Abstract
Deep learning is a branch of artificial intelligence where networks of simple interconnected units are used to extract patterns from data in order to solve complex problems. Deep learning algorithms have shown groundbreaking performance in a variety of sophisticated tasks, especially those related to images. They have often matched or exceeded human performance. Since the medical field of radiology mainly relies on extracting useful information from images, it is a very natural application area for deep learning, and research in this area has rapidly grown in recent years. In this article, we discuss the general context of radiology and opportunities for application of deep learning algorithms. We also introduce basic concepts of deep learning including convolutional neural networks. Then, we present a survey of the research in deep learning applied to radiology. We organize the studies by the types of specific tasks that that they attempt to solve and review a broad range of deep learning algorithms being utilized. Finally, we briefly discuss opportunities and challenges for incorporating deep learning in the radiology practice of the future.
Keywords: Deep learning, machine learning, artificial intelligence, convolutional neural networks, radiology, medical imaging
Introduction
The field of deep learning encompasses a group of artificial intelligence methods which employ a large number of simple interconnected units to perform complicated tasks. Deep learning algorithms, rather than using a set of pre-programmed instructions, are capable of learning from large amounts of data. The tasks solved by these algorithms include localizing and classifying objects in images, understanding language, playing games, and many others [1]. While the flagship of deep learning, convolutional neural networks, were first introduced decades ago, only in the last 5 years have astonishing success of these algorithms elevated their status from interesting but impractical ideas to the go-to algorithms in artificial intelligence. In recent years, not only have deep learning algorithms been able to surpass performance of other methods in artificial intelligence [2] but in some tasks, such as pneumonia recognition, they have shown performance superior to humans [3–5].
Arguably, the most well-known achievement of deep learning to date is its performance in the ImageNet competition. ImageNet is a database of more than 14,000,000 annotated natural images containing real world objects such as cars, animals, and buildings (http://www.image-net.org). One of the goals of the competition is to assign each image to one of 1000 predefined categories. When a deep learning-based algorithm first appeared in the competition in 2012, it dramatically improved the error rate from 0.258 in the previous year (http://image-net.org/challenges/LSVRC/2011/results) to 0.153 (http://image-net.org/challenges/LSVRC/2012/results.html). The error rate produced by a deep learning based methods dropped below that achieved by human observers in 2015 for the first time [5]. The performance of deep learning algorithms for image classification has been improving since then and is now considered to be comparable to or better than human performance for many tasks [6–8]. Other areas relevant to the topic of this article, where deep learning algorithms have seen impressive results, include the automatic generation of sophisticated captions for images that consist of full sentences [9] as well as localization and outlining of objects in images [10, 11].
There are likely three reasons for the recent success of deep learning algorithms: availability of data, increased processing power, and rapid development of algorithms. These are highly connected: availability of large datasets of images and computing power made it possible to demonstrate the strength of the basic concepts of deep learning, and these successes motivated the development of further datasets and algorithms. The availability of graphic processing units (GPUs), which can be used in a multi-core model for rapid data processing, has dramatically reduced computation times and enabled larger scientific and technical communities to become involved and to develop even more powerful algorithms which further advanced the field.
As the primary strength of deep learning has been in image analysis, the potential applications in radiology have become very quickly apparent. The development of algorithms for radiology has shown some inertia due to the time needed for acquisition of the appropriate expertise in the medical imaging community as well as limited availability of large medical imaging datasets. However, the last 2–3 years have seen remarkable productivity in the field. It is now well recognized by both researchers and clinicians that deep learning will play a significant role in radiology.
In this paper, we begin with a general overview of radiology as the application domain and consider where deep learning could have the most significant impact. Then, we introduce the general concepts of deep learning. This is followed by an overview of the recent work in the field, emphasizing developments related to MRI. The article closes with remarks regarding the future of deep learning in radiology.
The practice of radiology
Deep learning techniques (and artificial intelligence algorithms in general) have a tremendous potential to influence the practice of radiology. Unlike most other facets of medicine, nearly all of the primary data utilized in imaging as well as the outputs produced by radiologists (ie imaging reports) are digital, lending those data to analysis by artificial intelligence algorithms.
One of the most challenging tasks in the interpretation of images is that of disease detection, the rapid differentiation of abnormalities from normal background anatomy. For example, in the interpretation of mammography, each radiograph contains thousands of individual focal densities, regional densities, and geometric points and lines that must be interpreted to detect a small number of suspicious or abnormal findings. Fortunately, in order to be useful, a computer algorithm does not have to detect all objects of interest (e.g., abnormalities) and be perfectly specific (i.e., not mark any normal locations). For example, in screening mammography, approximately 80% of screening mammograms should be read as negative according to the ACR BI-RADS guideline. Of the 20% of examinations that trigger additional evaluation, many will ultimately be categorized as negative or benign [12]. An algorithm that could successfully categorize even half of screening mammograms as definitely negative would dramatically reduce the effort required to interpret a large batch of examinations.
disease management implications is undertaken. For focal masses generically, a large number of features must be integrated in order to decide how to appropriately manage the finding. These features can include size, location, signal intensity, borders, heterogeneity, change over time, and others. In some cases, simple criteria have been established and validated for the management of focal findings. For example, most focal lesions in the kidney can be characterized as either simple or minimally complex cysts, which almost uniformly do not require treatment. On the other hand, most lesions in the kidney that are solid are considered to have high malignant potential. Finally, a minority of focal kidney lesions are considered indeterminate and can be managed accordingly. Deep learning algorithms have the potential to assess a large number of features, including features previously not considered by radiologists, and to arrive at a repeatable conclusion in a fraction of the time required for a human interpreter.
While detection, diagnosis, and characterization of disease receive the primary attention among algorithm developers, another important area where artificial intelligence could contribute is in facilitating the workflow of the radiologists while interpreting images. With the near complete conversion from printed films to centralized digital Picture Archiving and Viewing Systems (PACS) as well as the availability of multi-planar, multi-contrast, and multi-phase MRI, radiologists have seen exponential growth in the size and complexity of image data to be analyzed. However, standard PACS systems are not able to reliably organize and present all relevant imaging data to the interpreter for a variety of reasons, including differences in sequence labeling, patient positioning, and anatomy between examinations, variability in modalities used to image the same portion of the anatomy, as well as other factors. In principle, an artificial intelligence algorithm could bring forward sequences from examinations that include the relevant body part(s), detect the image modality and contrast type, and determine the location of the area of interest within the relevant anatomy to reduce the radiologist’s effort in performing these relatively mundane tasks.
Finally, computer algorithms might be able to perform medical image interpretation tasks that radiologists do not perform on a regular basis. For example, the field of radiogenomics [13] aims to find relationships between imaging features of tumors and their genomic characteristics. Examples can be found in breast cancer [14], glioblastoma [15], low grade glioma [16], and kidney cancer [17]. However, due to its complexity, radiogenomics is not a part of the typical clinical practice of a radiologist. Another example is prediction of outcomes of cancer patients with applications in glioblastoma [15, 18], lower grade glioma, [16], and breast cancer [19]. While imaging features have a potential to predict patient outcomes, very few are currently used to guide oncological treatment. Deep learning could facilitate the process of incorporating more of the information available from imaging into the oncology practice.
An introduction to deep learning
Terminology
To understand deep learning, it is helpful to first understand the related concepts of artificial intelligence and machine learning. Artificial intelligence is the most generic of the three terms, comprising a set of computer algorithms that are able to perform complicated tasks or tasks that require intelligence when conducted by humans. Machine learning is a subset of artificial intelligence algorithms which, to perform these complicated tasks, are able to learn from provided data and do not require pre-defined rules of reasoning. The field of machine learning is very diverse and has already had notable applications in medical imaging [20]. Deep learning is a sub-discipline of machine learning that relies on networks of simple interconnected units. In deep learning models, these units are connected to form multiple layers that are capable of generating increasingly high level representations of the provided inputs (e.g. images). Below, in order to explain the architecture of deep learning models, we introduce the artificial neural network in general and one specific type: the convolutional neural network. Then, we detail the process of “learning” as applied to networks, which is the process of incorporating the patterns extracted from data into the deep neural networks.
Artificial Neural Networks
Artificial neural networks (ANNs) are machine learning models based on basic concepts dating as far as back as the 1940s, significant development in 1970s and 1980s and a period of notable popularity in 1990s and 2000s, followed by a period of being overshadowed by other machine learning algorithms. The ANN is based on a concept of an artificial neuron, which is a model of a nerve cell. While many neuron models have been proposed, a typical neuron simply multiplies each input by a certain weight, then adds all the products for all the inputs and applies a simple mathematical function referred to as an activation function, to produce a single output value. An illustration of a neuron and different activation functions is shown in Figs. 1A and 1B respectively. An ANN consists of a multitude of interconnected neurons, usually organized in layers. A simple ANN is illustrated in Fig. 1C. A traditional ANN typically used in the practice of machine learning contains 2 to 3 layers of neurons. Even though each neuron performs a very rudimentary calculation, the interconnected nature of the network allows for the performance of very sophisticated calculations and implementation of very complicated functions.
Fig. 1.
