Abstract
Investigating type I feline coronaviruses (FCoVs) in tissue culture is critical for understanding the basic virology, pathogenesis, and virus-host interactome of these important veterinary pathogens. This has been a perennial challenge as type I FCoV strains do not easily adapt to cell culture. Here we characterize replication kinetics and plaque formation of a model type I strain FIPV Black in Fcwf-4 cells established at Cornell University (Fcwf-4 CU). We determined that maximum virus titers (>107 pfu/mL) were recoverable from infected Fcwf-4 CU cell-free supernatant at 20 h post-infection. Type I FIPV Black and both biotypes of type II FCoV formed uniform and enumerable plaques on Fcwf-4 CU cells. Therefore, these cells were employable in a standardized plaque assay. Finally, we determined that the Fcwf-4 CU cells were morphologically distinct from feline bone marrow-derived macrophages and were less sensitive to exogenous type I interferon than were Fcwf-4 cells purchased from ATCC.
Keywords: Feline coronavirus, FIPV, Feline macrophage-like cell line, Plaque assay, Fcwf-4 cells, AK-D cells
1. Introduction
Feline coronaviruses (FCoVs) are members of the alphacoronavirus genus that infect cats and can cause the highly lethal disease known as feline infectious peritonitis (FIP) (Pedersen, 2009). FCoV infection is widespread among domestic feline populations, especially within multi-cat households and catteries, which can exhibit upwards of 96% seropositivity (Addie et al., 2003, Addie and Jarrett, 1992, Hohdatsu et al., 1992, Pedersen, 2009, Pedersen, 1976, Vennema et al., 1998). Despite the global burden of FCoV infection, there are no currently approved therapeutics to treat FIP; however, reports of direct inhibition of virus growth and treatment of individual cats using small molecule viral inhibitors have been promising (Kim et al., 2016, Kim et al., 2015, Kim et al., 2013, Kim et al., 2012, Murphy et al., 2018, Pedersen et al., 2017, St John et al., 2015).
FCoVs are typically grouped into two biotypes (or pathotypes), which have been classified as feline enteric coronavirus (FECV) and feline infectious peritonitis virus (FIPV), based on tissue tropism, disease progression, and genetic markers (reviewed in Kipar and Meli, 2014; Pedersen, 2014, Pedersen, 2009), although the range of disease signs and clinical outcomes are likely to extend beyond these two basic definitions. Endemic FECV causes mild enteritis associated with loose stool and diarrhea and commonly leads to an asymptomatic, persistent infection (Addie, 2011, Addie et al., 2003, Pedersen et al., 2008). A subset of these infections (3–10%) result in lethal FIP (Addie and Jarrett, 1992, Pedersen, 1976) arising from a shift in virus tropism and systemic infection of monocytes and macrophages. Perturbations of the host feline immune state leading to immune deficiency can allow virus replication to surge (Pedersen, 2009, Tekes and Thiel, 2016), resulting in the formation of a quasispecies and the genetic sampling required for progression of FECV to the second biotype, FIPV. The internal mutation theory proposes that, within an individual animal, FIPV arises directly from FECV due to accumulation of non-synonymous mutations in spike (S) (Licitra et al., 2014, Licitra et al., 2013, Rottier et al., 2005) and group-specific proteins (Chang et al., 2010, Herrewegh et al., 1995, Lin et al., 2009, Pedersen et al., 2012, Phillips et al., 2013, Poland et al., 1996, Vennema et al., 1998). The resulting infection of monocytes and macrophages by FIPV leads to systemic spread and development of immune-mediated FIP (Pedersen, 2009).
FCoV biotypes are further defined by their viral S protein. Classically, antigenicity of the S protein alone has been used to categorize FCoVs into two serotypes (type I and type II) (Hohdatsu et al., 1991, Pedersen et al., 1984), but a recent study proposes a more specific grouping of FCoVs into two clades via functionality-based S-protein sequencing (Whittaker et al., 2018). Type I viruses account for the bulk (80–90%) of natural infections in domestic cats, while type II FCoV, a naturally-occurring recombinant between the type I and canine coronavirus (CCoV) spike proteins (Herrewegh et al., 1998), is far less prevalent (< 10%) (Addie et al., 2003, Benetka et al., 2004, Hohdatsu et al., 1992, Kennedy et al., 2002). A commendable number of studies focus on type II FCoV as these viruses are much more easily propagated in cell culture. However, it is difficult to know how accurately type II laboratory strains reflect natural infections with type I viruses given that the bulk of what is known about type I is extrapolated from studies using a type II virus.
It is critical that we investigate type I FIPV in laboratory cell culture in order to understand the basic virology of natural infection, characterize type I clinical isolates, test novel therapeutics, and develop effective feline vaccines with broader coverage. However, this has been challenging because type I FCoVs cannot be easily adapted to laboratory cell culture; furthermore, the receptor for type I is not known (Cham et al., 2017, Dye et al., 2007, Hohdatsu et al., 1998), making the identification of highly permissive cell types difficult. Select type I isolates, such as the FIPV Black strain used in this study, have been adapted to growth in tissue culture at the cost of reduced in vivo virulence (Black, 1980, Pedersen, 2009, Tekes et al., 2007, Thiel et al., 2014). Feline airway epithelial (AK-D) cells propagate FIPV Black (Regan et al., 2012); however, these cells do not represent natural tropism for FIPV. Felis catus whole fetus 4 (Fcwf-4) cells are a more physiologically-relevant feline macrophage-like cell line (Jacobse-Geels and Horzinek, 1983), but these cells come with several technical drawbacks for studying type I FIPV. First, Fcwf-4 cell doubling time is slow (>31 h) (American Type Culture Collection, 2013) and cells do not grow to high density. Second, previous studies report that type I FIPV grows to low titers (< 105 pfu/mL) in these cells relative to type II (> 106 pfu/mL) (Tekes et al., 2012) and can be measured by determining the 50% tissue culture infectious dose (TCID50) (Ramakrishnan, 2016, Reed and Muench, 1938) or by plaque assay (Tekes et al., 2012, Tekes et al., 2010). Third, type I virus kinetics are variable in Fcwf-4 cells, requiring between 15 and 72 h to achieve maximum titer (Jacobse-Geels and Horzinek, 1983, Tekes et al., 2012, Tekes et al., 2007; This Study). Finally, some reports suggest that type I is highly cell-associated (Jacobse-Geels and Horzinek, 1983, Pedersen et al., 1984) and multiple freeze-thaw cycles may be required to recover virus. Together, these factors have made investigation of type I FIPV challenging.
