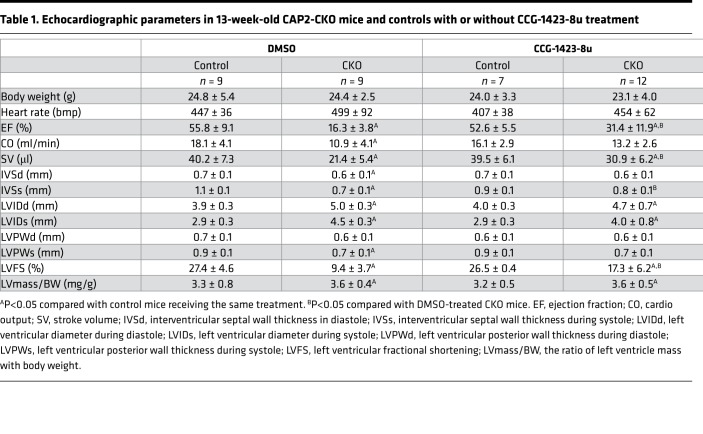
Table 1 shows the echocardiographic parameters for the CCG-1423-8u–treated CAP2-CKO mice. The CAP2-CKO mice had significantly increased LVIDd and LVIDs compared with control mice. The EF of CAP2-CKO mice at 13 weeks was 16.3% ± 3.8%, which was decreased 39.5% compared with control mice. CAP2-CKO mice treated with CCG-1423-8u had a reduction in the LVIDd and LVIDs and an EF of 31.4% ± 11.9% — an increase of 15.1% (P = 0.0002) compared with untreated CAP2-CKO mice. Overall, these results showed that CCG-1423-8u had positive effects on cardiac function during the progression of DCM in CAP2-CKO mice. The control mice treated with the same dosing schedule of CCG-1423-8u did not show any differences in echocardiographic parameters compared with untreated control mice, indicating that CCG-1423-8u does not cause any obvious cardiac toxicity or changes in baseline cardiac parameters.
Table 1. Echocardiographic parameters in 13-week-old CAP2-CKO mice and controls with or without CCG-1423-8u treatment.
Effects of CCG-1423-8u on survival of CAP2-CKO mice with heart failure.
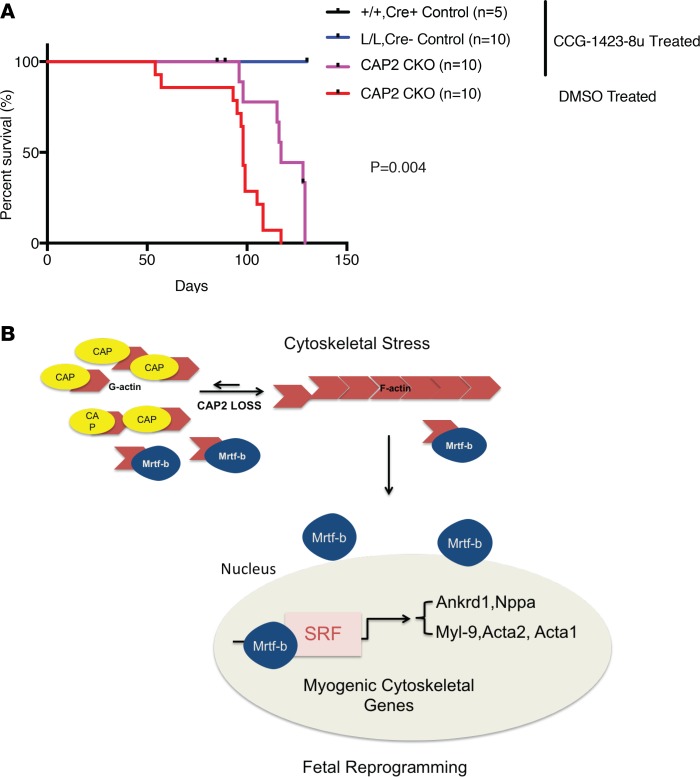
We next analyzed the effects of CCG-1423-8u treatment on CAP2-CKO survival. CAP2-CKO mice died between 8 and 15 weeks of age. We followed the same dosing schedule as described above; beginning at 6–7 weeks (45 ± 3 days) of age, CAP2-CKO mice were injected i.p. for 21 days with 0.15 mg/kg/d of CCG-1423-8u. DMSO-treated CAP2-CKO mice had a maximum survival of 117 days and a median survival of 98 days. CCG-1423-8u–treated CAP2-CKO mice had a significant increase in survival, with a median survival of 116 days (log rank P = 0.004), with some mice living up to 127 days of age (Figure 7A). Treatment had no effect on the survival of control mice. We repeated this experiment with a different treatment protocol, treating daily, beginning at 7 weeks of age. Again, mice lived for a maximum of 126 days with a median survival of 114 days (log rank P = 0.0005) (data not shown). Hence, suppressing SRF overactivation prolongs the lifespan of CAP2-CKO mice, consistent with its beneficial effects on cardiac fetal reprogramming, cardiac structure, and heart function.
