Abstract
β-2 Microglobulin (β2M) is a molecular chaperone for the major histocompatibility class I (MHC I) complex, hemochromatosis factor protein (HFE), and the neonatal Fc receptor (FcRn), but β2M may also have less understood chaperone-independent functions. Elevated plasma β2M has a direct role in neurocognitive decline and is a risk factor for adverse cardiovascular events. β2M mRNA is present in platelets at very high levels, and β2M is part of the activated platelet releasate. In addition to their more well-studied thrombotic functions, platelets are important immune regulatory cells that release inflammatory molecules and contribute to leukocyte trafficking, activation, and differentiation. We have now found that platelet-derived β2M is a mediator of monocyte proinflammatory differentiation through noncanonical TGFβ receptor signaling. Circulating monocytes from mice lacking β2M only in platelets (Plt-β2M–/–) had a more proreparative monocyte phenotype, in part dependent on increased platelet-derived TGFβ signaling in the absence of β2M. Using a mouse myocardial infarction (MI) model, Plt-β2M–/– mice had limited post-MI proinflammatory monocyte responses and, instead, demonstrated early proreparative monocyte differentiation, profibrotic myofibroblast responses, and a rapid decline in heart function compared with WT mice. These data demonstrate a potentially novel chaperone-independent, monocyte phenotype–regulatory function for platelet β2M and that platelet-derived 2M and TGFβ have opposing roles in monocyte differentiation that may be important in tissue injury responses.
Keywords: Vascular Biology
Keywords: Platelets
Platelet derived β-2 micro globulin and TGFβ have opposing roles in monocyte differentiation that may be important in tissue injury responses.
Introduction
The immune system must continually meet pathogen challenges, maintain tissue homeostasis, and facilitate sterile tissue injury repair. This requires the integrated responses of many cells, including platelets. Platelets have active roles in the regulation of innate and acquired immunity that are mediated in part by platelet secretion of immune effector molecules acting both locally and systemically, including PF4, RANTES, IL-1β, and TGFβ (1–3). The immune regulatory role for platelets includes the initiation or exacerbation of cardiovascular diseases such as atherosclerosis and myocardial infarction (MI) (4, 5). We have now discovered a potentially novel means by which platelets regulate circulating monocyte inflammatory phenotypes; platelet-derived β-2 microglobulin (β2M) exerts concentration-dependent, proinflammatory effects on monocytes that are opposed by TGFβ.
Platelets, like most cell types except RBCs, express major histocompatibility complex class I (MHC I). MHC I is composed of a 3-subunit MHCα chain and β2M. β2M is a chaperone for MHCα cell surface trafficking, and β2M stabilizes MHCα once at the membrane. β2M is neither covalently linked to MHCα, nor transmembrane bound, so it easily dissociates from the cell surface and is found at plasma concentrations of 1–2 μg/ml in healthy individuals (6). Plasma β2M has been described as a predictive biomarker for many vascular inflammatory diseases, including cardiovascular disease and age-associated neurocognitive decline (7–11), but a direct mechanistic role for β2M as an inflammatory molecule has not been demonstrated. β2M mRNA is the highest expressed platelet transcript (12, 13), and proteomic studies demonstrated that platelets contain abundant β2M protein (14, 15). We have now discovered that platelets are a major source of plasma β2M and that β2M has critical immune modulatory functions beyond its molecular chaperone roles.
Platelet-derived immune mediators have many direct and indirect effects on monocytes in disease-relevant conditions. Platelet interactions with an inflamed endothelial cell layer at sites of atherosclerotic lesion development lead to the arrest of monocytes at these sites and to the acceleration of atherosclerosis (16). The activation and differentiation of monocytes and macrophages are also directly affected by platelet-derived immune mediators (17, 18). Platelets and monocytes are both activated by MI and have important roles in MI outcomes (5, 19, 20). Immediately following an MI, there are elevated platelet-derived immune mediators and mobilization of proinflammatory monocytes that traffic into the infarct region, becoming M1-like proinflammatory macrophages. Following the early ischemic tissue injury responses, there is a decline in platelet activation and a shift in circulating monocytes to a proreparative phenotype; these monocytes traffic to become M2-like–reparative macrophages that promote infarct wound healing and fibrosis (21). We now demonstrate a direct mechanistic link between post-MI platelet activation, elevated platelet-derived plasma β2M, and platelet β2M–dependent regulation of monocyte phenotypes that affect cardiac injury responses to MI.
Results
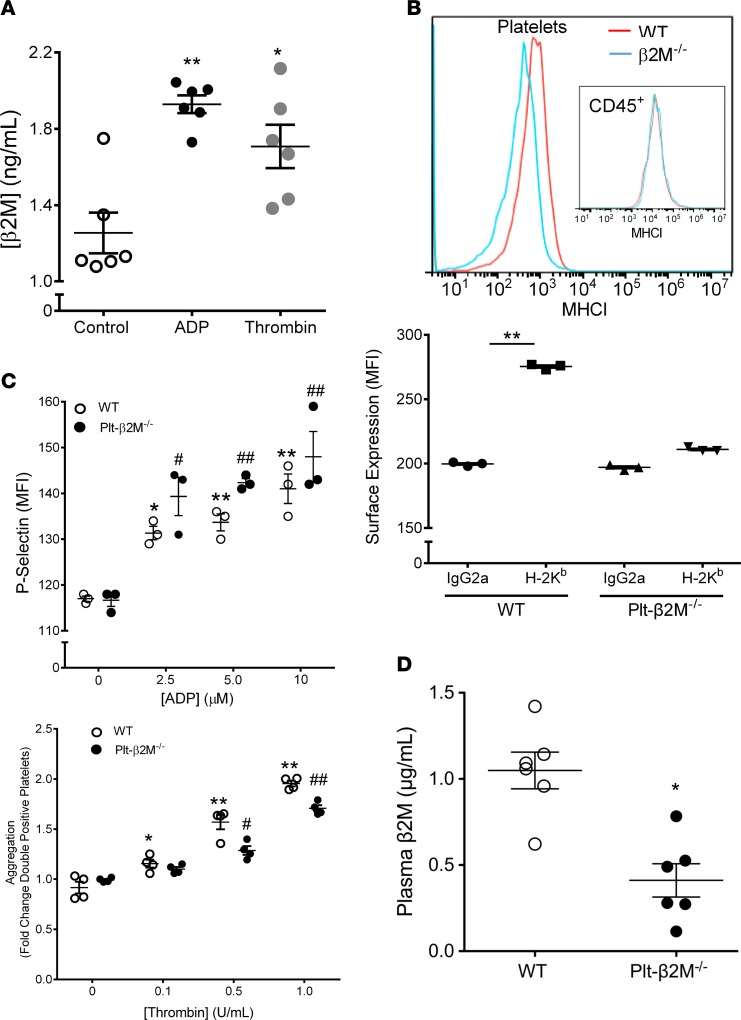
Platelet-derivedβ2M induces monocyte proinflammatory differentiation. Platelets have abundant B2m mRNA and MHC I, but the contribution of platelets to plasma β2M concentrations is not known. To first demonstrate that stimulated platelets release β2M, washed mouse platelets were activated with either (adenosine diphosphate) ADP or thrombin, and β2M release was quantified by ELISA. Activated platelets released β2M (Figure 1A). Global β2M–/– mice have many immune defects (22); therefore, to explore the role of platelet-derived β2M in immune function, we generated platelet-specific β2M-KO mice (PF4-Cre × β2Mfl/fl, Plt-β2M–/–; Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122943DS1). Washed platelets from WT and Plt-β2M–/– mice were isolated, and MHC I surface expression was determined. Platelets from Plt-β2M–/– mice had greatly reduced surface MHC I compared with WT platelets (Figure 1B), but MHC I expression did not differ on WBC, indicating platelet specificity in the β2M–/– mice (Figure 1B). Peripheral blood cell numbers, including CD8+ T cells, were similar in WT and Plt-β2M–/– mice (Supplemental Figure 2), indicating that, unlike complete β2M–/– mice, a lack of platelet β2M does not grossly affect immune development. A lack of β2M also did not affect platelet activation, as ADP and thrombin similarly activated platelets from WT and Plt-β2M–/– mice based on P-selectin surface expression (Figure 1C), platelet aggregation (Figure 1C), fibrinogen binding (Supplemental Figure 3), and activated platelet PF4 release (Supplemental Figure 3). Furthermore, we measured plasma β2M in age-matched WT and Plt-β2M–/– mice and found that plasma β2M in WT mice was very similar to reported human values (11), but plasma β2M was significantly reduced in Plt-β2M–/– mice (Figure 1D), with no difference in other major platelet-derived plasma molecules, including PF4 (Supplemental Figure 4). These data indicate that platelets are an important source of plasma β2M.
Figure 1. Platelets are a major source of plasma β2M.
(A) Activated platelets release β2M. Mouse platelets were isolated and treated with control buffer, ADP (10 μM), or thrombin (1 U/ml). β2M release was measured by ELISA (n = 4, *P < 0.05, **P < 0.01, 1-way ANOVA with Bonferroni correction). (B) Platelets, but not WBC, from Plt-β2M–/– mice lack MHC I surface expression. Anti–MHC I or control IgG was incubated with circulating platelets or CD45+ cells (insert) from WT and Plt-β2M–/–. MHC I was quantified by flow cytometry (n = 3, **P < 0.01, 1-way ANOVA with Bonferroni correction). (C) Platelets from WT and Plt-β2M–/– mice had similar activation and aggregation. Washed WT and Plt-β2M–/– platelets were stimulated with ADP, and surface P-selectin was measured. Platelets were also labeled with APC or PE antibodies, thrombin stimulated and platelet aggregates determined as double-positive cells by flow cytometry (n = 4 for both panels, 1-way ANOVA with Bonferroni correction). (D) Platelets are a major source of plasma β2M. Concentration of plasma β2M in 10-week-old WT and Plt-β2M–/– mice was determined by ELISA (n = 5, WT; n = 6, Plt-β2M–/–; *P < 0.05 vs. WT, unpaired 2-tailed t test with Welch’s correction).
