Abstract
Existing methods to improve detection of circulating tumor DNA (ctDNA) have focused on sensitivity for detecting genomic alterations but have rarely considered the biological properties of plasma cell-free DNA (cfDNA). We hypothesized that differences in fragment lengths of circulating DNA could be exploited to enhance sensitivity for detecting the presence of ctDNA and for non-invasive genomic analysis of cancer. We surveyed ctDNA fragment sizes in 344 plasma samples from 200 cancer patients using low-pass whole-genome sequencing (0.4×). To establish the size distribution of mutant ctDNA, tumor-guided personalized deep sequencing was performed in 19 patients. We detected enrichment of ctDNA in fragment sizes between 90–150 bp, and developed methods for in vitro and in silico size selection of these fragments. Selecting fragments between 90–150 bp improved detection of tumor DNA, with more than 2-fold median enrichment in >95% of cases, and more than 4-fold enrichment in >10% of cases. Analysis of size-selected cfDNA identified clinically actionable mutations and copy number alterations that were otherwise not detected. Identification of plasma samples from patients with advanced cancer was improved by predictive models integrating fragment length and copy number analysis of cfDNA, with AUC>0.99 compared to AUC<0.80 without fragmentation features. Increased identification of cfDNA from patients with glioma, renal, and pancreatic cancer was achieved with AUC>0.91, compared to AUC<0.5 without fragmentation features. Fragment size analysis and selective sequencing of specific fragment sizes can boost ctDNA detection and could complement or provide an alternative to deeper sequencing of cell-free DNA for clinical applications, earlier diagnosis and study of tumor biology.
Introduction
Blood plasma of cancer patients contains circulating tumor DNA (ctDNA), but this valuable source of information is diluted by much larger quantities of DNA of non-cancerous origins, such that ctDNA usually represents only a small fraction of the total cell-free DNA (cfDNA) (1, 2). High-depth targeted sequencing of selected genomic regions can be used to detect low amounts of ctDNA, but broader analysis with methods such as whole exome sequencing (WES) and shallow whole genome sequencing (sWGS) are only generally informative when ctDNA content is ~10% or greater (3–5). The concentration of ctDNA can exceed 10% of the total cfDNA in patients with advanced-stage cancers (6–8), but is much lower in patients with low tumor burden (9–12) and in patients with some cancer types such as gliomas and renal cancers (6). Current strategies to improve ctDNA detection rely on increasing depth of sequencing coupled with various error-correction methods (2, 13, 14). However, approaches that focus only on genomic alterations do not take advantage of the potential differences in chromatin organization or fragment sizes of ctDNA (15–17). Results of ever-deeper sequencing are also confounded by the likelihood of false positive results from detection of mutations from non-cancerous cells, clonal expansions in normal epithelia, or clonal hematopoiesis of indeterminate potential (CHIP) (13, 18, 19).
The cell of origin and the mechanism of cfDNA release into blood can mark cfDNA with specific fragmentation signatures, potentially providing precise information about cell type, gene expression, cell physiology or pathology, or action of treatment (15, 16, 20). cfDNA fragments commonly show a prominent mode at 167 bp, suggesting release from apoptotic caspase-dependent cleavage (21–24) (Fig. 1A). Circulating fetal DNA has been shown to be shorter than maternal DNA in plasma, and these size differences have been used to improve sensitivity of non-invasive prenatal diagnosis (22, 25–27). The size distribution of tumor-derived cfDNA has only been investigated in a few studies, encompassing a small number of cancer types and patients, and showed conflicting results (28–33). A limitation of previous studies is that determining the specific sizes of tumor-derived DNA fragments requires detailed characterization of matched tumor-derived alterations (30, 33), and the broader understanding and implications of potential biological differences have not previously been explored.
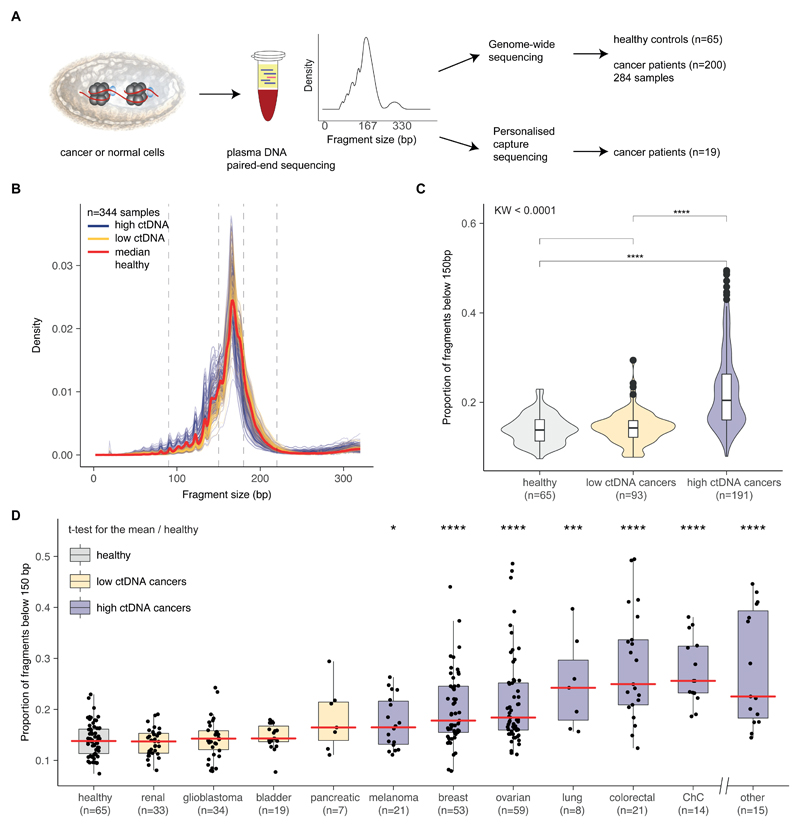
Figure 1. Survey of plasma DNA fragmentation with genome-wide sequencing on a pan-cancer scale.
A, The size profile of cfDNA can be determined by paired-end sequencing of plasma samples and reflects its organization around the nucleosome. cfDNA is released into the blood circulation by various means, each of which leaves a signature on the DNA fragment sizes. We inferred the size profile of cfDNA by analyzing with sWGS (n=344 plasma samples from 65 healthy controls and 200 cancer patients) and the size profile of mutant ctDNA by personalized capture sequencing (n=18 plasma samples). B, Fragment size distributions of 344 plasma samples from 200 cancer patients. Samples are split into two groups based on previous literature (6), with orange representing samples from patients with cancer types previously observed to have low amounts of ctDNA (renal, bladder, pancreatic, and glioma) and blue representing samples from patients with cancer types previously observed to have higher levels of ctDNA (breast, melanoma, ovarian, lung, colorectal, cholangiocarcinoma, and others, see table S1). C, Proportion of cfDNA fragments below 150 bp in those samples, grouped into cancer types as defined in B. The Kruskal-Wallis test for difference in size distributions indicated a significant difference between the group of samples from cancer types releasing high amounts of ctDNA and the group of samples from cancer types releasing low amounts, as well as the group of samples from healthy individuals (p<0.001). D, Proportion of cfDNA fragments below 150 bp by cancer type (all samples). Cancer types represented by fewer than 4 individuals are grouped in the “other” category. The red line indicates the median proportion for each cancer type. ChC=cholangiocarcinoma.
We hypothesized that we could improve the sensitivity for non-invasive cancer genomics by selective sequencing of ctDNA fragments and by leveraging differences in the biology that determine DNA fragmentation. To test this, we established a pan-cancer catalogue of cfDNA fragmentation features in plasma samples from patients with different cancer types and healthy individuals to identify biological features enriched in tumor-derived DNA. We developed methods for selecting specific sizes of cfDNA fragments prior to sequencing and investigated the impact of combining cfDNA size selection with genome-wide sequencing to improve the detection of ctDNA and the identification of clinically actionable genomic alterations.