Figure 7. SRF inhibition extends the lifespan of CAP2-CKO mice.
(A) Survival curves of CAP2-CKO mice treated with CCG-1423-8u (n = 10) or DMSO (n = 10). P value was determined by Log-rank (Mantel-Cox) test. (B) Schematic model of MRTF activated SRF signaling in CAP2-KO hearts.
Discussion
The SRF transcription factor stimulates expression of numerous sarcomere proteins. Knocking out SRF in the mouse heart causes a rapid onset of DCM, indicating that it is essential for both heart development and maintenance of cardiac homeostasis (5, 28, 29). However, hearts must balance SRF levels because overexpression of SRF also causes DCM (30, 31). SRF is overexpressed in many cardiac diseases, both ischemic and nonischemic, but it is not established if the overexpression is protective or pathological. Since we find beneficial effects of inhibiting SRF in CAP2-KO, we suggest that overexpression is pathological and that cardiomyocytes can overcompensate when they activate SRF in response to cytoskeletal stress (Figure 7B).
Cardiomyocyte transcriptional dedifferentiation, or fetal reprogramming, is common in both nonischemic and ischemic cardiomyopathy and is characterized by reexpression of fetal genes and isoforms. It is not known if fetal transcription factor activation is a protective, detrimental, or noncontributory response to the pathology of heart failure. There is some evidence that fetal reprogramming is a pathological contributor because β blocker–treated patients recovering from heart failure partially reverse the fetal gene reactivation, but the recovery of fetal gene expression was only correlative and is not seen in the partial recovery of patients on a LV assist device support (5, 32, 33). Indeed, Histone deacetylase (HDAC) inhibitors, which are classic dedifferentiation agents, show some effectiveness in experimental models of cardiomyopathy, but clinical use of multiple HDAC inhibitors to treat cancer is associated with adverse cardiac side effects (34–37). HDAC inhibitors are broad-acting epigenetic transcription regulators that affect the expression of many genes, suggesting that effective use of transcription modulators may require more specific drugs. SRF, which directly responds to cytoskeletal stress, is activated as part of the cardiac myoblast fetal expression program. Though transcription factors are generally poor drug targets, we find here that SRF can be successfully targeted with a small molecule inhibitor to delay DCM.
Fibrosis is commonly observed in DCM and ischemic heart damage. To initiate fibrosis, factors that activate fibroblasts, especially TGFβ, are released from damaged tissues. Activated fibroblasts, or myofibroblasts, express high levels of smooth muscle actin and secrete collagen (38). However, myofibroblasts are not the likely to be the target of CCG-1423-8u in our CAP2-KO animals, since we do not reliably observe high levels of collagen expression in qPCR assays or observe collagen deposits by Sirius Red staining (Supplemental Figure 4B). Moreover, we find that there are extremely high levels of Acta2 and Myl9 in isolated cardiomyocytes from CAP2-KO animals. There is some debate as to the ideal level of fibrosis in damaged hearts. While some level of fibrosis can assist tissue repair, excessive fibrosis compromises cardiac contraction. In any event, CCG-1423-8u inhibits SRF in fibroblasts and can reduce levels of lung fibrosis, suggesting that it may be beneficial in reducing excessive cardiac fibrosis, as well as acting on cardiomyocytes.
The optimal dose and dosing schedule of CCG-1423-8u has not been determined. Only a few structurally related SRF inhibitors have been tested in vivo before. Though they are metabolized relatively rapidly, they have been successfully used in multiple models of fibrosis. This is the first study to our knowledge to test an SRF inhibitor in animals with compromised cardiac function (25, 39). We did not observe any obvious discomfort in the control mice that were treated with CCG-1423-8u, suggesting that they will tolerate higher doses and more potent SRF inhibitors. We tested 2 dosing schedules, continuous dosing, and interrupted treatment, and we found that both extended survival to similar degrees. This suggests that there may be an early time window for treatment and that treating beyond the window does not improve efficacy.
