INTRODUCTION
Epidermal growth factor receptor (EGFR) mutation–positive lung cancers respond dramatically to EGFR tyrosine kinase inhibitors (TKIs),1–3 and repeat biopsies at acquired resistance can illuminate the critical molecular resistance mechanisms.4,5 Historically, resistance mutations were conceptualized as binomial variables (eg, cancers were either positive or negative for a given mutation); however, growing appreciation of intra- and intertumoral heterogeneity has shifted the paradigm toward resistant clones as dynamic populations, which shift in prevalence on the basis of the selective pressure of sequential therapies.6 This case report illustrates how serial molecular monitoring may provide unique insight into clonal evolution.
CASE REPORT
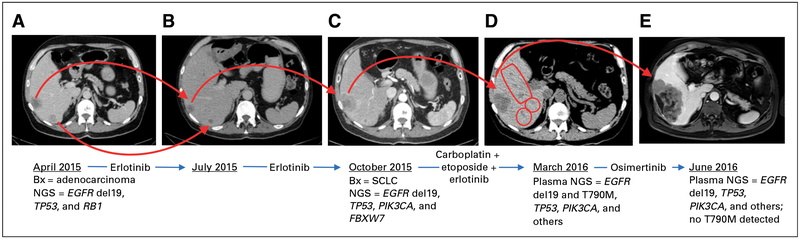
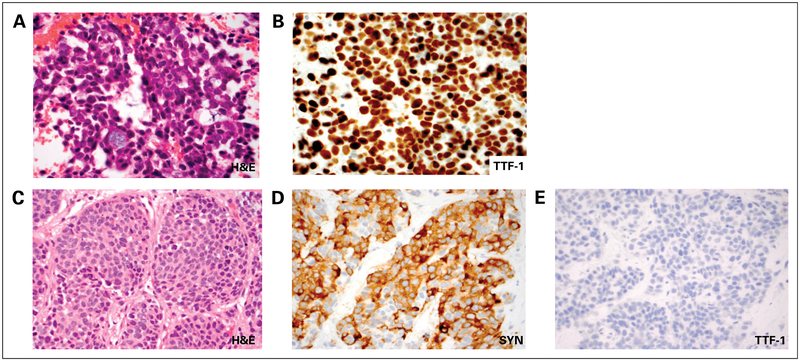
A 63-year-old man with back pain and a minimal smoking history presented for medical attention in April 2015. Lumbar spine magnetic resonance imaging (MRI) demonstrated multiple bone lesions, and computed tomography scans revealed a 4-cm right-sided hilar lung mass, regional thoracic lymphadenopathy, multiple hepatic metastases, a left-sided adrenal metastasis, and several osseous lesions(Fig 1A). Brain MRI visualized three asymptomatic brain metastases. Biopsies of the subcarinal lymph node and the left-side adrenal lesion were performed, which confirmed adenocarcinoma of lung origin (Figs 2A and 2B).
Fig 1.
Clinical course and representative radiographic images. (A to E) Selected radiographic images of the liver illustrate involvement with cancer. Treatments and key biopsy (Bx) results (tissue or liquid) are indicated underneath each image in chronologic order. NGS, next-generation sequencing; SCLC, small-cell lung cancer.
Fig 2.
Histopathologic findings. (A) Hematoxylin and eosin (H&E) stain and (B) thyroid transcription factor 1 (TTF-1) immunostain of fine-needle aspiration of a subcarinal lymph node at diagnosis show adenocarcinoma with diffuse TTF-1 expression consistent with a lung primary. (C) H&E stain and (D) synaptophysin (SYN) and (E) TTF-1immunostains of a liver core biopsy at the time of acquired resistance to erlotinib demonstrate small-cell lung cancer with solid nests of highly atypical epithelial cells with finely dispersed chromatin, inconspicuous nucleoli, and brisk mitotic activity. The tumor cells are positive for SYN and negative for TTF-1.
