Abstract
Psoriasis (PS) is a systemic, immune-mediated inflammatory disorder. However, the whole lymphocyte compartment and the potential pathologies of PS have not been fully characterized. In the present study, we examined whole lymphocyte subsets and signal transduction proteins using high-dimensional single-cell mass cytometry and a bioinformatics pipeline for an in-depth characterization of the immune cell subsets and protein profiles involved in pathways in the peripheral blood of patients with PS. We identified 15 major immune cell populations in T cell lineages and characterized various CD3+CD4+ Th and CD3+CD8+ T cytotoxic cell populations simultaneously across 24 leukocyte markers and 7 proteins related to the signal transduction pathways. High-dimensional analysis identified 3 new subsets that are abundant in PS peripheral blood, resembling CD3–CD4+ lymphoid tissue inducer cells, Tc17 cells, and CD8+CXCR3+ Tregs. We confirmed the CD3–CD4+ cells, and their features and functions, in an independent PS cohort. The use of single-cell mass cytometry allows systemic-level characterization of lymphocyte subpopulations and dysregulated signaling pathways in the blood of patients with PS, identifying abnormalities of different immune cell subsets. We validated that the CD3–CD4+ cells had elevated OX40 and decreased FRA2 expression, which were positively associated with the PS area and severity index.
Keywords: Autoimmunity, Inflammation
Keywords: Autoimmune diseases
Systems-level characterization of lymphocyte subpopulations in the blood of patients with psoriasis identifies abnormalities in immune cell subsets.
Introduction
Psoriasis (PS) is a chronic, immune-modulated inflammatory disease that affects patients far beyond the skin (1). About 2%−3% of individuals worldwide suffer from this debilitating skin disease (2), and about 30% of patients with PS also develop psoriatic arthritis. Besides skin and joint inflammation, there is now evidence of important systemic manifestations of PS (3, 4). In recent years, our growing knowledge of the PS pathogenic pathways and the advent of new technologies that are directly applicable to human biological substrates have spawned optimism that a cure for PS may be found (5). Targeted treatments with monoclonal antibodies were shown to be safe and efficacious in the short-term management of most patients with moderate to severe chronic plaque PS (6).
Despite these encouraging results, clinical outcomes remain highly variable, providing evidence that PS is a systemic and heterogeneous disease and highlighting the requirement to deepen our current understanding of the disease (7). Different subsets of Th cells have been reported to contribute to the pathogenesis of PS (1, 8, 9), such as Th1, Th9, Th17, and Th22 cells, whereas other contributors are skin-homing memory T cells (1) or Tregs (10). Therefore, complicated immune dysregulation may result in alterations in the frequency, function, or location of any one of these Th or T cytotoxic (Tc) subpopulations, which determine the severity of different pathologies (11, 12). There has been increasing interest in the role of the T cell costimulatory tumor necrosis factor receptor superfamily member 4 (TNFRSF4, also known as OX40) and its cognate ligand, OX40L, in immunoregulation of autoimmunity (13). OX40 plays a key role in maintaining late T cell proliferation and survival by suppressing apoptosis and by inducing T cell memory formation (14). OX40 (also known as CD134) is expressed in lesional, but not healthy, skin of patients with PS (15). OX40-OX40L, as an immune checkpoint, has been proposed as a potential therapeutic target to treat PS. In addition, studies at the transcription level have suggested that AP-1 proteins, comprising members of the FOS (C-FOS, FOSB, FRA-1, and FRA-2) and JUN families, acting as sequence-specific homodimeric and heterodimeric transcription factors, are abnormally expressed in skin biopsies of patients with inflammatory/proliferative skin diseases, such as PS (16, 17). Among the members, fos-related antigen 2 (FRA2) acts as a key regulator of terminal epidermal differentiation. Our previous study (18) found that FRA2 was involved in the pathogenesis of PS by regulating the IL-23 receptor (IL23R). Therefore, a comprehensive understanding of the nature of the dysregulated immune system in PS might identify potential novel targets to develop more efficacious therapies for PS.
However, the collective characterization of alterations in different subsets in immune-mediated diseases remains difficult because of the large phenotypic diversity across all lymphocyte populations (19, 20). In addition, there has been little research on the comprehensive analysis of Th cell lineages in patients with PS. Some recent reports have shown the use of single-cell mass cytometry to characterize Th populations simultaneously across 23 memory T cell–associated surface and intracellular molecules in healthy donors as well as to evaluate the predictive biomarkers in patients with cancer (20, 21). In addition, cytometry by time-of-flight (CyTOF) overcomes the limitations of fluorophore tags by detecting metal-tagged antibodies attached to single cells, which allows simultaneous multiparameter interrogation and even permits all samples in the same experiment group to be analyzed at the same time, which would diminish laboratory errors (22). In the present study, we used peripheral blood mononuclear cells (PBMCs) and lesional skin from patients with PS at disease onset without any treatments and health controls (23). High-dimensional single-cell mass cytometry and imaging mass cytometry were used, together with optimized immune marker panels and a customized, interactive bioinformatics pipeline, to generate a thorough analysis of the peripheral blood immune cells. We aimed to display the immune landscape of patients with PS, characterize the abnormalities of Th cell subsets, identify PS-associated pathogenic CD3–CD4+ cell populations, and preliminarily explore the potential role of CD3–CD4+ cells in the immune-mediated pathology of PS.
Results
Lymphocyte landscape of PS using mass cytometry.
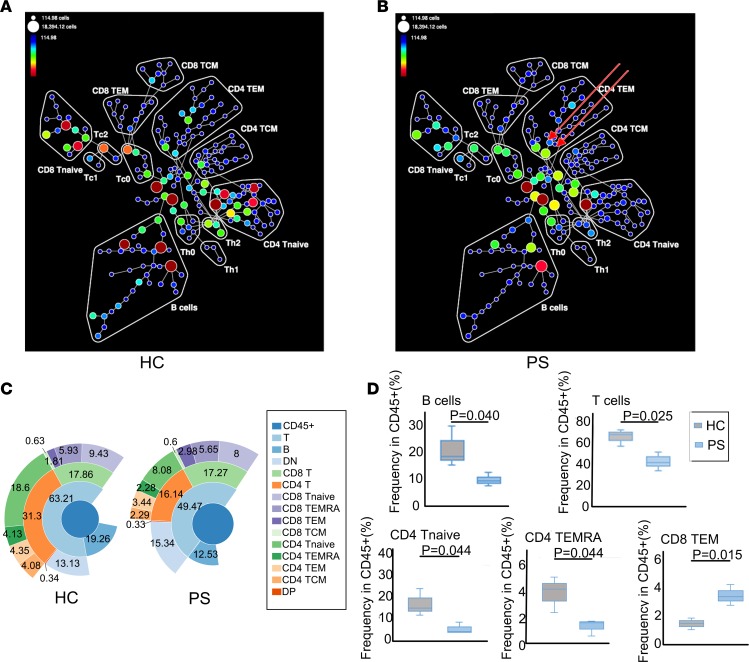
We analyzed PBMC samples isolated from the blood of 4 patients that were newly diagnosed with PS, without any treatment, as well as from 4 samples from age-and sex-matched healthy controls (HCs). Based on the combinations of surface markers, we used CyTOF to simultaneously analyze CD8 naive T, Tc0, Tc1, Tc2, CD8 central memory T (TCM), CD8 effective memory T (TEM), CD4 naive T, Th0, Th1, Th2, CD4 TCM, CD4 TEM, CD4+CD8+ T (double-positive T [DPT]), CD4–CD8– T (double-negative T [DNT]), and B cells within the PS and HC samples, respectively. The frequencies and compositions of those cell populations are presented in Figure 1, A and B. In particular, the frequencies of CD4 naive T, Th2, CD8 naive T, Tc0, Tc2, double-positive (DP), double-negative (DN), and B cells in PS samples were obviously lower compared with those in HC samples, while the proportion and constituents of CD4 TEM cells were more complicated. There were two subsets of CD4 TEM cells showing higher frequencies in patients with PS, which are indicated by arrows in Figure 1B. Then, we manually annotated lymphocytes into 15 major cell populations. Using the sunburst method, the frequencies of the 15 compartments are shown Figure 1C. The 15 compartments comprised CD45+ cells, T cells, B cells, DNT cells, CD8+ T cells, CD4+ T cells, CD8 naive T cells, CD8 terminally differentiated effector memory (TEMRA) cells, CD8 TEM cells, CD8 TCM cells, CD4 naive T cells, CD4 TEMRA cells, CD4 TEM cells, CD4 TCM cells, and DP cells. The frequencies of nearly all these compartments were lower in the PS samples, except for DNT (PS/HC fold change = 1.17) and CD8 TEM (1.64). Among those lower compartments, T cells (PS/HC fold change = 0.78), B cells (PS/HC fold change = 0.65), CD4+ T cells (PS/HC fold change = 0.52), CD4 naive T cells (PS/HC fold change = 0.43), CD4 TEMRA cells (PS/HC fold change = 0.55), CD4 TEM cells (PS/HC fold change = 0.79), and CD4 TCM cells (PS/HC fold change = 0.56) were abnormal. These data showed that the frequencies of total T and B cells were statistically significantly decreased in PS samples, especially CD4 naive T cells and CD4 TEMRA cells, while frequencies of CD8 TEM cells were increased (Figure 1D). This is the first study to our knowledge to demonstrate the adaptive immune landscape using mass cytometry of PS samples to show the overall dysregulation of the immune system.