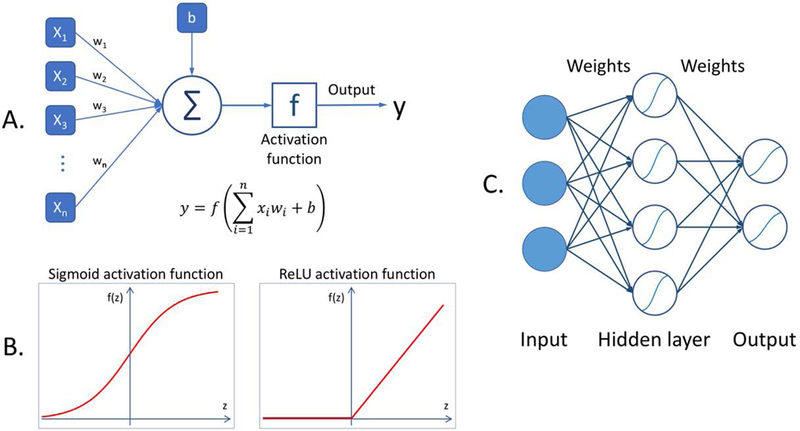
A diagram illustrating basic concepts of artificial neural network: (A) a model of a neuron where x1, …, xn are the network inputs, w1,…,wn are the weights, b is a bias, f is the activation function, and y is the neuron output, (B) two common activation functions, (C) a model of a simple neural network.
Convolutional Neural Networks
Deep neural networks are a special type of an ANN. The most common type of deep neural network is a deep convolutional neural network (CNN). Deep convolutional neural networks, while inheriting the properties of a generic ANN, also have their own specific features. First, they are “deep,” which is to say that they are typically comprised of 10–30 layer, and in extreme cases could exceed 1000 layers. Second, their neurons are connected such that multiple neurons share weights. This effectively allows the network to perform convolutions (or template matching) of the input image with the filters (defined by the weights) within the CNN. Another special feature of CNNs is that between some layers, they perform pooling operations (see Fig. 2) which make the network invariant to small changes in the input data. Finally, CNNs typically use a different nonlinear transformation when generating the output of a neuron as compared to traditional ANNs.
Fig. 2.
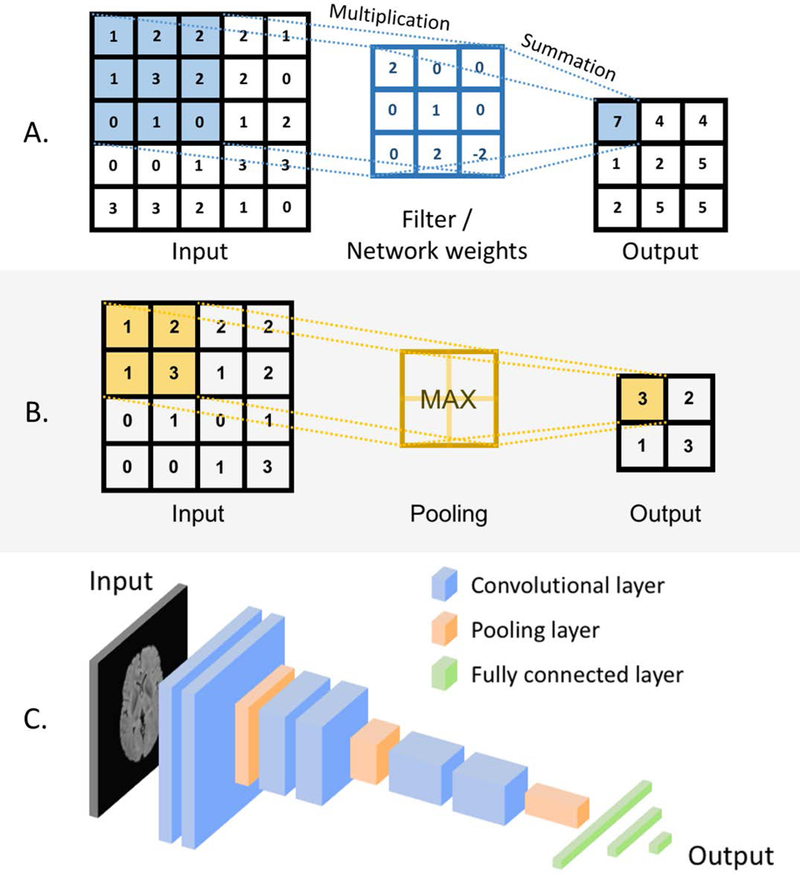
A diagram illustrating basic concepts of convolutional neural networks: (A) a convolutional layers: values in the convolutional filters implemented in the network weights (middle column) are multiplied by the pixel values and the products are summed up, (B) a max pooling layer: a maximum pixel value is taken in a given region, (C) an architecture of a simple convolutional neural network including convolutional, pooling, and fully connected layers.
Figure 2 illustrates key concepts for CNNs. Specifically, Fig. 2A demonstrates how a network performs a multiplication of its weights, organized in a matrix by the original pixels within an image. As this multiplication is repeated across different locations in the image this operation corresponds to filtering of an image where the filters (a.k.a. the convolutional kernels) are defined by the network weights. These layers are referred to as convolutional layers. Figure 2B shows the basic concept of a max pooling layer where a maximum value of multiple neighboring outputs of the previous layer is passed to the next layer. Convolutional layers, pooling layers, and fully connected layers (such as those in the multi-layer neural network in Fig. 1C) are the primary components of convolutional neural network. Figure 2C, shows an example of a small architecture for a typical CNN. A variety of deep learning architectures have been proposed, often driven by characteristics of the task at hand (e.g. fully convolutional neural networks for image segmentation). Some of these are described in more detail in the section of this paper that reviews the current state of the art.
The learning process for convolutional neural networks
Above, we described general characteristics of traditional neural networks and deep learning’s flagship, the convolutional neural network. Next, we will explore how to make those networks perform useful tasks. This is accomplished in the process referred to as learning or training. The learning process for a convolutional neural network simply consists of changing the weights of the individual neurons in response to the provided input data. In the most popular type of a learning process, called supervised learning, a training example contains an object of interest (e.g. a T2-weighted image of a tumor) and a label (e.g. the tumor’s pathology: benign or malignant). In our example, the image is presented to the network’s input, and the calculation is carried out within the network to produce a predicted summary value (such as a likelihood of malignancy) based on the current weights of the network. Then, the network’s prediction is compared to the actual label of the object (e.g. 0 for benign, 1 for malignant), and an error is calculated. A correction for the error is then propagated through the network to change the values of the network’s weights such that the next time the network analyzes this exact example, the error decreases. In practice, the correction of the weights is performed after a group of examples (a batch) are presented to the network. This process is called error backpropagation or stochastic gradient descent. Various modifications of the stochastic gradient descent algorithm have been developed [21]. In principle, this iterative process consists of calculations of error between the output of the model and the desired output and adjusting the weights in the direction where the error decreases.
The most straightforward way of training is to start with a random set of weights and train using available data specific to the problem being solved (training from scratch). However, given the large number of parameters (weights) in a network, often above 10 million, and a limited amount of training data for a specific task, a network may overtrain (a.k.a. overfit) to the available data (i.e. fitting to well to the training set and not generalizing well to test data), resulting in poor performance on test data. Two training methods have been developed to address this issue: transfer learning [22] and off-the-shelf features (a.k.a. deep features) [23]. There are many properties of the dataset used for pre-training that affect its usability, e.g. similarity of the structures present in the images, the size of the original dataset. However, quantitative effects of these factors are still a part of ongoing research on transfer learning methods. A diagram comparing training from scratch with transfer learning and off-the-shelf deep features is shown in Figure 3.
Fig. 3.
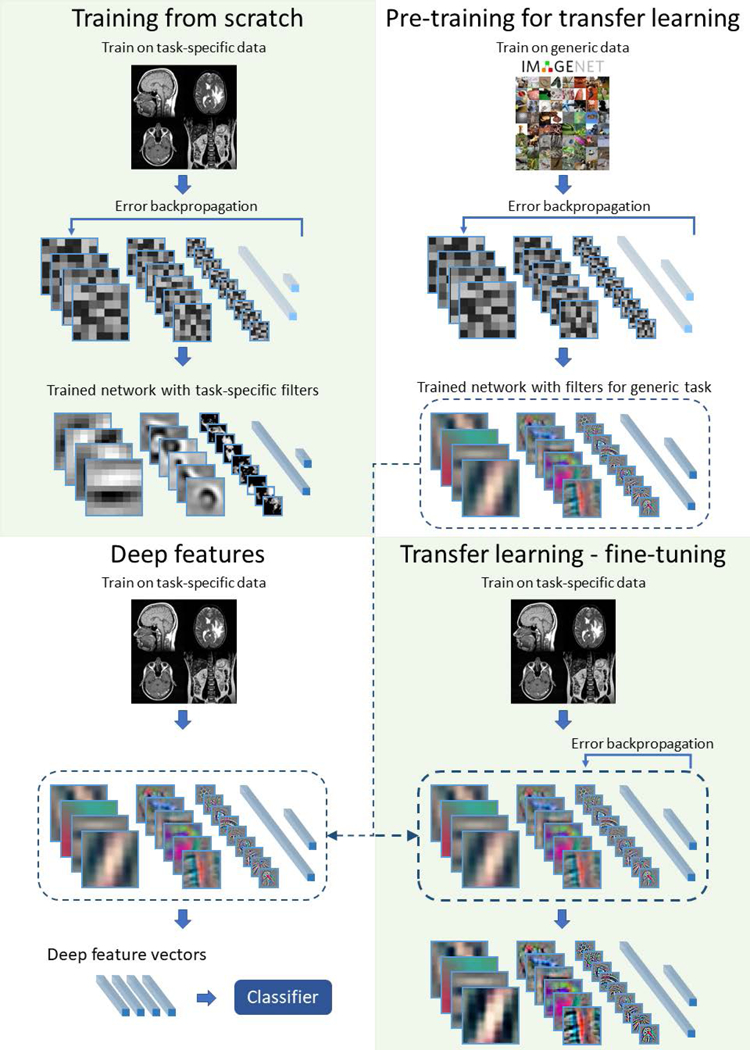
An illustration of different ways of training in deep neural networks: training from scratch, transfer learning, and deep features
In the transfer learning approach, the network is first trained using a different dataset, for example an ImageNet collection. Then, the network is “fine-tuned” through the addition of training data specific to the problem to be addressed. The idea behind this approach is that performing different visual tasks shares a certain level of processing such as recognition of edges or simple shapes. This approach has been shown successful in, for example, prediction of patient survival time from brain MRI in patients with glioblastoma [24] or in skin lesion classification [25]. Another approach that addresses the issue of limited training data is the deep “off-the-shelf” features approach which uses convolutional neural networks which have been trained on a different dataset to extract features from the images. This is done by using a pre-trained network and extracting outputs of layers prior to the network’s final layer. Those layers typically have hundreds or thousands of outputs. Then, these outputs are used as inputs to “traditional” classifiers such as linear discriminant analysis, support vector machines, or decision trees. This is similar to transfer learning (and is sometimes considered a part of transfer learning) with the difference being that the final layers of a CNN are replaced by a traditional classifier and the early layers are not additionally trained for the specific task at hand.
