Abstract
Hepatocellular carcinoma (HCC) is the world’s second leading cause of cancer death; 82.4% of patients die within 5 years. This grim prognosis is the consequence of a lack of effective early detection tools, limited treatment options, and the high frequency of HCC recurrence. Advances in the field of liquid biopsy hold great promise in improving early detection of HCC, advancing patient prognosis, and ultimately increasing the survival rate. In an effort to address the current challenges HCC screening and management, several studies have identified and evaluated liver–cancer-associated molecular signatures such as genetic alterations, methylation, and noncoding RNA expression in the form of circulating biomarkers in body fluids and circulating tumor cells of HCC patients. In this review, we summarize recent progress in HCC liquid biopsy, organized by the intended clinical application of the reported study.
INTRODUCTION
Cancer is one of the most challenging health care problems of our times, the second leading cause of death globally1 and in the United States.2,3 The number of new cases is expected to rise by about 70% over the next two decades.1 Technological advances have enhanced our understanding of carcinogenesis from a histo-pathological definition of tumorous cells to a molecular characterization as a disease of the genome and epigenome. This fundamental genetic understanding of cancer reveals its high heterogeneity and evolutionary dynamic (tumor evolution) as a function of time,4,5 particularly in response to treatment, thus not only highlighting the importance of genetics-based tumor analysis for precise management of cancer, but also providing the potential for patient selection in targeted drug development.
Liquid biopsy refers to the less- or noninvasive tests performed on blood or other body fluids, as opposed to surgical tumor biopsy, that provide genetic information about a patient’s tumor.6 The source of tumor material for liquid biopsy encompasses circulating cell-free tumor DNA (ctDNA), circulating tumor cells (CTC), and circulating exosomes from body fluids such as serum, plasma, urine, saliva, etc.
Among different cancer types, liver cancer, with hepatocellular carcinoma (HCC) as its primary form, is the second most common cause of cancer-related deaths worldwide and one of the fastest growing cancers in the United States.1,2,7 The high mortality rate of HCC is attributable to lack of effective early detection tools, limited treatment options, and high-frequency recurrence.8–10 Although liquid biopsies have shown promising applications already in clinic for several cancers such as colorectal carcinoma, breast cancer, and lung cancer11–16 in predicting response to therapy and monitoring relapse, their relevance in clinical application for liver cancer is limited. Due to the shedding of ctDNA into circulation by the microcirculation of discontinuous sinusoids (fenestrated capillaries with intercellular gaps and a fragmented basement membrane) in the liver, liver cancer should be particularly suitable for liquid biopsy for cancer genetics for precision medicine once more treatment options become available and for drug development. In the current review, we summarize recent developments in the field of liquid biopsy of HCC, organized by the intended clinical application of the reported study.
Screening and early diagnosis.
Early detection is critical for the effective treatment of HCC.7,9,10,17,18 Despite HCC surveillance programs for the high-risk population and well known, identifiable HCC-associated risk groups, such as those with chronic hepatitis B or C viral infection, alcoholism, or fatty liver disease, most HCC remains undetected until late stages (the 5-year survival rate remains less than a mere 10%)2,7,19–22 due to the lack of a sensitive and convenient screening method. The current, most used marker, serum alfa-fetoprotein (AFP), and its fucosylated glycoform, L3, are of limited value because of an overall sensitivity of only 50%. The remaining ~50% of HCC cases are considered to be AFP-negative HCC (less than 20 ng/mL, as suggested by American Association for the Study of Liver Diseases (AASLD) guideline for HCC screening9). Thus, there is an urgent need for a test that can be effective either alone or in combination with serum AFP to improve the early detection of HCC. Recent studies have suggested that liquid biopsy-based screening tests can be a promising and attractive option to detect cancer-causing genetic alterations.
Cell-free DNA integrity and quantification.
Although cell-free DNA (cfDNA) integrity as a marker for detection of HCC has been evaluated by number of studies,23–25 the results have not been conclusive. These studies generally evaluated DNA integrity by qPCR analysis of two differently sized amplicons of a gene of interest, such as Alu repeat or beta-actin. A recent study by the Dennis Lo group26 has taken a more comprehensive approach: using massive parallel sequencing to evaluate the size profile of plasma cfDNA and analyzing the z-score of the chromosome-arm level to distinguish tumor-derived DNA from the nontumoral DNA. Their analysis provides evidence that the tumor-derived DNA was shorter than the nontumor-derived DNA. Huang et al27 have also reported that cfDNA is more fragmentized in HCC patients than in patients with benign liver diseases and healthy individuals. It was also observed that this decrease in cfDNA integrity is reversed back to normal after resection of the HCC tumor. Interestingly, this shortened cfDNA feature has been reported for other cancers as well.28–31 Given the generalized nature of this marker quantification, more studies are needed to evaluate the value of cfDNA integrity decrease as a marker for HCC diagnosis or for cancer in general. Despite intriguing observation and the ease of performing this assay, the size and quantity of cfDNA as a marker for HCC and/or other cancers has fallen short of the desired specificity for cancer diagnosis.32–34
Somatic mutations.
The mutational landscape of HCC has been well characterized by multiple whole genome sequencing and whole exome sequencing based studies of tumor tissue.35–39 Briefly, TERT promoter activating mutations, found in 40%−60% HCC, and the mutually exclusive mutations of TP53 and CTNNB1 genes, together found in 30%−50% of HCC cases, are the three most frequently reported mutational events in HCC.35–39 Additional significantly mutated genes include the tumor suppressor genes AXIN1 (~8%) and RB1 (~4%), which were inactivated by mutation, and the chromatin remodeling genes ARID1A (~7%), ARID2 (~5%), and BAP1 (~5%). NFE2L2 and its interactor KEAP1, which are both instrumental in cellular antioxidant defenses, were also significantly mutated in ~3% and ~5% of HCC, respectively. Albumin (ALB) and APOB mutations were observed in ~13% and ~10% of tumors.
Among the dozens of reported mutations from tissue studies, the TP53 p.R249S, c.747G>T mutation is the most frequently reported HCC-associated somatic mutation in the peripheral body fluids, including serum, plasma, and urine, of HCC subjects.40–47 Although TP53 mutations are reported in almost all cancer types, the mutation at codon 249 of the TP53 gene is known to be highly specific for HCC. In case-control studies evaluating the performance of this marker as a tool for detection of HCC, a sensitivity of 15%−47% at a specificity of 46%−86% was obtained in plasma,44–46,48–55 as compared with a sensitivity of 4%–18% at a specificity of 83.3% in serum42,56,57 and 53% sensitivity at 75% specificity in the urine.41 This mutation is known to be more common in HCC subjects residing in regions with high prevalence of chronic hepatitis B infection and dietary aflatoxin exposure. Recent studies have reported the detection of TERT promoter and CTNNB1 mutation in the body fluids of HCC patients using digital PCR and Next generation sequencing (NGS) technologies.58–60 However, to the best of our knowledge, despite its high frequency found in HCC tumor, the TERT promoter mutation has also been reported in ~10% of cirrhosis cases, thus diminishing its specificity as a HCC biomarker.61–63 This could explain, at least in part, why TERT promoter mutations have not yet been reported as a biomarker for the early detection of HCC.
A recent experimental blood test called Cancer-SEEK, which combines genetic alterations and protein biomarkers in circulation for early detection of cancer, was able to detect liver cancer with an overall sensitivity of about 98% with 99% specificity in a case-control study with 44 HCC and 812 normal healthy controls.64 Encouragingly, the plasma of all 44 liver cancer cases was found to contain detectable level of at least one of the 16 gene mutations analyzed, suggesting the sufficient quantity of HCC ctDNA for sensitive genetic liquid biopsy. Of 44 HCC, TP53 mutation was detected in 59.09% (26/44) of plasma samples and CTNNB1 mutation was detected in 18.18% (8/44) of plasma samples. As anticipated, the detections of TP53 and CTNNB1 were in a mutually exclusive fashion, as previously suggested.36, 65 Mutations in CDKN2A (2/44), GNAS (¼4), KRAS (2/44), PI3KCA (2/44), and PTEN (3/4) genes were also detected in the HCC plasma. It is of note that the TP53 p.R249S, c.747G>T mutation was identified in 20.45% (9/44) HCC plasma samples, 3.22% (3/93) pancreatic cancer, 4.41% (3/68) stomach cancer, and none of the 812 healthy controls. The sensitivity of CancerSEEK was highest for liver cancer (100%) among the various stage 1 cancers evaluated. Although the results of this study demonstrate the promising future of HCC genetic liquid biopsy using ctDNA, as acknowledged by the authors, the high sensitivity of HCC detection would be compromised by specificity, since the controls in this study were limited to healthy individuals. Some of the markers included in the panel are reportedly related to inflammation and other diseases such as hepatitis and cirrhosis. As a result, false positives for HCC may be concerned when CancerSEEK blood test is implemented for HCC screening from at-risk population.