Figure 1. Immune cell distributions and compositions in PS and HC.
(A and B) The SPADE tree plots of all HC (n = 4) (A) and all PS patients (n = 4) (B) showing the different gated immune cell subpopulations. Node color and the size of each node in the tree indicate the frequency of cells. Red arrows indicate the abundant and heterogeneous pattern of CD4 TEM cells in PS. (C) Sunburst representation of the distributions and frequencies of the various cell subsets. Results are expressed as percentages. (D) Box plots comparing cell frequency (percentage of CD45+ cells) of B cells, T cells, CD4 naive T (Tnaive) cells, CD4 TEMRA cells, and CD8 TEM cells between HC and PS groups. The 5th and 95th percentile are shown in each box, and median (center line) is marked. *P < 0.05 by 2-tailed Student’s t test. PS, psoriasis; HC, healthy controls.
Expression levels of 24 immune markers on the major immune cell subsets of PS.
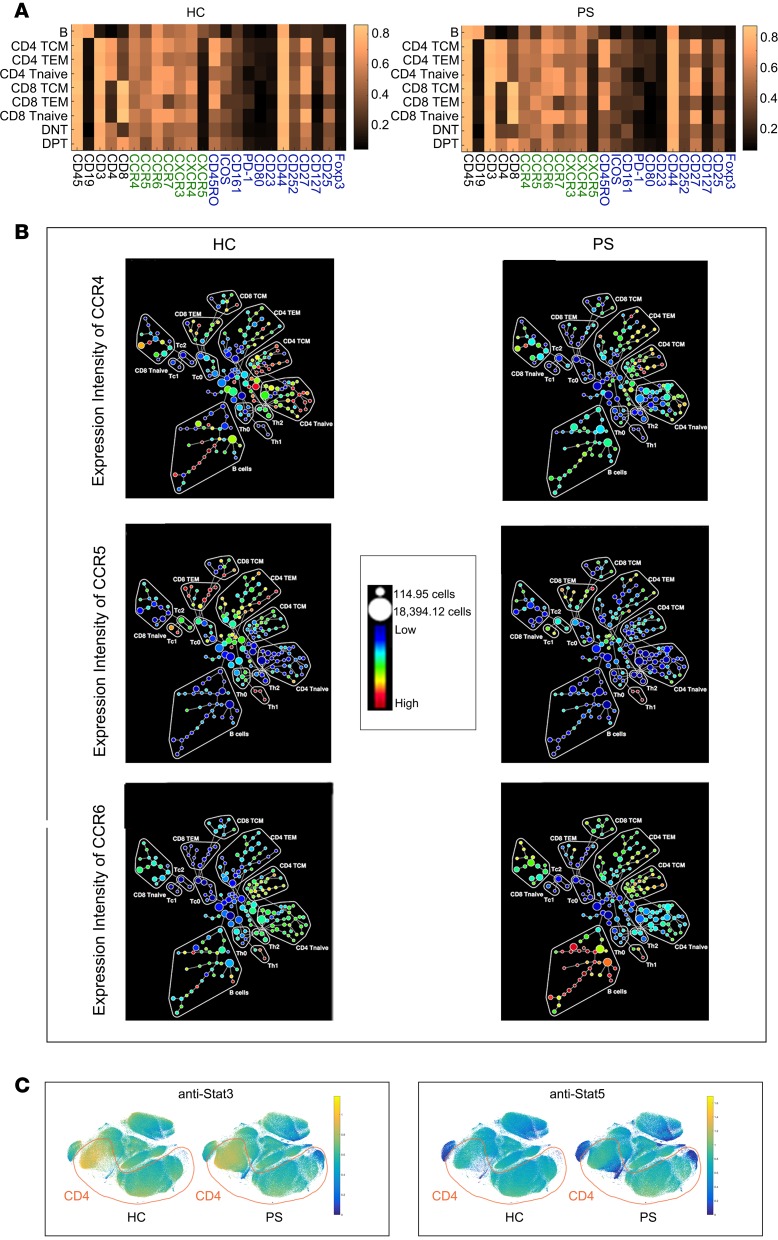
The adaptive immune landscape of PS is different to that of HC; therefore, we aimed to explore the detailed mechanism contributing to this difference. We constructed a heatmap to show the expression levels of 24 immune markers between the HC and PS groups (Figure 2, A and B). These markers could be divided into 3 groups: cluster differentiation antigens that define specific immune cells, activated markers, and chemokines. Generally, the body has a certain ability to maintain the immune balance, and there might be a little fluctuation of these immune markers in the major immune cell subsets. Our descriptive analysis showed that there were differences in the expression levels of the 24 immune markers on the major immune cell subsets of PS and HC samples; however, the fluctuations were not significant. Chemokines play an important role in attracting the immune cells to the skin, leading to the classic lesions of PS. We further focused on the expression of C-C motif chemokine receptor 4 (CCR4), CCR5, and CCR6 on the 15 lymphocyte compartments (Figure 2B). CCR4 and CCR6 were highly expressed on the CD4 TEM cells in the PS samples. In particular, we observed, for the first time to our knowledge that CCR6 levels were elevated on B cells. Compared with the HC samples, we observed CD4+ T cells with aberrant protein expression levels of STAT3 and STAT5 using viSNE analysis (Figure 2C). The descriptive data might provide more clues to explore the chemokines and signal transduction pathways of PS.
Figure 2. Phenotype fluctuations in PS.
(A) Heatmap depicting differential expression for coinhibitory or costimulatory population-defining markers and activation-associated and lymphoid homing–associated receptors across each subpopulation. (B) SPADE plot showing the median expression of CCR4, CCR5, and CCR6 in the 15 lymphocyte compartments of HC and PS. Node size indicates cell number; color coding represents median CCR4, CCR5, and CCR6 expression of each node. (C) viSNE map of PS and HC, illustrating color-coded cell populations that clustered based on cell surface marker expression. The relative location of CD4 was marked (red). viSNE analysis showing CD4+cells with aberrant proteins expressions of Stat3 and Stat5.
Altered subpopulations and signal transduction proteins of T cell lineages in PS.
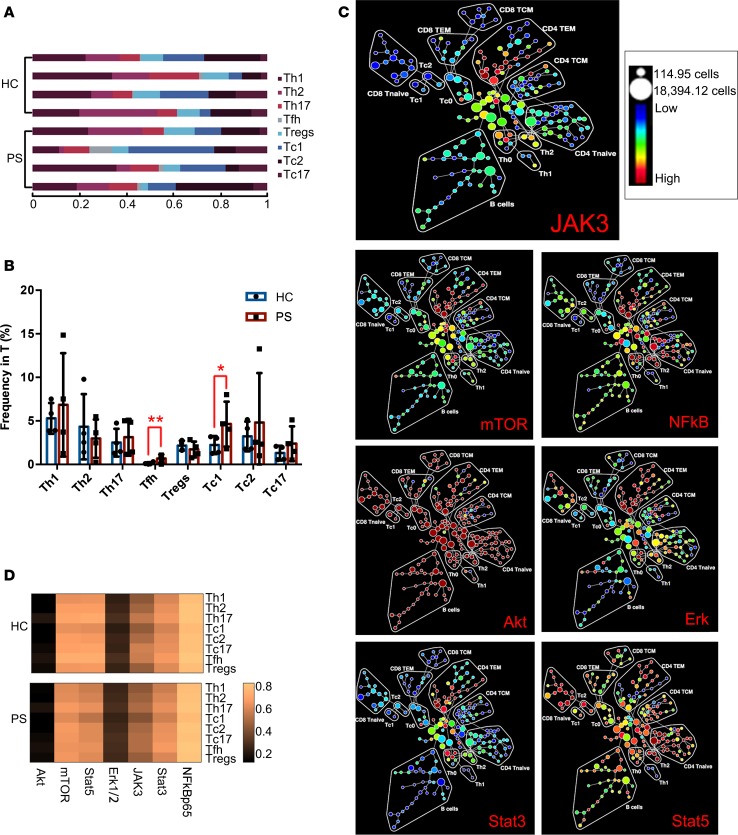
Differences in the frequencies and compositions of T cell lineages were observed in PS (Figure 1, B and D). Next, we sought to identify which subpopulations of T cells best described the differences between PS and HC in terms of cell frequency and activated markers. Subgroups of T cell lineages were gated (Figure 3, A and B) and the distributions of 7 key signal transduction proteins on T cell lineages are shown in Figure 3C. We subdivided CD4 TEM cells into Th1, Th2, Th17, and follicular Th (Tfh) cells and Tregs and divided the CD8 TEM cells into Tc1, Tc2, and Tc17 cells by manual annotation (Figure 3B). The frequencies of these subclusters between PS and HC were then compared. The limited samples meant that the frequencies of Th1 and Th17 cells were higher without significant differences in patients with PS. However, we observed that the frequencies of Tfh and Tc1 cells were significantly higher in PS (Figure 3B, P < 0.05). Meanwhile, Tc17 cells also showed an increased trend in the peripheral blood of patients with PS. Although the small sample size limited the significance of the statistical difference, the increasing trend of the Th subcluster was confirmed using flow cytometry in a follow-up experiment. These findings suggested that not only was there an imbalance of Th subpopulations but that there were also disordered CD8 TEM subclusters.