Deep learning vs “traditional” machine learning
Increasingly often we hear a distinction between deep learning and “traditional” machine learning (see Figure 4). The difference is very important, particularly in the context of medical imaging. In traditional machine learning, the first step is typically feature extraction. This means that to classify an object, one must decide for the algorithm which characteristics of an object will be important and implement algorithms that are able to capture these characteristics. A number of sophisticated algorithms in the field of computer vision have been proposed for this purpose and a variety of size, shape, texture, and other features have been extracted. This process is to a large extent arbitrary since the machine learning researcher or practitioner often must guess which features will be of use for a particular task and runs the risk of including useless and redundant features and, more importantly, not including truly useful features. In deep learning, the process of feature extraction and decision making are merged and trainable, and therefore no choices need to be made regarding which features should be extracted; this is decided by the network in the training process. However, the cost of allowing the neural network to select its wn features is a requirement for much larger training data sets.
Fig. 4.
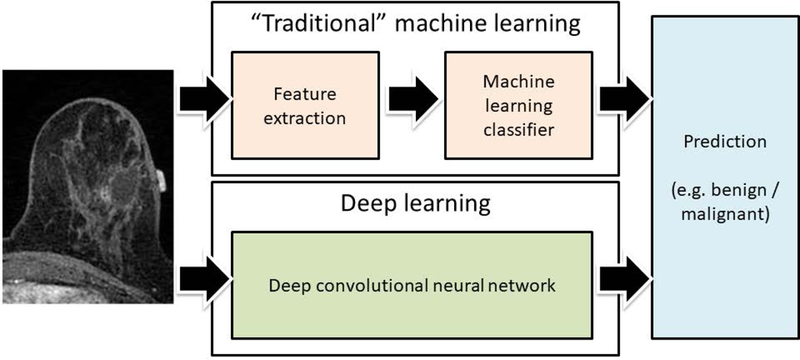
An illustration of difference between “traditional” machine learning and deep learning. In the “traditional” machine learning, a set of predefined features is extracted and used by a multivariate classifier. In deep learning the entire image is provided as an input to a neural network which outputs a decision.
Deep learning in radiology: state of the art
In this section, we give an overview of applications of deep learning in radiology. We organized this section by the tasks that the deep learning algorithms perform. Within each subsection, we describe different methods applied, and when possible, we systematically discuss the evolution of these methods in the recent years. Other recent reviews surveyed the applications of deep learning in broadly understood medical imaging (including pathology) [26] and specifically in brain segmentation in MRI [27].
Classification
In a classification task, an object is assigned to one of the predefined classes. A number of different classification tasks can be found in the domain of radiology such as: classification of an image or an examination to determine the presence or an absence of an abnormality; classification of abnormalities as benign or malignant; classification of cancerous lesions according to their histopathological and genomic features; prognostication; and classification for the purpose of organization radiological data.
Deep learning is becoming the methodology of choice for classifying radiological data. The majority of the available deep learning classifiers use convolutional neural networks with a varying number of convolutional layers followed by fully connected layers. The availability of radiological data is limited as compared to the natural image datasets which have driven the development of deep learning techniques over the last 5 years. Therefore, many applications of deep learning in medical image classification have resorted to techniques meant to alleviate this issue: off-the-shelf features and transfer learning [28] discussed in the previous section of this article. Off-the-shelf features have performed well in a variety of domains [23], and this technique has been successfully applied to medical imaging [29, 30]. In [29], the authors combined the deep off-the shelf features extracted from a pre-trained deep CNN network with hand-crafted features for determining malignancy of breast lesions in mammography, ultrasound, and MRI and achieved statistically significant improvements in performance compared to existing breast cancer computer-aided diagnosis methods. In [30], long-term and short term survival with improved (29%) accuracy was predicted for patients with lung carcinoma by combining off-the-shelf features with the traditional quantitative features. The other strategy, transfer learning, involves fine tuning of a network pre-trained on a different dataset. Transfer learning has been successfully applied to a variety of tasks such as classification of prostate MR images to distinguish patients with prostate cancer from patients with benign prostate conditions [31] using MRI. Most of the studies which apply the transfer learning strategy replace and retrain the deepest layer of a network, while the shallow layers are fixed after the initial training. A variant of the transfer learning strategy combines fine-tuning and deep features approaches. It fine-tunes a pre-trained network on a new dataset to obtain more task-specific deep feature representations. An ensemble of fine-tuned CNN classifiers was shown to outperform traditional CNNs in predicting radiological image modality on a test set of 4166 images [32]. A comparison of approaches using deep features and transfer learning with fine tuning has been shown useful for identifying radiogenomic relationships in breast cancer MRI [33]. Though, deep features performed better than transfer learning with the fine tuning approach, the method faced the issue of training on small dataset.
When sufficient data are available, an entire deep neural network can be trained from a random initialization (training from scratch). The size of the network to be trained depends on task and dataset characteristics. However, the commonly used architecture in medical imaging is based on AlexNet [2] and VGG [34] with modifications that have fewer layers and weights. Training from scratch has been applied to assessing for the presence of Alzheimer’s disease based on brain MRI using deep learning [35]. In this study using the publicly available ADNI cohort, sparse regression models were combined with deep neural networks to achieve higher classification performance compared with several non-deep learning based techniques in differentiating Alzheimer’s versus normal controls. Recent advances in the design of CNN architectures have made networks easier to train and more efficient. They have more layers and perform better (in terms of accuracy or area under the curve [AUC]) while having fewer trainable parameters, which reduces the likelihood of overtraining [36]. The most notable examples include Residual Networks (ResNets) [37] and the Inception architecture [38, 39]. A shift toward these more powerful networks has also taken place in applications of deep learning to radiology both for transfer learning and training from scratch. Three different ResNets were used to predict methylation of the O6-methylguanine methyltransferase gene status from brain tumor pre-surgical MRI [40] with an accuracy of 94.9%, which is better than conventional machine learning based techniques using MRI texture features. In [41], the InceptionV3 network was fine-tuned and served as a feature extractor instead of previously used GoogLeNet to classify wrist radiographs into two categories (with and without fracture). The authors leveraged the data augmentation to generate 11,112 training images from an initial set of 1389 images and obtained an AUC of 0.95 on the test set.
In another approach, auto-encoder [42] or stacked auto-encoder [43] networks have been trained from scratch, layer by layer, in an unsupervised way. A stacked denoising auto-encoder with backpropagation was used in [44] to determine the presence of Alzheimer’s disease. Auto-encoders and stacked auto-encoders can also be used to extract feature representations (similarly to the deep features approach) from hidden layers for further classification. Such feature representation has also been used in the identification of multiple sclerosis lesions in using MRI and myelin maps jointly [45].
Segmentation
In an image segmentation task, an image is divided into different regions in order to separate distinct parts or objects. In MRI, the common applications are segmentation of organs, substructures, or lesions, often as a preprocessing step for feature extraction and classification [46, 47]. Below, we discuss different types of deep learning approaches used in segmentation tasks in a variety of radiological images.
The most straightforward and still widely used method for image segmentation is classification of individual voxels based on small image patches (both 2-dimensional and 3-dimensional patches) extracted around the classified voxel. This approach has found use in various segmentation problems, for example brain tumor segmentation in [48–50], white matter segmentation in multiple sclerosis patients [51], segmentation of normal components of brain anatomy [52], and rectal cancer segmentation [53]. It allows for using the same network architectures and solutions that are known to work well for classification, however, there are some shortcomings of this method. The primary issue is that these methods are computationally inefficient, since they process overlapping parts of images multiple times. Another drawback is that each voxel is segmented based on a limited-size context window and ignores the wider context. In some cases, some global information, e.g. pixel location or relative position to other image parts, may be needed to correctly assign its label.