The patient was treated with radiation therapy to the painful vertebral metastasis and stereotactic radiosurgery to two brain lesions. Molecular testing of the subcarinal node through a next-generation sequencing (NGS) panel that covers > 200 genes (FoundationOne; Foundation Medicine, Cambridge, MA) revealed an EGFR exon 19 deletion (del19) mutation, a TP53V173L mutation, an EGFR amplification, and an RB1 loss of exons 18 and 19. Oral erlotinib 150 mg daily was initiated in May 2015, and uniform disease response was evident on restaging scans in July 2015 (Fig 1B). However, in October 2015, repeat imaging showed significant growth in a single liver lesion and a new 2.6-cm lesion in the spleen, with continued response elsewhere, including the brain (Fig 1C).
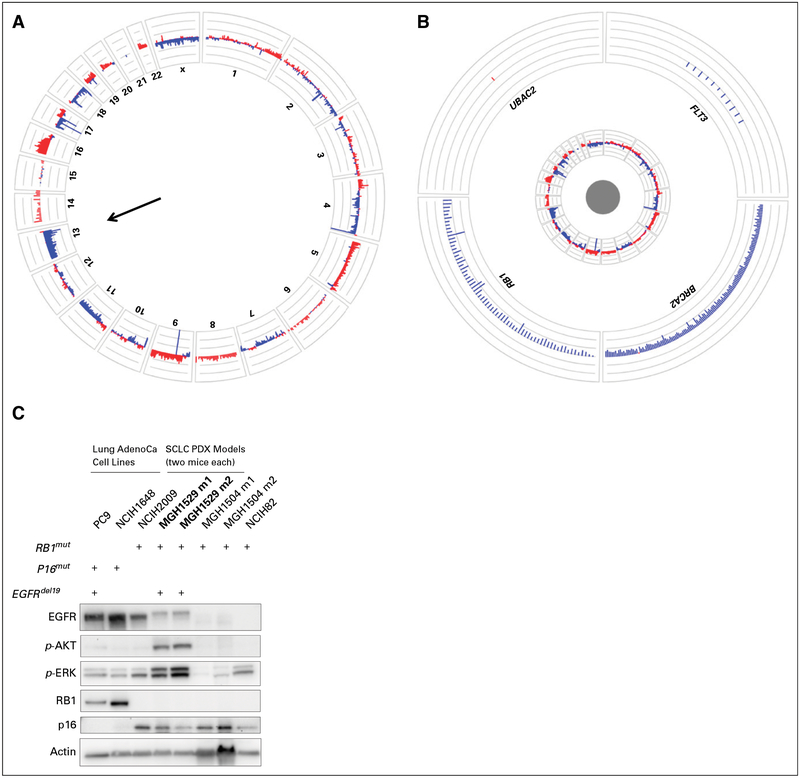
Repeat biopsy of the enlarging liver lesion in November 2015 revealed nests of highly atypical cells with finely dispersed chromatin, inconspicuous nucleoli, and abundant mitoses (Figs 2C to 2E). Immunohistochemical stains were positive for synaptophysin and chromogranin and negative for thyroid transcription factor 1 and napsin A. The protein encoded by the MKI67 gene labeling index was 80%. The overall features were consistent with small-cell lung cancer (SCLC) transformation.4 NGS that consisted of targeted hotspot evaluation in 39 genes (SNaPshot NGS; Massachusetts General Hospital, Boston, MA) confirmed the presence of the original EGFR del19 and TP53 mutations and showed additional mutations in PIK3CA (G545L), PIK3CA (G726L), ERBB3 (G337A), and FBXW7 (L8P). Biallelic loss of RB1 was detected (Figs 3A and 3B). A patient-derived xenograft generated from this biopsy specimen lacked RB protein expression but retained minimal EGFR expression and demonstrated activation of the mitogen-activated protein kinase and AKT pathways (Fig 3C), likely as a result of the presence of an activating PIK3CA mutation.
Fig 3.
