Abstract
2,4-Diacetylphloroglucinol (DAPG)-producing Pseudomonas spp. in the P. fluorescens complex are primarily responsible for a natural suppression of take-all of wheat known as take-all decline (TAD) in many fields in the United States. P. brassicacearum, the most common DAPG producer found in TAD soils in the Pacific Northwest (PNW) of the United States, has biological control, growth promoting and phytotoxic activities. In this study, we explored how the wheat cultivar affects the level of take-all suppression when grown in a TAD soil, and how cultivars respond to colonization by P. brassicacearum. Three cultivars (Tara, Finley, and Buchanan) supported similar rhizosphere population sizes of P. brassicacearum when grown in a TAD soil, however they developed significantly different amounts of take-all. Cultivars Tara and Buchanan developed the least and most take-all, respectively, and Finley showed an intermediate amount of disease. However, when grown in TAD soil that was pasteurized to eliminate both DAPG producers and take-all suppression, all three cultivars were equally susceptible to take-all. The three cultivars also responded differently to the colonization and phytotoxicity of P. brassicacearum strains Q8r1–96 and L5.1–96, which are characteristic of DAPG producers in PNW TAD soils. Compared with cultivar Tara, cultivar Buchanan showed significantly reduced seedling emergence and root growth when colonized by P. brassicacearum, and the response of Finley was intermediate. However, all cultivars emerged equally when treated with a DAPG-deficient mutant of Q8r1–96. Our results indicate that wheat cultivars grown in a TAD soil modulate both the robustness of take-all suppression and the potential phytotoxicity of the antibiotic DAPG.
Keywords: Gaeumannomyces graminis var. tritici
Crop plants often do not have genetic resistance to pathogenic soilborne fungi and nematodes, therefore they rely on individual or consortia of species of the soil or rhizosphere microbiota to defend their roots by directly inhibiting pathogens or inducing host resistance mechanisms (Bakker et al. 2013; Pieterse et al. 2014; Schlatter et al. 2017; Weller 2015; Weller et al. 2007). Suppressive soils (Expósito et al. 2017; Kyselková and Moënne-Loccoz 2012; Mendes et al. 2011; Raaijmakers and Mazzola 2016; Schlatter et al. 2017; Weller et al. 2002) are examples of natural microbial-based defense. Suppressive soils are “long standing” because a natural characteristic of the soil, which has unknown origins, limits a disease, or “induced” because the ability to suppress a disease is initiated by monoculture or growing a susceptible crop (Expósito et al. 2017; Schlatter et al. 2017; Weller 2015; Weller et al. 2002).
Take-all decline (TAD) is an example of induced suppression and controls take-all of wheat caused by Gaeumannomyces graminis var. tritici (Sanguin et al. 2009; Schlatter et al. 2017; Weller 2015; Weller et al. 2007). TAD is a spontaneous reduction in the severity of take-all, occurring during monoculture of wheat or barley (Hornby 1998; Schlatter et al. 2017; Weller 2015; Weller et al. 2002). In Pacific Northwest (PNW) wheat fields and other fields in the United States, TAD results from the enrichment of 2, 4-diacetylphloroglucinol (DAPG)-producing Pseudomonas spp. to population sizes of greater than 105 CFU per g of root, which is the threshold needed to suppress take-all (Raaijmakers and Weller 1998; Weller 2015; Weller et al. 2002). PNW TAD soils lose suppressiveness when DAPG-producing pseudomonads are eliminated, and nonsuppressive (conducive) soils gain suppressive activity when the bacteria are introduced (Raaijmakers and Weller 1998; Schlatter et al. 2017). DAPG is produced on wheat roots in TAD soils (Raaijmakersetal.1999) and the take-all pathogen is very sensitive to the antibiotic (Keel et al. 1992; Kwak et al. 2009), but it does not develop resistance to DAPG during wheat monoculture (Kwak et al. 2009). DAPG-deficient mutants of Pseudomonas lose the ability to control take-all (Weller 2015).
Pseudomonads that produce DAPG belong to the P. fluorescens complex (Loper et al. 2012) and they are genetically diverse (Frapolli et al. 2008; Weller et al. 2007), with 22 genotypes (designated A-T, PfY, and PfZ) described, but more are yet to be isolated (De La Fuente et al. 2006; Landa et al. 2002, 2006; Loper et al. 2012; Mavrodi et al. 2001; McSpadden Gardener et al. 2000; Weller et al. 2007). In PNW TAD soils, multiple genotypes occur, but D-genotype pseudomonads are primarily responsible for suppression of take-all, and these bacteria aggressively colonized wheat roots (Landa et al. 2003, 2006; Raaijmakers and Weller 2001; Weller et al. 2007). On the basis of whole genome sequencing, Loper et al. (2012) reported that the D-genotype P. fluorescens strain Q8r1–96 is related to P. brassicacearum NFM421. This finding prompted a new species designation of P. brassicacearum for D-genotype strains like Q8r1–96. DAPG-producers now occur in four species in the P. fluorescens complex: P. fluorescens, P. protegens, P. kilonensis, and P. brassicacearum (Loper et al. 2012; Vacheron et al. 2018). P. brassicacearum is especially notable because it has biological control, growth-promoting and pathogenic traits (Belimov et al. 2007) and it induces resistance in plants (Pieterse et al. 2014; Weller et al. 2012).
TAD occurs globally (Weller et al. 2002) and is used by PNW and other U.S. wheat farmers to manage take-all (Cook 2003). TAD development follows a similar pattern everywhere but the level of take-all suppression and the length of time before its onset (usually 4 to 6 years) varies among fields depending on the previous cropping history, local climate and soil factors (Cook 2007; Schlatter et al. 2017; Weller et al. 2002). Knowledge gaps about the variations in the length of time before the onset of TAD, and fluctuations in the intensity of disease suppression occurring among fields and years, still need to be filled (Cook and Veseth 1991; Hornby 1998; Weller 2015; Weller et al. 2002).
One factor that probably contributes to variations in the onset and intensity of TAD is the differences among wheat cultivars in ability to initiate and sustain take-all suppression. For example, wheat cultivars differ in their supportiveness of DAPG-producing pseudomonads (Kwak et al. 2012; Okubara et al. 2004), the genotypes that colonize them (Mazzola et al. 2004; Meyer et al. 2010), the amount of DAPG produced in the rhizosphere (Kwak et al. 2012; Okubara and Bonsall 2008), and expression of plant defense genes when colonized by DAPG producers (Maketon et al. 2012; Okubara et al. 2010). Kwaket al. (2012) demonstrated that cultivars Tara and Buchanan differ in the severity of take-all that develops when colonized by P. brassicacearum Q8r1–96. In a related study, Notz et al. (2001) showed that the expression of phlA in the 2,4-DAPG biosynthesis operon, differed significantly among populations of P. protegens CHA0 colonizing different corn cultivars. The basis for the differential responses of DAPG-producing pseudomonads to roots of different plants or cultivars is thought to be due to complex chemical cross-talk in the rhizosphere (Maurhofer et al. 2004).
The aim of our study was to determine whether the wheat cultivar affects the level of take-all suppression that occurs in a TAD soil. Our study builds on previous work showing that the interaction of P. brassicacearum strain Q8r1–96 with wheat is cultivar dependent (Kwak et al. 2012). We show that cultivar Tara supported a greater suppression of take-all in a TAD soil than did cultivar Buchanan, but the cultivar effect was lost when the soil was pasteurized. Furthermore, the supportiveness of each cultivar to take-all suppression correlated with the cultivar’s ability to tolerate phytotoxicity associated with colonization by P. brassicacearum. Collectively, our findings provide further insight into the basis of the relationship between cultivars of a crop and specific members of the rhizosphere microbial community that provide beneficial services to the plant.
MATERIALS AND METHODS
Soil and plants.
Wheat (Triticum aestivum L.) cultivars Tara (hard red spring), Finley (hard red winter), Buchanan (hard red winter), and Louise (soft white spring) were used in this study (Okubara et al. 2004). Tara was derived from the cross KODIAK/SPILLMAN/WPB00906, which was first made in 1987, and released commercially in 2001. Finley was derived from the cross WESTON/HATTON sib/TX69A450–1, which was made in 1989 and was released commercially in 1996. Buchanan was selected from an unknown field cross in 1982 and released commercially in 1991. Louise is a F4:5 head row selection derived from the cross Wakanz (PI506352)/Wawawai (PI574538), which was made in1992 and released commercially in 2006. Louise is currently the most widely grown soft white spring wheat in the PNW. These cultivars were selected for this study because they differ in the amount of DAPG produced on their roots (Kwak et al. 2012; Okubara and Bonsall 2008) and have different patterns of gene expression when exposed to P. brassicacearum Q8r1–96 (Maketon et al. 2012).