As part of this study, we characterized three feline cell lines– two from the American Type Culture Collection (ATCC) and one from Cornell University– and evaluated the replication kinetics, efficiency of plaque formation, and responsiveness of these cells to interferon (IFN) in order to identify the optimal cell culture conditions for type I FIPV Black. We found that an Fcwf-4 cell line established at Cornell University College of Veterinary Medicine, designated Fcwf-4 CU, propagates type I FIPV to significantly higher titers in cell-free supernatant and with more rapid kinetics compared to commercially available Fcwf-4 cells. We show that Fcwf-4 CU cells are less responsive to exogenous type I interferon than Fcwf-4 cells from the ATCC and are permissive to infection by both biotypes of type II FCoV. To facilitate quantitation of FIPV Black, we established a standardized plaque assay method using Fcwf-4 CU cells and commercially available AK-D cells and show that both cell types permit rapid and consistent quantitation of infectious titers of type I FIPV as well as type II FIPV and FECV from cell-free supernatants.
2. Results
2.1. Type I FIPV Black replication kinetics varies between feline cell types
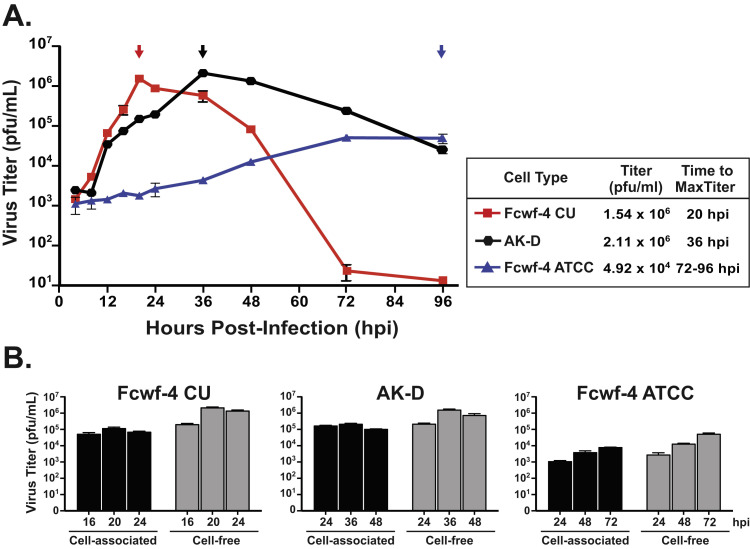
To determine the optimal cell type and conditions required to grow the type I FIPV Black strain, we evaluated virus growth kinetics using a standard infection time course. Cells were infected at a multiplicity of infection (MOI) of 0.1 and virus titer was determined by plaque assay from cell-free supernatants over 96 h. FIPV Black, a distinct type I lab strain that replicates in feline epithelial cells, replicated as expected in AK-D cells reaching a maximum titer > 106 pfu/mL at 36 h post-infection (hpi) ( Fig. 1 A). In our hands, using Fcwf-4 cells purchased from the ATCC, the replication of FIPV Black reached a maximum titer > 104 pfu/mL over 72–96 hpi ( Fig. 1 A). Strikingly, FIPV Black replication kinetics and maximum titer were drastically different in an Fcwf-4 cell line established at Cornell University College of Veterinary Medicine (Fcwf-4 CU). Using these cells, the virus reached a significantly higher maximum titer of > 106 pfu/mL at 20 hpi. In other words, nearly 100 times more virus was produced from the Fcwf-4 CU cells a full 2–3 days faster than in Fcwf-4 ATCC cells.
Fig. 1.
Fcwf-4 Cornell University (CU) cells produce high titers of cell-free type I FIPV with rapid growth kinetics. A) Virus growth kinetics measured from cell-free supernatants of AK-D (black), Fcwf-4 ATCC (blue), and Fcwf-4 CU (red) cells infected with FIPV Black (MOI=0.1) over 96 h. Arrows indicate time of peak titer presented as plaque-forming units (pfu)/mL. Titer determined by plaque assay on AK-D cells in triplicate; error bars ± SD. B) Cell-associated and cell-free virus titers were determined following infection of Fcwf-4 CU, AK-D, and Fcwf-4 ATCC cells with FIPV Black (MOI=0.1). Cell-free titer was determined from cell-clarified supernatants; cell-associated titer was determined from suspended cell monolayers following three freeze-thaw cycles alternating between −80 °C and 37 oC. Samples were taken at hours post-infection (hpi) just prior to, at, and following the maximum (max) virus titer for each cell type. Titers determined by plaque assay on AK-D cells in triplicate; error bars ± SD.
To address whether the differences in titer observed between AK-D, Fcwf-4 CU, and Fcwf-4 ATCC cells were due to differences in cell-free and cell-associated virus, we compared the cell-associated and cell-free virus titers from each cell type at the time points around the respective maximum titers. Surprisingly, the cell-free virus titers were higher than the cell-associated titers at all time points and in all cell types assayed ( Fig. 1 B). This indicates that FIPV Black virions are released into cell supernatant during infection of cell culture and freeze-thaw cycles are not necessary to obtain high virus titers.
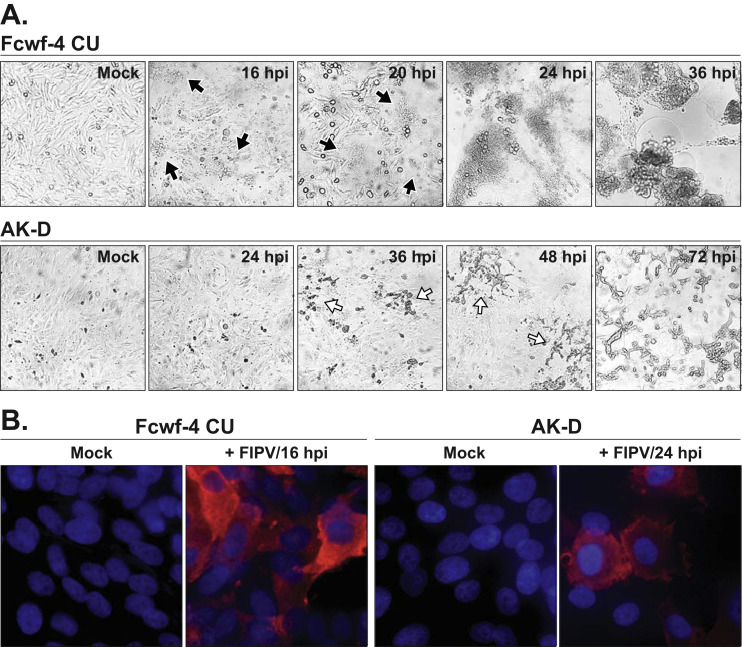
Although the maximum titers of FIPV Black were comparable between AK-D and Fcwf-4 CU cells, the progression of cell cytopathic effects (CPE) induced by the virus differed. FIPV Black formed large, uniform syncytia in Fcwf-4 CU cells, while individual cell-death-induced clearings were observed in infected AK-D cells ( Fig. 2 A). Of note, maximum titers from both cell types were obtained just prior to the appearance of major CPE, allowing a visual guide to virus collection. To further demonstrate this point, Fcwf-4 CU and AK-D cells infected with FIPV Black were labeled with an anti-nucleocapsid antibody (CCV2-2) (Poncelet et al., 2008) and visualized by immunofluorescence prior to the induction of major observable CPE. As expected, the majority of cells were positive for virus antigen ( Fig. 2 B) and the differences in CPE are clearly shown: note the syncytial membrane fusion in infected Fcwf-4 CU cells and the maintenance of distinct cell membranes in infected AK-D cells ( Fig. 2 B). Together, these results demonstrate the ability of the Fcwf-4 CU cells to rapidly produce high levels of type I FIPV Black in cell-free supernatants.