One approach that addresses the shortcomings of the voxel-based segmentation is a fully convolutional neural network (fCNN) [54]. Networks of this type process the entire image (or large portions of it) at the same time and output a 2-dimensional map of labels (i.e., a segmentation map) instead of a label for a single pixel. A very important advantage of fCNNs over the voxel-based approach is avoiding many repeated convolutions by analyzing a large portion of the image and providing the segmentation label for all the voxels at the same time. Example architectures that were successfully used in both natural images and radiology applications are encoder-decoder architectures such as U-Net [55–57] or Fully Convolutional DenseNet [58–60]. Various adjustments to these types of architectures have been developed that mainly focus on connections between the encoder and decoder parts of the networks, called skip connections. An fCNN was applied in [61] for in radiology include prostate gland segmentation in diffusion-weighted MRI. Although a relatively small dataset of over 100 cases was used, the segmentation quality as evaluated with Dice similarity coefficient was 0.89. In another study [62], a fCNN was used for segmentation of multiple sclerosis lesions and gliomas in MRI slices of axial, coronal and sagittal planes separately. In addition to differences in building blocks for fCNNs, different optimization functions have been explored that account for class imbalance (remarkable differences among the number of examples in each class), which is common in medical datasets [63]. In [64], weighted cross entropy loss was used for brain structure segmentation in MRI. The proposed method did not require any post-processing and offered on average 10 times faster processing of large MRI volumes comparing to other tested methods.
In order to segment 3-dimensional data, it is common to process data as 2-dimensional slices and then combine the 2-dimensional segmentation maps into a 3-dimensional map since 3D fCNNs are significantly larger in terms of trainable parameters and as a result require significantly larger amounts of data. Nevertheless, these obstacles can be overcome, and there are successful applications of 3D fCNNs in radiology, e.g. V-Net for prostate segmentation from MRI [65], 3D U-Net [66] for segmentation of the proximal femur for assessing osteoporosis [67], and brain glioma segmentation [68].
Finally, a deep learning approach that has found some application in medical imaging segmentation is recurrent neural networks. In [69], the authors applied a recurrent fully convolutional neural network for left-ventricle segmentation in multi-slice cardiac MRI to leverage inter-slice spatial dependences. Similarly, [70] used Long Short-Term Memory (LSTM) [71] type of recurrent neural network trained end-to-end together with fCNN to take advantage of 3D contextual information for pancreas segmentation in MR images. In addition, they proposed a novel loss function that directly optimizes a widely used segmentation metric, the Jaccard Index [72].
Detection
Detection is a task of localizing and pointing out (e.g., using a rectangular box) an object in an image. In radiology, detection is often an important step in the diagnostic process which identifies an abnormality (such as a mass or a nodule), an organ, an anatomical structure, or a region of interest for further classification or segmentation [73, 74]. Here, we discuss the common architectures used for various detection tasks in radiology along with example specific applications.
The most common approach to detection for 2-dimensional data is a 2-phase process that requires training of 2 models. The first phase identifies all suspicious regions that may contain the object of interest. The requirement for this phase is high sensitivity [75] and therefore it usually produces many false positives. A typical deep learning approach for this phase is a regression network for bounding box coordinates based on architectures used for classification [76, 77]. The second phase is simply classification of sub-images extracted in the previous step. In some applications, only one of the two steps uses deep learning. This strategy has been applied in cerebral microhemorrhage detection using a large dataset of 320 MRI volumes and achieved 93% sensitivity [78].
The classification step, when utilizing deep learning, is often performed using transfer learning. The models are often pre-trained on natural images, for example for thoraco-abdominal lymph node detection in [79] and pulmonary embolism detection in CT pulmonary angiogram images [28]. In other applications, models have been pre-trained using other medical imaging dataset to detect masses in digital breast tomosynthesis images [80]. The same network architectures can be used for the second phase as in a regular classification task (e.g. VGG [34], GoogLeNet [81], Inception [38], ResNet [37]) depending on the needs of a particular application.
While in the 2-phase detection process the models are trained separately for each phase, in the end-to-end approach one model encompassing both phases is trained. An end-to-end architecture that has proved to be successful in object detection in natural images, and was recently applied to medical imaging, is the Faster Region-based Convolutional Neural Network [10]. It uses a CNN to obtain a feature map which is shared between region proposal network that outputs bounding box candidates, and a classification network which predicts the category of each candidate. It was recently applied for intervertebral disc detection in X-ray images [82] and detection of colitis on CT images [83].
Another approach to detection is a single-phase detector that eliminates the first phase of region proposals. Examples of popular methods that were first developed for detection in natural images and rely on this approach are You Only Look Once (YOLO) [84], Single Shot MultiBox Detector [85] and RetinaNet [11]. In the context of radiology, a YOLO-based network called BC-DROID has been developed for region of interest detection in breast mammograms [86]. Single Shot MultiBox Detector has been employed for breast tumor detection in ultrasound images, outperforming other evaluated deep learning methods that were available at the time [87]. The authors of [88] applied the same network for detection of pulmonary lung nodules in CT images. The above-mentioned methods and architectures were widely adapted for natural images and some medical imagining modalities, e.g. CT, mammograms, X-rays, however, are still uncommonly applied in object detection using MRI.
In the examples above, 2-dimensional data have typically been used. For 3-dimensional imaging volumes, which are most commonly encountered in CT and MRI, results obtained from 2-dimensional processing can be combined to produce the final 3-dimensional bounding box. As an example, in [89] the authors performed detection of 3D anatomy in chest CT images by processing data slice by slice in one direction. Combining output from different planes was performed in several studies. Most of the them [90–92] used orthogonal planes of MRI and CT images performing detection in each direction separately. The results can then be combined in different ways, e.g. by an algorithm based on output probabilities [89] or using another machine learning method like random forest [88]. An alternative method for 3D detection has been proposed for automatic detection of lymph nodes by concatenating coronal, sagittal and axial views as a single 3-channel image [75].
Other Tasks in Radiology
While the majority of the applications of deep learning in radiology have been in classification, segmentation, and detection, other medical imaging-related problems have found some solutions in deep learning. Due the variety of those problems, there is no unifying methodological framework for these solutions. Therefore below, we organize the examples according to the problem that they attempt to address.
Image Registration:
In this task two or more images (often 3D volumes), typically of different types (e.g., T1-weighted and T2-weighted image sets) must be spatially aligned such that the same location in each image represents the same physical location in the depicted organ. Several approaches can be taken to address the problem. In one approach, similarity measures between image patches taken from the images of interest are calculated and used to register the image sets. The authors of [93] used deep learning to learn a similarity measure from T1-T2 MRI image pairs of the adult brain and tested it to register T1-T2 MRI interpatient images of the neonatal brain. This similarity measure performed better than the standard measure, called mutual information, which is widely used in registration [94]. In another deep learning-based approach to image registration, the deformation parameters between image pairs are directly learned using misaligned image pairs. A CNN-based network was trained to correct respiratory motion in 3D abdominal MR images [95] by predicting spatial transforms. All of these techniques are supervised regression techniques as they were trained using ground truth deformation information. In another approach [96], which was unsupervised, a CNN was trained end-to-end to generate a spatial transformation which minimized dissimilarity between misaligned image pairs.
Image generation/reconstruction:
Acquisition and hardware parameters can strongly affect the visual quality and detail of images obtained using the same modality. First, we discuss the applications that synthesize images generated using different acquisition parameters within the same modality. In [97], 7T like images were generated from 3T MR images by training a CNN with patches centered around voxels in the 3T MR images. Undersampled (in k-space) cardiac MRIs were reconstructed using a deep cascade of CNNs in [98]. A real-time method to reconstruct compressed sensed MRI using GAN has also been proposed [99]. In another approach [100] in order to synthesize brain MRI images based on other MRI sequences in the same patient, convolutional encoders were built to generate a latent representation of images. Then, based on that representation a sequence of interest was generated. Reconstruction of “normal-dose” CT images from low-dose CT images (which are degraded in comparison to normal-dose images) has been performed using patch-by-patch mapping of low-dose images to high-dose images using a shallow CNN [101]. In contrast, a deep CNN has been trained with low-dose abdominal CT images for reconstruction of normal-dose CT [102].
Deep learning has also been applied to synthesizing images of different modalities. For example, CT images have been generated using MR images by adopting an FCN to learn an end-to-end non-linear mapping between pelvic CT and MR images [103]. Synthetic CT images of the brain have also been generated from a single T1-weighted MR image set [104]. In another application to aid a classification framework for Alzheimer’s disease diagnosis with missing PET scans, PET patterns were predicted from MRI using CNN [105].
Image enhancement:
Image enhancement aims to improve different characteristics of the image such as resolution, signal-to-noise-ratio, and necessary anatomical structures (by suppressing unnecessary information) through various approaches such as super-resolution and denoising.
Super-resolution of images has particularly been explored in cardiac and lung imaging. Three dimensional near–isotropic cardiac and lung images often require long scan times in comparison to the time the subject can hold his or her breath. Thus, multiple thick 2D slices are acquired instead and the super-resolution methodology is applied to improve the through-plane resolution of the images. A deep cascade of CNNs has been shown to preserve anatomical structure up to 11 fold undersampling using cardiac MRI [106]. In another study [107] using CT, a single image super-resolution approach based on CNN was applied in a publicly available chest CT image dataset to generate high-resolution CT images which are preferred for interstitial lung disease detection. This method outperformed the traditional compressed sensing based approaches used in MR image reconstruction. In another study [108] to synthesize thin slice knee MRIs from thick slice knee MRIs, the proposed CNN based approach showed improved qualitative and quantitative performance over the state-of-the-art techniques in a test set of 17 patients.
