Abstract
Atopic dermatitis (AD) is a complex inflammatory skin disease mediated by immune cells of both adaptive and innate types. Among them, CD4+ Th cells are one of major players of AD pathogenesis. Although the pathogenic role of Th2 cells has been well characterized, Th17/Th22 cells are also implicated in the pathogenesis of AD. However, the molecular mechanisms underlying pathogenic immune responses in AD remain unclear. We sought to investigate how the defect in the AD susceptibility gene, Ets1, is involved in AD pathogenesis in human and mice and its clinical relevance in disease severity by identifying Ets1 target genes and binding partners. Consistent with the decrease in ETS1 levels in severe AD patients and the experimental AD-like skin inflammation model, T cell–specific Ets1-deficient mice (Ets1ΔdLck) developed severe AD-like symptoms with increased pathogenic Th cell responses. A T cell–intrinsic increase of gp130 expression upon Ets1 deficiency promotes the gp130-mediated IL-6 signaling pathway, thereby leading to the development of severe AD-like symptoms. Functional blocking of gp130 by selective inhibitor SC144 ameliorated the disease pathogenesis by reducing pathogenic Th cell responses. Our results reveal a protective role of Ets1 in restricting pathogenic Th cell responses and suggest a potential therapeutic target for AD treatment.
Keywords: Immunology
Keywords: Adaptive immunity, Allergy, T cells
Transcription factor Ets1 is a key regulator in the development and progression of atopic dermatitis by suppressing pathogenic Th cell responses.
Introduction
Atopic dermatitis (AD) is an inflammatory skin disorder with chronic, relapsing, pruritic, inflammatory eczematous eruption that usually develops in early childhood (1). The pathogenesis of AD is a complex interplay of genetic, environmental, skin barrier (including microbiota), and immunological factors (2), which mainly contribute to the development and progression of AD. Both innate and adaptive immune cells are involved in the complex immune network in cutaneous inflammation, yet CD4+ T cells are known to be a key population promoting AD pathogenesis (3, 4). Among the Th subsets, Th2 cells are the key pathogenic mediators of AD progression by producing IL-4, IL-5, IL-10, IL-13, and IL-31, which enhance IgE levels and development and survival of eosinophils (5). Th17 and Th22 immune responses are also reported in skin-related inflammatory diseases, including AD. Among different kinds of skin cells, keratinocytes play an important function in the regulation of inflammation, responding to environmental and proinflammatory stimuli such as the cytokines IL-17 and IL-22 released by Th17 or Th22 cells (6–11). However, the mechanisms occurring via the activity of cytokines such as IL-17 and IL-22 from pathogenic Th cells under conditions of AD are not fully understood.
Expression array technologies have identified a set of genes, including those encoding cytokines, chemokines, IgE receptor, proteases, and pattern recognition receptors (PRRs), that are relevant to AD pathogenesis (12). Recent genome-wide association studies (GWAS) have also updated the landscape of AD genetics, and for the most part, gene sets were related to immune signaling and T cell polarization (13). While not detracting from the importance of maintaining the skin barrier in the prevention and treatment of AD, the above findings led to new therapeutic approaches targeting immune modulation of AD. Recently, GWAS have identified 11 new loci for AD using a large number of cohorts (13). Among them, Ets1 was selected with the highest P value and further supported association with self-reported allergy (14). However, the method by which defective Ets1 leads to AD development is still unclear.
Ets1 is prototype of Ets family transcription factors, specifically binding to core GGAA/T element, and is shown to have versatile roles in various biological processes by regulating expression of multifarious target genes (15). Although expression of Ets1 is ubiquitous, high levels of Ets1 expression is strictly confined to the lymphoid organ (16, 17), indicating crucial roles of Ets1 for the development and functionality of lymphoid cells. Consistently, germline Ets1-KO mice (Ets1–/–) showed impaired development of NK, NKT, and Treg cells (18–21) and incomplete thymocyte development (22, 23), suggesting crucial roles of Ets1 in hematopoietic development. In T cells, Ets1 modulates various T cell–specific genes such as Tcra and Tcrb (24). Additionally, Ets1 has been shown to act as a positive regulator for Th1 differentiation (25) or a negative regulator for Th17 differentiation (26), suggesting that Ets1 mainly modulates effector function of Th cells. Although the various studies have shown crucial roles of Ets1 in the immune system, pathophysiological roles of Ets1 in CD4+ T cells are still under scrutiny in a variety of immune disorders.
In this study, we have examined the role of Ets1 in AD development and progression. We show that absence of Ets1 triggered spontaneous development of AD-like symptoms, and T cell–specific Ets1-deleted mice (Ets1ΔdLck) were more susceptible to experimental AD-like skin inflammation. In T cells, Ets1 directly regulates pathogenicity of CD4+ T cells by acting as a strong transcriptional repressor of multiple targets involved in AD development. Our results demonstrate an importance role of Ets1 as a key regulator in the development and progression of AD.
Results
Inverse correlation between Ets1 expression and pathogenesis of AD.