TAD is a field phenomenon but once established, the suppressiveness of the soil can be expressed and manipulated under controlled conditions in the greenhouse. Quincy take-all decline (Q-TAD) soil was collected in November 2011 and 2012 from the upper 30 cm of the soil profile from a field near Quincy, WA. The characteristics and disease suppressive activity of this TAD soil have been described (Raaijmakers and Weller 1998; Raaijmakers et al. 1997; Weller et al. 1997). The soil was sieved through a 0.5-cm-mesh screen, air dried and stored in plastic buckets at room temperature. To ensure that the population of DAPG producers was at a threshold density (>105 CFU g−1 of root fresh weight), which is required for take-all suppression, at the onset of each experiment, Q-TAD soil (2 kg) was placed in plastic pots (18 cm diameter × 25 cm deep) and 30 seeds of the spring wheat cultivar Louise were sown in each pot. Plants were maintained at 15 to 16°C in a growth room (12-h photoperiod), roots and shoots were harvested after 3 weeks and the remaining soil was mixed and again sown to cultivar Louise. After the second cycle of wheat, the population size of DAPG producers in the rhizosphere of individual seedlings was determined with the PCR-based endpoint-dilution assay (described below) followed by RFLP analysis of the terminal dilution (described below) to determine the dominant genotype of the DAPG producer present in the rhizosphere. One half of the cycled Q-TAD soil was pasteurized at 70°C for 2 h using compressed air mixed with steam (pasteurized Q-TAD) in order to eliminate both DAPG producers and take-all suppression (Raaijmakers and Weller 1998).
Fungal and bacterial strains and growth media.
G. graminis var. tritici strain R3–111a-1 was isolated from wheat in a field near Moses Lake, WA (K wak et al. 2009; Okagaki et al. 2015) and grown on 1/5-strength homemade potato dextrose agar (1/5× PDA) plates. Potato slices (40 g) were boiled in distilled water and the filtered extract was increased to a volume of 1 liter with distilled water. Dextrose (4 g liter−1) and agar (18 g liter−1) (Sigma Chemical Co., St. Louis, MO), were added, and the medium was adjustedtopH6.5 prior to autoclaving (Kwak et al. 2009). For long-term storage, strain R3–111a-1 was inoculated in the center of a plate of 1/5× PDA plus rifampicin (100 μg ml–1; Sigma Chemical Co.) and when the fungus covered half of the plate, it was sealed with parafilm and placed in the dark at 4°C.
Bacterial strains used in this study are shown in Table 1. DAPG-producing P. brassicacearum strains Q8r1–96 and L5.1–96 (formerly P. fluorescens D-genotype) were cultured on King’s medium B (KMB) (King et al. 1954) and stored in glycerol at −80°C. Strain Q8r1–96 was isolated from wheat grown in Q-TAD soil (Raaijmakers and Weller 2001) and its genome has been sequenced (Loper et al. 2012). Strain L5.1–96 was isolated from wheat grown in soil from a TAD field at the WSU Dryland Research Station, Lind, WA (Lind TAD) (Raaijmakers et al. 1997). Both strains have been well described for their biocontrol activity (Weller et al. 2007). Also used in the study, were three type III secretion system (T3SS) mutants of Q8r1–96; two of these, Q8r1–96ΔOPQR and Q8r1–96rscV, are mutated in genes encoding different conserved parts of the type III secretion apparatus (Mavrodi et al. 2011). The third mutant, Q8r1–96rspL, has a mutation in a gene encoding an alternative sigma factor that positively regulates transcription of T3SS genes in Q8r1–96 (Mavrodi et al. 2011).
TABLE 1.
Strains and mutants of Pseudomonas brassicacearum used in this study
| Strains | Characteristicsz | Source or reference |
|---|---|---|
| L5.1–96 | Wheat rhizosphere isolate from Lind, WA; DAPG+; Rifr | Landa et al. 2003 |
| Q8r1–96 | Wheat rhizosphere isolate from Quincy, WA; DAPG+; Rifr | Raaijmakers and Weller 1998 |
| Q8r1–96ΔOPQR | T3SS-deficient mutant; Δ(rsOP-rscQR)::aacC1; Rifr Gmr | Mavrodi et al. 2011 |
| Q8r1–96rspL | T3SS-deficient mutant; rspL::aph; Rifr Kanr | Mavrodi et al. 2011 |
| Q8r1–96rscV | T3SS-deficient mutant; rscv::aph; Rifr Kanr | Mavrodi et al. 2011 |
| Q8r1–96phlD | DAPG-deficient mutant; phlD::aph; Rifr Kanr | This study |
Rifr, rifampicin resistant; Kanr, kanamycin resistant; Gmr, gentamicin resistant; aph, aminoglycoside 3′-phosphotransferase; aacC1, gentamicin acetyltransferase 3–1; T3SS, type III protein secretion system; and DAPG, 2,4-diacetylphloroglucinol.
The Q8r1–96phlD mutant is deficient in the production of DAPG and was constructed as follows. Areas flanking the chosen Kmr cassette insertion site within phlD were amplified by PCR with primers sets phlDm1 (5′-TGC TTC TAA CTT TAT TGG CTT CTC G-5′)/phlDm2 (5′-GAT TTT GAG ACA CAA CGT GGG GAG ACG ATG AGG ACATGG-3′) and phlDm3 (5′-GGT TGT AAC ACT GGC AGA ATA CCG GGT CTT ACT TCC TG-3′)/phlDm4 (5′-GTG TTT TAT CGG GTC TGT CAG GC-3′). The 995-bp Km’ cassette of vector pUC4K (Vieira and Messing 1982) was amplified with primers Fkan (5′-CAC GTT GTG TCT CAA AAT CTC TGA TG-3′) and Rkan (5′-TCT GCC AGT GTT ACA ACC AAT TAA CC-3′). All amplicons were gel purified with a QIAEX II kit (Qiagen, Valencia, CA) and fused by overlap extension PCR with KOD Hot Start DNA polymerase (EMD Millipore, Billerica, MA) and primers phlDm1 and phlDm4. The resultant PCR fragment was cloned into the SmaI site of the gene replacement vector pEX18Tc (Hoang et al. 1998). The pEX18Tc-based plasmid was mobilized into Q8r1–96 from E. coli S17–1(λ-pir), and double crossover events were selected and verified by PCR as described by Mavrodi et al. (2006).
G. graminis var. tritici oat-kernel inoculum.
Inoculum of strain R3–111a-1 was prepared essentially as described by Ownley et al. (1992). Briefly, 250 ml of whole oat grains and 350 ml of distilled water were added to a 1-liter flask and autoclaved on two successive days. Sterile oat grains were inoculated with agar disks of G. graminis R3–111a-1 grown on 1/5× PDA. Flasks were incubated at room temperature, shaken after 10 days, and at 3 weeks after inoculation, the contents of each jar were air dried and stored at 4°C. Just prior to use, G. graminis var. tritici oat-kernel inoculum was ground in a blender and particles of 0.25 to 0.5 mm were collected and mixed into the soil (0.7%, wt/wt) (Ownley et al. 1992).
Effect of the wheat cultivar on the level of take-all suppression in TAD soil.
Nontreated wheat seeds were pregerminated on filter paper moistened with sterile water for one day before planting. Twelve seeds of cultivars Tara, Finley, or Buchanan were sown in square PVC pots (8 cm high × 7.5 cm wide) containing 200 g of Q-TAD or pasteurized Q-TAD soil amended with 0.7% (wt/wt) of freshly ground oat-kernel inoculum (particle sizes of 0.25 to 0.5 mm). Prior to planting, 50 ml of water supplemented with metalaxyl (Syngenta, Wilmington, DE) was added to each pot to prevent seed damping-off by indigenous Pythium spp. A 1-cm layer of nonamended soil was added over the seeds. Plants were incubated in a growth room (15 to 16°C, 12-h photoperiod). Pots received 50 ml of dilute 1/3× Hoagland’s solution (macro-elements only) once weekly. Each treatment consisted of five pots with each pot serving as a replicate. After 3 weeks of growth, one plant with rhizosphere soil attached to the roots was harvested from each treatment replicate to determine rhizosphere population sizes of DAPG producers and total heterotrophic aerobic culturable bacteria using the dilutionendpoint assay (described below). The severity of take-all on the remaining plants in each pot was determined using a 0 to 8 scale (0 = no disease and 8 = dead plant or nearly so) (Ownley et al. 1992).
Enumeration of rhizosphere bacterial populations and genotype determinations.