Comparison of the CAP2-KO and CAP2-CKO mice with human CAP2-DCM is in the early stages because only 2 patients have been described with homozygous CAP2 mutations (21). Their DCM was diagnosed at ages 5 and 12; the 12-year-old died shortly after diagnosis and was not completely phenotyped. The mutation that both patients had was at a splice site and generated a protein that lacked 2 exons but left most functional domains intact. Expression of the mutant protein was low, so it is not clear if the deleted regions reduced protein function or only affected stability. Phenotypically, the patients had DCM and conduction disease without the eye or height abnormalities observed in CAP2-KO, suggesting that they resembled CAP2-CKO more closely than CAP2-KO. However, we note that the CAP2-KO mice are a biased population because few mice survive infancy, so the survivors that we study may have less severe DCM than those that do not live long enough to be characterized.
Though this study utilized CAP2-KO mice to study DCM, many mutations seen in DCM are in genes for sarcomere proteins, including actin. Actin is difficult to target safely because actin-binding drugs, such as latrunculin A, cytochalasin D, phalloidin, and jasplakinolide, are not well tolerated, often causing cardiac toxicity (40–42). However, drugs that target signals to the actin cytoskeleton offer a less toxic alternative than those targeting actin itself. The most common signaling pathways to the cytoskeleton are through Rho family small GTPases. Druggable targets from the Rho pathway include Rho kinases JNK and p38. Inhibitors of these kinases, as well as ERK, have shown some effectiveness in DCM models caused by lamin A/C mutations, the most common mutation in DCM (3, 37, 43–48). Lamin A/C mutant mice develop DCM and conduction disease, with reduced levels of SRF activation, suggesting that a common element of lamin A/C and CAP2-KO DCM may be the misregulation of Rho signaling (49). Given the common pathway leading back to Rho and actin, treatment of DCM by SRF inhibition may benefit patients with DCM caused by mutations in genes other than CAP2 or even benefit patients with heart failure, where cytoskeletal stress and fetal reprogramming are also observed (5, 50).
Methods
Generation of mice used in this study.
CAP2-KO mice and CAP2-CKO mice were generated and genotyped as described (23). Mice were euthanized by CO2, and hearts were isolated from CAP2-KO mice and -CKO mice at the indicated ages. For all Western blots, RNA-seq, and real-time PCR experiments, CAP2-KO mice were compared directly with age- and sex-matched WT littermates taken from the litters of 6 different crosses between cap2+/– and cap2+/–. For CCG-1423-8u treatment experiments, CAP2-CKO mice and controls were taken from the litters of 10 different crosses between Myh6-Cre-cap2+/cap2+ or Myh6-Cre-cap2+/cap2loxp bred to cap2loxp/cap2loxp mice. Control cap2loxp/cap2loxp and Myh6Cre-cap2+/cap2+ were taken from each of the litters from which the CAP2-CKO mice were used.
CCG-1423-8u studies.
CCG-1423-8u was prepared as described (25). An analysis of the compound used in these studies is shown in Supplemental Figures 6 and 7. CCG-1423 was purchased from SelleckChem. For animal experiments, CCG-1423-8u was dissolved in DMSO at a concentration of 15 mg/ml, and then dissolved in PBS at a final concentration of 0.15 mg/ml before injection. Mice were injected i.p. with 0.1 ml/10 g for 14 days or 21 days at a dose of 0.15 mg/kg/d. Control mice were injected with 0.1 ml/10 g PBS containing 0.1% DMSO.
RNA isolation from whole hearts.
Total RNA was extracted using the RNeasy fibrous tissue kit (catalog 74704, Qiagen) according to the manufacturer’s instructions. The RNA concentration of each sample was measured by using Nanodrop 2000/2000c spectrophotometers (Thermo Fisher Scientific).
RNA-seq and analysis.
RNA-seq and analysis were performed by PrimBio Research Institute LLC, Next Generation Sequencing Core (University of Pennsylvania), and ITMAT Bioinformatics (University of Pennsylvania).
Library preparation and sequencing.