RB1 loss within the small-cell lung cancer (SCLC)–transformed tumor. Circos plots provide illustrative overviews of the next-generation sequencing (NGS) analyses of the liver biopsy specimens with SCLC transformation. (A) Summary of all evaluable probes across all chromosomes (red, genomic gains; blue, genomic losses) shows diffuse losses across chromosome 13, including the RB1 gene locus. (B) A magnified view of four specific genes on chromosome 13 shows that all examined loci of RB1 are lost, with only blue signals and complete absence of red signals. (C) Tissue obtained from the November 2015 liver biopsy (which shows SCLC) was used to generate a patient-derived xenograft (PDX) in an NSG mouse and subsequently passaged through additional NSG mice. The PDX tumor demonstrated SCLC histologic features consistent with the patient biopsy sample (not shown). A Western blot demonstrates relative protein levels of EGFR, p-AKT (S473), p-ERK, RB, p16, and actin (loading control) among PDX tumors (MGH1529; two tumors shown) with control lung adenocarcinoma (AdenoCa) and SCLC samples for comparison. Genetic characteristics of the various cell lines (RB1, p16, and EGFR exon 19 deletion [del19] mutations [mut]) are shown above the Western blot. Of note, the PDX tumor retained mild EGFR expression, although not as strongly as the AdenoCa controls, but had complete loss of RB1 expression similar to the de novo SCLC lines.
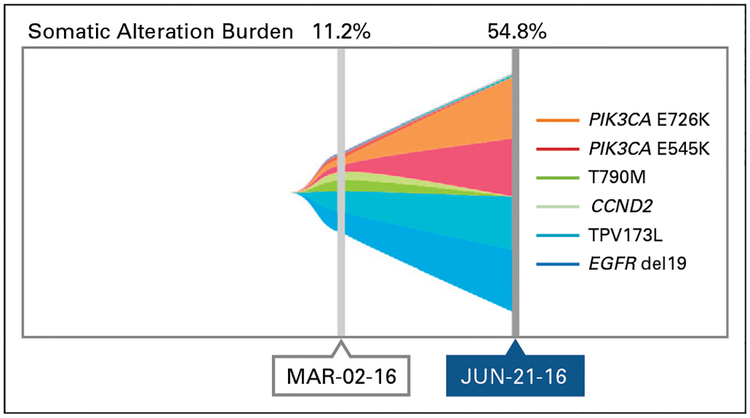
The patient was treated with carboplatin and etoposide chemotherapy and ongoing erlotinib. Scans after four cycles of chemotherapy showed a mixed response with slight regression in the previously biopsied (SCLC-transformed) liver metastasis, stable brain metastases, and multiple new distinct sites of hepatic progression (Fig 1D). Plasma-based cell-free circulating tumor DNA (ctDNA) genotyping (Guardant360; Guardant Health, Redwood City, CA) revealed the following genes (and mutant allele frequencies [MAFs]; Fig 4): EGFR del19 (11.1%), TP53 V173L (11.2%), CCND2 S271S(5%),EGFRT790M (3.5%), PIK3CA E726K (2.7%), PIK3CA E545K (2.6%), and NRAS V188V (1.4%). Of note, the Guardant360 assay can detect RB1 inactivating mutations but not allelic losses.
Fig 4.
Plasma genotyping. Graphic representation of the relative mutant allele frequencies detected in plasma circulating tumor DNA over time.
We interpreted the emergence of an EGFR T790M–positive clone as the most likely resistance mechanism within the growing liver nodules. The patient discontinued chemotherapy, and the T790M-specific TKI osimertinib7 was administered in March 2016. Restaging in June 2016 (performed with MRI because of renal dysfunction) revealed that the hepatic lesions that had most recently progressed on chemotherapy (presumed T790M positive) had stabilized, but the liver lesion biopsied in 2015 (SCLC histology at that time) had again enlarged with no other sites of systemic or intracranial progression (Fig 1E). We hypothesized that the SCLC-transformed clone was driving radiographic progression. A repeat plasma Guardant360 test in June 2016 confirmed that EGFR T790M was now undetectable, but increases were found in the MAFs of PIK3CA E726K (50.9%), PIK3CA E545K (54.3%), TP53 V173L (54.8%), and EGFR del19 (45.5%; Fig 4). In addition, new (compared with prior plasma testing) moderate-level amplification was noted in ERBB2 (HER2; 3.3-fold amplified in plasma), PIK3CA (3.3-fold), c-MYC (2.7-fold), and FGFR1 (2.6-fold). The patient was subsequently treated with docetaxel and then nivolumab without response. He died in November 2016. An autopsy was not pursued.