Rhizosphere bacterial population sizes were determined using the PCR-based dilution-endpoint assay as previously described (Mavrodi et al. 2012; McSpadden Gardener et al. 2001). Roots with rhizosphere soil from a single plant were placed into a 50-ml screw-cap plastic centrifuge tube containing 10 ml of sterile distilled water. The tube was vortexed for 1 min and then placed in an ultrasonic bath for 1 min to dislodge bacteria from the roots. Each root washing solution was then serially diluted in a 96-well plate (100 μl added into a well with 200 μl of water) and then 50 μl of each dilution was transferred to wells of a plate with 200 μl of 1/3× KMB broth supplemented with ampicillin (40 μg ml−1), chloramphenicol (13 μg ml−1), and cycloheximide (100 μg ml−1) (1/3× KMB+++). Population sizes of total culturable heterotrophic aerobic bacteria (total bacteria), were determined by transferring each dilution to a well with 200 μl of 1/10× tryptic soy broth supplemented with cycloheximide (100 μg ml−1) (1/10× TSB+). The inoculated plates were incubated at room temperature in the dark and the growth after 3 days was determined spectrophotometrically at 600 nm by using a Model 680 microplate reader (Bio-Rad, Hercules, CA). An OD > 0.1 was considered as positive for growth. Population sizes of DAPG-producing pseudomonads were calculated based on the final dilution that contained cells positive for phlD in the DAPG biosynthesis operon by PCR with the primers B2BF and BPR4 as previously described (Mavrodi et al. 2012; McSpadden Gardener et al. 2001). Population sizes of total bacteria were determined based on the final dilution with growth in the well with 1/10× TSB+.
The genotype of the DAPG-producing Pseudomonas spp. present in the terminal dilution of 96-well plates containing 1/3× KMB+++ was determined by restriction fragment length polymorphism (RFLP) analysis of phlD as previously described (Landa et al. 2002; McSpadden Gardener et al. 2001). Briefly, the 629-bp phlD fragment amplified with primers B2BF and BPR4 was cut with the restriction enzymes HaeIII, MspI, and TaqI and fragment patterns developing after gel electrophoresis were compared with patterns previously identified for each of 22 genotypes of DAPG-producing Pseudomonas spp. (De La Fuente et al. 2006; Landa et al. 2006; McSpadden Gardener et al. 2001).
Determination of the phytotoxicity of P. brassicacearum on wheat.
Wheat seeds were treated with a solution of 1% methyl cellulose (Sigma Chemical Co.) or suspensions of P. brassicacearum strains Q8r1–96 or L5.1–96 in 1% methyl cellulose to establish doses of 0, 102, 104, 106, or 108 CFU seed−1 essentially as described by Yang et al. (2011). Coated seeds were air-dried for 2 h in a laminar flow cabinet. Seeds were also treated at a dose of 108 CFU seed−1 with mutants of Q8r1–96 defective in the T3SS (Q8r1–96ΔOPQR, Q8r1–96rspL, and Q8r1–96rscV) and DAPG production (Q8r1–96phlD). The phytotoxicity of strains Q8r1–96 and L5.1–96 was tested on cultivars Tara, Finley, and Buchanan using a modification of the tube assay (Yang et al. 2011). Briefly, plastic tubes (2.5 cm diameter × 16.5 cm long) were hung in racks (200 cones/rack). Each tube was plugged with a cotton ball and filled with a 6.5-cm-thick column of sterile vermiculite. Three treated seeds were planted in each tube, covered with a 1.5-cm-thick layer of sterile vermiculite, and each tube received 10 ml of water. Racks of tubes were incubated at 15 to 16°C in a dark/light cycle of 12 h, and for the first 4 days after planting, each rack was covered with plastic. After the plastic was removed, each tube received 10 ml of diluted (1:3, vol/vol) Hoagland’s solution. All treatments were replicated five times and each replicate consisted of five tubes. Treatments were arranged in a randomized complete block design. Ten days after sowing, the number of shoots emerging was determined and the plants were removed from the tubes. The roots of each plant were washed free of vermiculite and the length of the shoots and roots was measured. The effects of pure DAPG on the growth of cultivars Tara, Finley, and Buchanan were determined as described by Kwak et al. (2012).
Data analysis.
Statistical analyses were performed by using appropriate parametric and nonparametric procedures with the STATISTIX 8.0 software (Analytical Software, St. Paul, MN). All population data were converted to log CFU g−1 fresh weight of root. Differences in population densities, germination, root disease rating, and root and shoot length among treatments were determined by standard analysis of variance. Mean comparisons among treatments were performed by using either Fisher’s protected least significant difference test (P = 0.05) or the Kruskal-Wallis all-pairwise comparison test (P = 0.05).
RESULTS
Impact of wheat cultivar on take-all suppression in TAD soil.
Two cycles of wheat (cultivar Louise) were grown in the Q-TAD soil in a growth room prior to use in order to enrich population sizes of DAPG producers to between 105 and 107 CFU g−1 of root. P. brassicacearum (genotype D) was the only genotype of DAPG producer detected in the rhizosphere of the wheat (data not shown). Pasteurization of the Q-TAD soil after the second cycle of wheat, reduced the population size of DAPG producers to below the detection limit (approximately 103 CFU g−1 of root fresh weigh) in the rhizosphere of cultivar Louise grown in that soil (data not shown).
The severity of take-all developing on cultivars Tara, Finley, and Buchanan was consistently less on wheat grown in Q-TAD compared with that in pasteurized Q-TAD soil in six separate experiments conducted over a period of 14 months in soils amended with inoculum of G. graminis var. tritici (Fig. 1; Table 2), indicating that pasteurization effectively reduced the suppressive activity of the soil. In pasteurized Q-TAD soil, the three cultivars showed no significant difference in take-all severity, indicating that they did not differ in sensitivity to the pathogen (Table 2). However, in Q-TAD soil, the severity of take-all on cultivar Tara was consistently and significantly less than that on cultivar Buchanan, suggesting a differential ability of cultivars to respond to TAD suppressiveness (Table 2; Fig. 2). The response of cultivar Finley was generally intermediate between Tara and Buchanan (Table 2).
Fig. 1.
Representative example of the effect of soil pasteurization on take-all suppression in Quincy take-all decline (Q-TAD) soil sown to cultivar Tara. A, Raw or B, pasteurized Q-TAD soil was amended with 0.7% (wt/wt) of oat-kernel inoculum of Gaeumannomyces graminis var. tritici. Twelve seeds were sown in each pot and plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Pots were watered 3 times a week. After 3 to 4 weeks of growth, the plants were harvested. Take-all severity was rated on a 0 to 8 scale (0 = healthy plant and 8 = dead or nearly so) and plant height was measured from the crown to the tip of the longest leaf. Representative data are shown in Table 2.
TABLE 2.
Suppression of take-all in raw and pasteurized Quincy take-all decline (Q-TAD) soil on wheat cultivars Tara, Finley, and Buchanan
| Experiment 1y |
Experiment 2 |
Experiment 3 |
Experiment 4 |
Experiment 5 |
Experiment 6 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cultivar | Soilx | Disease ratingz | Shoot heightz | Disease rating | Shoot height | Disease rating | Shoot height | Disease rating | Shoot height | Disease rating | Shoot height | Disease rating | Shoot height |
| Tara | Q-TAD | 6.1 B | 13.5 B | 5.7 B | 13.3 A | 5.4 C | 16.6 A | 4.3 C | 16.4 B | 4.6 B | 16.2 A | 4.7 C | 13.7 A |
| Finley | Q-TAD | 6.3 B | 15.0 A | 5.9 B | 13.0 A | 5.8 BC | 15.3 AB | 4.9 BC | 15.6 B | 4.8 B | 16.5 A | 4.8 BC | 14.2 A |
| Buchanan | Q-TAD | 7.3 A | 12.2 C | 6.4 A | 12.4 B | 6.2 B | 13.8 B | 4.8 BC | 14.8 B | 5.7 A | 13.1 B | 5.6 B | 12.9 A |
| Tara | TAD-P | – | – | – | – | 7.4 A | 10.1 C | 5.8 A | 15.9 B | – | – | 7.1 A | 10.0 B |
| Finley | TAD-P | – | – | – | – | 7.3 A | 11.1 C | 5.5 AB | 19.0 A | – | – | 7.0 A | 9.8 B |
| Buchanan | TAD-P | – | – | – | – | 7.4 A | 9.9 C | 5.2 AB | 19.0 A | – | – | 7.0 A | 9.7 B |
Q-TAD, raw (nonpasteurized) Quincy TAD soil; TAD-P, Q-TAD soil that was pasteurized for 2 h at 70°C.
