Abstract
The tumor microenvironment presents physical, immunologic, and metabolic barriers to durable immunotherapy responses. We have recently described roles for both T cell metabolic insufficiency as well as tumor hypoxia as inhibitory mechanisms that prevent T cell activity in murine tumors, but whether intratumoral T cell activity or response to immunotherapy varies between patients as a function of distinct metabolic profiles in tumor cells remains unclear. Here, we show that metabolic derangement can vary widely in both degree and type in patient-derived cell lines and in ex vivo analysis of patient samples, such that some cells demonstrate solely deregulated oxidative or glycolytic metabolism. Further, deregulated oxidative, but not glycolytic, metabolism was associated with increased generation of hypoxia upon implantation into immunodeficient animals. Generation of murine single-cell melanoma cell lines that lacked either oxidative or glycolytic metabolism showed that elevated tumor oxygen consumption was associated with increased T cell exhaustion and decreased immune activity. Moreover, melanoma lines lacking oxidative metabolism were solely responsive to anti–PD-1 therapy among those tested. Prospective analysis of patient sample immunotherapy revealed that oxidative, but not glycolytic, metabolism was associated with progression on PD-1 blockade. Our data highlight a role for oxygen as a crucial metabolite required for the tumor-infiltrating T cells to differentiate appropriately upon PD-1 blockade, and suggest that tumor oxidative metabolism may be a target to improve immunotherapeutic response.
Keywords: Immunology
Keywords: Cancer immunotherapy, Glucose metabolism, T cells
Tumor cells’ intrinsic ability to consume oxygen represents a barrier for checkpoint blockade immunotherapy.
Introduction
The treatment of advanced melanoma has been revolutionized by immunotherapy, most prominently the antibody-mediated blockade of the coinhibitory molecule programmed death 1 (PD-1) (1). In recent years, it has become clear that PD-1 blockade can result in rapid and durable antitumor responses, leading to FDA approval of PD-1 inhibitors for melanoma in the first-line metastatic and, more recently, adjuvant settings (2). However, it has also become clear that most melanoma patients that receive immunotherapy will not receive its life-saving benefits, as the response rate is 30%–40%.
Unlike targeted therapies, in which expression of BRAF predicts response, CTLA-4 and PD-1 represent general checkpoints in all T cell immunity, with no validated predictive marker (2). While the expression of the ligand for PD-1, PD-L1, has been proposed as a potential biomarker of response, PD-L1–low patients can still respond to checkpoint blockade and there is still a high degree of resistance in PD-L1–high patients (3). While these data might suggest that there are other T cell–surface immune checkpoints that continue to inhibit T cell function in the tumor, it is also likely the case that other, T cell–extrinsic immunoregulatory factors exist that inhibit T cell function in addition to PD-1 signaling.
Much study has been aimed at identifying these non-checkpoint means of T cell inhibition, many of which relate to factors in the tumor microenvironment. Barriers to T cell infiltration, defective antigen cross-presentation, and expansion/recruitment of suppressive cell types like myeloid-derived suppressor cells or regulatory T cells are each associated with both identification of patients that may not respond to immunotherapy and targets for improving its efficacy (4).
However, in addition to the distinct population of cells that comprise the tumor microenvironment, it is generally appreciated that the tumor microenvironment has a metabolic landscape distinct among tissues, and that this too may serve as a barrier for antitumor immunity (5, 6). Tumors have areas of hypoxia and acidosis, depleted essential amino acids like glutamine and arginine, and have a unique lipid profile among tissues (7, 8). The effects of these individual factors on T cell function have been rigorously studied, and much recent work has gone into identifying what the consequences are for tumor-infiltrating T cells experiencing these metabolic stressors in aggregate. Indeed, recent work has examined the role of tumor glucose metabolism as a central barrier for antitumor immunity; glucose has been found to be limiting in some tumors, and thus T cell competition for this vital nutrient becomes a regulatory factor in the tumor microenvironment (9, 10). Indeed, a recent report suggests that cell lines derived from adoptive cell therapy–resistant melanoma patients are more glycolytic than those that would go on to respond (11). Importantly, glucose deprivation is not a common phenotype of all tumors, and (for instance) renal cell carcinoma has almost no glucose deprivation compared with normal tissue (12). These data suggest that the competition for glucose in the tumor microenvironment may not fully explain the metabolic insufficiency underlying T cell dysfunction in cancer.
We have recently shown that T cells that infiltrate tumors exhibit major defects in oxidative metabolism. Tumor-infiltrating T cells show defects in mitochondrial biogenesis, fusion, and oxidative function (12–14). Further, mitochondrial dysfunction occurs concomitantly with the development of T cell exhaustion but independently of PD-1 signaling (14). Boosting T cell mitochondrial metabolism through metabolic reprogramming (14), costimulatory immunotherapy (13, 15), or mitigation of tumor cell oxidative metabolism (16) results in increased antitumor immunity and response to PD-1 blockade immunotherapy, suggesting that the oxidative axis is also an important metabolic consideration in cancer immunity.
It has long been appreciated that tumors can have variable degrees of hypoxia (7, 17), and that there is vast metabolic heterogeneity between tumor types, patients, and even within the same tumor and different tumor metastases from the same patient (18). However, how those patient-to-patient variabilities relate to the response to immunotherapy remains unclear. Part of this lack of clarity relates to the fact that metabolic profiling was both laborious and required a large amount of tissue, or relied on various indirect readouts. Extracellular flux analysis (enabled by Seahorse technology) provides real-time profiling of oxidative versus glycolytic metabolism from relatively small numbers of cells (19). Thus, we sought to use this tool to profile tumor cell metabolism directly from melanoma patients, as a means to infer the metabolic status of the tumor microenvironment.