Image enhancement through denoising application of using deep has also been described in [109] where the authors performed denoising of DCE-MRI images of a brain (for stroke and brain tumors) by training an ensemble of deep auto-encoders using synthesized data. Removal of Rician noise in real and synthetic three-dimensional MR images using a deep convolutional neural network aided with residual learning can be performed by excluding the traditional steps of optimization and estimation of the noise level parameter [110]. An encoder-decoder CNN architecture [111] was used to denoise the noisy uptake signal between a pre-contrast MR sequence (zero gadolinium dose for contrast) and a 10% low-dose post-contrast MR sequence of brain. With the help of this model, full contrast high quality post-contrast sequences were reconstructed from sequences with 10-fold reduction in contrast dose for different pathologies (including glioma) in brain for 50 patients.
Content-based image retrieval:
In the most typical version of this task, the algorithm, given a query image, finds the most similar images in a given database. To accomplish this task, a deep CNN can first be trained to distinguish between different organs [112]. Then, features from the three fully connected layers in the network are extracted for the images in the set from which the images were retrieved (evaluation dataset). The same features can be then extracted from the query image and compared with those of the evaluation dataset to retrieve the image.
Objective image quality assessment:
Objective quality assessment measures of medical images aim to classify an image to be of satisfactory or unsatisfactory quality for subsequent tasks. Objective quality measures of medical images diagnosis and aid in better treatment [113]. Image quality of fetal ultrasound has recently been predicted using CNN [114]. Another study attempted to reduce the data acquisition variability in echocardiograms using a CNN trained on the quality scores assigned by an expert radiologist [115]. As another example, simple CNN architecture has been reported for classifying T2-weighted liver MR images as diagnostic or non-diagnostic quality by CNN [116].
Main challenges and pitfalls in development of deep learning algorithms
While applications of deep learning in medical imaging show tremendous promise, there are some challenges and potential pitfalls, and caution should be exercised in research on the topic. One of the principal challenges is availability of data. While millions of training examples are available for problems related to natural images, the datasets for medical images are typically much smaller, with a typical number of patients in the hundreds range. This, combined with the large number of parameters in a deep neural network that require optimization, results in a high risk of overtraining and subsequent low performance on data that were not used in the training process. Some solutions that can help alleviate this issue are pre-training of the models with other datasets, use of smaller models, and augmentation of the data by including slight alterations of the original images in the dataset. A related issue is often a very small number of cases with a disease (e.g., cancer) as compared to healthy patients. This issue, referred to as class imbalance, can lead to highly diminished performance [118]. Some solutions have been proposed to this issue such as higher rate of sampling of the examples from the minority class for training [118]. However, despite the solutions that have been proposed to address small data set size and class imbalance, these remain important challenges to the use of deep learning in radiology.
Given the high risk of overtraining, there is a high likelihood of reporting performance that does not reflect the true ability of a model to classify/predict/segment when the validation is not conducted properly. Though evaluation of models through splitting datasets into training dataset which is used for the development of the model and test set used for estimating the model’s performance as well as crossvalidation (splitting the dataset into training and test sets and combining the evaluation results) can provide fairly accurate estimate of generalization performance, these methods also have limitations. Therefore, sharing the developed models for testing by other institutions can facilitate further development by testing reproducibility and can increase the confidence of the scientific community in these models.
Finally, when aiming to develop deep learning models that could be used clinically, one must ensure a proper validation setup in the experimental validation of the models. This is highly challenging and sometimes overlooked. It requires not only posing a clinical question that is of significance and answers that change clinical decision making but also careful curation of the dataset to include only those patients that are relevant to the question and precise definitions of other non-imaging variables such as pathology/genomic markers and patient outcomes. This requires a close and continuous collaboration with clinicians and/or other experts in a given application field at many stages of the development as well as a strong understanding of the clinical reality of the problem by the technical expert. While those and other issues pose challenges in the development of deep learning models, none of them are insurmountable.
Future of deep learning in radiology
There is a general agreement that deep learning will play a role in the future practice of radiology and MRI specifically. Some predict that deep learning algorithms will conduct mundane tasks, leaving radiologists with more time to focus on intellectually demanding challenges. Others believe that radiologists and deep learning algorithms will work hand-in-hand to deliver performance superior to either alone. Finally, some predict that deep learning algorithms will replace radiologists (at least in their image interpretation capacity) altogether.
Incorporation of deep learning in radiology will be associated with multiple challenges. First, and currently foremost, is the technological challenge. While deep learning has shown extraordinary promise in other image-related tasks, the results in radiology are still far from showing that deep learning algorithms will replace a radiologist in the entire scope of their diagnostic work. Some recent studies suggest performance of these algorithms comparable to expert humans in narrowly defined tasks, but these results are only applicable to a very small minority of the tasks that radiologists perform [4, 119–125] . This is likely to change in upcoming years given the rapid progress in implementing the deep learning algorithms in the realm of radiology.
Implementation of deep learning in radiology practice also poses legal and ethical challenges. Primarily: who will be responsible for the mistakes that a computer will make? While this is a difficult question, similar questions have been posed and resolved when other technologies were introduced, for example elevators and cars. Since artificial intelligence penetrates various areas of human activity, questions of this type will likely be studied and answers proposed in the coming years.
Other challenges will include patient acceptance or non-acceptance of a human’s not being involved in the process of interpreting their images (regardless of the performance) as well as regulatory issues. Finally, an important practical issue is how to incorporate deep learning algorithms into the radiology workflow in order to improve, rather than disrupt, the radiology practice.
Conclusion
In summary, in this paper we have discussed the principles of deep learning as well as the current practice of radiology to elucidate how these new algorithms may be incorporated into radiologists’ workflow. We have discussed the progress and state of art in the field. Finally, we have discussed some challenges and questions related to implementation of deep learning in the current practice of imaging. All signs show that deep learning will play a significant role in radiology. The next 5 years will be a very exciting time in the field that may see many questions stated in this article answered through a collaboration of machine learning scientists and radiologists.
Fig. 5.
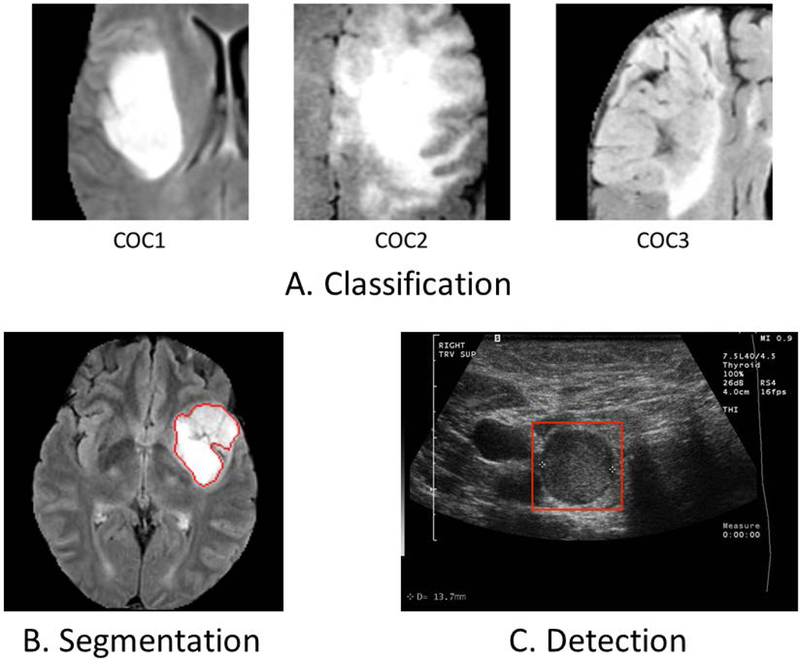
Examples of applications of deep neural network to medical images in our laboratory: (A) A classification task in which a CNN was designed to distinguish between different genomic subtypes (cluster of clusters) of lower grade gliomas in MRI, (B) An automatic segmentation of low grade glioma tumors in MRI, (C) A detection of thyroid nodules in ultrasound
Table 1.