Wheat seeds were pregerminated for one day before sowing. Twelve seeds were sown in square PVC pots (8 cm high, 7.5 cm wide) containing 200 g of sieved Q-TAD or TAD-P soil amended with 0.7% (wt/wt) of ground oat-kernel inoculum of the take-all pathogen (Gaeumannomyces graminis var. tritici isolate R3-111a-1). A 1-cm layer of the soil was spread on top of the seeds. Plants were grown in a controlled-environment chamber at 15 to 16°C with a 12-h photoperiod. Pots received 50 ml of dilute (1:3, vol/vol) Hoaglund’s solution (macro-elements only) once a week. After 3 to 4 weeks of growth, the plants were harvested. Six plants with rhizosphere soil were used from each treatment to determine the rhizosphere bacterial populations of naturally occurring 2,4-diacetylphloroglucinol producers (Table 3). Take-all severity was rated on a 0 to 8 scale where 0 = healthy plant and 8 = dead or nearly so. For each soil, five replicates were used. Plant height was measured in centimeters from the crown to the tip of the longest leaf.
Statistical analyses were performed by using appropriate parametric and nonparametric procedures with the STATISTIX 8.0 software (Analytical Software, St. Paul, MN). Mean in the same column followed by the same letter are not significantly different at P = 0.05. Comparisons among treatments were performed by using Fisher’s protected least significant difference test (P = 0.05) or the Kruskal-Wallis all-pairwise comparison test.
Fig. 2.
Representative example of suppression of take-all in Quincy take-all decline (Q-TAD) soil on wheat cultivars Tara, Finley, and Buchanan. Six experiments like these were conducted over a period of 14 months with Q-TAD soil collected from the field in 2011 and 2012 and showed similar results. Twelve seeds were sown in each pot containing Q-TAD soil amended with 0.7% (wt/wt) of oat-kernel inoculum of Gaeumannomyces graminis var. tritici. Plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Pots were watered three times a week. After 3 to 4 weeks of growth, the plants were harvested. Take-all severity was rated on a 0 to 8 scale (0 = healthy plant and 8 = dead or nearly so) and plant height was measured from the crown to the tip of the longest leaf. Representative data are shown in Table 2.
The population sizes of DAPG producers in the rhizospheres of the three cultivars in Q-TAD soil were not significantly different (Table 3), and in most cases were above 106 CFU g−1 of root. P. brassicacearum (genotype D) was the only genotype of DAPG producers detected on all three cultivars. The three cultivars also supported similar population sizes of total bacteria. These findings indicate that the differential response of the three cultivars in TAD soil was not due to differences in population sizes or genotype of DAPG producers or total bacteria in the rhizosphere.
TABLE 3.
Population density (log CFU g−1) of 2,4-DAPG-producing pseudomonads and total culturable bacteria in the rhizosphere of wheat grown in Quincy take-all decline soil
| Experiment 1z |
Experiment 2 |
Experiment 3 |
Experiment 4 |
|||||
|---|---|---|---|---|---|---|---|---|
| Cultivar | DAPG+ | Total | DAPG+ | Total | DAPG+ | Total | DAPG+ | Total |
| Tara | 6.7 A | 9.3 A | 7.5 A | 9.6 A | 7.1 A | 8.7 A | 6.5 A | 9.3 A |
| Finley | 7.5 AB | 9.5 A | 7.4 A | 9.4 A | 6.9 A | 8.9 A | 6.1 A | 8.9 AB |
| Buchanan | 7.5 AB | 9.7 A | 7.6 A | 9.4 A | 7.0 A | 8.9 A | 5.7 A | 8.6 B |
Experiments 1 to 4 refer to bacterial populations in the rhizosphere of cultivars shown in Table 2. DAPG+, populations of 2,4-DAPG-producing Pseudomonas spp.; Total, total culturable heterotrophic aerobic bacteria. Bacterial populations are expressed as log CFU g−1 of root fresh weight. Means in the same column followed by the same letter are not significantly different at P = 0.05 according to the Fisher’s protected least significant difference test.
Wheat cultivars differ in their response to the phytotoxicity of P. brassicacearum.
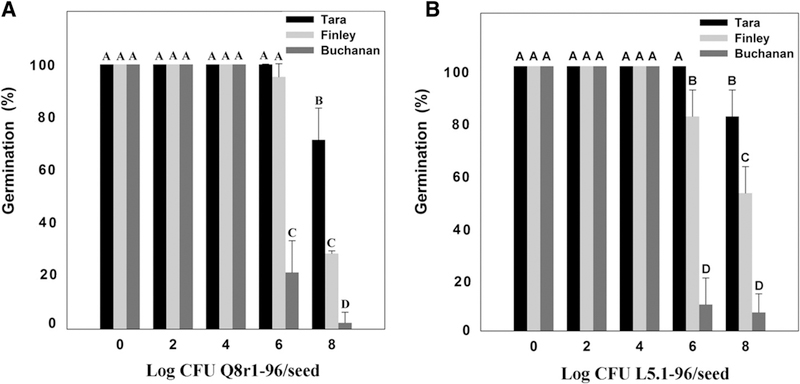
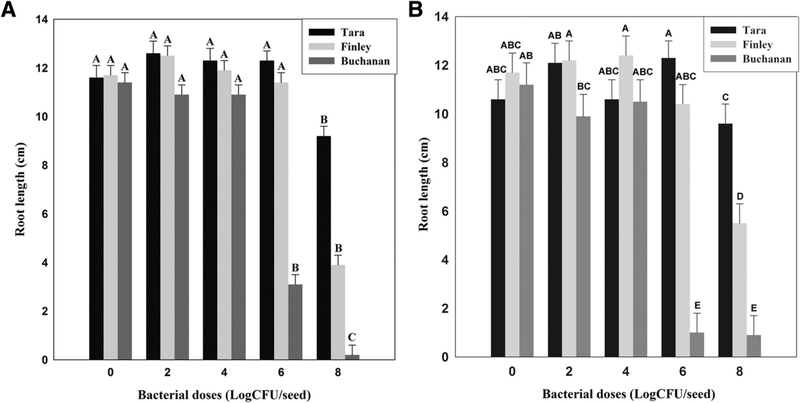
P. brassicacearum can be a minor pathogen on some crops (Belimov et al. 2007; Brazelton et al. 2008; Kwak et al. 2012); therefore, we determined whether the cultivars differ in sensitivity to the phytotoxicity of P. brassicacearum. Nontreated seed of Tara, Finley, and Buchanan and seed treated with strains Q8r1–96 or L5.1–96 at doses of 102 or 104 CFU seed−1 had 100% germination and emergence, indicating that there were no differences in seed viability among the three cultivars at low bacterial doses. However, at doses of 106 or 108 CFU seed−1, the three cultivars differed significantly in germination and emergence, with Tara showing the greatest emergence, Buchanan the least, and Finley intermediate (Fig. 3). For example, strain Q8r1–96 at a dose of 106 CFU seed−1 significantly reduced the germination and emergence of cultivar Buchanan down to about 20% compared with 100 and 97 % for cultivars Tara and Finley, respectively (Fig. 3). Strain L5.1–96 had a similar effect on the germination and emergence of the three cultivars as Q8r1–96. For example, at a dose of 106 CFU of L5.1–96 seed−1, emergence of Tara, Finley, and Buchanan was 100, 81, and 9%, respectively (Fig. 3). The effects of strains Q8r1–96 and L5.1–96 on the root length (Fig. 4) and shoot height (data not shown) of the three cultivars were similar to those seen with measurements of germination and emergence.
Fig. 3.
Effect of Pseudomonas brassicacearum strains A, Q8r1–96 and B, L5.1–96 on seed germination and emergence of the wheat cultivars Tara, Finley, and Buchanan. Bacteria were applied at doses ranging from 102 to 108 CFU seed−1 in 1% methyl cellulose. Seeds with the dose of 0 CFU were coated with only 1% methyl cellulose. Plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Emergence was measured 10 days after the seeds were sown. Bars with the same letter are not significantly different according to Fisher’s protected least significant difference test (P = 0.05).
Fig. 4.
Effect of Pseudomonas brassicacearum A, Q8r1–96 and B, L5.1–96 on root length of wheat cultivars Tara, Finley, and Buchanan. Bacteria were applied at doses ranging from 102 to 108 CFU seed−1 in 1% methyl cellulose. Seeds with the dose of 0 CFU were coated with only 1% methyl cellulose. Plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Root length was measured 10 days after the seeds were sown. Bars with the same letter are not significantly different according to Fisher’s protected least significant difference test (P = 0.05).