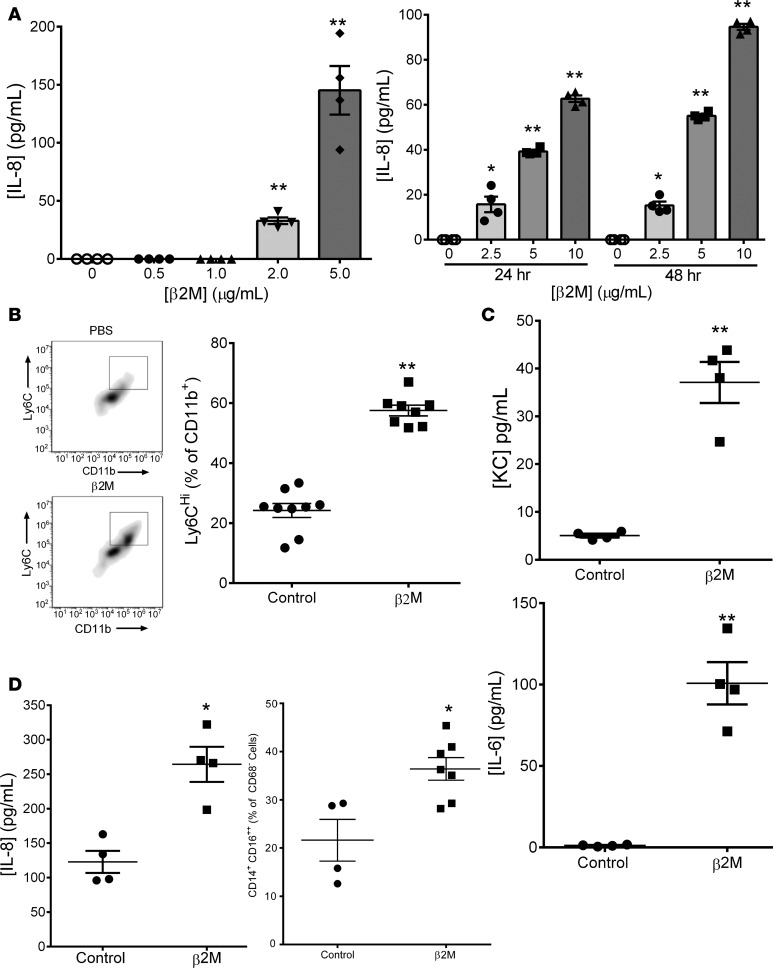
Because β2M has chaperone-independent functions (23), and because platelets have important roles in monocyte responses, we asked whether β2M induced monocyte activation. The human THP-1 monocyte cell line was treated with control buffer or recombinant human β2M, and IL-8 release was determined in a time- and dose-dependent manner. β2M induced IL-8 release beginning at concentrations consistent with those associated with increased cardiovascular event risk (Figure 2A) (6, 11). Mouse monocytes are broadly classified as proinflammatory (Ly6Chi, IL-6, KC [mouse IL-8 homologue] secreting) or proreparative (LyC6lo and IL-10 secreting). To determine whether β2M induced a primary mouse monocyte proinflammatory response, isolated mouse BM monocytes were incubated with β2M for 48 hours, and their inflammatory phenotype was determined by Ly6C expression and cytokine release. Recombinant β2M increased the number of Ly6Chi mouse monocytes and the secretion of both KC and IL-6 (Figure 2, B and C). Human peripheral blood monocytes were also isolated and incubated with control buffer or recombinant β2M. Characteristic of proinflammatory human monocytes (24–26), β2M increased both the number of CD16++ monocytes and monocyte IL-8 secretion (Figure 2D; confirmed with THP-1 cells in Supplemental Figure 5). These data demonstrated that β2M induced a proinflammatory monocyte phenotype.
Figure 2. β2M induced monocyte inflammatory responses.
(A) THP-1 cells were treated with β2M, and IL-8 release was determined by ELISA. β2M induced inflammatory molecule production in a time- and dose-dependent manner (n = 4, *P < 0.05, **P < 0.01 vs. 0, 1-way ANOVA with Bonferroni correction). (B and C) β2M induced a primary mouse monocyte proinflammatory phenotype. Mouse BM monocytes were isolated and treated with control PBS or β2M (5 μg/ml). (B) Forty-eight hours later, Ly6C surface expression was measured by flow cytometry (n = 9, control; n = 8, β2M; **P < 0.01, unpaired 2-tailed t test with Welch’s correction). (C) KC and IL-6 release determined by ELISA (n = 9, control; n = 8, β2M; **P < 0.01, unpaired 2-tailed t test with Welch’s correction). (D) β2M induced a human monocyte inflammatory phenotype. Human peripheral blood monocytes were isolated and treated with control PBS or β2M (10 μg/ml). CD16 surface expression was measured by flow cytometry (right panel, n = 4, *P < 0.05, unpaired 2-tailed t test with Welch’s correction) and IL-8 release by ELISA (left panel, n = 4 control, n = 7 β2M, *P < 0.05, unpaired 2-tailed t test with Welch’s correction). All graphs represent experiments that were repeated at least twice.
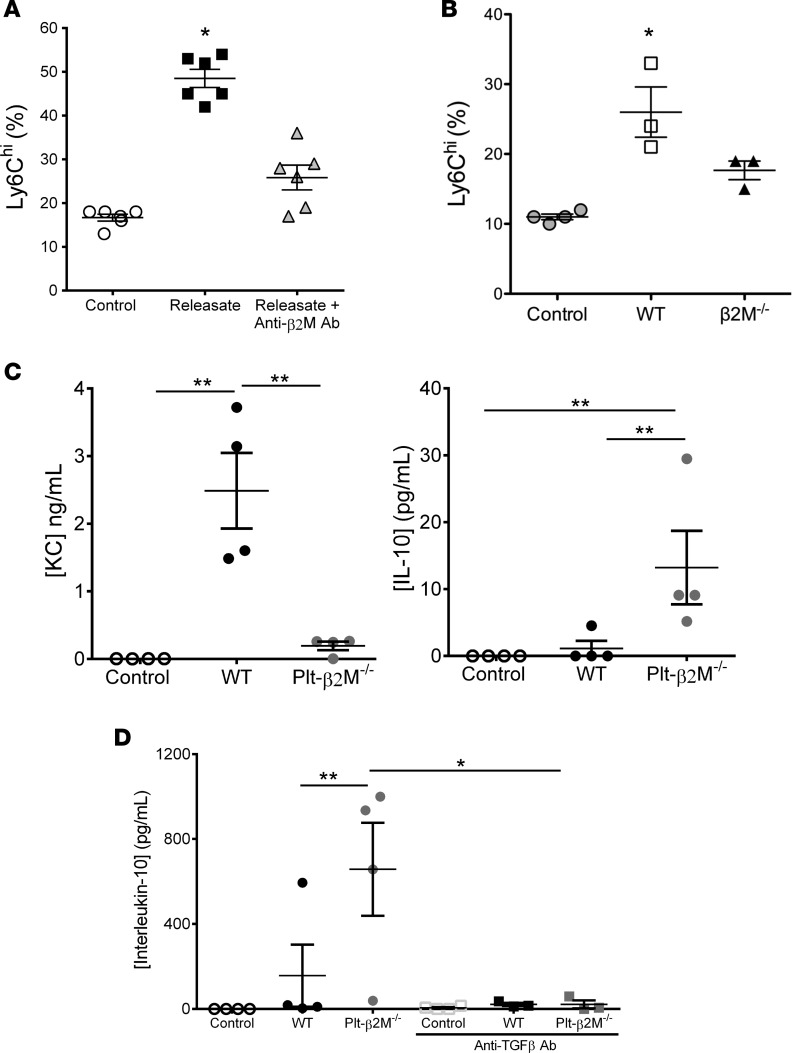
Platelets have well-described roles in both monocyte trafficking and activation (1), but platelet-mediated monocyte effector differentiation is not as well understood. To determine the role of platelet-derived β2M in monocyte effector responses, mouse monocytes were incubated with control buffer, the releasate from activated washed mouse platelets, or platelet releasate and anti-β2M antibody (platelets were thrombin activated and thrombin neutralized with hirudin; monocytes were incubated with platelet releasate at a ratio of 1 monocyte to releasate from 10 platelets). Consistent with a platelet β2M–mediated proinflammatory phenotype, platelet releasate increased Ly6Chi monocytes and KC secretion, but platelet-induced monocyte polarization was attenuated by anti-β2M blocking antibody (Figure 3A and Supplemental Figure 6). Mouse monocytes were also treated with the releasate from WT or Plt-β2M–/– platelets. In contrast to WT platelets, the releasate from Plt-β2M–/– platelets did not induce a Ly6Chi phenotype or KC secretion (Figure 3, B and C). Plt-β2M–/– releasate instead increased monocyte IL-10 release, typical of a proreparative monocyte phenotype (Figure 3C). Similar results were noted using platelet releasates from both male and female mice (Supplemental Figure 7). These findings indicate that, in the absence of β2M, platelet activation promotes proreparative monocyte differentiation.
Figure 3. Platelets induce a monocyte proinflammatory phenotype in a β2M-dependent manner.
(A–B) Mouse monocytes were incubated with control buffer, the releasate from WT platelets ± anti-β2M antibody (A), or Plt-β2M–/– mouse platelet releasate (B) (10:1 platelet/monocyte ratio). Control WT platelet releasate induced Ly6Chi monocytes more than anti-β2M antibody (n = 4 control, n = 6 releasate ± anti-β2M Ab, *P < 0.05 vs. control, 1-way ANOVA with Bonferroni correction) (A) or Plt-β2M–/– mouse platelet releasate (B) (n = 4, control; n = 3, WT; Plt-β2M–/–; *P < 0.05 vs. control, 1-way ANOVA with Bonferroni correction). (C) Releasate from β2M–/– platelets did not induce mouse monocyte KC, but instead induced IL-10 release (both panels, n = 4; *P < 0.01 vs. control, 1-way ANOVA with Bonferroni correction). (D) Plt-β2M–/– releasate–induced IL-10 is TGFβ dependent. Anti-TGFβ antibody (10 μg/ml) blocked Plt-β2M–/– releasate–induced IL-10 production (n = 4; *P < 0.05, 1-way ANOVA with Bonferroni correction). All graphs and tables are representative from experiments repeated at least twice.
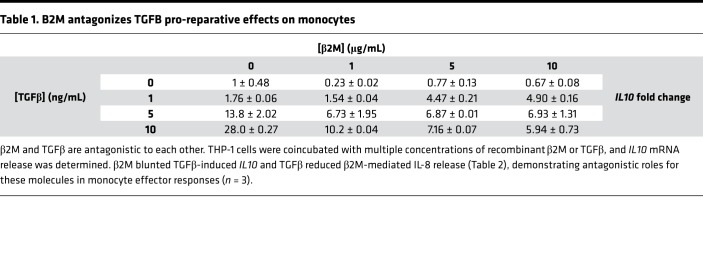
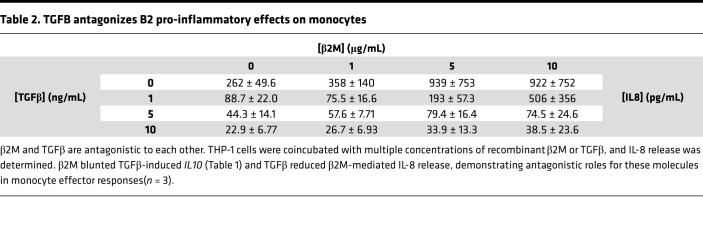
Platelets contain and secrete both β2M and TGFβ (27, 28), but TGFβ is associated with inducing a proreparative/profibrotic tissue environment and monocyte phenotype. TGFβ and β2M have been noted to be antagonistic in other cellular environments (29, 30), so to determine their effects on monocytes, THP-1 cells were treated with recombinant β2M and TGFβ1 at multiple concentration ratios, and monocyte responses were determined after 48 hours. β2M reduced TGFβ1-mediated IL10 expression (proreparative cytokine), and TGFβ1 limited β2M-induced monocyte inflammatory cytokine production (IL-8) (Tables 1 and 2). We therefore determined whether the proreparative phenotype induced by β2M–/– platelet releasate was due to increased TGFβ signaling in the absence of β2M. Monocytes were incubated with WT or Plt-β2M–/– releasate in the presence of anti-TGFβ blocking antibody. Blocking TGFβ blunted Plt-β2M–/– releasate–induced IL-10 production (Figure 3D) and increased Plt-β2M–/– releasate–induced proinflammatory responses (Supplemental Figure 8). There was no difference in platelet releasate TGFβ1 concentration between WT and Plt-β2M–/– mice (Supplemental Figure 9). These data indicate that platelet-derived β2M and TGFβ1 have antagonistic effects on monocyte polarization.
Table 1. B2M antagonizes TGFB pro-reparative effects on monocytes.
Table 2. TGFB antagonizes B2 pro-inflammatory effects on monocytes.