Fig. 2.
FIPV Black-induced cell cytopathic effect varies between infected cell types. A) Progression of cell cytopathic effects (CPE) induced by FIPV Black over time in Fcwf-4 and AK-D cells (MOI=0.1). Localized cell syncytia formation (black, closed arrows) and discrete cell death (white, open arrows) indicated on brightfield images taken at 20x magnification. B) Immunofluorescence staining of FIPV Black-infected Fcwf-4 CU and AK-D cells (MOI=0.1) at indicated hours post-infection (hpi). Nucleocapsid protein staining (Red; CCV2-2) indicates infected cells; nuclei stained in blue (Hoesch3342). Images taken at 60x magnification.
2.2. AK-D and Fcwf-4 CU cells facilitate rapid and consistent plaque assays for FIPV Black
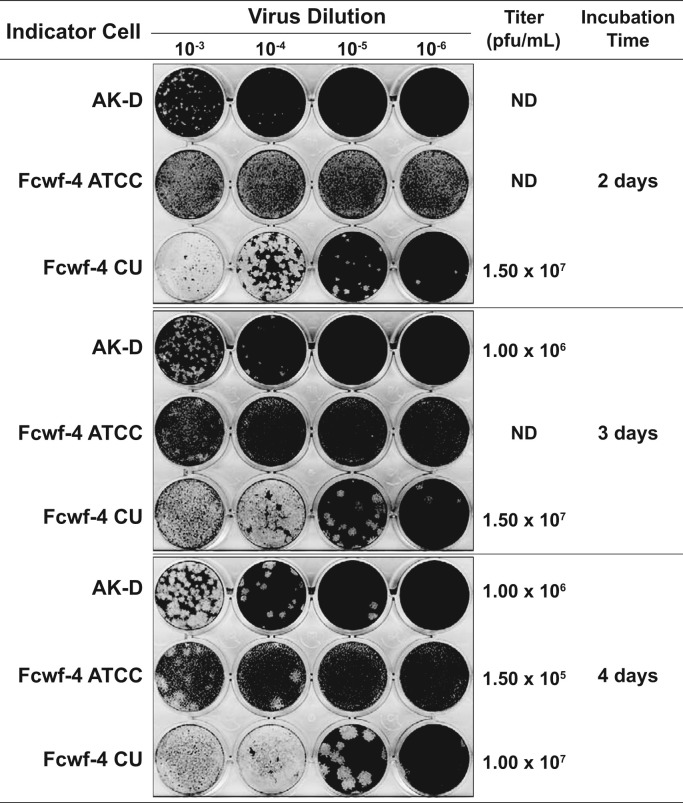
After observing the rapid and uniform development of CPE and release of virus into cell supernatants during infection of AK-D and Fcwf-4 CU cells, we reasoned that these cells would be employable in a standardized plaque assay to consistently determine FIPV Black titer. To this end, we calculated the endpoint titer and compared the size, uniformity, and timing of virus plaque development over time in AK-D, Fcwf-4 ATCC, and Fcwf-4 CU cells following infection with 10-fold dilutions of the same FIPV Black virus stock initially grown on Fcwf-4 CU cells. A detailed description of the plaque assay is provided in the Materials and Methods; we note here that Oxoid agar is critical for visualizing clear plaques. At 2 days post-infection (dpi), FIPV Black formed enumerable plaques on both AK-D and Fcwf-4 CU cells with the latter cells producing more numerous and larger plaques at higher dilutions ( Fig. 3 top). Plaques were not observed in Fcwf-4 ATCC cells at 2 dpi ( Fig. 3 top). Plaques were detected at 3 dpi ( Fig. 3 middle); and were more clear at 4 dpi in Fcwf-4 ATCC cells ( Fig. 3 bottom). Calculated titers overall were higher in Fcwf-4 CU (>107 pfu/mL) than in AK-D cells (≥106 pfu/mL); however, we report that both cell types are useful for determining virus titer, whereas the Fcwf-4 ATCC cells are not ideal for use in this assay.
Fig. 3.
Evaluating FIPV Black plaque development and titer over time in different feline cell lines. Tenfold serial dilutions (10-3-10-6) of virus inoculum, derived from 24 h cell-free supernatant of Fcwf-4 CU cells infected with FIPV Black (MOI=0.1), were applied to AK-D, Fcwf-4 ATCC, and Fcwf-4 CU indicator cells. Following Oxoid agar/media overlay, plates were incubated for 2 (top), 3 (middle), or 4 (bottom) days before cells were fixed with 3.7% formaldehyde and stained with 0.1% crystal violet. Titers were calculated by plaque counts and are presented as plaque forming units (pfu)/mL. ND indicates that titers were not determined.
