Abstract
Purpose of Review
Epigenetic variations have been shown to reveal vulnerability to diabetes and its complications. Although it has become clear that metabolic derangements, especially hyperglycemia, can impose a long-term metabolic memory that predisposes to diabetic complications, the underlying mechanisms remain to be understood. It has been suggested that epigenetics (e.g. histone modification, DNA methylation, and non-coding RNAs) help link metabolic disruption to aberrancies related to diabetic kidney disease (DKD). In this review we discuss the key findings and advances made in the epigenetic risk profile of DKD and provide perspectives on the emerging topics that implicate epigenetics in DKD.
Recent Findings
Epigenetic profiles can be profoundly altered in patients with diabetes, in circulating blood cells as well as in renal tissues. These changes provide useful insight into the mechanisms of diabetic kidney injury and progressive kidney dysfunction.
Summary
Increasing evidence supports the role of epigenetic regulation in DKD. More studies are needed to elucidate the mechanism and importance of epigenetic changes in the initiation and progression of DKD and to further explore their diagnostic and therapeutic potential in the clinical management of patients with diabetes who have a high risk for DKD.
Keywords: Diabetic nephropathy, diabetic kidney disease, epigenetic regulation
Introduction
Diabetic kidney disease (DKD), also known as diabetic nephropathy, is one of the major microvascular complications of diabetes, affecting nearly 50% of all patients with diabetes [1]. With the increase of diabetic population worldwide, DKD has been recognized as the leading cause of end-stage renal disease (ESRD) and is strongly associated with mortality in diabetic patients [2].
DKD is characterized by albuminuria and/or an initially increased glomerular filtration rate (GFR) that decreases in the middle to end stage, and is typically accompanied by hypertension [3-5]. The most common histological changes of DKD include glomerular hypertrophy, glomerular and tubular basement membrane thickening, mesangial matrix expansion, glomerulosclerosis, podocyte effacement, and arteriolar wall thickening [6].
Epigenetic regulation, defined as heritable changes in gene expression and function that occur without an alteration in the DNA sequence [7,8] has been identified as an important mechanism underlying diabetes and its complications, such as DKD [9]. Metabolic derangements, such as hyperglycemia, can cause profound epigenetic alterations (e.g. histone modification and DNA methylation) in various renal cells, which in turn may compromise kidney function. A “metabolic memory” imposed by hyperglycemia has been suggested to contribute to the increased risk of diabetic complications, including DKD [10]. Here we review the recent advances in epigenetic risk profile of DKD and discuss the novel insights into the role of epigenetics in progressive kidney dysfunction and pathologic changes in the kidney of high-risk patients with diabetes.
Metabolic memory and epigenetic changes in diabetes
Longitudinal epidemiologic studies and clinical trials of patients with either type 1 or type 2 diabetes consistently demonstrate improved clinical outcomes resulting from intensive glycemic control vs. conventional insulin treatment [11,13,16,17]. After the intervention period, participants in the intensive glycemic control arm continued to have lower risk of vascular complications compared to diabetic patients who received conventional treatment. In particular, the risk of diabetic nephropathy remained significantly higher in the conventional treatment group compared to the intensive control arm [11-17]. Such a phenomenon that early hyperglycemia has persistent and enduring effects in diabetes vascular complications has been described as “metabolic memory” or “legacy effect” [18-22].
In the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Study (DCCT/EDIC), participants with type 1 diabetes in the intensive glycemic control arm had a 39% reduction in microalbuminuria and 54% reduction in albuminuria compared with with those in the conventional treatment in the initial DCCT phase [11]. Despite conversion to intensive control for almost a decade those originally assigned to conventional therapy experienced a higher prevalence of microalbuminuria and albuminuria compared to those consistently managed with intensive treatment [12,13,18]. Over a median follow-up of 22 years, the risk of impairment in GFR was still significantly lower in the intensive treatment group than it in the conventional treatment group [14]. The UK Prospective Diabetes Study (UKPDS) and the ADVANCE collaborative group, which have conducted analogous studies in participants with type 2 diabetes, also reported similar phenomena [15-17].
To understand the mechanisms underlying metabolic memory, researchers have begun to investigate epigenetics using samples collected from the DCCT/EDIC trial. Miao et al. profiled the histone modifications in the blood monocytes and lymphocytes obtained from two groups of participants with type 1 diabetes at year 16-17 of EDIC: participants randomized to the DCCT conventional treatment group who had progression of retinopathy or nephropathy in EDIC (case subjects), and participants randomized to the DCCT intensive treatment group who had no progression of retinopathy or nephropathy (controls subjects). The authors found that case subjects had greater number of promoter regions with enrichment in H3K9Ac (an active histone mark), as compared with control subjects. Importantly, the H3K9Ac levels were positively and significantly associated with glycated hemoglobin (HbA1c) levels in all subjects at all time periods (P< 2.2×10−16). Furthermore, among the top genes differentially acetylated were those related to the nuclear factor-κB (NF-κB) inflammatory pathway well known to be associated with diabetes complications, with over 15 genes in this pathway depicting hyperacetylated promoters [23]. These findings provide the first direct evidence of a relationship between epigenetic modifications and metabolic memory in diabetes.
More recently, Chen et al. have analyzed DNA methylation profiles in similar samples from the DCCT/EDIC studies. In addition to the blood monocytes collected from the case subjects and controls subjects as noted above (i.e. samples collected by EDIC year 16-17), the DNA methylation profile in whole blood DNA collected and archived at the end of DCCT ~1993 (and beginning of EDIC) was also analyzed. While hundreds of differentially methylated loci (DML) were identified in both monocytes and whole blood between the two groups, the 3’UTR region of thioredoxin-interacting protein (TXNIP), a gene known to be associated with hyperglycemia and diabetic complications, was found to be significantly hypomethylated in the case group. More importantly, a set of these DML exhibited similar differences that persist for about 17 years from the DCCT into the EDIC Study, again supporting an epigenetic explanation for the metabolic memory linking hyperglycemia to diabetic complications [24].