Results
Surveying the fragmentation features of tumor cfDNA
We generated a catalogue of cfDNA fragmentation features (Fig. 1A) from 344 plasma samples from 200 patients with 18 different cancer types, and additional 65 plasma samples from healthy controls (Fig. 1B, fig. S1, table S1, and table S2). The size distribution of cfDNA fragments in cancer patients differed in the size ranges of 90–150 bp, 180–220 bp, and 250–320 bp compared to healthy individuals (Fig. 1B and fig. S2). cfDNA fragment sizes in plasma of healthy individuals and in plasma of patients with late stage glioma, renal, pancreatic, and bladder cancers, were significantly longer than in other late stage cancer types including breast, ovarian, lung, melanoma, colorectal, and cholangiocarcinoma (p<0.001, Kruskal-Wallis; Fig. 1C). Sorting the 18 cancer types according to the proportion of cfDNA fragments in the size range 20–150 bp resulted in an order very similar to that obtained by Bettegowda et al. based on the concentrations of ctDNA measured by individual mutation assays (Fig. 1D) (6). In contrast to previous reports (6, 34), this sorting was performed without any analysis or prior knowledge of the presence of mutations or somatic copy number alterations (SCNAs), yet allowed the investigation of ctDNA content in different cancers.
Sizing up mutant ctDNA
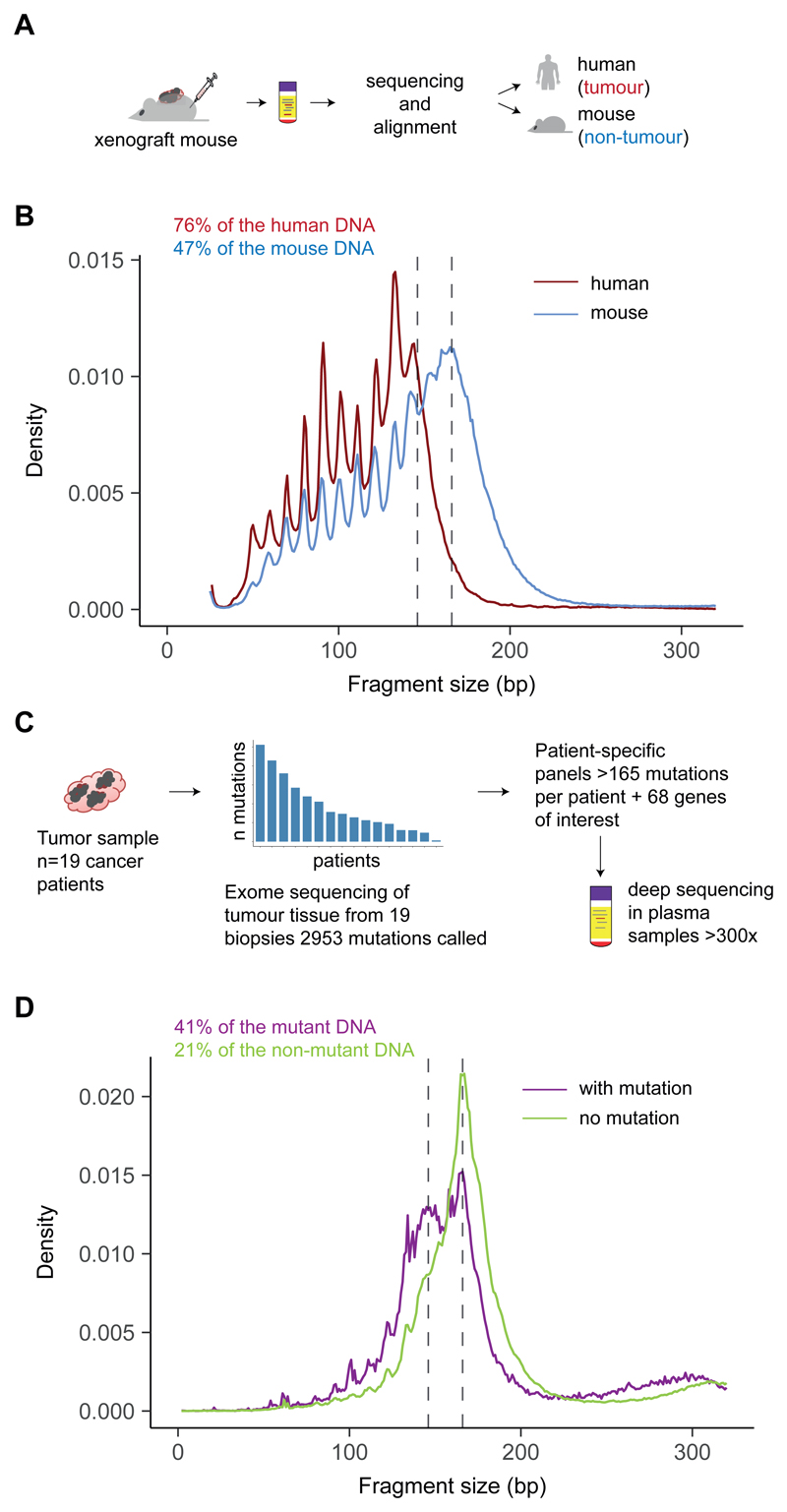
We determined the size profile of mutant ctDNA in plasma using two high-specificity approaches. First, we inferred the specific size profile of ctDNA and non-tumor cfDNA with sWGS from the plasma of mice bearing human ovarian cancer xenografts (Fig. 2A). We observed a shift in ctDNA fragment sizes to less than 167 bp (Fig. 2B). Second, the size profile of mutant ctDNA was determined in plasma from 19 cancer patients, using deep sequencing with patient-specific hybrid-capture panels developed from whole-exome profiling of matched tumor samples (Fig. 2C). By sequencing hundreds of mutations at a depth >300× in cfDNA, allele-specific reads from mutant and normal DNA were obtained. Enrichment of DNA fragments carrying tumor-mutated alleles was observed in fragments ~20–40 bp shorter than nucleosomal DNA sizes (multiples of 167 bp) (Fig. 2D). We determined that mutant ctDNA is generally more fragmented than non-mutant cfDNA, with a maximum enrichment of ctDNA in fragments between 90 and 150 bp (fig. S3), as well as enrichment in the size range 250–320 bp. These data also indicated that mutant DNA in plasma of patients with advanced cancer (pre-treatment) is consistently shorter than predicted mono-, and di-nucleosomal DNA fragment lengths (Fig. 2D).
Figure 2. Determining the size profile of mutant ctDNA with animal models and personalized capture sequencing.
A, A mouse model with xenografted human tumor cells enabled the discrimination of DNA fragments released by cancer cells (reads aligning to the human genome) from the DNA released by healthy cells (reads aligning to the mouse genome), with the use of sWGS. B, Fragment size distribution from the plasma extracted from a mouse xenografted with a human ovarian tumor, showing ctDNA originating from tumor cells (red) and cfDNA from non-cancerous cells (blue). Two vertical lines indicate 145 bp and 167 bp. The fraction of reads shorter than 150 bp is indicated. C, Design of personalized hybrid-capture sequencing panels developed to specifically determine the size profiles of mutant DNA and non-mutant DNA in plasma from 19 patients with late-stage cancers. Capture panels included somatic mutations identified in tumor tissue by WES. A mean of 165 mutations per patient was then analyzed from matched plasma samples. Reads were aligned and separated into fragments carrying either the reference or the mutant sequence. Fragment sizes for paired-end reads were calculated. D, Size profiles of mutant DNA and non-mutant DNA in plasma from 19 patients with late stage cancers were determined by tumor-guided capture sequencing. The fraction of reads shorter than 150 bp is indicated.