Results
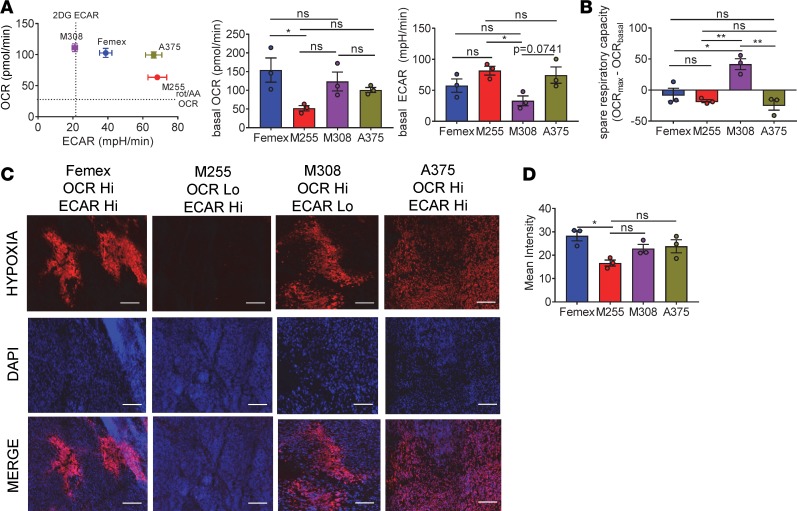
We first employed Seahorse analysis to examine various melanoma cell lines that are used in human cancer biology. Plotting the basal metabolism of these cell lines on a 2-dimensional plot of oxidative metabolism (oxygen consumption rate, OCR) versus glycolytic metabolism (extracellular acidification rate, ECAR) revealed that there were distinct metabolic profiles between cell lines that have been cultured in vitro for years (Figure 1A). There were tumor cell lines that were extremely energetic (A375, Femex), utilizing both glycolytic and oxidative metabolism, as well as cell lines that performed heightened oxidative (M308) or glycolytic (M255) metabolism, but not both (Figure 1A). Stimulation of maximal oxidative metabolism using uncouplers (FCCP) showed that even when stressed, some cells would solely use one type of metabolic pathway (Figure 1B). These results suggested that tumor cells do not solely exist on a single axis from quiescent to energetic, but that there could also be distinctions in the type of metabolism that is deregulated, and which pathway is deregulated may be part of the tumor cell identity.
Figure 1. Melanoma cell lines are metabolically heterogeneous and produce various degrees of hypoxia in vivo.
(A) Representative OCR versus ECAR of human melanoma cell lines (left) and tabulated baseline OCR and ECAR (right) from multiple experiments. (B) Tabulated respiratory capacity (maximal, uncoupled respiration by FCCP stimulation) from cells as in A. (C) Pimonidazole staining of full tumor sections from NSG mice bearing human melanoma cell line tumors measuring 5 mm across. Scale bars: 100 μm. (D) Tabulated results of the internal hypoxyprobe intensity from set brightness normalized for each day of imaging. Data represent 3 independent experiments. *P < 0.05, **P < 0.01 by 1-way ANOVA. ns, not significant. Error bars indicate SEM.
As we and others have previously shown that heightened oxidative metabolism of tumor cells is associated with the development of intratumoral hypoxia (16), we wanted to confirm how these metabolically distinct cell lines generated hypoxic environments in vivo. Using pimonidazole to mark areas of low oxygen tension (20), we determined the extent of tumor microenvironment hypoxia resulting from implantation of these cell lines in immunodeficient (NOD.Cg-PrkdcscidIl2rgnull or NSG) animals, analyzing when each tumor reached 5- to 6-mm diameters. Patient-derived melanoma cell lines that bore a high degree of oxidative metabolism in metabolic flux assays also generated substantially more intratumoral hypoxia, suggesting that the ability of tumors to generate hypoxic environments is mechanistically linked to their utilization of mitochondrial oxidative phosphorylation pathways (Figure 1, C and D). Importantly, tumors formed from the solely glycolytic M255 cell line produced less hypoxia in vivo, suggesting that tumor cell oxidative metabolism is a central driver of intratumoral hypoxia.
We next sought to determine how these metabolic changes and the generation of intratumoral hypoxia might impact antitumor immunity and the response to immunotherapy. As these human melanoma cell lines only grew in immunodeficient animals, we instead sought to reverse translate these findings into a mouse model in which only the metabolic pathways were modified. Single cells were sorted from melanoma tumors arising in the Ptenfl/flBrafLSL-V600ETyrCre.ER genetically engineered mouse model (21) to generate implantable melanoma cell lines (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124989DS1). These were favorable over the more broadly used B16 melanoma, as they carry mutations consistent with human disease. Metabolic profiling of the resulting lines facilitated our selection of clone 24, which exhibited extraordinarily high oxidative and glycolytic metabolism compared with a C57BL/6J normal melanocyte cell line (Melan-A) (Supplemental Figure 1B). This cell line could be injected intradermally into C57BL/6J mice and form extremely aggressive tumors with a poor tumor infiltrate (Supplemental Figure 1C). Importantly, this tumor model is completely resistant to anti–PD-1 immunotherapy (Supplemental Figure 1D). Thus, this model, based on a single energetic tumor cell, could allow the dissection of contributions from a single metabolic pathway to immune dysfunction.