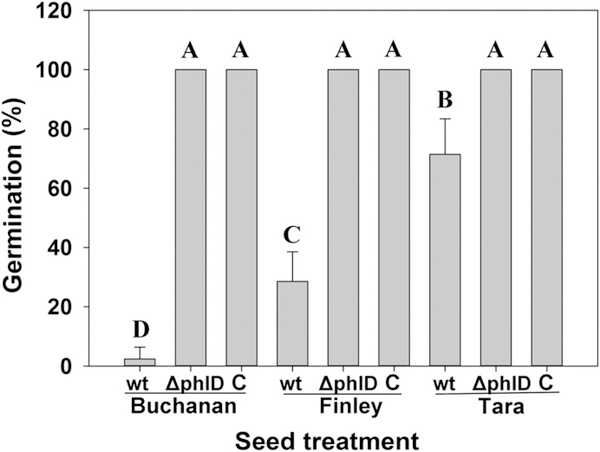
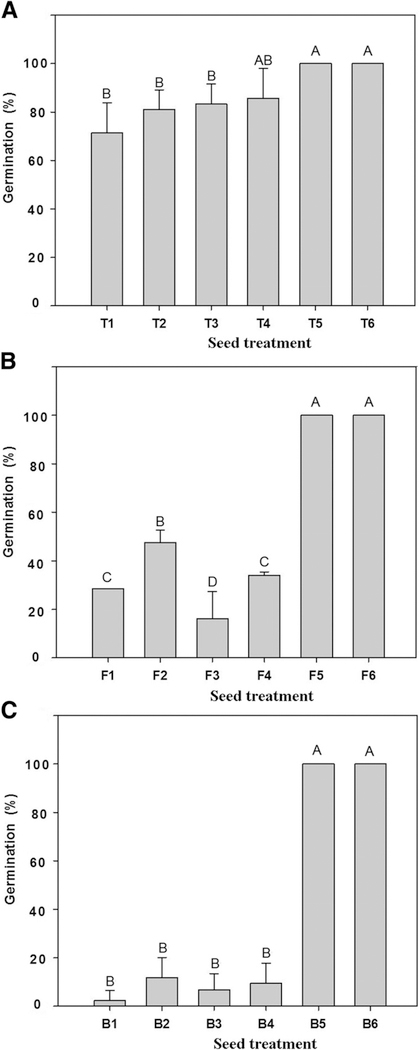
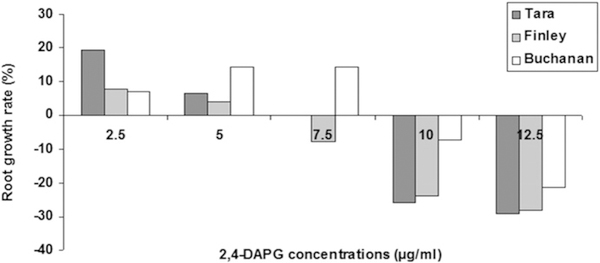
DAPG interferes with auxin signaling in roots, and at high concentrations it damages root tissues (Brazelton et al. 2008; Kwak et al. 2012). T3SS are present in both beneficial and pathogenic plant-associated bacteria, and in phytopathogens, T3SS and effectors play central roles in the capacity to overcome plant defense and cause disease on susceptible hosts. Hence, we tested DAPG and T3SS mutants of Q8r1–96 for phytotoxicity on the three cultivars. The DAPG-deficient mutant Q8r1–96phlD completely lost phytotoxicity; it had no effect on germination or emergence of the three cultivars, and the cultivars did not differ from the nontreated control (Figs. 5 and 6). In contrast, the three T3SS mutants showed no consistent loss in phytotoxicity on the three cultivars because the emergence of seed treated with the mutants was generally comparable to emergence of seed treated with the wild-type strain (Fig. 6). However, only Finley seeds treated with Q8r1–96ΔOPQR had greater emergence than Q8r1–96. Root (Fig. 7) and shoot measurements (data not shown) of the mutant treatments mirrored the measurements of emergence. Collectively, these results indicated that DAPG production is a key factor in reducing the germination and emergence of Tara, Finley, and Buchanan. However, DAPG is not the only factor responsible for the differential response of the cultivars to P. brassicacearum because roots of the three cultivars growing on agar with concentrations of 2.5 to 12.5 μg ml−1 responded fairly similarly to the antibiotic except at a dose of 7.5 μg ml−1 (Fig. 8). In fact, Buchanan was least inhibited at the higher doses of DAPG.
Fig. 5.
Germination and emergence of cultivars Buchanan, Finley, and Tara treated with Pseudomonas brassicacearum Q8r1–96 (wt) and 2,4-diacetylphloroglucinol (DAPG)-deficient mutant Q8r1–96phlD (ΔphlD) at a dose of 108 seed−1 in 1% methyl cellulose. The nontreated control (C) was coated with only 1% methyl cellulose. Plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Emergence was measured 10 days after the seeds were sown. Bars with the same letter are not significantly different according to Fisher’s protected least significant difference test (P = 0.05).
Fig. 6.
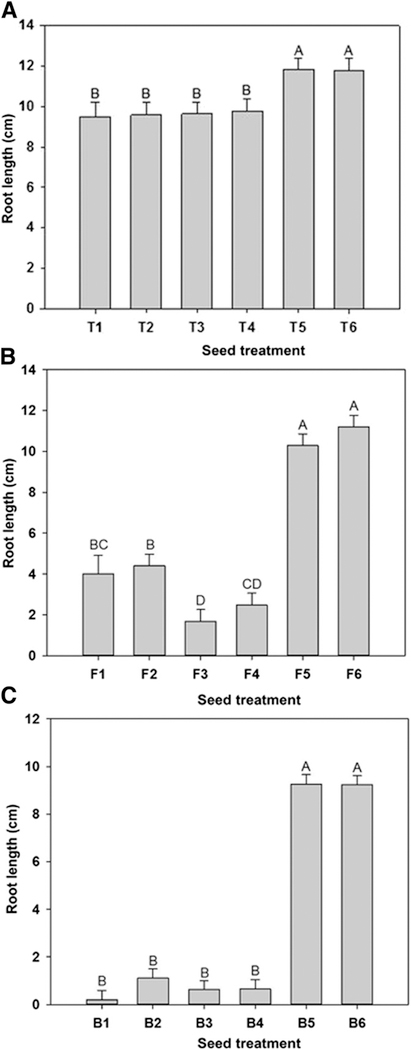
Effect of Pseudomonas brassicacearum Q8r1–96 and its mutants on germination and emergence of cultivars A, Tara, B, Finley, and C, Buchanan. Wheat seeds were treated with: Q8r1–96 (T1, F1, B1); Q8r1–96ΔOPQR (T2, F2, B2); Q8r1–96rspL (T3, F3, B3); Q8r1–96rscV (T4, F4, B4); and Q8r1–96phlD (T5, F5, B5). Bacteria were applied at a dose of 108 CFU seed−1 in 1% methyl cellulose. The nontreated control (T6, F6, B6) was coated with 1% methyl cellulose. Plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Emergence was measured 10 days after seeding. Bars with the same letter are not significantly different according to the Fisher’s protected least significant difference test (P = 0.05).
Fig. 7.
Effect of Pseudomonas brassicacearum Q8r1–96 and its mutants on root length of cultivars A, Tara, B, Finley, and C, Buchanan. Wheat seeds were treated with: Q8r1–96 (T1, F1, B1); Q8r1–96ΔOPQR (T2, F2, B2); Q8r1–96rspL (T3, F3, B3); Q8r1–96rscV (T4, F4, B4); and Q8r1–96phlD (T5, F5, B5). Bacteria were applied at a dose of 108 CFU seed−1 in 1% methyl cellulose. The nontreated control (T6, F6, B6) was coated with 1% methyl cellulose. Plants were incubated in a growth room (15 to 16°C; 12-h photoperiod). Root length was measured 10 days after seeding. Bars with the same letter are not significantly different according to Fisher’s protected least significant difference test (P = 0.05).
Fig. 8.
Effect of 2,4-diacetylphloroglucinol (DAPG) on the growth of cultivars Tara, Finley, and Buchanan. Two-day-old seedlings were transferred to plates containing DAPG at concentrations ranging from 0 to 12.5 μg/ml as described by Kwak et al. (2012). The seedlings were incubated for 24 h at room temperature in the dark, and then the roots of 30 seedlings were measured. Root growth rate (%) = (root length of seedling on plate with DAPG - root length of control)/root length of control × 100%.