Selecting tumor-derived DNA fragments
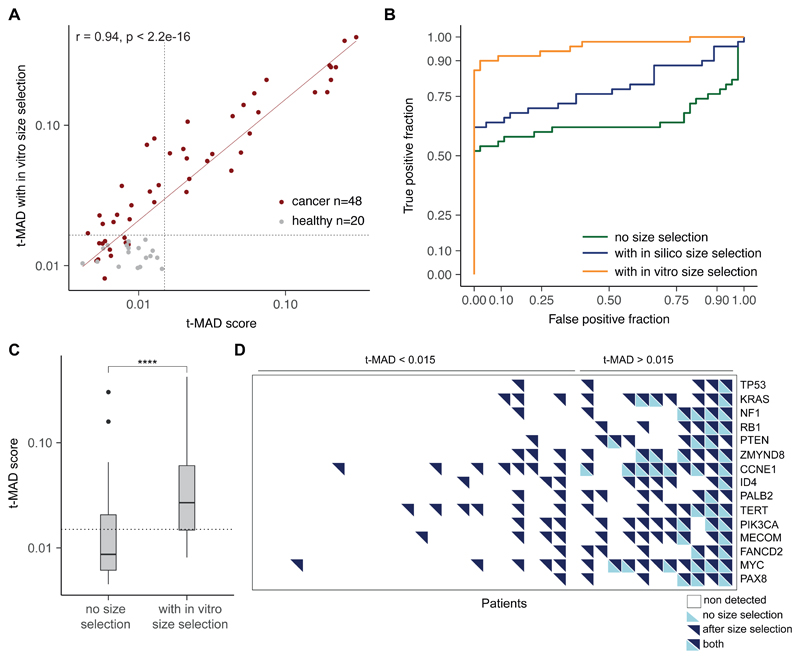
We evaluated whether the shorter cfDNA fragments in plasma can be harnessed to improve ctDNA detection. We determined the feasibility of selective sequencing of shorter fragments using in vitro size selection with a bench-top microfluidic device followed by sWGS, in 48 plasma samples from 35 patients with high-grade serous ovarian cancer (HGSOC) (Fig. 3A, fig. S4, and fig. S5). We assessed the accuracy and quality of the size selection with the plasma from 20 healthy individuals (Fig. 3B and fig. S6). We also explored the utility of in silico size selection of fragmented DNA using read-pair positioning from unprocessed sWGS data (Fig. 3A). In silico size selection was performed once reads were aligned to the genome reference, by selecting the paired-end reads that corresponded to the fragment lengths in a 90–150 bp size range. Fig. 3C, Fig. 3D, and Fig. 3E illustrate the effect of in vitro size selection for one HGSOC case (see all 5 samples in fig. S7 and fig. S8). First, we identified SCNAs in plasma cfDNA before treatment, when the concentration of ctDNA was high (Fig. 3C). Only a small number of focal SCNAs were observed in the subsequent plasma sample collected 3 weeks after initiation of chemotherapy (without size selection, Fig. 3D). In vitro size selection of the same post-treatment plasma sample showed a median increase of 6.4 times in the amplitude of detectable SCNAs without size selection. Selective sequencing of shorter fragments in this sample resulted in the detection of multiple other SCNAs that were not observed without size selection (Fig. 3E), and a genome-wide copy-number profile that was similar to that obtained before treatment when ctDNA concentrations were 4 times higher, with additional copy-number alterations identified in this sample despite the lower initial concentration of ctDNA (Fig. 3C). In silico size selection also enriched ctDNA but to a lower extent than using in vitro size selection (fig. S7). We concluded that selecting short DNA fragments in plasma can enrich tumor content on a genome-wide scale.
Figure 3. Enhancing the tumor fraction from plasma sequencing with size selection.
A, Plasma samples collected from ovarian cancer patients were analyzed in parallel without size selection or using either in silico or in vitro size selection. B, accuracy of the in vitro and in silico size selection determined on a cohort of 20 healthy controls. The size distribution before size selection is shown in green, after in silico size selection (with sharp cutoff at 90 and 150 bp) in blue, and after in vitro size selection in orange. Vertical lines indicate 90 bp and 150 bp. C, SCNA analysis with sWGS from plasma DNA of an ovarian cancer patient collected before initiation of treatment, when ctDNA MAF was 0.271 for a TP53 mutation as determined by TAm-Seq. Inferred amplifications are shown in blue and deletions in orange. Copy number neutral regions are in gray. D, SCNA analysis of a plasma sample from the same patient as in panel C, collected three weeks after treatment start. The MAF for the TP53 mutation at this time point was 0.068, and sWGS revealed only limited evidence of copy number alterations (before size selection). E, Analysis of the same plasma sample as in D after in vitro size selection of fragments between 90 bp and 150 bp in length. The MAF for the TP53 mutation increased to 0.402 after in vitro size selection, and SCNAs were clearly apparent by sWGS. More SCNAs were detected in comparison to C and D (for example in chr2, chr9, chr10). SCNAs were also detected in this sample after in silico size selection (fig. S7).
Quantifying the impact of size selection
To quantitatively assess the enrichment after size selection on a genome-wide scale, we developed a metric from sWGS data (<0.4× coverage) called t-MAD (trimmed Median Absolute Deviation from copy-number neutrality, see Fig. 4A). All sWGS data were downsampled to 10 million sequencing reads for comparison. To define the detection threshold, we measured the t-MAD score for sWGS data from 65 plasma samples from 46 healthy individuals and took the maximal value (median=0.01, range 0.004–0.015). We compared t-MAD to the mutant allele fraction (MAF) in the high ctDNA cancer types as assessed by digital PCR (dPCR) or WES in 97 samples. We observed a high correlation (Pearson correlation, r=0.80) between t-MAD and MAF (Fig. 4B), for samples with t-MAD greater than the detection threshold (0.015), or with MAF>0.025. fig. S9 shows that the slope of t-MAD versus MAF fit lines differed between cancer types (range 0.17–1.12), reflecting likely differences in the extent of SCNAs. We estimated the sensitivity of t-MAD for detecting low amounts of ctDNA using a spike-in dilution of DNA from a patient with a TP53 mutation into DNA from a pool of 7 healthy individuals (fig. S10), which confirmed that the t-MAD score was linear with ctDNA fraction down to MAF of ~0.01. In addition, t-MAD scores greater than the detection threshold (0.015) for samples were present even in samples with MAF as low as 0.004. t-MAD was also strongly correlated with tumor volume determined by RECIST1.1 (Pearson correlation, r=0.6, p<0.0001, n=35) (fig. S11).
Figure 4. Quantifying the ctDNA enrichment by sWGS with in silico size selection and t-MAD.