There are a few technical challenges that need to be overcome in order to develop somatic mutations for a liquid biopsy for HCC screening and early detection. These include development of sophisticated technology that is capable of detecting such rare mutations (up to 0.01%) in a background of normal DNA. Also, mutations vary based on the etiological factor that is responsible for hepatocarcinogenesis. Given that HCC has multifactorial risk factors, such as viral infections, alcoholic liver disease, nonalcoholic fatty liver disease, etc., a panel that integrates multiple genes and multiple locations within a given gene will be needed to optimize performance. Additionally, combining genetic alterations with either DNA methylation markers and/ or circulating RNA-based markers and/or protein markers could also be a possibility.
DNA methylation.
Studies have indicated that DNA methylation of tumor suppressor genes such as CDKN2A, RASSF1A, and GSTP1 are early events of carcinogenesis and hence are promising markers for developing an HCC screening test,66–69 in which sensitivity is more important than specificity, to identify positive patients for more sophisticated imaging diagnosis. Similar to cancer-associated genetic mutations, cancer-related methylation events can also be detected in the circulation of the patients with cancer, including HCC.53,70–72 In fact, the first blood-based colorectal screening test that has been approved by the United States Food and Drug Administration (USFDA), Epi proColon, is a test for methylated SEPT9 DNA in plasma.73 Several studies have reported on the performance of methylated markers, both individually and in panels, for the diagnosis of HCC.74–76
Xu et al77 have developed a new diagnostic and prognostic blood test for early detection of HCC. An HCC-specific methylation marker panel was identified by comparing HCC tissue and normal peripheral blood mononuclear cells (PBMC) methylation to generate a diagnostic prediction model and was tested using cfDNA samples from plasma of 1098 HCC patients and 835 normal controls. Application of this model yielded a sensitivity of 85.7% and specificity of 94.3% for HCC in a training data set of 715 HCC and 560 normal samples and a sensitivity of 83.3% and specificity of 90.5% in a validation data set of 383 HCC and 275 normal samples. The combined diagnostic score of the model was able to differentiate liver diseases (Hepatitis B virus (HBV) and/or HCV infection, fatty liver) and HCC similar to normal healthy controls. Similar to CancerSEEK, no cirrhosis controls were used in this study, raising concern about the specificity of the test performance.
HCC is one of the few cancers with an identified high-risk population and an implemented surveillance program.10 Given its incidence, a general population screening is not warranted. Hence, HCC screening performance must be evaluated against this HCC high-risk population as controls instead of healthy controls. This includes patients with hepatitis B, and cirrhosis of any origin. This is especially true in the case of methylation biomarkers since they represent very early events of carcinogenesis and are often detectable in precancerous conditions such as liver cirrhosis.67 For example, Dong et al78 reported the detection of methylated RASSF1A in the serum of 52.04% (51/75) of patients with HBV-HCC, 13.33% (10/75) with liver cirrhosis, 4.44% (4/90) with chronic hepatitis B, and 3.75% (3/80) healthy controls. Similar trends were also seen in the methylated APC, BVES, TIMP3, GSTP1, and HOXA9 genes that were also evaluated in this study as a multigene panel for HCC diagnosis. In another study, methylated RASSF1A was detected in 10% of the healthy controls (2/20), 62.5% of the hepatitis C group (25/40), and in 90% of the HCC group (36/40).79 A cut-off determination comparing the quantity of methylation between HCC and at-risk populations could be one approach to include these methylation markers for HCC screening.69 The challenges that researchers face in translating methylation markers from the bench to the bedside include the impact of the HCC-specific methylation site locations, nontumor-associated methylation, technology-related obstacles as discussed in this review article,69 and the inclusion of age-matched high-risk groups, such as patients with cirrhosis, in the study design for marker development since, DNA methylation is an early event in tumorigenesis and is related to age as well.
Circulating tumor cells.
One main pathway for metastasis is through tumor cell in the systemic circulation and similar to other solid tumors,80,81 CTC positivity has been shown to a promising tool for HCC diagnosis.82 It is estimated that approximately 106 cells/g of tumor are shed into the circulation each day; however, the short half-life of CTCs results in approximately 1 CTC in 1 billion blood cells being present in the circulation at a given time.83 Various techniques have been developed to detect these rare CTCs, based either on the physical (size, density, or charge) or cell surface expression, properties of CTCs. To separate the enriched CTCs with leukocytes, specific antibody-based enrichment techniques have been incorporated such immunomagnetic bead separation using epithelial surface antigen markers such as Epithelial cell adhesion molecule (EpCAM).84 However, only about 35% of HCC cases express EpCAM. Other methods that are independent of epithelial antigen expression have been studied, including size-based filtration method, flow cytometry,82 and RT-PCR-based HCC-specific RNA quantification85 or sequencing for identification of HCC mutations.86 A recent meta-analysis also demonstrated a more robust diagnosis accuracy for HCC with nonmagnetic isolation methods over magnetic methods dependent on epithelial antigen expression. Based on data of 20 studies on CTCs for HCC with 998 eligible study subjects, a pooled sensitivity of CTC detection was 67% at a pooled specificity of 98% with liver and tumorous disease and 100% with healthy controls.87
Several new developments that combine genomic analysis to CTC enrichment have improved the specificity of CTC detection. Kalinich et al used a microfluidic chip device (iChip), which depletes hematopoietic cells from blood by size-based exclusion of red blood cells, platelets, and plasma, followed by magnetic deflection of white blood cells, and combined with RNA-based digital PCR to detect CTC-derived signatures.88 Based on this test, 56% (9 out of 16) untreated HCC cases had detectable CTCs including early stage HCC. It also showed much lower detection in patients with “no-evidence of disease” after curative-intent treatment, demonstrating a high degree of specificity of the test. Guo et al85 obtained a sensitivity of 72.5% at 95% specificity by identifying the subpopulations of CTC with stem cell phenotypes and constructing a qRT-PCR-based RNA marker diagnostic CTC panel in a well-designed training (200 HCC, 101 CHB/LC and 100 healthy) and validation sets (195 HCC, 100 CHB/LC, 100 benign liver lesions, and 110 healthy). The panel was also accurate for early stage and AFP-negative HCC.
Circulating RNA.
Several microRNAs (miRNA) and long noncoding RNAs (lncRNA) have been reported as potential biomarkers for liver cancer with great promise. MicroRNAs are small noncoding RNAs involved in gene expression. Specific miRNAs, such as miR-21, miR-200a, miR-122, miR-223, let-7f, miR-155, etc., have been demonstrated to be associated with HCC.75 MicroRNAs are released in the circulation by either cell lysis or secretion. In the plasma, microRNAs bind to certain proteins such as Argonaute 2 and high-density lipoprotein or are packaged into exosomes, which protect them from degradation by RNAse in the circulation.89 Circulating miRNAs have been extensively investigated for their potential as biomarkers for HCC detection, both individually and in combination as a panel.75,76 Also, similar to methylated DNA markers, some miRNAs had a lower specificity for HCC when comparing HCC in chronic liver disease populations tothat of normal healthy controls.76,90–94 A panel of miR-122 and let-7,95 panel of miR-122, miR-885–5p, miR-221, and miR-22 with AFP,96 miR-143 and miR-224 with AFP97 have been reported to have a good sensitivity for HCC diagnosis.95 A meta-analysis of 24 studies by Ding et al found expression levels of miR-21, miR-122, and miR-192 to be highly selective for HCC diagnosis.98 Some miRNAs such as miR-16, and miR30e and miR223 were found to be downregulated in the serum of HCC patients as compared with chronic liver diseases and healthy controls.99,100 Although promising, most of these miRNA biomarkers are in their early phases and need to be thoroughly evaluated in the five-phase format recommended by the Early Detection Research Network101 for HCC diagnosis.
lncRNAs are greater than 200 bp transcripts that are not translated. Their expression has been involved in the regulation of multiple carcinogenic processes such as proliferation, apoptosis, invasion, and metastasis. Similar to other biomarkers, the lncRNAs provide the best accuracy when combined in a panel of lncRNA markers with either other microRNAs and/or AFP due to the high heterogeneity of HCC. For instance, HULC and Linc00152 have been shown to be associated with HCC in a case-control study (66 HCC, 32 chronic hepatitis, and 53 healthy controls).102 In this study, lncRNAs LINC00152, RP11–160H22.5, and XLOC014172 along with serum AFP had a promising performance (AUROC of 0.985 and 0.986) discriminating HCC (n = 100) development from both cirrhotic (n= 100) and healthy patients (n= 100), respectively. Four RNA-based biomarker panels [lncRNA-C terminal binding protein, androgen responsive (lncRNACTBP), microRNA-16–2 (miR-16–2), microRNA-21–5-P (miR-21–5p), and LAMP2] had a positive predictive value of 87% and a negative predictive value of 80% in a validation study of 100 HCC and 100 chronic hepatitis patients.103 Similar to miRNA markers, most of these markers, although promising, are in need for a thorough development for HCC diagnosis.