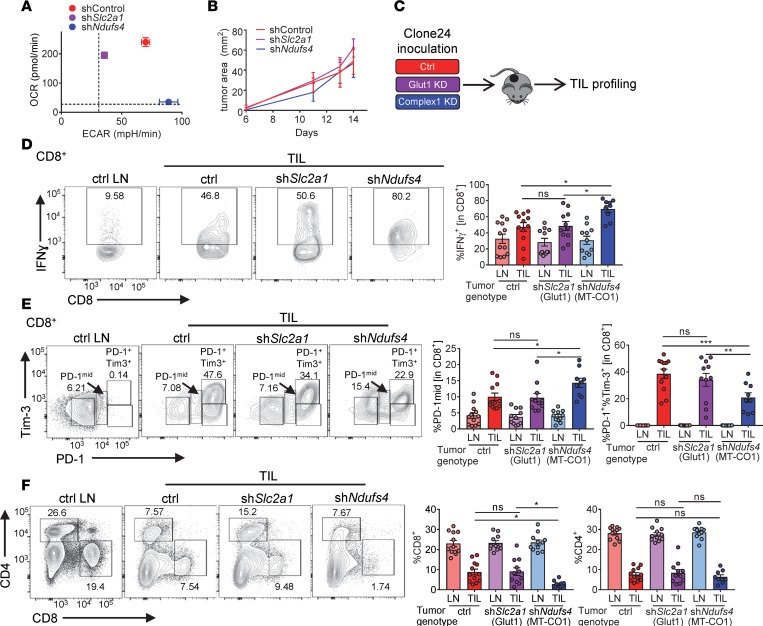
In order to interrogate the role of glycolysis versus oxidative metabolism in immunotherapy resistance, we generated clone 24 lines stably expressing RNA interference constructs targeting either glucose uptake (shSlc2a1, targeting the glucose transporter GLUT1) or mitochondrial oxidative phosphorylation (shNdufs4, targeting a subunit of mitochondrial complex I). These lines efficiently knocked down their target protein (Supplemental Figure 2A) compared with scrambled RNA–expressing lines. When these cell lines were metabolically profiled, GLUT1-knockdown tumors had impaired glycolytic metabolism, while complex I–knockdown tumors had impaired oxidative phosphorylation (Figure 2A and Supplemental Figure 2B), essentially mirroring the metabolic phenotype of the human cell lines we analyzed previously. These tumor lines proliferated in vitro and formed tumors of similar sizes when implanted into B6 mice (Figure 2B and Supplemental Figure 2C). However, examination of the immune infiltrate and immune cell function revealed substantial differences; T cells infiltrating control (both oxidative and glycolytic) tumors showed high degrees of T cell dysfunction: defective cytokine production upon restimulation and high coinhibitory molecule expression (Figure 2, C–E). Near-complete loss of glycolytic activity in clone 24 tumors (GLUT1-knockdown tumors) did little to affect the phenotype of tumor-infiltrating T cells, but the loss of oxidative metabolism in tumor (complex I–knockdown tumors) resulted in significant improvements in T cell status: increased cytokine production and a shift from terminally exhausted PD-1hiTim-3+ cells to PD-1int cells (Figure 2E). Interestingly, however, we observed a significant decrease in CD8+ T cell numbers when tumor cells lacked oxidative metabolism and favored glycolysis (complex 1–knockdown tumors) (Figure 2F), despite being functionally superior (Figure 2, D and E). Thus, changing only oxidative metabolism was sufficient to alter the status of the tumor microenvironment such that steady-state T cell dysfunction was reduced.
Figure 2. Oxidative metabolism in tumor cells promotes increased T cell dysfunction.
(A) Representative OCR versus ECAR of clone 24 melanoma cells transfected with scrambled control, Slc2a1 (Glut1), or Ndufs4 (complex I) shRNA. (B) Tumor growth curves of C57BL/6J inoculated with 250,000 of clone 24 melanoma cells as in A intradermally. (C) Schematic of TIL profiling from mice bearing clone 24 tumors. (D) Representative flow cytogram (left) and tabulated data from multiple experiments (right) of PMA- and ionomycin-induced IFN-γ from CD8+ T cells from clone 24–bearing mice. LN, lymph node; TIL, tumor-infiltrating lymphocyte (n = 12 per group). (E) Representative flow cytogram (left) and tabulated data from multiple experiments (right) of PD-1 and Tim-3 expression as in D. (F) Representative flow cytogram (left) and tabulated data from multiple experiments (right) of CD4+ and CD8+ T cells as in D. Data represent 5 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by 2-way ANOVA (A) or 1-way ANOVA (C–E). ns, not significant. Error bars indicate SEM.
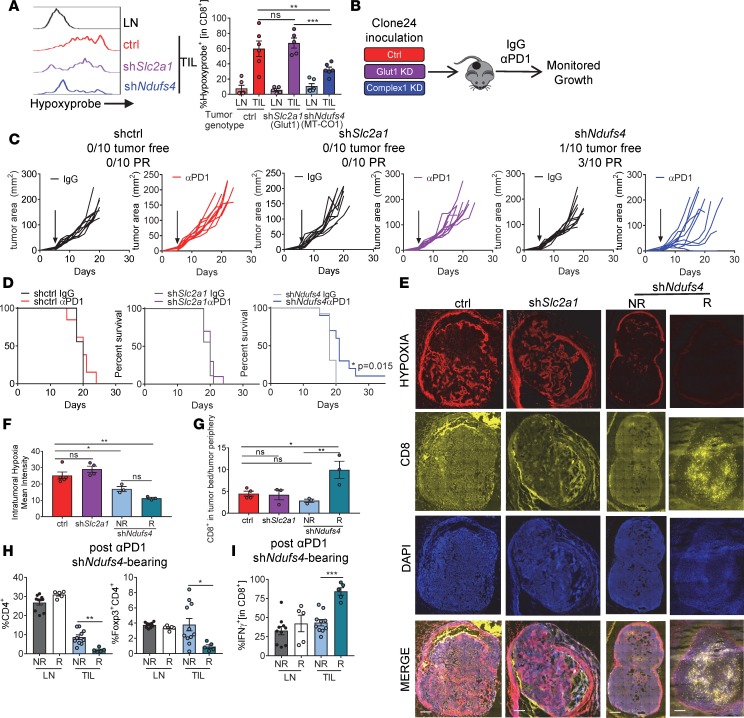
Pimonidazole pulsing of tumor-bearing animals revealed that CD8+ T cells isolated from tumors formed by complex I–knockdown tumors experienced less hypoxia, while tumor-infiltrating lymphocytes (TILs) isolated from control and less glycolytic tumors (GLUT1-knockdown) experienced similar, high levels of hypoxia (Figure 3A). Hypoxia can be inhibitory to T cell proliferation and function (22), and a hypoxia-related transcriptional profile is associated with adaptive resistance to checkpoint blockade immunotherapy (23). We thus asked whether clone 24 tumors lacking these specific metabolic pathways were more sensitive to immunotherapy, which we initiated when tumors were palpable (measuring 1–3 mm in any direction) (Figure 3B). Energetic control– and glycolysis-deficient Slc2a1-knockdown tumors had no response to anti–PD-1 immunotherapy, similar to their parental line (Figure 3C). However, mice bearing mitochondrial complex 1–knockdown tumors, having dramatically reduced oxidative metabolism, were significantly more sensitive to checkpoint blockade, with 40% of the mice experiencing a partial or complete response as well as increased overall survival (Figure 3, C and D). Notably however, complex I deficiency did not result in response to anti–PD-1 in all mice; 60% of the animals implanted with complex I–knockdown tumors did not respond whatsoever to anti–PD-1 therapy.