A, Workflow to quantify tumor fraction from SCNA as a genome-wide score named t-MAD. B, Correlation between the MAF of SNVs determined by digital PCR or hybrid-capture sequencing and t-MAD score determined by sWGS. Data included 97 samples from patients of multiple cancer types with matched MAF measurements and t-MAD scores. Pearson correlation (coefficient r) between MAF and t-MAD scores was calculated for all cases with MAF>0.025 and t-MAD>0.015. Linear regression indicated a fit with a slope of 0.44 (purple solid line). C, Comparison of t-MAD scores determined from sWGS between healthy samples, samples collected from patients with cancer types that exhibit low amounts of ctDNA, and from patients with cancer types that exhibit high amounts of ctDNA (as in Fig. 1). All samples for which t-MAD could be calculated have been included. D, ROC analysis comparing the classification of these plasma samples from high ctDNA cancer samples (n=189) and plasma samples from healthy controls (n=65) using t-MAD had an area under curve (AUC) of 0.69 without size selection (black solid curve). After applying in silico size selection to the samples from the cancer patients, we observed an AUC of 0.90 (black dashed curve). E, Determination of t-MAD from longitudinal plasma samples of a colorectal cancer patient. t-MAD was analyzed before and after in silico size selection of the DNA fragments 90-150 bp, and then compared to the RECIST status for this patient. F, Application of in silico size selection to 6 patients with long-term follow-up. t-MAD score was determined before and after in silico size selection of the short DNA fragments. Dark blue circles indicate samples in which ctDNA was detected both with and without in silico size selection. Light blue circles indicate samples where ctDNA was detected only after in silico size selection. Empty circles indicate samples where ctDNA was not detected by either analysis. Times when RECIST status was assessed are indicated by a red bar for progression, or an orange bar for regression or stable disease.
Using t-MAD, we detected ctDNA from 69% (130/189) of the samples from cancer types where ctDNA concentrations were shown to be high (Fig. 4C). From cancer types for which ctDNA concentrations are suspected to be low (glioma, renal, bladder, pancreatic), we detected ctDNA in 17% (10/57) of the cases (Fig. 4C). We used in silico size selection of the DNA fragments between 90–150 bp from the high ctDNA cancers (n=189) and healthy controls (n=65) to improve the sensitivity for detecting t-MAD (Fig. 4D). Receiver operating characteristic (ROC) analysis comparing the t-MAD score for the samples revealed an area under the curve (AUC) of 0.90 after in silico size selection, against an AUC of 0.69 without size selection (Fig. 4D).
We explored whether size selected sequencing could improve the detection of response or disease progression. We used sWGS of longitudinal plasma samples from six cancer patients (Fig. 4E, F) and in silico size selection of the cfDNA fragments between 90–150 bp. In two patients, size-selected samples indicated tumor progression 60 and 87 days before detection by imaging or unselected t-MAD analysis (Fig. 4E, F). Other longitudinal samples exhibited improvements in the detection of ctDNA with t-MAD and size selection (Fig. 4F).
Identifying more clinically relevant genomic alterations with size selection
We next tested whether size selection could increase the sensitivity for detecting cancer genomic alterations in cfDNA. To test effects on copy number aberrations, we studied 35 patients with HGSOC as the archetypal copy-number driven cancer (35). t-MAD was used to quantify the enrichment of ctDNA with in vitro size selection in 48 plasma samples, including samples collected before and after initiation of chemotherapy treatment. In vitro size selection resulted in an increase in the calculated t-MAD score from the sWGS data for 47/48 of the plasma samples (98%, t-test, p=0.06) with a mean 2.5 and median 2.1-fold increase (Fig. 5A and table S3). We compared the t-MAD scores against those obtained by sWGS for the plasma samples from healthy individuals. 39 of the 48 size-selected HGSOC plasma samples (82%) had a t-MAD score greater than the highest t-MAD value determined in the in vitro size selected healthy plasma samples (Fig. 5A, fig. S12 and fig. S6), compared to 24 out of 48 without size selection (50%). ROC analysis comparing the t-MAD score for the samples from the cancer patients (pre- and post-treatment initiation, n=48) and healthy controls (n=46) revealed an AUC of 0.97 after in vitro size selection, with maximal sensitivity and specificity of 90% and 98%, respectively. This was superior to detection by sWGS without size selection (AUC=0.64) (Fig. 5B).
Figure 5. Quantifying the ctDNA enrichment by sWGS with in vitro size selection.
A, The effect of in vitro size selection on the t-MAD score. For each of 48 plasma samples collected from 35 patients, the t-MAD score was determined from the sWGS after in vitro size selection (y axis) and without size selection (x axis). In vitro size selection increased the t-MAD score for nearly all samples, with a median increase of 2.1-fold (range from 1.1 to 6.4 fold). t-MAD scores determined from sWGS for 46 samples from healthy individuals were all <0.015 both before and after in vitro size selection. B, ROC analysis comparing the classification of plasma samples from cancer patients (n=48) and plasma samples from healthy controls (n=46) using t-MAD had an area under curve (AUC) of 0.64 without size selection (green curve). After applying in silico size selection to the samples from the patients and controls, we observed an AUC of 0.78 (blue curve), and after in vitro size selection, an AUC of 0.97 (orange curve). C, Comparison of t-MAD scores determined from sWGS between matched ovarian cancer samples with and without in vitro size selection. The t-test for the difference in means indicates a significant increase in tumor fraction (measured by t-MAD) with in vitro size selection (p<0.0001). D, Detection of SCNAs across 15 genes frequently mutated in recurrent ovarian cancer, measured in plasma samples collected during treatment for 35 patients. Patients were ranked from left to right by increasing tumor fraction as quantified by t-MAD (before in vitro size selection). SCNAs are labeled as detected for a gene if the mean log2 ratio in that region was greater than 0.05. Empty squares represent copy number neutral regions, bottom left triangles in light blue indicate that SCNAs were detected without size selection, and top right triangles in dark blue represent SCNAs detected after in vitro size selection.
We then determined if this improved sensitivity resulted in the detection of SCNAs with potential clinical value. Across the genome, t-MAD scores evaluating SCNAs were higher after size selection in 33/35 (94%) HGSOC patients, and the magnitude of the copy number (log2 ratio) values significantly increased after in vitro size selection (t-test for the means, p=0.003) (Fig. 5C). We compared the relative copy number values for 15 genes frequently altered in HGSOC (table S4). Analysis of plasma cfDNA after size selection revealed a large number of SCNAs that were not observed in the same samples without size selection (Fig. 5D), including amplifications in key genes such as NF1, TERT, and MYC (fig. S13).
We also tested whether similar enrichment was seen for substitutions, to exclude the possibility that size selection might only increase the sensitivity for sWGS analysis. We performed whole exome sequencing of plasma cfDNA from 23 patients with 7 cancer types (fig. S1). We used the WES data to compare the size distributions of fragments carrying mutant or non-mutant alleles (Fig. 6A), and to test whether size selection could identify additional mutations. We first selected 6 patients with HGSOC and performed WES of plasma DNA with and without in vitro size selection in the 90–150 bp range, analyzing time points before and after initiation of treatment (36). In addition, in silico size selection for the same range of fragment sizes was performed (Fig. 6A). Analysis of the MAF of SNVs revealed statistically significant enrichment of the tumor fraction with both in vitro size selection (mean 4.19-fold, median 4.27-fold increase, t-test, p<0.001) and in silico size selection (mean 2.20-fold, median 2.25-fold increase, t-test, p<0.001) (Fig. 6A and fig. S14). Three weeks after initiation of treatment, ctDNA fractions are often lower (36), and therefore we further analyzed post-treatment plasma samples using Tagged-Amplicon Deep Sequencing (TAm-Seq) (37). We observed enrichment of MAFs by in vitro size selection between 0.9 and 11 times (mean 2.1 times, median 1.5 times), with one outlier sample exhibiting a relative enrichment of 118 times, compared to the same samples without size selection (fig. S15).