For the CAP2-KO RNA-seq samples, cDNA libraries were constructed using Ion Total RNA-Seq Kit v2 from Invitrogen (catalog 4479789) and manufacturer-recommended protocol. The prepared libraries were run on the Agilent 2100 Bioanalyzer to determine the yield and size distribution of each library. Approximately 100 pM of pooled barcoded libraries were used for templating using Invitrogen Ion Chef 200 kit (catalog 4488377) and manufacturer-recommended protocol. The loaded chip was then placed into an Ion Proton sequencer, and the run was started using an Ion Torrent RNA-seq run plan that was configured based on type of library, species, number of run flows required, type of plug-in required, and adapter trimming, as well as other parameters specific to the transcriptome run.
For the CKO RNA-seq samples, total RNA quantity and quality were assayed with an Agilent 2100 Bioanalyzer instrument using the RNA 6000 Nano Kit (Agilent Technologies). Libraries were prepared using TruSeq Stranded mRNA HT Sample Prep Kit (Illumina) as per standard protocol in the kit’s sample preparation guide. Libraries were assayed for size with D1000 ScreenTape kit of Agilent 2200 TapeStation System (Agilent Technologies) and quantified using KAPA Library Quantification Kit for Illumina platforms (KAPABiosystems). Single-read sequencing (100 bp) of multiplexed samples was performed on an Illumina HiSeq 4000 sequencer. Illumina’s bcl2fastq version 2.20.0.422 software was used to convert bcl to fastq files.
Alignment and data analysis.
For the KO experiment, the raw sequence files were aligned to the mouse genome (mm10) reference sequence by the alignment plugin tool in Ion Torrent Suite software using the default parameters. Quality control, quantification, and normalization was performed by the StrandNGS program using its DEseq algorithm. Differential expression was analyzed using limma-voom. For the CKO experiment, the raw sequence files were aligned to the mouse genome (mm10) using STAR version 2.6.0c. One sample, a male CKO, had an abnormally low number of reads and was removed from further analysis. The remaining samples had their reads quantified and normalized using PORT 0.8.5d (https://github.com/itmat/Normalization; c482d07aa8b03236004f47f2c80ecf2332870da4). Differential expression between WT and CKO was analyzed by limma-voom with a sex cofactor. For both experiments, pathway analysis was performed with Ingenuity IPA software using the top genes by differential expression with an FDR <0.2 and fold-change of at least 1.2. The RNA-seq data for all the samples in both experiments were deposited on Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE125603.
Cardiomyocyte isolation.
Cardiomyocytes were isolated by the Langendorff method, using retrograde perfusion through the aorta with enzyme-containing solutions, as described (51). Briefly, mice were anesthetized with an i.p. injection of ketamine/xylazine, 100 mg/kg per 12.5 mg/kg with 100 IU of Heparin, respectively. After prepping with antiseptic, the chest was opened, and a cardiectomy was performed. The heart was mounted on a perfusion apparatus and perfused with a calcium-free Krebs-Henseleit buffer followed by a collagenase-based enzyme solution to digest the extracellular matrix through the aorta. After digestion, myocytes were dispersed into a single-cell suspension in Krebs- Henseleit buffer.
qPCR.
Immediately after RNA was extracted, 100 μg of RNA was used to synthesize cDNA, using the High-Capacity RNA-to-cDNA Kit (catalog 4387406, Thermo Fisher Scientific). Predesigned SYBR Green primers were obtained from Integrated DNA Technologies (IDT) for the following genes: Myl9 (Mn.PT.58.41726289), Nppa (Mm.PT.58.12973594.g), Acta1 (Mm.PT.58.7312945), Acta2 (Mm.PT.58.163206644), cFOS (Mm.PT.58.29977214), Ankrd1 (Mm.PT.58.11977470), Gapdh (Mm.PT.39a.1), Casq1 (Mm.PT.58.42990820), and Bmp10 (Mm.PT.58.5708693). Cap2 primers (Mm00482645) were obtained from Thermo Fisher Scientific. SYBR Green PCR Master Mix was purchased from Thermo Fisher Scientific (catalog 4309155). The real-time PCR reaction contained 5l mastermix, 100 nM of each primer, and 0.5 μl of template in a 10-μl reaction volume. Amplification was carried out on a ViiA7 Real-Time PCR system from Applied Biosystems. Relative levels of mRNA expression were calculated according to the 2–ΔΔCT method. Individual expression values were normalized by comparison with Gapdh mRNA.
Subcellular protein fraction extractions from hearts and Western blot analysis.