Epigenetic modifications
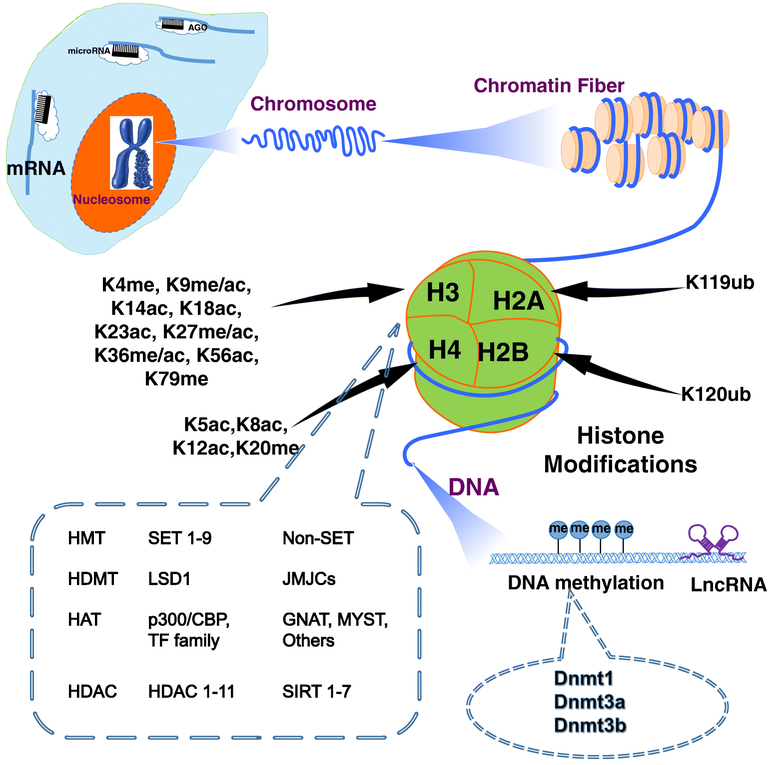
Many types of epigenetic processes have been identified and discussed extensively in several excellent reviews [25-27], and the three major types to be discussed here are DNA methylation, histone modification, and non-coding RNA (ncRNA)-associated gene regulation. We will first provide a brief introduction of these epigenetic regulation and the key enzymes that have been investigated in the context of DKD (Fig. 1), before we cover their profile in DKD.
Figure 1. Overview of epigenetic modifications and mediators in DKD.
Chromatins are subjected to epigenetic regulations, including DNA methylation, posttranslational histone modifications, and microRNA (miRNA) and long noncoding RNAs (lncRNAs)-mediated gene regulation. miRNAs typically affect mRNA expression at post-transcriptional level by targeting the 3’-untranslated region of mRNA in the cytoplasm. DNA methylation and histone modification influence gene regulation in the nucleus. These molecular processes are intricately regulated by various epigenetic enzymes as depicted. LncRNAs can regulate gene expression through either nuclear or cytoplasmic mechanism. DNMT, DNA methyltransferases; HMT, histone methyltransferases; HDM, histone demethylase; HAT, histone acetyltransferases; and histone deacetylases (HDACs).
DNA methylation involves the addition of methyl group to DNA molecules, mostly on cytosines [28]. In mammals, DNA methylation is almost exclusively found in cytosine-phosphate-guanine (CpG) dinucleotides [29]. Methylation in the promoter regions typically leads to gene silencing whereas methylation in the gene bodies allows for active transcription [30,31]. Transfer of methyl groups to DNA is catalyzed by DNA methyltransferases (DNMTs) [32,33].
Histone modifications mainly occur in the exposed histone amino-terminal tails in histone octamers comprising two copies of histone H2A, H2B, H3, or H4. Among over 60 different types of histone modifications, methylation and acetylation of lysine (K) and arginine (R) are the most extensively studied to date [34]. These histone modifications are tightly regulated by enzymes responsible for adding or removing the epigenetic marks, often referred to as “writers”, (i.e., histone acetylases and methyltransferases) and “erasers”, (i.e., deacetylases and demethylases) [35].
Histone methylation on H3K4, H3K9, and H3K36 typically leads to transcriptional activation. In contrast, methylation on H3K27 by enhancer of zeste homolog 2 (EZH2) often denotes transcriptional suppression. These methylation events can be reversed by by demethylases, such as lysine specific demethylases (LSDs) and Jumonji C-domain containing demethylases [36-39].
Histone acetylation or hyperacetylation (e.g. H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac) is typically associated with transcriptional activation, as the acetyl group can decrease the negative charge of DNA, making chromatin more accessible to transcription factors (TFs) and their coactivators. In contrast, hypoacetylation usually results in a more compact chromatin conformation, leading to transcriptional repression. Histone acetylation and deacetylation are regulated by histone acetyltransferases (HATs) and deacetyltransferases (HDAC) respectively [42]. Among HATs, P300 and cAMP response element binding protein (CREB)-binding protein (CBP) are the best characterized in DKD [40,41]. HDACs, a large protein family consists of many members have been investigated extensively for their role in a variety of disease conditions, including DKD [43,44].
Non-coding RNAs are encoded by and transcribed from the non-protein-coding regions in the genome. While microRNA (miRs) have been intensively studied in DKD in the past decade (please refer to references 45-48 for review), long noncoding RNAs (lncRNAs) have recently emerged to be key regulators implicated in diabetes complications. LncRNAs, with transcript length >200 nucleotides, can epigenetically regulate gene expression through diverse molecular mechanisms [49,50], as we will discuss in the next section using several examples related to DKD.
Epigenetic profile in DKD patients and potential mechanistic link
In this section, we will review current studies performed with different type of samples (i.e. body fluids vs renal tissues) for epigenetic profiling, which likely reveal systemic vs local epigenetic changes underlying DKD.
Epigenetic profile in body fluids
Useful epigenetic information carried in body fluids (e.g., peripheral blood, urine, saliva) of human subjects can be collected non-invasively. Changes in the urine are the most common manifestation of DKD. However, although DNA can be extracted from human urine and DNA methylation has been detected [51-53], to date there is a lack of evidence that DNA methylation or histone modification can be detected in the urine informative of underlying disease process and risk of adverse outcomes in people with DKD .
