Abstract
Treg differentiation, maintenance, and function are controlled by the transcription factor FoxP3, which can be destabilized under inflammatory or other pathological conditions. Tregs can be destabilized under inflammatory or other pathological conditions, but the underlying mechanisms are not fully defined. Herein, we show that inflammatory cytokines induce ER stress response, which destabilizes Tregs by suppressing FoxP3 expression, suggesting a critical role of the ER stress response in maintaining Treg stability. Indeed, genetic deletion of Hrd1, an E3 ligase critical in suppressing the ER stress response, leads to elevated expression of ER stress–responsive genes in Treg and largely diminishes Treg suppressive functions under inflammatory condition. Mice with Treg-specific ablation of Hrd1 displayed massive multiorgan lymphocyte infiltration, body weight loss, and the development of severe small intestine inflammation with aging. At the molecular level, the deletion of Hrd1 led to the activation of both the ER stress sensor IRE1α and its downstream MAPK p38. Pharmacological suppression of IRE1α kinase, but not its endoribonuclease activity, diminished the elevated p38 activation and fully rescued the stability of Hrd1-null Tregs. Taken together, our studies reveal ER stress response as a previously unappreciated mechanism underlying Treg instability and that Hrd1 is crucial for maintaining Treg stability and functions through suppressing the IRE1α-mediated ER stress response.
Keywords: Immunology
Keywords: Molecular biology, T cells, Ubiquitin-proteosome system
The ubiquitin ligase HRD1 maintains regulatory T cell integrity by suppressing inflammatory cytokine-induced IRE1a-p38 activation.
Introduction
During inflammation, multiple levels of physiological stress are encountered within the microenvironment, such as hypoxia, low pH, nutrient deprivation, microbial products, and inflammatory cytokines. The notion that such stresses can perturb the function of the ER and trigger ER stress has long been proposed (1, 2). In turn, ER stress is a major contributor to inflammatory diseases, such as Crohn’s disease and type 2 diabetes (3). However, it is not clear which immune cell types are involved in ER stress–induced inflammation. Tregs represent a distinct cell lineage among functional CD4+ T cell subsets and perform immune-suppressive rather than immune-effector actions (4). Modulation of Treg function may be a potential mechanism underlying ER stress–induced inflammation.
Expression of the transcription factor FoxP3 was determined to be the master regulator and marker of Tregs (5). CD4+FoxP3+ T cells derived from CD4+CD8+ in mature T cells in thymus are the natural Tregs (nTregs), and the peripheral conventional naive CD4+FoxP3– T cells can differentiate into CD4+FoxP3+ induced Tregs (iTregs) (6). Although nTregs and iTregs are generated at different sites, both have suppressive functions and are required to maintain immune homeostasis (4). Both the expression level and stability of the FoxP3 protein are essential for the differentiation and function of Tregs (7, 8). Diminished FoxP3 expression leads to Treg deficiency, which results in activation and expansion of autoreactive T cells and other innate immune cells, as well as increased production of IL-2 and a wide range of proinflammatory Th1, Th2, and Th17 cytokines (9, 10). It has been shown that proinflammatory cytokines and TLR agonist stimuli suppress FoxP3 expression to impair Treg suppressive functions during autoimmune inflammatory disease (11–13). However, the molecular mechanisms underlying how the proinflammatory cytokines and TLR agonist stimuli trigger the Treg instability are still not fully defined.
Hrd1 is an E3 ubiquitin ligase that localizes to the ER membrane. The best-characterized function of Hrd1 is to catalyze the degradation of misfolded/unfolded proteins, a process called ER-associated degradation (ERAD) (14, 15). Perturbations in ERAD leads to accumulation of misfolded/unfolded proteins within the ER, which triggers the unfolded protein response (UPR) and ER stress. Three major ER transmembrane proteins are involved in UPR pathways: PKR-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1α (IRE1α), and activating transcription factor 6 (ATF6) (16–18). IRE1α is the most highly conserved UPR sensor and functions as a protein kinase and endoribonuclease (RNase) (19, 20). Studies have shown important roles of Hrd1 in regulating both innate and adaptive immune responses both in physiological and pathological settings (21–23), but the role of Hrd1 in regulating the suppressive activities of Tregs is unknown.
We and others have shown that inflammatory cytokines and TLR signaling induces the ER stress response during inflammatory disease (1, 24). Together with the fact that both the inflammatory cytokines induce Treg instability (11–13), we speculated whether the inflammatory cytokines destabilize Tregs at least partially through ER stress response. In this study, we found that a lack of Hrd1 in Tregs triggered ER stress responses and reduced their capacity to execute their suppressive functions. At molecular level, the deletion of Hrd1 led to upregulation of the ER stress sensor IRE1α, and impaired FoxP3 expression caused by Hrd1 deletion was largely rescued by an ER stress inhibitor that was specific to IREα kinase activity. In conclusion, our findings indicate that Hrd1 is crucial for maintaining Treg properties by antagonizing the IRE1α-mediated ER stress.
Results
ER stress destabilizes FoxP3 in Tregs.
To investigate the effect of ER stress on Treg stability, we treated TGF-β–converted mouse iTregs with tunicamycin, a pharmacological ER stress inducer. After 3 days, FoxP3 expression was significantly reduced in the tunicamycin-treated group, and the loss of FoxP3 could be rescued by adding tauroursodeoxycholate (TUDCA), an ER stress inhibitor (Figure 1, A and B). The loss of FoxP3+ Tregs was not likely due to increased apoptosis because tunicamycin treatment did not further increase the fraction of fixable viability dye–positive (FVD+) cells (Figure 1C), suggesting that ER stress plays a critical role in maintaining FoxP3 stability.
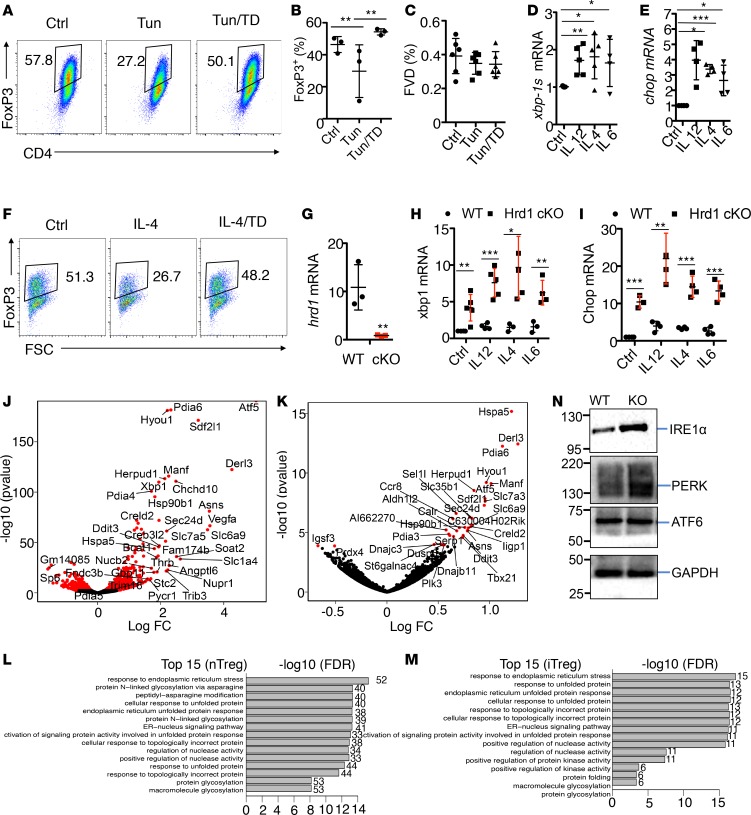
Figure 1. Hrd1 suppresses the ER stress response in Tregs.