2.3. Fcwf-4 CU cells are distinct in morphology and IFN-responsiveness
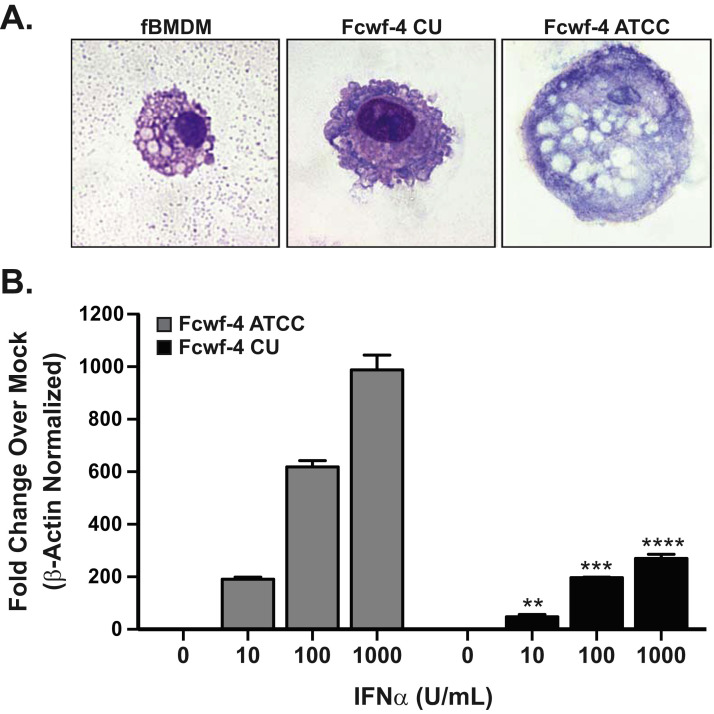
The apparent differences in production and kinetics of FIPV Black virus in two Fcwf-4 cell lines led us to ask if there are morphologic or functional differences between the two cell types. To answer this question, we first compared the single-cell morphologies of Fcwf-4 ATCC, Fcwf-4 CU cells, and primary feline bone marrow-derived macrophages (fBMDMs) by Wright-Giemsa staining. The typical morphologic characteristics of feline macrophages (large cytoplasmic inclusions, a non-dominant nucleus, a non-ruffled cell membrane) (Bienzle et al., 2003) were observed for the fBMDMs ( Fig. 4 A). Comparison of the two Fcwf-4 cell lines revealed stark differences in morphology. Fcwf-4 ATCC cells were large with a smooth cell membrane and had large and abundant cytoplasmic inclusions comparable to the fBMDMs ( Fig. 4 A). Fcwf-4 CU cells were more similar in size to fBMDMs; however, the Fcwf-4 CU cell line exhibited fewer cytoplasmic inclusions and more cell membrane ruffling ( Fig. 4 A) than either the fBMDMs or Fcwf-4 ATCC cells. Neither Fcwf-4 cell line had a “true” macrophage morphology further corroborating their original “macrophage-like” description (Jacobse-Geels and Horzinek, 1983). As macrophages are innate immune cells that restrict virus replication through production of interferon-stimulated genes (ISGs) in response to type I IFN, we reasoned that differences in virus replication may be due to variation in cell IFN-responsiveness. Therefore, we asked if Fcwf-4 ATCC and CU cell lines differed in responsiveness to exogenous type I IFN by measuring the resulting ISG54 transcript production following treatment with IFN. Remarkably, Fcwf-4 ATCC cells produced significantly higher ISG54 transcripts in response to IFN stimulation compared to Fcwf-4 CU cells ( Fig. 4 B), suggesting that the Fcwf-4 CU cells are much less responsive to IFN. This is not to say, however, that Fcwf-4 CU cells are insensitive to IFN since they also exhibit significant, dose-dependent ISG54 transcript production. Together, these data highlight the distinct morphology of the Fcwf-4 CU cells and suggest that enhanced virus replication in these cells may be due, at least in part, to reduced IFN-responsiveness.
Fig. 4.
Fcwf-4 CU cells differ in morphology and IFN-responsiveness from Fcwf-4 ATCC. A) Single-cell morphology of feline bone marrow-derived macrophages (fBMDMs), Fcwf-4 CU, and Fcwf-4 ATCC cells determined by Wright-Giemsa stain following cytospin preparation. Images were taken at 100 × . B) Interferon stimulated gene (ISG) 54 transcript levels from Fcwf-4 ATCC (grey) and Fcwf-4 CU (black) cell lines stimulated with increasing concentrations of feline type I interferon (IFNα). After 6 h, total RNA was extracted and analyzed by qPCR for ISG54 and feline beta-actin. ISG54 mRNA expression was normalized to β-actin via the delta Ct method (2-ΔCt[ΔCt=Ct(gene of interest)-Ct(β-actin)]), then presented as relative expression over corresponding mock (basal) expression. Data are representative of three independent experiments performed in triplicate and presented as means ± SD. Values were analyzed by unpaired t-test. * * P < 0.01; * ** P < 0.001; * ** * P < 0.0001.
2.4. Fcwf-4 CU cells replicate both biotypes of type II FCoV
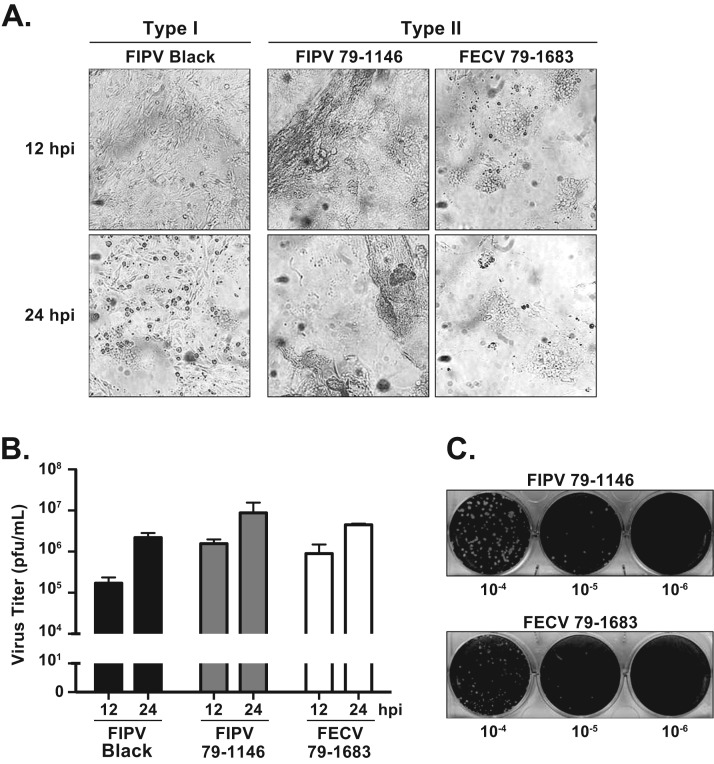
Due to the high titer and rapid kinetics of type I FIPV Black replication in the Fcwf-4 CU cells, we next addressed whether these cells are permissive to type II FCoV infection. Fcwf-4 CU cells were infected with type I FIPV Black or one of two type II viruses, FIPV 79–1146 or FECV 79–1683. Cell cytopathic effects were observed over time in these cells, with the viruses all forming similar, large syncytia ( Fig. 5 A). Cell-free virus titers were determined at 12 and 24 hpi. All three viruses accumulated to titers > 106 pfu/mL in supernatants by 24 hpi ( Fig. 5 B), with FIPV 79–1146 reaching the highest titer. The kinetics of the type II viruses were faster than the type I FIPV Black strain, producing higher titers by 12 hpi ( Fig. 5 B) and inducing more substantial syncytial CPE by 16 hpi ( Fig. 5 A). Next, we asked if the Fcwf-4 CU cells could be utilized in a plaque assay for determining the titer of type II FCoV. Indeed, both FIPV 79–1146 and FECV 79–1683 formed clear, uniform, enumerable plaques at 24 hpi ( Fig. 5 C). Thus, we have demonstrated that the Fcwf-4 CU cells replicate both FCoV types and biotypes to high titers and are employable in a plaque assay to consistently determine the titers of all viruses assayed.
Fig. 5.