Attempt has also been made with DNA extracted from saliva. Sapienza et al. profiled DNA methylation in saliva collected from African American and Hispanic participants with diabetes and ESRD undergoing hemodialysis and that in participants with diabetes but without nephropathy. Among 187 genes that were differentially methylated between the two groups of participants, 39 were involved in kidney development or diabetic nephropathy, or have been associated with dialysis-induced changes in gene expression in peripheral blood cells [54]. With only one report to date, it is yet difficult to evaluate the potential use of saliva DNA methylation in the evaluation of DKD risk.
Compared with epigenetic profiling using urine and saliva, tests in peripheral blood seem to be more informative. In addition to the two elegant studies mentioned earlier [23,24] in profiling DNA methylation and histone modification in blood samples collected from DCCT/EDIC, several other studies have also reported the association between epigenetic profiles measured in peripheral blood samples and DKD. Most of these studies have focused on DNA methylation.
Gautier et al. profiled the genome-wide DNA methylation in leukocytes from non-diabetic offspring of mothers with type 1 diabetes (case group) in comparison with offspring of fathers with type 1 diabetes (control group). Among 87 CpG sites differently methylated, DNMT1 (the key enzyme for the maintenance of DNA methylation) was under-methylated in cases, and the 74 under-methylated sites were correlated with GFR in cases and controls [55]. Wing et al. studied the genome-wide DNA methylation pattern in whole blood samples associated with the decline in kidney function among 40 participants in the Chronic Renal Insufficiency (CRIC) study comparing the highest and lowest rates of decline in eGFR. Their results showed that 80% of CpG sites were hypermethylated in individuals with stable kidney function compared with 16% of CpG sites in rapid progressors. Specifically, methylation in CpG islands of NPHP4, IQSEC1 and TCF3, genes involved in pathways known to promote the epithelial to mesenchymal transition, were associated with rapid loss of kidney function [56]. In a more recent well-powered epigenome-wide association study (EWAS), Chu et al. profiled DNA methylation in peripheral blood leukocyte samples from 2,264 (586 cases with diabetes) African Americans participants in the Atherosclerosis Risk in Communities (ARIC) study and 2595 (394 cases with diabetes) in the Framingham Heart Study (FHS). Of 19 CpG sites significantly associated with eGFR/CKD, five were also associated with renal fibrosis in biopsies from participants with CKD and showed concordant DNA methylation changes in kidney cortex. The study further reported that methylation at PTPN6/PHB2 cg19942083 in kidney cortex associates with lower renal PTPN6 expression, higher eGFR, and less renal fibrosis; these regions are likely enriched with TF binding sites [57]. Given that PTPN6 encodes protein tyrosine phosphatase non-receptor type 6, aka Src homology-2 domain-containing phosphatase-1 (SHP-1) and that increased renal SHP-1 expression has been implicated in kidney disease and vascular complications in the setting of diabetes, the dysregulation of methylation at this site may reveal an epigenetic mechanism underlying DKD.
In addition to these genome-wide profiling of DNA methylation, a number of studies have revealed the association of DNA methylation at select gene loci with DKD risk. One example is the let-7a, which is known to decrease collagen (Col) and fibronectin (FN) expression induced by high glucose along with suppression of expression of the target gene ubituitin-like, containing PHD and RING finger domains 1 (UHRF1) essential for DNMT1 activity activity [59,60]. Peng et al. found that the methylation of let-7a promoter in the blood of DKD group was significantly higher than that in the control groups (including both healthy control and diabetic patients without DKD), whereas its level was lower in the plasma of people with DKD. The average let-7a methylation rate was 96.2% in the DKD group, 76.6% in the diabetes without nephropathy group, and 63.2% in healthy controls [58]. It is possible that the link between let-7a methylation and DKD is not only associative but may also be a mechanism in the pathogesis of DKD.
Epigenetic changes in people with DKD have also been associated with the manifestation of clinical phenotypes. Maghbooli et al. found that in comparing subjects with diabetes and with or without albuminuria, global DNA methylation in peripheral blood monocytes was significantly higher in those with albuminuria. This finding implicates DNA hypermethylation as an independent risk factor for albuminuria in patients with diabetes [61].
Other studies have suggested that methylation levels in select genes or select regions can also be associated with different manifestation of DKD. For example, methylation states of the tissure inhibitor of metalloproteinase 2 (TIMP-2) and aldo-keto reductase family 1, member B1 (AKR1B1) genes in peripheral blood was negatively correlated with albuminuria of people with DKD [62]. In another study, methylation in gene promoter of connective tissue growth factor (CTGF), a strong pro-fibrotic factor was significantly decreased in the peripheral blood of participants with diabetic nephropathy compared with participants with diabetes but without nephropathy and with non-diabetic controls. The degree of methylation in the CTGF gene is positively associated with CTGF concentration in serum, which is positively correlated with the urinary albumin-to-creatinine ratio, blood urine nitrogen and serum creatinine, but negatively correlated with eGFR [63]. Swan et al. examined the methylation patterns in genes that affect mitochondrial function and found that that the methylation levels of CpG in the TAMM41 (encoding TAM41 mitochondrial translocator assembly and maintenance homolog) and COX6A1 (gene encoding cytochrome c oxidase subunit 6A1) in peripheral blood were significantly different between healthy controls and persons with DKD [64]. Given the role of these genes in mitochondrial function, it is possible that the altered DNA methylation in part mediates the disruption of mitochondrial metabolism in DKD.
Collectively, these efforts in profiling epigenetic states using circulating blood samples strongly suggest the association between epigenetic changes and DKD risk, both in incidence and disease progression. These reports also implicate genes with significant variations in DNA methylation as promising candidates for future experimental studies to illuminate the underlying regulatory mechanisms contributing to the incidence and progression of DKD.
Researchers have also begun to test the changes in lncRNAs using blood samples. Several lncRNAs have been detected in the blood from patients with DKD. For example, the level of lncRNA NR_033515 was significantly higher in the serum of patients with DKD than normal controls, and it was correlated with the severity of DKD and positively associated with diagnostic markers of DKD such as KIM-1 [65]. Plasmacytoma variant translocation (PVT1), a lncRNA significantly upregulated in human mesangial cells by high glucose, has been identified to be a candidate gene for ESRD in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism analysis in DNA samples from the participants of Gila River Indian Commnity [66]. Using the Genetics of Kidneys Diabetes (GoKinD) trial database, variants in the PVT1 gene have been associated with ESRD in type 1 diabetes [67].