An overview of papers presented in the review split by task and organ.
| Task | Site | Reference |
|---|---|---|
| Classification of breast mass lesions | Breast | (Antropova, Huynh, and Giger 2017) |
| Classification of survival groups | Lung | (Paul R, Hawkins SH, Balagurunathan Y, Schabath MB, Gillies RJ, Hall LO 2016) |
| Classification of prostate cancer | Prostate | (Wang et al. 2017) |
| Classification of image modality | Multiple | (Kumar et al. 2017) |
| Classification of genomic subtypes | Breast | (Zhu et al. 2017) |
| Classification for Alzheimer’s Disease | Brain | (Suk, Lee, and Shen 2017) |
| Classification of O6-methylguanine methyltransferase gene status | Brain | (Korfiatis et al. 2017) |
| Classification of fracture | Wrist | (Kim and Mackinnon 2017) |
| Classification for Alzheimer’s Disease | Brain | (Ortiz et al. 2017) |
| Classification for multiple sclerosis | Brain | (Yoo et al. 2018) |
| Classification of genomic subtypes | Brain | (Akkus, Ali, et al. 2017) |
| Classification of genomic subtypes | Brain | (Wachinger, Reuter, and Klein 2017) |
| Segmentation of rectal cancer | Rectum | (Trebeschi et al. 2017) |
| Segmentation of brain in fetal US | Fetal brain | (Salehi et al. 2017) |
| Segmentation of liver and hepatic lesions | Liver | (Christ et al. 2017) |
| Segmentation of liver tumor | Liver | (X. Li et al. 2017) |
| Segmentation of prostate gland | Prostate | (Clark et al. 2017) |
| Segmentation of sclerosis lesions and gliomas | Brain | (McKinley et al. 2016) |
| Segmentation of brain structure | Brain | (Mehta and Sivaswamy 2017) |
| Segmentation of prostate | Prostate | (Milletari, Navab, and Ahmadi 2016) |
| Segmentation of proximal femur | Proximal femur | (Deniz et al. 2017) |
| Segmentation of gliomas | Brain | (Shen and Anderson, n.d.) |
| Segmentation of left-ventricle | Brain | (Poudel, Lamata, and Montana 2016) |
| Segmentation of pancreas | Pancreas | (Cai et al. 2017) |
| Segmentation of left-ventricle | Heart | (Avendi, Kheradvar, and Jafarkhani 2016) |
| Detection of sclerosis | Brain | (Rey et al. 2002) |
| Detection of lymph nodes | Lymph nodes | (Roth et al. 2014) |
| Detection of cerebral microhemorrhage | Brain | (Dou et al. 2016) |
| Detection of Thoraco-abdominal lymph nodes | Lymph nodes | (Shin et al. 2016) |
| Detection of pulmonary embolism | Lung | (Tajbakhsh et al. 2016) |
| Detection of masses | Breast | (Samala et al. 2016) |
| Detection of intervertebral disc | Spine | (Sa et al. 2017) |
| Detection of colitis | Colon | (J. Liu et al. 2017) |
| Detection of breast cancer | Breast | (Platania et al. 2017) |
| Detection of breast tumor | Breast | (Cao et al. 2017) |
| Detection of pulmonary lung nodules | Lung | (N. Li et al. 2017) |
| Detection of 3D anatomy in chest | Chest | (de Vos et al. 2016) |
| Detection of knee cartilage | Knee | (Prasoon et al. 2013) |
| Detection of sclerotic metastases | Brain | (Roth, Lu, et al. 2016) |
| Detection of lymph nodes | Lymph nodes | (Roth, Lu, et al. 2016) |
| Detection of colonic polyp | Colon | (Roth, Lu, et al. 2016) |
| Detection of fractures on spine | Spine | (Roth, Wang, et al. 2016) |
| Registration of T1-T2 MRI of the neonatal brain | Brain | (Simonovsky et al. 2016) |
| Registration of brain MRI | Brain | (Maes et al. 1997) |
| Correction of respiratory motion | Abdomen | (Lv et al. 2017) |
| Registration of cardiac cine MRI | Heard | (de Vos et al. 2017) |
| Reconstruction of 7T-like MRI | Brain | (Bahrami et al. 2016) |
| Reconstruction of MRI | Heart | (Schlemper et al. 2017) |
| Reconstruction of compressed sensed MRI | Abdomen | (Yang et al. 2017) |
| Synthesis of MRI | Brain | (Chartsias et al. 2017) |
| Reconstruction of CT images from low-dose CT images | Multiple | (H. Chen et al. 2017) |
| Reconstruction of CT images from low-dose CT images | Abdomen | (Kang, Min, and Ye 2017) |
| Generation of CT images from MRI | Pelvis | (Nie et al. 2016) |
| Generation of CT images from MRI | Brain | (Han 2017) |
| Prediction of PET pattern from MRI | Brain | (R. Li et al. 2014) |
| Super-resolution in MRI | Heart | (Oktay et al. 2016) |
| Enhancement of DCE-MRI | Brain | (Benou et al. 2017) |
| Denoising of 3D MRI | Brain | (Jiang et al. 2017) |
| Content-based image retrieval | Multiple | (Qayyum et al. 2017) |
| Automatic objective image quality assessment | Fetal | (Wu Lingyun, Cheng Jie-Zhi, Li Shengli, Lei Baiying, Wang Tianfu 2017) |
| Automatic objective image quality assessment | Heart | (Abdi AH, Luong C, Tsang T, Allan G, Nouranian S, Jue J, Hawley D, Fleming S, Gin K, Swift J 2017) |
| Diagnostic quality assessment of MRI | Liver | (Esses et al. 2017) |
Acknowledgments:
The authors would like to thank Gemini Janas for reviewing and editing this article.
Grant Support:
The authors would like to acknowledge funding from the National Institutes of Biomedical Imaging and Bioengineering grant 5 R01 EB021360.
BIBLIOGRAPHY
- 1.Liu W, Wang Z, Liu X, Zeng N, Liu Y, Alsaadi FE: A survey of deep neural network architectures and their applications. Neurocomputing 2017; 234:11–26. [Google Scholar]
- 2.Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. In Adv Neural Inf Process Syst; 2012:1097–1105. [Google Scholar]
- 3.Dodge S, Karam L: A Study and Comparison of Human and Deep Learning Recognition Performance Under Visual Distortions. arXiv Prepr arXiv170502498 2017. [Google Scholar]
- 4.Rajpurkar P, Irvin J, Zhu K, et al. : CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv Prepr arXiv171105225 2017. [Google Scholar]
- 5.He K, Zhang X, Ren S, Sun J: Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proc IEEE Int Conf Comput Vis; 2015:1026–1034. [Google Scholar]
- 6.Taigman Y, Yang M, Ranzato M, Wolf L: Deepface: Closing the gap to human-level performance in face verification. In Proc IEEE Conf Comput Vis pattern Recognit; 2014:1701–1708. [Google Scholar]
- 7.Wu R, Yan S, Shan Y, Dang Q, Sun G: Deep image: Scaling up image recognition. arXiv Prepr arXiv150102876 2015. [Google Scholar]
- 8.Chung JS, Senior AW, Vinyals O, Zisserman A: Lip Reading Sentences in the Wild. In CVPR; 2017:3444–3453. [Google Scholar]
- 9.Karpathy A, Fei-Fei L: Deep visual-semantic alignments for generating image descriptions. In Proc IEEE Conf Comput Vis Pattern Recognit; 2015:3128–3137. [DOI] [PubMed] [Google Scholar]
- 10.Ren S, He K, Girshick R, Sun J: Faster R-CNN: Towards real-time object detection with region proposal networks. In Adv Neural Inf Process Syst; 2015:91–99. [DOI] [PubMed] [Google Scholar]
- 11.Lin T-Y, Goyal P, Girshick R, He K, Dollár P: Focal loss for dense object detection. arXiv Prepr arXiv170802002 2017. [DOI] [PubMed] [Google Scholar]
- 12.Ghate S V, Soo MS, Baker JA, Walsh R, Gimenez EI, Rosen EL: Comparison of recall and cancer detection rates for immediate versus batch interpretation of screening mammograms. Radiology 2005; 235:31–35. [DOI] [PubMed] [Google Scholar]
- 13.Mazurowski MA: Radiogenomics: What It Is and Why It Is Important. J Am Coll Radiol 2015; 12:862–866. [DOI] [PubMed] [Google Scholar]
- 14.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI: Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014; 273:365–372. [DOI] [PubMed] [Google Scholar]
- 15.Gutman DA, Cooper LA, Hwang SN, et al. : MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013; 267:560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazurowski MA, Clark K, Czarnek NM, Shamsesfandabadi P, Peters KB, Saha A: Radiogenomics of lower-grade glioma: algorithmically-assessed tumor shape is associated with tumor genomic subtypes and patient outcomes in a multi-institutional study with The Cancer Genome Atlas data. J Neurooncol 2017:1–9. [DOI] [PubMed] [Google Scholar]
- 17.Karlo CA, Di Paolo PL, Chaim J, et al. : Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology 2014; 270:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazurowski MA, Desjardins A, Malof JM: Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro Oncol 2013; 15:1389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazurowski MA, Grimm LJ, Zhang J, et al. : Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. Eur J Radiol 2015; 84:2117–2122. [DOI] [PubMed] [Google Scholar]
- 20.Erickson BJ, Korfiatis P, Akkus Z, Kline TL: Machine Learning for Medical Imaging. RadioGraphics 2017; 37:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruder S: An overview of gradient descent optimization algorithms. 2016. [Google Scholar]
- 22.Yosinski J, Clune J, Bengio Y, Lipson H: How transferable are features in deep neural networks? In Adv Neural Inf Process Syst; 2014:3320–3328. [Google Scholar]
- 23.Sharif Razavian A, Azizpour H, Sullivan J, Carlsson S: CNN features off-the-shelf: an astounding baseline for recognition. In Proc IEEE Conf Comput Vis pattern Recognit Work; 2014:806–813. [Google Scholar]
- 24.Ahmed KB, Hall LO, Goldgof DB, Liu R, Gatenby RA: Fine-tuning convolutional deep features for MRI based brain tumor classification. In Med Imaging 2017 Comput Diagnosis. Volume 10134; 2017:101342E. [Google Scholar]
- 25.Esteva A, Kuprel B, Novoa R, Ko J, Nature SS-, 2017 undefined: Dermatologist-level classification of skin cancer with deep neural networks. nature.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litjens G, Kooi T, Bejnordi BE, et al. : A Survey on Deep Learning in Medical Image Analysis. Med Image Anal 2017; 42:60–88. [DOI] [PubMed] [Google Scholar]