Figure 3. Compositional alterations and functional changes in effector T cells of PS (n = 4) and HC (n = 4).
(A) Stacked bars depicted T cell subtypes in each individual sample. Cell abundance was expressed as normalized percentage of T cells. (B) Bar plots comparing cell frequency (percentage of T cells) of Th1 cells, Th2 cells, Th17 cells, Tfh cells, Tregs, Tc1 cells, Tc2 cells, and Tc17 cells. Error bars represent SD. *P < 0.05; ** P < 0.01 by 2-tailed Student’s t test. (C) SPADE maps depicting the fold change of JAK3, mTOR, Stat3, Stat5, Akt, Erk, and NF-κB expression in PS compared with HC groups. Signaling fold changes for each node in the SPADE diagram were calculated as the difference of arcsine median intensity of the PS patients compared with the HC. (D) Profiles in signal transduction proteins of Th1, Th2, Th17, Tfh, Treg, Tc1, Tc2, and Tc17 subsets in HC and PS groups.
Based on the above abnormalities in the frequency, composition, and functional markers of T cells in PS, we further determined the potential pathogenic mechanism underlying the systemic inflammatory status. We use spanning-tree progression analysis of density-normalized events (SPADE) mapping to demonstrate the expression changes of the 7 key proteins and visualized the distribution of these proteins in the 15 population phenotypes between PS and HC. The 7 proteins consist of mTOR, STAT5A/B, STAT3, JAK3, ERK1/2, NF-κB p65, and AKT serine/threonine kinase 1/2/3 (AKT1/2/3), which are central orchestrators of immune responses (24). We observed for the first time to our knowledge that mTOR, STAT3, and JAK3 were more predominantly expressed in CD4 TEM cells, and their expression levels were higher in samples from patients with PS compared with those from HC. This suggested that JAK3/STAT3 is important pathway for the inflammatory response in patients with PS. At the same time, the STAT5A/B, AKT1/2/3, ERK1/2, and NF-κB p65 responses were shared across the immune systems of patients with PS without specificity (Figure 3C), which suggested that complicated upstream mechanisms may be involved in the pathogenesis of PS, leading to dysregulated signaling pathways. CD4 TEM cells would be a potential focus of research. However, we need to pinpoint the subsets of CD4 TEM cells that contribute to the dysregulation of PS.
Eight clusters of TEM cells were segregated based on combinations of cluster differentiation antigens and chemokines (25). The eight clusters included Tregs and Th1, Th2, Th17, Tfh, Tc1, Tc2, and Tc17 cells. We then identified that the signal transduction proteins were expressed in the 8 clusters between PS and HC without obvious fluctuations (Figure 3D). This might have been caused by the small sample size. Thus, the phosphorylation status of these proteins should also be defined.
Identification of CD3–CD4+ subsets in patients with PS using the viSNE algorithm.
viSNE analysis could allow us to identify and characterize new cell subpopulations. We ran viSNE analysis on cells of all PS and HC samples using 31 staining markers and identified 4 populations with an obviously different abundance in PS compared with that in HC (Figure 4A). Three of them have not been reported before to our knowledge. In terms of marker expression, we found that the 4 detected populations (P1, P2, P3, and P4 in Figure 4A) contained a core signature of CD45+, CD44+, and CCR7+ markers. Backprojection of those 4 populations to the ACCENSE density map showed that they corresponded to subpopulations 3, 5, 22, and 23, respectively (Figure 4B). On further view of all the markers, P2 expressed CD45+, CD44+, CCR7+, CD3–, and CD4+. The detailed features of the P2 subpopulation are illustrated in Figure 4C. The features of P1, P3, and P4 are provided in the Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125306DS1). Based on the combinations of surface markers, the P2 subset (CD3–CD4+ cells) resembled lymphoid tissue inducer (26) cells. In addition, signal-transducing proteins, such as NF-κB p65, mTOR, and STAT3, were highly expressed in the CD3–CD4+ population, which suggested its potentially proinflammatory role in PS. STAT5 and ERK1/2 levels were decreased in P2. When compared with the CD3+CD4+ T cells of PS and HC samples (Figure 4, C and D), there were obviously differences between CD3–CD4+ cells and CD3+CD4+ cells, for example, lower FOXP3 expression in CD3–CD4+ cells from the PS samples. During data processing, we noticed an interesting phenomenon: OX40L (also known as CD252) was hardly expressed in the P2 population (Figure 4C). Further detailed study of subset P2 and exploration of the underlying mechanism of its involvement with PS might be helpful to develop precise targeted treatment.
Figure 4. Four suspect populations in the red dotted ellipse regions composed almost entirely of cells from PS patients were identified as a potential target of treatment.
(A) viSNE map of PS (n = 4) and HC (n = 4) illustrating color-coded cell populations that clustered based on cell surface marker expression. viSNE analysis identifies 4 subpopulations that appear dominantly in PS. (B) ACCENSE density map of immune cells defined in A. Subpopulations 1−23 identified as local maxima in the density map are marked. The color scale signifies cell densities of similar marker phenotypes. (C) Expression signatures of the marked subpopulations P2 in A. Bar heights indicate the median expression for each marker within the subpopulation, estimated from cells sampled close to the locations of the subpopulation peaks. The horizontal line for each marker corresponds to its median expression level across all the cells in the data set. (D) Expression signatures of CD3+CD4+cells in HC and PS. Bar heights indicate the median expression for each marker within the subpopulation.
CD3–CD4+ subsets in skin lesions of patients with PS using imaging mass cytometry.
Imaging mass cytometry (IMC) uses laser ablation to generate plumes of particles that are carried to the mass cytometer by a stream of inert gas. IMC has a resolution comparable to light microscopy, with the high content of mass cytometry enabled through the use of isotopically labeled probes and ICP-MS detection (27). Two samples of PS lesions were obtained and stained for the above-mentioned 31 immune markers. Figure 5A shows that a predominantly lymphocytic infiltrate was present in the dermis of the PS skin, including CD3, CD4, and CD8 T cells. The images also showed that CD3 (red) colocalized with both CD4 (green) and CD8 (white). The CD3–CD4+ cells are clearly delineated in green. We also determined the distribution pattern of various label combinations, such as CD3, CD4, CD8, JAK3, and DNA; CD3, CD4, CD8, mTOR, and DNA; CD3, CD4, CD8, STAT3, and DNA; CD3, CD4, CD8, CCR4, and DNA; CD3, CD4, CD8, CXCR3, and DNA; and CD3, CD4, CD8, CXCR4, and DNA (Figure 5, B and C). These results were consistent with the results for peripheral blood as analyzed using CyTOF.
Figure 5. Representative mass cytometry images of skin tissue samples from HC and PS patients.
(A) Overlay of CD3 (red), CD4 (green), CD8 (white), and DNA (blue). (B) Top: overlay of CD3 (red), CD4 (green), CD8 (white), JAK3 (cyan), and DNA (blue). Bottom left: overlay of CD3 (red), CD4 (green), CD8 (white), mTOR (cyan), and DNA (blue). Bottom right: overlay of CD3 (red), CD4 (green), CD8 (white), Stat5 (cyan), and DNA (blue). (C) Top: overlay of CD3 (red), CD4 (green), CD8 (white), CCR4 (cyan), and DNA (blue). Bottom left: overlay of CD3 (red), CD4 (green), CD8 (white), CXCR3 (cyan), and DNA (blue). Bottom right: overlay of CD3 (red), CD4 (green), CD8 (white), CXCR4 (cyan), and DNA (blue). Scale bars: 100 μm. Data are representative of results of 2 experiments.