Epigenetic profile in renal tissue
Compared to epigenetic profiling using body fluids, it is more difficult to probe epigenetic changes in renal tissues, which are not easily accessible. Nevertheless, investigators have focused on this organ as the relevant site of the disease process. Wang et al. has recently analyzed the gene expression omnibus (GEO) public database archiving data from kidney and reported 121 genes with hypermethylated sites and 579 genes with hypomethylated sites in the kidney tissue. Among these genes with differential methylation states, two hypomethylated genes [Peroxisome proliferator-activated receptor alpha (PPARA) and Glutaminase (GLS)] were specific for tubular cells and one hypermethylated gene (PIK3C2B) were specific for glomeruli [68]. Because tubular cells have abundant mitochondria and are important for acid-base regulation through ammoniagenesis, these findings may suggest epigenetic modulation of mitochondrial biogenesis (PGC-1α) and acid-base regulation (glutaminase) that may contribute to the disease process of DKD.
Hayashi et al. found that KLF4 expression is decreased in proteinuric states in human renal tissues. Restoration of KLF4 expression in diseased glomeruli in vivo resulted in restoration of nephrin expression and the attenuation of proteinuria. To explain the mechanism, the authors suggested that KLF4 overexpression leads to demethylation in the nephrin promoter region and consequently increases nephrin promoter activity. In contrast, KLF4 overexpression caused an increase of DNA methylation in the vimentin promoter, which in turn suppresses the expression of vimentin [69].
The association of histone methylation and acetylation in DKD progression have also been examined in renal tissue or cells [70-72]. H3K9ac levels were significantly increased in renal biopsies from patients with DKD, especially in the podocytes [70]. The change of histone methylation/acetylation in different renal cells has been suggested to induce the expression of potent profibrotic factors such as TGF-β1, CTGF, PAI-1 and Col-1A [41,73,74], which have been correlated with increased renal fibrosis and DKD progress to ESRD [75].
HDAC 2/4/5/9 have been shown to be upregulated in kidney biopsy tissue obtained from patients with diabetes. The mRNA levels of HDAC2/4/5 in patients with DKD were negatively correlated with eGFR. Inhibition of HDACs reduced albuminuria and mesangial expansion, ameliorated podocyte injury, attenuated glomerulosclerosis and decreased production of proinflammatory mediators in diabetic animals [76-79]. The mRNA and protein levels of TFEB, an important regulator of the autophagy-lysosome pathway, were reduced in the renal parenchyma of patients with DKD in comparison to individuals without diabetes or kidney disease. TFEB acetylation was increased by HDAC6 inhibitor in tubular cell. Treating right kidney-nephrectomized rats with an HDAC6 inhibitor attenuated proteinuria, tubule epithelial cell death, and tubulointerstitial collagen IV protein accumulation, concomitantly with an increase in renal protein levels of the TFEB target gene beclin 1 [80]. All of the above indicates that HDAC changes influence progression of DKD, suggesting that HDAC inhibition might be a potential treatment for DKD.
Belonging to Class III HDAC, sirtuins, including SIRT1-7, have been implicated in aging, transcription, apoptosis, and inflammation. The levels of Sirt1, Sirt3, Sirt4, and Sirt6 were reduced in kidneys from patients with diabetic nephropathy. Moreover, Sirt1 expression in proximal tubular and glomerular regions were lower in the kidneys of patients with heavy proteinuria compared with those with moderate proteinuria. Diabetic mice with Sirt6 deletion developed marked albuminuria, significant mesangial matrix expansion, glomerular basement membrane thickening, and podocyte foot process broadening and effacement by increased H3K9ac and activating Notch signaling pathways [70]. Pharmacological targeting of Sirt6-mediated Notch signaling pathways at multiple levels may provide a novel approach for the treatment of proteinuric kidney disease.
The studies about lncRNAs in DKD are limited, but there has been rising interest in their role in the pathogenesis of DKD. In one of the earliest studies, a key lncRNA, lnc-megacluster (Inc-MGC) was shown to promote mesangial cell extracellular matrix accumulation and hypertrophy related to early DKD. Lnc-MGC is a host non-coding transcript to a megacluster of nearly 40 miRNAs (miR-379 cluster), which are coordinately increased by HG and TGFβ1 in cultured mouse and human mesangial cells, and in glomeruli of mouse models of type1 and type 2 diabetes. Notably, knockdown of Lnc-MGC in diabetic mice with a modified antisense oligonucleotide (GapmeR) reduced glomerular hypertrophy and other features of early diabetic nephropathy, demonstrating the translational potential of targeting renal lncRNAs [81]. LncRNA taurine upregulated gene 1 (Tug 1) was the first lncRNA identified to regulate podocyte function, and through RNA sequencing its transcript levels were shown to be decreased in DKD compared with controls. Lower expression levels of TUG1 was correlated with reduced levels of eGFR in patients with DKD. Podocyte-specific diabetic Tug1 transgenic (TugPodTg ) mice showed a significant reduction in albuminuria. This was associated with a reduction in mesangial matrix expansion, improvement in effacement of podocyte foot process and glomerular basement membrane thickening. The effect of Tug1 overexpression on DKD was shown to regulate the PGC1α and influence mitochondrial bioenergetics [82]. A recent study has identified another lncRNA, Erbb4-IR, to be a potential key regulator in DKD. Kidney-specific silencing of Erbb4-IR could protect against the development of type 2 DKD, such as elevated microalbuminuria, serum creatinine, and progressive renal fibrosis in db/db mice. Inhibition of Erbb4-IR in mouse tubular epithelial cells suppressed TGFβ1-induced collagen I and α-SMA expression inhibiting tubular epithelial mesenchymal transition, extracellular matrix accumulation and significantly reducing the severity of tubulointerstitial fibrosis [83,84]. In another study, Linc01619 was found to be downregulated in renal biopsy tissues collected from patients with DKD. It was negatively correlated with serum creatinine and proteinuria, and positively correlated with eGFR [85]. Additional lncRNAs reported to have functions related to DKD in different renal cell types [86-98] are summarized in Table 1.