- 27.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ: Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017; 30:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tajbakhsh N, Shin JY, Gurudu SR, et al. : Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Med Imaging 2016; 35:1299–1312. [DOI] [PubMed] [Google Scholar]
- 29.Antropova N, Huynh BQ, Giger ML: A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017; 44:5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deep Feature Transfer Learning in Combination with Traditional Features Predicts Survival Among Patients with Lung Adenocarcinoma. Tomography 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Yang W, Weinreb J, et al. : Searching for prostate cancer by fully automated magnetic resonance imaging classification: Deep learning versus non-deep learning. Sci Rep 2017; 7:15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A, Kim J, Lyndon D, Fulham M, Feng D: An Ensemble of Fine-Tuned Convolutional Neural Networks for Medical Image Classification. IEEE J Biomed Heal Informatics 2017; 21:31–40. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Z, Albadawy E, Saha A, Zhang J, Harowicz MR, Mazurowski MA: Deep Learning for identifying radiogenomic associations in breast cancer. arXiv Prepr arXiv171111097 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv Prepr arXiv14091556 2014. [Google Scholar]
- 35.Suk H-I, Lee S- W, Shen D: Deep ensemble learning of sparse regression models for brain disease diagnosis. Med Image Anal 2017; 37:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canziani A, Paszke A, Culurciello E: An analysis of deep neural network models for practical applications. arXiv Prepr arXiv160507678 2016. [Google Scholar]
- 37.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In Proc IEEE Conf Comput Vis Pattern Recognit; 2016:770–778. [Google Scholar]
- 38.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA: Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In AAAI; 2017:4278–4284. [Google Scholar]
- 39.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z: Rethinking the inception architecture for computer vision. In Proc IEEE Conf Comput Vis Pattern Recognit; 2016:2818–2826. [Google Scholar]
- 40.Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ: Residual Deep Convolutional Neural Network Predicts MGMT Methylation Status. J Digit Imaging 2017; 30:622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DH, Mackinnon T: Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol 2017. [DOI] [PubMed] [Google Scholar]
- 42.Hinton GE, Salakhutdinov RR: Reducing the dimensionality of data with neural networks. Science (80- ) 2006; 313:504–507. [DOI] [PubMed] [Google Scholar]
- 43.Bengio Y, Lamblin P, Popovici D, Larochelle H: Greedy layer-wise training of deep networks. In Adv Neural Inf Process Syst; 2007:153–160. [Google Scholar]
- 44.Ortiz A, Munilla J, Martínez-Murcia FJ, Górriz JM, Ramírez J: Learning longitudinal MRI patterns by SICE and deep learning: Assessing the Alzheimer’s disease progression. In Commun Comput Inf Sci. Volume 723; 2017:413–424. [Google Scholar]
- 45.Yoo Y, Tang LYW, Brosch T, et al. : Deep learning of joint myelin and T1w MRI features in normal-appearing brain tissue to distinguish between multiple sclerosis patients and healthy controls. NeuroImage Clin 2018; 17:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akkus Z, Ali I, Sedlář J, et al. : Predicting Deletion of Chromosomal Arms 1p/19q in Low-Grade Gliomas from MR Images Using Machine Intelligence. J Digit Imaging 2017; 30:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Wang Y, Yu J, Guo Y, Cao W: Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep 2017; 7:5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Havaei M, Davy A, Warde-Farley D, et al. : Brain tumor segmentation with deep neural networks. Med Image Anal 2017; 35:18–31. [DOI] [PubMed] [Google Scholar]
- 49.Milletari F, Ahmadi S-A, Kroll C, et al. : Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput Vis Image Underst 2017. [Google Scholar]
- 50.Hussain S, Anwar SM, Majid M: Brain tumor segmentation using cascaded deep convolutional neural network. In Eng Med Biol Soc (EMBC), 2017 39th Annu Int Conf IEEE; 2017:1998–2001. [DOI] [PubMed] [Google Scholar]
- 51.Valverde S, Cabezas M, Roura E, et al. : Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. Neuroimage 2017; 155:159–168. [DOI] [PubMed] [Google Scholar]
- 52.Wachinger C, Reuter M, Klein T: DeepNAT: Deep convolutional neural network for segmenting neuroanatomy. Neuroimage 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trebeschi S, van Griethuysen JJM, Lambregts DMJ, et al. : Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep 2017; 7:5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation. In Proc IEEE Conf Comput Vis Pattern Recognit; 2015:3431–3440. [DOI] [PubMed] [Google Scholar]
- 55.Ronneberger O, Fischer P, Brox T: U-net: Convolutional networks for biomedical image segmentation. In Int Conf Med Image Comput Comput Interv; 2015:234–241. [Google Scholar]
- 56.Salehi SSM, Hashemi SR, Velasco-Annis C, et al. : Real-Time Automatic Fetal Brain Extraction in Fetal MRI by Deep Learning. arXiv Prepr arXiv171009338 2017. [Google Scholar]
- 57.Christ PF, Ettlinger F, Grün F, et al. : Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks. arXiv Prepr arXiv170205970 2017. [Google Scholar]
- 58.Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y: The one hundred layers tiramisu: Fully convolutional densenets for semantic segmentation. In Comput Vis Pattern Recognit Work (CVPRW), 2017 IEEE Conf; 2017:1175–1183. [Google Scholar]
- 59.Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng PA: H-DenseUNet: Hybrid densely connected UNet for liver and liver tumor segmentation from CT volumes. arXiv Prepr arXiv170907330 2017. [DOI] [PubMed] [Google Scholar]
- 60.Chen L, Wu Y, DSouza AM, Abidin AZ, Xu C, Wismüller A: MRI tumor segmentation with densely connected 3D CNN. . [Google Scholar]
- 61.Clark T, Wong A, Haider MA, Khalvati F: Fully deep convolutional neural networks for segmentation of the prostate gland in diffusion-weighted MR images. In Int Conf Image Anal Recognit; 2017:97–104. [Google Scholar]
- 62.McKinley R, Wepfer R, Gundersen T, et al. : Nabla-net: A Deep Dag-Like Convolutional Architecture for Biomedical Image Segmentation. In Int Work Brainlesion Glioma, Mult Sclerosis, Stroke Trauma Brain Inj; 2016:119–128. [Google Scholar]
- 63.Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ: Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations In Deep Learn Med Image Anal Multimodal Learn Clin Decis Support. Springer; 2017:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta R, Sivaswamy J: M-net: A Convolutional Neural Network for deep brain structure segmentation. In Biomed Imaging (ISBI 2017), 2017 IEEE 14th Int Symp; 2017:437–440. [Google Scholar]
- 65.Milletari F, Navab N, Ahmadi S-A: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In 3D Vis (3DV), 2016 Fourth Int Conf; 2016:565–571. [Google Scholar]
- 66.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In Int Conf Med Image Comput Comput Interv; 2016:424–432. [Google Scholar]
- 67.Deniz CM, Hallyburton S, Welbeck A, Honig S, Cho K, Chang G: Segmentation of the Proximal Femur from MR Images using Deep Convolutional Neural Networks. arXiv Prepr arXiv170406176 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen L, Anderson T: Multimodal Brain MRI Tumor Segmentation via Convolutional Neural Networks. . [Google Scholar]
- 69.Poudel RPK, Lamata P, Montana G: Recurrent fully convolutional neural networks for multi-slice mri cardiac segmentation. In Int Work Reconstr Anal Mov Body Organs; 2016:83–94. [Google Scholar]
- 70.Cai J, Lu L, Xie Y, Xing F, Yang L: Improving deep pancreas segmentation in ct and mri images via recurrent neural contextual learning and direct loss function. arXiv Prepr arXiv170704912 2017. [Google Scholar]
- 71.Hochreiter S, Schmidhuber J: Long short-term memory. Neural Comput 1997; 9:1735–1780. [DOI] [PubMed] [Google Scholar]
- 72.Jaccard P: The distribution of the flora in the alpine zone. New Phytol 1912; 11:37–50. [Google Scholar]
- 73.Avendi MR, Kheradvar A, Jafarkhani H: A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 2016; 30:108–119. [DOI] [PubMed] [Google Scholar]
- 74.Rey D, Subsol G, Delingette H, Ayache N: Automatic detection and segmentation of evolving processes in 3D medical images: Application to multiple sclerosis. Med Image Anal 2002; 6:163–179. [DOI] [PubMed] [Google Scholar]
- 75.Roth HR, Lu L, Seff A, et al. : A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. In Int Conf Med Image Comput Comput Interv; 2014:520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szegedy C, Reed S, Erhan D, Anguelov D, Ioffe S: Scalable, high-quality object detection. arXiv Prepr arXiv14121441 2014. [Google Scholar]
- 77.Erhan D, Szegedy C, Toshev A, Anguelov D: Scalable object detection using deep neural networks. In Proc IEEE Conf Comput Vis Pattern Recognit; 2014:2147–2154. [Google Scholar]
- 78.Dou Q, Chen H, Yu L, et al. : Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 2016; 35:1182–1195. [DOI] [PubMed] [Google Scholar]