Fcwf-4 CU cells and are permissive to both biotypes of type II FCoV. A) Cell cytopathic effect at 12 and 24 h post-infection (hpi) of Fcwf-4 CU cells infected with indicated FCoV strains (MOI=0.1). B) FCoV growth kinetics in Fcwf-4 CU cells (MOI=0.1) determined by C) plaque assay on Fcwf-4 CU indicator cells. Tenfold serial dilutions of cell-free virus inoculum were applied to cells. Following Oxoid agar/media overlay, cells were fixed in 3.7% formaldehyde and stained using 0.1% crystal violet after 24 h. FCoV used were the type I strain FIPV Black and the type II strains FIPV WSU 79–1146 and FECV WSU 79–1683. Plaque assay performed in triplicate; error bars ± SD.
3. Discussion
Since the isolation of the FIPV Black strain in 1980 (Black, 1980) it has remained a predominant model of type I FIPV because it is cultivatable in commercially available Fcwf-4 cells. However, different groups have reported major variations in the growth kinetics, maximum obtainable titer, and techniques for recovery of this virus from tissue culture (Jacobse-Geels and Horzinek, 1983, Pedersen et al., 1984, Tekes et al., 2012, Tekes et al., 2007, Thiel et al., 2014). For example, maximal titers measured by determining the TCID50 can range between > 103 (Jacobse-Geels and Horzinek, 1983) and 104 TCID50/mL (Takano et al., 2015) at 24–48 hpi, or measured by plaque assay between > 104 (36 hpi) (Tekes et al., 2012, Tekes et al., 2007) and > 107 pfu/mL (20 hpi) (This Report). This suggests either high variability in the Fcwf-4 cell lines used or co-adaptation between a particular virus and Fcwf-4 cell line used during laboratory cultivation. Indeed, we report significant differences in FIPV Black replication properties between an Fcwf-4 cell line that was newly purchased from the ATCC, and the Fcwf-4 cells that were established at Cornell University. The enhanced rate of FIPV Black virus growth and increased maximum titer (>107 pfu/mL by 20 hpi) obtained from infected Fcwf-4 CU cells, however, may not be due to co-adaptation with our particular strain of FIPV Black, as these cells also replicated both biotypes of type II viruses. Instead, the Fcwf-4 CU cells may be highly susceptible to FCoV infection in general and therefore may be particularly useful in generating highly-permissive cell types to isolate and grow clinical type I FCoVs. The increased virus infection of Fcwf-4 CU cells could be due to any number of cellular factors; however, it is tempting to speculate that the reduced IFN-responsiveness of the Fcwf-4 CU cells relative to the Fcwf-4 ATCC cells may significantly enhance infection in the former. Further, these Fcwf-4 CU cells may express a higher density of the yet unknown type I virus receptor (Cham et al., 2017, Dye et al., 2007) and therefore may be critical in identifying the receptor or other cellular characteristics that allow for enhanced type I virus replication.
FIPV Black infection of Fcwf-4 cells has also been reported to be highly cell-associated (Jacobse-Geels and Horzinek, 1983, Pedersen et al., 1984), requiring suspension and freeze-thaw cycling to release infectious virus. In contrast, we found significantly higher titers of virus in cell-free supernatants and speculate that multiple freeze-thaw cycles may actually decrease virus titer by damaging the virus envelope. The release of virus into cell supernatants and the uniform CPE observed in AK-D and Fcwf-4 CU cells were critical in establishing a standardized plaque assay using either cell type. Further, we report that the stage of CPE development can be used as an indicator of when maximal virus titers can be recovered.
One possible explanation for the varied reports on titer of FIPV Black is that laboratory lines of Fcwf-4 cells have deviated from the original ATCC stock. This is likely what occurred at Cornell University to produce the Fcwf-4 CU cell line, given that Fcwf-4 cells were obtained from the ATCC and then passaged for many years. To our knowledge, this is the first report describing the phenotypic differences between the original Fcwf-4 cells available from the ATCC and a distinct lineage that was derived from the original cells.
Many challenges are still associated with the growth of type I FCoV in tissue culture, including the lack of a known cell-entry receptor and no highly permissive cell type that rapidly grows clinical samples of these viruses. However, our studies with the Fcwf-4 CU cell line demonstrate that rapid, high titers of type I FIPV Black can be recovered from cell-free supernatants and enumerated using a standardized plaque assay. It is our hope that the Fcwf-4 CU cells will alleviate some of the technical hardships associated with the growth of type I FCoV and expedite investigation of a wider range of type I FCoV strains. The Fcwf-4 CU cells, due to their distinct growth kinetics and enhanced replication of FIPV Black virus, will be deposited at the ATCC to facilitate their distribution to the research community.
4. Materials and methods
4.1. Viruses
Feline coronavirus strains including type I feline infectious peritonitis virus (FIPV) Black (TN-406), type II FIPV WSU 79–1146, and type II feline enteric coronavirus (FECV) WSU 79–1683 were kindly provided by Dr. Fred Scott, Cornell University College of Veterinary Medicine, Ithaca, NY.
4.2. Cell Lines
Feline airway epithelial (AK-D) cells were purchased from the American Type Culture Collection (ATCC) (ATCC® CCL-150™) and maintained in Dulbecco's Modified Eagle Medium (DMEM; Gibco, #12100-046) containing 10% fetal bovine serum (FBS) (Atlanta Biologicals, #S11150), supplemented with 2.2 g/L of sodium bicarbonate (Sigma, #S5761), 1% non-essential amino acids (HyClone, #SH30238.01), 1% HEPES (HyClone, #SH30237.01), 1% sodium pyruvate (Corning, #25-000-CI), 1% L-glutamine (HyClone, #SH30034.01), and 1% penicillin/ streptomycin (Corning, #30-002-CI). When cells grew to a confluent monolayer, the medium was removed and the monolayer was rinsed with PBS. The cells were removed by addition of 2 mL of 0.25% trypsin (Gibco, #15090-046) in versene solution (0.48 mM EDTA in PBS) for 1–2 min at room temperature. For routine passaging, approximately 5.0 × 105 - 1.0 × 106 cells were transferred (1:5 split) to a new T-75 flask every 3 days. Felis catus whole fetus (Fcwf-4) cells were purchased from the ATCC (ATCC® CRL-2787™), designated Fcwf-4 ATCC cells. Fcwf-4 ATCC cells were maintained in Minimal Essential Medium Eagle (EMEM) (Sigma, #M0268) containing 10% FBS, supplemented with 1.5 g/L sodium bicarbonate, 1% non-essential amino acids, 1% HEPES, 1% sodium pyruvate, 1% L-glutamine and 1% penicillin/streptomycin. As described by the ATCC, the doubling time for these cells is > 31 h. When cells were confluent in T-25 flask, the monolayer was washed with PBS, then cells were removed by addition of 1.5 mL 0.25% trypsin-versene solution for 2–3 min at room temperature. For routine passaging, approximately 1.0 × 105 - 5.0 × 105 cells were transferred (1:3 split) to a new T-25 flask every 3 days. A second source of Felis catus whole fetus cells were provided by Dr. Edward J. Dubovi, Cornell University College of Veterinary Medicine, Ithaca, NY, designated Fcwf-4 CU, and maintained in the same medium as the Fcwf-4 ATCC cells. When cells were confluent, the monolayer was washed with PBS, then cells were removed by addition of 2 mL 0.25% trypsin-versene solution for 1–2 min at room temperature. For routine passaging, approximately 5.0 × 105 - 1.0 × 106 cells were transferred (1:10 split) to a new T-75 flask every 3 days. All cells used in this study were monitored for mycoplasma contamination using a PCR-based assay. Cell culture supernatants were routinely collected after 3 days of culture and then heat-inactivated at 95 °C for 10 min. PCR amplification for mycoplasma detection was performed using a forward primer: 5’- GGC GAA TGG GTG AGT AAC ACG -3’ and a reverse primer: 5’- CGG ATA ACG CTT GCG ACC TAT G -3’. Thermocycler settings were as follows: initial denaturation at 95 °C for 10 min; 35 cycles consisting of denaturation at 95 °C for 45 s, annealing at 60 °C for 45 s, and extension at 72 °C for 60 s; and a final extension at 72 °C for 10 min. The PCR products were analyzed on 1% (w/v) agarose gel. DNA fragments were visualized with a UV transilluminator after being stained with ethidium bromide. If mycoplasma was detected, the cells were treated for 7 days with 0.5 μg/mL of mycoplasma removal agent (Bio-Rad, #BUF035). All results shown here are from mycoplasma negative cells.