(A–C) Mouse CD4 T cells were polarized into iTreg for 5 days and then treated with the ER stress inducer tunicamycin or tunicamycin/TUDCA in medium supplemented with 1 ng/ml IL-2. FoxP3 expression was analyzed by flow cytometry (n = 6–9 per group). Representative images (A) and statistical analysis of FoxP3 expression (B) and fixable viability dye (C) in Treg are shown. (D and E) CD4+FoxP3+ Tregs were sorted from the SPL and pLN by YFP expression. The cells then were treated with or without cytokines, including IL-12 (10 ng/ml), IL-4 (10 ng/ml), or IL-6 (50 ng/ml) for 10 hours. The mRNA levels of xbp-1s (D) and chop (E) were evaluated by qPCR analysis after 10 hours of treatment (n is at least 4 biological samples per group). (F) Tregs were cultivated with IL-4 or further with the ER stress inhibitor TD for 2 days; the FoxP3 expression levels were determined. (G) CD4+YFP+ Tregs were sorted, and Hrd1 mRNA in the Tregs from WT and Hrd1fl/flFoxP3Cre mice were measured (n = 5 per group). (H and I) Sorted WT and Hrd1fl/fl-FoxP3cre CD4+YFP+ Tregs were treated with anti-CD3 or anti-CD3 plus IL-12, IL-4, or IL-6 for 10 hours. The mRNA expression level of xbp1 (H) and chop (I) were analyzed by qPCR (n = 3–4 per group). (J and K) Volcano plot comparing the P value versus sorted CD4+YFP+ Tregs (J) and polarized CD4+YFP+ iTregs (K) from WT and Hrd1fl/fl-FoxP3cre mice. Genes labeled in red are ER stress–associated genes significantly differentially expressed between WT and Hrd1fl/fl-FoxP3cre Tregs. (L and M) GSEA was performed using the Gorilla bioinformatics tool for significantly upregulated genes in Hrd1fl/fl-FoxP3cre sorted YFP+ nTregs and Hrd1fl/fl-FoxP3cre polarized iTregs relative to WT. (N) CD4+ T cells were polarized into iTregs in vitro for 5 days. Protein expression levels in WT and Hrd1fl/fl-FoxP3cre iTregs were then analyzed by immunoblotting. Data shown as mean ± SD in B–E and G–I. *P < 0.05, **P < 0.01, and ***P < 0.005 by 2-tailed Student’s t test.
Since ER stress resulted in significant loss of FoxP3 protein expression and the notion that inflammatory cytokines can regulate Treg stability has long been proposed (25–27), we explored the impact of inflammatory cytokines on ER stress responses in Tregs. Indeed, the ER stress responsive genes including spliced form of Xbp-1 (Xbp1s, controlled by the ER stress sensor IRE1α) and CHOP (also known as Ddit3, controlled by PERK) (19, 20, 28–30) are both dramatically increased in Tregs when exposed to inflammatory cytokine treatment (Figure 1, D and E), suggesting that proinflammatory cytokines induce the ER stress response in Tregs. To support this conclusion, we further demonstrated that the ER stress inhibitor TUDCA (31, 32) largely rescued Tregs from the inflammatory cytokine IL-4–induced Treg depolarization (Figure 1F). Similarly, ER stress inhibitor TUDCA treatment largely diminished the inflammatory cytokine–induced expression of ER stress–responsive genes, including Hrd1 (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121887DS1). Together with the fact that the pharmacological ER stress inducer destabilized both human and mouse Tregs, our results suggest an important role of ER stress response in inflammatory cytokine–induced Treg destabilization.
Deletion of Hrd1 triggers the ER stress response in Tregs.
To further characterize the effect of ER stress on Treg function, we generated Treg-specific Hrd1-KO (Hrd1fl/fl-FoxP3cre) mice by breeding Hrd1 floxed mice with FoxP3YFP–Cre (FoxP3cre) transgenic mice. Quantitative PCR (qPCR) analysis confirmed the complete elimination of Hrd1 mRNA expression in Tregs from the Hrd1fl/fl-FoxP3cre mice (Figure 1G). As expected, genetic suppression of Hrd1 resulted in a dramatic upregulation of ER stress–responsive genes in Tregs (Figure 1, H and I) compared with WT Tregs when cocultivated with inflammatory cytokines including IL-12, IL-4, and IL-6. Further analysis indicated that the inflammatory cytokine treatment significantly induced ER stress–responsive genes, including Xbp-1s, Chop, and Hrd1 (Supplemental Figure 1B), indicating that the inflammatory cytokines induces ER stress response in Tregs and suggesting that the upregulated Hrd1 functions as a negative regulator to protect Tregs from inflammatory cytokine–induced instability. However, Hrd1 is not differentially expressed in each T cell subset (Supplemental Figure 1C). Together with our recent report that Hrd1 is required for optimal production of Th1 and Th17 cytokines, these results indicate that Hrd1 plays a diverted role in T cell immunity, including in maintaining Treg stability.
Our data show that Hrd1 is critical to suppress this upregulated ER stress response. To support this conclusion, our genome-wide transcriptome analysis further showed that, in both nTregs and iTregs from the Hrd1fl/fl-FoxP3cre mice, the most dramatic changes in gene expression were seen in ER stress–related genes (Figure 1, J and K). We then performed gene-set enrichment analysis to identify key networks regulated by Hrd1. Notably, all of the top 15 upregulated gene sets in nTregs and iTregs from Hrd1fl/fl-FoxP3cre mice were associated with ER stress pathways (Figure 1, L and M). ER stress induces the activation of 3 ER stress sensors: ATF6, PERK, and IRE1α. We found that the protein levels of PERK and IRE1α, but not ATF6, are upregulated in Hrd1fl/fl-FoxP3cre iTregs (Figure 1N). These results indicate that the Hrd1 suppresses the ER stress response in Tregs.
Treg-specific deletion of Hrd1 precipitates inflammatory disease in aged mice.
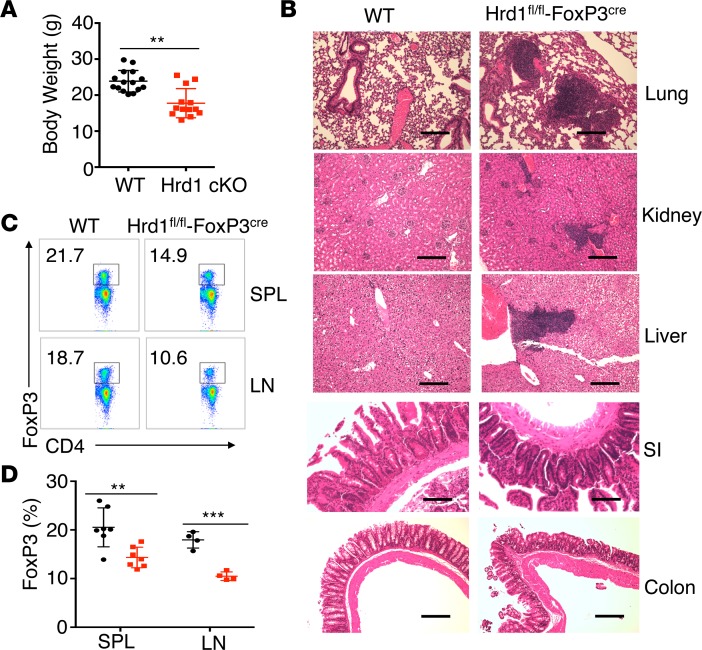
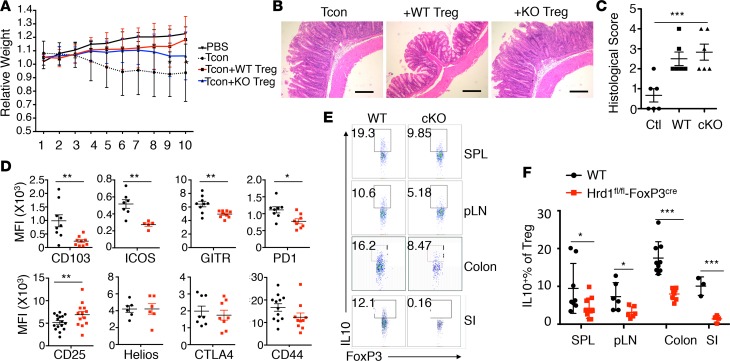
While Treg-specific Hrd1 gene deletion did not affect the growth of mice at younger ages, Hrd1fl/fl-FoxP3cre mice showed a markedly lower body weight compared with WT starting at 4–5 months of age (Figure 2A), which is likely due to the development of spontaneously inflammatory disease as characterized by increased lymphocyte infiltration in multiple organs — including lung, kidney, liver, and colon — because the increased lymphocytes infiltration was observed in the tissue sections from Hrd1-null but not WT mice (Figure 2B). Analysis of the spleen (SPL) and peripheral lymph nodes (pLN) of old Hrd1fl/fl-FoxP3cre mice showed a notable decrease in the percentage of CD4+FoxP3+ Tregs (Figure 2, C and D). Therefore, loss of Hrd1 in Tregs resulted in multiorgan inflammation in aged mice, indicating that Hrd1 is required for maintaining the Treg stability and suppressive functions to protect mice from inflammatory disease.