Development of multimarker models and/or algorithms for hepatocellular carcinoma prediction.
As discussed above, a panel of multiple biomarkers derived from different cancer pathways is needed for an HCC screening test to attain sufficient sensitivity and robustness and104–106 to overcome cancer’s high heterogeneity. Combining data from several different types of liquid biopsies (CTCs, other types of circulating cells, ctDNA or tumor-derived extracellular vesicles) or other forms of biomarkers (proteins, miRNAs, metabolites, etc.) may provide complementary information, resulting in more accurate and sensitive early detection methods. To analyze multiple variables and generate algorithms for classification, many different multivariate models can be applied (eg k-nearest neighbor and Bayesian classifiers, etc.107,108 Among these, logistic regression (LR) is most commonly used, and classification and regression trees (CART) have also become popular. However, LR and CART may lack the robustness necessary to serve as effective algorithms for cancer screening because of increasing numbers of variables. In addition, biomarkers maybe also needed to tailor these models for various etiologies. Machine learning techniques have recently been used in the field of classification, showing promise in predictive accuracy and robustness in various heterogeneous classification settings, for example, the human gut microbiome and detection of cancers such as ovarian, lung, and breast.109,110
We, Wang et al, had applied the machine learning algorithm, random forest (RF), and proposed the novel statistical algorithms fixed sequential (FS) and two-step (TS) for biomarker development for HCC screening.111 These two novel statistical algorithm, compared with both the commonly used multivariate techniques LR and CART, performed significant better in both sensitivity and robustness, as models for the development of HCC screening test using multiple biomarkers. The two models FS and TS using RF machine learning techniques provided a substantial improvement in performance over the commonly used models LR and CART within the iterative crossvalidation experiment. Various other machine learning techniques such as neural networks and support vector machines112 have also been employed in cancer biomarker development. It would be of interest to examine the performance of such techniques for HCC screening.
Liquid biopsy for hepatocellular carcinoma management.
Diagnosis of HCC is usually confirmed by radiology such as multiphasic computed tomography (CTscan) or magnetic resonance imaging (MRI) with agents that characterize the blood flow in the liver. Tissue biopsy, if performed, has value only if it is positive for HCC. It is not routinely performed because of the risk of bleeding, tumor seeding, and inability to rule out HCC in the event of a negative biopsy.10 Liquid biopsy of HCC ctDNA can be a superior alternative because it can provide confirmation of HCC diagnosis in such indeterminate cases, as opposed to the “wait and watch” approach that is currently been employed for such early stage HCC. It also provides HCC genetics for precision medicine when treatment options for targeted therapy become available. Current treatment options for HCC are broadly classified into two categories: either curative (surgical resection or liver transplantation) or palliative, based on the size and number of the tumor nodules, assessment of the liver function based on the Child Pugh score. Intermediate stage tumors are treated by transcatheter arterial chemoembolization (TACE) and advanced stage tumors are currently treated with multikinase inhibitors such as Sorafenib or regorafenib and immunotherapy with Nivolumanb.113 At the present time, no liquid biopsy or genetic markers have been applied to the decision-making of any of the current treatment plans. Recent progress in the applications of ctDNA in HCC precision medicine is discussed in more detail in companion diagnostic tool for hepatocellular carcinoma-targeted therapy section, which covers recent progress in the development of the companion diagnosis tool for HCC target therapy and immune-therapy below.
As mentioned earlier, in addition to difficulties in early detection and limited treatment options, a high recurrence rate also contributes to the high mortality rate of HCC. Rates of recurrence range from 15% for transplant to near 100% for surgery or ablation.17,114–122 Recurrence is most common within 2 years of treatment. The high HCC recurrence rate is attributed to (1) incomplete treatment, (2) micrometastases within the liver, and (3) de novo lesions.117 Currently, HCC recurrence is monitored by serum AFP or other serum proteins and serial imaging. Notably, there are no specific guidelines addressing how HCC recurrence should be monitored. This is likely due to the limited sensitivity of the available methods. MRI and/or CT imaging is the gold standard for diagnosis, although it is expensive and has limited utility in the detection of small tumors (<2 cm) and tumors in the presence of previously treated lesions (especially from local ablation), cirrhosis, obesity, and dysplastic nodules.121–123 Liquid biopsy-based methods, on the other hand, are unaffected by the abovementioned limitations of imaging, and can thus be a great option for evaluating the response to treatment and monitoring of HCC recurrence.
Somatic mutations and methylation markers for recurrence and prognosis.
In additional to its potential in the early detection, ctDNA biomarkers have also been reported as good prognosis markers and indicators of HCC progression. RASSF1A methylation levels in plasma have been shown to be a prognostic factor for overall survival,71 and can be taken into account with tumor size68 and LINE-1 hypomethylation to correlate early recurrence and poor prognosis in sera of patients after curative resection.124 The combined diagnostic score based on 10 selected methylation markers developed by Xu et al77 associated with tumor stage, tumor burden, detectable residual tumor post-treatment, disease progression, and development of HCC recurrence. Wong et al125 demonstrated association between p15 methylation and liver cancer recurrence or metastasis.
Recent strategies have focused on expanding the panel of cfDNA alterations for detection, assessment of HCC molecular information, intratumoral heterogeneity and monitoring of HCC recurrence. Cai et al126 extensively analyzed the mutation profiles of 574 cancer-related genes known to harbor actionable mutations from the plasma of four HCC patients. The circulating DNA captured more than 98% of subclonal mutations detected in the matching tissue. Circulating levels of 61.64%–94.12% of subclonal mutations correlated to patients’ tumor burden in samples collected at preoperation, postoperation, and follow-up at recurrence time points. In this study, one of the patients displayed increasing circulating levels of somatic mutations prior to imaging diagnosis and increase of AFP levels, suggesting the promising utility of liquid biopsy as a tool for monitoring recurrence. This elevation of DNA markers in cfDNA before MRI imaging diagnosis was also observed in a pilot study (n= 10) by Hann et al, which is the first study to combine both mutational and methylation markers in one panel.40 In this study, urine samples were collected prospectively from HCC patients (when available) after curative treatment at follow-up visits. Five patients developed recurrence during the study. The samples were retrospectively analyzed in a blinded fashion for HCC DNA bio-markers (TP53 R249S mutation, methylation of RASSF1A, and GSTP1). These markers were elevated in the urine of four patients up to 9 months before or at the time of diagnosis of HCC recurrence by MRI imaging. Although MRI and/or CT imaging is the current gold standard for diagnosis of recurrent HCC, it has difficulty in detecting early recurrence in the previously treated areas (especially after local ablation). This study demonstrates not only the efficacy of using ctDNA for monitoring HCC recurrence but also the applications of urine as a noninvasive body fluid for HCC liquid biopsy.
Circulating RNA.
Kim et al127 recently demonstrated that combination of plasma miR-21, −26a, and −29a-3p expression could predict early TACE refractoriness in patients (n = 198) with history of TACE-treated HCC. Interestingly, Lu et al identified hypermethylated regions encoding for miRNAs in the plasma of HCC subjects and demonstrated their diagnostic and prognostic potential.128 Increased expression of serum let-7f has been shown to correlate with tumor size (>5 cm) and with early HCC recurrence.129 HCC-associated circulating miRNAs levels of miR-224 and miR-500 decreased following surgery thus reflecting tumor dynamics.76,130 Conversely, circulating miR-30e, miR223, and miR-125a-5p are downregulated in HCC patients.99, 131 Lnc00974, lncRNA MALAT1, and SPRY-IT1 have been shown to be detectable in the circulation of HCC patients, and their levels correlate with disease severity and prognosis.132–134
Circulating tumor cells.