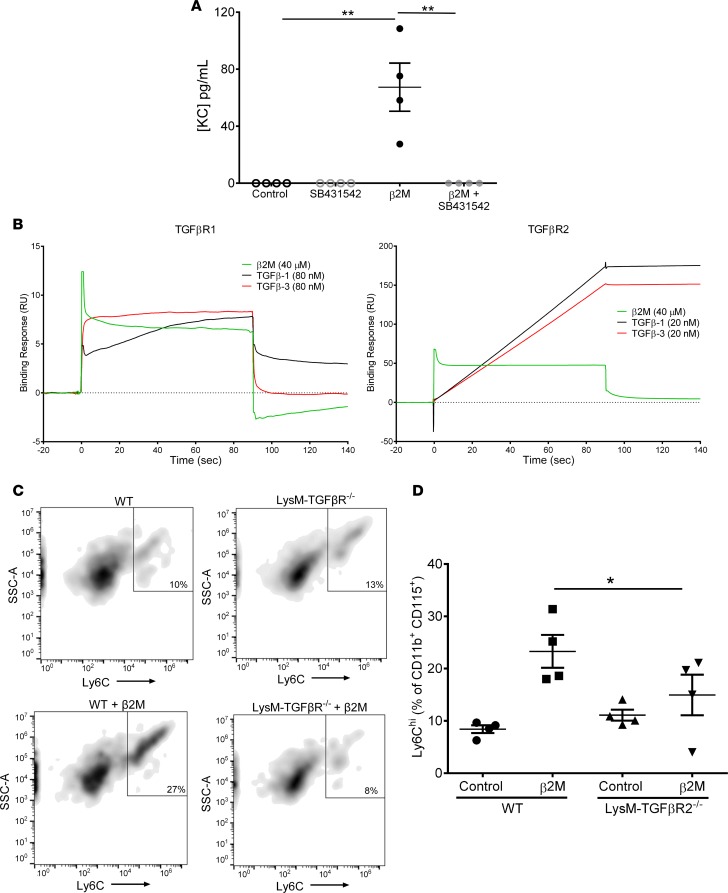
β2M signals through a noncanonical TGFβreceptor pathway. We next sought to determine whether β2M and TGFβ signal through the same receptor but with different downstream outcomes. To do so, we first pharmacologically inhibited mouse monocyte TGFβR1/TGFβR2 receptor complex activity using the TGFβR1 kinase inhibitor SB431542 (31). TGFβR inhibition blocked β2M-induced KC release (Figure 4A). Treatment of THP-1 cells with SB431542 also blocked β2M-induced upregulation of surface CD16 and IL8 mRNA expression, as well as TGFβ1-induced IL10 expression (Supplemental Figure 10). As further proof that β2M interacts with TGFβR, we performed surface plasma resonance (SPR). TGFβR1 and TGFβR2 were covalently bound to a sensorchip, and either recombinant β2M, TGFβ1, TGFβ3, BSA, or FBS were used as the flow-through ligands. β2M, TGFβ1, and TGFβ3 bound to the TGFβR1 at ratios similar to their signaling concentrations (Figure 4B, left), consistent with the physiologic observation that β2M signals monocytes at μg/ml concentrations compared with TGFβ signaling at ng/ml concentrations. We confirmed β2M binding to TGFβR2 but at a weaker ratio compared with TGFβ1 and TGFβ3 (Figure 4B, right). Negative controls BSA and FBS both had negative binding responses, indicating that β2M did not nonspecifically bind TGFβR (Supplemental Figure 11). As further proof of TGFβR-dependent β2M monocyte activation, we isolated BM monocytes from WT or myeloid cell–specific TGFβR2–/– mice (LysMCre-TGFβR2fl/fl) (32), treated the monocytes with control buffer or β2M, and determined Ly6C expression 48 hours later. β2M increased Ly6C expression on WT, but not TGFβR2–/–, monocytes (Figure 4, C and D).
Figure 4. β2M proinflammatory phenotype signaling is through noncanonical TGFβ receptor signaling.
(A) Inhibition of TGFβR signaling ameliorated β2M-induced monocyte activation. Mouse monocytes were incubated with control buffer, β2M, or β2M and a TGFβR1 kinase inhibitor (SB431542). KC production was determined 48 hours later (n = 4; **P < 0.01, 1-way ANOVA with Bonferroni correction). (B) β2M binds to TGFβR1 and TGFβR2. Fc-TGFβR1 or Fc-TGFβR2 were immobilized on a sensor chip, and β2M, TGFβ1, or TGFβ3 binding was determined by SPR. (C and D) WT and TGFβR2–/– monocytes were incubated with control buffer or β2M. (C) β2M induced WT, but not TGFβR–/– monocyte, Ly6Chi phenotype. (D) Quantification of C (n = 4; *P < 0.05, 1-way ANOVA with Bonferroni correction).
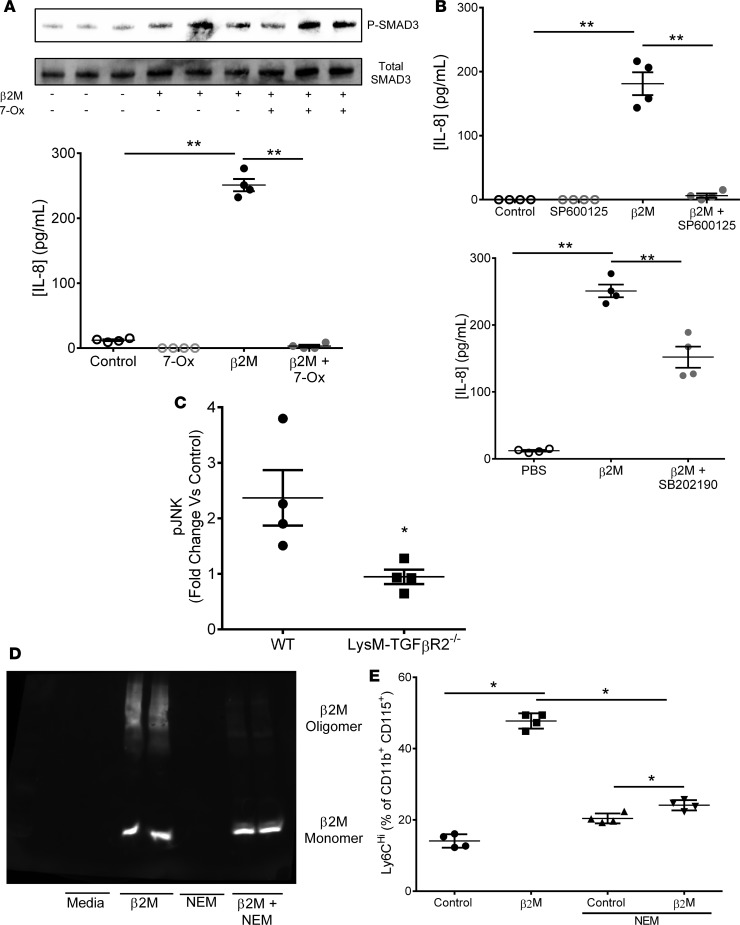
TGFβR signaling can either be antiinflammatory (canonical, Smad2/3 dependent) (28), or proinflammatory (noncanonical, TAK1/p38/JNK dependent) (33). To explore downstream β2M-TGFβR signaling, THP-1 cells were either treated with a TAK1 (noncanonical pathway) inhibitor or a SMAD3 (canonical pathway) inhibitor prior to β2M treatment. The TAK1 inhibitor completely blocked β2M-induced IL-8 secretion, despite no change in β2M-induced SMAD3 phosphorylation (Figure 5A; phosphorylated-TAK1 and phosphorylated-SMAD3 quantified in Supplemental Figure 12). SMAD3 inhibition only partially blocked β2M-induced IL-8 release (Supplemental Figure 13), further indicating TAK1-dominant β2M signaling. To explore downstream TAK1 signaling, THP-1 cells were treated with JNK or p38 inhibitors. JNK inhibition strongly blocked β2M signaling, while p38 inhibition only partially blocked β2M inflammatory responses (Figure 5B). Similar results were found using primary mouse monocytes and SB431542, SIS, 7-Ox, and SP600125 (Supplemental Figure 14). Monocytes treated with 2M from WT, but not TGFβR2–/– mice, had increased P-JNK as measured by the change in intracellular P-JNK compared with nontreated controls using intracellular flow cytometry (Figure 5C; TGFβ1 treated negative controls in Supplemental Figure 15). Similar to the immunoblot data, β2M increased intracellular P-JNK and P-SMAD2/3 in primary mouse monocytes treated with β2M, but this was blocked by the TAK1 and JNK inhibitors (Supplemental Figure 15). These data indicate that β2M downstream signaling is, in part, dependent on TAK1 and JNK.
Figure 5. β2M signals through a noncanonical TGFβ receptor signaling mechanism.
(A) Inhibition of TAK1 (7-ox, 1 μM) blocked β2M-induced THP-1 IL-8 but had no effect on β2M-induced SMAD3 activation (P-SMAD3). THP-1 cells were treated with TAK1 inhibitor or control buffer prior to β2M. IL-8 was determined by ELISA (n = 4; **P < 0.01 vs. control, 1-way ANOVA with Bonferroni correction), and P-SMAD3 and total SMAD3 were determined by immunoblot (representative image quantified in Supplemental Figure 12 and 13). (B) JNK inhibitor (SP600125, 10 μM) greatly attenuated THP-1 IL-8 production, but p38 inhibitor (SB202190, 10 μM) only partially attenuated IL-8 (both panels, n = 4, **P < 0.01 vs. control, 1-way ANOVA with Bonferroni correction). (C) β2M-induced WT but not TGFβR2–/– monocyte p-JNK. WT and TGFβR2–/– monocytes were incubated with control buffer or β2M, and intracellular p-JNK was determined by flow cytometry (n = 4, *P < 0.05, unpaired 2-tailed t test with Welch’s correction). (D and E) Blocking β2M oligomer formation with NEM inhibited β2M induced monocyte inflammatory phenotype. β2M was pretreated with buffer or NEM and then added to mouse monocytes for 48 hours. (D) NEM reduced β2M oligomer formation (nonreducing gel), and (E) Ly6Chi monocytes (n = 4, *P < 0.05, 1-way ANOVA with Bonferroni correction).
β2M forms oligomeric complexes in a concentration-dependent manner through cysteine sulphydryl group cross-linking (34). To determine whether β2M signaling is dependent on oligomer formation, we pretreated β2M with N-ethylmaleimide (NEM) to irreversibly block cysteine-dependent oligomer formation and confirmed this by immunoblot after running control and treated β2M on a nonreducing gel (Figure 5D). Control or NEM-treated β2M was then incubated with mouse monocytes. NEM-treated, monomeric β2M did not significantly increase Ly6Chi monocytes compared with control β2M (Figure 5E). Treatment of recombinant β2M with NEM did not affect the measurement of β2M by ELISA (Supplemental Figure 16). These data indicate that β2M signaling may be dependent on β2M in its oligomeric conformation.