4.3. Generation of feline bone marrow-derived macrophages
Feline femurs were harvested by Cornell University Veterinary Biobank from specific pathogen free cats euthanized according to IACUC approved protocols. Total bone marrow content was collected. Red blood cells and fatty tissue were removed by lysis in ACK lysis buffer and straining through a 0.70 µm filter (Falcon). The remaining cells, predominantly hematopoietic stem cells, were cryopreserved at 5.0 × 107 cells/mL in 90% FBS and 10% DMSO. Feline bone marrow derived macrophages (fBMDMs) were differentiated as previously reported (Gow et al., 2013) with slight modification. Briefly, 5.0 × 107 bone marrow cells were plated in 100 × 26 mm petri dishes (VWR, #25387-030) in DMEM (Corning, #10–017-CV) supplemented with 20% FBS, 10,000 IU/mL recombinant human (rh) M-CSF (PeproTech, #300-25) and 50 μM β-mercaptoethanol then incubated at 37 °C and 5% CO2. At day 3, supernatant was removed, clarified of cells, diluted 1:1 with fresh DMEM supplemented with 20% FCS and 10,000 IU/mL rhM-CSF, and returned to cells. fBMDMs were recovered on day 6 by gentle aspiration in PBS following 30 min, 4 °C incubation in PBS.
4.4. Plaque assay
The plaque assay technique was established using both AK-D and Fcwf-4 CU cells. 6.5 × 105 cells per well were plated in 6-well plates or 3.0 × 105 cells per well were plated in 12-well plates. Cells were infected with 10-fold serial dilutions of viral samples for 1 h at 37 °C, followed by overlaying with a 0.5% Oxoid agar (Oxoid LTD, #LP0028)-DMEM containing 1% FBS mixture. Plates were incubated at 37 °C for 48 h (or the indicated time) and fixed using 3.7% formaldehyde-PBS solution for 30 min. Viral plaques were visualized by staining with 0.1% crystal violet for 30 min and photographed. We note plaques were clearly evident when we used Oxoid agar, but not if we used Noble agar.
4.5. Viral growth kinetics
We analyzed the growth kinetics of type I FIPV Black in three cell lines: AK-D, Fcwf-4 CU, and Fcwf-4 ATCC. Type II FCoV titers were evaluated in Fcwf-4 CU cells. For all cell types, 1.5 × 105 cells were plated in 24-well plates or 3.0 × 105 cells in 12-well plates. After incubating for 24 h, cells were infected with FIPV in serum-free media at a multiplicity of infection (MOI) of 0.1 at 37 °C. After a 1 h incubation, the infectious media were replaced with fresh media containing 2% FBS. At the indicated time points, cell-free supernatants and/or infected cells were harvested and used for titration by plaque assay on AK-D (FIPV Black) or Fcwf-4 CU (type II FCoVs) cells.
4.6. Evaluating virus titers from cell-free supernatant versus cell-associated samples
The supernatant was collected from the cultures at the time indicated and subjected to centrifugation at 2200 ×g for 10 min at 4 °C to remove any dead cells. This cell free supernatant was aliquoted and frozen at −80 °C until use. To prepare the cell-associated sample, 0.5 mL of medium was added to the infected cells in the 24-well plate, and the entire plate was frozen at −80 °C. The infected cells were then subjected to two additional rounds of freezing and thawing (37 °C for 1–2 min). After the third thaw, the cells and medium were transferred to a centrifuge tube and centrifuged at 2200 ×g for 10 min at 4 °C to remove cell debris. The supernatant containing the viruses released from the cells during the freeze-thaw process was designated the cell-associated virus sample.
4.7. Immunofluorescence detection of FIPV nucleocapsid protein
Monolayers of 1.0 × 105 AK-D and Fcwf-4 CU cells were cultured in 8-well chamber slides (Nalge Nunc International, #177445) at 37 °C for 24 h. Cells were infected with FIPV Black at a MOI of 0.1 for 1 h at 37 °C. At times indicated, the infected cells were fixed with 3.7% formaldehyde-PBS solution for 30 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and then incubated with blocking solution containing 5% normal goat serum and 0.1% Triton X-100 in PBS at 4 °C overnight. For immunofluorescence staining of FIPV Black-infected cells, the cells were incubated with mouse monoclonal anti-FIPV nucleocapsid (CCV2-2) (Poncelet et al., 2008) (Bio-Rad, #MCA2594B) as a primary antibody at a dilution of 1:500 at room temperature for 1 h. Cells were then incubated with a secondary antibody, Alexa Fluor 568-conjugated goat anti-mouse IgG (Thermo Fisher Scientific, #A11004) at a dilution of 1:1000 in the presence of Hoechst33342 (Thermo Fisher Scientific, #H1399) at a dilution of 1:1000 for nucleus stain. After 30 min incubation with the secondary antibody at room temperature, the cells were then washed with PBS, mounted, and examined under a fluorescence microscope.