Validation of CD3–CD4+ cells in an independent PS cohort using flow cytometry and confocal microscopy.
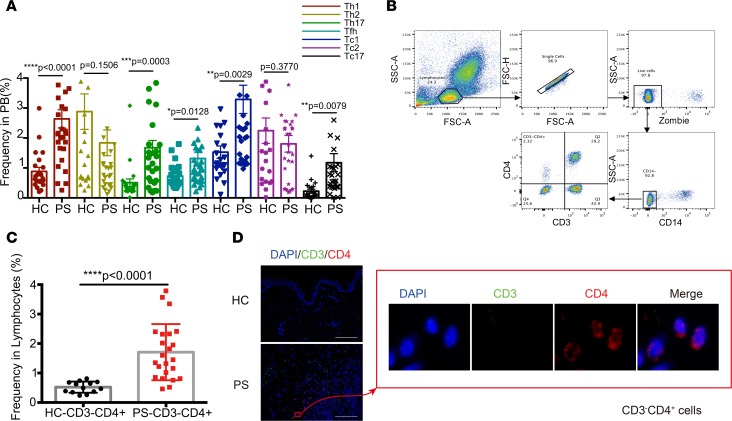
To independently validate the computational results, a blinded validation was performed on peripheral blood from a second independent cohort of 29 patients with PS and 23 age-and sex-matched HCs. We designed two flow cytometry–based validation panels using a reduced number of markers. One panel was used to confirm the Th cell lineages using 7 immunofluorescent markers. The results confirmed the increased proportion of Th1 and Th17 cells (Figure 6A), which had been reported before (8). We further discovered that Tfh, Tc1, and Tc17 cell numbers were significantly elevated in samples for patients with PS compared with those in samples from the HC group. Another panel was used to identify CD3–CD4+ cells in the 23 PS and 13 HC samples. In the CyTOF panel, CD161 (28) was used to preliminarily exclude the sources of NK cells. We used Zombie and CD14 to exclude nonspecific staining and CD4+ monocytes, respectively (Figure 6B). There was a distinct CD3–CD4+ subset observed in Figure 6B. Circulating CD3–CD4+ cells (0.52% ± 0.05%) were detected in HCs at a lower frequency that in the PS group, with a high degree of uniformity (Figure 6C). Significantly higher numbers of circulating CD3–CD4+ cells (1.7% ± 0.20%) were validated with varying frequency in samples from different patients with PS. viSNE analysis showed that a wide variety of chemokines were expressed on the CD3–CD4+ cells, such as CCR4, CCR5, CCR6, CCR7, CXCR3, and CXCR4 (Figure 4C), which might attract CD3–CD4+ cells to cutaneous psoriatic lesions. Therefore, we determined whether there were CD3–CD4+ cells in the lesional areas of PS samples using confocal microscopy. The confocal microscopy showed that there were many CD3+ cells in the lesions (Figure 6D), which was in accord with the IMC results. The resulting image (Figure 6D) also showed clearly the relative location of CD3–CD4+ cells. These results highlighted a potential role of CD3–CD4+ cell subsets in PS.
Figure 6. Validation CD3–CD4+ cells and Th subsets in an independent PS cohort.
(A) Percentages of Th cell subsets, including Tfh, Th1, Th2, Th17, Tc1, Tc2, and Tc17 cells in PBMCs, in HC (n = 23) and PS patients (n = 29). (B) CD3–CD4+ cells separating from lymphocytes by sequential gates. (C) The percentage of CD3–CD4+ cells in lymphocytes was compared between PS (n = 23) and HC (n = 13). (D) Confocal images showing immunofluorescence localization of CD3 (green), CD4 (red), and nuclear (blue). Data are representative of results of 3 experiments. Scale bars: 50 mm. *P < 0.05; ***P < 0.0005; ****P < 0.0001 as assessed by the 2-tailed Student’s t test. Data are presented as mean ± SD.
Characterization of CD3–CD4+ cells in PS.
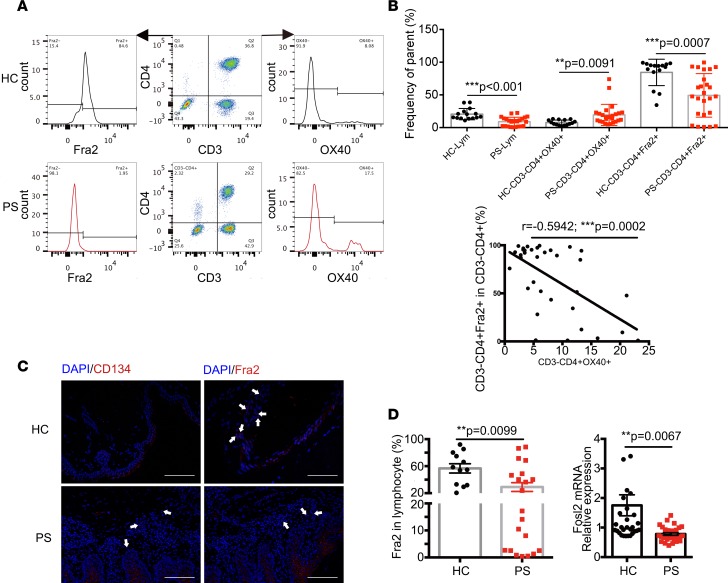
OX40L was rarely expressed on CD3–CD4+ cells from patients with PS, and there was explicit dysregulation of signal-transducing proteins. A previous study (15) showed that OX40 was expressed on the lesional skin of PS patients. In addition, the OX40-OX40L interaction provides a specific costimulatory signal to activated effector T cells. Thus, we wondered how those higher circulating CD3–CD4+ cells in PS are maintained. Therefore, we assumed that OX40 might be involved in the higher levels of circulating CD3–CD4+ cells of PS samples. Our previous study showed that FRA2 is involved in the pathology of PS by regulating IL23R (18). Thus, we further explored OX40 and FRA2 in CD3–CD4+ cells using flow cytometry (Figure 7A). We found that OX40 levels were upregulated on the CD3–CD4+ population (Figure 7B). By contrast, we observed significantly decreased levels of the nuclear transcription factor FRA2 on this population of cells. Notably, there was a negative correlation between OX40 and FRA2 levels (Figure 7B). Additionally, confocal imaging showed higher levels of OX40 and lower amounts of FRA2 in the dermis of PS skin lesions compared with skin from the HC group (Figure 7C). The percentage of CD3–CD4+ cells in lymphocytes was limited. Therefore, it is difficult to obtain sufficient quantities of these cells for functional analysis. We next assessed mRNA and protein levels of FRA2 in PBMCs from the PS and HC groups using quantitative real-time PCR (qPCR) and flow cytometry, respectively. We observed a lower expression of FRA2 at both the mRNA and protein levels in patients with PS (Figure 7D).
Figure 7. Characterization of CD3–CD4+cells in PS and HC.
(A) Flow cytometry showing the OX40 and FRA2 expression in CD3–CD4+cells of PS (n = 23) and HC (n = 13). (B) Percentages of CD3–CD4+OX40+ and CD3–CD4+Fra2+ cells in HC (n = 13) and PS patients (n = 23) were compared by 2-tailed Student’s t test, and the correlation of CD3–CD4+OX40+ and CD3–CD4+Fra2+ cells was analyzed by Pearson correlation analysis. (C) Confocal images showing immunofluorescence localization of CD134 (otherwise known as OX40) and Fra2 in the skin of PS and HC. White arrows indicate the positive expression of OX40 and Fra2 staining in dermis. The experiment was repeated 3 times. Scale bars: 50 mm. (D) The relative Fra2 expression was measured by flow cytometry (left, 13 HCs and 23 PS patients) and quantified using real-time PCR (right, 30 HCs and 32 PS patients). The fosl2 transcript was normalized to the copies of β-actin mRNA from the same sample.**P < 0.005; ***P < 0.0005, as assessed by the 2-tailed Student’s t test.
Function assay of CD3–CD4+ T cells in PS.