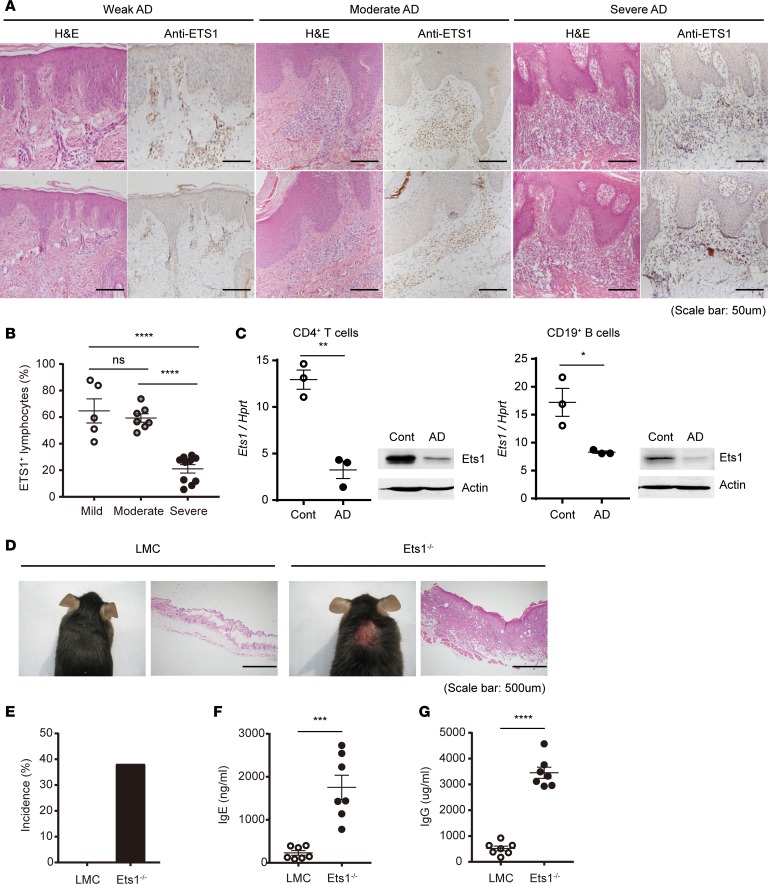
In order to investigate the clinical relevance of Ets1 expression in AD pathogenesis, we first analyzed the expression level of ETS1 in skin residual lymphocyte from AD patients with varying disease severity (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124202DS1). In weak and moderate AD patients diagnosed by their clinical symptoms, approximately 60% of tissue-infiltrated lymphocytes were shown to express ETS1 (weak, 444 ETS1+ cells among 721 cells; moderate, 1,400 ETS1+ cells among 2,271 cells). However, in the case of severe AD patients, ETS1 expression in tissue-infiltrated lymphocytes was significantly reduced to 20% (2,137 ETS1+ cells among 10,209 cells) (Figure 1, A and B) suggesting that reduced ETS1 level is highly correlated with severe AD. To corroborate these findings in mice, we analyzed Ets1 level in an experimental AD-like skin inflammation model induced by alternative application of hapten and house dust mite (HDM) extract in BALB/c mice (27) (Supplemental Figure 2A). Upon induction of the disease, mice showed clinical and molecular facets of AD-like symptoms, including destruction of ear tissues (Supplemental Figure 2B), increased ear thickness (Supplemental Figure 2C), elevated total and antigen-specific IgE (Supplemental Figure 2, D and E), and altered skin barrier integrity (Supplemental Figure 2F). Ets1 expression was decreased in lymphocytes (CD4+ T cells and CD19+ B cells) from the skin-draining lymph nodes (dLNs) upon induction of AD-like skin inflammation (Figure 1C), substantiating the notion that reduced Ets1 level is highly correlated with severe AD-like inflammation. In addition, we found that Ets1 germline–KO mice (Ets1–/– in C57BL/6 genetic background) bred under conventional conditions developed AD-like pruritic and erosive skin inflammation (Figure 1D). The incidence of AD-like skin inflammation was around 40% in Ets1-deficient mice (Figure 1E), with enhanced serum IgE and IgG levels (Figure 1, F and G). Collectively, these data suggest the role of Ets1 as a protective regulator of AD pathogenesis.
Figure 1. Ets1 expression is significantly reduced in skin lesion of severe AD patients and experimental animal model.
(A) H&E staining of the human skin biopsies confirmed the clinical diagnoses of weak, moderate, and severe atopic dermatitis (AD). Ets1+ lymphocytes were analyzed by IHC in the same tissues. (B) Frequency of Ets1+ lymphocytes from AD patient group with different disease severity is summarized in the graph. The data are expressed as mean ± SEM. ****P ≤ 0.0001; 1-way ANOVA. (C) After 4 weeks of AD induction in BALB/c mice, expression level of Ets1 was analyzed in peripheral CD4+ T and CD19+ B cells from skin-draining LNs by qPCR or immunoblotting. Quantification represents 3 independent experiments. (D) Representative phenotype of spontaneous AD-like skin inflammation developed in Ets1–/– mice (in C57BL/6 genetic background) under conventional conditions. (E) Incidence of AD-like skin inflammation among age-matched littermate controls (LMC) and Ets1–/– mice was calculated (n = 43 of LMC and 21 of Ets1–/– mice). (F and G) The total IgE and IgG levels in serum from mice groups at 12–16 weeks kept in conventional conditions were measured by ELISA (data comes from the analysis of 7 individual mice in each group). Error bars represent the mean ± SEM. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; ****P ≤ 0.0001; Student’s t test.
Ets1 deficiency increases the susceptibility to experimental AD.