Table 1.
Long non-coding RNAs involved in diabetic nephropathy
| Cell type | LncRNA | Cell function | Potential targets | ref |
|---|---|---|---|---|
| mouse mesangial cell | CYP4B1-PS1-001 | overexpression of CYP4B1-PS1-001 inhibited proliferation and fibrosis of mesangial cells | PCNA,cyclin-D1,Col 1,Fibronectin | 86 |
| ENSMUST-00000147869 | Reduce cell proliferation, decreased the synthesis of fibronectin and collagen I | PCNA,cyclin-D1,Col 1, Fibronectin | 87 | |
| Gm4419 | Regulate the pro-inflammatory and fibrosis cytokines and mesangial cell proliferation | NF-kB/NRLP3 | 88 | |
| lincRNA 1700020I24Rik | affect cell proliferation and expressions of renal fibrosis markers | miR-34a-5p/Sirt1/HIF-1α | 89 | |
| human mesangial cell (HMC) | Lnc-MGC | inhibit the expression of key cluster miRNAs in the kidney, and regulate TGF-β signaling, cellular hypertrophy, extracellular matrix synthesis and ER stress. | PTEN,CUGBP2,PUM2, TNRC6B,CPEB4, BHC80,EDEM3, ATF3 and others | 81 |
| ASncmtRNA-2 | TGF-b and FN expression were up regulated by HG-induced ASncmtRNA-2 in HMC | TGF-b | 90 | |
| PVT1 | FN1, COL4A,TGFB1,PAI-1 expression were decreased in HMC treated with high glucose | FN1, COL4A,TGFB1,PAI-1 | 91 | |
| podocyte | Lnc TUG1 | modulates mitochondrial bioenergetics through PGC-1a in podocyte | peroxisome proliferator–activated receptor g coactivator a (PGC-1 a) | 82 |
| Linc01619 | regulates miR-27a/FOXO1 mediated ER stress and podocyte apoptosis | miR 27a/FOXO1 | 85 | |
| ENSRNOG00000037522 | inhibit the high glucose-induced podocyte epithelial mesenchymal transition | PODXL-1,nephrin | 92 | |
| MALAT1 | MALAT1 regulated by b-catenin, MALAT1 knock-down rectified podocyte damage via down-regulating SRSF1 overexpression | SRSF1 | 93 | |
| Gm5524 Gm15645 |
Affect autophagy and apoptosis of podocyte | Atg5,Atg7,Bcl2 | 98 | |
| HK2 | MALAT1 | Down regulated MALAT1 decreased the expression of the pro-inflammatory cytokines IL-6 and TNF-α | ELAVL1, NLRP3, Caspase-1, TNF-a, IL-1β ,IL-6 | 94,95 |
| MIAT | regulated HK2 cell viability via stabilizing Nrf2 expression | Nrf2 | 96 | |
| Lnc ZEB1-AS1 | Inhibition of lnc ZEB1-AS1 may increase HG-induced ECM accumulation by downregulating ZEB1 expression | ZEB1 | 97 | |
| tubular epithelial cell (TEC)/mouse mesangial cell (MMC) | Erbb4-IR | Regulate the collagen expression in TEC and MMC through miR29b | miR29b | 83 |
Conclusion
Although insulin and a variety of newer medications have been used to control hyperglycemia, microvascular complications such as DKD are still inevitable in many patients with diabetes. Furthermore, many patients with DKD develop ESRD, a devastating outcome of CKD. Currently, there is still a lack of effective measures to predict the risk for DKD development and progression. Epigenetic changes including DNA methylation, histone modification, and noncoding RNAs, are increasingly recognized features of DKD and recent evidence suggests that epigenetic mechanisms may influence the pathogenesis and progression of DKD. Given the rapid progress in epigenetics and epigenomics due to increasingly affordable high-throughput technology and advances in bioinformatics software, more data addressing the epigenetics in DKD can be obtained in a much more cost-effective manner. These advances may reveal additional epigenetic signatures or enhance profiling in the DKD population.
Further investigation is necessary to delineate any causal relationship of epigenetic modification with DKD pathogenesis in light of recent advances in the understanding of basic molecular and cellular mechanisms. In this regard, integration of these data with emerging genome-wide association results for DKD could help place the activation or repression of specific genes on causal pathways. This could enable the application of current knowledge of epigenetics to the evaluation of DKD risk, and facilitate the discovery of new treatment to manage this debilitating disease. A major challenging to the translation of knowledge to the evaluation of patients with DKD is the limited availability of samples and the heterogeneity of renal tissues with multiple cell types. Moreover, epigenetic data generated from patients with DKD in different studies are often affected by heterogeneity in the selection of study population, the nature of the samples collected, and the profiling methods employed. More consistency in study protocols and experimental procedures is needed to promote the reproducibility and comparability of various studies.
Although microRNAs have been showed to be a useful marker in the diagnosis and severity of albuminuria as well as markers of pathological change in the body fluids of patients with DKD, whether DNA methylation, histone modification, and lncRNA levels in urine or serum can be reliable markers to evaluate risks for DKD remains largely unknown. In particular, the role of lncRNAs in the context of DKD is a relative new area for exploration. More research is warranted to elucidate mechanisms by which these non-coding transcripts alter molecular and cellular processes related to DKD. Several small molecule inhibitors that target epigenetic regulation have been administered in DKD animal models and show promise in attenuating the pathological manifestations of DKD. However, none of them have been tested in patients with DKD. It would be of great interest to investigate whether these therapeutic strategies designed to influence epigenetic profile could delay or prevent the development of DKD and/or even reverse the progression of this condition.
Acknowledgment
This work was supported by the National Natural Science Fund of China 81570609 and 81770667 (to X. L.), and US PHS NIH research grants K99/R00HL122368 (to Z.C.) and R01 DK065073, R01 DK081705, R01DK58191, R01 HL106089 (to R.N.). The authors would also like to acknowledge the support from Ella Fitzgerald Foundation. We apologize to colleagues whose important primary studies we were unable to cite due to space constraints.