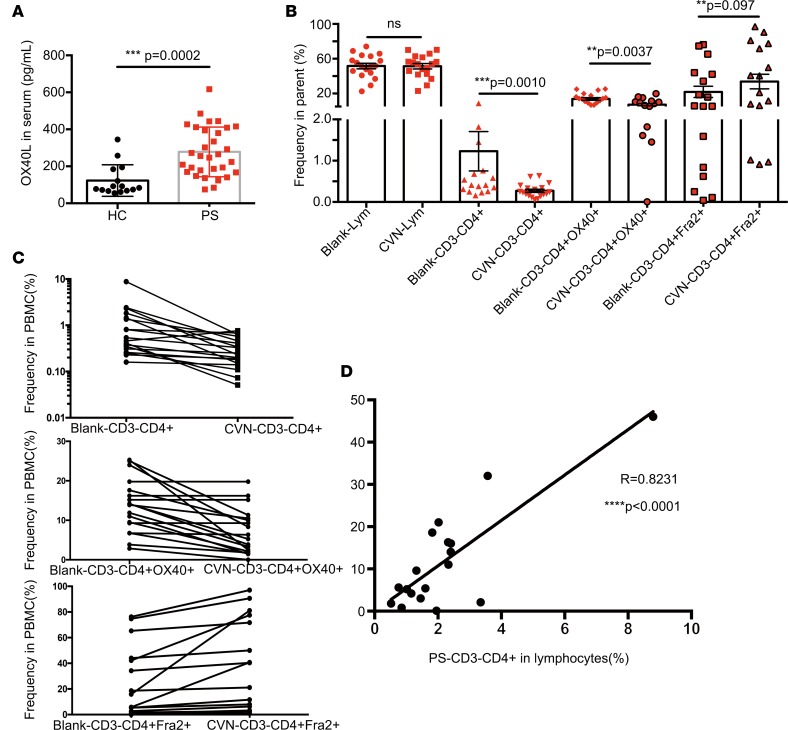
The OX40-OX40L interaction mainly promotes the expansion and survival of effector T cells. Therefore, ELISA was used to measure the serum OX40L level in PS and HC samples. The OX40L level was significantly higher in the PS group than in the HC group (Figure 8A, P = 0.0002). We further isolated PBMCs from the blood of an independent cohort of 18 patients with PS. Freshly isolated PBMCs were cultured in vitro with the OX40-OX40L inhibitor, chlorazol violet N (CVN), for 36 hours to probe the changes of OX40 and FRA2 and the numbers of CD3–CD4+ cells. Interestingly, inhibition of the OX40-OX40L costimulatory interaction significantly reduced the numbers of CD3–CD4+ cells (Figure 8B). The inhibitor of the OX40-OX40L interaction simultaneously affected the expression levels of OX40 and FRA2. CD3–CD4+OX40+ cells numbers obviously decreased and CD3–CD4+Fra2+ cell numbers were elevated in PBMCs treated with CVN (Figure 8C). We next sought to determine whether this population in the blood of patients with PS might reflect the disease activity. We collected and analyzed clinical data of the 18 patients with PS. A significant correlation was found between the frequency of CD3–CD4+ cells and the PS area and severity index (PASI) (Figure 8D). These analyses indicated that the potentially newly discovered CD3–CD4+ cells in PS might be developed as a biomarker to assess disease activity.
Figure 8. Function assay of CD3–CD4+ cells in PS.
(A) Serum concentrations of OX40L in HC (n = 15) and PS patients (n = 31). The data are illustrated as the mean ± SD and analyzed by 2-tailed Student’s t test. (B) The changes of CD3–CD4+cells, CD3–CD4+OX40+, and CD3–CD4+Fra2+cells in PBMCs with CVN treatment in PS (n = 18) were analyzed by 2-tailed Student’s t test. (C) After 36 hours of treatment with CVN, the trend of the CD3–CD4+, CD3–CD4+OX40+, and CD3–CD4+Fra2+ cell percentage in PBMCs was shown (n = 18). (D) There was positive correlation between CD3–CD4+cells and PASI before CVN treatment (n = 18) by Pearson correlation analysis. Correlation analysis was performed using the Pearson’s correlation. **P < 0.005; ***P < 0.0005; ****P < 0.0001.
Discussion
PS is a multifaceted, chronic skin disorder with many associated comorbidities; it imposes both a physical and psychological burden on patients. Growing evidence shows that targeting a single cytokine secreted by Th cells, such as IL-12, IL-17, IL-22, IL-23, TNF-α, and so on, could benefit patients with PS (29, 30). However, patients with PS experience highly variable clinical outcomes, and dysregulated or excessive activity of anticytokines in related pathways might promote inflammation (31). Thus, there is an urgent need to comprehensively understand the dysregulated immune system in PS and to optimize current therapy.
Mass cytometry offers the opportunity to simultaneously analyze dozens of single-cell markers in complex cellular samples, resulting in highly complex data sets (32). Conventional approaches for flow cytometry data analysis are not suitable for such data sets, because they suffer from individual user bias and require prior knowledge of the cell type of interest. In the current study, we initially used 31 antibodies to detect heterogeneity within the major adaptive immune cells in patients with PS. SPADE analysis showed that PBMCs from the PS and HC group could be classified into different clusters (Figure 1, A and B). Two abundant subpopulations of CD4 TEM cells were identified in the PS group (Figure 1B), which were important in the initiation and amplification phase of cutaneous inflammation (8). The significantly lower level of B cells might contribute to the lack of autoantibodies in PS; however, this requires further study.
We then used sunburst analysis to show the proportion of the 15 major immune subsets (Figure 1C). The proportions and functional markers of CD4 naive T cells, CD4 TEM cells, and CD8 TEM cells were altered in the PS group (Figure 2A). This analysis showed that, although PS was driven by T cell activation, in general, there were no obvious differences in the steady state of the overall cellular immunity reached by the PS and HC groups. However, this result might be related with our limited number of samples. Similar to previous studies (33, 34), the fraction containing CD4 and CD8 T cells expressed higher levels of skin-homing receptors, such as CCR4, CCR5, and CCR6 (Figure 2B), which mediate lymphocyte retention in dermis of patients with PS (34). We further observed that CD4 TEM cells and CD8 TEM cells predominant expressed CCR4 and CCR6 in PS (Figure 2B).
Th1, Th2, and Th17 cell and Treg populations have been identified as major players in determining the type of immune response in PS (10, 30). Therefore, the identification of these subsets, combined with expanded phenotyping for Tfh, Tc1, Tc2, and Tc17 cells, was performed on gated T cells (Figure 3A). Consistent with previous studies (8, 30), our data showed that levels of circulating Th1 and Th17 cells were nonsignificantly increased in PS. The lack of statistical significance may be explained by the limited sample size, as the variances between individuals were clearly demonstrated. Moreover, we also identified markedly elevated levels of circulating Tfh, Tc1, and Tc17 cells in PS, which were validated in an independent cohort of patients with PS (Figure 6A). These findings of abundant subpopulations in PS identified abnormal CD8 and CD4 autoreactive T cells levels in PS, which provided an opportunity to explore therapeutic targets and develop personalized and combined medicines.
However, precisely how these various subpopulations of lymphocytes affect the immune system in patients with PS should be determined. Therefore, we used SPADE analysis to study the expression levels of mTOR, STAT5A/B, STAT3, JAK3, ERK1/2, NF-κB p65, and AKT1/2/3, which are key players in immune responses. Among these protein profiles, we found the elevated levels mTOR, STAT3, and JAK3 were mainly restrictedly to CD4 TEM cells, which play a vital role in PS (35). In the PS group, JAK3 levels were markedly higher in CD4 T cells, especially CD4 TEM cells, and Th2 cells and were higher than those in the HC group (Figure 3C). Tofacitinib, formerly designated CP-690,550, is the most widely studied JAK inhibitor in humans. It inhibits JAK3 and JAK1 and to a lesser extent JAK2. Importantly, tofacitinib is under investigation for the treatment of PS (11). Our study indicated that a more specific inhibitor of JAK3 might more beneficial to treat PS. Similarly, in patients with PS, higher levels of mTOR and STAT3 were also predominantly expressed in CD4 TEM cells, suggesting these proteins as potential therapeutic targets. STAT5A/B, AKT1/2/3, ERK, and NF-κB p65 might not be suitable for inhibition, because they were widely expressed in the inflammatory status of PS (Figure 3C). In the present study, for the first time to our knowledge, we visualize the 7 key signaling proteins in the PS immune system and precisely outlined their profiles in the 15 lymphocyte compartments. However, the phosphorylation status of these proteins will be determined in a future study.
To discover the complexity of immunology phenotypes in patients with PS, we use the CyTOF technique to achieve a system-level characterization. Our in-depth characterization of lymphocyte compartments revealed three new subsets that were present in higher amounts in the peripheral blood from patients with PS compared with that in the HC group: CXCR3+CCR5+CD8+ Tregs, CD3–CD4+LTi cells, and Tc17 cells (Figure 4, A and B). We also labeled 31 markers in the lesional skin of PS by IMC, which preliminarily confirmed the presence of the CD3–CD4+ subset and depicted the distribution and expression of key proteins related to signal transduction pathways, such JAK3, STAT5, and mTOR, in skin biopsy of PS (Figure 5). This was the first visualization of the compositions and protein profiles in those subclusters of the immune system in PS to our knowledge.