Discussion
EGFR mutation–positive lung adenocarcinomas have been observed to transform to an SCLC phenotype as a resistance mechanism to frontline EGFR inhibitors in 5% of patients.4,5,8 This particular case of SCLC transformation illustrates the complexities of clonal evolution in acquired resistance and, importantly, demonstrates how serial genotyping through plasma and tissue may help us to follow the various clones clinically and to prioritize therapeutics.
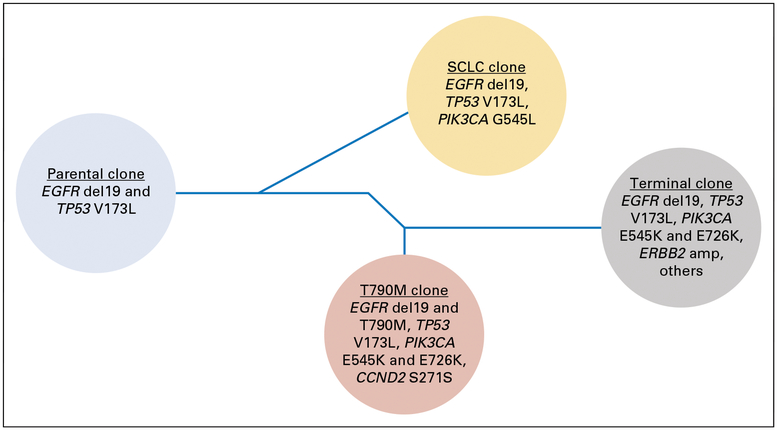
We hypothesize that all tumor cells in this patient carried common founder mutations in EGFR del19 and TP53 V173L (Fig 5). However, one resistant clone with SCLC morphology emerged clinically in October 2015, and genotyping confirmed an additional private PIK3CA E545L mutation, which often is seen in SCLC-transformed EGFR mutant clones.4 During chemotherapy, we believe that the SCLC clone diminished, whereas another clone that harbored an acquired EGFR T790M mutation and perhaps two other distinct PI K3CA mutations (E545K and E726K) emerged as observed in plasma ctDNA in March 2016. Although a tissue biopsy specimen could not be obtained at that time, on the basis of our prior observation that SCLC and adenocarcinoma populations can oscillate in response to specific treatment4 and that T790M is rarely seen in SCLC-transformed tissue biopsy specimens, we hypothesize that the T790M-positive clone detected in plasma maintained an adenocarcinoma phenotype. The T790M clone was no longer detectable by June 2016 after treatment with osimertinib, but one or more other clones became dominant with increasing MAFs, and subsequently, the disease became refractory to therapy (Fig 4). This elevation in ctDNA and subsequent clinical decline mirror data that demonstrated the correlation of increased MAFs and decreased overall survival.9–11
Fig 5.
Clonal evolution schematic. Dendrogram of a hypothetical phylogenetic evolution of subclones in this patient. Each circle represents a hypothesized clone and its key genetic features. The timing of the branch points is illustrative and not meant to convey exact data. Not all documented molecular changes are included in the illustration. SCLC, small-cell lung cancer.
In addition, the patient had a tumor with baseline RB1 mutation that was expected to lead to loss of function and is believed to play an essential role in the histologic transformation to SCLC among EGFR mutant cancers. We previously demonstrated that RB1 is uniersally lost in SCLC-transformed cancers, although not sufficiently for transformation.12 Recent work has demonstrated that baseline RB1 loss among EGFR mutant tumors is a strong predictor for subsequent SCLC transformation.13 As the clinical use of NGS panels increases and baseline inactivating RB1 mutations are more frequently detected, more data to understand the implications will be required, including a better understanding of the critical steps that lead to the lineage shift, so that we can develop more-effective treatment strategies.14
Finally, this case report illustrates the potential utility of longitudinal molecular profiling during targeted therapy. At present, two mutation-specific Food and Drug Administration–approved plasma tests may be used to select EGFR TKIs.15,16 Clinical practice is rapidly evolving, but on the basis of current evidence, it is reasonable to offer plasma genotyping upon progression with frontline EGFR TKIs to evaluate for T790M. If a T790M clone is detected, initiation of osimertinib is standard; however, if plasma testing is negative for T790M, reflexive tissue biopsy should be performed because approximately 30% of ctDNA test results will be false negative.17 The exact role of liquid biopsies for serial monitoring requires more rigorous evaluation, but a key appeal is that tumor biopsy findings may underestimate the full spectrum of resistant clones present at the time of progression.6,18,19 In practice, we commonly see the type of mixed radiographic response as observed in this patient with regression of some sitesbut continued growth in others. Heterogeneity among distinct cancer subclones may explain such disparate responses, and longitudinal plasma testing might be a valuable adjunct to tissue biopsies to understand the dynamic evolution of various clones.