Figure 2. Hrd1fl/fl-FoxP3cre mice developed age-related spontaneous multiorgan inflammation.
(A) Body weights of WT and Hrd1fl/fl-FoxP3cre mice at the age of 4–6 months (n = 13–15 per group). (B) Representative images of H&E staining of the lung, kidney, liver, small intestine (SI), and colon in WT and Hrd1fl/fl-FoxP3cre mice. Data are representative of 3 independent experiments with 2 pairs of WT and Hrd1fl/fl-FoxP3cre mice for each experiment. Scale bars: 100 μm. (C and D) Representative image (C) and frequencies of FoxP3 (D) in Tregs from the SPL and pLN in the aged WT and Hrd1fl/fl-FoxP3cre mice (n = 4–7 per group). Data are shown as mean ± SD in A and D. **P < 0.01 and ***P < 0.005 by 2-tailed Student’s t test.
Decreased FoxP3+ Treg frequency and stability in Hrd1fl/fl-FoxP3Cre mice.
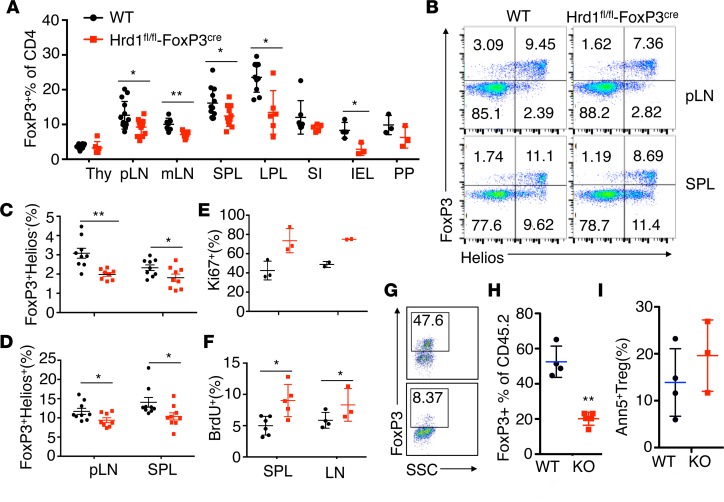
The fact that Hrd1fl/fl-FoxP3cre mice develop inflammatory disease with aging is likely due to the reduced Treg frequency in mice. To further confirm this, we analyzed the Treg frequency in 8- to 10-week-old WT mice and Hrd1fl/fl-FoxP3cre mice. Consistent with our observation in aged mice, the percentages of Tregs are significantly reduced in the pLN, mesenteric lymph nodes (mLN), SPL, large intestine lamina propria, and small intestine intraepithelial cells of the 8-week old Hrd1fl/fl-FoxP3cre mice (Figure 3, A and B). This result indicated that Hrd1 is required to maintain Treg homeostasis and/or stability. Using Helios as an intracellular marker to distinguish nTregs from iTregs (33), we found a lower frequency of both FoxP3+Helios– iTregs and FoxP3+Helios+ nTregs in the SPL and pLN of Hrd1fl/fl-FoxP3cre mice compared with WT mice (Figure 3, B–D). Neurophilin 1 (Nrp1) is another marker that can distinguish between nTregs and iTregs. Similarly, we observed that both Nrp1+FoxP3+ and Nrp1–FoxP3+ Tregs were slightly but significantly decreased in Hrd1-deficient Tregs compared with WT Tregs (Supplemental Figure 2, A and B), suggesting that Hrd1 plays an essential role in maintaining nTreg and iTreg populations.
Figure 3. Decreased FoxP3+ Treg frequencies and reduced FoxP3 stability in Hrd1fl/fl-FoxP3cre mice.
(A) Flow cytometry analysis and frequency of FoxP3+ Tregs in WT and Hrd1fl/fl-FoxP3cre young mice (n = 3–14 per group). Thy, thyroid; pLN, popliteal lymph node; mLN, mesenteric lymph node; SPL, spleen; LPL, lamina propria lymphocytes; SI-IEL, small intestinal intraepithelial lymphocytes; PP, Peyer’s patches. (B–D) Representative images and statistical analysis of Helios expression in the CD4+ cells from the pLN and SPL in WT and Hrd1fl/fl-FoxP3cre mice (n = 8–9 per group). (E and F) BrdU staining 24 hours after injection of BrdU (n = 4–6 per group) and Ki67 staining (n = 2–3 per group) in Tregs (CD4+FoxP3+) from the SPL and pLN of WT and Hrd1fl/fl-FoxP3cre mice. (G–I) CD4+CD25+YFP+ Tregs from CD45.2+ WT and Hrd1-cKO mice were sorted, mixed with CD4+ CD45.1 naive T cells, and adoptively transferred into RAG1-null mice. Eight weeks later, CD45.2+CD4+ Tregs were gated, and the expression of FoxP3 were analyzed (G and H). SSC, side scatter The survival of CD45.2 cells were analyzed by Annexin V staining (I) (n = 4 per group). Data are shown as mean ± SD. *P < 0.05 and **P < 0.01 by 2-tailed Student’s t test.
We previously observed that Hrd1-null conventional T cells showed markedly lower proliferation (23), suggesting a possibility that Hrd1-deficency limits Treg expansion. However, Tregs from WT and Hrd1fl/fl-FoxP3cre mice showed similar or even larger populations of Ki67+ cells and a significantly higher BrdU incorporation (Figure 3, E and F), indicating that the observed reduction in Tregs was not due to a defect in proliferation. We then used an adoptive transfer approach to determine whether Hrd1 is required to maintain FoxP3 expression. Naive CD4+CD25–CD44loCD62Lhi T cells were sorted and transferred to Rag1–/– mice alone or in combination with sorted Hrd1fl/fl-FoxP3cre Tregs or littermate control WT Tregs. Most Hrd1-deficient Tregs lost FoxP3 expression at 10 weeks after adoptive transfer (Figure 3, G and H). In contrast, the survival of Tregs in vivo was unaltered by Hrd1 deficiency (Figure 3I), indicating that Hrd1 is critical for maintaining FoxP3 stability, as well as suppressing their functions. This may contribute to the lower Treg populations observed in the SPL and pLN of Hrd1fl/fl-FoxP3cre mice (Figure 3, A and B), suggesting that Hrd1 is required to main FoxP3 expression under inflammatory conditions and consequently protect mice from inflammatory diseases.
However, when CD4+CD25+YFP+ Tregs were sorted from WT and Hrd1-null mice and cultivated with naive CD4 T cells, Hrd1 deficiency appears to have little effect on Treg suppressive functions (Supplemental Figure 3A), suggesting that Hrd1 maintains Treg stability under inflammatory conditions. Indeed, when the inflammatory cytokine IL-4 was added to the coculture, the suppressive activity of Hrd1-null Tregs was significantly reduced (Supplemental Figure 3, B and C). To further support this conclusion, in contrast to the fact that most Hrd1-null Tregs lost FoxP3 when adoptively transferred into lethally irradiated WT mice with naive CD4 T cells that develop colitis (Figure 3, G and H), cotransfer WT and Hrd1-null Tregs without naive CD4 T cells largely rescued Hrd1-null Tregs from losing FoxP3 expression (Supplemental Figure 3, D–G).
Increased T cell activation and altered immune homeostasis in Hrd1fl/flFoxP3Cre mice.