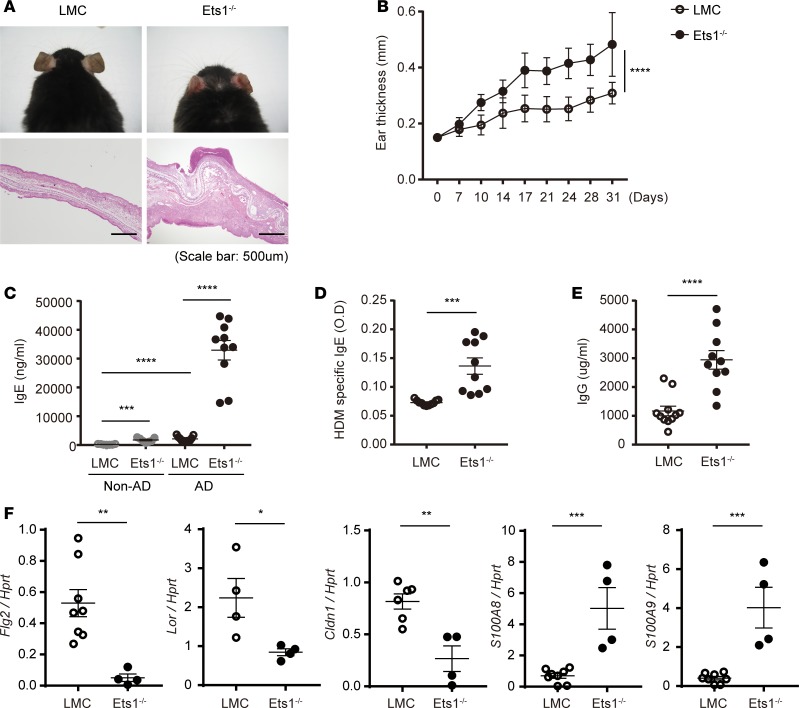
We also confirmed that Ets1–/– mice are more susceptible to induction of AD-like skin inflammation in specific pathogen free (SPF) conditions. Compared with littermate control (LMC), Ets1–/– mice showed significantly increased ear thickness and clinical symptoms, including edema, erosion, erythema, and heavy infiltration of mononuclear cells (Figure 2, A and B). Moreover, Ets1–/– mice showed much higher level of total serum IgE (Figure 2C), HDM-specific IgE (Figure 2D), and total serum IgG (Figure 2E). Consistent with histopathology shown in Figure 2A, significantly increased infiltration of immune cells such as CD4+ T cells and CD19+ B cells were observed in inflamed tissue of Ets1–/– mice (Supplemental Figure 3, A–C), leading to an alteration in expression levels of skin-barrier integrity–related molecules (Figure 2F). Ets1–/– mice showed much lower levels in filaggrin, claudin, loricrin, S100A8, and S100A9 compared with LMC mice. These data suggest that deficiency of Ets1 makes mice more susceptible for the development of AD-like symptoms, implying the vigilant roles of Ets1 in AD pathogenesis.
Figure 2. Ets1 deficiency promotes experimental AD pathogenesis under SPF conditions.
(A) Representative photographs of mouse ears from each group (upper) and H&E staining of the ear biopsies (lower) confirmed clinical symptoms of AD. (B) Ear thickness during the course of AD was measured 24 hours after DNCB or HDM extract application by using a dial thickness gauge. The data are expressed as mean ± SD. ****P ≤ 0.0001 (from day 17–31); 2-way ANOVA. (C–E) Total IgE (C), HDM allergen-specific IgE (D), and total IgG levels (E) in serum from the mouse groups were measured by ELISA. (F) RNA was collected from the cells of ear tissues, and the relative levels of filaggrin, claudin, loricrin, S100A8, and S100A9 were evaluated by qPCR after normalization with Hprt level. Data represent results from 3 independent experiments. Error bars represent the mean ± SEM. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; ****P ≤ 0.0001; Student’s t test.
T cell–specific Ets1 deletion enhances the susceptibility to AD induction.