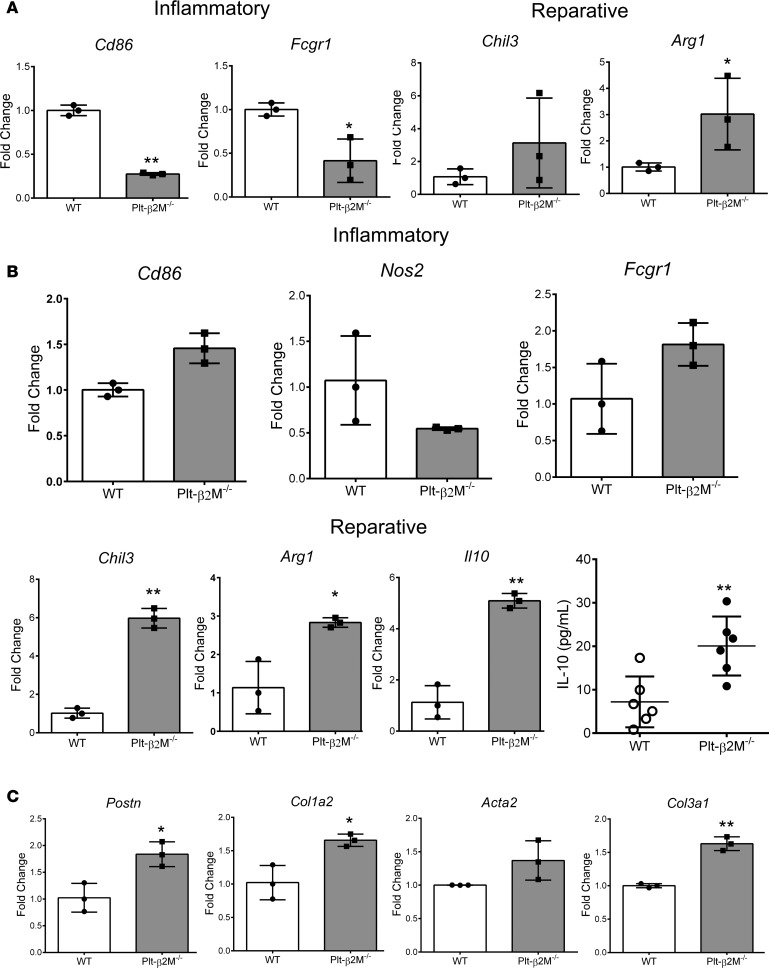
Plateletβ2M regulates monocyte responses to tissue injury. Upon tissue trafficking, monocytes polarize and differentiate to macrophage subsets that are most simply defined as proinflammatory (M1-like) or proreparative (M2-like). Macrophage polarization and differentiation are, in part, dependent on the trafficking monocyte phenotype. Ly6Chi monocytes tend to differentiate to M1-like macrophages that are inflammatory and phagocytic, whereas Ly6Clo reparative monocytes differentiate primarily to profibrotic M2-like macrophages that promote fibroblast extracellular matrix (ECM) production (35, 36). To determine whether platelet-derived β2M regulates basal circulating monocyte phenotypes, peripheral blood monocytes were isolated from WT and Plt-β2M–/– mice by negative selection, and markers of a proinflammatory or proreparative differentiation were quantified by quantitative PCR (qPCR). Compared with WT monocytes, Plt-β2M–/– monocytes were skewed to a proreparative phenotype (Figure 6A). Monocytes isolated from the BM of WT and Plt-2M–/– mice had similar Ly6C expression and responses to WT platelet releasates, indicating that β2M exerts its effects after BM development (Supplemental Figure 17). To determine whether monocytes from Plt-β2M–/– mice are skewed to become profibrotic macrophages, peripheral blood monocytes from WT and Plt-β2M–/– mice were isolated and cocultured with primary adult cardiac fibroblasts for 72 hours. When cocultured with fibroblasts, monocytes isolated from Plt-β2M–/– mice expressed gene markers indicative of reparative M2-like macrophages and had increased IL-10 secretion compared with WT mouse–derived monocytes cocultured with fibroblasts (Figure 6B). There was also increased myofibroblast activation when fibroblasts were cocultured with monocytes from Plt-β2M–/– mice (Figure 6C). These data indicate that platelet-derived β2M has a major role in monocyte to macrophage differentiation that affects their profibrotic potential.
Figure 6. Platelet β2M regulates circulating monocyte differentiation.
(A) Circulating monocytes were isolated from WT and Plt-β2M–/– mice, and proinflammatory and proreparative monocyte gene markers were quantified by qPCR. Plt-β2M–/– mouse monocytes had proreparative monocyte gene expression (all panels, n = 3; *P < 0.05, unpaired 2-tailed t test with Welch’s correction). (B and C) Monocytes from Plt-β2M–/– mice differentiate to proreparative, fibroblast activating, macrophages in vitro. Peripheral blood monocytes from WT and Plt-β2M–/– mice were coincubated with cardiac fibroblasts for 72 hours and macrophage differentiation (B) and fibroblast activation (C) were determined by qPCR, and IL-10 secretion determined by ELISA. Plt-β2M–/––derived monocytes had increased proreparative differentiation and IL-10 secretion and induced more fibroblast activation compared with WT mouse monocytes (B and C, n = 4; *P < 0.05, **P < 0.01 vs. WT, unpaired 2-tailed t test with Welch’s correction).
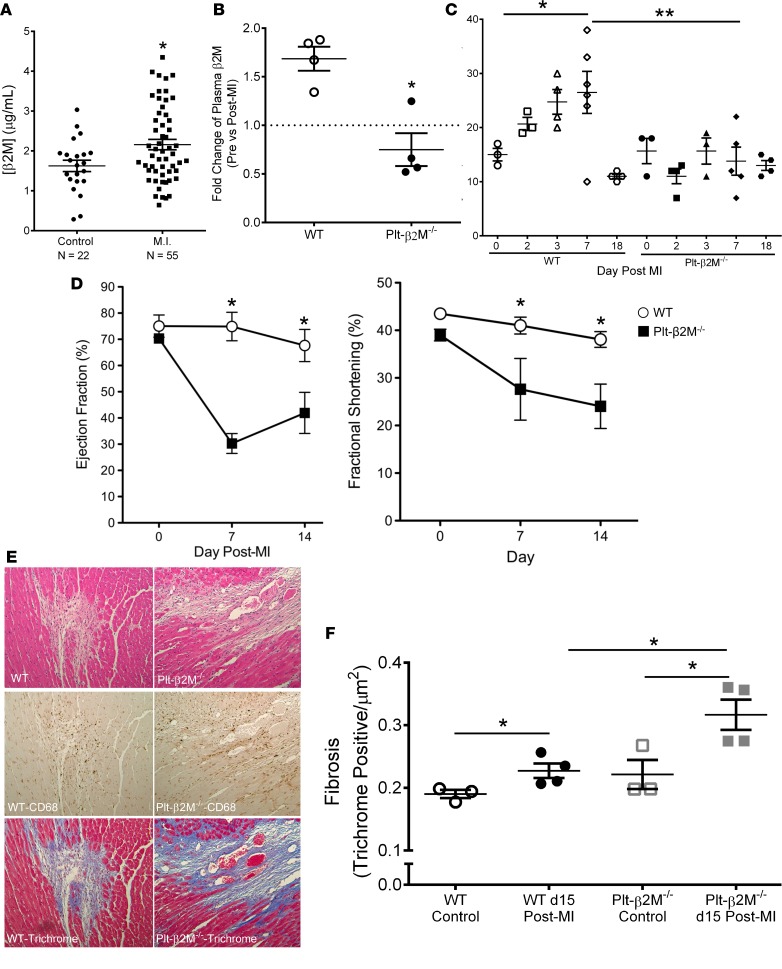
Platelets and monocytes are systemically activated and recruited to the heart by acute MI (37). At the time of emergency room presentation, plasma β2M was increased in confirmed MI patients compared with healthy controls, demonstrating the association of elevated β2M with acute MI events (Figure 7A). To explore the role of platelet-derived β2M in monocyte responses to MI, we used a mouse permanent ligation of the left anterior descending (LAD) artery model but induced relatively small MIs to better explore the inflammatory component. WT and Plt-β2M–/– mice had similar tissue perfusion (Supplemental Figure 18), and like human MI patients, WT mice had significantly elevated post-MI plasma β2M; however, Plt-β2M–/– mice did not (Figure 7B). This demonstrates that platelets are a major source of the increased plasma β2M after MI; there was no significant difference in plasma TGFβ1 between WT and Plt-β2M–/– mice either before or after MI (Supplemental Figure 19).
Figure 7. Platelet-derived β2M mediates monocyte inflammatory responses to myocardial infarction.
(A) Plasma β2M is elevated in humans after MI. Plasma was isolated from healthy subjects and confirmed MI patients. β2M was measured by ELISA (n = 22, control; n = 55, MI; *P < 0.05 vs. control, unpaired 2-tailed t test with Welch’s correction). (B) WT, but not Plt-β2M–/–, mice had increased post-MI plasma β2M. β2M measured before and 1 day after MI by ELISA (n = 5; *P < 0.05 vs. Plt-β2M–/–, 1-way ANOVA with Bonferroni correction). (C) WT mice had increased circulating Ly6Chi monocytes after MI, but Plt-β2M–/– mice had no change from day 0 (n = 3, WT at 0 and 2 days, and Plt-β2M–/– at 0, 2, and 3 days; n = 4, WT at 3 and 18 days, and Plt-β2M–/– at 18 days; n = 5, Plt-β2M–/– at 7 days; n = 6, WT at 7 days; *P < 0.05, **P < 0.01, 1-way ANOVA with Bonferroni correction). (D) Plt-β2M–/– mice had a rapid post-MI decline in heart function compared with WT mice (n = 5, *P < 0.05 vs. Plt-β2M–/–, unpaired 2-tailed t test with Welch’s correction). (E) WT and Plt-β2M–/– mice had similar post-MI monocyte lineage infiltrates (CD68+), but monocytes are associated with areas of ECM deposition (trichrome) in Plt-β2M–/– mice. Representative images 20× images, from day 15 after MI. (F) Fibrosis quantification (n = 3, control; n = 4, day 15 after MI; *P < 0.05, 1-way ANOVA with Bonferroni correction).
There are 2 phases of mouse monocyte responses to MI: (a) an initial Ly6Chi proinflammatory monocyte response, followed 4–7 days later by (b) Ly6Clo proreparative monocytes that promote wound healing and fibrosis (20, 21). To characterize post-MI monocyte responses in WT and Plt-β2M–/– mice, blood was collected on multiple days, and circulating monocyte Ly6C expression was determined (gating strategy, Supplemental Figure 20). WT mice had an expected early Ly6Chi monocyte response that peaked at about day 7 before declining (Figure 7C). However, Plt-β2M–/– mice had no change in post-MI Ly6Chi monocytes compared with baseline (Figure 7C). Cardiac function was assessed by echocardiography. Somewhat surprisingly, compared with WT mice, Plt-β2M–/– mice had a rapid decline in post-MI heart function (Figure 7D, Supplemental Figure 21, and Supplemental Figure 22). By day 28 after MI, Plt-β2M–/– mice still had worse heart function compared with WT mice, but the ejection fraction (EF) and fractional shortening (FS) began to converge (Supplemental Figure 23). Histologic examination on day 15 demonstrated that WT and Plt-β2M–/– mice had similar CD68+ (monocyte lineage marker) infiltrates (Figure 7E), but the CD68+ cells tended to be focused in areas of fibrosis (trichrome stain) in Plt-β2M–/– mice compared with WT mice (representative images, Figure 7E). The area of fibrosis was significantly higher in Plt-β2M–/– mice on day 15 compared WT mice (Figure 7F). These data are consistent with a proreparative skewing of monocytes in Plt-β2M–/– mice, leading to an early proreparative M2-like macrophage response and increased myofibroblast activation.