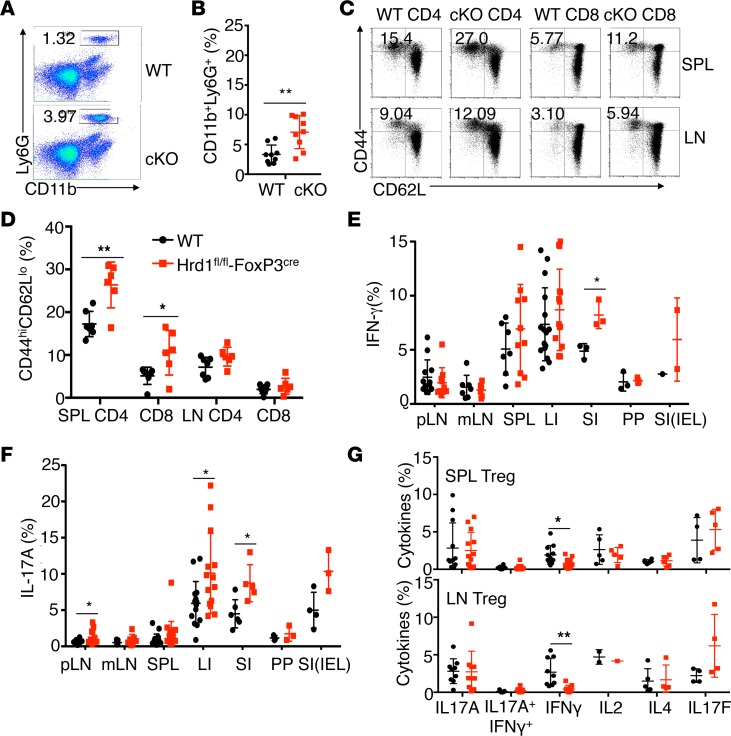
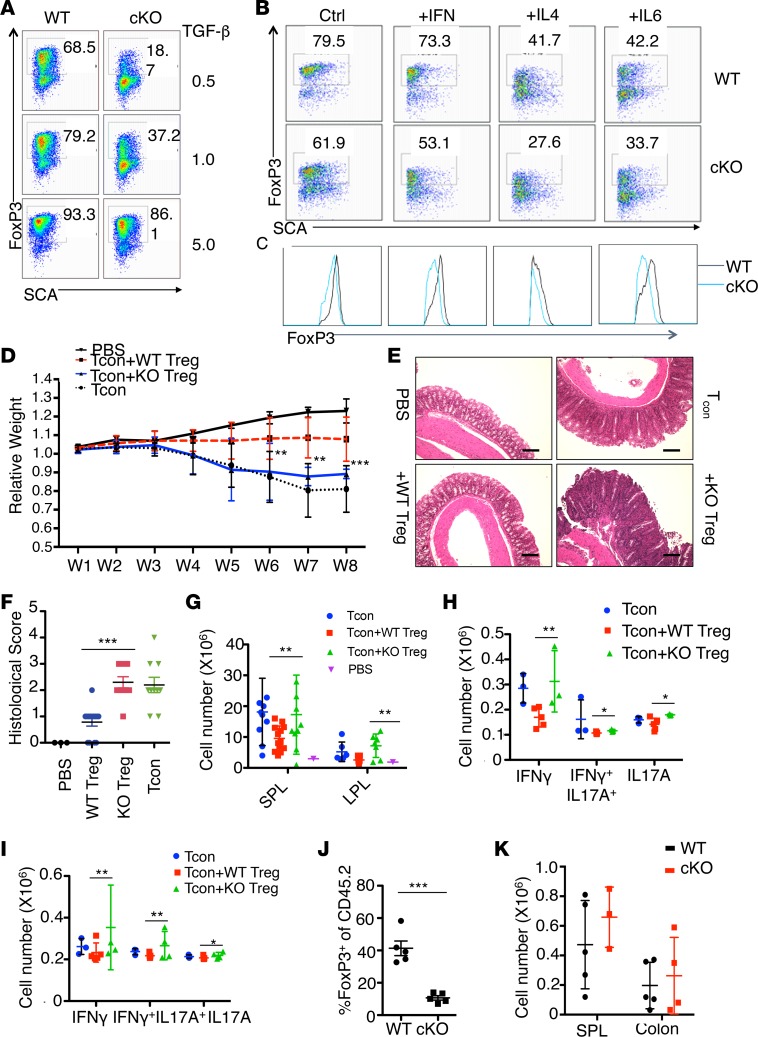
The reduction of FoxP3+ Tregs upon Hrd1 gene deletion prompted us to examine whether immune homeostasis was altered in Hrd1fl/fl-FoxP3cre mice. Analysis of the thymocytes of Hrd1fl/fl-FoxP3cre and WT mice revealed comparable percentages and numbers of CD4+CD8–, CD4–CD8+, and CD4+CD8+ thymocytes (Supplemental Figure 2C). The percentages of CD3+, CD4+, and CD8+ T cells; B (B220+) cells; and conventional DCs (CD11c+MHCII+) in the peripheral lymphoid organs, as well as serum antibody levels, were largely similar between WT and Hrd1fl/fl-FoxP3cre mice (Supplemental Figure 2, D–G). However, Hrd1fl/fl-FoxP3cre mice exhibited a higher percentage of neutrophils (CD11b+Ly6G+) in the SPL (Figure 4, A and B). In addition, compared with WT mice, Hrd1fl/fl-FoxP3cre mice had a significantly higher percentage of CD44hiCD62Llo cells in the CD4+ and CD8+ compartments (Figure 4, C and D) in the SPL, suggesting that T cells from Hrd1fl/fl-FoxP3cre mice were spontaneously activated in vivo. Moreover, CD4+ T cells from the small intestine lamina propria of Hrd1fl/fl-FoxP3cre mice produced significantly larger amounts of IFN-γ. More IL-17A–producing CD4 T cells were detected in the pLN, large and small intestine lamina propria, and intraepithelial lymphocytes in Hrd1fl/fl-FoxP3cre mice compared with WT counterparts (Figure 4, E and F). These results suggest that Hrd1-deficient FoxP3+ Tregs are functionally defective, thereby disrupting immune homeostasis and permitting inflammation. Examination of cytokine production by CD4+FoxP3+ Tregs in the SPL and pLN of Hrd1fl/fl-FoxP3cre mice revealed no increase in the production of IL-17A and IL-4, and an even lower IFN-γ production compared with WT Tregs (Figure 4G), indicating that Hrd1-deficient Tregs do not convert into T effector cells.
Figure 4. Increased T cell activation and altered immune homeostasis in Hrd1fl/fl-FoxP3cre mice.
(A and B) Representative images (A) and quantification (B) of neutrophils in the SPL in WT and Hrd1fl/fl-FoxP3cre mice (n = 8–9 per group). (C and D) Representative images (C) and quantification (D) of the expression of CD62L and CD44 on WT and Hrd1fl/fl-FoxP3cre splenic and LN CD4+ and CD8+ T cells (n = 6 per group). (E and F) Expression of IFN-γ+(E) and IL-17A+ (F) in CD4+ T cells from WT and Hrd1fl/fl-FoxP3cre mice (n = 3–16 per group). (G) Expression of Th subset cytokines in the SPL and LN Tregs from WT and Hrd1fl/fl-FoxP3cre (n = 2–13 per group). Data are shown as mean ± SD. *P < 0.05 and **P < 0.01 by 2-tailed Student’s t test.
Loss of Treg properties in Hrd1fl/fl FoxP3Cre mice.
Our in vitro suppressive assay shows that the suppressive functions of Hrd1-null Tregs are largely intact, at least within a short cocultivation period (3 days), but are largely diminished when inflammatory cytokines were added (Supplemental Figure 3), suggesting that Hrd1 is critical to maintain Treg suppressive functions under inflammatory conditions. To investigate the effect of Hrd1-deficiency on the suppressive function of Tregs specifically, we performed an in vivo suppressive assay. Naive CD4+CD25–CD44loCD62Lhi T cells were sorted and transferred to Rag1–/– mice alone or in combination with an equal number of sorted CD4+CD25+YFP+ Tregs from either Hrd1fl/fl-FoxP3cre mice or their littermate controls. Transfer of naive CD4+CD25–CD44loCD62Lhi alone to Rag1–/– mice induced T cell–mediated colitis. Hrd1-deficient Tregs failed to prevent the loss of body weight caused by naive T cells in Rag1–/– mice (Figure 5A). Histological examination of the large intestine indicated that lymphocyte infiltrates were increased and showed mucosal hyperplasia in mice with adoptively transferred Hrd1-deficient Tregs, similar to what was observed in mice that received naive T cells (Figure 5, B and C).
Figure 5. Reduced Treg suppressive function and altered Treg signature profiles Hrd1fl/fl-FoxP3cre mice.