Figure 3. Inhibiting oxidative metabolism in tumor cells reduces intratumoral hypoxia and increases sensitivity to PD-1 blockade therapy.
(A) Representative pimonidazole staining and tabulated data from multiple experiments of CD8+ T cells isolated from clone 24 knockdown tumors (n = 6 per group). (B) Schematic of C57BL/6J mouse inoculated with 250,000 of clone 24 knockdown cells intradermally, and then treated with 200 μg anti–PD-1 or isotype controls thrice weekly when tumors reached 1–3 mm. (C) Tumor growth from mice treated as in B. Tumor-free indicates a complete regression. PR indicates mice that showed tumor regression for at least 2 measurements. Each line represents 1 animal. (D) Survival curve of mice treated as in B. (E) Pimonidazole, CD8, and DAPI staining of full tumor sections from mice bearing clone 24 knockdown tumors (left). Scale bars: 500 μm. (F) Tabulated results of the internal hypoxyprobe intensity from a set brightness normalized for each day of imaging (n = 4 per group). (G) Ratio of T cell counts in tumor bed versus periphery of tumor (n = 4 per group). (H) Tabulated flow cytometry data from multiple experiments of CD4+ and CD4+Foxp3+ T cells from clone 24 Ndufs4–knockdown mice treated with 200 μg anti–PD-1 (n = 5–10 per group). (I) Tabulated flow cytometry data from multiple experiments of IFN-γ in CD8+ T cells from mice as in H. Data represent 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA (A–C and F–I) or log-rank test (D). ns, not significant. Error bars indicate SEM.
We next wanted to examine how T cells infiltrate hypoxic tumor regions in our model, using pimonidazole imaging and T cell stains by immunofluorescence. We found that in both control and Slc2a1-knockdown tumors (GLUT1-knockdown), hypoxic regions were evident in the tumor bed and that T cells were excluded from these regions. However, this imaging revealed that T cells infiltrating the less oxidative tumors were more likely to infiltrate the tumor bed rather than be excluded to the tumor periphery (Figure 3, E–G). We also profiled the infiltrate of shNdufs4 tumors shortly after receiving checkpoint blockade immunotherapy based on whether that mouse was responding (defined as 2 sequential daily measurements showing tumor regression). Analysis of the infiltrate when tumors were still similarly sized from Ndufs4-knockdown tumor–bearing mice showed that there still existed some animal-to-animal variability in terms of the functional phenotypes of the tumor microenvironment. TILs from nonresponding shNdufs4 tumors experienced more hypoxia and were characterized by a more immunosuppressive tumor microenvironment (higher CD4+ and regulatory T cell infiltrate) (Figure 3H). We also profiled the tumor-infiltrating T cell effector function and saw significantly more IFN-γ in responding shNdufs4 tumors (Figure 3I). Thus, the oxidative axis represents an important barrier to effective immunotherapy, and inhibition of oxidative metabolism results in increased penetrance of anti–PD-1 treatment.
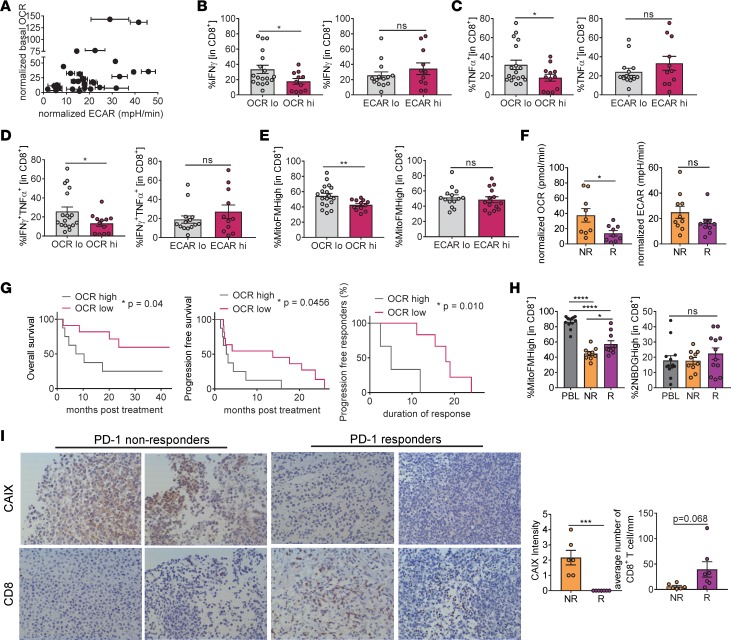
As previously mentioned, extracellular flux analysis can be conducted on exceedingly small numbers of tumor cells (5,000–10,000 cells), facilitating the metabolic profiling of tumor cells from human patient biopsy samples from our IRB-approved blood and tissue melanoma bank. Metabolic profiling of CD45– tumor samples revealed similarly distinct metabolic profiles compared with what we observed in the panel of melanoma cell lines; some patients had relatively quiescent tumor cells, while others had tumor cells with deranged glycolytic and oxidative metabolic pathways. Further, there were also patients that predominantly upregulated either ECAR or OCR activity (Figure 4A). Evaluation of the tumor infiltrate from each of these patient samples revealed that oxidative, but not glycolytic, metabolism in the tumor cells was associated with repressed T cell function and metabolism (Figure 4, B–E, and Supplemental Figure 3, A and B).