Furthermore, we focused on the potentially novel p2 population (Figure 6B), which resembled LTi cells, which are CD4+ but CD3– and maintain the integral organization of lymphoid tissue, but might also function as antigen-presenting cells in certain circumstances (36, 37). LTi cells are an innate lymphoid cell (ILC) subset that appears to be closely related to ILC3s (38), which were first identified as CD4+CD3− cells scattered among fetal and neonatal lymph nodes. In human tissue, LTi cells are characterized as CD7+CD161+CD4–CD94–RORγt+ (38). However, circulating CD3–CD4+ cells in the present study have not been reported in patients with PS before, and those cells are CD161–. This potentially novel population is different from the LTi cells in tissues and the CD3+CD4+ population of PS and HC (Figure 4D). Many inflammatory chemokine receptors, such as CCR4, CCR7, and CXCR3, are upregulated on CD3–CD4+ cells, and the IMC results confirmed those chemokines in the PS lesional skin (Figure 5). These data suggested that the involvement of CD3–CD4+ cells in PS might be guided by their responding chemokine ligands. Therefore, we validated the abundant CD3–CD4+ cells in the whole blood and skin lesion of another PS cohort (Figure 6, B–D). Our previous study (18) showed that FRA2 is an important transcription factor in the development of PS via its regulation of the expression of IL23R. FRA2 is known to regulate the production of inflammatory cytokines and thus may have an important role in PS pathogenesis (39). In addition, recent studies showed that OX40 or OX40L may be expressed on a number of other cell types that control of immune function (36). Notably, an anti-OX40 monoclonal antibody (KHK4083) has been administered to patients with PS in a phase I clinical trial with improved effects (15). Our study showed that CD3–CD4+ cells rarely expressed OX40L (Figure 4C). The OX40-OX40L interaction regulates T cell tolerance, peripheral T cell homeostasis, and T cell–mediated inflammatory diseases (14). Thus, we further explored OX40 and FRA2 within the abundant CD3–CD4+ cells.
The CD3–CD4+ cells had higher OX40 and lower FRA2 levels, when compared with those in the HC group (Figure 7, A and B), and there was a negative correlation between OX40 and FRA2 levels. We also confirmed the presence of these molecules in the lesional skin of patients with PS (Figure 7C). Similar to previous studies (15, 40), OX40 was highly expressed in the lesional skin of PS samples and anti-OX40 treatment in PS indicated an improvement in the PASI score (15). The limited amounts of CD3–CD4+ cells in peripheral blood were not enough to explore the mechanism. Therefore, the significantly lower expression of FRA2 at the protein and mRNA levels was confirmed using flow cytometry and qPCR in PBMCs from patients with PS (Figure 7D). These data indirectly support the view that the abnormal OX40 and FRA2 levels on CD3–CD4+ cells have an important role in PS.
OX40 mainly promotes the expansion and survival of effector T cells (14) and was likely to be predominantly expressed on activated T cells. Therefore, OX40-OX40L, as a checkpoint between adaptive and innate immunity, might be potential candidate for PS treatment. The OX40-OX40L interaction also controls the phosphorylation of PI3K and AKT, which are upstream activators of mTOR (41). OX40 also plays a key role in maintaining T cell proliferation and survival by inducing T cell memory formation (15). The low levels of CD3–CD4+ cells in circulation make it difficult to perform functional assays. Therefore, we used ELISA to test OX40L levels in PS serum, which showed an obviously higher level of OX40L in patients’ serum compared with that in HC serum (Figure 8A). The OX40+CD3–CD4+ cells might interact with high soluble OX40L in serum to mediate PS development. We next isolated PBMCs from PS and treated them with CVN (C32H22N6Na2O8S2), a specific inhibitor of the OX40-OX40L interaction (42). CVN has been confirmed in ex vivo T cell polarization assays, where it successfully mimicked the effects of an agonistic anti-OX40 antibody (43). After 36 hours of in vitro incubation, we observed reduced numbers of circulating CD3–CD4+ cells and increased FRA2 expression. In addition, the level of CD3–CD4+ cells positively correlated with the PASI score before CVN treatment. Thus, the CD3–CD4+ cells might be a novel biomarker for PS disease activity.
Our study contained some limitations. Mass cytometry and IMC are advanced techniques involving expensive reagents; however, they do allow simultaneous evaluation of multiple parameters. Similar to that during gene sequencing, we could obtain big data in one sample using 31 markers by mass cytometry. Therefore, we used small numbers of samples to initially screen the potential abnormality of the adaptive immune system in patients with PS. Much of the data based on the mass cytometry was descriptive, and the tendency for the increased expression of some molecules or subsets did not show statistical significance, which requires further validation. However, we ultimately observed certain obvious and potentially novel subsets based on these advanced techniques and bioinformatic analyses and validated the CD3–CD4+ subset in an independent cohort. To our knowledge this was the first comprehensive visualization of the adaptive immune system of PS, which identified 3 possibly new subsets, which should be validated in further studies.
Future studies involving the systematic collection of blood from patients with PS at different disease statuses, in larger multicenter cohorts, are needed to confirm the involvement of OX40-OX40L, Tfh cells, Tc17 cells, CXCR3+CD8+ Tregs, and CD3–CD4+ cells and to determine the underlying mechanisms involving these subpopulations. A comprehensive understanding of the nature of PS would help to develop more effective therapies.
Methods
Reagents.
Millipore filtered deionized water was used as sample carrier and to prepare chemicals. Cell staining buffer consisted of PBS with 0.5% (w/v) BSA and 0.02% (w/v) sodium azide. Unlabeled carrier protein–free antibodies and fluorescent antibodies for flow cytometry validation were purchased from the indicated vendors (Supplemental Table 1). In-house antibody conjugation with lanthanide isotopes was carried out using Maxpar X8 antibody labeling kits (Fluidigm Sciences), as per the manufacturer’s instructions. Antibodies staining panel used for mass cytometry are summarized in Supplemental Table 1.
Subjects.
The study was conducted according to the Declaration of Helsinki. Human peripheral blood specimens of PS patients with PS vulgaris (total n = 38) without any treatment at the disease onset were sampled from Renji Hospital. PASI was determined, which is a standard measure of PS clinical disease activity. The symptoms and biopsy results of PS patients were recorded (Supplemental Figure 2, A–C). Healthy donors (total n = 30) with were recruited from the Medical Examination Center of Renji Hospital. Patients characteristics are shown in Supplemental Table 2.
Sample processing and storage.
PBMCs were isolated from sodium heparin vacutainer blood samples by Ficoll-Paque PLUS (GE Healthcare Biosciences) density gradient separation within 4 hours of collection. According to the standard experimental protocol provided by Fluidigm company, the PBMC count of each sample was controlled to be approximately 1 × 106 to 3 × 106. During data acquiring on mass cytometry, the PBMC number in each sample was controlled to be 1.5 × 106. Cells were then stained with cisplatin for 5 minutes at 37°C for live-dead cell discrimination and processed further for long-term storage at –80°C until analysis, as described in ref. 44. Serum was isolated from blood by centrifuging the tubes at 300 g for 10 minutes and stored at –80˚C.
Barcoding, antibody staining, and data acquisition.
Mass tag barcoding (45) was performed according to the instructions. For antibody staining, pooled cells were first incubated with TruStain fcX (10 minutes, room temperature) for FC blocking, washed twice with Cell staining buffer, and then stained (30 minutes, room temperature) with a metal-tagged surface-stain antibody cocktail using predetermined concentrations detailed in Supplemental Table 1. Cells were washed twice and permeabilized using Intracellular Fixation & Permeabilization Buffer Set (eBiosciences) and then stained with intracellular antibodies (Supplemental Table 2). Cell pellets were then diluted in distilled water to the appropriate concentration to achieve an acquisition rate of 500 events/s on the CyTOF instrument (46). All other settings were either default settings or optimized with tuning solution, as instructed by DVS Sciences.
Data analysis.
Data collected from the Helios instrument in.fcs file format were normalized using Four Element EQ Beads in the CyTOF software as described in ref. 47 for intra- and interfile signal drift and then analyzed on Cytobank (https://www.cytobank.org/). Sequential gating (21) was performed to discern living intact singlets from debris, cell aggregates, and dead cells.