(A) Colitis progression was assessed by body weight loss (n = 10 per group). (B and C) Representative images of the large intestine after H&E staining (B) and histological analysis (C) 10 weeks after adoptive transfer. Scale bars: 100 μm. Data are representative of 2 independent experiments with 5 mice per group in each experiment. (D) Expression of Treg-specific surface markers in thymic Tregs from WT and Hrd1fl/fl-FoxP3cre mice (n = 5–13 per group). (E and F) Representative images and quantification of IL-10–producing Tregs from WT and Hrd1fl/fl-FoxP3cre mice (n = 4–10 per group). Data are shown as mean ± SD. *P < 0.05; **P < 0.01; and ***P < 0.005 by 2-tailed Student’s t test.
To gain the insight into the reduced immune suppressive function of Hrd1-deficient Tregs in vivo, we first examined the expression of Treg-specific molecules. We found that the expression of CD103, ICOS, GITR, and PD1 were significantly reduced in the thymic Tregs (Figure 5D). Interestingly, expression of the Treg marker CD25 was upregulated in Hrd1fl/fl-FoxP3cre thymic Tregs. CD4+FoxP3+Tregs in the SPL and pLN in Hrd1fl/fl-FoxP3cre mice also showed higher expression of CD25 but not other markers (Supplemental Figures 4 and 5). These observations suggested that CD4+FoxP3+ Tregs have an activated phenotype in the absence of Hrd1, with a corresponding reduction in the FoxP3+CD25– population (Supplemental Figure 6). One of the crucial mechanisms by which Tregs exert their immune suppressive functions is through upregulation of immunomodulatory cytokine production, such as IL-10 and TGF-β (34). Analysis of cytokine production by CD4+FoxP3+ Tregs from Hrd1fl/fl-FoxP3cre mice showed a notable decrease in IL-10 production (Figure 5, E and F), but not TGF-β, in the SPL, pLN, and small and large intestine lamina propria. Thus, these data indicate that ablation of Hrd1 results in activation of CD4+FoxP3+ Tregs and decreased IL-10 production, and they indicate that Hrd1 is essential for maintaining Treg immune suppressive functions.
Hrd1 is required for TGF-β–mediated conversion of iTregs and their stability and function. To further understand the effect of Hrd1 deficiency on iTregs, we examined FoxP3 expression in both WT and Hrd1fl/fl-FoxP3cre naive CD4+CD25– T cells after incubation with different concentrations of TGF-β. Lower concentrations of TGF-β were unable to induce FoxP3 expression in Hrd1fl/fl-FoxP3cre iTregs; however, increasing the concentration of TGF-β rescued FoxP3 expression (Figure 6A), suggesting an important role of Hrd1 in TGF-β–mediated Treg differentiation. We next examined the stability of the in vitro converted iTregs with higher concentration of TGF-β (5 ng/ml) by adding the proinflammatory cytokines to the culture medium. Addition of IL-12, IL-4, and IL-6 severely compromised FoxP3 expression in Hrd1fl/fl-FoxP3cre Tregs by significantly reducing both the percentages of FoxP3+ Tregs and FoxP3 mean fluorescent intensity (MFI) (Figure 6, B and C), whereas in WT Tregs, only a modest loss of FoxP3 was observed in the presence of these cytokines. Thus, Hrd1 protected Tregs from inflammatory cytokine–induced depolarization.
Figure 6. Hrd1fl/fl-FoxP3cre mice exhibit impaired generation and function of TGF-β–converted iTregs.
(A) CD4+ T cells were isolated from WT and Hrd1fl/fl-FoxP3cre mice and polarized into iTreg in vitro with 0.5, 1, or 5 ng/ml of TGF-β for 5 days. The expression of FoxP3 was analyzed by flow cytometry. (B and C) The TGF-β–converted Tregs from WT and Hrd1fl/fl-FoxP3cre mice were treated with complete medium supplemented with 5 ng/ml IL-2, with/without inflammatory cytokines IFN-γ (100 ng/ml), IL-4 (10 ng/ml), and IL-6 (50 ng/ml) for 3 days. The percentages (B) and mean fluorescent intensity (MFI) (C) of FoxP3 were measured by flow cytometry. Data are representative of 3 independent experiments. (D) Colitis progress was assessed by body weight loss (n = 6–8 per group). (E and F) Representative images of large intestine after H&E staining (E) and histological analysis (F) 10 weeks after adoptive transfer (n = 3–8 per group). Scale bars: 100 μm. (G) The total number of CD45.1+ T cells in the SPL and large intestine were measured (n = 6–10 per group). (H and I) The absolute number of IFN-γ+IL-17A–, IFN-γ+IL-17A+, and IFN-γ–IL-17A+ cells differentiated from transferred naive cells (CD45.1+) in the large intestine and SPL were calculated (n = 3–5 per group). (J) Expression level of FoxP3 from transferred Tregs (CD45.2+) in the SPL was analyzed by intracellular staining (n = 3–5 per group). (K) The absolute number of CD45.2+ cells in the SPL and large intestine were calculated (n = 3–5 per group). Data are shown as mean ± SD. *P < 0.05; **P < 0.01; and ***P < 0.005 by 2-tailed Student’s t test.
To further investigate the effect of Hrd1 deficiency on the biological function of iTregs, we performed an in vivo suppressive assay. We used Hrd1-deficient iTregs by sorting TGF-β–polarized (5 ng/ml) CD4+YFP+ cells, transferred with T naive cells into Rag1-deficient mice. In contrast to the full protective effect of WT iTregs, Hrd1-deficient iTregs failed to prevent the loss of body weight and led to increased gut lymphocyte infiltrates in mice after adoptive transfer (Figure 6, D–F). Further analysis showed that Hrd1-deficient Tregs were unable to inhibit the proliferation of naive T cells and induction of cytokine-producing T cells, particularly IFN-γ+, IL-17A+, and IFN-γ+L-17A+ cytokines in the SPL and large intestine (Figure 6, G–I), indicating that Hrd1-null Tregs failed to suppress the gut inflammation in mice.
Notably, in contrast to the fact that about 43% of WT Tregs were able to maintain FoxP3 expression at 10 weeks after adoptive transfer, less than 7% Hrd1-null Tregs were FoxP3+ (Figure 6J). Importantly, the numbers of CD45.2+ cells were similar in the recipients that received either WT or Hrd1-null CD45.2+ Tregs (Figure 6K). Together with the fact that nether ER stress inducer nor Hrd1 gene deletion enhanced Treg apoptosis (Figure 1C and Figure 3I), our results demonstrated that Hrd1-deficient Tregs have defects in suppressive activity in vivo due to loss of FoxP3 expression rather than cell apoptosis.