DISCUSSION
In this study, we showed for the first time that wheat cultivars differ significantly in their ability to support suppression of take-all when grown in a TAD soil. Our study was precipitated by a discussion with a Washington grower/collaborator whose farm has hosted many research plots over the last 25 years. He only plants cultivar Tara in fields with a long history of wheat monoculture because of its consistent performance compared with other cultivars. His fields in wheat monoculture were previously shown to support threshold densities of DAPG producers (Raaijmakers and Weller 1998) and to be in the state of TAD. We selected Finley and Buchanan to compare with Tara because these three cultivars have been compared in previous studies (Kwak et al. 2012; Okubara and Bonsall 2008; Okubara et al. 2004) and Tara and Buchanan showed different patterns of gene expression when colonized by P. brassicacearum strain Q8r1–96 (Maketon et al. 2012).
Suppression of take-all in the Q-TAD soil was significantly greater on Tara compared with Buchanan, and Finley was intermediate in its response. Our findings of differential wheat cultivar responses in TAD soil are supported by those of Kwak et al. (2012), who demonstrated that seed treatments with P. brassicacearum Q8r1–96 also provided greater biocontrol against take-all on Tara than on Buchanan. We think that our findings are relevant to the field setting because it is well known that TAD suppressiveness can be transferred, manipulated and eliminated similarly under both field and controlled conditions (Hornby 1998; Schlatter et al. 2017; Weller et al. 2002).
We concluded that the differences among the three cultivars in supporting suppression of take-all were not related to their differential sensitivity to G. graminis var. tritici because in pasteurized Q-TAD soil, all of the cultivars were equally diseased. Also, it is well documented that no commercial wheat cultivar has resistance to take-all (Cook 2003; Freeman and Ward 2004; Hornby 1998). Secondly, cultivar differences were not due to variations in rhizosphere population densities of P. brassicacearum because all three cultivars were colonized to the same extent in the Q-TAD soil. Finally, the populations of total culturable heterotrophic aerobic bacteria in the rhizospheres of the three cultivars did not differ consistently.
Previous reports that DAPG can negatively impact the growth of wheat (Kwak et al. 2012) and tomato (Brazelton et al. 2008), then prompted our evaluation of the impact of P. brassicacearum on the three cultivars as a possible factor in their differential response to take-all in Q-TAD soil. Surprisingly, P. brassicacearum strains Q8r1–96 and L5.1–96 were significantly more deleterious to the germination, emergence, and root and shoot growth of cultivar Buchanan than cultivars Tara and Finley. This relationship between the cultivar’s emergences when treated with P. brassicacearum and response to take-all in TAD soil is currently being expanded to more cultivars. The wide range of responses shown by the three PNW cultivars tested suggests that there is probably considerable variability among the 100+ commercial cultivars currently grown in the PNW in their interaction with P. brassicacearum.
We think that P. brassicacearum strains isolated from TAD soils are best characterized as minor pathogens (Schlatter et al. 2017) because their beneficial or deleterious activity on a wheat cultivar is clearly dose dependent, with low and moderate doses being beneficial to the plant, and higher doses being deleterious. A population density of 105 CFU DAPG producers g−1 of root is the threshold required for take-all suppression (Raaijmakers and Weller 1998), and this dose applied as a seed treatment did not reduce the germination and root growth of wheat, but densities of 106 to 107 (or greater) CFU g−1 of root are not uncommon on wheat roots in TAD soil (Weller et al. 2007). Strains Q8r1–96 and L5.1–96, which are from two different TAD fields, inhibited the growth of the three cultivars similarly, suggesting that this is a common trait of P. brassicacearum strains from PNW TAD soils. Belimov et al. (2007) reported that P. brassicacearum strains Am3R and 520–1 showed the ability to cause necrosis when injected into tomato fruits. Interestingly, strain Q8r1–96 also caused a similar necrosis when injected into immature tomato fruits (M. Yang and D. M. Weller, unpublished data). We are currently testing the ability of P. brassicacearum isolates from other TAD soils to cause necrosis in tomato fruits and stems.
The finding that P. brassicacearum strains from Washington State TAD soils are minor pathogens is especially notable and fits an emerging picture seen in examples of disease-suppressive soils. The microbial agent(s) responsible for specific suppression often share some characteristics of a pathogen (Kyselková and Moënne-Loccoz 2012) and thus considerable niche overlap with the target pathogen would be expected. For example, different formae speciales of Fusarium oxysporum cause Fusarium wilt on many crop species. In many examples of Fusarium wilt suppressive soils, nonpathogenic or mildly pathogenic F. oxysporum strains contribute to the suppression of wilt diseases (Kyselková and Moënne-Loccoz 2012; Weller et al. 2002) either through competition with the pathogen for nutrients and infection courts or by induction of host resistance mechanisms (Alabouvette 1986; Expósito et al. 2017; Larkin and Fravel 1999: Weller et al. 2002). In addition, common scab, caused by Streptomyces scabies and other Streptomyces spp., is a chronic problem in potato production, but continuous monoculture of potato leads to potato scab decline through a build-up of antibiotic-producing nonpathogenic Streptomyces species or weakly pathogenic S. scabies (Expósito et al. 2017; Liu et al. 1996; Lorang et al. 1995; Kyselková and Moënne-Loccoz 2012). Finally, crown gall, caused by Agrobacterium tumefaciens, affects many dicotyledonous crop species. On almond, crown gall is naturally suppressed when the population of nonpathogenic agrobacteria reaches a threshold density (New and Kerr 1972).
The antibiotic DAPG inhibits a broad spectrum of organisms including bacteria, fungi, protists, and nematodes (Keel et al. 1992; Weller et al. 2007) and it targets basic cellular processes in ascomycetes (Kwak et al. 2011). It also inhibits the growth and seed germination of a wide variety of plant species (Keel et al. 1992; Maurhofer et al. 1995) and causes distinct necrotic lesions on roots of wheat and tomato (Brazelton et al. 2008; Kwak et al. 2012). The herbicidal activity of DAPG resembles that of 2,4-dichlorophe-nonxyacetic acid (2,4-D) and generally dicots are more sensitive to DAPG than monocots (Keel et al. 1992). Brazelton et al. (2008) showed that DAPG can inhibit primary root growth and stimulate lateral root production in tomato seedlings, and the effects require a DGT-dependent auxin-signaling pathway. They also showed that DAPG failed to stimulate the activity of the GH3 promoter, which is 2,4-D responsive, indicating that DAPG and 2,4-D do not act via the same signal transduction pathway. We showed that a key determinant in the phytotoxicity of Q8r1–96 to wheat was production of DAPG, given that the DAPG-deficient mutant of Q8r1–96, Q8r1–96phlD, failed to reduce germination, emergence, and root and shoot length when applied to Tara, Finley and Buchanan and the mutant did not differ from the noninoculated control. However, the antibiotic was not the sole determinant responsible for the differential responses to seed treatment by Q8r1–96 because all three cultivars responded similarly when grown on media amended with DAPG (Kwak et al. 2012). In addition, in a soil-free assay in a Petri dish, Tara was previously shown to accumulate significantly more DAPG on the roots than Finley or Buchanan when inoculated with strain Q8r1–96 (Okubara and Bonsall 2008), yet in raw soil Q8r1–96 produced less DAPG on Tara than it did on Buchanan (Kwak et al. 2012). Given the importance of the T3SS in plant-microbe interactions, we then hypothesized a role for the T3SS of P. brassicacearum in the differential inhibition of cultivar growth. However, T3SS-deficient mutants of Q8r1–96 were no different that the wild type in their effect on emergence and root growth of the three cultivars. We currently are focused on determining the additional traits and genes that work in concert with DAPG production to modulate the differential effect of P. brassicacearum on Tara, Finley, and Buchanan.
A lingering question about the role of P. brassicacearum in TAD is how a single species of Pseudomonas within the milieu of microbes in the soil and rhizosphere microbiomes can be responsible for such powerful natural disease suppression (Weller 2015; Weller et al. 2007). Strains of P. brassicacearum and wheat have an exceptional mutual affinity (Landa et al. 2006; Raaijmakers and Weller 2001) that allows the bacterium to maintain population sizes above the threshold bacterial density (105 CFU g−1 of root) required to suppress take-all throughout the growing season. Because of this affinity, Landa et al. (2006) initially suggested that these bacteria may border on being mutualistic symbionts (Andrews and Harris 2000), with wheat benefiting from a highly effective line of defense against take-all, against which it has no resistance, and the P. brassicacearum benefiting from a substantial increase in population size in a highly competitive rhizosphere environment (Weller et al. 2007). Our finding that P. brassicacearum strains from TAD soils are minor pathogens also provides new insight and clues to the exceptional ability of this bacterium to colonize wheat and suppress take-all. However, the genes and traits in P. brassicacearum and wheat responsible for this mutual affinity remain unknown and a topic of intense investigation.