Figure 4. Oxidative metabolism of tumor cells and hypoxia are associated with decreased antitumor immunity and poor clinical response to PD-1 blockade therapy in patients.
(A) Normalized OCR versus normalized ECAR of isolated tumor cells from melanoma patient biopsies. Values normalized based on rotenone/antimycin A for OCR and 2DG for ECAR (n = 30). (B) Tabulated flow cytometry data of IFN-γ production from CD8+ T cells isolated as in A as a function of high OCR or ECAR. (C) Tabulated flow cytometry data of TNF-α from CD8+ T cells as in B. (D) Tabulated flow cytometry data of IFN-γ and TNF-α from CD8+ T cells as in B. (E) Tabulated flow cytometry data of MitoTracker FM from CD8+ T cells as in B. (F) Tabulated normalized OCR and tabulated normalized ECAR of isolated tumor cells from melanoma patients that progressed on (NR, nonresponder) or responded to (R, responder defined as stable disease, partial or complete response) (n = 19). (G) Overall survival, progression-free survival, and duration of response of patients treated with either pembrolizumab or nivolumab monotherapy based on tumor cell oxidative metabolism. (H) Representative flow cytogram (left) and tabulated data from multiple experiments (right) of MitoTracker FM and 2NBDG of CD8+ T cells from patients as in F. (I) Representative immunohistochemistry at ×20 magnification (left) and tabulated CAIX scoring and CD8+ T cell numbers of FFPE sections from patients as in F. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001 by unpaired t test (B–F, H, and I) or Wilcoxon’s test (G). ns, not significant. Error bars indicate SEM.
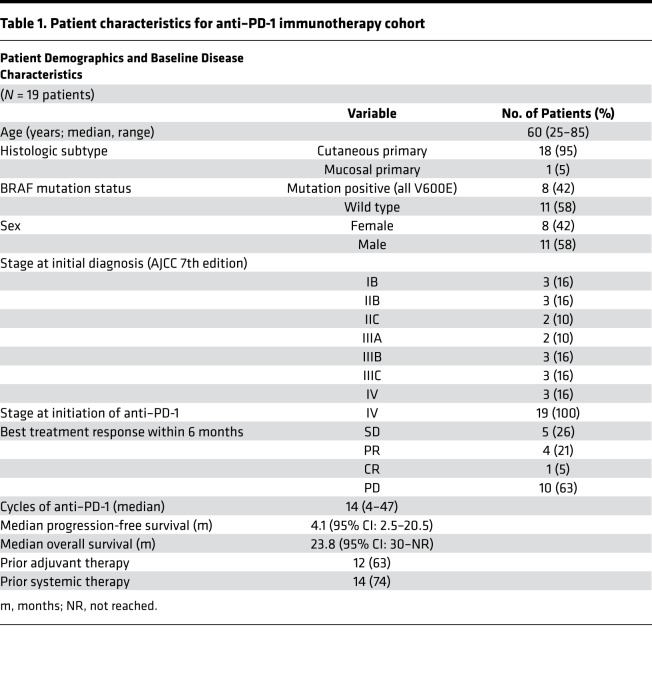
As our preclinical data suggested that response to anti–PD-1 immunotherapy hinged critically on the availability of oxygen in the tumor microenvironment, we also asked whether metabolic profiling of melanoma patient tumor cells might predict the response to nivolumab or pembrolizumab monotherapy. We evaluated a cohort of 19 patients (Table 1) who had received anti–PD-1 monotherapy for metastatic melanoma: 1 patient was on concomitant vemurafenib (on clinical trial), 14 patients were biopsied prior to initiation of treatment, and 5 were biopsied on treatment. When evaluating this cohort of 19 patients, we defined a “responder” as a patient who had stable disease, or a partial or complete response to immunotherapy with a duration of at least 6 months. Indeed, patients who did not respond to anti–PD-1 checkpoint blockade (n = 10) had tumor cells characterized by increased oxidative metabolism, while glycolytic metabolism had no predictive value (Figure 4F). Also, high oxidative tumor cell metabolism was associated with poorer overall survival after anti–PD-1 immunotherapy (Figure 4G). Patients who had low tumor oxygen consumption had greater progression-free survival and duration of response (Figure 4G).
Table 1. Patient characteristics for anti–PD-1 immunotherapy cohort.