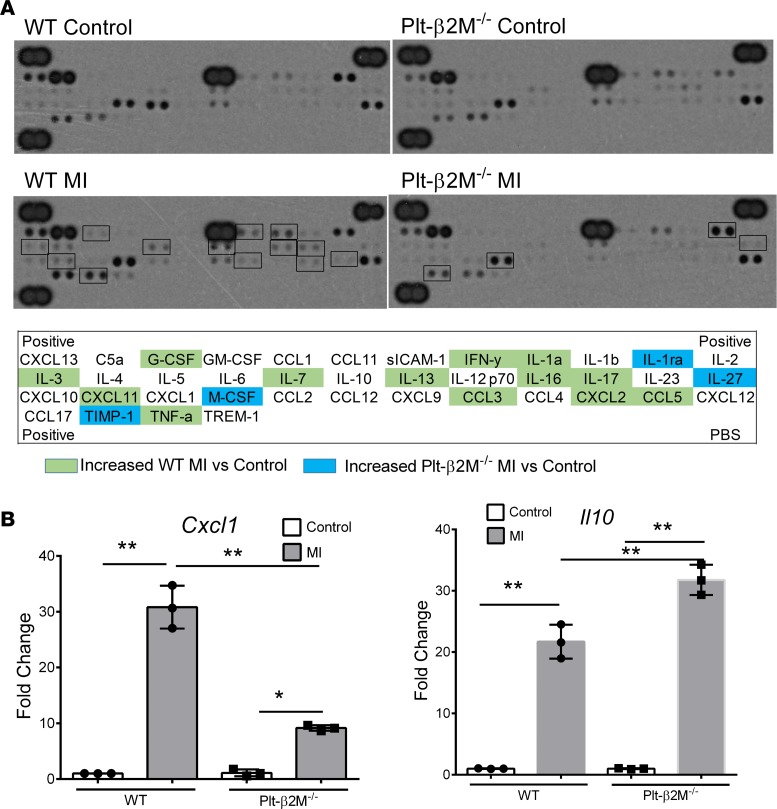
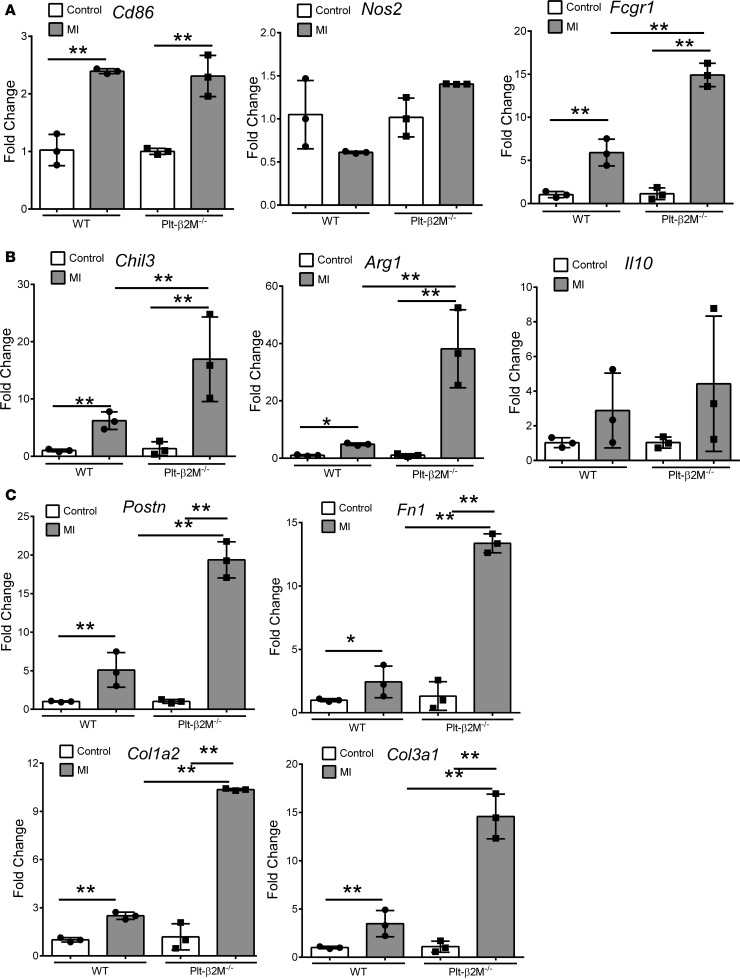
To begin to define the early immune responses to MI in WT and Plt-β2M–/– mice, we performed a protein array using pooled plasma from mice on days 0 and 3 after MI. WT mice had an expected post-MI increase in multiple inflammatory molecules, including IFNγ, IL-1, and TNFα (Figure 8A). The plasma cytokines elevated in Plt-β2M–/– mice were more indicative of proreparative responses, including elevated M-CSF and TIMP1 (Figure 8A). WT and Plt-β2M–/– mice had very similar pre- and post-MI peripheral blood counts, including monocytes (Supplemental Figure 24), as well as very similar T cell and neutrophil responses to MI (Supplemental Figure 25). To better define the early post-MI circulating monocyte differentiation, monocytes were isolated on days 0 and 3, and inflammatory cytokines were quantified by qPCR relative to their genotype controls. As expected, WT monocytes had increased post-MI expression of Cxcl1, while Plt-β2M–/– monocytes had increased expression of proreparative Il10 (Figure 8B), further demonstrating that platelet β2M promotes a proinflammatory monocyte response to tissue injury, and in its absence, a rapid proreparative response occurs. To assess the early post-MI macrophage infiltrates in the heart, markers of macrophage phenotype and myofibroblast activation were determined by qPCR on day 3 after MI. Plt-β2M–/– and WT mice had similarly increased proinflammatory macrophage markers (Figure 9A), but only Plt-β2M–/– mice had greatly increased expression of proreparative macrophage gene markers (Figure 9B). WT mice also had more M1-like MHC IIhi macrophages in the heart compared with Plt-β2M–/– (Supplemental Figure 26). We further validated the difference in heart macrophages by histological staining for Arginase-1 (Arg1), a marker for M2-like macrophages. There was a significant increase in post-MI Arg1+ cells in Plt-β2M–/– mice compared with WT mice (Supplemental Figure 27). Myofibroblast activation markers were also increased in Plt-β2M–/– mouse hearts at this early time point (Figure 9C), suggesting that proreparative monocyte responses and M2-like macrophage differentiation in Plt-β2M–/– mice leads to early profibrotic responses to myocardial injury and an important role for platelets in post-MI monocyte/macrophage responses.
Figure 8. Plt-β2M–/– mice have an early proreparative skewed monocyte response to MI.
(A) Plasma inflammatory protein array. WT mice had a day 3 post-MI increase in proinflammatory plasma proteins, whereas Plt-β2M–/– mice had a greater increase in proreparative plasma proteins. (B) Circulating monocytes were isolated from WT and Plt-β2M–/– mice before and 3 days after MI, and qPCR was performed for Cxcl1 and Il10. Monocytes from Plt-β2M–/– mice had less Cxcl1 and increased Il10 expression after MI compared with WT mice (n = 4, **P < 0.01, 1-way ANOVA with Bonferroni correction).
Figure 9. Plt-β2M–/– mice have an early reparative and fibrotic response to MI.
Cardiac mRNA was isolated from control and day-3 post-MI hearts. (A and B) Plt-β2M–/– mouse hearts had increased post-MI markers of proreparative macrophage phenotype (A) (n = 3, normalized to genotype control; **P < 0.01, 1-way ANOVA with Bonferroni correction) and increased fibroblast activation compared with WT mice (C) (n = 3, normalized to genotype control; **P < 0.01, 1-way ANOVA with Bonferroni correction).
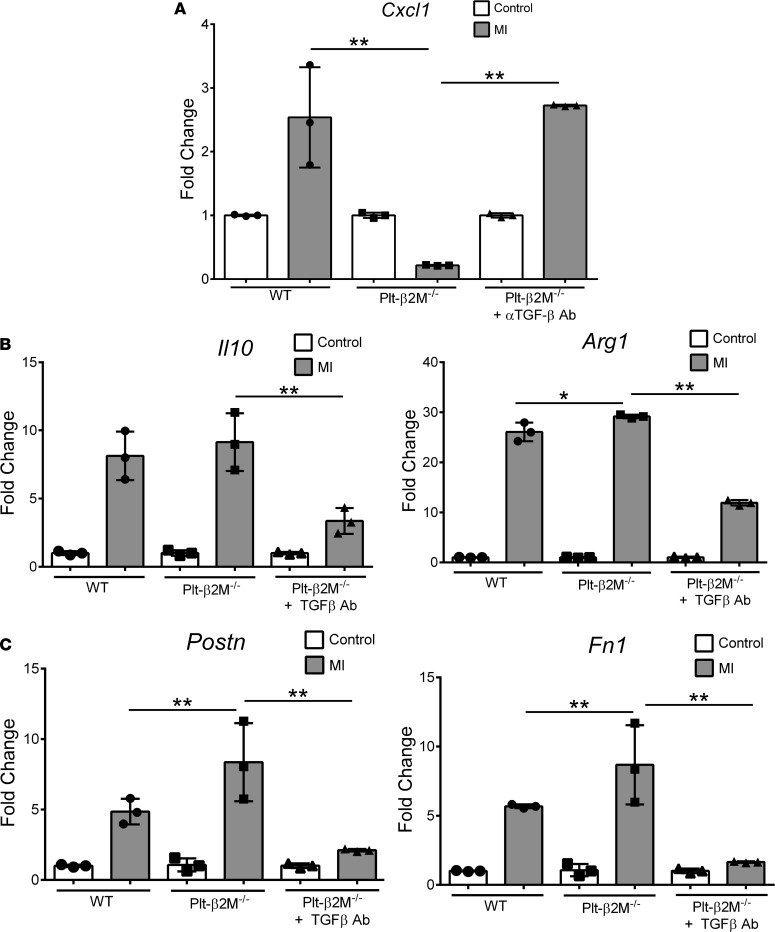
Because platelet-derived β2M and TGFβ have opposing monocyte differentiation effects in vitro, we determined whether the in vivo Plt-β2M–/– proreparative changes are due to enhanced TGFβ signaling. Plt-β2M–/– mice were treated with a TGFβ blocking antibody to reduce plasma TGFβ1 (Supplemental Figure 28) and subjected to ligation of the LAD coronary artery. On day 4 after MI, Plt-β2M–/– mice treated with TGFβ blocking antibody had similar monocyte proinflammatory Cxcl1 expression as WT mice (Figure 10A). Markers of cardiac macrophage differentiation and myofibroblast activation were also analyzed; increased M2-like profibrotic macrophage and myofibroblast activation markers were attenuated in Plt-β2M–/– mice treated with anti-TGFβ antibody (Figure 10, B and C). WT mice treated anti-TGFβ antibody had no significant change in macrophage or myofibroblast activation markers at day 4 after MI compared with genotype controls (Supplemental Figure 29), indicating that, at this early time point, TGFβ signaling is only dominant in Plt-β2M–/– mice. These in vivo data indicate that, while platelet-derived β2M polarized monocytes toward proinflammatory responses, TGFβ acts to promote monocyte profibrotic responses in response to MI.
Figure 10. Plt-β2M–/– early proreparative responses to MI are at least in part TGFβ dependent.
Plt-β2M–/– mice were treated with anti-TGFβ antibody prior to MI. On day 4 after MI, monocyte and macrophage inflammatory phenotypes and myofibroblast activation were determined. (A–C) Blocking TGFβ in Plt-β2M–/– mice increased circulating monocyte inflammatory cytokine expression (n = 3, **P < 0.01, 1-way ANOVA with Bonferroni correction) (A), decreased cardiac proreparative macrophages (n = 3; *P < 0.05, **P < 0.01, 1-way ANOVA with Bonferroni correction) (B), and decreased myofibroblast activation compared with control Plt-β2M–/– mice (n = 3, **P < 0.01, 1-way ANOVA with Bonferroni correction) (C).
Discussion
Taken together, our data demonstrate that platelet-derived β2M has a direct role as a central regulator of monocyte inflammatory responses. We have found that β2M and TGFβ may both signal monocytes through the same receptor but with different downstream signaling pathways and physiologic outcomes. This implies that robust platelet activation, such as occurs after MI, leads to elevated concentrations of plasma β2M that promote proinflammatory monocyte responses. However, after the immediate infarct event, the amount of ongoing platelet activation, and thus plasma β2M concentrations, eventually decline below the β2M signaling threshold. There is then a shift to proreparative monocytes that may be facilitated by a decline in β2M, allowing for TGFβ signaling that occurs at much lower concentrations than β2M. Latent TGFβ also becomes activated after MI, perhaps accounting for a delay in TGFβ-dominant signaling. Our results point to the important role of platelets as master regulators of monocyte differentiation responses in ischemic tissue injury, including platelet regulation of monocyte activation and inflammatory phenotype differentiation in a time after tissue injury–dependent manner. How β2M and TGFβ both signal through a common receptor, but with different outcomes, is not clear. TAK1 signaling is, in part, ubiquitination dependent (38, 39), but ubiquitination of SMAD4 can limit SMAD2/3 nuclear trafficking (40). It is possible that ubiquitination-dependent events downstream of β2M and TGFβ receptor binding may regulate the different signaling outcomes, but more work is needed to demonstrate this.