Collectively, our findings reveal yet another layer of complexity in the interactions among G. graminis var. tritici, wheat, and P. brassicacearum that leads to the natural suppression of take-all in the soils of the PNW, United States. Historically, in the field of biological control of plant pathogens, it has often been thought that more is better, and many studies have sought to increase the production of biocontrol traits such as antibiotic or lytic enzyme production in order to enhance disease suppression. In the case of TAD, we think that wheat cultivars differentially modulate their interactions with P. brassicacearum to ensure that DAPG production in the rhizosphere remains close to the “Goldilocks Zone,” where sufficient antibiotic is produced to suppress G. graminis var. tritici but not so much as to stunt root growth. Given that wheat lacks genetic resistance to take-all, one key defensive strategy used by wheat to cope with take-all is proliferation of secondary roots to outgrow and avoid the pathogen. G. graminis var. tritici requires high moisture for optimal infection, and drying of the soil dramatically retards its growth, thus allowing new young and emerging roots to avoid the pathogen (Cook 2003). As a result, any factor that retards root growth and vigor will increase the severity of take-all because it will keep susceptible young root tissue in contact with take-all inoculum for a longer period of time. In order for wheat roots to reap the benefits of protection by P. brassicacearum during TAD, root growth cannot be slowed by DAPG, as clearly can occur with cultivar Buchanan.
In conclusion, our results suggest that growers have the potential to enhance the robustness of take-all suppression in their TAD fields by selecting a cultivar which has the ability to optimally modulate its interactions with P. brassicacearum. Indeed, analysis by next-generation sequencing of the rhizosphere microbiota of wheat (Mahoney et al. 2017) and other crops (Mendes et al. 2018), clearly shows how cultivars differentially shape the microbiome to their benefit or detriment. Finally, given that it is now thought that roots are the key to the next green revolution, plant breeding programs need to place much greater emphasis on developing cultivars of wheat and other crops that are more supportive of disease-suppressive and growth-promoting microbiota (Wissuwa et al. 2009).
Acknowledgments
Funding: This research was supported by a grant from the O. A. Vogel Wheat Research Fund of Washington State University, Pullman, Washington.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Contributor Information
Mingming Yang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Agronomy, Northwest A&F University, Yangling 712100, P.R. China.
Dmitri V. Mavrodi, Department of Cell and Molecular Biology, The University of Southern Mississippi, Hattiesburg 39406
Linda S. Thomashow, U.S. Department of Agriculture, Agricultural Research Service, Wheat Health, Genetics and Quality Research Unit, Pullman, WA 99164-6430.
David M. Weller, U.S. Department of Agriculture, Agricultural Research Service, Wheat Health, Genetics and Quality Research Unit, Pullman, WA 99164-6430..
LITERATURE CITED
- Alabouvette C 1986. Fusarium-wilt suppressive soils from the Châteaurenard region: Review of a 10-year study. Agronomie 6:273–284. [Google Scholar]
- Andrews JH, and Harris RF 2000. The ecology and biogeography of microorganisms on plant surfaces. Annu. Rev. Phytopathol 38:145–180. [DOI] [PubMed] [Google Scholar]
- Bakker PAHM, Berendsen RL, Doornbos RF, Wintermans PCA, and Pieterse CMJ 2013. The rhizosphere revisited: Root microbiomics. Front. Plant Sci 4:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belimov AA, Dodd IC, Safronova VI, Hontzeas N, and Davies WJ 2007. Pseudomonas brassicacearum strain Am3 containing 1-aminocyclopropane-1-carboxylate deaminase can show both pathogenic and growth-promoting properties in its interaction with tomato. J. Exp. Bot 58:1485–1495. [DOI] [PubMed] [Google Scholar]
- Brazelton JN, Pfeufer EE, Sweat TA, Gardener BBM, and Coenen C 2008. 2,4-Diacetylphloroglucinol alters plant root development. Mol. Plant-Microbe Interact 21:1349–1358. [DOI] [PubMed] [Google Scholar]
- Cook RJ 2003. Take-all of wheat. Physiol. Mol. Plant Pathol 62:73–86. [Google Scholar]
- Cook RJ 2007. Take-all decline: model system in the science of biological control and clue to the success of intensive cropping Pages 399–414 in: Biological Control: A Global Perspective. Vincent C, Goettel MS, and Lazarovits G, eds. CAB International, Wallingford, UK. [Google Scholar]
- Cook RJ, and Veseth RJ 1991. Page 151 in: Wheat Health Management. American Phytopathological Society, St. Paul, MN. [Google Scholar]
- De La Fuente L, Mavrodi DV, Landa BB, Thomashow LS, and Weller DM 2006. phlD-based genetic diversity and detection of genotypes of 2,4-diacetylpholorogucinol-producing Pseudomonas fluorescens. FEMS Microbiol. Ecol 56:64–78. [DOI] [PubMed] [Google Scholar]
- Expósito RG, de Bruijn I, Postma J, and Raaijmakers JM 2017. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol 8:2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frapolli M, Moënne-Loccoz Y, Meyer J, and Défago D 2008. A new DGGE protocol targeting 2,4-diacetylphloroglucinol biosynthetic gene phlD from phylogenetically contrasted biocontrol pseudomonads for assessment of disease-suppressive soils. FEMS Microbiol. Ecol 64:468–481. [DOI] [PubMed] [Google Scholar]
- Freeman J, and Ward E 2004. Gaeumannomyces graminis, the take-all fungus and its relatives. Mol. Plant Pathol 5:235–252. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, and Schweizer HP 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
- Hornby D 1998. Take-All of Cereals: A Regional Perspective. CAB International, Wallingford, UK. [Google Scholar]
- Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, Wirthner P, Haas D, and Défago G 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: Importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant-Microbe Interact 5:4–13. [Google Scholar]
- King EO, Ward MK, and Raney DE 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med 44:301–307. [PubMed] [Google Scholar]
- Kwak YS, Bakker PAHM, Glandorf DCM, Rice JT, Paulitz TC, and Weller DM 2009. Diversity, virulence, and 2,4-diacetylphloroglucinol sensitivity of Gaeumannomyces graminis var. tritici isolates from Washington State. Phytopathology 99:472–479. [DOI] [PubMed] [Google Scholar]
- Kwak YS, Bonsall RF, Okubara PA, Paulitz TC, Thomashow LS, and Weller DM 2012. Factors impacting the activity of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens against take-all of wheat. Soil Biol. Biochem 54:48–56. [Google Scholar]
- Kwak YS, Han S, Thomashow LS, Rice JT, Paulitz TC, Kim D, and Weller DM 2011. Saccharomyces cerevisiae genome-wide mutant screen for sensitivity to 2,4-diacetylphloroglucinol, an antibiotic produced by Pseudomonas fluorescens. Appl. Environ. Microbiol 77:1770–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyselková M, and Moënne-Loccoz Y 2012. Pseudomonas and other microbes in disease-suppressive soils Pages 93–140 in: Organic Fertilisation, Soil Quality and Human Health.Lichtfouse E, ed. Sustainable Agriculture Reviews. Vol. 9 Springer, Dordrecht. [Google Scholar]
- Landa BB, Mavrodi DV, Thomshow LS, and Weller DM 2003. Interactions between strains of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens in the rhizosphere of wheat. Phytopathology 93:982–994. [DOI] [PubMed] [Google Scholar]
- Landa BB, Mavrodi OV, Schroeder KL, Allende-Molar R, and Weller DM 2006. Enrichment and genotypic diversity of phlD-containing fluorescent Pseudomonas spp. in two soils after a century of wheat and flax monoculture. FEMS Microbiol. Ecol 55:351–368. [DOI] [PubMed] [Google Scholar]
- Landa BB, Mavrodi VO, Raaijmakers MJ, McSpadden Gardener BB, Thomashow LS, and Weller DM 2002. Differential ability of genotypes of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens strains to colonize the roots of pea plants. Appl. Environ. Microbiol 68: 3226–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RP, and Fravel DR 1999. Mechanisms of action and dose-response relationships governing biological control of Fusarium wilt of tomato by nonpathogenic Fusarium spp. Phytopathology 89:1152–1161. [DOI] [PubMed] [Google Scholar]
- Liu D, Anderson NA, and Kinkel LL 1996. Selection and characterization of strains of Streptomyces suppressive to the potato scab pathogen. Can. J. Microbiol 42:487–502. [Google Scholar]
- Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LDH, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van Mortel J, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin SA, Kluepfel DA, Wechter PW, Anderson AJ, Kim YC, Pierson LS, Pierson EA, Lindow SE, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, and Paulsen IT 2012. Comparative genomics of plant-associated Pseudomonas spp: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang JM, Liu D, Erson NA, and Schottel JL 1995. Identification of potato scab inducing and suppressive species of Streptomyces. Phytopathology 85:261–268. [Google Scholar]
- Mahoney AK, Yin C, and Hulbert SH 2017. Community structure, species variation, and potential function of different winter wheat (Triticum aestivum) cultivars. Front. Plant Sci 8:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maketon C, Fortuna AM, and Okubara PA 2012. Cultivar dependent transcript accumulation in wheat roots colonized by Pseudomonas fluorescens Q8r1–96 wild type and mutant strains. Biol. Control 60:216–224. [Google Scholar]
- Maurhofer M, Baehler E, Notz R, Martinez V, and Keel C 2004. Cross talk between 2,4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Appl. Environ. Microbiol 70:1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurhofer M, Keel C, Haas D, and Défago G 1995. Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathol. 44:40–50. [Google Scholar]
- Mavrodi DV, Joe A, Mavrodi OV, Hassan KA, Weller DM, Paulsen IT, Loper JE, Alfano JR, and Thomashow LS 2011. Structural and functional analysis of the type III secretion system from Pseudomonas fluorescens Q8r1–96. J. Bacteriol 193:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi OV, Mavrodi DV, Parejko JA, Thomashow LS, and Weller DM 2012. Irrigation differentially impacts populations of indigenous antibiotic-producing Pseudomonas spp. in the rhizosphere of wheat. Appl. Environ. Microbiol 78:3214–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi OV, Mavrodi DV, Park AA, Weller DM, and Thomashow LS 2006. The role of dsbA in colonization of the wheat rhizosphere by Pseudomonas fluorescens Q8r1–96. Microbiology 152:863–872. [DOI] [PubMed] [Google Scholar]
- Mavrodi OV, McSpadden Gardener BB, Mavrodi DV, Bonsall RF, Weller DM, and Thomashow LS 2001. Genetic diversity of phlD from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas species. Phytopathology 91:35–43. [DOI] [PubMed] [Google Scholar]
- Mazzola M, Funnell DL, and Raaijmakers JM 2004. Wheat cultivar-specific selection of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas species from resident soil populations. Microb. Ecol 48:338–348. [DOI] [PubMed] [Google Scholar]
- McSpadden Gardener BB, Mavrodi DV, Thomashow LS, and Weller DM 2001. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology 91:44–54. [DOI] [PubMed] [Google Scholar]
- McSpadden Gardener BB, Schroeder KL, Kalloger SE, Raaijmakers JM, Thomashow LS, and Weller DM 2000. Genotypic and phenotypic diversity of phlD-containing Pseudomonas isolated from the rhizosphere of wheat. Appl. Environ. Microbiol 66:1939–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, and Tsai SM 2018. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, Van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, and Raaijmakers JM 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. [DOI] [PubMed] [Google Scholar]
- Meyer JB, Lutz MP, Frapolli M, Pechy-Tarr M, Rochat L, Keel C, Défago G, and Maurhofer M 2010. Interplay between wheat cultivars, biocontrol pseudomonads, and soil. Appl. Environ. Microbiol 76:6196–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New PB, and Kerr A 1972. Biological control of crown gall: Field measurements and glasshouse experiments. J. Appl. Bact 35:279–287. [Google Scholar]
- Notz R, Maurhofer M, Schnider-Keel U, Duffy B, Haas D, and Défago G 2001. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91:873–881. [DOI] [PubMed] [Google Scholar]
- Okagaki LH, Nunes CC, Sailsbery J, Clay B, Brown D, John T, Oh Y, Young N, Fitzgerald F, Hass BJ, Zeng Q, Young S, Adiconis X, Fan L, Levin JZ, Mitchell TK, Okubara PA, Farman ML, Kohn LM, Birren B, Ma L, and Dean RA 2015. Genome sequences of three phytopathogenic species of the Magnaporthaceae family of fungi. Gene Genomics Genet. 5:2539–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubara PA, and Bonsall RF 2008. Accumulation of Pseudomonas-derived 2,4-diacetylphloroglucinol on wheat seedling roots is influenced by host cultivar. Biol. Control 46:322–331. [Google Scholar]
- Okubara PA, Call DR, Kwak Y-S, and Skinner DZ 2010. Induction of defense gene homologues in wheat roots during interactions with Pseudomonas fluorescens. Biol. Control 55:118–125. [Google Scholar]
- Okubara PA, Kornoely JP, and Landa BB 2004. Rhizosphere colonization of hexaploid wheat by Pseudomonas fluorescens strains Q8r1–96 and Q2–87 is cultivar-variable and associated with changes in gross root morphology. Biol. Control 30:392–403. [Google Scholar]
- Ownley BH, Weller DM, and Thomashow LS 1992. Influence of in situ and in vitro pH on suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens 2–79. Phytopathology 82:178–184. [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, and Bakker PAHM 2014. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol 52:347–375. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, Bonsall RF, and Weller DM 1999. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology 89:470–475. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, and Mazzola M 2016. Soil immune responses. Science 352:1392–1393. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JM, and Weller DM 1998. Natural plant protection by 2,4-diacetylphloroglucinol producing Pseudomonas spp. in take-all decline soils. Mol. Plant-Microbe Interact 11:144–152. [Google Scholar]
- Raaijmakers JM, and Weller DM 2001. Exploiting genetic diversity of 2,4-diacetylphlorogluciol-producing Pseudomonas spp.: Characterization of superior root-colonizing P. fluorescens strain Q8r1–96. Appl. Environ. Microbiol 67:2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, Weller DM, and Thomashow LS 1997. Frequency of antibiotic producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol 63:881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguin H, Sarniguet A, Gazengel K, Moënne-Loccoz Y, and Grundmann GL 2009. Rhizosphere bacterial communities associated with disease suppressive stages of take-all decline in wheat monoculture. New Phytol. 184:694–707. [DOI] [PubMed] [Google Scholar]
- Schlatter D, Kinkel L, Thomashow L, Weller D, and Paulitz T 2017. Disease suppressive soils: New insights from the soil microbiome. Phytopathology 107:1284–1297. [DOI] [PubMed] [Google Scholar]
- Vacheron J, Desbrosses G, Renoud S, Padilla R, Walker V, Muller D, and Prigent-Cambaret C 2018. Differential contribution of plant-beneficial functions from Pseudomonas kilonensis F113 to root system architecture alterations in Arabidopsis thaliana and Zea mays. Mol. Plant-Microbe Interact 31:212–223. [DOI] [PubMed] [Google Scholar]
- Vieira J, and Messing J 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268. [DOI] [PubMed] [Google Scholar]
- Weller DM 2015. Take-all decline and beneficial pseudomonads Pages 363–370 in: Principles of Plant-Microbe Interactions—Microbes for Sustainable Agriculture. Lugtenberg B, ed. Springer, Switzerland. [Google Scholar]
- Weller DM, Landa BB, Mavrodi OV, Schroeder KL, De La Fuente L, Bankhead SB, Allende-Molar R, Bonsall RF, Mavrodi DV, and Thomashow LS 2007. Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol. 9: 4–20. [DOI] [PubMed] [Google Scholar]
- Weller DM, Mavrodi DV, van Pelt JA, Pieterse CM, van Loon LC, and Bakker PAHM 2012. Induced systemic resistance (ISR) in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 102:403–412. [DOI] [PubMed] [Google Scholar]
- Weller DM, Raaijmakers JM, McSpadden Gardener BB, and Thomashow LS 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol 40:309–348. [DOI] [PubMed] [Google Scholar]
- Weller DM, Raaijmakers JM, and Thomashow LS 1997. The rhizosphere ecology of antibiotic-producing pseudomonads and their role in take-all decline Pages 58–64 in: Plant Growth-Promoting Rhizobacteria-Present Status and Future Prospects. Ogoshi A, Kobayashi K, Homma Y, Kodama F, Kondo N, and Akino S, eds. Nakanishi Printing, Sapporo. [Google Scholar]
- Wissuwa M, Mazzola M, and Picard C 2009. Novel approaches in plant breeding for rhizosphere-related traits. Plant Soil 321:409–430. [Google Scholar]
- Yang MM, Mavrodi DV, Mavrodi OV, Bonsall RF, Parejko JA, Paulitz TC, Thomashow LS, Yang HT, Weller DM, and Guo JH 2011. Biological control of take-all by fluorescent Pseudomonas spp. from Chinese wheat fields. Phytopathology 101:1481–1491. [DOI] [PubMed] [Google Scholar]