- 79.Shin H-C, Roth HR, Gao M, et al. : Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016; 35:1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samala RK, Chan H-P, Hadjiiski L, Helvie MA, Wei J, Cha K: Mass detection in digital breast tomosynthesis: Deep convolutional neural network with transfer learning from mammography. Med Phys 2016; 43:6654–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szegedy C, Liu W, Jia Y, et al. : Going deeper with convolutions. In Proc IEEE Conf Comput Vis pattern Recognit; 2015:1–9. [Google Scholar]
- 82.Sa R, Owens W, Wiegand R, et al. : Intervertebral disc detection in X-ray images using faster R-CNN. In Eng Med Biol Soc (EMBC), 2017 39th Annu Int Conf IEEE; 2017:564–567. [DOI] [PubMed] [Google Scholar]
- 83.Liu J, Wang D, Lu L, et al. : detection and diagnosis of colitis on computed tomography using deep convolutional neural networks. Med Phys 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Redmon J, Divvala S, Girshick R, Farhadi A: You only look once: Unified, real-time object detection. In Proc IEEE Conf Comput Vis Pattern Recognit; 2016:779–788. [Google Scholar]
- 85.Liu W, Anguelov D, Erhan D, et al. : Ssd: Single shot multibox detector. In Eur Conf Comput Vis; 2016:21–37. [Google Scholar]
- 86.Platania R, Shams S, Yang S, Zhang J, Lee K, Park S-J: Automated Breast Cancer Diagnosis Using Deep Learning and Region of Interest Detection (BC-DROID). In Proc 8th ACM Int Conf Bioinformatics, Comput Biol Heal Informatics; 2017:536–543. [Google Scholar]
- 87.Cao Z, Duan L, Yang G, et al. : Breast Tumor Detection in Ultrasound Images Using Deep Learning. In Int Work Patch-based Tech Med Imaging; 2017:121–128. [Google Scholar]
- 88.Li N, Liu H, Qiu B, et al. : Detection and Attention: Diagnosing Pulmonary Lung Cancer from CT by Imitating Physicians. arXiv Prepr arXiv171205114 2017. [Google Scholar]
- 89.de Vos BD, Wolterink JM, de Jong PA, Viergever MA, Isgum I: 2D image classification for 3D anatomy localization: employing deep convolutional neural networks. In Med Imaging Image Process; 2016:97841Y. [Google Scholar]
- 90.Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M: Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In Int Conf Med image Comput Comput Interv; 2013:246–253. [DOI] [PubMed] [Google Scholar]
- 91.Roth HR, Lu L, Liu J, et al. : Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging 2016; 35:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roth HR, Wang Y, Yao J, Lu L, Burns JE, Summers RM: Deep convolutional networks for automated detection of posterior-element fractures on spine CT. arXiv Prepr arXiv160200020 2016. [Google Scholar]
- 93.Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N, Komodakis N: A deep metric for multimodal registration In Int Conf Med Image Comput Comput Interv. Springer; 2016:10–18. [Google Scholar]
- 94.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P: Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging 1997; 16. [DOI] [PubMed] [Google Scholar]
- 95.Lv J, Yang M, Zhang J, Wang X: Respiratory motion correction for free-breathing 3D abdominal MRI using CNN based image registration: a feasibility study. Br J Radiol 2017:20170788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Vos BD, Berendsen FF, Viergever MA, Staring M, Išgum I: End-to-End Unsupervised Deformable Image Registration with a Convolutional Neural Network BT - Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support : Third International Workshop, DLMIA 2017, and 7th International Edited by Cardoso MJ, Arbel T, Carneiro G, et al. Cham: Springer International Publishing; 2017:204–212. [Google Scholar]
- 97.Bahrami K, Shi F, Zong X, Shin HW, An H, Shen D: Reconstruction of 7T-Like Images from 3T MRI. IEEE Trans Med Imaging 2016; 35:2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schlemper J, Caballero J, Hajnal J V, Price A, Rueckert D: A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging 2017; PP:1. [DOI] [PubMed] [Google Scholar]
- 99.Yang G, Yu S, Dong H, et al. : DAGAN: Deep De-Aliasing Generative Adversarial Networks for Fast Compressed Sensing MRI Reconstruction. IEEE Trans Med Imaging 2017:1–1. [DOI] [PubMed] [Google Scholar]
- 100.Chartsias A, Joyce T, Giuffrida MV, Tsaftaris SA: Multimodal MR Synthesis via Modality-Invariant Latent Representation. IEEE Trans Med Imaging 2017; 0062(January):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen H, Zhang Y, Zhang W, et al. : Low-dose CT via convolutional neural network. Biomed Opt Express 2017; 8:679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kang E, Min J, Ye JC: A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys 2017; 44:e360–e375. [DOI] [PubMed] [Google Scholar]
- 103.Nie D, Cao X, Gao Y, Wang L, Shen D: Estimating CT Image from MRI Data Using 3D Fully Convolutional Networks BT - Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICC Edited by Carneiro G, Mateus D, Peter L, et al. Cham: Springer International Publishing; 2016:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han X: MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 2017; 44:1408–1419. [DOI] [PubMed] [Google Scholar]
- 105.Li R, Zhang W, Suk H-I, et al. : Deep Learning Based Imaging Data Completion for Improved Brain Disease Diagnosis. Med Image Comput Comput Assist Interv 2014; 17(03):305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oktay O, Bai W, Lee M, et al. : Multi-input Cardiac Image Super-Resolution Using Convolutional Neural Networks. In Med Image Comput Comput Interv -- MICCAI 2016 19th Int Conf Athens, Greece, Oct 17–21, 2016, Proceedings, Part III; 2016:246–254. [Google Scholar]
- 107.Umehara K, Ota J, Ishida T: Application of Super-Resolution Convolutional Neural Network for Enhancing Image Resolution in Chest CT. J Digit Imaging 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chaudhari AS, Fang Z, Kogan F, et al. : Super‐resolution musculoskeletal MRI using deep learning. Magn Reson Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Benou A, Veksler R, Friedman A, Riklin Raviv T: Ensemble of expert deep neural networks for spatio-temporal denoising of contrast-enhanced MRI sequences. Med Image Anal 2017; 42:145–159. [DOI] [PubMed] [Google Scholar]
- 110.Jiang D, Dou W, Vosters L, Xu X, Sun Y, Tan T: Denoising of 3D magnetic resonance images with multi-channel residual learning of convolutional neural network. arxiv.org 2017. [DOI] [PubMed] [Google Scholar]
- 111.Gong E, Pauly JM, Wintermark M, Zaharchuk G: Deep learning enables reduced gadolinium dose for contrast‐enhanced brain MRI. J Magn Reson Imaging 2018. [DOI] [PubMed] [Google Scholar]
- 112.Qayyum A, Anwar SM, Awais M, Majid M: Medical image retrieval using deep convolutional neural network. Neurocomputing 2017; 266:8–20. [Google Scholar]
- 113.Chow LS, Paramesran R: Review of medical image quality assessment. Biomed Signal Process Control 2016; 27:145–154. [Google Scholar]
- 114.Wu Lingyun, Cheng Jie-Zhi, Li Shengli, Lei Baiying, Wang Tianfu ND: FUIQA: Fetal ultrasound image quality assessment with deep convolutional networks. IEEE Trans Cybern 2017; 45:1336–1349. [DOI] [PubMed] [Google Scholar]
- 115.Abdi AH, Luong C, Tsang T, Allan G, Nouranian S, Jue J, Hawley D, Fleming S, Gin K, Swift J RR: Automatic Quality Assessment of Echocardiograms Using Convolutional Neural Networks: Feasibility on the Apical Four-Chamber View. IEEE Trans Med Imaging 2017; 36:1221–1230. [DOI] [PubMed] [Google Scholar]
- 116.Esses SJ, Lu X, Zhao T, et al. : Automated image quality evaluation of T 2 -weighted liver MRI utilizing deep learning architecture. J Magn Reson Imaging 2017. [DOI] [PubMed] [Google Scholar]
- 117.Kim DH, Mackinnon T: Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Elsevier; 2017. [DOI] [PubMed] [Google Scholar]
- 118.Buda M, Maki A, Mazurowski MA: A systematic study of the class imbalance problem in convolutional neural networks. arXiv Prepr arXiv171005381 2017. [DOI] [PubMed] [Google Scholar]
- 119.Kooi T, Litjens G, van Ginneken B, et al. : Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 2017; 35:303–312. [DOI] [PubMed] [Google Scholar]
- 120.Grewal M, Srivastava MM, Kumar P, Varadarajan S: RADNET: Radiologist Level Accuracy using Deep Learning for HEMORRHAGE detection in CT Scans. arXiv Prepr arXiv171004934 2017. [Google Scholar]
- 121.Gale W, Oakden-Rayner L, Carneiro G, Bradley AP, Palmer LJ: Detecting hip fractures with radiologist-level performance using deep neural networks. arXiv Prepr arXiv171106504 2017. [Google Scholar]
- 122.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP: Performance of a Deep-Learning Neural Network Model in Assessing Skeletal Maturity on Pediatric Hand Radiographs. Radiology 2017:170236. [DOI] [PubMed] [Google Scholar]
- 123.Olczak J, Fahlberg N, Maki A, et al. : Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop 2017; 88:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jamaludin A, Lootus M, Kadir T, et al. : Automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J 2017; 26:1374–1383. [DOI] [PubMed] [Google Scholar]
- 125.Merkow J, Luftkin R, Nguyen K, Soatto S, Tu Z, Vedaldi A: DeepRadiologyNet: Radiologist Level Pathology Detection in CT Head Images. arXiv Prepr arXiv171109313 2017. [Google Scholar]