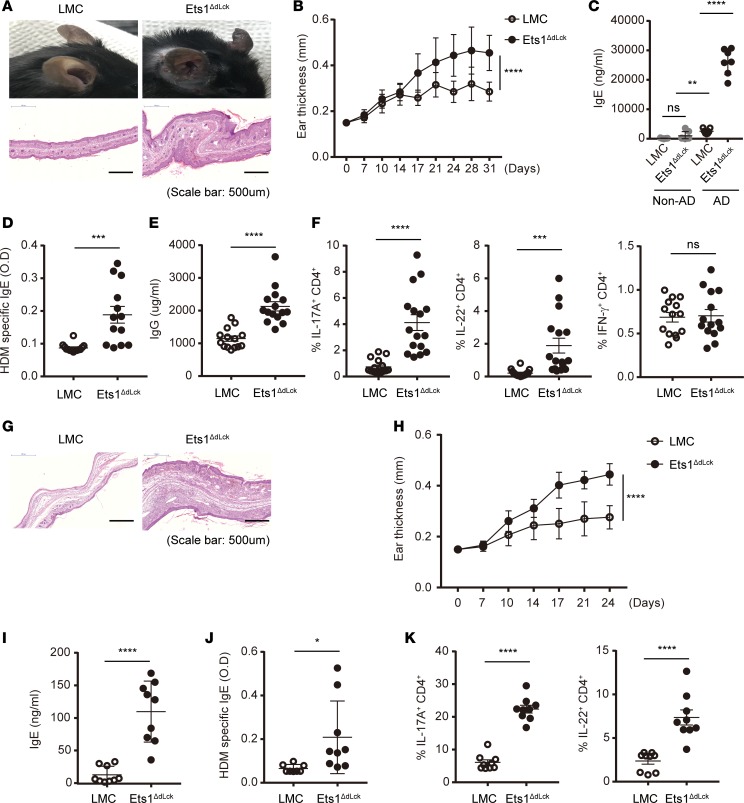
Next, we tested whether CD4+ T cells are involved in AD pathogenesis. Since systemic deletion of Ets1 leads to profound immune cell dysregulation (18–21, 25, 26), including defective T cell development (Supplemental Figure 4), we decided to delete Ets1 expression in mature CD4+ T cells. We generated Ets1 floxed mice (Ets1fl/fl) that have the exon 7 DNA binding regions flanked by 2 loxp sites (28). Ets1fl/fl mice were crossed with distal Lck-Cre mice, not with CD4-Cre mice, to avoid a developmental defect of T cell due to deletion of Ets1 in the thymus (28, 29). Indeed, distal Lck-Cre Ets1fl/fl mice (Ets1ΔdLck) showed specific deletion of Ets1 in peripheral CD4+ T cells (Supplemental Figure 5A). Ets1ΔdLck mice showed no defect in thymic T cell development (Supplemental Figure 5B) and thymic Treg development (Supplemental Figure 5C), and they showed normal peripheral CD4+ T cells proportion (Supplemental Figure 5, D and E) and overall intact cellularity in lymphoid tissues (Supplemental Figure 5F). In contrast to germline Ets1–/– mice, Ets1ΔdLck mice did not show any signs of autoimmune diseases. Next, we tested whether Ets1ΔdLck mice are susceptible to induction of AD-like skin inflammation compared with LMC mice. Indeed, Ets1ΔdLck mice were more susceptible to the disease induction and showed AD-like pathophysiology similar to germline Ets1–/– mice, including increased ear thickness (Figure 3, A and B), serum total IgE, antigen-specific IgE and IgG levels (Figure 3, C–E), and massive mononuclear cells inflation (Supplemental Figure 6A), and they showed an alteration in expression levels of skin-barrier integrity–related molecules (Supplemental Figure 6B). dLN CD4+ T cells isolated from Ets1ΔdLck mice at the peak of the disease showed a massive increase in cellularity (Supplemental Figure 6C) and an increase in the levels of Th2 cytokines (IL-4, IL-10, and IL-13) (Supplemental Figure 6D). Moreover, we also observed significant elevation of IL-17A and IL-22, but not IFN-γ, production in Ets1ΔdLck mice (Figure 3F). We also tested whether enhanced AD-like skin inflammation in Ets1ΔdLck mice is due to an alteration of Tregs under homeostatic and disease conditions. No significant differences were observed in Treg frequency in homeostatic conditions between the LMC and Ets1ΔdLck mice (Supplemental Figure 5, C and D). Ets1ΔdLck mice also did not show any defect in Treg frequency, even in AD-like disease conditions (Supplemental Figure 7A). We also analyzed frequency of Helios+ (Supplemental Figure 7B) and Ki-67+ (Supplemental Figure 7C) among CD4+Foxp3+ Tregs, as they are functional markers of suppressive activity of Tregs (30, 31). No significant differences were observed between LMC and Ets1ΔdLck mice under disease conditions. To further confirm that the causality of Ets1-dependent intrinsic defects in CD4+ T cells is the key mechanism for AD pathogenesis in vivo, we performed adoptive transfer experiments. CD45.2+CD4+ T cells from LMC or Ets1ΔdLck mice, together with WT B cells (CD45.1+), were adoptively transferred into Rag1–/– mice; then, the mice were induced to develop experimental AD-like skin inflammation by alternative application of hapten and HDM extract. The recipient mice reconstituted with Ets1ΔdLck CD4+ T cells developed severe AD-like symptoms, such as severe tissue destruction (Figure 3G), increased ear thickness (Figure 3H), and higher levels of IgE (Figure 3, I and J). Furthermore, recipient mice reconstituted with CD4+ T cells from Ets1ΔdLck mice showed significantly higher frequencies of IL-17A+ and IL-22+ populations than the recipient mice reconstituted with LMC CD4+ T cells (Figure 3K). These results collectively suggest that Ets1 in CD4+ T cells has an intrinsic role to inhibit the differentiation of IL-17A/IL-22–producing pathogenic Th cells in the context of AD pathogenesis.
Figure 3. Specific deletion of Ets1 in mature T cells (Ets1ΔdLck) promotes AD pathogenesis by enhancing Th17 and Th2 responses.