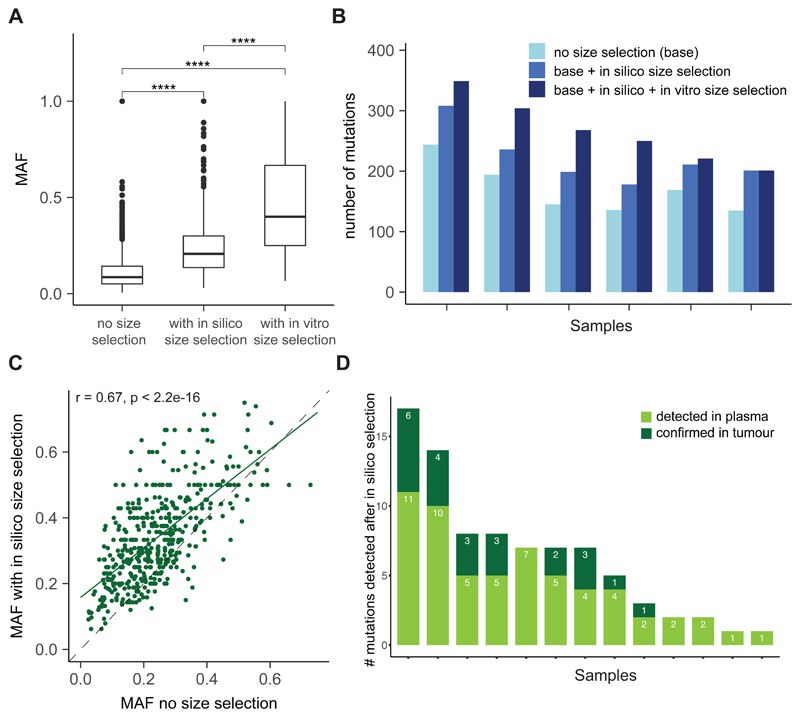
Figure 6. Improving the detection of somatic alterations by WES in multiple cancer types with size selection.
A, Analysis of the MAF of mutations detected by WES in 6 patients with HGSOC without size selection and with either in vitro or in silico size selection. B, Comparison of size-selected WES data with non-selected WES data to assess the number of mutations detected in plasma samples from 6 patients with HGSOC. For each patient, the first bar in light blue shows the number of mutations called without size selection, the second bar quantifies the number of mutations called after the addition of those identified with in silico size selection, and the third, dark blue bar shows the number of mutations called after addition of mutations called after in vitro size selection. C, Patients (n=16) were retrospectively selected from a cohort with different cancer types (colorectal, cholangiocarcinoma, pancreatic, prostate) enrolled in early phase clinical trials. Matched tumor tissue DNA was available for each plasma sample, and 2 patients also had a biopsy collected at relapse. WES was performed on tumor tissue DNA and plasma DNA samples, and in silico size selection was applied to the data. 2061/2133, 97% of the shared mutations detected by WES showed higher MAF after in silico size selection. D, Mutations detected only after in silico selection of WES data from 16 patients (as in C) compared to mutations called by WES of the matched tumor tissue. Three of 16 patients had no additional mutations identified after in silico size selection. Of the 82 mutations detected in plasma after in silico size selection, 23 (28%) had low signal in tumor WES data and were not identified in those samples without size selection.
Size selection with both in vitro and in silico methods increased the number of mutations detected by WES by an average of 53% compared to no size selection (Fig. 6B). We identified a total of 1023 mutations in the non-size-selected samples. An additional 260 mutations were detected by in vitro size selection, and an additional 310 mutations were called after in silico size selection (Fig. 6B and table S5). To exclude the possibility that the improved sensitivity for mutation detection was a result of sequencing artefacts, we validated whether new mutations were also detectable in tumor specimens. We used in silico size selection in an independent cohort of 16 patients for whom matched tumor tissue DNA was available (table S6). In silico size selection enriched the MAF for nearly all mutations (2061/2133, 97%), with an average increase of MAF of ×1.7 (Fig. 6C). For 13 of 16 patients (81%), we identified additional mutations in plasma after in silico size selection. Of these 82 additional mutations, 23 (28%) were confirmed to be present in the matched tumor tissue DNA (Fig. 6D). Notably, this included mutations in key cancer genes including BRAF, ARID1A, and NF1 (fig. S16).
Detecting cancer by supervised machine learning combining cfDNA fragmentation and somatic alteration analysis
It is important to note that although in vitro and in silico size selection increase the sensitivity of detection, they also result in a loss of cfDNA for analysis. In analysis of ctDNA based on genomic signals, potentially-informative data is lost since regions of the cancer genome which are not mutated or altered do not contribute to detection (fig. S17). We hypothesized that leveraging other biological properties of the cfDNA fragmentation profile could enhance the detection of ctDNA.
We defined other cfDNA fragmentation features from sWGS data including (1) the proportion of fragments in multiple size ranges, (2) the ratios of proportions of fragments in different sizes, and (3) the amplitude of oscillations in fragment size density with 10 bp periodicity (see Materials and Methods and Fig. 7A). These fragmentation features were compared between cancer patients and healthy individuals (fig. S18), and the feature representing the proportion (P) of fragments between 20–150 bp exhibited the highest AUC (0.819). Principal component analysis (PCA) of the samples represented by t-MAD and fragmentation features showed a separation between healthy samples and samples from cancer patients and identified fragment features that were aligned (in PCA analysis) with t-MAD scores (Fig. 7B).
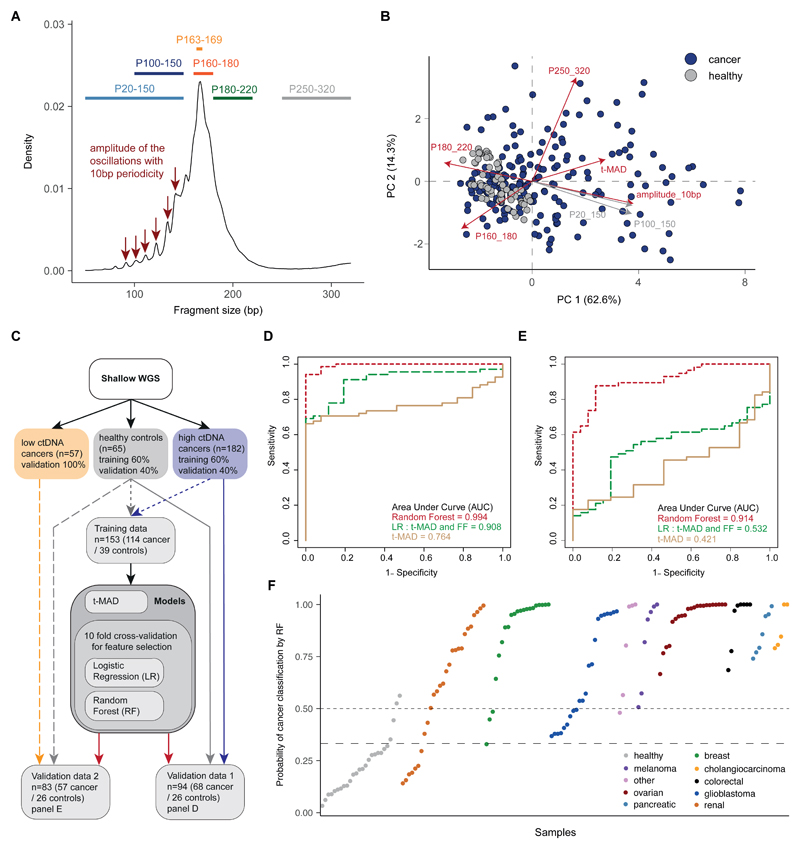
Figure 7. Enhancing the potential for ctDNA detection by combining SCNAs and fragment-size features.