We also explored how the tumor microenvironment metabolism might relate to the metabolic sufficiency of the tumor-infiltrating CD8+ T cells. While, as we have previously shown, most tumor-infiltrating T cells showed defects in mitochondrial staining by MitoTracker FM, metabolic insufficiency was more severe in pretreatment TILs from patients that subsequently progressed on anti–PD-1 (Figure 4H). The capacity to take up the glucose tracer 2NBDG appeared to be similar between responders and nonresponders (Figure 4H). As increased oxidative metabolism is linked to increased hypoxia, we sought to determine whether tumor hypoxia might be associated with immunotherapy resistance. True hypoxia imaging via pimonidazole requires a prebiopsy pulse with this tracer, but hypoxia can be inferred using staining for carbonic anhydrase IX (CAIX) (20). Indeed, tumor sections from immunotherapy-resistant patients were significantly more hypoxic than those that responded (Figure 4I). Thus, oxidative metabolism in tumor cells and consequent hypoxia forms a barrier to T cell activity and proper differentiation in the tumor microenvironment and thus clinical response to checkpoint blockade immunotherapy.
Discussion
In this study, we explored the effects of tumor cell metabolism on the generation of a harsh metabolic microenvironment, and how it contributed to both steady-state T cell dysfunction and response to immunotherapy. Our data place tumor mitochondrial oxidative consumption as a barrier to response to PD-1 blockade. Further, our data suggest that the degree of oxidative metabolism varies within the patient population, and that the tumor cell metabolic profile may be related to the likelihood of response to anti–PD-1 immunotherapy.
One of the primary consequences of deranged tumor cell metabolism is the generation of hypoxia. While exposure to hypoxia alone can have both positive and detrimental effects on T cell fate and function (24–26), hypoxia is certainly considered inhibitory to T cells in vivo (22, 27). Interestingly, transcriptional profiling tumors in anti–PD-1 responders versus nonresponders revealed a dominant hypoxia signature in immunotherapy-resistant patients (10, 23). Our data support previous work implicating hypoxia and hypoxic signaling in the metabolic and functional reprogramming of T cells (24–26), and suggest that hypoxia may divert differentiation into a dysfunctional exhausted phenotype rather than one associated with an effector-memory state. Further, we have previously shown in preclinical models that more oxidative tumors tend to be resistant to checkpoint blockade immunotherapy (16). Our data in nonresponders versus responders support the notion that tumor hypoxia correlates to resistance to anti–PD-1–based immunotherapy. Some studies have previously suggested that it is the capacity of tumor cells to outcompete T cells for glucose that limits T cell function in the tumor microenvironment (9), and that cell lines derived from nonresponders to adoptive cell therapy were more glycolytic (11). Our study found no role for increased glucose metabolism in tumor cells or repressed T cell glucose uptake in the response to checkpoint blockade immunotherapy, and identifies oxygen as a key metabolite at the core of anti–PD-1 immunotherapy sensitivity. Future studies will seek to determine whether adoptive cell therapies versus checkpoint blockade are metabolically distinct therapies, preferentially utilizing different fuel sources.
Oxygen is required for several cellular processes. Indeed, oxygen is required for oxidative phosphorylation of ATP, but it is also a key player in the generation of reactive oxygen species, the decarboxylation reactions of the TCA cycle, and the demethylation reactions of DNA and histones. Given that the epigenetic stability of exhausted T cells represents a barrier to PD-1 blockade in chronic viral infection and cancer (28, 29), the effects of hypoxia on T cells may not simply be energetic but may prevent any epigenetic reprogramming induced by PD-1 blockade and other factors.
One intriguing observation in this study is that while the less-oxidative shNdufs4 melanomas were more sensitive to PD-1 blockade immunotherapy, there were significantly fewer CD8+ T cells in these tumors. While it is clear from both preclinical and patient data that T cell infiltration is an important prerequisite for response to PD-1 blockade immunotherapy (30, 31), our data refine this observation and suggest that, provided there is some T cell infiltration, it is the quality of the infiltrating T cells that dictate response, and potentially apply a metabolic component to the immune-excluded phenotype. Possessing tumor-infiltrating T cells that are less hypoxic and more metabolically sufficient correlated with increased effector function and response to immunotherapy. This is an important consideration for future work exploring the importance of T cell infiltration as a means to predict responses to anti–PD-1.
Finally, our data support the use of metabolic modulators, especially those that may target oxidative metabolism, to bolster immunotherapeutic response. Re-normalizing the oxygen tension of the tumor microenvironment represents an attractive modality to improve the efficacy of various therapies, especially immunotherapy. Low-dose VEGF inhibitors may improve the quality of the tumor vasculature (32), elimination of hypoxic tumor cells can potentially improve response to checkpoint blockade (33), and respiratory hyperoxia can re-oxygenate the environment and improve response to immunotherapy (34). At our institution, we recently opened a phase I/II randomized clinical trial of pembrolizumab versus pembrolizumab plus the type II diabetes drug metformin (which inhibits mitochondrial complex I, of which Ndufs4 is a subunit) in patients with advanced unresectable melanoma (NCT03311308), based on our preclinical work showing that metformin treatment can reduce tumor hypoxia and synergize with PD-1 blockade (16). These therapeutic efforts seek to remodel the tumor microenvironment such that it is metabolically permissive to T cell function, providing an opportunity to improve the efficacy of immunotherapeutic treatments of cancer.
Methods
Metabolic assays.
Tumor cell metabolic output was measured by Seahorse technology as previously described (16). Tumor cells were seeded in XFe96 plates at 10,000–40,000 cells per well in complete RPMI. Cells were washed in minimal unbuffered assay media containing glucose, pyruvate, and glutamine 1 hour before the assay. Cells received sequential injections of 2 μM oligomycin, 2 μM FCCP, 10 mM 2-deoxyglucose (2DG), and 0.5 μM rotenone/antimycin A. Normalized OCR was calculated by subtracting OCR measurements after rotenone/antimycin A injection from basal OCR. Normalized ECAR was calculated by subtracting ECAR measurements after 2DG injection from maximum ECAR measurements.