(A) Representative photographs of mouse ears from each group (upper) and H&E staining of the ear biopsies (lower) confirmed the clinical symptoms of AD. (B) Ear thickness during the course of AD was measured. The data are expressed as mean ± SD. ****P ≤ 0.0001 (from day 17–31); 2-way ANOVA. (C–E) Total IgE (C), HDM allergen-specific IgE (D), and total IgG levels (E) in serum from the mouse groups were measured by ELISA. (F) Ex vivo isolated total lymphocytes from skin-draining LNs under AD were given PMA and ionomycin stimulation with GolgiStop or GolgiPlug for 4 hours. Intracellular staining of IL-17, IL-22, and IFN-γ by CD4-gated T cells were analyzed. (G) Naive CD4+ T cells were isolated from LMC and Ets1ΔdLck mice by means of fluorescence-activated cell sorting and transferred i.v. into Rag1−/− mice (5 × 105/mouse) with WT CD45.1+CD19+ B cells (2 × 106/mouse). AD was induced the day after cell transfer. H&E staining of the ear biopsies confirmed clinical symptoms of AD. (H) Ear thickness during the course of AD was measured. The data are expressed as mean ± SD. **P ≤ 0.005; ***P ≤ 0.0005; ****P ≤ 0.0001 (from day 17–24); 2-way ANOVA. (I and J) The total IgE (J) and HDM allergen-specific IgE levels (J) in serum from the mouse groups were measured by ELISA. (K) Intracellular staining of IL-17 and IL-22 by CD4-gated T cells were analyzed. Data represent results from 3–4 independent experiments. Error bars represent the mean ± SEM. *P ≤ 0.05; ****P ≤ 0.0001; Student’s t test.
Ets1 deficiency enhances pathogenic Th cell response in AD development.
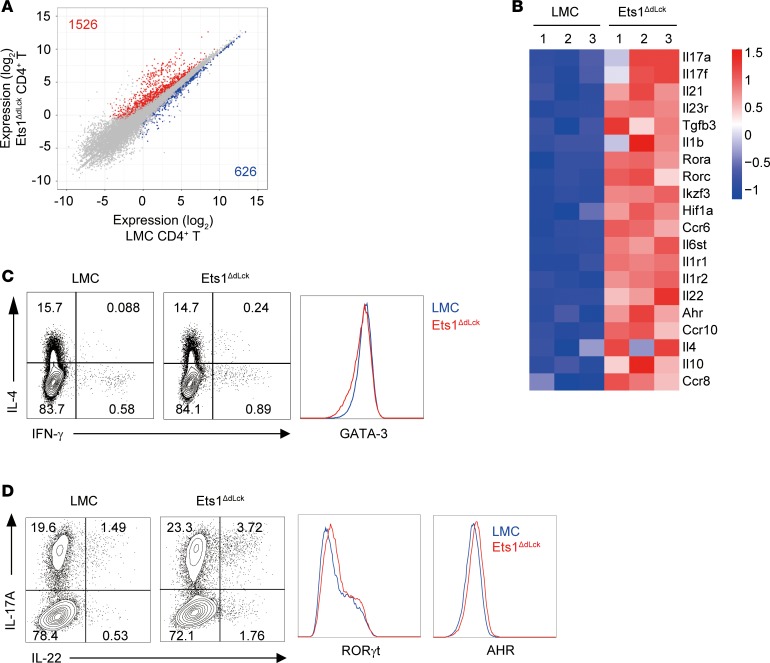
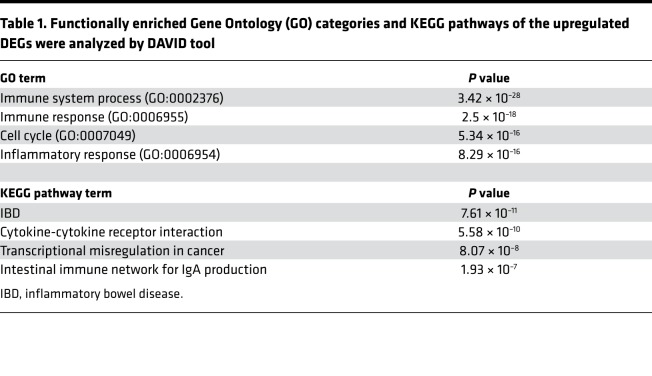
To identify the molecular mechanisms responsible for enhanced AD-like pathogenesis upon Ets1 deficiency in CD4+ T cells, we compared transcriptome profile between CD4+ T cells from LMC and Ets1ΔdLck mice at the peak of disease progression by performing RNA sequencing (RNA-seq). Among the genes expressed in the CD4+ T cells (~13,113), 1,526 genes were upregulated and 626 genes were downregulated in Ets1-deficient CD4+ T cells (defined as genes with at least 1.5-fold change, adjusted P ≤ 0.05, and expression above a minimal threshold based on the distribution of all expressed genes) relative to their expression in LMC CD4+ T cells (Figure 4A). Various inflammatory pathways such as inflammatory responses (in Gene Ontology [GO] terms) (http://geneontology.org) and cytokine-to-cytokine receptor interaction (in Kyoto Encyclopedia of Genes and Genomes [KEGG] pathways) (https://www.genome.jp/kegg/) were significantly enriched in Ets1-deficient CD4+ T cells (Table 1). Interestingly, genes related with pathogenic Th17 immune response (Il17a/f, Il22, Rora, and Rorc) were significantly upregulated in Ets1-deficient CD4+ T cells, together with some of the Th2-related genes (Il4, Il10, and Ccr8) (Figure 4B). Next, we tested whether Ets1 deficiency leads to dysregulation of Th cell differentiation in non-AD conditions. Naive CD4+ T cells isolated from LMC or Ets1ΔdLck mice were differentiated into Th1, Th2, Th17, or pathogenic Th17 cells in vitro (32); then, levels of cytokines were analyzed. In line with a previous report in germline Ets1–/– mice (25), CD4+ T cells from the Ets1ΔdLck mice showed reduced IFN-γ production but similar T-bet expression under in vitro Th1 differentiation conditions (Supplemental Figure 8A). Likewise in germline Ets1–/– mice (26), CD4+ T cells from the Ets1ΔdLck mice showed increased IL-17A production under in vitro Th17 differentiation (Supplemental Figure 8B). Ets1-deficient T cells from Ets1ΔdLck mice showed a similar frequency of IL-4–producing Th2 cells with comparable GATA-3 expression (Figure 4C). Interestingly, although we could still detect comparable expression of RORγt and AHR, a significant increase in IL-17 and IL-22 production was observed in the Ets1-deficient CD4+ T cells under pathogenic Th17 differentiation conditions (Figure 4D). Ex vivo CD4+ T cells from Ets1ΔdLck mice also showed significantly enhanced production of IL-17A and IL-22 (Supplemental Figure 8, C–E). No significant differences were observed in Th2-related cytokines (Supplemental Figure 8, C–E). These results suggest that Ets1 deficiency may enhance pathogenic Th17 (IL-17A and IL-22) production, which mediates AD-like skin inflammation.