A, Schematic illustrating the selection of different size ranges and features in the distribution of fragment sizes. For each sample, fragmentation features included the proportion (P) of fragments in specific size ranges, the ratio between certain ranges, and a quantification of the amplitude of the 10 bp oscillations in the 90-145 bp size range calculated from the periodic “peaks” and “valleys”. B, Principal Component Analysis (PCA) comparing cancer and healthy samples using data from t-MAD scores and the fragmentation features. Red colored arrows indicate features that were selected as informative by the predictive analysis. C, Workflow for the predictive analysis combining SCNAs and fragment size features. sWGS data from 182 plasma samples from patients with cancer types with high amounts of ctDNA (colorectal, cholangiocarcinoma, lung, ovarian, breast) were split into a training set (60% of samples) and a validation set (Validation data 1, together with the healthy individual validation set). A further dataset of sWGS from 57 samples of cancer types exhibiting low amounts of ctDNA (glioma, renal, pancreatic) was used as Validation data 2, together with the healthy individual validation set. Plasma DNA sWGS data from healthy controls were split into a training set (60% of samples) and a validation set (used in both Validation data 1 and Validation data 2). D, ROC curves for Validation data 1 (samples from cancer patients with high ctDNA amounts=68, healthy=26) for 3 predictive models built on the pan-cancer training cohort (cancer=114, healthy=39). The beige curve represents the ROC curve for classification with t-MAD only, the long dashed green line represents the logistic regression model combining the top 5 features based on recursive feature elimination (t-MAD score, 10 bp amplitude, P(160-180), P(180-220), and P(250-320)), and the dashed red line shows the result for a random forest classifier trained on the combination of the same 5 features, independently chosen for the best RF predictive model. E, ROC curves for Validation data 2 (samples from cancer patients with low ctDNA amounts=57, healthy=26) for the same 3 classifiers as in D. The beige curve represents the model using t-MAD only, the long-dashed green curve represents the logistic regression model combining the top 5 features (t-MAD score, 10 bp amplitude, P(160-180), P(180-220), and P(250-320)), and the dashed red curve shows the result for a random forest classifier trained on the combination of same 5 predictive features. F, Plot representing the probability of classification as cancer with the RF model for all samples in both validation datasets. Samples are separated by cancer type and sorted within each by the RF probability of classification as cancer. The dashed horizontal line indicates 50% probability (achieving specificity of 24/26=92.3%), and the long-dashed line indicates 33% probability (achieving specificity of 22/26=84.6%).
We next explored the potential of fragmentation features to enhance the detection of tumor DNA in plasma samples. A predictive analysis was performed using the t-MAD score and 9 fragmentation features across 304 samples (239 from cancer patients and 65 from healthy controls) (Fig. 7C, fig. S19, and table S2). The 9 fragmentation features determined from sWGS included five features based on the proportion (P) of fragments in defined size ranges: P(20–150), P(100–150), P(160–180), P(180–220), P(250–320); three features based on ratios of those proportions: P(20–150)/P(160–180), P(100–150)/P(163–169), P(20–150)/P(180–220); and a further feature based on the amplitude of the oscillations having 10 bp periodicity observed below 150 bp.
Variable selection and the classification of samples as “healthy” or “cancer” were performed using logistic regression (LR) and random forests (RF) trained on 153 samples and validated on two datasets of 94 and 83 independent samples (Fig. 7C). The best feature set for the LR model included t-MAD, 10 bp amplitude, P(160–180), P(180–220), and P(250–320). The same five variables were independently identified using the RF model (with some differences in their ranking). Fig. S20 shows performance metrics for the different algorithms on training set data using cross-validation. Using t-MAD alone in the validation pan-cancer dataset (Fig. 7D and fig. S19), we could distinguish cancer samples from healthy individuals with AUC=0.764. Using the LR model improved the classification of the samples to AUC=0.908. The RF model (trained on the 153-sample training set) could distinguish cancer from healthy individuals even more accurately in the validation data set (n=94) with AUC=0.994. On the second validation dataset containing low-ctDNA cancer samples (n=83) (Fig. 7E), t-MAD alone or the LR performed less well, with AUC values of 0.421 and 0.532, respectively. However, the RF model was still able to distinguish low-ctDNA cancer samples from healthy controls with AUC=0.914. At a specificity of 95%, the RF model correctly classified as cancer 64/68 (94%) of the samples from high-ctDNA cancers (colorectal, cholangiocarcinoma, ovarian, breast, melanoma) and 37/57 (65%) of the samples from low-ctDNA cancers (pancreatic, renal, glioma) (Fig. 7F). In a second iteration of model training, we omitted t-MAD, using only the 4 fragmentation features (fig. S21). The RF model could still distinguish cancer from healthy controls, albeit with slightly reduced AUCs (0.989 for cancer types with high amounts of ctDNA and 0.891 for cancer types with low amounts of ctDNA), suggesting that the cfDNA fragmentation pattern is the most important predictive component.
Discussion
Our results indicate that exploiting fundamental properties of cfDNA with fragment-specific analyses can allow more sensitive evaluation of ctDNA. We based the fragment size selection criteria on a biological observation that ctDNA fragment size distribution is shifted from non-cancerous cfDNA. Our work builds on a comprehensive survey of plasma cfDNA fragmentation patterns across 200 patients with multiple cancer types and 65 healthy individuals. We identified features that could determine the presence and amount of ctDNA in plasma samples, without a priori knowledge of somatic aberrations. We caution that this catalog is limited to double-stranded DNA from plasma samples and is subject to potential biases incurred by the DNA extraction and sequencing methods we used. Additional biological effects could contribute to further selective analysis of cfDNA. Other bodily fluids (urine, cerebrospinal fluid, saliva), different nucleic acids and structures, altered mechanisms of release into circulation, or sample processing methods could exhibit varying fragment size signatures and could offer additional exploitable biological patterns for selective sequencing.
Previous work has reported the size distributions of mutant ctDNA, but only considered limited genomic loci, cancer types, or cases (30, 32, 33). We identified the size differences between mutant and non-mutant DNA on a genome-wide and pan-cancer scale. We developed a method to size mutant ctDNA without using high-depth WGS. By sequencing >150 mutations per patient at high depth, we obtained large numbers of reads that could be unequivocally identified as tumor-derived, and thus determined the size distribution of mutant ctDNA and non-mutant cfDNA in cancer patients. A potential limitation of our approach is that capture-based sequencing is biased by probe capture efficiency and therefore our data may not accurately reflect ctDNA fragments <100 bp or >300 bp.
Our work provides strong evidence that the modal size of ctDNA for many cancer types is less than 167 bp, which is the length of DNA wrapped around the chromatosome. In addition, our work also shows that there is enrichment of mutant DNA fragments at sizes greater than 167 bp, notably in the range 250–320 bp. These longer fragments may explain previous observations that longer ctDNA can be detected in the plasma of cancer patients (29, 32). The origin of these long fragments is still unknown, and their observation could be linked to technical factors. However, it is likely that mechanisms of compaction and release of cfDNA into circulation, which may differ depending on its origin, will be reflected by different fragment sizes (38). Improving the characterization of these fragments will be important, especially for future work combining analysis of ctDNA with that of other entities in blood such as microvesicles and tumor-educated platelets (39, 40). Fragment-specific analyses not only increase the sensitivity for detection of rare mutations, but could be used to track modifications in the size distribution of ctDNA. Future work should address whether this approach could be used to elucidate mechanistic effects of treatment on tumor cells, for example by distinguishing between necrosis and apoptosis based on fragment size (41).
Genome-wide and exome sequencing of plasma DNA at multiple time points during cancer treatment have been proposed as non-invasive means to study cancer evolution and for the identification of possible mechanisms of resistance to treatment (3). However, WGS and WES approaches are costly and have thus far been applicable only in samples for which the tumor DNA fraction was >5%–10% (3–5, 42). We demonstrated that we could exploit the differences in fragment lengths using in vitro and in silico size selection to enrich for tumor content in plasma samples, which improved mutation and SCNA detection in sWGS and WES data. We demonstrated that size selection improved the detection of mutations that are present in plasma at low allelic fractions, while maintaining low sequencing depth by sWGS and WES. Size selection can be achieved with simple means and at low cost and is compatible with a wide range of downstream genome-wide and targeted genomic analyses, greatly increasing the potential value and utility of liquid biopsies as well as the cost-effectiveness of cfDNA sequencing.