Subcellular protein fractions were extracted from whole hearts using a Subcellular Protein Fractionation Kit for Tissues (catalog 87790, Thermo Fisher Scientific). Protein concentration was quantified with a Bradford Protein Assay Kit (catalog 23246, Thermo Fisher Scientific). 30 μg of protein was resolved by SDS-PAGE (4-12% gel, Invitrogen). The proteins were transferred to PVDF (Invitrogen) membranes and probed with an anti-MRTFA antibody (catalog sc-21558, Santa Cruz Biotechnology Inc., 1:1000), an anti-MRTFB antibody (catalog TE2565871, Invitrogen, 1:500) or an anti-SRF antibody (catalog sc-335, Santa Cruz Biotechnology Inc., 1:1000). Antibodies against Lamin-B (catalog sc-374015, Santa Cruz Biotechnology Inc., 1:1000), or GAPDH (catalog sc-25778, Santa Cruz Biotechnology) were used as loading controls. MRTF is ~150 kDa, and SRF is ~67 kDa. Immunoreactivity was visualized by use of ECL according to manufacturer’s protocol (catalog 32106, Thermo Fisher Scientific). Band intensity was measured and quantified using ImageJ (NIH).
Histopathology analysis.
Sections (4μm) of paraffin-embedded hearts stained with H&E were examined using light microscopy. Myocyte cross-sectional area was measured using ImageJ. For each heart, the mean cross-sectional area was determined from 120 myocytes in 3 different visuals, measured using ImageJ. Heart size was calculated using the weight after sacrifice of the heart, corrected for the length of tibia and body weight, weighed before sacrificing.
Echocardiograph.
Transthoracic echocardiography was performed as described previously (23). Mice were anesthetized by inhalation of 4% isoflurane in a glass chamber and then maintained during procedures in 1.5% isoflurane via nose cones. Echocardiography was performed using a Vevo 770 (VisualSonics) with a linear 30-MHz probe (RMV 707B). LV parameters were obtained from M-mode recordings in short-axis view. Interventricular septal wall thickness in diastole (IVSd), posterior wall thickness in diastole (PWd), LVEDD, and LVESD diameters were calculated from the mean of at least 3 separate cardiac cycles. LV EF was calculated by dividing stroke volume by the end diastolic volume (EDV). LV mass was calculated with following formula: LV mass(g) = 1.05 × (5/6 [epicardial × long axes (epicardial) – (endocardial – long axes endocardial)]) and corrected for body weight.
Multiple-hypothesis testing consideration.
A total of 62 two-sample tests were performed, all with a significance threshold of 0.05. A total of 31 of the 62 P values are significant at the 0.05 level. We therefore would conservatively expect 0.05 × 62 = 3.1 false positives from 62 tests if all null hypotheses were true. Thus, we (conservatively) expect approximately 28 of our rejected null hypotheses to be correctly rejected. With an expectation of 28 true positives, we can recalculate the expected number of falsely rejected null hypotheses to conservatively be 0.05 × 34 = 1.7. Therefore, we expect approximately 2 false positives from the 31 rejected null hypotheses. This number is conservative since many of the observed P values are in fact much smaller than 0.05. The overall conclusions of this paper are robust to such a small number of false positives among all rejected null hypotheses, particularly among the hypotheses with marginal P values.
Statistics.
Differences between groups were compared using 2-tailed Student’s t test, 2-way ANOVA with Bonferroni’s correction, or 2-tailed paired t test. Differences in survival of groups were compared using Log-rank (Mantel-Cox) test using GraphPad Prism software (version 7.0). Data are expressed as means ± SEM. P values are provided in the figures or in the legends. P < 0.05 was considered statistically significant.
Study approval.
All animal studies were reviewed and approved by the IACUC at the University of Pennsylvania, and studies were carried out in accordance with the approved guidelines.
Author contributions
YX, KB, BKA, and KW designed and executed experiments; SB designed synthesis and testing of SRF inhibitors; TGB executed bioinformatics; KBM and JF contributed project supervision and manuscript preparation.
Supplementary Material
Acknowledgments
The authors thank Gregory R. Grant (University of Pennsylvania) for insightful discussion on the bioinformatics and statistical analysis in this manuscript. This project was supported by NIH R01HL134923 to JF.
Version 1. 02/14/2019
In-Press Preview
Version 2. 03/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(6):e124629. https://doi.org/10.1172/jci.insight.124629.
Contributor Information
Yao Xiong, Email: xiongyao@pennmedicine.upenn.edu.