Figure 4. RNA-seq analysis of CD4+ T cells from LMC and Ets1ΔdLck mice under AD condition.
RNA was collected from FACS-sorted cells of skin-draining LNs at the peak of the disease onset (day 31). (A) Plot of gene expression (as log2 normalized read count) in LMC vs. Ets1ΔdLck-derived CD4+ T cells. Significantly down- and upregulated genes (defined as genes with at least 1.5-fold change, adjusted P ≤ 0.05, and expression above a minimal threshold based on the distribution of all expressed genes) are colored blue or red, respectively, and their numbers are shown. (B) Heatmap of selected genes were depicted. Three replicates are shown in order. (C and D) Naive CD4+ T cells from LMC and Ets1ΔdLck mice were cultured 3–4 days with plate-bound anti-CD3 and anti-CD28 in the presence of skewing cytokines IL-4 and anti–IL-12 for Th2 and IL-1β and IL-6 and IL-23 for pathogenic Th17. Expression of the indicated cytokines and transcription factors were analyzed by flow cytometry. Representative plot comes from the analysis of 4–6 individual mice.
Table 1. Functionally enriched Gene Ontology (GO) categories and KEGG pathways of the upregulated DEGs were analyzed by DAVID tool.
Elevated gp130 mediates pathogenic Th17 cell differentiation in Ets1-deficient T cells.
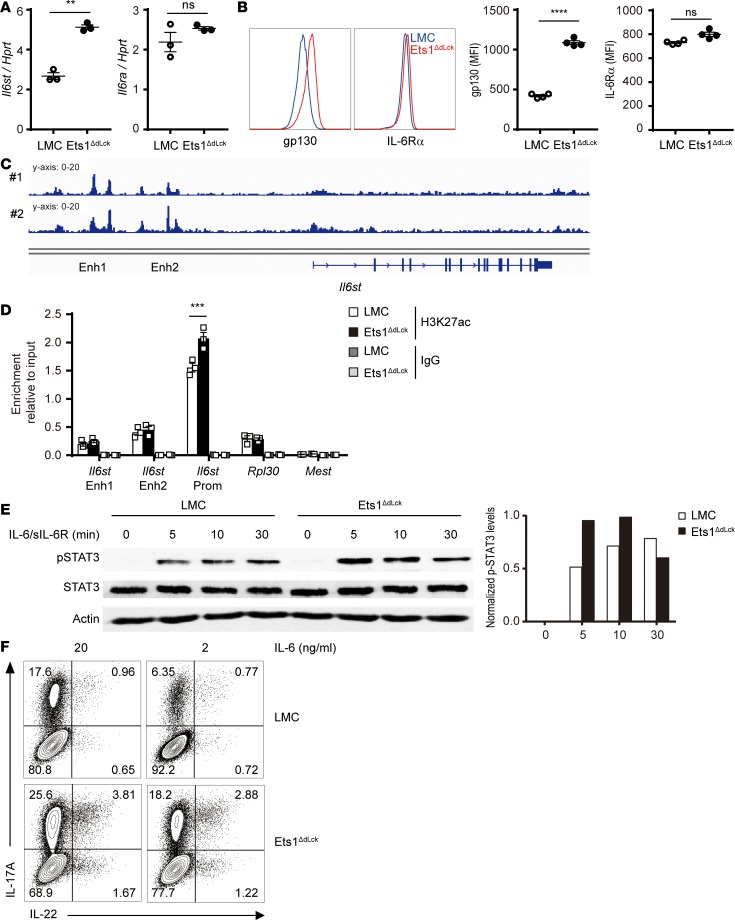
Since Ets1-deficient CD4+ T cells did not show a significant alteration in key regulators of Th17 such as RORγt and AHR under ex vivo and in vitro differentiation, we hypothesized that upstream signaling molecules involved in differentiation of Th17 cells might be affected in Ets1-deficient CD4+ T cells. Indeed, among various pathogenic Th17 genes analyzed in the disease condition, we identified Il6st (encodes gp130, a signaling subunit for the IL-6 receptor complex) as a key target gene (Figure 4B). Il6st showed a significant elevation in Ets1-deficient CD4+ T cells, while Il6ra level was not altered (Figure 5, A and B) under homeostatic conditions. To further confirm the intrinsic role of Ets1 in regulation of gp130, a mixed BM chimera experiment was performed. BM cells from LMC (CD45.2 Ets1fl/fl) or Ets1-deficient mice (CD45.2 Ets1ΔdLck) were mixed with BM cells from congenic CD45.1 WT mice at a 1:1 ratio and injected into sublethally irradiated Rag1–/– mice. Indeed, we could clearly observe increased gp130 levels in the Ets1-deficient BM transferred group, implying that Ets1 has a cell-intrinsic role as a transcriptional repressor for gp130 expression (Supplemental Figure 9A). To further dissect molecular mechanisms of Ets1 mediated Il6st expression, we reanalyzed previously reported Ets1 ChIP-seq data using ex vivo CD4+ T cells from the WT mice (33, 