Several studies have shown that CTCs are significantly associated with HCC recurrence and poor prognosis. Fan et al first reported using CTC with stem-like characteristics (CD90+ and CD44+) as a prognostic marker for HCC recurrence after hepatectomy.135,136 Using a qRT-PCR-based RNA platform for CTC detection has also demonstrated the use of CTCs not only as a diagnostic test but also as risk prediction tools for HCC recurrence after surgery.85 In this multicenter cohort study, patients with persistently high CTC load had a higher propensity for recurrence. This panel incorporated EpCAM, CD90, CD133, and CK19, which had better correlation to prognosis than AFP or EpCAM alone. While these have been largely focused on Asian population with chronic HBV and cirrhosis, findings are promising for the use of CTCs for disease prognostication of HCC. Further studies with incorporation of patients from other geographic areas, with various etiology of HCC including cirrhosis from nonalcoholic fatty liver disease or aflatoxin exposure should be explored.
Similarly, accumulating evidence shows that a subset of CTCs has an epithelial-mesenchymal transition phenotype that is associated with more metastaticspread by vascular invasion137–139 One study evaluated 46 patients with HCC in which CTCs with mesenchymal features with twist and vimentin expression were detected in 39 (84.8%) and 37 (80.4%) patients, respectively. This significantly correlated with portal vein tumor thrombus.140 There are also emerging data on heterogeneity of the epithelial-mesenchymal transition status in CTCs across different vascular compartments of the circulation.141 While studies need more validation with larger patient cohort, there’s a promising evidence for use of CTCs for disease progression in HCC. The challenges in bringing CTCs for disease management are lacking of a technology platform that is tailored for capture HCC CTC.
Companion diagnostic tool for hepatocellular carcinoma-targeted therapy.
Currently, no targeted therapy has been approved for liver cancer. As a result, no companion test is needed for HCC treatment. As our understanding of the HCC genetic landscape and HCC drivers increases, precision medicine for liver cancer could become feasible in the near future.
Table 1 summarizes the potential actionable (drug-gable) genetic biomarkers and the status of drug development for HCC therapy. For instance, at least one potentially druggable mutation was identified in 88.5% (23/26) of HCC patients in a recent study that investigated ctDNA analysis in advanced HCC patients.142 Drugs targeting the TERT pathway, either by telomerase enzyme inhibition or telomerase active immunotherapy (GX301, Imetelstat, and GV1001), and the Wnt and/or CTNNB1 pathway (pRI-724 and XAV939) are currently in various stages of clinical development, ranging Ph I–III studies for non-HCC cancers.143 TP53 genomic alterations correlate with increased VEGF-A expression and such tumors can potentially be targeted by antiangiogenic drugs such as Bevacizumab.144 A retrospective study suggests that patients with TP53 mutations had longer progression free survival with bevacizumab-containing therapies when compared with nonbevacizumab containing regimen (median 11.0 vs 4.0 months [P < 0.0001]).145 Another report indicates that TP53 mutations predict sensitivity to VEGF and/or VEGFR inhibitors and are associated with improvement of the disease outcome.146 Finally, TP53 mutations have been associated with better outcomes in sarcoma patients treated with the VEGFR inhibitor pazopanib.147 TP53 may also be targetable by WEE1 inhibitors.148 Other genomic alterations with possible targeted treatments include ARID1, BRAF, CCNE1, CDK4, CDK6, CDKN2A, CTNNB1, EGFR, ERBB2, FGFR1, KRAS, MET, MYC, PI3KCA, and PTEN.142 An overview of the current research on HCC-related biomarker selection and potential future personalized drug testing for HCC has been discussed in detail in recent review articles.143,149
Table 1.
Summary of potentially targetable genetic biomarkers for HCC treatment and the stage in development of their corresponding drugs
| Pathway | Candidate marker | Drug | Drug mechanism | Status |
|---|---|---|---|---|
| Telomerase maintenance | TERT | GX301 | hTERT multipeptide therapeutic vaccine | Ph II, prostate cancer |
| Imetelstat | Antitelomerase antisense oligonucleotides | Ph I-III, hepatoblastoma, myelodysplastic syndrome, breast and nonsmall cell lung cancer, etc. | ||
| GV1001 | Telomerase-derived anticancer pep-tide vaccine | Ph II, HCC; Ph III, pancreatic cancer | ||
| Wnt pathway | CTNNB1 | PRI-724 | beta-catenin antagonist | Ph I, HCV cirrhosis; Ph I-III, pancreatic, colorec-tal, and myeloid cancers |
| AXIN1 | XAV939 Galunisertib | Wnt inhibitor TGF-β inhibitor | Preclinical Ph I-II, HCC; PhI-II, pancreatic, prostate, breast, ovarian, etc., cancers | |
| Cell cycle | TP53 | Bevacizumab | Recombinant humanized monoclonal antibody that blocks angiogenesis by inhibiting VEGFA | PH I-III, HCC; FDA approved, colon, non-small cell lung, glioblastoma multiforme, ovarian, and cervical cancers |
| Pazopanib | Multitargeted receptor tyrosine kinase inhibitor | Ph I-III, HCC; FDA-approved (Renal cell carcinoma and Sarcoma) | ||
| CCND1 | ON 013105 | Cyclin D modulator | Ph I, Lymphoma, ALL and advanced solid tumors | |
| RBI | Palbociclib | CDK4 and 6 inhibitor | Ph II, HCC, FDA approved for breast cancer | |
| CDKN2a | llorasertib | Aurora and VEGFR inhibitor | ph I in CDKN2a deficient solid tumors | |
| Chromatin remodeling | ARID1 | GSK126 | EZH2 methyltransferase inhibitor | Preclinical |
| MAP kinase | FAK | GSK2256098 | FAK inhibitor | Ph I-II, meningioma and pancreatic cancer |
| RPS6KA3 | Refametinib | MEK inhibitor | Ph II, HCC; Ph I-II, pancreatic and advanced solid tumors | |
| RAS/MAPK, PI3K/Akt/mTOR | cMET expression | Tivantinib | MET inhibitor | Failed Ph III, HCC; Ph I-II, nonsmall cell lung and esophageal cancer |
| AFP | Ramucirumab | fully human monoclonal antibody(IgGi) | Ph II-III HCC, FDA approved for gastric or gas-tro-esophageal junction adenocarcinoma, colorectal and nonsmall cell lung cancer | |
| FGF19 amplification | JNJ-42756493 (erdafltinib) | Pan-Fibroblast Growth Factor Receptor (FGFR) Tyrosine Kinase Inhibitor | Ph I, HCC; Ph I-II urothelial, nonsmall cell lung, esophageal, breast cancer, lymphoma, and cholangiocarcinoma | |
| FGFR4/klothβ expression | FGF401 | FGFR4 inhibitor | Ph I/II HCC | |
| PI3KCA | Taselisib | PI3K inhibitor | Ph I-III in breast cancer, other advanced cancer |
Abbreviation: HCC, hepatocellular carcinoma.
The USFDA has granted accelerated approval to Nivolumab, a programmed cell death protein-1 (PD-1) immune checkpoint inhibitor for the treatment of patients with HCC who have been previously treated with Sorafenib, based on the CheckMate-040 study, where it had an objective response rate of 20%.113 Pembrolizumab, a humanized antibody targeting the PD-1 receptor, elicited promising progression-free survival and overall survival results in patients with advanced HCC who received previous treatment with sorafenib, according to phase II findings.150 Tumor mutational burden (TMB; total number of mutations per coding area of tumor genome), measured in tumor tissue by whole exome sequencing, is a biomarker for predicting response to immunotherapy.151,152 Recent studies have shown that TMB can be accurately measured in smaller gene panels limited to several hundred genes instead of the whole exome.153,154 Since a tumor biopsy may not capture tumor heterogeneity, emerging evidence indicates that it may be feasible and more effectively to asses TMB in ctDNA.155 Thus, liquid biopsy has the potential to be the companion diagnostic tool for identification of responders to HCC immunotherapy.
Technology platform in detection of circulating cell-free tumor DNA and circulating tumor cells for hepatocellular carcinoma liquid biopsy.