4.8. Quantification of ISG54 transcript production by RT-qPCR following stimulation with feline IFN
Cells (1.0 ×105 in a 24-well plate) were treated with 10, 100, or 1000 U/mL of purified feline IFN-alpha (PBL Assay Science, #15100-1) for 6 h. To determine ISG54 and feline β-actin mRNA production, total RNA was extracted and an equal amount of RNA (1000 ng) was used for cDNA synthesis using RT2 HT First-Strand Kit (Qiagen, #330401). qPCR was performed with specific primers for feline β-actin transcript (FWD 5’- CAA CCG TGA GAA GAT GAC TCA GA -3’; REV 5’- CCC AGA GTC CAT GAC AAT ACC A -3’) or ISG54 transcript (FWD 5’- CCT GAG CTG CAG CCT TTC AGA ACA G -3'; REV 5'- CAC GTG AAA TGG CAT TTA AGT TGC CGC AG -3') using RT2 SYBR Green qPCR Mastermix (Qiagen, #330502). A Bio-Rad CFX96 thermocycler was set as follows: one step at 95 °C (10 min); 40 cycles of 95 °C (15 s), 60 °C (1 min), and plate read; one step at 95 °C (10 s); and a melt curve from 65 °C to 95 °C at increments of 0.5 °C/0.05 s. Samples were evaluated in triplicate and data are representative of three independent experiments. The levels of mRNA are reported relative to β-actin mRNA and expressed as 2−ΔCT [ΔC T = C T(gene of interest) −C T(β-actin)].
4.9. Morphologic staining
Feline bone marrow-derived macrophages, Fcwf-4 CU, or Fcwf-4 ATCC cells, grown for 24 h in 100 × 26 mm petri dishes (VWR, #25387-030), were washed with PBS, incubated in PBS for 30 min at 4 °C, then collected with gentle pipetting. Using a Cytospin (Shandon), 200 μL of cells (10,000 cells/mL) suspended in PBS supplemented with 2% BSA were spun onto glass coverslips (pre-treated with 2% BSA in PBS) at 115 xg for 6 min. Cells were dried, rinsed in PBS, then fixed in absolute methanol for 1 min. Wright-Giemsa (Thermo Fisher Scientific, #9990710) staining was performed per the manufacturer's instructions using a 3 min primary stain and a 2 min counter stain, and washed with 1 mL rinse solution. Slides were dried and imaged under oil immersion at 100x magnification.
Acknowledgements
We thank Matthew Hackbart, Dr. Xufang Deng, Dr. Jean K. Millet and Javier Jaimes for helpful discussions. We also thank Dr. Fred Scott and Dr. Edward J. Dubovi for the provision of reagents. Tissue samples and associated phenotypic data were provided by the Cornell Veterinary Biobank, a resource built with the support of National Institutes of Health (NIH) grant R24 GM082910 and the Cornell University College of Veterinary Medicine. This work was supported by a Pilot Project grant issued to S.C.B. by Loyola University of Chicago and a research grant from the Winn Feline Foundation Bria Fund (#MTW17–022 to S.C.B. and G.R.W.). R.C.M. is supported by the (NIH) T32 Training Grant for Experimental Immunology (#AI007508) and the Arthur J. Schmitt Dissertation Fellowship in Leadership and Service (Arthur J. Schmitt Foundation). G.R.W. is supported by research grants from the Cornell Feline Health Center and the Winn Feline Foundation.
References
- Addie D.D. Feline coronaviral infections. In: Greene C., editor. Infectious Diseases of the Dog and Cat. Saunders; 2011. pp. 92–108. [Google Scholar]
- Addie D.D., Jarrett O. A study of naturally occurring feline coronavirus infections in kittens. Vet. Rec. 1992;130:133–137. doi: 10.1136/vr.130.7.133. [DOI] [PubMed] [Google Scholar]
- Addie D.D., Schaap I.A.T., Nicolson L., Jarrett O. Persistence and transmission of natural type I feline coronavirus infection. J. Gen. Virol. 2003;84:2735–2744. doi: 10.1099/vir.0.19129-0. [DOI] [PubMed] [Google Scholar]
- American Type Culture Collection, 2013. Fcwf4 [Fcwf] (ATCC ® CRL 2787 ™) Product Sheet.
- Benetka V., Kübber-Heiss A., Kolodziejek J., Nowotny N., Hofmann-Parisot M., Möstl K. Prevalence of feline coronavirus types I and II in cats with histopathologically verified feline infectious peritonitis. Vet. Microbiol. 2004;99:31–42. doi: 10.1016/j.vetmic.2003.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienzle D., Reggeti F., Clark M.E., Chow C. Immunophenotype and functional properties of feline dendritic cells derived from blood and bone marrow. Vet. Immunol. Immunopathol. 2003;96:19–30. doi: 10.1016/S0165-2427(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Black J.W. Recovery and in vitro cultivation of a coronavirus from laboratory-induced cases of feline infectious peritonitis (FIP) Vet. Med. Small Anim. Clin. 1980;75:811–814. [PubMed] [Google Scholar]
- Cham T.C., Chang Y.C., Tsai P.S., Wu C.H., Chen H.W., Jeng C.R., Pang V.F., Chang H.W. Determination of the cell tropism of serotype 1 feline infectious peritonitis virus using the spike affinity histochemistry in paraffin-embedded tissues. Microbiol. Immunol. 2017;61:318–327. doi: 10.1111/1348-0421.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.W., de Groot R.J., Egberink H.F., Rottier P.J.M. Feline infectious peritonitis: insights into feline coronavirus pathobiogenesis and epidemiology based on genetic analysis of the viral 3c gene. J. Gen. Virol. 2010;91:415–420. doi: 10.1099/vir.0.016485-0. [DOI] [PubMed] [Google Scholar]
- Dye C., Temperton N., Siddell S.G. Type I feline coronavirus spike glycoprotien fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J. Gen. Virol. 2007;88:1753–1760. doi: 10.1099/vir.0.82666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow D.J., Garceau V., Pridans C., Gow A.G., Simpson K.E., Gunn-Moore D., Hume D.A. Cloning and expression of feline colony stimulating factor receptor (CSF-1R) and analysis of the species specificity of stimulation by colony stimulating factor-1 (CSF-1) and interleukin-34 (IL-34) Cytokine. 2013;61:630–638. doi: 10.1016/j.cyto.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., de Groot R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Vennema H., Horzinek M.C., Rottier P.J.M., de Groot R.J. The molecular genetics of feline coronaviruses: comparitive sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212:622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohdatsu T., Izumiya Y., Yokoyama Y., Kida K., Koyama H. Differences in virus receptor for type I and type II feline infectious peritonitis virus. Arch. Virol. 1998;143:839–850. doi: 10.1007/s007050050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohdatsu T., Okada S., Ishizuka Y., Yamada H., Koyama H. The prevalence of types I and II feline coronavirus infections in cats. J. Vet. Med. Sci. 1992;54:557–562. doi: 10.1292/jvms.54.557. [DOI] [PubMed] [Google Scholar]
- Hohdatsu T., Okada S., Koyama H. Characterization of monoclonal antibodies against feline infectious peritonitis virus type II and antigenic relationship between feline, porcine, and canine coronaviruses. Arch. Virol. 1991;117:85–95. doi: 10.1007/BF01310494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobse-Geels H.E., Horzinek M.C. Expression of feline infectious peritonitis coronavirus antigens on the surface of feline macrophage-like cells. J. Gen. Virol. 1983;64:1859–1866. doi: 10.1099/0022-1317-64-9-1859. [DOI] [PubMed] [Google Scholar]
- Kennedy M., Citino S., McNabb A.H., Moffatt A.S., Gertz K., Kania S. Detection of feline coronavirus in captive Felidae in the USA. J. Vet. Diagn. Investig. 2002;14:520–522. doi: 10.1177/104063870201400615. [DOI] [PubMed] [Google Scholar]
- Kim Y., Liu H., Galasiti Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.O., Pedersen N.C. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12:e1005531. doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lovell S., Tiew K.-C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.-O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Mandadapu S., Groutas W., Chang K. Potent inhibition of feline coronaviruses with peptidyl compounds targeting coronavirus 3C-like protease. Antivir. Res. 2013;97:161–168. doi: 10.1016/j.antiviral.2012.11.005.Potent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Shivanna V., Narayanan S., Prior A.M., Weerasekara S., Hua D.H., Kankanamalage A.C., Groutas W.C., Chang K.O. Broad-spectrum inhibitors against 3C-like proteases of feline coronaviruses and feline caliciviruses. J. Virol. 2015;89:4942–4950. doi: 10.1128/JVI.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A., Meli M.L. Feline infectious peritonitis: still an enigma? Vet. Pathol. 2014;51:505–526. doi: 10.1177/0300985814522077. [DOI] [PubMed] [Google Scholar]
- Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19:1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra, B.N., Sams, K.L., Lee, D.W., Whittaker, G.R., 2014. Feline coronaviruses associated with feline infectious peritonitis have modifications to spike protein activation sites at two discrete positions. arXiv:1412.4034v1 (q-bio.GN).