It is interesting to note that, while platelets have been associated with promoting inflammatory effects in the context of MI and other acute tissue injury models, platelet-derived TGFβ has been shown to contribute to fibrosis in models of cardiac hypertrophy using the mouse transaortic constriction (TAC) model (27, 41). TAC promotes cardiac remodeling and fibrosis by producing pressure overload, and it results in less platelet activation than does MI. Mice lacking TGFβ only in platelets had less TAC-induced cardiac fibrosis (27), and mice depleted of platelets after TAC had a reduction in the proreparative cytokine IL-10 compared with control TAC mice (41). This indicates that, in more chronic vascular disease models, TGFβ signaling may dominate and produce a proreparative monocyte phenotype due to its lower signaling threshold compared with β2M. Therefore, injury size and tissue damage may dictate the degree of platelet activation response, which in turn — through β2M and TGFβ concentrations — may direct monocyte differentiation responses.
Although this study examined β2M in the context of MI, the results may also be extended to other disease contexts. Elevated β2M is a biomarker of both age- and HIV-associated neurocognitive decline (11, 42), although the β2M cell source and its role in the disease process beyond a simple biomarker is not known. Platelet activation is also linked to HIV-associated neurocognitive decline (43), but more research must be conducted to evaluate if this occurs through a β2M-dependent mechanism. In addition, TGFβR is expressed by many types of cells. Our studies only examined β2M-TGFβR signaling in monocytes. Similar interactions may occur in other cell types, but more work is needed to determine whether our findings are monocyte specific, likely extending these studies into many other fields of biologic research.
Here, we demonstrated a potentially novel mechanism of platelet-mediated monocyte responses and differentiation. Platelet-derived β2M and TGFβ function in opposition to regulate monocyte proinflammatory vs. proreparative differentiation. In the absence of either immune mediator, the inflammatory response balance is disrupted and adverse outcomes may occur. Therefore, as therapeutic strategies targeting platelets in an inflammatory context are contemplated, these counter-balancing effects must be considered along the posttissue time continuum, emphasizing the complex and still poorly understood roles for platelets in immune responses.
Methods
Reagents.
Anti–mouse APC MHC I [H-2Db] (catalog 17-5999-80, clone 28-14-8), –mouse PE-Cy5 MHC II [I-A/I-E] (catalog 50-141-32, clone [M5/114.15.2]), –mouse APC CD4 (catalog 17-0041-82, clone GK1.5), –mouse APC CD9 (catalog 17-0091-82, clone KMC8), and –mouse PE CD9 (catalog 12-00-91-81, clone KMC8) antibodies were purchased from eBioscience. Antibodies to mouse PE F4/80 (catalog 123110, clone BM8), mouse FITC CD3 (catalog 100306, clone 145-2C11), mouse FITC Ly6C (catalog 128006, clone HK1.4), mouse/human APC CD11b (catalog 101212, clone [M1/70]), mouse PerCP/Cy5.5 CD115 (catalog 135526, clone AFS98), mouse PE CD182 (catalog 149303, clone SA044G4), mouse PerCP/Cy5.5 CD184 (catalog 146510, clone L276F12), human BV421 CD14 (catalog 325627, clone HCD14), human APC CD16 (catalog 302012, clone 3G8), human APC/Cy7 CD68 (catalog 333821, clone [Y1/82A]), human BV605 CD163 (catalog 333615, clone [GHI/61]), and mouse APC IgG2a (catalog 400219, clone MOPC-173) were purchased from BioLegend. Mouse FITC P-selectin/CD62P (catalog 553744, clone RB40.34), mouse FITC CD45 (catalog 553080, clone 30-F11), mouse PE CD8a (catalog 553033, clone 53-6.7), mouse FITC Ly6G (catalog 551460, clone 1A8), human BV786 CD83 (catalog 565336, clone HB15e), mouse AlexaFluor 647 phospho-JNK (catalog 562481, clone N9-66), and mouse PE phospho-SMAD2/3 (catalog 562586, clone O72-670) antibodies were purchased from BD Biosciences. Human fibrinogen with Oregon Green 488 Conjugate (catalog F-13192) was purchased from Thermo Fisher Scientific. Rabbit anti-CD68 (catalog ab125212) for IHC, rabbit anti-TGFβ (catalog ab92486) and rabbit AlexFluor 488 anti-β2M (catalog ab195298) antibodies for immunoblot were purchased from Abcam. Rabbit monoclonal antibodies to phospho-Smad3 (catalog 9520S, clone C25A9), total Smad3 (catalog 9513S), phospho-Tak1 (catalog 4531S), total Tak1 (catalog 5206S), and Arg1 (catalog 93668S, clone D4E3M) were purchased from Cell Signaling Technology. ELISA kits for human IL-8 (catalog DY208), human IL-6 (catalog DY206), mouse IL-6 (catalog DY406), mouse KC (catalog DY453), mouse IL-10 (catalog M1000B), mouse TGFβ1 (catalog DY1679), mouse PF4 (catalog DY595), and human β2M (catalog KGE019) were purchased from R&D Systems. Proteome profiler mouse cytokine array kit, panel A (catalog ARY006) was purchased from R&D Systems and performed according to manufacturer instructions. ELISA kit for mouse β2M (catalog LS-F14141) was purchased from LifeSpan BioSciences. LPS (catalog L6529) from E. coli O55:B5 was purchased from MilliporeSigma. Human recombinant β2M (catalog BDB551089) was purchased from BD Biosciences. Human β2M isolated from human urine (PRO-553) was purchased from ProSpec. Human recombinant TGFβ1 (catalog 78067) and TGFβ3 (catalog 78131) was purchased from Stemcell Technologies. Recombinant mouse TGFβ1 (catalog 14-8342-62) was purchased from Thermo Fisher Scientific. 2-Methyltioadenosine diphosphate trisodium salt (ADP, catalog 1624) and TAK1 inhibitor (5Z)-7-Oxozeaenol (catalog 3604) were purchased from Tocris Bioscience. ALK-5 inhibitor SB431542 (catalog 61-646-15MG) was purchased from Thermo Fisher Scientific. P38 inhibitor SB202190 and JNK inhibitor SP600125 were purchased from Thermo Fisher Scientific. Human thrombin and Smad3 inhibitor SIS3 were purchased from Cayman Chemical. Neutralizing antibody to TGFβ and β2M were purchased from Bio X Cell. Recombinant human TGFβ receptor 1 and human TGFβ receptor 2 Fc chimera proteins were purchases from R&D Systems. Series S CM5 chip for surface plasmon resonance (SPR) was purchased through Thermo Fisher Scientific.
Mouse studies.
All mice used in this study were on a C57BL/6J background. After verification that male and female WT and β2M–/– platelets induced similar monocyte responses (Supplemental Figure 7), male mice were used for all studies. Platelet-specific β2M-KO mice were generated by crossing PF4-Cre mice (The Jackson Laboratory) with β2Mfl/fl mice (Cyagen Biosciences) to obtain a PF4-Cre-β2Mfl/fl. The B2m gene (GenBank accession no. NM_009735, Ensembl: ENSMUSG00000060802) is located on mouse chromosome 2. Exon 1 contains the translation initiation codon. Exon 3 contains the stop codon. Exons 2–3 were selected as the conditional KO region flanked by LoxP sites (Supplemental Figure 1). The targeting vector was generated using BAC clones from the C57BL/6J library and will be transfected into the C57BL/6 ES cell line. The conditional KO allele was obtained after Flp-mediated removal of the Neo selection marker. Cre mediated recombination deleted exons 2–3 of B2m. Myeloid TGFβR2–/– and control WT mice were provided by Julie Ann Sterling (Vanderbilt University, Nashville, Tennessee, USA) (32).
MI was induced by ligation of the LAD coronary artery, and echocardiography was performed (VisualSonics) by standard transsternal approach, as we have described in past studies (5). Mice were perfused with methylene blue, and hearts were collected and sectioned to indirectly quantify size of infarcted zone.
Complete blood counts (CBCs) were performed using an Abaxis VetScan HM5 on mouse blood obtained by retroorbital collection into EDTA coated tubes (Greiner Bio One). Mouse plasma was isolated from blood collected in EDTA tubes and stored at –20°C.
Mouse hearts were collected at day 3 after MI and immediately placed into digestion buffer at 37°C for 1 hour. Digestion buffer consisted of 1× PBS (Corning), 305 U/mg collagenase II (Worthington), and 100 mM CaCl2 (MilliporeSigma). The digested hearts passed through a 100-μm mesh nylon strainer and were then centrifuged at 350 g for 5 minutes. The pellet was resuspended into PBS and stained for flow cytometry.
Mice treated with anti-TGFβ antibody (clone 1D11.16.8, BioXcell) received initial injection (300 μg) the day before LAD ligation. A second injection (100 μg) was given on day 2 after MI. Mouse hearts and peripheral blood were collected at day 4 as previously described.
IHC.
WT and PF4Cre+-β2Mfl/fl mouse hearts were collected and immediately placed into fixative (60% methanol, 10% acetic acid, 30% dH2O). Hearts were cross sectioned, paraffin embedded, and sectioned onto slides at a thickness of 5 μM.
For immunostaining, slides were deparaffinized and rehydrated and placed into 3% H2O2 for 15 minutes. Slides were washed with TBS 3 times. In a pressure cooker, slides were incubated in Dako Target Retrieval Solution (catalog S1699) for 15 minutes, washed in PBS, and then incubated in Dako Protein Block (catalog X0909) for 30 minutes. Anti-CD68 (Abcam, catalog 125212) was diluted 1:500 into Dako Antibody Diluent (catalog S0809), anti-arg1 was diluted 1:500 (Cell Signaling Technology, catalog 93668S) and incubated overnight at 4°C. Slides were rinsed in PBS and biotinylated anti-rabbit antibody (Vector Laboratories, BA-1000, 1:250 in Dako Antibody Diluent) and were incubated for 30 minutes at room temperature. Slides were rinsed and incubated with VECTASTAIN Elite ABC-HRP Kit (Vector Laboratories, catalog PK-6100) for 30 minutes, and DAB peroxidase (HRP) substrate (Vector Laboratories, SK-4100) was added for 5 minutes. Slides were washed in dH20 for 5 minutes, counterstained, and coverslip were added. As a negative control, rat IgG2b (catalog I-1000, Vector Labs) was used in the primary antibody step.
Paraffin embedded mouse heart sections were mounted and stained with Masson’s trichrome reagent. Slides were deparaffinized, rehydrated, and placed in Bouin’s Fix Solution (Thermo Fisher Scientific, catalog 11-201) in the oven for 15 minutes. Slides were washed until the yellow color disappeared. Equal parts of Welgerts Hematoxylin A and B (Thermo Fisher Scientific, catalogs 50-317-75 and 50-317-79) were mixed, and solution was added to each section for 5 minutes. Slides were rinsed with tap water for 5 minutes and then quickly rinsed with dH20. Belbrich Scarlet Fuchsin (MilliporeSigma, catalog HT151) was added to samples for 5 minutes. Slides were rinsed with dH20 for 5 minutes. One part Phosphtungstic Acid (MilliporeSigma, catalog HT152), 1 part Phosphomolybdic Acid (MilliporeSigma, catalog HT153), and 2 parts dH20 were mixed and the phosphtungstic/phosphomolybdic solution added to each slide for 5 minutes. The solution was tapped off, and Aniline Blue (MilliporeSigma, catalog HT154) was added to each slide for 5 minutes. Slides were then rinsed with 1% acetic acid (diluted in dH20) for 1 minute, dehydrated to xylene, and mounted with a coverslip.