34). Indeed, we could observe a significant enrichment of Ets1 to the promoter region of Il6st and upstream regulatory elements, as well (Figure 5C). We also analyzed histone modifications status in the promoter region of Il6st (e.g, histone H3 Lys 27 acetylation, H3K27ac) as a parameter for the permissive chromatin state (35). ChIP-PCR analysis was performed with anti-H3K27ac antibody on the CD4+ T cells isolated from LMC and Ets1ΔdLck mice. Indeed, we found an increased enrichment of H3K27ac in the promoter region of Il6st (Figure 5D) in Ets1-deficient CD4+ T cells. Rpl30 and Mest were used as positive and negative controls, respectively. We also analyzed the levels of phospho-STAT3, which is the key factor as a downstream signal involved in gp130-mediated trans-signaling pathways for Th17 response (36, 37). Indeed, Ets1-deficient CD4+ T cells showed stronger phosphorylation of STAT3 (Figure 5E) upon IL-6/sIL-6R complex stimulation and showed higher IL-17A and IL-22 levels, even at limited doses of IL-6 (Figure 5F). These results enlightened the role of Ets1 as the transcriptional repressor for Il6st expression to inhibit hyperactivation of the STAT3-mediated pathogenic Th17 response.
Figure 5. Effect of Ets1 deficiency on gp130 expression, IL-17A and IL-22 production, and IL-6–mediated STAT3 activation.
(A) RNA was collected from the naive CD4+ T cells from LMC and Ets1ΔdLck mice, and relative expression of Il6st and Il6ra were evaluated by qPCR (n = 3). (B) Protein levels of Il6st (gp130) and Il6ra (IL-6Rα) were examined by surface staining. Mean fluorescence intensity (MFI) is presented (n = 4). (C) Ets1 ChIP-seq confirmed binding of Ets1 on regulatory elements of Il6st locus. Numbers represent 2 different sets of data. (D) Histone modification (H3K27ac) of Il6st locus was analyzed by ChIP-qPCR. Quantification represents 3 independent experiments. (E) Naive CD4+ T cells from LMC and Ets1ΔdLck mice were stimulated with IL-6/sIL-6R complex for the indicated times, and STAT3, phospho-STAT3, and actin expression was examined by immunoblotting. The blots were scanned, and the levels of p-STAT3 were normalized to the total amount of STAT3 and depicted as a graph. (F) Naive CD4+ T cells from LMC and Ets1ΔdLck mice were cultured 3–4 days with plate-bound anti-CD3 and anti-CD28 in the presence of skewing cytokines IL-1β, IL-6, and IL-23. IL-6 was given under optimal (20 ng/ml) or limited (2 ng/ml) conditions. Expressions of the indicated cytokines were analyzed by flow cytometry. Representative plot comes from the analysis of 4–6 individual mice. Error bars represent the mean ± SEM. **P ≤ 0.005; ***P ≤ 0.0005; ****P ≤ 0.0001; Student’s t test.