Footnotes
Conflict of Interest
Lixia Xu, Rama Natarajan, and Zhen Chen declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Jones CA, Krolewski AS, Rogus J, Xue JL, Collins A, Warram JH. Epidemic of end-stage renal disease in people with diabetes in the United States population: do we know the cause? Kidney Int 2005;67(5): 1684–91 [DOI] [PubMed] [Google Scholar]
- 2.Sarnak MJ, Levey AS, Schoolwerth AC, Coresh J, Culleton B, Hamm LL, et al. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disease, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation 2003; 108(17): 1054–69 [DOI] [PubMed] [Google Scholar]
- 3.Schultz CJ, Neil HA, Dalton RN, Dunger DB; Oxforn Regional Prospective Study Group. Risk of nephropathy can be detected before the onset of microalbuminuria during the early years after diagnosis of type 1 diabetes. Diabetes Care 2000;23(12): 1811–5 [DOI] [PubMed] [Google Scholar]
- 4.Tonneijck L, Muskiet MH, Smits MM, van Bommel EJ, Heerspink HJ, van Raalte DH, et al. Glomerular Hyperfiltration in Diabetes: Mechanisms, Clinical Significance, and Treatment. J Am Soc Nephrol 2017;28(4):1023–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hovind P, Rossing P, Tarnow L, Smidt UM, Parving HH. Progression of diabetic nephropathy. Kidney Int 2001;59(2):702–9 [DOI] [PubMed] [Google Scholar]
- 6.Najafian B, Mauer M. Morphologic features of declining renal function in type 1 diabetes. Semin Nephrol 2012;32(5):415–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2014; 429(6990):457–63 [DOI] [PubMed] [Google Scholar]
- 8.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science 2010; 330(6004):612–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen ED, Kaestner KH, Natarajan R, Patti ME, Sallari R, Sander M, et al. Epigenetics and Epigenomics: Implications for Diabetes and Obesity. Diabetes 2018;67(10):1923–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceriello A, Ihnat MA, Thorpe JE. Clinical review 2: The "metabolic memory": is more than just tight glucose control necessary to prevent diabetic complications? J Clin Endocrinol Metab 2009;94(2):410–5 [DOI] [PubMed] [Google Scholar]
- 11.Diabetes Control and Complications Trial Research Group, Nathan DM, Genuth S, Lachin J, Cleary P, Crofford O, Davis M, et al. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993; 329(14):977–86 [DOI] [PubMed] [Google Scholar]
- 12.Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group, Lachin JM, Genuth S, Cleary PA, Davis MD, Nathan DM. Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. N Engl J Med 2000; 342(6):381–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Writing team for DCCT/EDIC Research Group. Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: The Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA 2003; 290(16):2159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DCCT/EDIC Research Group, de Boer IH, Sun W, Cleary PA, Lachin JM, Molitch ME, Steffes MW, et al. Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med 2011; 365(25):2366–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UKPDS Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet 1998; 352(9131):837–53 [PubMed] [Google Scholar]
- 16.Retnakaran R, Cull CA, Thorne KI, Adler AI, Holman RR; UKPDS Study Group. Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective Diabetes Study 74. Diabetes 2006; 55(6):1832–9 [DOI] [PubMed] [Google Scholar]
- 17.ADVANCE Collaborative Group, Patel A, MacMahon S, Chalmers J, Neal B, Billot L, Woodward M, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med 2008; 358(24):2560–72 [DOI] [PubMed] [Google Scholar]
- 18.Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med 2005; 353(25):2643–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceriello. Hypothesis: the "metabolic memory", the new challenge of diabetes. Diabetes Res Clin Pract 2009; 86 Suppl: S2–6 [DOI] [PubMed] [Google Scholar]
- 20.Bianchi C, Miccoli R, Del Prato S. Hyperglycemia and vascular metabolic memory: truth or fiction? Curr Diab Rep 2013; 13(3):403–10 [DOI] [PubMed] [Google Scholar]
- 21.Chamers J, Cooper ME. UKPDS and the Legacy Effect. N Engl J Med 2008;359(15): 1618–20 [DOI] [PubMed] [Google Scholar]
- 22.Kuzhively J, Tahsin B, Hart P, Fogelfeld L. Legacy effect in combined diabetic-renal multifactorial intervention in patients with advanced diabetic nephropathy. J Diabetes Complications 2018;32(5):474–9 [DOI] [PubMed] [Google Scholar]
- 23.Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014; 63(5):1748–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.••.Chen Z, Miao F, Paterson AD, Lachin JM, Zhang L, Schones DE, et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci USA 2016; 113(21): E3002–11This study reports that the DNA-methylation differences during the DCCT persist at certain loci associated with glycemia for years during the EDIC Study and highlights an epigenetic explanation for metabolic memory.