The SPADE algorithm (48) was performed on gated live single cells to generate immune landscape for each sample and each group. In SPADE, the color gradient indicates the median expression level of the chosen marker. The SPADE settings were as follows: 5 clustering markers (CD19, CD4, CD8a, CCR7, CD45RO), 200 nodes, and equal down sampling of 10,000 events for each sample. 5000 cells from each sample (after arcsine transformation) were chosen randomly and subjected to t-SNE dimension reduction with 12 surface markers to generate viSNE maps (49). Due to the large number of cells in a single t-SNE running, a fast Barnes-Hut t-SNE method was adopted, reducing dimension from 19 to 2. The 2D t-SNE maps were then fed to an automatic clustering algorithm ACCENSE (50).
Manual gating on bivariate plots and SPADE or viSNE maps allowed for identification of populations of interest on the basis of canonical marker expression patterns (21). The relative frequency and median marker expression levels (arcsine ratio) of each defined population were exported for further analysis and plotting. Heatmaps generated using MATLAB (MathWorks) were used to visualize median marker expression levels scaled to each marker for each population.
IMC.
To assess the spatial distribution of T cell subpopulations in skin tissues, IMC (Fluidigm) (51) was employed to analyze regions of interest of skin frozen section from PS patients. Tissue sections were collected and prepared as previously described (52). Briefly, the slides were firstly blocked with 3% BSA in PBS and then incubated with a mixture of metal-conjugated antibodies overnight at 4°C in a wet chamber, followed by washing twice with PBST for 10 minutes. The slides were then counterstained with freshly prepared Ir working solution for DNA staining. Images were acquired using the Hyperion Laser Scanning Module coupled to a Helios mass cytometer. An area of 400 × 5 pixels for each metal spot was ablated at a frequency of 200 Hz.
Flow cytometry validation.
Established panels (Supplemental Table 1) and protocols were used as described previously (53). Briefly, cells were blocked with Fc block in staining buffer for 10 minutes at room temperature, incubated with antibodies in staining buffer for 30 minutes at 4°C, and then washed twice with staining buffer. Samples were resuspended in 400 μl of 1% PFA in staining buffer for 10 minutes and washed with staining buffer. Then samples were resuspended in 300 μl PBS and analyzed by flow cytometry in an Aria analyzer (BD Biosciences). FlowJo software (Treestar Inc.) was used for data analysis. The analysis strategy for PBMCs by flow cytometry was provided in Supplemental Figure 3.
Immunofluorescence staining.
Fresh human skin samples were fixed in formalin and embedded in paraffin. Protocols were employed as described previously (54). Briefly, paraffin-embedded 5-μm sections from human PS skin were stained for immunological assessment of Fra2, OX40, CD3, or CD4 (Abcam) expression.
Rabbit/mouse anti-human antibody in conjugation with goat anti-rabbit FITC or goat anti-mouse PE (Abcam) were used. Negative controls were run simultaneously for all assays by omitting primary antibody. Stained sections were examined and photographed using a Carl Zeiss LSM 800 confocal microscope.
qPCR.
Protocols were employed as described previously (53). Total RNA from human PBMCs was isolated using TRIzol according to the manufacturer’s instructions (Invitrogen). An equal amount of total RNA from each individual sample was used as a template in cDNA preparation with Superscript II (Invitrogen). cDNA amplification was performed using the Brilliant SYBR Green QPCR Master Mix kit (Stratagene) according to the instructions recommended by the manufacturer. qPCR was performed on an Applied Biosystems Quantstudio 7 Flex (Applied Biosystems). Sequences of primers used in this study are shown in Supplemental Table 3. Calculations for relative mRNA transcript levels were performed using the comparative CT method (2–ΔΔT).
ELISAs.
According to the ELISA protocol, a microplate was coated with diluted capture antibody and incubated overnight at 4°C. After washing 3 times with 0.05% Tween-20 in PBS, the plate was blocked with 1% BSA in PBS at room temperature for 2 hours. The plate was then washed 3 times, and diluted samples and standards were then added in duplicate to the corresponding wells for 2 hours at room temperature. After washing, captured cytokines reacted with the detection antibody, and the plate was incubated 2 hours at room temperature. Streptavidin-HRP was then added to each well for 20 minutes at room temperature. Color was developed by incubating with substrate solution for 20 minutes at room temperature. The reaction was terminated with stop solution, and absorbance was read using a microplate reader set to 450 nm.
Cell isolation and culture.
PBMCs were separated from the peripheral blood of PS samples by density gradient centrifugation (GE Healthcare). Then, trypan blue exclusion demonstrated that the viability was >96% for all preparations. PBMCs were plated in 6-well plates and allowed to settle 30 minutes in a 5% carbon dioxide atmosphere at 37°C. PBMCs were then incubated with CVN (TCI Chemical) for 36 hours (42). After 36 hours culture, cells were collected for flow cytometry experiment.
Statistics.
Statistical analyses were done using GraphPad Prism v7. All the continuous variables were presented as mean ± SD and were analyzed by 2-tailed Student’s t test, unless specified otherwise. Correlation analysis was performed using the Pearson’s correlation. P < 0.05 was considered statistically significant.
Study approval.
The Renji Hospital, Shanghai Jiao Tong University School of Medicine, approved the acquisition of blood and tissue from patients evaluated. Written informed consent was obtained from patients prior to their enrollment in the study.
Author contributions
RG designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. TZ designed research studies, conducted experiments, acquired data, analyzed data, and provided reagents. XM conducted experiments and analyzed data. ZL conducted experiments and analyzed data. JL conducted experiments and provided reagents. YG acquired data and provided reagents. XL conducted experiments and analyzed data. YY conducted experiments and analyzed data. GZ conducted experiments and analyzed data. XD designed research studies, analyzed data, wrote the manuscript, and provided mentorship of TZ. XC designed research studies, analyzed data, and wrote the manuscript. LL designed research studies, analyzed data, wrote the manuscript, and provided mentorship of RG.
Supplementary Material
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFC0909002) and the National Science Foundation of China (grant 81373209). The authors would like to thank Bruce Kirkham (Guy’s & St Thomas’ NHS Foundation Trust, London, United Kingdom) for his guidance on the language and logic of this study.
Version 1. 02/12/2019
In-Press Preview
Version 2. 03/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(6):e125306. https://doi.org/10.1172/jci.insight.125306.
Contributor Information
Ruru Guo, Email: loisen@163.com.
Ting Zhang, Email: T.Zhang@sjtu.edu.cn.
Xinyu Meng, Email: mengxy17@foxmail.com.
Zhen Lin, Email: 281657520@qq.com.
Jinran Lin, Email: aminolin@163.com.
Yu Gong, Email: gongyu8521@sina.cn.
Xuesong Liu, Email: cedarsky@163.com.
Yuetian Yu, Email: fishyyt@sina.com.
Guilin Zhao, Email: 258047199@qq.com.
Xianting Ding, Email: dingxianting@sjtu.edu.cn.
Xiaoxiang Chen, Email: xiaoxiang0721@gmail.com.
Liangjing Lu, Email: lu_liangjing@163.com.