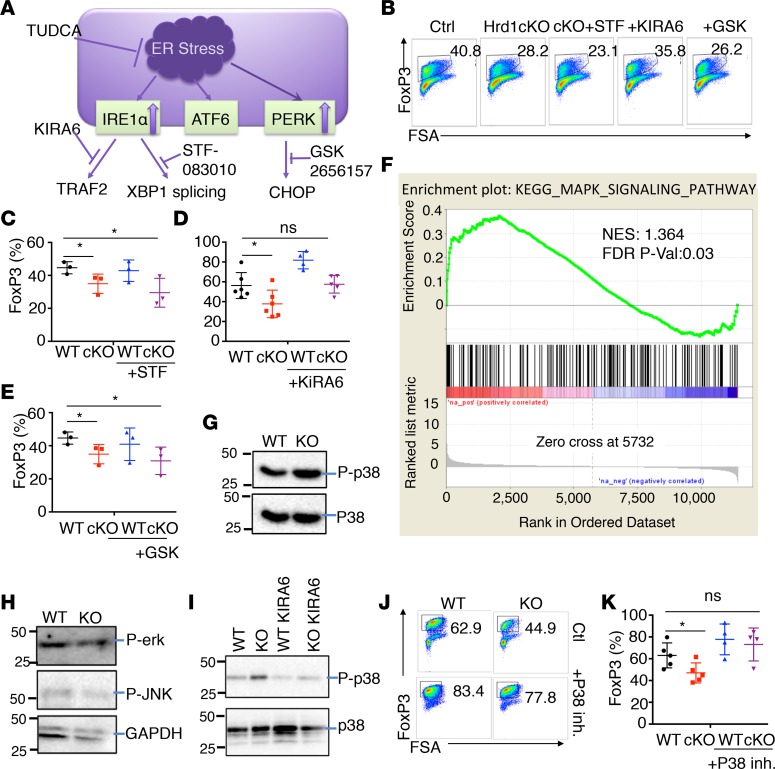
The IRE1αkinase pathway of the ER stress response is most important for FoxP3 stability. Our immunoblotting analysis revealed that Hrd1 deficiency led to the upregulation of IRE1α and PERK, but not ATF6, ER stress pathways (Figure 1M). To investigate whether elevated IRE1α and PERK activation contribute to the loss of TGF-β–mediated expression of FoxP3 in Hrd1fl/fl-FoxP3cre Tregs (Figure 7A), we treated Hrd1-deficient Tregs with the IRE1α kinase inhibitor KIRA6 (35), the IRE1α RNase inhibitor STF-083010 (N-[(2-hydroxynaphthalen-1-yl) methylidene] thiophene-2-sulfonamide) (36), or the PERK inhibitor GSK2656157 (37). The impairment in TGF-β–induced (1ng/ml) FoxP3 expression in Hrd1-deficient Tregs was rescued by treatment with KIRA6, but not STF-083010 or GSK2656157 (Figure 7, B–E), indicating that the IRE1α kinase — but not its RNase activity — during the ER stress response is involved in inducing FoxP3 expression in Tregs. The IRE1α kinase domain is important for TRAF2 binding and further activates mitogen-activated protein kinase kinase kinase (MKKK) apoptosis signal regulating kinase 1 (ASK1) (38, 39), while the IRE1α RNase domain splices a 26-nucleotide intronic sequence from the xbp1 mRNA (40). Signaling pathways downstream of ASK1 have been shown to involve the phosphorylation and activation of MKK4/MKK7 and MKK3/MKK6, the upstream kinases for JNK and p38, respectively (41). Gene set enrichment analysis (GSEA) revealed significantly upregulated MAPK signaling (Figure 7F) but unaltered NF-κB activation (Supplemental Figure 7) in Hrd1fl/fl-FoxP3cre Tregs, suggesting that the MAPK pathway triggered by ER stress might contribute to the observed phenotypes in Hrd1fl/fl-FoxP3cre Tregs. Analysis of the activation of downstream MAPKs, p-Erk, p-JNK, and p-p38 revealed that p-p38, but not its total protein expression, was significantly upregulated in Hrd1fl/fl-FoxP3cre Tregs compared with WT Tregs. In contrast, the levels of p-Erk and p-JNK were not altered by Hrd1 deficiency in Tregs (Figure 7, G and H), indicating that the increase in p38 activation is likely responsible for impaired Treg differentiation upon Hrd1 gene deletion. Hrd1 deletion–induced p-p38 upregulation could be blunted by treatment with the IRE1α kinase inhibitor KIRA6 in iTregs (Figure 7I), suggesting that increased IRE1α kinase activity is responsible for activation of p-p38. To support this notion, either shRNA-mediated p38 knockdown or addition of a p38-specific inhibitor largely rescued the loss of FoxP3 in Hrd1fl/fl-FoxP3cre Tregs (Figure 7, J and K, and Supplemental Figure 8). In contrast, neither JNK- nor Erk-specific inhibitor treatment rescued Hrd1-null Tregs (Supplemental Figure 9). Collectively, our results indicate that Hrd1 suppresses the IRE1α/p38 pathway to promote FoxP3 expression in Tregs.
Figure 7. The IRE1α kinase branch of the ER stress response was most important for expression of FoxP3.
(A) Schematic of ER stress and unfolded protein responses. (B–E) Under TGF-β 1 ng/ml Treg polarization, we added TUDCA (100 μM), Tunicamycin (0.5 M), IRE1α kinase inhibitor KIRA6 (1 μM), IRE1α endonuclease inhibitor STF-083010 (10 μM), PERK inhibitor GSK2656157 (0.5 μM), and (J and K) p38 MAPK inhibitor SB230580 (10 M) for the first 2 days and then changed to normal medium on day 3. Representative image and statistical analysis of FoxP3 expression after adding ER stress inhibitors into Treg polarization medium (n = 3–6 per group). (F) MAPK pathway GSEA of WT and Hrd1fl/fl-FoxP3cre TGF-β–converted iTregs. (G and H) The expression levels of p-p38, p-Erk, and p-JNK were analyzed by immunoblotting. Data are representative of 3 independent experiments. (I) The expression levels of p-p38 were analyzed after adding IRE1α kinase inhibitor KIRA6. (J and K) Representative image (J) and statistical analysis (K) of FoxP3 expression after adding p38 protein inhibitor into Treg medium (n = 4–5 per group). Data are shown as mean ± SD. *P < 0.05 by Student’s 2-tailed t test.
Discussion
It is well established that Tregs can often be destabilized under inflammatory or other pathological conditions, leading to diminished suppressive functions, but the molecular mechanism underlying how the inflammatory cytokine stimuli triggers Treg instability remain unclear. Our results define a critical role of Hrd1 in suppressing ER stress–induced Treg depolarization under inflammation conditions. This conclusion is supported by the following observations: (a) inflammatory cytokines trigger ER stress response in Tregs, which is further enhanced by genetic Hrd1 deletion; (b) pharmacological ER stress inducer facilitates Treg depolarization; (c) mice with Treg-specific Hrd1 gene deletion develop multiple organ inflammation; (d) Hrd1 deletion in Tregs leads to the loss of FoxP3 expression and suppressive functions in vivo; (e) Hrd1 deletion results in the elevated IRE1α and PERK expression; and (f) Hrd1-null naive CD4 T cells are resistant to TGF-β–induced Treg polarization, which is largely rescued by IRE1α kinase inhibitor but not the inhibitors to IRE1α endoribonuclease activity or PERK.
While our genome-wide analysis detected a significant increase in a variety of ER stress–responsive genes, Hrd1-mediated suppression of IRE1α kinase but not its endoribonuclease activation plays a dominant role in maintaining Treg stability. Previous work by our laboratory indicated that Hrd1 targets IRE1α for ubiquitination and degradation in the synovial fibroblasts (42). We speculate that this specific targeted suppression of IRE1α function might also occur in Tregs. Indeed, a significant increase in IRE1α protein expression was detected in Hrd1-null Tregs. The elevated IRE1α in Hrd1-null Tregs appears to facilitate Treg instability through activating the downstream MAP kinase p3, because IRE1α kinase inhibitor largely diminished the increased p38 activation. More importantly, the p38-specific inhibitor or shRNA-mediated p38 knockdown fully rescued FoxP3 expression in Hrd1-null Tregs.
Given the fact that T cells from Hrd1-deficient mice were hyporesponsive to TGF-β treatment and that TGF-β–converted Hrd1fl/fl-FoxP3cre iTregs were less suppressive under inflammation conditions, we suspect that TGF-β signaling might be compromised upon Hrd1 deletion. We did not observe a change in TGF-β target gene expression by RNA sequencing (RNA-seq) analysis (Supplemental Figure 5), suggesting that Hrd1 might regulate TGF-β signaling in a noncanonical way. In addition, p38 signaling has been reported to associate with TGF-β–mediated conversion of CD4+CD25– T cells into Tregs and that loss of p38 redundant isoforms in T cells upregulates FoxP3 expression (43). Similarly, our findings suggest that the signal strength of the p38 signaling pathway is critical for TGF-β–induced FoxP3 expression.
We demonstrated that both Helios+ nTreg and Helios– iTreg frequencies were reduced significantly in Hrd1-deficient mice, indicating that Hrd1 is crucial for maintaining not only TGF-β–derived iTregs, but also thymus-derived nTregs, though we did not see a reduction in the Treg population in the thymus. As expression of CD103 is downregulated while CD62L is upregulated in Hrd1fl/fl-FoxP3cre Tregs, it is possible that Hrd1-deficient Tregs fail to appropriately migrate and accumulate in the SPL, LN, and gut. We also observed a reduction in the suppressive capabilities in both Hrd1fl/fl-FoxP3cre nTregs and iTregs. Significantly reduced antiinflammatory IL-10 production might contribute to the decrease in immune suppressive function of Tregs from Hrd1fl/fl-FoxP3cre mice. A previous study showed that IL-10 produced by FoxP3+ Tregs controls mature Th17 and Th17+Th1+ cells but not Th1 cells (44). Consistent with this, we found increased IL-17A, but not IFN-γ, production in the pLN and small and large intestines in Hrd1fl/fl-FoxP3cre mice. However, under T cell–mediated colitis setting, we observed that Hrd1fl/fl-FoxP3cre Tregs could not control inflammation in colitis with increased IL-17A+–, IL-17A+IFN-γ+–, and IFN-γ+–producing cells (Figure 7, G and H). These data highlight the importance of accessing Treg functionality outside of the intact steady state in animals.