Size selection can be applied in silico, which incurs no added costs, or in vitro, which adds a simple and low-cost intermediate step that can be applied to either the extracted DNA or the libraries created from it. This approach, applied prospectively to new studies, could boost the clinical utility of ctDNA detection and analysis and creates an opportunity for re-analysis of large volumes of existing data (4, 34, 43). The limitation of this technique is a potential loss of material and information, since some of the informative fragments may be found in size ranges that are filtered out or de-prioritized in the analysis. This may be particularly problematic if only a few copies of the fragments of interest are present in the plasma. Despite potential loss of material, we demonstrated that classification algorithms can learn from cfDNA fragmentation features and SCNA analysis and improve the detection of ctDNA with a cheap sequencing approach. Moreover, the cfDNA fragmentation features alone can be leveraged to classify cancer and healthy samples with a high accuracy (AUC=0.989 for high ctDNA cancers, and AUC=0.891 for low ctDNA cancers).
Analysis of fragment sizes could provide improvements in other applications. Introducing fragment size information on each read could enhance mutation-calling algorithms from high-depth sequencing, to distinguish tumor-derived mutations from other sources such as somatic variants or background sequencing noise. In addition, cfDNA from patients analyzed with CHIP is likely to be structurally different from ctDNA released during tumor cell proliferation (18, 19). Thus, fragmentation analysis or selective sequencing strategies could be applied to distinguish clinically relevant tumor mutations from those present in clonal expansions of normal cells. This will be critical for the development of cfDNA-based methods for identification of patients with early stage cancer.
Size selection could also have an impact on the detection of other types of DNA in body fluids or enrichment of signals from circulating bacterial or pathogen DNA and mitochondrial DNA. These DNA fragments are not associated with nucleosomes and are often highly fragmented below 100 bp. Filtering or selection of such fragments may prove to be important in light of the recently established link between the microbiome and treatment efficiency (17, 44). Moreover, recent work highlights a stronger correlation of ctDNA detection with cellular proliferation than with cell death (45). We hypothesize that the mode of the distribution of ctDNA fragment sizes at 145 bp could reflect cfDNA released during cell proliferation, and the fragments at 167 bp may reflect cfDNA released by apoptosis or maturation/turnover of blood cells. The effect of other cancer hallmarks (46) on ctDNA biology, structure, concentration, and release is yet unknown.
In summary, ctDNA fragment size analysis, via size selection and machine learning approaches, boosts non-invasive genomic analysis of tumor DNA. Size selection of shorter plasma DNA fragments enriches ctDNA and assists in the identification of a greater number of genomic alterations with both targeted and untargeted sequencing at minimal additional cost. Combining cfDNA fragment size analysis and the detection of SCNAs with a non-linear classification algorithm improved the discrimination between samples from cancer patients and those from healthy individuals. Because the analysis of fragment sizes is based on the structural properties of ctDNA, size selection could be used with any downstream sequencing applications. Our work could help overcome current limitations of sensitivity for liquid biopsy, supporting expanded clinical and research applications. Our results indicate that exploiting the endogenous biological properties of cfDNA provides an alternative paradigm to deeper sequencing of ctDNA.
Materials and Methods
Study design
344 plasma samples from 200 patients with multiple cancer types were collected along with plasma from 65 healthy controls. Among the patients, 172 individuals, and notably the OV04 samples, were recruited through prospective clinical studies at Addenbrooke’s Hospital, Cambridge, UK, approved by the local research ethics committee (REC reference numbers: 07/Q0106/63; and NRES Committee East of England - Cambridge Central 03/018). Written informed consent was obtained from all patients, and blood samples were collected before and after initiation of treatment with surgery or chemotherapeutic agents. DNA was extracted from 2 mL of plasma using the QIAamp circulating nucleic acid kit (Qiagen) or QIAsymphony (Qiagen) according to the manufacturer’s instructions. In addition, 28 patients were recruited as part of the Copenhagen Prospective Personalized Oncology (CoPPO) program (Ref: PMID: 25046202) at Rigshospitalet, Copenhagen, Denmark, approved by the local research ethics committee. Baseline tumor tissue biopsies were available from all 28 patients, together with re-biopsies collected at relapse from two patients, and matched plasma samples. Brain tumor patients were recruited at Addenbrooke’s Hospital, Cambridge, UK, as part of the BLING study (REC – 15/EE/0094). Bladder cancer patients were recruited at the Netherlands Cancer Institute, Amsterdam, The Netherlands, and approval according to national guidelines was obtained (N13KCM/CFMPB250) (47). 65 plasma samples were obtained from healthy control individuals using a similar collection protocol (Seralab). Plasma samples have not been freeze-thawed more than 2 times to reduce artifactual fragmentation of cfDNA. A flowchart of the study is presented in fig. S1.
Supplementary Material
One sentence summary.
Selective sequencing or in silico analysis for differences in DNA fragment size can improve the detection of circulating tumor DNA
Acknowledgments
The authors would like to thank all members of the Rosenfeld Lab and Brenton Lab for their help and constructive discussion, in particular Mareike Thompson, Andrea Ruiz-Valdepanas, Jenny P.Y. Chan, and Anja Lisa Riediger. The authors would like to also thank the Cancer Research UK Cambridge Institute core facilities for their support, in particular the genomics, bioinformatics and biorepository facilities. Support is also acknowledged from the Cancer Research UK Cambridge Cancer Centre, the Cambridge Experimental Cancer Medicine Centre (ECMC), Cancer Molecular Diagnostics Laboratory (CMDL) and NIHR Biomedical Research Centre (BRC). We would like to acknowledge our patients and caregivers, and the help and support of the research nurses, trial staff and the staff at Addenbrooke’s Hospital and Rigshospitalet. In particular, we would like to acknowledge Charlotte Hodgkin, Heather Biggs and Karen Hosking. We would like to thank Hedley Carr and AstraZeneca for support for the CALIBRATE study.
Funding: We would like to acknowledge the support of The University of Cambridge, Cancer Research UK and the EPSRC (CRUK grant numbers A11906 (NR), A20240 (NR), A22905 (JDB), A15601 (JDB), A25177 (CRUK Cancer Centre Cambridge), A17242 (KMB), A16465 (CRUK-EPSRC Imaging Centre in Cambridge and Manchester)). The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013) / ERC Grant Agreement n. 337905. The research was supported by the National Institute for Health Research Cambridge, National Cancer Research Network, Cambridge Experimental Cancer Medicine Centre and Hutchison Whampoa Limited. This research is also supported by Target Ovarian Cancer and the Medical Research Council through their Joint Clinical Research Training Fellowship for Dr Moore. The CALIBRATE study was supported by funding from AstraZeneca. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions: FM, AMP, DC, EM, JDB and NR conceptualized and designed the study; FM, AMP, EM, LBA, KH, CGS, JCMW, DG, RM, TG, AS, IG, OO, CAP, MMS, IH, KP, WNC performed experiments and collected data; FM, AMP, DC, EM and CGS conceptualized the size selection approach; FM, AMP and EM designed and performed in vitro size selection; FM and DC conceptualized and designed the fragmentation feature analysis, with input from F. Marass and NR; DC conceptualized and designed the t-MAD index with input from FM; FM and DC carried out bioinformatics analysis of SCNAs from sWGS; JM performed bioinformatics analysis of TAm-Seq; FM and LBA designed the tailored captured sequencing and performed WES; FM and JM performed bioinformatics analysis of the capture sequencing and WES; ME developed and optimized mutation calling algorithms; RM, KB and SR designed the animal model; JCG, SP, RB, MMS, GDS, JB, SM, PC, CW, RM, MvdH have collected human samples; MJL and J. Burge performed histopathology revision; FM, DC, AMP, EM, JDB and NR wrote the manuscript; all co-authors have critically reviewed the manuscript; FM, AMP, DC, JDB and NR supervised the study; FM coordinated the study.