All IHC images analyzed were taken under 10× magnification, using a BX41 microscope, and imaged with SPOT camera and SPOT Basic imaging software. Trichrome slides were quantified using a previously published method (44) that utilizes ImageJ (NIH) with the color deconvolution (version 1.5) plug-in. The Masson trichrome vector was selected, and the green component was used for analysis. Each sample’s threshold was adjusted to upper slider 0, lower slider 200 to distinguish areas of collagen deposition. The images were measured with pixel intensity and normalized to surface area of tissue for each section. Slides stained for Arg1 were analyzed using ImageJ (NIH) and were converted to 8-bit; threshold was adjusted to upper slider 0, lower slider 160 to differentiate between positive cells (brown) and negative (blue). The adjusted images were measured for pixel intensity and normalized to tissue surface area.
Platelet studies.
Mouse platelets were obtained by retro-orbital bleed into heparinized Tyrodes solution. Washed WT and PF4-Cre-β2Mfl/fl mouse platelets were isolated as previously described (45). For activation, platelets were incubated with thrombin or ADP for 30 minutes. Surface CD62P, MHC I, and fibrinogen binding were analyzed by FACS. Washed platelets were resuspended in the monocyte culture media and stimulated with 1 U/mL of thrombin for 20 minutes; thrombin was neutralized with hirudin (1 U/ml). Platelets were pelleted by centrifugation, and the supernatant was collected to treat primary mouse cells in culture. TGFβ1 and PF4 release were measured by ELISA. The ratio of platelets to monocytes was at a 100:1 or 10:1 ratio, respective to the experiment.
For platelet aggregation, whole mouse blood was centrifuged for 15 minutes at 270 g, platelet rich plasma (PRP) was isolated, and samples were washed and resuspended in plasma at a concentration of 50 × 107 platelets/ml. Samples were incubated with a 1:100 dilution of anti- CD9-APC or CD9-PE (Abcam) antibodies for 15 minutes. Labeled platelets were mixed 1:1 and agonist-stimulated at 37°C while shaking, as we have published (46). Double-positive platelets were then quantified by flow cytometry.
Human plasma was collected from MI patients under the University of Rochester School of Medicine IRB approved protocol RSRB00061784, and normal healthy controls under IRB protocol RSRB00028659.
Cell culture.
THP-1 cells (ATCC) were seeded in 24-well plates with RPMI media and 1× glutamax (Thermo Fisher Scientific), 2% penicillin/streptomycin (Invitrogen), 10% FBS (Invitrogen), 1× Sodium Pyruvate (Invitrogen), 1× nonessential amino acids (Invitrogen), and 1× minimum essential medium vitamins (Invitrogen). Cells were incubated with SB431542 (10 μM, Thermo Fisher Scientific), (5Z)-7-Oxozeaenol (1 μM, Tocris Bioscience), SIS3 (10 μM, Cayman Chemical), or SP600125 (10 μM, Thermo Fisher Scientific) 1 hour prior to treatment with β2M (5 μg/ml) or TGFβ1 (10 ng/ml). CD16 surface expression was measured through FACS. IL-8 release was determined through ELISA (DuoSet, R&D Systems). Transcripts for Il8 and Il10 were analyzed by qPCR. THP-1 was treated with β2M (0, 1, 2, 5, 10 μg/ml) and/or TGFβ1 (0, 1, 5, 10 μg/ml) for 48 hours. IL-8 release was measured through ELISA and Il10 transcripts through qPCR.
Primary mouse monocytes were isolated from femurs and tibias of mice. The BM was flushed with isolation buffer (1× PBS, 2% FBS, 1 mM EDTA; Thermo Fisher Scientific) using a 20-gauge needle (BD Biosciences), and RBCs were lysed with ACK Lysis Buffer (Thermo Fisher Scientific). Cell suspension was passed through a 100-μm mesh nylon strainer, and monocytes were isolated using EasySep Mouse Monocyte Isolation Kit (Stemcell Technologies). Circulating monocytes were collected by retro-orbital bleed into EDTA-coated tubes and spun to collect a WBC-rich buffy coat. RBCs were lysed and monocytes were isolated by the described negative selection kit (StemCell Technologies, EasySep Mouse Monocyte Isolation Kit, catalog 19861). Isolated monocytes were cultured in DMEM with 1× Glutamax (Thermo Fisher Scientific), 10% FBS, and 2% penicillin/streptomyocin. Primary mouse monocytes were treated with recombinant β2M (5 μg/ml) or recombinant TGFβ1 (10 ng/ml) for 48 hours. Isolated mouse monocytes treated with platelet releasate were pretreated with control buffer, anti-TGFβ antibody (10 μg/ml), or anti-β2M antibody (10 μg/ml) for 1 hour prior to treatment with platelet releasate. Recombinant β2M (5 μg/ml) or PBS was pretreated overnight at 37°C with NEM (Thermo Fisher Scientific, 10 μM) or control buffer under constant rocking. Primary mouse monocytes were then treated with the coincubated PBS or β2M ± NEM for 48 hours. Ly6C surface expression was measured through FACS.
Human peripheral blood was obtained from the New York Blood Center (New York, New York, USA). The buffy coat was isolated from whole blood using Ficoll-Paque Plus (GE Healthcare) density centrifugation with SepMate-50 (Stemcell Technologies) according to the manufacturer instructions. From the buffy coat, primary human monocytes were isolated using EasySep Human Monocyte Isolation Kit (Stemcell Technologies). Monocytes were incubated in the RPMI media with 10% FBS and 2% penicillin/streptomyocin. Human monocytes were treated with purified β2M derived from human urine (10 μg/ml).
Adult cardiac fibroblasts were isolated from C57/Bl6 mice hearts. Hearts were excised from the mouse chest, and the aorta and atria were removed prior to cutting the ventricular myocardium into 1-mm pieces in 1× PBS. Tissue was then digested in 10 mg of Collagenase II (Worthington) diluted in 10 ml of PBS using a gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec). Digested tissue was then placed through a 100-mm strainer with full media (high glucose DMEM media containing 10% FBS and 1% penicillin/streptomycin) and centrifuged at 270 g for 10 minutes. Pelleted cells were then plated on plastic culture dishes in full media for 5 hours to allow for the adherence of fibroblasts. After 5 hours, media was removed and adherent fibroblasts were washed twice with PBS before replacing cells with fresh full media. Media changes occurred once a day until fibroblasts reached 80% confluency. Isolated peripheral monocytes were then added and cocultured for 72 hours.
SPR.
SPR measurements were performed on a General Electric (GE) Biacore T200 instrument, utilizing a CM5 sensor chip. TGFβR1/Alk-5 was covalently surface immobilized to the CM5 sensor chip at a concentration of 30 μM/ml. Experiments were performed using a running buffer of 10 mM HEPES (pH 7.4), 150 mM NaCl, 0.005 % Tween 20, and 1% DMSO that was of 0.2 μm filtered and degassed. Kinetics/affinity binding experiments for TGFβ1 (80 nM), TGFβ3 (80 nM), β2M (40 μM), BSA (7 μM), and FBS (10%) were run at a flow rate of 30 μl/min. Injections had a contact time of 90 seconds and a dissociation time of 120 seconds, and binding was measured in relative response units (RU) that were calculated based on the difference between the immobilized protein flow cell and control flow cell to subtracted blank injections of buffer alone. The regeneration buffer consisted of 2 M NaCl and was injected after each ligand was flowed. Regeneration buffer had a contact time of 30 seconds, a flow rate of 30 μl/min, and a stabilization period of 60 seconds.
TGFβR2 was covalently surface immobilized to the CM5 sensor chip at a concentration of 60 μM/ml. Affinity binding experiments for TGFβ1 (20 nM), TGFβ3 (20 nM), β2M (40 μM), BSA (7 μM), and FBS (10%) were run at a flow rate of 30 μl/min. Injections had a contact time of 60 seconds and a dissociation time of 120 seconds, and binding was measured in relative RU that were calculated based on the difference between the immobilized protein flow cell and control flow cell to subtracted blank injections of buffer alone. The regeneration buffer consisted of glycine pH 2.0 (GE Healthcare) and was injected after each ligand was flowed. Regeneration buffer had a contact time of 30 seconds, a flow rate of 30 μl/min, and a stabilization period of 60 seconds.
Analysis of results were performed using GE Biacore T200 evaluation software version 3.0. Results were graphed in GraphPad Prism as an average of replicates.
qPCR.
Hearts from mice were collected day 0 or day 3 after MI and placed into RNAlater RNA Stabilization Reagent (Qiagen). Isolated hearts were homogenized using a Tissue-Tearor (BioSpec). Isolated primary mouse monocytes from peripheral blood, whole blood platelets, and/or primary heart fibroblasts were pelleted and resuspended in RLT lysis buffer. Cultured THP-1 cells were pelleted and resuspended in RLT lysis buffer. RNA was extracted using RNeasy Mini Kit (Qiagen). Concentration of RNA was determined using Nanodrop (Thermo Fisher Scientific). The isolated RNA was turned into cDNA using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). Real-time PCR analysis was performed using the TaqMan Gene Expression Master Mix Protocol on a Bio-Rad iCycler. TaqMan Gene expression primers were purchased from Thermo Fisher Scientific.
Immunoblotting.
Cells were lysed with 1× cell lysis buffer (Cell Signaling Technology) mixed with protease inhibitor cocktail (MilliporeSigma) per manufacture instructions. Protein samples were mixed 1:1 with 2× Laemmli buffer and loaded into Mini-PROTEAN TGX Gels (Bio-Rad). The gels were run at 100 V, in 1× Tris/Glycine/SDS Buffer. Protein was transferred from the SDS-PAGE gel to a nitrocellulose membrane (Bio-Rad) in transfer buffer at 110 V for 1 hour with an ice pack. Blots were blocked in blocking buffer consisting of 3% BSA (MilliporeSigma) dissolved in Tris-buffered saline (Thermo Fisher Scientific) with 0.1% Tween-20 (TBS-T) for 1 hour at room temperature. Primary antibody was diluted 1:1000 in blocking buffer and incubated overnight at 4°C. Anti–rabbit HRP was used as the secondary antibody (GE Healthcare) and diluted 1:10,000 in 5% milk for 1 hours at room temperature under constant rocking. Immunoblots were developed with Supersignal West Pico (Thermo Fisher Scientific) using the Bio-Rad ChemiDoc MP chemiluminescence settings.
Nonreducing gel electrophoresis.
Recombinant β2M (10 μg/ml) was pretreated overnight at 37°C with NEM (10 μM) or control buffer under constant rocking. Samples were mixed 1:4 into 4× non-reducing loading buffer (bromophenol blue, xylene cyanol, glycerol in H2O). Samples were loaded into Mini-PROTEAN TGX Gels (Bio-Rad). The blots were run at 100 V, in 1× Tris/glycine buffer. Protein was transferred from the nonreducing gel to a nitrocellulose membrane (Bio-Rad) in transfer buffer at 100 V for 1 hour with an ice pack. Blots were blocked in blocking buffer consisting of 3% BSA (MilliporeSigma) dissolved in Tris-buffered saline (Thermo Fisher Scientific) with 0.1% Tween-20 (TBS-T) for 1 hour at room temperature. FITC-tagged β2M antibody was diluted 1:1000 in blocking buffer and incubated overnight at 4°C. Blots were developed using Bio-Rad ChemiDoc MP Alexa 488 setting.
Data analysis.
Flow cytometers were either Accuri C6 or BD LSR II. All FACS samples were analyzed using FlowJo version 7.6. Proteome profiler mouse cytokine array kit, panel A was analyzed using ImageJ version 1.50i. Real-time PCR was analyzed in Microsoft Excel using calculations for fold change 2(–ΔΔCT) using Gapdh as a reference gene and normalized to genotype controls. Relative protein expression was determined using Bio-Rad Image Lab version 6.0.