For example, although tissue biopsy may offer critical information about the histology and molecular alterations of a specific progressing lesion, it may lack information about other sites of disease. Conversely, ctDNA genotyping could paint a more-complete picture of the competing resistance clones within a patient, although precise determination of which molecular alterations coexist within one clone or in one anatomic site are not currently possible. Indeed, other studies have demonstrated that multiple resistance mechanisms can be detected within plasma,18 and we and others have observed that longitudinal ctDNA analyses can track the rise and fall of distinct subclones.6,20
In summary, this case report illustrates that the relative magnitude of resistant subclones can fluctuate in response to therapy, that liquid biopsies hold great potential to detect and monitor distinct genetic subpopulations within a patient, and that the presence of a baseline RB1 mutation in an EGFR mutant cancer and subsequent SCLC transformation raises important questions about monitoring such patients. For those with EGFR mutant lung cancers, both tissue and liquid biopsy specimens can yield critical information to elucidate dominant clones that drive cancer growth at various time points. As our appreciation of the complexities of resistance and cancer heterogeneity grows, longitudinal plasma testing likely will play an increasing clinical role.
Footnotes
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
REFERENCES
- 1.Sequist LV, Yang JC, Yamamoto N, et al. : Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 31:3327–3334, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, et al. : Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361:947–957, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Carcereny E, Gervais R, et al. : Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13:239–246, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Sequist LV, Waltman BA, Dias-Santagata D, et al. : Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3:75ra26, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu HA, Arcila ME, Rekhtman N, et al. : Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 19:2240–2247, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piotrowska Z, Niederst MJ, Karlovich CA, et al. : Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov 5:713–722, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jänne PA, Yang JC, Kim DW, et al. : AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 372:1689–1699, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Camidge DR, Pao W, Sequist LV: Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat Rev Clin Oncol 11:473–481, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Bettegowda C, Sausen M, Leary RJ, et al. : Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:224ra24, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwaederlé MC, Patel SP, Husain H, et al. : Utility of genomic assessment of blood-derived circulating tumor DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clin Cancer Res 23:5101–5111, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Messaoudi S, Mouliere F, Du Manoir S, et al. : Circulating DNA as a strong multimarker prognostic tool for metastatic colorectal cancer patient management care. Clin Cancer Res 22:3067–3077, 2016 [DOI] [PubMed] [Google Scholar]
- 12.Niederst MJ, Sequist LV, Poirier JT, et al. : RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 6:6377, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JK LJ, Lee J, Kim S, et al. : Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol 35:3065–3074, 2017 [DOI] [PubMed] [Google Scholar]
- 14.Farago AF, Piotrowska Z, Sequist LV. Unlocking the mystery of small-cell lung cancer transformation in EGFR mutant adenocarcinoma. J Clin Oncol 35:2987–2988, 2017 [DOI] [PubMed] [Google Scholar]
- 15.Food and Drug Administration: FDA approves first blood test to detect gene mutation associated with non-small cell lung cancer, 2016. https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm504488.htm
- 16.Kwapisz D: The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med 5:46, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxnard GR, Thress KS, Alden RS, et al. : Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non–small-cell lung cancer. J Clin Oncol 34:3375–3382, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chabon JJ, Simmons AD, Lovejoy AF, et al. : Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 7:11815, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suda K, Murakami I, Sakai K, et al. : Small cell lung cancer transformation and T790M mutation: Complimentary roles in acquired resistance to kinase inhibitors in lung cancer. Sci Rep 5:14447, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo M, Siravegna G, Blaszkowsky LS, et al. : Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov 6:147–153, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]