Several E3 ubiquitin ligases have been found to regulate the differentiation and function of Tregs — for example, Grail, Cbl-b, Itch, Stub1, and Vhl (25, 45–48). Here, we found an additional E3 ligase, Hrd1, that is crucial for maintaining the stability and function of Tregs. Several proteins have been identified as substrates for Hrd1, with p27 as the most important in T cells (23). We previously showed that Hrd1 appears to positively regulate T cell clonal expansion by targeting p27, the cyclin-dependent kinase inhibitor, for ubiquitination and degradation (23). Given that Hrd1 is upregulated upon TCR stimulation and T conventional cells and Tregs develop from the same pool of thymocyte progenitor cells, it will be important to determine how engagement of the same E3 ligase in these 2 related cell types execute such different mechanisms. In addition, Hrd1 is not differentially expressed in each T cell subset, including Th1, Th2, Th17, and Tregs. Together with our recent report that Hrd1 is required for optimal production of Th1 and Th17 cytokines, these results indicate that Hrd1 plays a diverted role in T cell immunity, including in maintaining Treg stability. Therefore, it is likely that Hrd1 functions in a cell type–specific manner and the microenvironment of each cell type determines the specificity of Hrd1 target proteins and their interactions.
In short, our study uncovered a key role of the E3 ubiquitin ligase Hrd1 in mediating Treg expression, function, and stability. We demonstrated that Hrd1 deletion leads to upregulation of the UPR and leads to reduced Treg stability and function. Our findings reveal a new link between Hrd1 and inflammatory diseases involving ER stress, such as Crohn’s disease, ulcerative colitis, obesity, and type 2 diabetes.
Methods
Animals.
The Hrd1-floxed mice were used as reported (21) and the Treg-specific Hrd1-deficient mice (Hrd1fl/fl-FoxP3cre) were generated by breeding Hrd1-floxed mice with FoxP3YFP-Cre mice, whose YFP expression under the FoxP3 promoter allows us to sort CD4+CD25+YFP+ Tregs (The Jackson Laboratory, 016959). The littermates of either Cre-positive Hrd1+/+ or Hrd1fl/+ were used as WT controls. Mice were maintained at the Northwestern University mouse facility under pathogen-free conditions according to institutional guidelines.
Real-time RT PCR analysis.
Tregs were treated with or without recombinant inflammatory cytokines including IL-12 (catalog 577002), IL-4 (catalog 574302), and IL-6 (catalog 575702) (BioLegend) or further with inhibitors including ER stress inhibitor sodium TUDCA (catalog T-0682, MilliporeSigma), IRE1α inhibitors KIRA6 (catalog 6166) and STF-083010 (catalog 4509) (Tocris Bioscience), PERK inhibitor GSK2656157 (catalog 5046510, MilliporeSigma), p38 inhibitor SB203580 (catalog 559389), JNK inhibitor SP600125 (catalog 420119), and Erk inhibitor II FR180204 (catalog 328007) (MilliporeSigma). Total RNA from the treated Tregs were isolated using Trizol (catalog 15596018, Thermo Fisher Scientific). The levels of each indicated genes were determined by real-time RT-PCR as recently reported(49–51).
Western blotting analysis.
WT and Hrd1-null Tregs, ether sorted or polarized, were lyzed with RIPA cell lysis buffer (Bio-Rad), which was freshly added with protease inhibitor cocktail and then were boiled at 95°C in 20 μl of 2× Laemmli’s buffer. Samples were analyzed on an 8–12% SDS-PAGE gel and electrotransferred onto polyvinylidene difluoride membranes (MilliporeSigma). After blocking with 4% milk dissolved in TBST (tris-buffered saline, 0.1% Tween 20), membranes were probed with the appropriate primary antibodies. After washing twice with TBST, membranes were incubated with horseradish peroxidase–conjugated secondary antibodies, including IRE1 (catalog ab37073), PERK (catalog ab65142), p38 (catalog ab170099), phosphor-p38 (catalog ab16587), JNK1 (catalog ab199380), p-JNK (catalog ab47337), Erk (catalog ab32537), p-Erk (catalog ab176660), and GAPDH (catalog ab181602) (all from Abcam). After washing twice with TBST, membranes were treated with enhanced chemiluminescence (ECL) detection system (GE Healthcare) and exposed. If necessary, membranes were stripped with stripping buffer (Bio-Rad), washed, blocked, and then reprobed with other antibodies.
Flow cytometry.
Single-cell suspensions were used for staining using fluorescent-labeled specific antibodies against CD3 (clone 17A2), CD4 (clone GK 1.5), CD8 (clone 53-6.7), CD25 (clone PC61), CD44 (clone IM7), CTLA4 (clone UC10-4B9), PD-1 (clone 29F or 1A12), CD103 (clone 2E7), ICOS (clone 7E or 17G9), Helios (clone 22F6), Nrp1 (clone 3E12) from Biolegend as well as FoxP3 (clone FJK-16s) and CD62L (clone MEL-14) from eBioscience. For intracellular cytokine staining, cells were stimulated with PMA (10 ng/ml, Sigma-Aldrich), ionomycin (1 μg/ml, Sigma-Aldrich), and Golgi-Blocker Monensin (10 μg/ml, Biolegend) for 4–6 hours.
Mouse in vitro Treg differentiation.
Naive CD4 T cells were isolated and stimulated with plate-bound anti-CD3 (3 μg/ml) plus anti-CD28 (5 μg/ml) and polarizing cytokines (Tregs; TGF-β 0.1, 1, or 5 ng/ml; IL-2 5 ng/ml; anti–IFN-γ 2 ng/ml; and anti–IL-4 2 ng/ml) for 5 days. The expression of FoxP3 was analyzed by intracellular staining and flow cytometry.
Histology.
Mouse tissues were fixed in 10% formalin overnight and washed with 70% ethanol the next day. After the 70% ethanol wash, samples were immediately sent to Northwestern University Histology Core facility and stained with H&E. Scores were assigned according to the infiltrate lymphocytes by blinded assessment of at least 3 sections of each tissue per mouse.
RNA-seq.
FoxP3+CD4+ Tregs from WT and Hrd1fl/fl-FoxP3cre mice were sorted by YFP expression. After 5 days of in vitro Treg polarization, iTregs were sorted by YFP expression and antibody staining with anti-CD4. Three biological replicates of each population were sorted. Total RNA was extracted with RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen) and sent to NUseq Core (Northwestern University) for RNA-seq analysis.
Treg in vivo suppressive assay.
Naive CD4 T cells were sorted in the Northwestern flow cytometry center, with the marker (CD4+CD25–CD44loCD62Lhi) from SJL mice, which have congenic marker CD45.1. Tregs from the C57/BL6 genetic background (CD45.2) WT and Hrd1fl/fl-FoxP3cre mice were sorted by CD4+FoxP3+ YFP+. For in vitro polarized Tregs, YFP+ cells were sorted after 5 days polarization. Naive T cells (0.5 million) and Tregs (0.25 million) were mixed and adoptively transferred into Rag–/– mice through i.p. injection. Mice were then monitored on a weekly basis for weight changes and signs of disease.
Apoptosis and proliferation assay.
For analysis of apoptosis, splenocytes from WT and Hrd1fl/fl-FoxP3cre mice were stained with apoptosis marker Annexin 5 (BioLegend). In order to assess Treg proliferation in vivo, WT and Hrd1fl/fl-FoxP3cre mice were administered 2 mg BrdU by i.p. injection 24 hours before analysis, following the protocol provided by the manufacturers from eBioscience. Cells then were stained with anti-BrdU and further analyzed by flow cytometry to detect the incorporation of BrdU.
Statistics.