We are entering a new era of precision medicine led by technical advances in analysis of nucleic acids and other biological molecules and generation of targeted therapeutic agents. Precision medicine is driven by accurate evaluation of pathologic tumor markers, specifically in this review, markers measured in bodily fluids. Because the amount of ctDNA is very limited (as low as 0.001%), a highly sensitive technology platform and robust sample preparation are both essential to accurately identify ctDNA markers and characterize CTCs once isolated. This was highlighted in a recent study with a less than satisfactory congruence obtained from patient-paired blood samples between two commercial liquid biopsy tests.156 Thus, a positive detection of ctDNA markers indicates the existence of such markers in the circulation, but a negative result only indicates ctDNA markers were nondetectable at the time of biopsy using the methods of detection and sample processing. More than one sampling may be needed to efficiently detect the ctDNA markers.
Mutations in the TERT promoter, CTNNB1, and TP53 genes are the three most frequently altered genes in HCC. Huang et al used digital PCR assays with a limit of detection of 0.01% to evaluate the four gene loci, TP53 (R249S), CTNNB1 (T41A, S45P), and TERT (c.1–124C>T) in 41 HCC patients.27 At least one kind of circulating mutant was found in 27 patients (56.3%, 27/48), with the mutant allele frequency ranging from 0.33% to 23.7%. Although PCR technology has a desirable sensitivity for detecting ctDNA markers, it is restricted by its ability to assess only a limited number of markers of interest from a given sample. Technology with a higher throughput for number of markers multiplexed, such as targeted next-generation sequencing technology, has gained more attention for profiling HCC ctDNA markers. Initial attempts to use next-generation sequencing technology, such as the Miseq platform, in evaluating the TERT, CTNNB1, and TP53 hotspots with limited coverage (453 bp) resulted in an unsatisfactory sensitivity of 20% (8/41) for at least one detectable marker in the plasma of HCC patients.157 A recent study59 focused on a 58-gene panel that included HCC driver gene and a candidate drug-able mutation (JAK1) by targeting ultradeep NGS in the paired plasma, serum, PBMC, and tissues from eight HCC patients. Of the 21 mutations identified in the tissue with deep sequencing, 9 (43%) were confidently detected in both plasma and serum. A subset of the tissue and plasma mutations detected by NGS were tested for validation by Sanger and/or digital PCR and confirmed. This is the first study to compare the mutation detection performance in paired plasma and serum from five HCC patients. Interestingly, in this small sample size study, no significant differences were noted in the DNA recovery rates, median DNA fragment size, and mutation detection rate from both sources. This is surprising because plasma is considered superior to serum as a source of circulating cell-free DNA.158, 159 A digital ctDNA sequencing study targeting a panel of 68 genes detected at least one genetic alteration in 100% of advanced HCC plasma samples (n=14).142 Mutations were detected in TP53 (8/14), CTNNB1 (4/14), PTEN (1/14), CDKN2A (1/14), ARID1A (1/14), and MET (1/14) genes.
Although the CTC is not currently being used for HCC detection or management, the CTC has been used in other cancers for prognosis or management, including breast cancer.160,161 The only technology that is approved by the FDA to capture CTC is CellSearch, which uses antibodies to capture EpCAM-positive cells. Unfortunately, only 35% of HCC CTCs express EpCAM, thus this technology platform does not have the desired sensitivity to capture HCC CTC. Incorporation of more HCC-specific surface antigens into a capture platform may improve the sensitivity of the capture. Other technology platforms that use microfluidic chips (CTC-iChip), size-selection, or combinations of technologies, as discussed in circulating tumor cells section, are under development to improve the sensitivity and specificity of HCC CTC captures for the subsequent downstream characterization of captured cells.
CONCLUSIONS
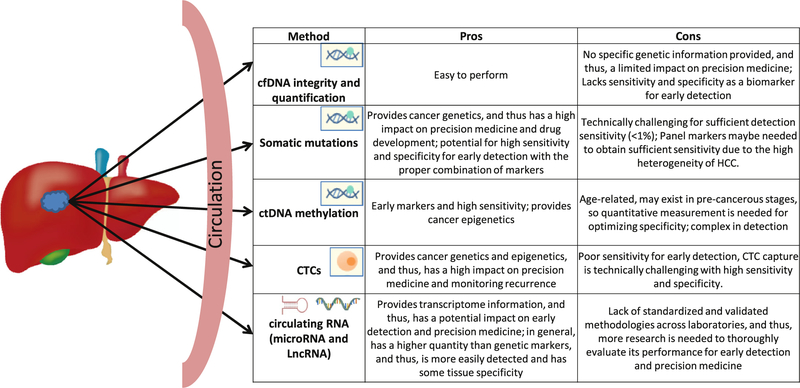
All cancers, including HCC, result from accumulation of genomic and epigenomic modifications and thus consequentially orchestrate aberrant expression or forms of its downstream molecules, such as RNA and proteins. Identification of such modifications (markers) underlying the development of HCC should permit unambiguous screening and early detection if such modifications can be detected in a less or noninvasive liquid biopsy and provide characterization of the tumor, allowing for a more precise treatment plan. Anatomically, highly vascularized liver cancer should shed significant amounts of tumor-derived molecules in the body fluid for liquid biopsy. Although—unlike colorectal cancer, breast cancer, and lung cancer—liquid biopsy has not been used in HCC clinical applications, the exciting progress of its applications, as discussed, in HCC early detection, prognosis prediction, and monitoring recurrence highlights great promise in near future. Fig 1 summarizes the pros and cons of the categories of different liquid biopsy markers for HCC early detection and precision medicine.
Fig 1.
Approaches for HCC liquid biopsy and a summary of the pros and cons of each category of markers. HCC, hepatocellular carcinoma.
Given the high heterogeneity of HCC, we envision a panel of genetic and epigenetic DNA, RNA, or protein markers with other clinical variables such as patient risk factors (age, gender, hepatitis B and/or C, alcohol, Non-alcoholic steatohepatitis (NASH), etc.), clinical laboratory information (AFP, liver function), and radiographic studies (ultra-sound), to be integrated into an algorithm for both screening and early detection. As our understanding of the HCC genetic landscape expands and more treatment options become available, liquid biopsy tests for HCC management should also take place in near future. Simultaneously, efforts should also be made for the standardization and quality control of technology platform. Additionally, attention should be drawn to study designs for developing liquid biopsy tests for the screening and early detection of HCC to ensure the inclusion of high-risk populations as controls in order to better define test performance for HCC detection, and thus facilitating the translation of experimental liquid biopsy tests to clinical applications.
ACKNOWLEDGMENTS
This work is supported by R01CA202769 (YHS), R43CA213610 (SJ), and R44CA165312 (SJ). We would like to thank Amy Lu and Gianni Aranoff for their assistance in manuscript preparation. All authors have read the journal’s policy on disclosure of potential conflicts of interest. Ying-Hsiu Su and Amy K. Kim have no conflicts to disclose. Surbhi Jain is an employee of JBS Science, Inc. All authors have read the journal’s authorship statement, and the manuscript has been reviewed by and approved by all named authors.
Abbreviations:
- AASLD
American Association for the Study of Liver Diseases
- AFP
alfa-fetoprotein
- CART
classification and regression trees
- cfDNA
circulating cell-free DNA
- CTC
circulating tumor cells
- ctDNA
circulating cell-free tumor DNA
- CTscan
computed tomography
- EpCAM
epithelial cell adhesion molecule
- FS
fixed sequential
- HCC
hepatocellular carcinoma
- lncRNA
long noncoding RNA
- LR
logistic regression
- miRNA
microRNA
- MRI
magnetic resonance imaging
- PBMC
peripheral blood mononuclear cells
- qPCR
quantitative polymerase chain reaction
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- RF
random forest
- TACE
transcatheter arterial chemoembolization
- TMB
tumor mutational burden
- TS
two-step
- USFDA
United states Food and Drug Administration
REFERENCES
- 1.Ferlay JSI, Ervik M, Dikshit R, et al. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available at: http://globocan.iarc.fr. Accessed March 14, 2017. [Google Scholar]
- 2.Howlader NNA, Krapcho M, Miller D, et al. (eds). SEER cancer statistics review, 1975–2013, Bethesda, National Cancer Institute, http://seer.cancer.gov/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site, April 2016. 2016. [Google Scholar]
- 3.Kohler BA, Ward E, McCarthy BJ, et al. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst 2011;103:714–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. [DOI] [PubMed] [Google Scholar]
- 6.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472. [DOI] [PubMed] [Google Scholar]
- 7.American Cancer Society. Cancer facts and figures 2017.