Statistics.
All statistical tests were done using GraphPad Prism. Standard 2-tailed Student’s t test was used for when 2 independent samples were compared, with P < 0.05 considered significant. One-way ANOVA with Bonferroni correction was used when comparing more than 2 independent groups. *P < 0.05; **P < 0.01. All data represents mean ± SEM.
Study approval.
All animal studies were approved by the University of Rochester IACUC under protocol no. 2009-022. Human studies were approved by the University of Rochester Institutional Review Board under protocol number RSRB00061784 and normal healthy controls under IRB protocol RSRB00028659. All subjects provided informed consent prior to their participation.
Author contributions
ZTH and CNM planned, performed, and analyzed experiments and wrote the manuscript. DNP, SKT, AM, PQ, AAA, and ALJ all performed and analyzed experiments. JP, KEM, SJC, MRE, JLJ, JAS, ARM, and EMS all provided experimental input, reagents, and analyzed data.
Supplementary Material
Acknowledgments
CNM was supported by NIH grants HL124018 and HL141106, AHA grant 13EIA14250023, University of Rochester Center for AIDS Research P30AI078498, and University of Rochester CTSA award no. UL1 TR002001. ZTH was supported by NIH grants 5T32AI049815-17, University of Rochester CTSA funding GR500380, and AHA 18PRE3403016. DP was supported by T32 AI 7285-30
Version 1. 01/31/2019
In-Press Preview
Version 2. 03/07/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(5):e122943. https://doi.org/10.1172/jci.insight.122943.
Contributor Information
Zachary T. Hilt, Email: Zachary_hilt@urmc.rochester.edu.
Daphne N. Pariser, Email: daphne_pariser@urmc.rochester.edu.
Sara K. Ture, Email: Sara_Ture@URMC.Rochester.edu.
Amy Mohan, Email: Amy_Mohan@urmc.rochester.edu.
Pearl Quijada, Email: pearl_quijada@urmc.rochester.edu.
Scott J. Cameron, Email: scott_Cameron@URMC.Rochester.edu.
Julie A. Sterling, Email: julie.sterling@Vanderbilt.edu.
Alyssa R. Merkel, Email: alyssa.r.merkel@vanderbilt.edu.
Andrew L. Johanson, Email: ajohans5@jhmi.edu.
Jermaine L. Jenkins, Email: jermaine_jenkins@urmc.rochester.edu.
Eric M. Small, Email: Eric_Small@urmc.rochester.edu.
Kathleen E. McGrath, Email: KathleenE_McGrath@urmc.rochester.edu.
James Palis, Email: James_Palis@urmc.rochester.edu.
Michael R. Elliott, Email: michael_elliott@urmc.rochester.edu.
Craig N. Morrell, Email: Craig_Morrell@urmc.rochester.edu.
References
- 1.Gawaz M, Langer H, May AE. Platelets in inflammation and atherogenesis. J Clin Invest. 2005;115(12):3378–3384. doi: 10.1172/JCI27196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrell CN, Aggrey AA, Chapman LM, Modjeski KL. Emerging roles for platelets as immune and inflammatory cells. Blood. 2014;123(18):2759–2767. doi: 10.1182/blood-2013-11-462432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi G, et al. Platelet factor 4 limits Th17 differentiation and cardiac allograft rejection. J Clin Invest. 2014;124(2):543–552. doi: 10.1172/JCI71858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Hundelshausen P, et al. RANTES deposition by platelets triggers monocyte arrest on inflamed and atherosclerotic endothelium. Circulation. 2001;103(13):1772–1777. doi: 10.1161/01.CIR.103.13.1772. [DOI] [PubMed] [Google Scholar]
- 5.Cameron SJ, et al. Platelet Extracellular Regulated Protein Kinase 5 Is a Redox Switch and Triggers Maladaptive Platelet Responses and Myocardial Infarct Expansion. Circulation. 2015;132(1):47–58. doi: 10.1161/CIRCULATIONAHA.115.015656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson AM, et al. Beta2-microglobulin as a biomarker in peripheral arterial disease: proteomic profiling and clinical studies. Circulation. 2007;116(12):1396–1403. doi: 10.1161/CIRCULATIONAHA.106.683722. [DOI] [PubMed] [Google Scholar]
- 7.Kim MK, et al. Clinical utility of serum beta-2-microglobulin as a predictor of diabetic complications in patients with type 2 diabetes without renal impairment. Diabetes Metab. 2014;40(6):459–465. doi: 10.1016/j.diabet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Vianello A, et al. β2-Microglobulin and TIMP1 Are Linked Together in Cardiorenal Remodeling and Failure. Cardiorenal Med. 2015;5(1):1–11. doi: 10.1159/000369260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prentice RL, et al. Proteomic risk markers for coronary heart disease and stroke: validation and mediation of randomized trial hormone therapy effects on these diseases. Genome Med. 2013;5(12):112. doi: 10.1186/gm517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nead KT, et al. Usefulness of the addition of beta-2-microglobulin, cystatin C and C-reactive protein to an established risk factors model to improve mortality risk prediction in patients undergoing coronary angiography. Am J Cardiol. 2013;111(6):851–856. doi: 10.1016/j.amjcard.2012.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith LK, et al. β2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat Med. 2015;21(8):932–937. doi: 10.1038/nm.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy L, Beaulieu LM, Tanriverdi K, Freedman JE. The role of RNA uptake in platelet heterogeneity. Thromb Haemost. 2017;117(5):948–961. doi: 10.1160/TH16-11-0873. [DOI] [PubMed] [Google Scholar]
- 13.Rowley JW, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118(14):e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkhart JM, et al. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120(15):e73–e82. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- 15.Coppinger JA, et al. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood. 2004;103(6):2096–2104. doi: 10.1182/blood-2003-08-2804. [DOI] [PubMed] [Google Scholar]
- 16.Huo Y, et al. Circulating activated platelets exacerbate atherosclerosis in mice deficient in apolipoprotein E. Nat Med. 2003;9(1):61–67. doi: 10.1038/nm810. [DOI] [PubMed] [Google Scholar]
- 17.Gleissner CA, Shaked I, Erbel C, Böckler D, Katus HA, Ley K. CXCL4 downregulates the atheroprotective hemoglobin receptor CD163 in human macrophages. Circ Res. 2010;106(1):203–211. doi: 10.1161/CIRCRESAHA.109.199505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleissner CA, Shaked I, Little KM, Ley K. CXC chemokine ligand 4 induces a unique transcriptome in monocyte-derived macrophages. J Immunol. 2010;184(9):4810–4818. doi: 10.4049/jimmunol.0901368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sager HB, et al. Proliferation and Recruitment Contribute to Myocardial Macrophage Expansion in Chronic Heart Failure. Circ Res. 2016;119(7):853–864. doi: 10.1161/CIRCRESAHA.116.309001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nahrendorf M, Pittet MJ, Swirski FK. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121(22):2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nahrendorf M, et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204(12):3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science. 1990;248(4960):1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Yi Q. Beta2-microglobulin as a potential initiator of inflammatory responses. Trends Immunol. 2003;24(5):228–229. doi: 10.1016/S1471-4906(03)00076-0. [DOI] [PubMed] [Google Scholar]
- 24.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11(11):762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Zhang L, Yu C, Yang XF, Wang H. Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark Res. 2014;2(1):1. doi: 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee R, Kanti Barman P, Kumar Thatoi P, Tripathy R, Kumar Das B, Ravindran B. Non-Classical monocytes display inflammatory features: Validation in Sepsis and Systemic Lupus Erythematous. Sci Rep. 2015;5:13886. doi: 10.1038/srep13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer A, et al. Platelet TGF-β1 contributions to plasma TGF-β1, cardiac fibrosis, and systolic dysfunction in a mouse model of pressure overload. Blood. 2012;119(4):1064–1074. doi: 10.1182/blood-2011-09-377648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F, et al. TGF-β induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget. 2016;7(32):52294–52306. doi: 10.18632/oncotarget.10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowley DR, Dang TD, McBride L, Gerdes MJ, Lu B, Larsen M. Beta-2 microglobulin is mitogenic to PC-3 prostatic carcinoma cells and antagonistic to transforming growth factor beta 1 action. Cancer Res. 1995;55(4):781–786. [PubMed] [Google Scholar]
- 30.Sun W, et al. Human epithelial-type ovarian tumour marker beta-2-microglobulin is regulated by the TGF-β signaling pathway. J Transl Med. 2016;14:75. doi: 10.1186/s12967-016-0832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 32.Buenrostro D, et al. Early TGF-β inhibition in mice reduces the incidence of breast cancer induced bone disease in a myeloid dependent manner. Bone. 2018;113:77–88. doi: 10.1016/j.bone.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, et al. LPS-induced up-regulation of TGF-beta receptor 1 is associated with TNF-alpha expression in human monocyte-derived macrophages. J Leukoc Biol. 2008;83(5):1165–1173. doi: 10.1189/jlb.0807521. [DOI] [PubMed] [Google Scholar]
- 34.Santambrogio C, et al. DE-loop mutations affect beta2 microglobulin stability, oligomerization, and the low-pH unfolded form. Protein Sci. 2010;19(7):1386–1394. doi: 10.1002/pro.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Linthout S, Miteva K, Tschöpe C. Crosstalk between fibroblasts and inflammatory cells. Cardiovasc Res. 2014;102(2):258–269. doi: 10.1093/cvr/cvu062. [DOI] [PubMed] [Google Scholar]
- 36.Mills CD, Harris RA, Ley K. Macrophage Polarization: Decisions That Affect Health. J Clin Cell Immunol. 2015;6(5):364. doi: 10.4172/2155-9899.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao XM, et al. Splenic release of platelets contributes to increased circulating platelet size and inflammation after myocardial infarction. Clin Sci. 2016;130(13):1089–1104. doi: 10.1042/CS20160234. [DOI] [PubMed] [Google Scholar]
- 38.Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26(22):3214–3226. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 39.Chen IT, Hsu PH, Hsu WC, Chen NJ, Tseng PH. Polyubiquitination of Transforming Growth Factor β-activated Kinase 1 (TAK1) at Lysine 562 Residue Regulates TLR4-mediated JNK and p38 MAPK Activation. Sci Rep. 2015;5:12300. doi: 10.1038/srep12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heldin CH, Moustakas A. Role of Smads in TGFβ signaling. Cell Tissue Res. 2012;347(1):21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 41.Yang F, et al. Coronary artery remodeling in a model of left ventricular pressure overload is influenced by platelets and inflammatory cells. PLoS One. 2012;7(8):e40196. doi: 10.1371/journal.pone.0040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enting RH, et al. Cerebrospinal fluid beta2-microglobulin, monocyte chemotactic protein-1, and soluble tumour necrosis factor alpha receptors before and after treatment with lamivudine plus zidovudine or stavudine. J Neuroimmunol. 2000;102(2):216–221. doi: 10.1016/S0165-5728(99)00219-2. [DOI] [PubMed] [Google Scholar]
- 43.Singh MV, et al. Characterization of platelet-monocyte complexes in HIV-1-infected individuals: possible role in HIV-associated neuroinflammation. J Immunol. 2014;192(10):4674–4684. doi: 10.4049/jimmunol.1302318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Yu Q, Xu C-B. A convenient method for quantifying collagen fibers in atherosclerotic lesions by ImageJ software. Int J Clin Exp Med. 2017;10(10):14927–14935. [Google Scholar]
- 45.Matsushita K, et al. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115(2):139–150. doi: 10.1016/S0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modjeski KL, Ture SK, Field DJ, Cameron SJ, Morrell CN. Glutamate Receptor Interacting Protein 1 Mediates Platelet Adhesion and Thrombus Formation. PLoS One. 2016;11(9):e0160638. doi: 10.1371/journal.pone.0160638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.