For flow cytometric assays, cells were pulsed with 50 μM 2NBDG (Cayman Chemical) in vitro for 30 minutes at 37°C. Cells were then surface stained at 4°C in the presence of 20 nM MitoTracker Deep Red FM for 15 minutes, then washed and assayed by flow cytometry.
Immunoblot analysis.
Cells were lysed in 1% NP-40 lysis buffer as previously described (35). Lysates were separated by SDS-PAGE in 4%–12% Bolt or Bio-Rad gels, transferred to PVDF membranes, and blocked in 3% milk in Tris-buffered saline with 0.1% Tween 20. Primary antibodies against Glut1 (Abcam, catalog ab115730, clone EPR3915), Ndufs4 (Santa Cruz Biotechnology, catalog sc-100567, clone 1-E-4), and actin (Santa Cruz Biotechnology, catalog sc-47778, clone C4) were added in 3% milk/TBST overnight at 4°C. Secondary antibodies (anti-rabbit or anti-mouse linked to horseradish peroxidase, Jackson ImmunoResearch) were added in 3% milk/TBST for 1–2 hours at room temperature. Enhanced chemiluminescence was visualized using Western Lightning (PerkinElmer). Digitally captured films were analyzed densitometrically by ImageJ software (NIH).
Retroviral RNA interference.
shRNA for Slc2a1 and Ndufs4 was subcloned into pMKO.1 (originally generated by William Hahn and obtained from Addgene, plasmid 10676). This vector was then transiently transfected into clone 24 melanoma cells. shRNA knockdown for each experiment was confirmed by immunoblot.
Immunotherapy and tumor models.
C57BL/6J mice were treated with 200 μg anti–PD-1 (clone J43, Bio X Cell, catalog BE0033-2) or respective hamster isotype controls intraperitoneally 3 times per week. For tumor growth experiments, mice were injected intradermally with 250,000 clone 24 melanoma cells. When tumors reached 3 mm in any direction (typically day 5), mice began receiving immunotherapy.
Histology.
In some experiments, after tumor growth, PD-1 treatment, and pimonidazole pulsing, tumors were dissected and frozen at –80°C in Optimal Cutting Temperature Compound (OCT) (Tissue-Tek) and sectioned (Cryostat microtome). Tissue was fixed in histology-grade acetone (Thermo Fisher Scientific) at –20°C, then rehydrated in staining buffer, stained with hypoxyprobe (Hypoxyprobe, catalog HP7-100Kit), anti-CD8–Alexa Fluor 647 (Biolegend, catalog 100727, clone 53-6.7), and DAPI (Life Technologies), and mounted with ProLong Diamond Antifade Mountant (Life Technologies). Sections were imaged with an Olympus IX83 microscope and analyzed with ImageJ and NIS-Elements Imaging Software (Nikon).
Immunohistochemistry.
Antigen retrieval was performed using Diva Decloaker (Biocare Medical) in a decloaking chamber (Biocare Medical). Tissues were incubated at room temperature with primary antibody (CAIX, Novus Biologicals, catalog nb100-417; or CD8, Dako Cytomation, clone C8/144B) at 1:500 or 1:100 for 30 minutes. Visualization and detection of primary antibody was achieved using the HiDef Detection HRP Polymer System (Cell Marque) and DAB Substrate Kit (Cell Marque). Tissues were counterstained using Harris Hematoxylin. CAIX scoring was determined by percentage of total staining area on tumor cells and CD8+ T cell counts were quantified using ImageJ software.
Human peripheral blood lymphocyte, TIL, and tumor cell isolation.
Tumor digests and isolation of human peripheral blood lymphocytes, TILs, and tumors were performed as previously described (14). Cells were rested at 37°C for 30 minutes prior to flow cytometric assays or CD45-negative isolation.
Statistics.
All experiments were performed with at least 3 replicates and analyzed using GraphPad Prism software. All graphs include the mean with SEM and differences were considered significant at a P value of less than 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001; ns indicates not significant). Two-tailed unpaired t test was used when comparing 2 groups, 1- or 2-way ANOVA was used for multiple comparisons, and either Wilcoxon’s or log-ranked test was used for survival analysis.
Study approval.
Animal work in this study was approved by the University of Pittsburgh Institutional Animal Care and Use Committee, accredited by the AAALAC. Procedures were performed under their guidelines. C57BL/6J and BRafCA PtenloxP Tyr::CreERT2 mice were obtained originally from the Jackson Laboratories and bred in house. Biobanked peripheral blood lymphocytes and unseparated tumor from melanoma patients were utilized in accordance with our IRB-approved (University of Pittsburgh Cancer Institute [UPCI]) tissue banking protocol UPCI 96-099, and metabolically profiled utilizing IRB-approved protocol UPCI 17-127.
Author contributions
YGN provided clinical expertise, collected and analyzed clinical data, obtained research funding, oversaw translational research, and helped write the manuscript. AVM performed the majority of the research, analyzed data, and helped write the manuscript. CS procured patient samples. UR and AK conducted pathological analysis. RB helped collect clinical data. SZ assisted with statistical analysis. JMK provided clinical and scientific insight. GMD conceived of and oversaw research, analyzed data, obtained research funding, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the UPMC Hillman Cancer Center Melanoma and Skin Cancer SPOREs (P50CA121973-09 to YGN, JMK, and GMD); Stand Up To Cancer — American Association for Cancer Research (SU2C-AACR-IRG-04-16 to GMD); an NIH Director’s New Innovator Award (DP2AI136598 to GMD); Young Investigator Award from Alliance for Cancer Gene Therapy/Swim Across America (to GMD); the Hillman Fellows for Innovative Cancer Research Program funded by the Henry L. Hillman Foundation; Cancer Center Support Grant P30CA047904 (to YGN); and the UPMC Hillman Cancer Center (start-up funds to GMD). This work utilized the UPMC Hillman Cancer Center Flow Cytometry and Animal Facilities, supported in part by P30CA047904. The authors would also like to thank the UPMC Pathology Department and Biospecimen Core, especially Tony Green and Elaine Isherwood.