- 8.Daher S, Massarwa M, Benson AA, Khoury T. Current and future treatment of hepatocellular carcinoma: an updated comprehensive review. J Clin Transl Hepatol 2018;6:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heimbach J, Kulik LM, Finn R, et al. Aasld guidelines for the treatment of hepatocellular carcinoma. Hepatology 2017;67(1):358–80. [DOI] [PubMed] [Google Scholar]
- 11.Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol 2017;23:5086–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hench IB, Hench J, Tolnay M. Liquid biopsy in clinical management of breast, lung, and colorectal cancer. Front Med 2018;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison GJ, Goldkorn A. Development and application ofliquid biopsies in metastatic prostate cancer. Curr Oncol Rep 2018;20:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malapelle U, Sirera R, Jantus-Lewintre E, et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn 2017;17:209–15. [DOI] [PubMed] [Google Scholar]
- 15.Chung JH, Pavlick D, Hartmaier R, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with estrogen receptor-positive metastatic breast cancer. Ann Oncol 2017;28:2866–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Y, Ji JS, Jin JG, Kuo WP, Kang H. Cancer Liquid Biopsy: Is It Ready for Clinic? IEEE Pulse 2017;8:23–7. [DOI] [PubMed] [Google Scholar]
- 17.Marrero JA, Pelletier S. Hepatocellular carcinoma. Clin Liver Dis 2006;10:339–51. [DOI] [PubMed] [Google Scholar]
- 18.Davis GL, Dempster J, Meler JD, et al. Hepatocellular carcinoma: management of an increasingly common problem. Proc (Bayl Univ Med Cent) 2008;21:266–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrero JA. Hepatocellular carcinoma. Curr Opin Gastroenterol 2006;22:248–53. [DOI] [PubMed] [Google Scholar]
- 20.Seki S, Sakaguchi H, Kitada T, et al. Outcomes of dysplastic nodules in human cirrhotic liver: a clinicopathological study. Clin Cancer Res 2000;6:3469–73. [PubMed] [Google Scholar]
- 21.Sherman M Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis 2005;25:143–54. [DOI] [PubMed] [Google Scholar]
- 22.Hoofnagle JH. Hepatocellular carcinoma: summary and recommendations. Gastroenterology 2004;127:S319–23. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Sun L-y, Zheng H-q, Zhang Q-f, Jin X-m. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology 2012;44:318–24. [DOI] [PubMed] [Google Scholar]
- 24.El-Shazly SF. Evaluation of serum DNA integrity as a screening and prognostic tool in patients with hepatitis C virus-related hepatocellular carcinoma. Int J Biol Markers 2010;25:79–86. [DOI] [PubMed] [Google Scholar]
- 25.Wang BG, Huang H-Y, Chen Y-C, et al. Increased plasma DNA integrity in cancer patients. Cancer Res 2003;63:3966. [PubMed] [Google Scholar]
- 26.Jiang P, Chan CWM, Chan KCA, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317–E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang A, Zhang X, Zhou S-L, et al. Plasma circulating cell-free DNA integrity as a promising biomarker for diagnosis and surveillance in patients with hepatocellular carcinoma. J Cancer 2016;7:1798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobhani N, Generali D, Zanconati F, Bortul M, Scaggiante B. Cell-free DNA integrity for the monitoring of breast cancer: future perspectives? World J Clin Oncol 2018;9:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumari S, Husain N, Agarwal A, et al. Diagnostic value of circulating free dna integrity and global methylation status in gall bladder carcinoma. Pathol Oncol Res 2018: 1–12. [DOI] [PubMed] [Google Scholar]
- 30.Chiara B, Vittoria EM, Paola DB, Salvatore P, Donato N, Marco A. Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int J Cancer 2017;140:1888–98. [DOI] [PubMed] [Google Scholar]
- 31.Salvianti F, Giuliani C, Petrone L, et al. Integrity and quantity of total cell-free dna in the diagnosis of thyroid cancer: correlation with cytological classification. Int J Mol Sci 2017;18:1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piciocchi M, Cardin R, Vitale A, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int 2013;7:1050–7. [DOI] [PubMed] [Google Scholar]
- 33.Truszewska A, Foroncewicz B, Paczek L. The role and diagnostic value of cell-free DNA in systemic lupus erythematosus. Clin Exp Rheumatol 2017;35:330–6. [PubMed] [Google Scholar]
- 34.Zhang S, Lu X, Shu X, et al. Elevated plasma cfdna may be associated with active lupus nephritis and partially attributed to abnormal regulation of neutrophil extracellular traps (NETs) in patients with systemic lupus erythematosus. Intern Med 2014;53:2763–71. [DOI] [PubMed] [Google Scholar]
- 35.Nault J-C, Zucman-Rossi J. Genetics of hepatocellular carcinoma: the next generation. J Hepatol 2014;60:224–6. [DOI] [PubMed] [Google Scholar]
- 36.Ozen C, Yildiz G, Dagcan AT, et al. Genetics and epigenetics of liver cancer. New Biotechnol 2013;30:381–4. [DOI] [PubMed] [Google Scholar]
- 37.Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760–4. [DOI] [PubMed] [Google Scholar]
- 39.Ally A, Balasundaram M, Carlsen R, et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–41.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hann H-W, Jain S, Park G, Steffen JD, Song W, Su Y-H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res 2017;3:105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SY, Dhillon V, Jain S, et al. A locked nucleic acid clamp-mediated PCR assay for detection of a p53 codon 249 hotspot mutation in urine. J Mol Diagn 2011;13:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosny G, Farahat N, Tayel H, Hainaut P. Ser-249 TP53 and CTNNB1 mutations in circulating free DNA of Egyptian patients with hepatocellular carcinoma versus chronic liver diseases. CancerLett 2008;264:201–8. [DOI] [PubMed] [Google Scholar]
- 43.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene 2007;26:2166,76. [DOI] [PubMed] [Google Scholar]
- 44.Kuang SY, Lekawanvijit S, Maneekarn N, et al. Hepatitis B 1762T/1764A mutations, hepatitis C infection, and Codon 249 p53 mutations in hepatocellular carcinomas from Thailand. Cancer Epidemiol Biomarkers Prev 2005;14:380–4. [DOI] [PubMed] [Google Scholar]
- 45.Lleonart ME, Kirk GD, Villar S, et al. Quantitative analysis of plasma TP53 249Ser-mutated DNA by electrospray ionization mass spectrometry. Cancer Epidemiol, Biomarkers Prev 2005;14:2956–62. [DOI] [PubMed] [Google Scholar]
- 46.Kirk GD, Lesi OA, Mendy M, et al. 249ser TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene 2005;24:5858–67. [DOI] [PubMed] [Google Scholar]
- 47.Szymańska K, Lesi OA, Kirk GD, et al. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int J Cancer 2004;110:374–9. [DOI] [PubMed] [Google Scholar]
- 48.Kirk GD. Ser-249 p53 mutations in plasma DNA of patients with hepatocellular carcinoma from The Gambia. J Natl Cancer Inst 2000;92:148–53. [DOI] [PubMed] [Google Scholar]
- 49.Jackson PE. Specific p53 mutations detected in plasma and tumors of hepatocellular carcinoma patients by electrospray ionization mass spectrometry. Cancer Res 2001;61:33–5. [PubMed] [Google Scholar]
- 50.Zhang YJ, Rossner P ?Jr., Chen Y, et al. Aflatoxin B 1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int J Cancer 2006;119:985–91. [DOI] [PubMed] [Google Scholar]
- 51.Igetei R, Otegbayo J, Ndububa D, et al. Detection of p53 codon 249 mutation in Nigerian patients with hepatocellular carcinoma using a novel evaluation of cell-free DNA. Ann Hepatol 2008;7:339–44. [PubMed] [Google Scholar]
- 52.Kuang SY, Jackson PE, Wang JB, et al. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl AcadSci 2014;101(10):3575–80, 0308232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su Y-H, Lin SY, Song W, Jain S. DNA markers in molecular diagnostics for hepatocellular carcinoma. Expert Rev Mol Diagn 2014;14:803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villar S, Ortiz-Cuaran S, Abedi-Ardekani B, et al. Aflatoxin-induced TP53 R249S mutation in hepatocellular carcinoma in Thailand: association with tumors developing in the absence of liver cirrhosis. PLoS One 2012;7:e37707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X-H, Sun L-H, Lu D-D, et al. Codon 249 mutation in exon 7 of p53 gene in plasma DNA: maybe a new early diagnostic marker of hepatocellular carcinoma in Qidong risk area, China. World J Gastroenterol. 2003;9:692–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimbi GC, Kew MC, Yu MC, Arakawa K, Hodkinson J. 249serp53 mutation in the serum of black southern African patients with hepatocellular carcinoma. J Gastroenterol Hepatol 2005;20:1185–90. [DOI] [PubMed] [Google Scholar]
- 57.Mah Y-H, Hsu C-S, Liu C-H, et al. Serum p53 gene polymorphisms and severity ofhepatitis B or C-related chronic liver diseases in Taiwan. Hepatol Int 2011;5:814–21. [DOI] [PubMed] [Google Scholar]