Competing interests: NR, JDB and DG are cofounders, shareholders and officers/consultants of Inivata Ltd, a cancer genomics company that commercializes ctDNA analysis. Inivata Ltd had no role in the conceptualization, study design, data collection and analysis, decision to publish or preparation of the manuscript. JDB received research funding from Aprea and NCI, and has received advisory board fees from Astra-Zeneca. F. Marass and NR are co-inventors of patent WO/2016/009224 on “A method for detecting a genetic variant”. F. Mouliere, JW, KH, CM, CS, NR and other authors may be listed as co-inventors on patent application number 1803596.4 on “Improvements in variant detection” and other potential patents describing methods for the analysis of DNA fragments and applications of circulating tumor DNA. IG is currently an employee of Novartis AG, a relationship that started after all his work contributing to this manuscript had been completed. Novartis had no role in the work presented in this manuscript. Other co-authors declare that they have no competing interests.
Data and materials availability: Sequencing data for this study are deposited in the EGA database, accession number EGAS00001003258. Other data associated with this study are present in the paper or supplementary materials.
References
- 1.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017 doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 2.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 3.Murtaza M, Dawson S-J, Tsui DWY, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin S-F, Kingsbury Z, Wong ASC, Marass F, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 4.Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, Gydush G, Reed SC, Rotem D, Rhoades J, Loginov D, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, Benezeder T, Auer M, Pischler C, Mannweiler S, Pichler M, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med. 2014;6:224ra24–224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Goodman SN, David KA, Juhl H, Kinzler KW, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 9.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong H-L, Christie M, Kosmider S, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL, Zhou L, Scherer F, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque IS, Elemento O. Challenges in Using ctDNA to Achieve Early Detection of Cancer. bioRxiv. 2017 237578. [Google Scholar]
- 14.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, Zhou L, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulz P, Thallinger GG, Auer M, Graf R, Kashofer K, Jahn SW, Abete L, Pristauz G, Petru E, Geigl JB, et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet. 2016;48:1273–1278. doi: 10.1038/ng.3648. [DOI] [PubMed] [Google Scholar]
- 16.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, De Vlaminck I. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep. 2016;6:27859. doi: 10.1038/srep27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Ulrich B, Supplee J, Kuang Y, Lizotte PH, Feeney N, Guibert N, Awad MM, Wong K-K, Janne PA, Paweletz CP, et al. False positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-18-0143. clincanres.0143.2018. [DOI] [PubMed] [Google Scholar]
- 20.Bronkhorst AJ, Wentzel JF, Aucamp J, van Dyk E, du Plessis L, Pretorius PJ. Characterization of the cell-free DNA released by cultured cancer cells. Biochim Biophys Acta - Mol Cell Res. 2016;1863:157–165. doi: 10.1016/j.bbamcr.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–65. [PubMed] [Google Scholar]
- 22.Lo YMD, Chan KCA, Sun H, Chen EZ, Jiang P, Lun FMF, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RWK. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 23.Chandrananda D, Thorne NP, Bahlo M, Tam L-S, Liao G, Li E. High-resolution characterization of sequence signatures due to non-random cleavage of cell-free DNA. BMC Med Genomics. 2015;8:29. doi: 10.1186/s12920-015-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang P, Lo YMD. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016;32:360–371. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Yu SCY, Chan KCA, Zheng YWL, Jiang P, Liao GJW, Sun H, Akolekar R, Leung TY, Go ATJI, van Vugt JMG, Minekawa R, et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci U S A. 2014;111:8583–8. doi: 10.1073/pnas.1406103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lun FMF, Tsui NBY, Chan KCA, Leung TY, Lau TK, Charoenkwan P, Chow KCK, Lo WYW, Wanapirak C, Sanguansermsri T, Cantor CR, et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:19920–5. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minarik G, Repiska G, Hyblova M, Nagyova E, Soltys K, Budis J, Duris F, Sysak R, Gerykova Bujalkova M, Vlkova-Izrael B, Biro O, et al. Utilization of Benchtop Next Generation Sequencing Platforms Ion Torrent PGM and MiSeq in Noninvasive Prenatal Testing for Chromosome 21 Trisomy and Testing of Impact of In Silico and Physical Size Selection on Its Analytical Performance. PLoS One. 2015;10:e0144811. doi: 10.1371/journal.pone.0144811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-Free DNA in Human Blood Plasma. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, Hoon DSB. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–6. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 30.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, Gongora C, Thierry AR, Lee T, editors. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS One. 2011;6:e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouliere F, El Messaoudi S, Pang D, Dritschilo A, Thierry AR. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol. 2014;8:927–941. doi: 10.1016/j.molonc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang P, Chan CWM, Chan KCA, Cheng SH, Wong J, Wong VW-S, Wong GLH, Chan SL, Mok TSK, Chan HLY, Lai PBS, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112:E1317–25. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J, Kwiatkowski DJ, editors. Fragment Length of Circulating Tumor DNA. PLOS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles JV, Mokhtari R, Gandara DR, Mack PC, Odegaard JI, Nagy RJ, Baca AM, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-17-3837. clincanres.3837.2017. [DOI] [PubMed] [Google Scholar]
- 35.Macintyre G, Goranova TE, De Silva D, Ennis D, Piskorz AM, Eldridge M, Sie D, Lewsley L-A, Hanif A, Wilson C, Dowson S, et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet. 2018;1 doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, Freeman S, Moyle P, Sala E, Sayal K, Hosking K, et al., editors. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLOS Med. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DWY, Kaper F, Dawson S-J, Piskorz AM, Jimenez-Linan M, Bentley D, Hadfield J, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 38.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Best MG, Sol N, Tannous BA, Wesseling P, Wurdinger T. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell. 2015;28:666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Best MG, Sol N, In’t Veld SGJG, Vancura A, Muller M, Niemeijer A-LN, Fejes AV, Tjon Kon Fat L-A, Huis In ’t Veld AE, Leurs C, Le Large TY, et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell. 2017;32:238–252.e9. doi: 10.1016/j.ccell.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riediger AL, Dietz S, Schirmer U, Meister M, Heinzmann-Groth I, Schneider M, Muley T, Thomas M, Sültmann H. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep. 2016;6 doi: 10.1038/srep33505. 33505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belic J, Koch M, Ulz P, Auer M, Gerhalter T, Mohan S, Fischereder K, Petru E, Bauernhofer T, Geigl JB, Speicher MR, et al. Rapid Identification of Plasma DNA Samples with Increased ctDNA Levels by a Modified FAST-SeqS Approach. Clin Chem. 2015;61:838–849. doi: 10.1373/clinchem.2014.234286. [DOI] [PubMed] [Google Scholar]
- 43.Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, Choudhury AD, Gydush G, Reed SC, Rhoades J, Rotem D, et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations With Survival in Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2018;36:543–553. doi: 10.1200/JCO.2017.76.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 45.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, Marafioti T, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 47.Patel KM, van der Vos KE, Smith CG, Mouliere F, Tsui D, Morris J, Chandrananda D, Marass F, van den Broek D, Neal DE, Gnanapragasam VJ, et al. Association Of Plasma And Urinary Mutant DNA With Clinical Outcomes In Muscle Invasive Bladder Cancer. Sci Rep. 2017;7 doi: 10.1038/s41598-017-05623-3. 5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, van Essen HF, Eijk PP, Rustenburg F, Meijer GA, Reijneveld JC, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.