- 25.Feinberg AP. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N Engl J Med 2018; 378(14): 1323–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Q, Vijayakumar A, Kahn BB. Metabolites as regulators of insulin sensitivity and metabolism. Nat Rev Mol Cell Biol 2018; 19(10):654–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Egervari G, Wang Y, Berger SL, Lu Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat Rev Mol Cell Biol 2018; doi: 10.1038/s41580-018-0029-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016; 534(7609):652–7 [DOI] [PubMed] [Google Scholar]
- 29.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462(7271):315–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS genetic 2010; 6(9): e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 2010; 463(7284):1101–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindstrom VM Jr, Flynn J, Reich NO. Reconciling structure and function in HhaI DNA cytosine-C-5 methyltransferase. J Biol Chem 2000. ;275(7):4912–9 [DOI] [PubMed] [Google Scholar]
- 33.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci USA 1983; 80(18):5559–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet 2016; 32(1):42–56 [DOI] [PubMed] [Google Scholar]
- 35.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014; 13(9):673–91 [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004. ; 119(7):941–53 [DOI] [PubMed] [Google Scholar]
- 37.Kozub MM, Carr RM, Lomberk GL, Fernandez-Zapico ME. LSD1, a double-edged sword, confers dynamic chromatin regulation but commonly promotes aberrant cell growth. F1000 Res 2017; 6:2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439(7078):811–6 [DOI] [PubMed] [Google Scholar]
- 39.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298(5595): 1039–43 [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Li X, He K, Li B, Liu K, Qi J, et al. C-peptide prevents NF-κB from recruiting p300 and binding to the inos promoter in diabetic nephropathy. FASEB J 2018; 32(4):2269–79 [DOI] [PubMed] [Google Scholar]
- 41.Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-β1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol 2012; 304(5):F601–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 2007;26(37):5395–407 [DOI] [PubMed] [Google Scholar]
- 43.Seto E, Yoshida M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb Perspect Biol 2014; 6(4):a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J, Wang Y, Cui W, Lou Y, Sun G, Zhang D, et al. Role of Epigenetic Histone Modifications in Diabetic Kidney Disease Involving Renal Fibrosis. J Diabetes Res 2017; 2017:7242384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanner N, Bechtel-Walz W. Epigenetics of kidney disease. Cell Tissue Res 2017; 369(1):75–92 [DOI] [PubMed] [Google Scholar]
- 46.Assmann TS, Recamonde-Mendoza M, de Souza BM, Bauer AC, Crispim D. MicroRNAs and diabetic kidney disease: Systematic review and bioinformatic analysis. Mol Cell Endocrine 2018; 477:90–102 [DOI] [PubMed] [Google Scholar]
- 47.Ichii O, Horino T. MicroRNAs associated with the development of kidney diseases in humans and animals. J Toxicol Pathol 2018; 31(1):23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dewanjee S, Bhattachearjee N. MicroRNA: A new generation therapeutic target in diabetic nephropathy. Biochem Pharmacol 2018; 155:32–47 [DOI] [PubMed] [Google Scholar]
- 49.Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172(3):393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157(1):77–94 [DOI] [PubMed] [Google Scholar]
- 51.Leicamwasam A, Sexton-Oates A, Carmody J, Ekinci EI, Dwyer KM, Saffery R. DNA methylation profiling of genomic DNA isolated from urine in diabetic chronic kidney disease: A pilot study. PLoS one 2018; 13(2): e0190280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.EI Bali L, Diman A, Bernard A, Roosens NH, De Keersmaecker SC. Comparative study of seven commercial kits for human DNA extraction from urine samples suitable for DNA biomarker-based public health studies. J Biolmol Tech 2014; 25(4):96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokota M, Tatsumi N, Tsuda I, Takubo T, Hiyoshi M. DNA extraction from human urinary sediment. J Clin Lab Anal 1998; 12(2):88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sapienza C, Lee J, Powell J, Erinle O, Yafai F, Reichert J, et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 2011; 6(1):20–8 [DOI] [PubMed] [Google Scholar]
- 55.Gautier JF, Porcher R, Abi Khalil C, Bellili-Munoz N, Fetita LS, Travert F, et al. Kidney Dysfunction in Adult Offspring Exposed In Utero to Type 1 Diabetes Is Associated with Alterations in Genome-Wide DNA Methylation. PLoS ONE 2015;10(8): e0134654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wing MR, Devaney JM, Joffe MM, Xie D, Feldman HI, Dominic EA, et al. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant 2014; 29(4):864–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.•.Chu AY, Tin A, Schlosser P, Ko YA, Qiu C, Yao C, et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun 2017;8(1):1286.This well-powered EWAS identified differential DNA methylation associated with kidney function and CKD. It further demonstrated methylation at PTPN6/PHB2 in kidney cortex was associated with lower renal PTPN6 expression, higher eGFR, and less renal fibrosis.
- 58.Peng R, Liu H, Peng H, Zhou J, Zha H, Chen X, et al. Promoter hypermethylation of let-7a-3 is relevant to its down-expression in diabetic nephropathy by targeting UHRF1. Gene 2015; 570(1):57–63 [DOI] [PubMed] [Google Scholar]
- 59.Yan N, Wen L, Peng R, Li H, Liu H, Peng H, et al. Naringenin Ameliorated Kidney Injury through Let-7a/TGFBR1 Signaling in Diabetic Nephropathy. J Diabetes Res 2016;2016: 8738760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bashtrykov P, Jankevicius G, Jurkowska RZ, Ragnozin S, Jeltsch A. The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J Biol Chem 2014; 289(7):4106–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maghbooli Z, Larijani B, Emamgholipour S, Amini M, Keshtkar A, Pasalar P. Aberrant DNA methylation patterns in diabetic nephropathy. J Diabetes Metab Disord 2014; 13:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aldemir O, Turgut F, Gokce C. The association between methylation levels of targeted genes and albuminuria in patients with early diabetic kidney disease. Renal Failure 2017; 39(1):597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H, Cail X, Yi B, Huang J, Wang J, Sun J. Correlation of CTGF gene promoter methylation with CTGF expression in type 2 diabetes mellitus with or without nephropathy. Mol Med Rep 2014; 9(6):2138–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swan EJ, Maxwell AP, McKnight AJ. Distinct methylation patterns in genes that affect mitochondrial function are associated with kidney disease in blood-derived DNA from individuals with Type 1 diabetes. Diabet Med. 2015;32(8):1110–5 [DOI] [PubMed] [Google Scholar]