- Lin C.N., Su B.L., Huang H.P., Lee J.J., Hsieh M.W., Chueh L.L. Field strain feline coronaviruses with small deletions in ORF7b associated with both enteric infection and feline infectious peritonitis. J. Feline Med. Surg. 2009;11:413–419. doi: 10.1016/j.jfms.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B.G., Perron M., Murakami E., Bauer K., Park Y., Eckstrand C., Liepnieks M., Pedersen N.C. The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Vet. Microbiol. 2018;219:226–233. doi: 10.1016/J.VETMIC.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. An update on feline infectious peritonitis: virology and immunopathogenesis. Vet. J. 2014;201:123–132. doi: 10.1016/j.tvjl.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline Med. Surg. 2009;11:225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. Serologic studies of naturally occurring feline infectious peritonitis. Am. J. Vet. Res. 1976;37:1449–1453. [PubMed] [Google Scholar]
- Pedersen N.C., Allen C.E., Lyons L.A. Pathogenesis of feline enteric coronavirus infection. J. Feline Med. Surg. 2008;10:529–541. doi: 10.1016/j.jfms.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Black J.W., Boyle J.F., Evermann J.F., McKeirnan A.J., Ott R.L. Molecular Biology and Pathogenesis of Coronaviruses. Springer; Boston, MA: 1984. Pathogenic differences between various feline coronavirus isolates; pp. 365–380. [DOI] [PubMed] [Google Scholar]
- Pedersen N.C., Kim Y., Liu H., Galasiti Kankanamalage A.C., Eckstrand C., Groutas W.C., Bannasch M., Meadows J.M., Chang K.-O. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J. Feline Med. Surg. 2017;20:378–392. doi: 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Liu H., Scarlett J., Leutenegger C.M., Golovko L., Kennedy H., Kamal F.M. Feline infectious peritonitis: role of the feline coronavirus 3c gene in intestinal tropism and pathogenicity based upon isolates from resident and adopted shelter cats. Virus Res. 2012;165:17–28. doi: 10.1016/j.virusres.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.E., Hilt D.A., Jackwood M.W. Comparative sequence analysis of full-length genome of FIPV at different tissue passage levels. Virus Genes. 2013;47:490–497. doi: 10.1007/s11262-013-0972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A.M., Vennema H., Foley J.E., Pedersen N.C. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J. Clin. Microbiol. 1996;34:3180–3184. doi: 10.1128/jcm.34.12.3180-3184.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncelet L., Coppens A., Peeters D., Bianchi E., Grant C.K., Kadhim H. Detection of antigenic heterogeneity in feline coronavirus nucleocapsid in feline pyogranulomatous meningoencephalitis. Vet. Pathol. 2008;45:140–153. doi: 10.1354/vp.45-2-140. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Regan A.D., Millet J.K., Tse L.P.V., Chillag Z., Rinaldi V.D., Licitra B.N., Dubovi E.J., Town C.D., Whittaker G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J.M., Nakamura K., Schellen P., Volders H., Haijema B.J. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005;79:14122–14130. doi: 10.1128/JVI.79.22.14122-14130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John S.E., Therkelsen M.D., Nyalapatla P.R., Osswald H.L., Ghosh A.K., Mesecar A.D. X-ray structure and inhibition of the feline infectious peritonitis virus 3C-like protease: Structural implications for drug design. Bioorg. Med. Chem. Lett. 2015;25:5072–5077. doi: 10.1016/j.bmcl.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Nakano K., Doki T., Hohdatsu T. Differential effects of viroporin inhibitors against feline infectious peritonitis virus serotypes I and II. Arch. Virol. 2015;160:1163–1170. doi: 10.1007/s00705-015-2370-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Bank-Wolf B., Maier R., Thiel H.-J., Thiel V. Chimeric feline coronaviruses that encode type II spike protein on type I genetic background display accelerated viral growth and altered receptor usage. J. Virol. 2010;84:1326–1333. doi: 10.1128/jvi.01568-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Stallkamp I., Thiel V., Thiel H.-J. Genome organization and reverse genetic analysis of a type I feline coronavirus. J. Virol. 2007;82:1851–1859. doi: 10.1128/JVI.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Spies D., Bank-Wolf B., Thiel V., Thiel H.-J. A reverse genetics approach to study feline infectious peritonitis. J. Virol. 2012;86:6994–6998. doi: 10.1128/JVI.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Thiel H.J. Advances in Virus Research. Academic Press; 2016. Feline coronaviruses: pathogenesis of feline infectious peritonitis; pp. 193–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Thiel H.-J., Tekes G. Tackling feline infectious peritonitis via reverse genetics. Bioengineered. 2014;5:396–400. doi: 10.4161/bioe.32133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker G.R., André N.M., Millet J.K. Improving virus taxonomy by recontextualizing sequence-based classification with biologically relevant data: the case of the alphacoronavirus 1 species. mSphere. 2018;3:1–8. doi: 10.1128/mSphere.00463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]