Simon Berritt, Email: simon.berritt@pfizer.com.
Bennette K. Attipoe, Email: Bennette.Attipoe@pennmedicine.upenn.edu.
Thomas G. Brooks, Email: thobr@sas.upenn.edu.
Kevin Wang, Email: kaiwenw@seas.upenn.edu.
Kenneth B. Margulies, Email: Kenneth.Margulies@uphs.upenn.edu.
Jeffrey Field, Email: jfield@upenn.edu.
References
- 1.Deo R, Albert CM. Epidemiology and genetics of sudden cardiac death. Circulation. 2012;125(4):620–637. doi: 10.1161/CIRCULATIONAHA.111.023838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280(5364):750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 3.McNally EM, Mestroni L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ Res. 2017;121(7):731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey PA, Leinwand LA. The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol. 2011;194(3):355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dirkx E, da Costa Martins PA, De Windt LJ. Regulation of fetal gene expression in heart failure. Biochim Biophys Acta. 2013;1832(12):2414–2424. doi: 10.1016/j.bbadis.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98(2):159–169. doi: 10.1016/S0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 7.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16(11):588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Field J, et al. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990;61(2):319–327. doi: 10.1016/0092-8674(90)90812-S. [DOI] [PubMed] [Google Scholar]
- 9.Fedor-Chaiken M, Deschenes RJ, Broach JR. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990;61(2):329–340. doi: 10.1016/0092-8674(90)90813-T. [DOI] [PubMed] [Google Scholar]
- 10.Shima F, et al. Effect of association with adenylyl cyclase-associated protein on the interaction of yeast adenylyl cyclase with Ras protein. Mol Cell Biol. 1997;17(3):1057–1064. doi: 10.1128/MCB.17.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Cyclase-associated protein 1 (CAP1) is a prenyl-binding partner of Rap1 GTPase. J Biol Chem. 2018;293(20):7659–7673. doi: 10.1074/jbc.RA118.001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston AB, Collins A, Goode BL. High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat Cell Biol. 2015;17(11):1504–1511. doi: 10.1038/ncb3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotila T, et al. Structural basis of actin monomer re-charging by cyclase-associated protein. Nat Commun. 2018;9(1):1892. doi: 10.1038/s41467-018-04231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gourlay CW, Ayscough KR. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26(17):6487–6501. doi: 10.1128/MCB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono S. The role of cyclase-associated protein in regulating actin filament dynamics - more than a monomer-sequestration factor. J Cell Sci. 2013;126(Pt 15):3249–3258. doi: 10.1242/jcs.128231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simino J, Shi G, Arnett D, Broeckel U, Hunt SC, Rao DC. Variants on chromosome 6p22.3 associated with blood pressure in the HyperGEN study: follow-up of FBPP quantitative trait loci. Am J Hypertens. 2011;24(11):1227–1233. doi: 10.1038/ajh.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42(2):117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 19.Arking DE, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46(8):826–836. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celestino-Soper PB, et al. Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol Cytogenet. 2012;5:17. doi: 10.1186/1755-8166-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. doi: 10.1136/jmedgenet-2018-105498. Aspit L, et al. CAP2 mutation leads to impaired actin dynamics associates with supraventricular tachycardia dilated cardiomyopathy [published online ahead of print (December 5, 20180). J Med Genet. https://jmg.bmj.com/content/early/2018/12/04/jmedgenet-2018-105498. [DOI] [PubMed] [Google Scholar]
- 22.Peche VS, et al. Ablation of cyclase-associated protein 2 (CAP2) leads to cardiomyopathy. Cell Mol Life Sci. 2013;70(3):527–543. doi: 10.1007/s00018-012-1142-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Field J, et al. CAP2 in cardiac conduction, sudden cardiac death and eye development. Sci Rep. 2015;5:17256. doi: 10.1038/srep17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evelyn CR, et al. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6(8):2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- 25.Bell JL, Haak AJ, Wade SM, Kirchhoff PD, Neubig RR, Larsen SD. Optimization of novel nipecotic bis(amide) inhibitors of the Rho/MKL1/SRF transcriptional pathway as potential anti-metastasis agents. Bioorg Med Chem Lett. 2013;23(13):3826–3832. doi: 10.1016/j.bmcl.2013.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lexow J, Poggioli T, Sarathchandra P, Santini MP, Rosenthal N. Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis Model Mech. 2013;6(6):1470–1476. doi: 10.1242/dmm.010470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bersell K, et al. Moderate and high amounts of tamoxifen in αMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis Model Mech. 2013;6(6):1459–1469. doi: 10.1242/dmm.