Version 1. 02/05/2019
In-Press Preview
Version 2. 03/07/2019
Electronic publication
Footnotes
Conflict of interest: The authors declare that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(5):e124989. https://doi.org/10.1172/jci.insight.124989.
Contributor Information
Yana G. Najjar, Email: najjaryg@upmc.edu.
Ashley V. Menk, Email: avm9@pitt.edu.
Cindy Sander, Email: sandca@UPMC.EDU.
Uma Rao, Email: raounm@upmc.edu.
Arivarasan Karunamurthy, Email: karunamurthya@upmc.edu.
Roma Bhatia, Email: bhatiar@upmc.edu.
Shuyan Zhai, Email: shuyan.zhai@pitt.edu.
John M. Kirkwood, Email: kirkwoodjm@upmc.edu.
Greg M. Delgoffe, Email: delgoffeg@upmc.edu.
References
- 1.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozeman EA, Dekker TJA, Haanen JBAG, Blank CU. Advanced melanoma: current treatment options, biomarkers, and future perspectives. Am J Clin Dermatol. 2018;19(3):303–317. doi: 10.1007/s40257-017-0325-6. [DOI] [PubMed] [Google Scholar]
- 3.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 4.Munn DH, Bronte V. Immune suppressive mechanisms in the tumor microenvironment. Curr Opin Immunol. 2016;39:1–6. doi: 10.1016/j.coi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wegiel B, Vuerich M, Daneshmandi S, Seth P. Metabolic switch in the tumor microenvironment determines immune responses to anti-cancer therapy. Front Oncol. 2018;8:284. doi: 10.3389/fonc.2018.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scharping NE, Delgoffe GM. Tumor microenvironment metabolism: a new checkpoint for anti-tumor immunity. Vaccines (Basel) 2016;4(4) doi: 10.3390/vaccines4040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24(8):472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing Y, Zhao S, Zhou BP, Mi J. Metabolic reprogramming of the tumour microenvironment. FEBS J. 2015;282(20):3892–3898. doi: 10.1111/febs.13402. [DOI] [PubMed] [Google Scholar]
- 9.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho PC, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor t cell responses. Cell. 2015;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascone T, et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 2018;27(5):977–987.e4. doi: 10.1016/j.cmet.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siska PJ, et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight. 2017;2(12):e93411. doi: 10.1172/jci.insight.93411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menk AV, et al. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J Exp Med. 2018;215(4):1091–1100. doi: 10.1084/jem.20171068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scharping NE, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity. 2016;45(2):374–388. doi: 10.1016/j.immuni.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teijeira A, et al. Mitochondrial morphological and functional reprogramming following CD137 (4-1BB) costimulation. Cancer Immunol Res. 2018;6(7):798–811. doi: 10.1158/2326-6066.CIR-17-0767. [DOI] [PubMed] [Google Scholar]
- 16.Scharping NE, Menk AV, Whetstone RD, Zeng X, Delgoffe GM. Efficacy of PD-1 blockade is potentiated by metformin-induced reduction of tumor hypoxia. Cancer Immunol Res. 2017;5(1):9–16. doi: 10.1158/2326-6066.CIR-16-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morfoisse F, Renaud E, Hantelys F, Prats AC, Garmy-Susini B. Role of hypoxia and vascular endothelial growth factors in lymphangiogenesis. Mol Cell Oncol. 2015;2(4):e1024821. doi: 10.1080/23723556.2015.1024821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warmoes MO, Locasale JW. Heterogeneity of glycolysis in cancers and therapeutic opportunities. Biochem Pharmacol. 2014;92(1):12–21. doi: 10.1016/j.bcp.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pike Winer LS, Wu M. Rapid analysis of glycolytic and oxidative substrate flux of cancer cells in a microplate. PLoS ONE. 2014;9(10):e109916. doi: 10.1371/journal.pone.0109916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young RJ, Möller A. Immunohistochemical detection of tumour hypoxia. Methods Mol Biol. 2010;611:151–159. doi: 10.1007/978-1-60327-345-9_12. [DOI] [PubMed] [Google Scholar]
- 21.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Ertl HC. Starved and asphyxiated: how can CD8(+) T cells within a tumor microenvironment prevent tumor progression. Front Immunol. 2016;7:32. doi: 10.3389/fimmu.2016.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clever D, et al. Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell. 2016;166(5):1117–1131.e14. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14(11):1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phan AT, Goldrath AW. Hypoxia-inducible factors regulate T cell metabolism and function. Mol Immunol. 2015;68(2 Pt C):527–535. doi: 10.1016/j.molimm.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Patel SP, Roszik J, Qin Y. Hypoxia-driven immunosuppressive metabolites in the tumor microenvironment: new approaches for combinational immunotherapy. Front Immunol. 2018;9:1591. doi: 10.3389/fimmu.2018.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariathasan S, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turan T, et al. Immune oncology, immune responsiveness and the theory of everything. J Immunother Cancer. 2018;6(1):50. doi: 10.1186/s40425-018-0355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiffmann LM, et al. A combination of low-dose bevacizumab and imatinib enhances vascular normalisation without inducing extracellular matrix deposition. Br J Cancer. 2017;116(5):600–608. doi: 10.1038/bjc.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamieson SM, et al. Evofosfamide for the treatment of human papillomavirus-negative head and neck squamous cell carcinoma. JCI Insight. 2018;3(16):e122204. doi: 10.1172/jci.insight.122204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatfield SM, et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med. 2015;7(277):277ra30. doi: 10.1126/scitranslmed.aaa1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delgoffe GM, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501(7466):252–256. doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.