- 65.Gao J, Wang W, Wang F, Guo C. LncRNA-NR_033515 promotes proliferation, fibrogenesis and epithelial-to-mesenchymal transition by targeting miR-743b-5p in diabetic nephropathy. Biomed Pharmacother 2018; 106:543–52 [DOI] [PubMed] [Google Scholar]
- 66.Hadson RL, Craig DW, Millis MP, Yeatts KA, Kobes S, Pearson JV, et al. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study Diabetes 2007;56(4):975–83 [DOI] [PubMed] [Google Scholar]
- 67.Milles MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 2007;56(12):3027–32 [DOI] [PubMed] [Google Scholar]
- 68.Wang YZ, Xu WW, Zhu DY, Zhang N, Wang YL, Ding M, et al. Specific expression network analysis of diabetic nephropathy kidney tissue revealed key methylated sites. J Cell Physiol 2018; 233(10):7139–47 [DOI] [PubMed] [Google Scholar]
- 69.Hayashi K, Sasamura H, Nakamura M, Azegami T, Oguchi H, Sakamaki Y, et al. KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. J Clin Invest 2014; 124(6):2523–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z, et al. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun 2017; 8(1):413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S, et al. Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant 2010; 25(6):1811–7. [DOI] [PubMed] [Google Scholar]
- 72.De Marinis Y, Cai M, Bompada P, Atac D, Kotoa O, Johansson ME, et al. Epigenetic regulation of the thioredoxin-interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney Int 2016; 89(2):342–53 [DOI] [PubMed] [Google Scholar]
- 73.Xu H, Wu X, Qin H, Tian W, Chen J, Sun L, et al. Myocardin-Related Transcription Factor A Epigenetically Regulates Renal Fibrosis in Diabetic Nephropathy. J Am Soc Nephrol 2015; 26(7):1648–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan H, Reddy MA, Deshpande S, Jia Y, Park JT, Lanting LL, et al. Profibrotic Gene Regulation by 12/15-Lipoxygenase and Its Oxidized Lipid Products in Diabetic Nephropathy. Antioxid Redox Signal 2016;24(7):361–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulated fibrotic gene expression. J Am Soc Nephrol. 2010;21(12):2069–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Noh H, Oh EY, Seo JY, Yu MR, Kinm YO, Ha H, et al. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-beta1-induced renal injury.Am J Physiol Renal Physiol 2009; 297(3):F729–39 [DOI] [PubMed] [Google Scholar]
- 77.Lin CL, Lee PH, Hsu YC, Lei CC, Ko JY, Chuang PC, et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction.J Am Soc Nephrol 2014; 25(8):1698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X, Liu J, Zhen J, Zhang C, Wan Q, Liu G, et al. , Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int 2014; 86(4):712–25 [DOI] [PubMed] [Google Scholar]
- 79.Liu F, Zong M, Wen X, Li X, Wang J, Wang Y, et al. Silencing of Histone Deacetylase 9 Expression in Podocytes Attenuates Kidney Injury in Diabetic Nephropathy. Sci Rep 2016; 6:33676. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Brijmohan AS, Batchu SN, Majumder S, Alghamdi TA, Thieme K, McGaugh S, et al. HDAC6 Inhibition Promotes Transcription Factor EB Activation and Is Protective in Experimental Kidney Disease. Front Pharmacol 2018; 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.•.Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 2016; 7:12864.This study provides a thorough description of Lnc-MGC, a lncRNA megacluster regulated by endothplasmic reticulum stress and its relevance to human DKD.
- 82.•.Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016; 126(11):4205–18This study provides extensive evidence for an important regulatory crosstalk between lncRNAs and mitochondria mitochondrial bioenergetics in podocytes in the diabetic milieu.
- 83.Sun SF, Tang PMK, Feng M, Xiao J, Huang XR, Li P, et al. Novel lncRNA Erbb4-IR Promotes Diabetic Kidney Injury in db/db Mice by Targeting miR-29b. Diabetes 2018;67(4):731–44 [DOI] [PubMed] [Google Scholar]
- 84.Feng M, Tang PM, Huang XR, Sun SF, You YK, Xiao J, et al. TGF-β Mediates Renal Fibrosis via the Smad3-Erbb4-IR Long Noncoding RNA Axis. Mol Ther. 2018;26(1): 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bai X, Geng J, Li X, Wan J, Liu J, Zhou Z, et al. Long Noncoding RNA LINC01619 Regulates MicroRNA-27a/Forkhead Box Protein O1 and Endoplasmic Reticulum Stress-Mediated Podocyte Injury in Diabetic Nephropathy. Antioxid Redox Signal. 2018;29(4):355–76 [DOI] [PubMed] [Google Scholar]
- 86.Wang M, Wang S, Yao D, Yan Q, Lu W. A novel long non-coding RNA CYP4B1-PS1-1 regulates proliferation and fibrosis in diabetic nephropathy. Mol Cell Endocrinol 2016; 426:136–45 [DOI] [PubMed] [Google Scholar]
- 87.Wang M, Yao D, Wang S, Yan Q, Lu W. Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine 2016;54(1):81–92 [DOI] [PubMed] [Google Scholar]
- 88.Yi H, Peng R, Zhang LY, Sun Y, Peng HM, Liu HD, et al. LincRNA-Gm4419 knockdown ameliorates NFκB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis 2017; 8(2):e2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li A, Peng R, Sun Y, Liu H, Peng H, Zhang Z. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell Death Dis 2018; 9(5):461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao Y, Chen ZY, Wang Y, Liu Y, Ma JX, Li YK. Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp Ther Med 2017; 13(2):581–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE 2011; 6(4):e18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ling L, Tan Z, Zhang C, Gui S, Hu Y, Chen L. Long noncoding RNA ENSRNOG00000037522 is involved in the podocyte epithelial-mesenchymal transition in diabetic rats. Int J Mol Med 2018; 41(5):2704–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, et al. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J Cell Mol Med. 2017; 21(11):2732–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu D, Cheng YG, Huang X, Zhong MW, Liu SZ, Hu SY. Downregulation of lncRNA MALAT1 contributes to renal functional improvement after duodenal-jejunal bypass in a diabetic rat model. J Physiol Biochem 2018. ; 74(3):431–9 [DOI] [PubMed] [Google Scholar]
- 95.Li X, Zeng L, Cao C, Lu C, Lian W, Han J, et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res 2017. ; 350(2)327–35 [DOI] [PubMed] [Google Scholar]
- 96.Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun 2015; 468(4):726–32 [DOI] [PubMed] [Google Scholar]
- 97.Wang J, Pang J, Li H, Long J, Fang F, Chen J, et al. lncRNA ZEB1-AS1 Was Suppressed by p53 for Renal Fibrosis in Diabetic Nephropathy Mol Ther Nucleic Acid 2018; 12:741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feng Y, Chen S, Xu J, Zhu Q, Ye X, Ding D, et al. Dysregulation of lncRNAs GM5524 and GM15645 involved in high-glucose induced podocyte apoptosis and autophagy in diabetic nephropathy. Mol Med Rep 2018. ; 18(4) 3657–64 [DOI] [PMC free article] [PubMed] [Google Scholar]