- 58.Huang A, Zhang X, Zhou S-L, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital pcr is feasible and reflects intratumoral heterogeneity. J Cancer 2016;7:1907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labgaa I, Villacorta-Martin C, D’Avola D, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 2018;37:3740–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao W, Yang H, Xu H, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepato-cellular carcinoma patients by targeted deep sequencing. Oncotarget 2016;7:40481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013;4:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charles NJ, Julien C, Luca DT, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014;60:1983–92. [DOI] [PubMed] [Google Scholar]
- 63.Pinyol R, Tovar V, Llovet JM. TERT promoter mutations: gate-keeper and driver of hepatocellular carcinoma. J Hepatol 2014;61:685–7. [DOI] [PubMed] [Google Scholar]
- 64.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyer P, Zekri A-R, Hung C-W, et al. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol 2009;88:107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain S, Xie L, Boldbaatar B, et al. Differential methylation of the promoter and first exon of the RASSF1A gene in hepatocarcinogenesis. Hepatol Res 2015;45:1110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeo W, Wong N, Wong W-L, Lai PBS, Zhong S, Johnson PJ. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver International: Munksgaard International Publishers; 2005. p. 266–72. [DOI] [PubMed] [Google Scholar]
- 69.Jain S, Wojdacz TK, Su H-Y. Challenges for the application of DNA methylation biomarkers in molecular diagnostic testing for cancer. Expert Rev Mol Diagn 2013;13:283–94. [DOI] [PubMed] [Google Scholar]
- 70.Hua D, Hu Y, Wu Y-Y, et al. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol 2011;91:455–60. [DOI] [PubMed] [Google Scholar]
- 71.Huang Z-H, Hu Y, Hua D, Wu Y-Y, Song M-X, Cheng Z-H. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol 2011;91:702–7. [DOI] [PubMed] [Google Scholar]
- 72.Liu J-B, Zhang Y-X, Zhou S-H, et al. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol 2011;17:4718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lamb YN. 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther 2017;21:225–32. [DOI] [PubMed] [Google Scholar]
- 74.Yin C-Q, Yuan C-H, Qu Z, Guan Q, Chen H, Wang F-B. Liquid biopsy of hepatocellular carcinoma: circulating tumor-derived biomarkers. Dis Markers 2016;2016:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pezzuto F, Buonaguro L, Buonaguro MF, Tornesello LM. The role of circulating free DNA and microRNA in non-invasive diagnosis of HBV- and HCV-related hepatocellular carcinoma. Int J Mol Sci 2018;19:1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okajima W, Komatsu S, Ichikawa D, et al. Liquid biopsy in patients with hepatocellular carcinoma: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol 2017;23:5650–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu R-h, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155. [DOI] [PubMed] [Google Scholar]
- 78.Dong X, Hou Q, Chen Y, Wang X. Diagnostic value of the methylation of multiple gene promoters in serum in hepatitis B virus-related hepatocellular carcinoma. Dis Markers 2017;2017:2929381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohamed NA, Swify EM, Amin NF, Soliman MM, Tag-Eldin LM, Elsherbiny NM. Is serum level of methylated RASSF1A valuable in diagnosing hepatocellular carcinoma in patients with chronic viral hepatitis C? Arab J Gastroenterol 2012;13:111–5. [DOI] [PubMed] [Google Scholar]
- 80.Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Satelli A, Batth I, Brownlee Z, et al. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget 2017;8:49329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yan L, Jin G, Chaofeng W, et al. Circulation times of prostate cancer and hepatocellular carcinoma cells by in vivo flow cytometry. Cytometry A 2011;79A:848–54. [DOI] [PubMed] [Google Scholar]
- 83.Hesketh RL, Zhu AX, Oklu R. Hepatocellular carcinoma: can circulating tumor cells and radiogenomics deliver personalized care? Am J Clin Oncol 2015;38:431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamashita T, Forgues M, Wang W, et al. EpCAM and a-feto-protein expression defines novel prognostic subtypes of hepato-cellular carcinoma. Cancer Res 2008;68:1451–61. [DOI] [PubMed] [Google Scholar]
- 85.Guo W, Sun Y-F, Shen M-N, et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res 2018;24:2203–13. [DOI] [PubMed] [Google Scholar]
- 86.Kelley RK, Magbanua MJM, Butler TM, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer 2015;15:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun C, Liao W, Deng Z, et al. The diagnostic value of assays for circulating tumor cells in hepatocellular carcinoma: a meta-analysis. Medicine 2017;96:e7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kalinich M, Bhan I, Kwan TT, et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. PNAS 2017;114:1123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. PNAS 2011;108:5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gui J, Tian Y, Wen X, et al. Serum microRNA characterization identifies miR-885–5p as a potential marker for detecting liver pathologies. Clin Sci 2011;120:183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qi P, Cheng S-q, Wang H, Li N, Chen Y-f, Gao C-f. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One 2011;6:e28486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.El-Garem H, Ammer A, Shehab H, et al. Circulating micro-RNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol 2014;6:818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shanshan C, Hao C, Shanshan G, et al. Differential expression of plasma microRNA-125b in hepatitis B virus–related liver diseases and diagnostic potential for hepatitis B virus–induced hepatocellular carcinoma. Hepatol Res 2017;47:312–20. [DOI] [PubMed] [Google Scholar]
- 94.Sohn W, Kim J, Kang SH, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med 2015;47:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chao-Hung H, Tsung-Hui H, Sheng-Nan L, et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer 2016;138:714–20. [DOI] [PubMed] [Google Scholar]
- 96.Zekri A-RN, Youssef ASE-D, El-Desouky ED, et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumor Biol 2016;37:12273–86. [DOI] [PubMed] [Google Scholar]
- 97.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 2011;45:355–60. [DOI] [PubMed] [Google Scholar]
- 98.Ding Y, Yan J-L, Fang A-N, Zhou W-F, Huang L. Circulating miRNAs as novel diagnostic biomarkers in hepatocellular carcinoma detection: a meta-analysis based on 24 articles. Oncotarget 2017;8:66402–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhattacharya S, Steele R, Shrivastava S, Chakraborty S, Di Bisceglie AM, Ray RB. Serum miR-30e and miR-223 as novel noninvasive biomarkers for hepatocellular carcinoma. Am J Pathol 2016;186:242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei Ge, Yu D-c, Li Q-g, Chen X, Zhang C-y, Ding Y-t. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin Lab 2014;60:427–34. [DOI] [PubMed] [Google Scholar]
- 101.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. JNCI Cancer Spectrum 2001;93:1054–61. [DOI] [PubMed] [Google Scholar]
- 102.Yuan W, Sun Y, Liu L, Zhou B, Wang S, Gu D. Circulating LncRNAs serve as diagnostic markers for hepatocellular carcinoma. Cell Physiol Biochem 2017;44:125–32. [DOI] [PubMed] [Google Scholar]
- 103.El-tawdi A, Matboly M, Hussein Shehata H, et al. Evaluation of circulatory RNA-based biomarker panel in hepatocellular carcinoma 2016;20:265–77. [DOI] [PubMed] [Google Scholar]
- 104.Jia H-L, Ye Q-H, Qin L-X, et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res 2007;13:1133. [DOI] [PubMed] [Google Scholar]
- 105.Wang M, Long RE, Comunale MA, et al. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 2009;18:1914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 2008;68:645–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karabatak M A new classifier for breast cancer detection based on Naïve Bayesian. Measurement 2015;72:32–6. [Google Scholar]
- 108.Li C, Zhang S, Zhang H, et al. Using the K-nearest neighbor algorithm for the classification of lymph node metastasis in gastric cancer. Comput Math Methods Med 2012;2012:876545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 2015;13:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dietterich TG. Ensemble methods in machine learning In: In: Proceedings of the First international workshop on multiple classifier systems, Part of the Lecture Notes in Computer Science book series (LNCS, volume 1857): Springer-Verlag; 2000:1–15. [Google Scholar]