References
- 1.Elder JT, et al. Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol. 2010;130(5):1213–1226. doi: 10.1038/jid.2009.319. [DOI] [PubMed] [Google Scholar]
- 2.Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386(9997):983–994. doi: 10.1016/S0140-6736(14)61909-7. [DOI] [PubMed] [Google Scholar]
- 3.Golden JB, McCormick TS, Ward NL. IL-17 in psoriasis: implications for therapy and cardiovascular co-morbidities. Cytokine. 2013;62(2):195–201. doi: 10.1016/j.cyto.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361(5):496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 5.Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: A comprehensive review. J Autoimmun. 2015;64:66–73. doi: 10.1016/j.jaut.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad C, Gilliet M. Psoriasis: from pathogenesis to targeted therapies. Clin Rev Allergy Immunol. 2018;54(1):102–113. doi: 10.1007/s12016-018-8668-1. [DOI] [PubMed] [Google Scholar]
- 7.Reich K. The concept of psoriasis as a systemic inflammation: implications for disease management. J Eur Acad Dermatol Venereol. 2012;26(Suppl 2):3–11. doi: 10.1111/j.1468-3083.2011.04410.x. [DOI] [PubMed] [Google Scholar]
- 8.Diani M, Altomare G, Reali E. T helper cell subsets in clinical manifestations of psoriasis. J Immunol Res. 2016;2016:7692024. doi: 10.1155/2016/7692024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong AW, Voyles SV, Armstrong EJ, Fuller EN, Rutledge JC. A tale of two plaques: convergent mechanisms of T-cell-mediated inflammation in psoriasis and atherosclerosis. Exp Dermatol. 2011;20(7):544–549. doi: 10.1111/j.1600-0625.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 10.Bovenschen HJ, van de Kerkhof PC, van Erp PE, Woestenenk R, Joosten I, Koenen HJ. Foxp3+ regulatory T cells of psoriasis patients easily differentiate into IL-17A-producing cells and are found in lesional skin. J Invest Dermatol. 2011;131(9):1853–1860. doi: 10.1038/jid.2011.139. [DOI] [PubMed] [Google Scholar]
- 11.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36(4):542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354(6):610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 13.Webb GJ, Hirschfield GM, Lane PJ. OX40, OX40L and autoimmunity: a comprehensive review. Clin Rev Allergy Immunol. 2016;50(3):312–332. doi: 10.1007/s12016-015-8498-3. [DOI] [PubMed] [Google Scholar]
- 14.Ishii N, Takahashi T, Soroosh P, Sugamura K. OX40-OX40 ligand interaction in T-cell-mediated immunity and immunopathology. Adv Immunol. 2010;105:63–98. doi: 10.1016/S0065-2776(10)05003-0. [DOI] [PubMed] [Google Scholar]
- 15.Papp KA, Gooderham MJ, Girard G, Raman M, Strout V. Phase I randomized study of KHK4083, an anti-OX40 monoclonal antibody, in patients with mild to moderate plaque psoriasis. J Eur Acad Dermatol Venereol. 2017;31(8):1324–1332. doi: 10.1111/jdv.14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wurm S, et al. Terminal epidermal differentiation is regulated by the interaction of Fra-2/AP-1 with Ezh2 and ERK1/2. Genes Dev. 2015;29(2):144–156. doi: 10.1101/gad.249748.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr TM, Wheaton JD, Houtz GM, Ciofani M. JunB promotes Th17 cell identity and restrains alternative CD4+ T-cell programs during inflammation. Nat Commun. 2017;8(1):301. doi: 10.1038/s41467-017-00380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhen L, Ruru G, Liangjing L, Xiaoxiang C. The study of Fra2 in the pathology of psoriasis by regulating IL23R. J Diagnostics Concepts Practice. 2018;(3):254-259. http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CJFQ&dbname=CJFDLAST2018&filename=ZDLS201803006&v=MDMxMDFyQ1VSTE9mWk9kdEZ5RG5VN3pLUHluSGZiRzRIOW5Nckk5RllvUjhlWDFMdXhZUzdEaDFUM3FUcldNMUY= Accessed on March 12, 2019.
- 19.Eberl G. Immunity by equilibrium. Nat Rev Immunol. 2016;16(8):524–532. doi: 10.1038/nri.2016.75. [DOI] [PubMed] [Google Scholar]
- 20.Kunicki MA, Amaya Hernandez LC, Davis KL, Bacchetta R, Roncarolo MG. Identity and diversity of human peripheral Th and T regulatory cells defined by single-cell mass cytometry. J Immunol. 2018;200(1):336–346. doi: 10.4049/jimmunol.1701025. [DOI] [PubMed] [Google Scholar]
- 21.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165(4):780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths CEM, et al. The global state of psoriasis disease epidemiology: a workshop report. Br J Dermatol. 2017;177(1):e4–e7. doi: 10.1111/bjd.15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadhouders R, Lubberts E, Hendriks RW. A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J Autoimmun. 2018;87:1–15. doi: 10.1016/j.jaut.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efe C, Ozaslan E, Nasiroglu N, Tunca H, Purnak T, Altiparmak E. The development of autoimmune hepatitis and primary biliary cirrhosis overlap syndrome during the course of connective tissue diseases: report of three cases and review of the literature. Dig Dis Sci. 2010;55(8):2417–2421. doi: 10.1007/s10620-009-0996-9. [DOI] [PubMed] [Google Scholar]
- 27.Chang Q, Ornatsky OI, Siddiqui I, Loboda A, Baranov VI, Hedley DW. Imaging mass cytometry. Cytometry A. 2017;91(2):160–169. doi: 10.1002/cyto.a.23053. [DOI] [PubMed] [Google Scholar]
- 28.Povoleri GAM, et al. Human retinoic acid-regulated CD161+ regulatory T cells support wound repair in intestinal mucosa. Nat Immunol. 2018;19(12):1403–1414. doi: 10.1038/s41590-018-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kupetsky EA, Mathers AR, Ferris LK. Anti-cytokine therapy in the treatment of psoriasis. Cytokine. 2013;61(3):704–712. doi: 10.1016/j.cyto.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 30.Kagami S, Rizzo HL, Lee JJ, Koguchi Y, Blauvelt A. Circulating Th17, Th22, and Th1 cells are increased in psoriasis. J Invest Dermatol. 2010;130(5):1373–1383. doi: 10.1038/jid.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mease P. New pathways of treatment for psoriatic arthritis. Lancet. 2017;389(10086):2268–2270. doi: 10.1016/S0140-6736(17)31427-7. [DOI] [PubMed] [Google Scholar]
- 32.van Unen V, et al. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity. 2016;44(5):1227–1239. doi: 10.1016/j.immuni.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Sgambelluri F, et al. A role for CCR5(+)CD4 T cells in cutaneous psoriasis and for CD103(+) CCR4(+) CD8 Teff cells in the associated systemic inflammation. J Autoimmun. 2016;70:80–90. doi: 10.1016/j.jaut.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Iijima N, Iwasaki A. Tissue instruction for migration and retention of TRM cells. Trends Immunol. 2015;36(9):556–564. doi: 10.1016/j.it.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diani M, et al. Blood to skin recirculation of CD4+ memory T cells associates with cutaneous and systemic manifestations of psoriatic disease. Clin Immunol. 2017;180:84–94. doi: 10.1016/j.clim.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Croft M. Control of immunity by the TNFR-related molecule OX40 (CD134) Annu Rev Immunol. 2010;28:57–78. doi: 10.1146/annurev-immunol-030409-101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MY, et al. CD4(+)CD3(-) accessory cells costimulate primed CD4 T cells through OX40 and CD30 at sites where T cells collaborate with B cells. Immunity. 2003;18(5):643–654. doi: 10.1016/S1074-7613(03)00110-9. [DOI] [PubMed] [Google Scholar]
- 38.Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13(2):75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 39.Zenz R, et al. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res Ther. 2008;10(1):201. doi: 10.1186/ar2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumura Y, et al. Expression of CD134 and CD134 ligand in lesional and nonlesional psoriatic skin. Arch Dermatol Res. 2003;294(12):563–566. doi: 10.1007/s00403-002-0363-6. [DOI] [PubMed] [Google Scholar]
- 41.Song J, Salek-Ardakani S, Rogers PR, Cheng M, Van Parijs L, Croft M. The costimulation-regulated duration of PKB activation controls T cell longevity. Nat Immunol. 2004;5(2):150–158. doi: 10.1038/ni1030. [DOI] [PubMed] [Google Scholar]
- 42.Song Y, Margolles-Clark E, Bayer A, Buchwald P. Small-molecule modulators of the OX40-OX40 ligand co-stimulatory protein-protein interaction. Br J Pharmacol. 2014;171(21):4955–4969. doi: 10.1111/bph.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song Y. Small-Molecule Probes for the Modulation of Ligand-Receptor Interactions within the TNF Superfamily. Scholarly Repository. University of Miami. https://scholarlyrepository.miami.edu/oa_dissertations/1398/ Published April 29, 2015. Accessed February 25, 2019.
- 44.Brodie TM, Tosevski V. High-dimensional single-cell analysis with mass cytometry. Curr Protoc Immunol. 2017;118:5.11.1–5.11.25. doi: 10.1002/cpim.31. [DOI] [PubMed] [Google Scholar]
- 45.Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81(16):6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 47.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83(5):483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anchang B, et al. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protoc. 2016;11(7):1264–1279. doi: 10.1038/nprot.2016.066. [DOI] [PubMed] [Google Scholar]
- 49.Amir el-AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proc Natl Acad Sci U S A. 2014;111(1):202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 52.Chang Q, Ornatsky O, Hedley D. Staining of frozen and formalin-fixed, paraffin-embedded tissues with metal-labeled antibodies for imaging mass cytometry analysis. Curr Protoc Cytom. 2017;82:12.47.1–12.47.8. doi: 10.1002/cpcy.29. [DOI] [PubMed] [Google Scholar]
- 53.Guo R, et al. A role for receptor-interacting protein kinase-1 in neutrophil extracellular trap formation in patients with systemic lupus erythematosus: a preliminary study. Cell Physiol Biochem. 2018;45(6):2317–2328. doi: 10.1159/000488179. [DOI] [PubMed] [Google Scholar]
- 54.Li H, et al. Cyr61/CCN1 induces CCL20 production by keratinocyte via activating p38 and JNK/AP-1 pathway in psoriasis. J Dermatol Sci. 2017;88(1):46–56. doi: 10.1016/j.jdermsci.2017.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.