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parlakian A, et al. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol. 2004;24(12):5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parlakian A, et al. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112(19):2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001;280(4):H1782–H1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 31.Angelini A, Li Z, Mericskay M, Decaux JF. Regulation of Connective Tissue Growth Factor and Cardiac Fibrosis by an SRF/MicroRNA-133a Axis. PLoS ONE. 2015;10(10):e0139858. doi: 10.1371/journal.pone.0139858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowes BD, et al. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346(18):1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 33.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96(5):592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 34.Antos CL, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278(31):28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 35.Xu Q, Patel D, Zhang X, Veenstra RD. Changes in cardiac Nav1.5 expression, function, and acetylation by pan-histone deacetylase inhibitors. Am J Physiol Heart Circ Physiol. 2016;311(5):H1139–H1149. doi: 10.1152/ajpheart.00156.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seki M, et al. Class I Histone Deacetylase Inhibition for the Treatment of Sustained Atrial Fibrillation. J Pharmacol Exp Ther. 2016;358(3):441–449. doi: 10.1124/jpet.116.234591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arabacilar P, Marber M. The case for inhibiting p38 mitogen-activated protein kinase in heart failure. Front Pharmacol. 2015;6:102. doi: 10.3389/fphar.2015.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haak AJ, et al. Targeting the myofibroblast genetic switch: inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J Pharmacol Exp Ther. 2014;349(3):480–486. doi: 10.1124/jpet.114.213520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutchings KM, et al. Pharmacokinetic optimitzation of CCG-203971: Novel inhibitors of the Rho/MRTF/SRF transcriptional pathway as potential antifibrotic therapeutics for systemic scleroderma. Bioorg Med Chem Lett. 2017;27(8):1744–1749. doi: 10.1016/j.bmcl.2017.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manasek FJ, Burnside B, Stroman J. The sensitivity of developing cardiac myofibrils to cytochalasin-B (electron microscopy-polarized light-Z-bands-heartbeat) Proc Natl Acad Sci USA. 1972;69(2):308–312. doi: 10.1073/pnas.69.2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott VR, Boehme R, Matthews TR. New class of antifungal agents: jasplakinolide, a cyclodepsipeptide from the marine sponge, Jaspis species. Antimicrob Agents Chemother. 1988;32(8):1154–1157. doi: 10.1128/AAC.32.8.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandalovičová A, et al. Migrastatics-Anti-metastatic and Anti-invasion Drugs: Promises and Challenges. Trends Cancer. 2017;3(6):391–406. doi: 10.1016/j.trecan.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muchir A, et al. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J Clin Invest. 2007;117(5):1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu W, Shan J, Bonne G, Worman HJ, Muchir A. Pharmacological inhibition of c-Jun N-terminal kinase signaling prevents cardiomyopathy caused by mutation in LMNA gene. Biochim Biophys Acta. 2010;1802(7-8):632–638. doi: 10.1016/j.bbadis.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muchir A, et al. Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc Res. 2012;93(2):311–319. doi: 10.1093/cvr/cvr301. [DOI] [PubMed] [Google Scholar]
- 46.Wu W, Muchir A, Shan J, Bonne G, Worman HJ. Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation. 2011;123(1):53–61. doi: 10.1161/CIRCULATIONAHA.110.970673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu JT, Muchir A, Nagy PL, Worman HJ. LMNA cardiomyopathy: cell biology and genetics meet clinical medicine. Dis Model Mech. 2011;4(5):562–568. doi: 10.1242/dmm.006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waddingham MT, et al. Chronic Rho-kinase inhibition improves left ventricular contractile dysfunction in early type-1 diabetes by increasing myosin cross-bridge extension. Cardiovasc Diabetol. 2015;14:92. doi: 10.1186/s12933-015-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497(7450):507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burke MA, et al. Molecular profiling of dilated cardiomyopathy that progresses to heart failure. JCI Insight. 2016;1(6):e86898. doi: 10.1172/jci.insight.86898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li D, Wu J, Bai Y, Zhao X, Liu L. Isolation and culture of adult mouse cardiomyocytes for cell signaling and in vitro cardiac hypertrophy. J Vis Exp. 2014;87(87):e51357. doi: 10.3791/51357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.