All data are indicated as mean ± SD. The Student’s two-tailed t test at a setting of “Unpaired t test with Welch’s correction. Do not assume equal SDs” was used for the statistical analysis. P < 0.05 was considered as a significant difference. *P < 0.05, **P < 0.01, and ***P < 0.001 (unpaired t test). Mean ± SD.
Study approval.
All the studies using animal study proposals were approved by the IACUC at Northwestern University. Both sexes of the mice were used, and littermates were used as controls.
Author contributions
YX, ZS, SML, and DF contributed to the design and implementation of all experiments and writing of the manuscript. JMC and HJ contributed to RNA-seq experiments and bioinformatics analyses. Yana Zhang, BG, JW, EM, IG, and Yusi Zhang contributed to genotyping and immunoblot experiments.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 grants (AI079056, AI108634, and AR006634) to DF, as well as The Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility (supported by the grants CA060553 and NIH 1S10OD011996-01) and the NUseq Core and Northwestern Mouse Histology and Phenotyping Laboratory (MHPL) (supported by NCI P30-CA060553). This work was also partially supported by the Basic Science Research Program (grant no. 2015R1D1A1A01057465) through the National Research Foundation funded by the Korean government to SML. We thank Liang Zhou’s lab for the reagents and technical expertise in gut lymphocyte isolation. We also thank Melissa Brown, Stephen Miller, and Chyung-Ru Wang for providing reagents, critical reading of the manuscript, and constructive suggestions during our research.
Version 1. 03/07/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(5):e121887. https://doi.org/10.1172/jci.insight.121887.
Contributor Information
Yuanming Xu, Email: T8J8Q6@u.northwestern.edu.
Johanna Melo-Cardenas, Email: jmelo@u.northwestern.edu.
Yana Zhang, Email: yana.zhang@northwestern.edu.
Isabella Gau, Email: IsabellaGau2017@u.northwestern.edu.
Juncheng Wei, Email: juncheng.wei@northwestern.edu.
Elena Montauti, Email: ElenaMontauti2015@u.northwestern.edu.
Yusi Zhang, Email: zhangyusi_scu@163.com.
Beixue Gao, Email: gaob@northwestern.edu.
Hongjian Jin, Email: hongjianjin@gmail.com.
Zhaolin Sun, Email: fangdavid68@gmail.com.
Sang-Myeong Lee, Email: leesangm@jbnu.ac.kr.
Deyu Fang, Email: fangd@northwestern.edu.
References
- 1.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettigole SE, Glimcher LH. Endoplasmic reticulum stress in immunity. Annu Rev Immunol. 2015;33:107–138. doi: 10.1146/annurev-immunol-032414-112116. [DOI] [PubMed] [Google Scholar]
- 3.Garg AD, Kaczmarek A, Krysko O, Vandenabeele P, Krysko DV, Agostinis P. ER stress-induced inflammation: does it aid or impede disease progression? Trends Mol Med. 2012;18(10):589–598. doi: 10.1016/j.molmed.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Kim JM, Rudensky A. The role of the transcription factor Foxp3 in the development of regulatory T cells. Immunol Rev. 2006;212:86–98. doi: 10.1111/j.0105-2896.2006.00426.x. [DOI] [PubMed] [Google Scholar]
- 5.Rudensky A. Foxp3 and dominant tolerance. Philos Trans R Soc Lond B Biol Sci. 2005;360(1461):1645–1646. doi: 10.1098/rstb.2005.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12(3):157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 7.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445(7129):766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 8.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8(3):277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 9.Kanangat S, et al. Disease in the scurfy (sf) mouse is associated with overexpression of cytokine genes. Eur J Immunol. 1996;26(1):161–165. doi: 10.1002/eji.1830260125. [DOI] [PubMed] [Google Scholar]
- 10.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 11.Valencia X, Stephens G, Goldbach-Mansky R, Wilson M, Shevach EM, Lipsky PE. TNF downmodulates the function of human CD4+CD25hi T-regulatory cells. Blood. 2006;108(1):253–261. doi: 10.1182/blood-2005-11-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurence A, et al. STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity. 2012;37(2):209–222. doi: 10.1016/j.immuni.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y, et al. Inflammation negatively regulates FOXP3 and regulatory T-cell function via DBC1. Proc Natl Acad Sci USA. 2015;112(25):E3246–E3254. doi: 10.1073/pnas.1421463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner RG, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151(1):69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2(7):379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 16.Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66(2 Suppl 1):S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J Clin Invest. 2009;119(10):3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73(6):1197–1206. doi: 10.1016/0092-8674(93)90648-A. [DOI] [PubMed] [Google Scholar]
- 20.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74(4):743–756. doi: 10.1016/0092-8674(93)90521-Q. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Qiu Q, Gao B, Kong S, Lin Z, Fang D. Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic cell MHCII expression for CD4 T cell priming during inflammation. J Exp Med. 2014;211(12):2467–2479. doi: 10.1084/jem.20140283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue T, Tsai B. The Grp170 nucleotide exchange factor executes a key role during ERAD of cellular misfolded clients. Mol Biol Cell. 2016;27(10):1650–1662. doi: 10.1091/mbc.E16-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, et al. The ER membrane-anchored ubiquitin ligase Hrd1 is a positive regulator of T-cell immunity. Nat Commun. 2016;7:12073. doi: 10.1038/ncomms12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu Q, et al. Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 2013;32(18):2477–2490. doi: 10.1038/emboj.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, et al. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity. 2013;39(2):272–285. doi: 10.1016/j.immuni.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329(5999):1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10(9):1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park JS, et al. Isolation, characterization and chromosomal localization of the human GADD153 gene. Gene. 1992;116(2):259–267. doi: 10.1016/0378-1119(92)90523-R. [DOI] [PubMed] [Google Scholar]
- 29.Aman P, et al. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11) Genes Chromosomes Cancer. 1992;5(4):278–285. doi: 10.1002/gcc.2870050403. [DOI] [PubMed] [Google Scholar]
- 30.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6(3):439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 31.Miller SD, et al. Tauroursodeoxycholic acid inhibits apoptosis induced by Z alpha-1 antitrypsin via inhibition of Bad. Hepatology. 2007;46(2):496–503. doi: 10.1002/hep.21689. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe N, Lam E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem. 2008;283(6):3200–3210. doi: 10.1074/jbc.M706659200. [DOI] [PubMed] [Google Scholar]
- 33.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30(5):616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh R, et al. Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158(3):534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papandreou I, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkins C, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73(6):1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 38.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 39.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16(11):1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 41.Ichijo H, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275(5296):90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 42.Gao B, et al. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 2008;9(5):480–485. doi: 10.1038/embor.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayakawa M, et al. Loss of Functionally Redundant p38 Isoforms in T Cells Enhances Regulatory T Cell Induction. J Biol Chem. 2017;292(5):1762–1772. doi: 10.1074/jbc.M116.764548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber S, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3+ and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34(4):554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nurieva RI, et al. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity. 2010;32(5):670–680. doi: 10.1016/j.immuni.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiao G, et al. T cell activation threshold regulated by E3 ubiquitin ligase Cbl-b determines fate of inducible regulatory T cells. J Immunol. 2013;191(2):632–639. doi: 10.4049/jimmunol.1202068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin HS, Park Y, Elly C, Liu YC. Itch expression by Treg cells controls Th2 inflammatory responses. J Clin Invest. 2013;123(11):4923–4934. doi: 10.1172/JCI69355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH, Elly C, Park Y, Liu YC. E3 Ubiquitin Ligase VHL Regulates Hypoxia-Inducible Factor-1α to Maintain Regulatory T Cell Stability and Suppressive Capacity. Immunity. 2015;42(6):1062–1074. doi: 10.1016/j.immuni.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei J, et al. HRD1-ERAD controls production of the hepatokine FGF21 through CREBH polyubiquitination. EMBO J. 2018;37(22):e98942. doi: 10.15252/embj.201898942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, et al. The endoplasmic reticulum-resident E3 ubiquitin ligase Hrd1 controls a critical checkpoint in B cell development in mice. J Biol Chem. 2018;293(33):12934–12944. doi: 10.1074/jbc.RA117.001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei J, et al. ER-associated ubiquitin ligase HRD1 programs liver metabolism by targeting multiple metabolic enzymes. Nat Commun. 2018;9(1):3659. doi: 10.1038/s41467-018-06091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.