- 111.Wang J, Jain S, Chen D, Song W, Hu C-T, Su Y-H. Development and evaluation of novel statistical methods in urine bio-marker-based hepatocellular carcinoma screening. Sci Rep 2018;8:3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform 2006;2:59–77. [PMC free article] [PubMed] [Google Scholar]
- 113.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet North Am Ed 2017;389:2492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hung IF-N, Wong DK-H, Poon RT-P, et al. Risk factors and post-resection independent predictive score for the recurrence of hepatitis b-related hepatocellular carcinoma. PLoS One 2016;11:e0148493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chong CC Lee KF Ip PC, et al. Pre-operative predictors of post-hepatectomy recurrence of hepatocellular carcinoma: can we predict earlier? Surgeon 2012;10:260–6. [DOI] [PubMed] [Google Scholar]
- 116.Poon RT-P, Fan S-T, Ng IO-L, Lo C-M, Liu C-L, Wong J. Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer 2000;89:500–7. [PubMed] [Google Scholar]
- 117.Sherman M Recurrence of hepatocellular carcinoma. N Engl J Med 2008;359:2045–7. [DOI] [PubMed] [Google Scholar]
- 118.Lok A, McMahon B. Chronic hepatitis B. Hepatology 2001;34:1225–41. [DOI] [PubMed] [Google Scholar]
- 119.Portolani N, Coniglio A, Ghidoni S, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg 2006;243:229–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kamiyama T, Nakanishi K, Yokoo H, et al. Recurrence patterns after hepatectomy of hepatocellular carcinoma: implication of milan criteria utilization. Ann Surg Oncol 2009;16:1560–71. [DOI] [PubMed] [Google Scholar]
- 121.Minami Y, Nishida N, Kudo M. Therapeutic response assessment of RFA for HCC: contrast-enhanced US, CT and MRI. World J Gastroenterol 2014;20:4160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Minami Y, Kudo M. Imaging modalities for assessment of treatment response to nonsurgical hepatocellular carcinoma therapy: contrast-enhanced US, CT, and MRI. Liver Cancer 2015;4:106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Willatt JM, Hussain HK, Adusumilli S, Marrero JA. MR imaging of hepatocellular carcinoma in the cirrhotic liver: challenges and controversies. Radiology 2008;247:311–30. [DOI] [PubMed] [Google Scholar]
- 124.Liu ZJ, Huang Y, Wei L, et al. Combination ofLINE-1 hypomethylation and RASSF1A promoter hypermethylation in serum DNA is a non-invasion prognostic biomarker for early recurrence of hepatocellular carcinoma after curative resection. Neoplasma 2017;64:795–802. [DOI] [PubMed] [Google Scholar]
- 125.Wong IHN, Dennis Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res 2000;6:3516. [PubMed] [Google Scholar]
- 126.Zhi-Xiong C, Geng C, Yong-Yi Z, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer 2017;141:977–85. [DOI] [PubMed] [Google Scholar]
- 127.Kim SS, Cho HJ, Nam JS, et al. Plasma microRNA-21, 26a, and 29a-3p as predictive markers for treatment response following transarterial chemoembolization in patients with hepatocellular carcinoma. J Korean Med Sci 2018;33:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu C-Y, Chen S-Y, Peng H-L, Kan P-Y, Chang W-C, Yen C-J. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget 2017;8: 6406–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giray BG, Emekdas G, Tezcan S, et al. Profiles of serum microRNAs; miR-125b-5p and miR223–3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep 2014;41:4513–9. [DOI] [PubMed] [Google Scholar]
- 130.Yamamoto Y, Kosaka N, Tanaka M, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Bio-markers 2009;14:529–38. [DOI] [PubMed] [Google Scholar]
- 131.Zheng J, Zhou Z, Xu Z, et al. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol Med Rep 2015;12:1584–90. [DOI] [PubMed] [Google Scholar]
- 132.Tang J, Zhuo H, Zhang X, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis 2014;5:e1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou M, Zhang X-Y, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother 2017;85:348–54. [DOI] [PubMed] [Google Scholar]
- 134.Konishi H, Ichikawa D, Yamamoto Y, et al. Plasma level of metastasis—associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci 2016;107:149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fan S, Yang Z, Ho D, Ng M, Yu W, Wong J. Prediction of post-hepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: a prospective study. Ann Surg 2011;254:569–76. [DOI] [PubMed] [Google Scholar]
- 136.Yun-Fan S, Yang X, Xin-Rong Y, et al. Circulating stem cell-like epithelial cell adhesion molecule—positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013;57:1458–68. [DOI] [PubMed] [Google Scholar]
- 137.Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012;148:349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lawson DA, Bhakta NR, Kessenbrock K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015;526:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015;6:10697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li YM, Xu SC, Li J, et al. Epithelial–mesenchymal transition markers expressed in circulating tumor cells in hepatocellular carcinoma patients with different stages of disease. Cell Death Dis 2013;4:e831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun Y, Guo W, Xu Y, et al. Circulating tumors cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res 2017;24:547. [DOI] [PubMed] [Google Scholar]
- 142.Ikeda S, Lim JS, Kurzrock R. Analysis of tissue and circulating tumor DNA by next-generation sequencing of hepatocellular carcinoma: implications for targeted therapeutics. Mol Cancer Ther 2018;17:1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chan SL, Wong AM, Lee K, Wong N, Chan AKC. Personalized therapy for hepatocellular carcinoma: where are we now? Cancer Treat Rev. 2016;45:77–86. [DOI] [PubMed] [Google Scholar]
- 144.Schwaederlé M, Lazar V, Validire P, et al. VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: implications for antiangiogenesis therapy. Cancer Res 2015;75:1187. [DOI] [PubMed] [Google Scholar]
- 145.Said R, Hong DS, Warneke CL, et al. P53 mutations in advanced cancers: clinical characteristics, outcomes, and correlation between progression-free survival and bevacizumab-containing therapy. Oncotarget 2013;4:705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wheler JJ, Janku F, Naing A, et al. TP53 alterations correlate with response to VEGF/VEGFR inhibitors: implications for targeted therapeutics. Mol Cancer Ther 2016;15:2475. [DOI] [PubMed] [Google Scholar]
- 147.Koehler K, Liebner D, Chen JL. TP53 mutational status is predictive of pazopanib response in advanced sarcomas. Ann Oncol 2016;27:539–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clausse V, Goloudina AR, Uyanik B, et al. Wee1 inhibition potentiates Wip1-dependent p53-negative tumor cell death during chemotherapy. Cell Death Dis 2016;7:e2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang B, Finn RS. Personalized clinical trials in hepatocellular carcinoma based on biomarker selection. Liver Cancer 2016;5:221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Andrew X Zhu RSF, Cattan Stéphane, Edeline Julien, et al. KEYNOTE-224: pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib Oral presentation at: 2018 gastrointestinal cancers symposium; January 18–20, 2018; San Francisco, CA: 2018. [Google Scholar]
- 151.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med 2016;14:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Khagi Y, Goodman AM, Daniels GA, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res 2017;23:5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol 2017;4:868–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016;7:65364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wong FCK, Sun K, Jiang P, Cheng YKY, Chan KCA, Leung TY, et al. Cell-free DNA in maternal plasma and serum: a comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin Biochem 2016;49:1379–86. [DOI] [PubMed] [Google Scholar]
- 159.Veronika K, David V, Lenka S, Lenka B, Sabina S. Cell–free DNA—minimally invasive marker of hematological malignancies. Eur J Haematol 2017;99:291–9. [DOI] [PubMed] [Google Scholar]
- 160.Riethdorf S, O’Flaherty L, Hille C, Pantel K. Clinical applications of the CellSearch platform in cancer patients. Adv Drug Deliv Rev 2018;125:102–21. [DOI] [PubMed] [Google Scholar]
- 161.Beije N, Jager A, Sleijfer S. Circulating tumor cell enumeration by the CellSearch system: the clinician’s guide to breast cancer treatment? Cancer Treat Rev 2015;41:144–50. [DOI] [PubMed] [Google Scholar]