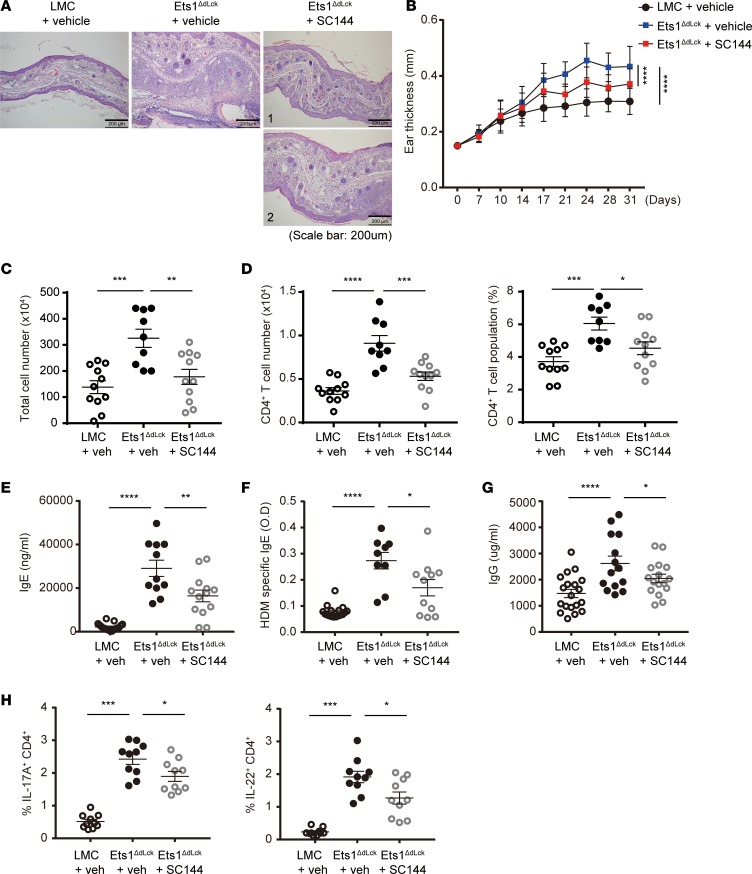
Administration of gp130 inhibitor suppresses AD pathogenesis of Ets1ΔdLckmice. To further validate the functional importance of gp130, we determined the effect of gp130 inhibition on AD-like pathogenesis of Ets1ΔdLck mice. For this purpose, we employed SC144 as a specific inhibitor of gp130. SC144 has been known to induce gp130 phosphorylation (S782) and deglycosylation to reduce STAT3 phosphorylation and its nuclear translocation, which subsequently inhibits the expression of downstream target genes (38). Administration of mock (vehicle) or SC144 was initiated a week after AD-like disease induction and continued every day (weekdays) by i.p. injection until the end of experiment. Compared with the vehicle-treated group, SC144 treatment significantly inhibited the AD-like symptoms in Ets1ΔdLck mice, characterized by reduced pathophysiology and ear thickness (Figure 6, A and B). This was associated with reduced lymphocytes infiltration (Figure 6, C and D) and reduced serum IgE (Figure 6, E and F) and IgG level (Figure 6G). Moreover, SC144 treatment significantly reduced IL-17A and IL-22 production in CD4+ T cells from the dLNs compared with vehicle controls (Figure 6H). These results indicate the causal function of gp130 in Ets1ΔdLck CD4+ T cells for the exaggerated AD-like pathology, and they are suggestive of a potential therapeutic remedy for AD upon gp130 blockade.
Figure 6. The gp130 inhibitor SC144 suppresses pathophysiology of AD in Ets1ΔdLck mice.
Experimental AD was induced in LMC and Ets1ΔdLck mice. The gp130 inhibitor, SC144, was administered via i.p. routes a week after the initial sensitization (5 times during weekdays for 3 weeks) and continued until the end of experiment. (A) H&E staining of the ear biopsies confirmed the clinical symptoms of AD. (B) Ear thickness during the course of AD was measured. The data are expressed as mean ± SD. ****P ≤ 0.0001 (from day 21–31); 2-way ANOVA. (C and D) Cells from the ear tissues were collected from each group of mice under AD (C) and absolute number and frequency of CD4+ T cells were calculated (D). (E–G) The total IgE (E), HDM allergen-specific IgE (F), and total IgG levels (G) in serum from the mouse groups were measured by ELISA. (H) Intracellular staining of IL-17 and IL-22 by CD4-gated T cells were analyzed. Data represent results from 3 independent experiments. Error bars represent the mean ± SEM. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; ****P ≤ 0.0001; Student’s t test.
Discussion
In this study, we revealed an important role of Ets1 as the key transcriptional repressor to preclude the differentiation of pathogenic Th17 cells, thereby suppressing the development and progression of AD-like skin inflammation. Furthermore, we identified Il6st (gp130) as the key factor involved in exaggerated AD-like pathology in the absence of Ets1 expression.
Previously, Ets1 has shown its causal roles in various inflammatory diseases. Various GWAS have identified ETS1 as candidate factor responsible for the susceptibility to various human autoimmune diseases (39–45), including systemic lupus erythematosus (SLE) (46). Intriguingly, a recent study has identified 11 new genetic loci associated with AD susceptibility that includes ETS1 with the highest P value (13). In this study, we defined an important role of Ets1 in AD pathogenesis. We could observe a strong negative correlation of ETS1 expression in lymphocytes with disease severity among AD patients and spontaneous AD-like skin inflammation in germline Ets1–/– mice. Because of versatile roles of Ets1 in various cell types such as survival of T cells (47), keratinocyte differentiation (48), and ILC2 development (49), it was hard to decipher which types of cells are mainly responsible for development of AD-like skin inflammation in the germline Ets1–/– system. To overcome this issue, we generated Ets1ΔdLck mice (details described below) to study the T cell–specific role of Ets1 in AD pathophysiology. Even though we could not find any immunological abnormality, such as naive/memory T cell ratio and Treg frequency, Ets1ΔdLck mice recapitulated overall AD-like symptoms comparable with the symptom of germline Ets1–/– mice, indicating that T cells are the causal population to induce AD pathogenesis in the absence of Ets1. Our study provides cellular details how the dysfunction of the transcriptional regulator Ets1 promotes pathogenic Th cell responses leading to AD pathogenesis.
Though a previous study has suggested the negative function of Ets1 in Th17 differentiation (26), the molecular action mechanisms of Ets1 for Th17 differentiation and pathophysiological evidence remain elusive. How does the deficiency of Ets1 in CD4+ T cells trigger AD pathogenesis? Multiple mechanisms might be involved, including the enhanced population of IL-17A– and IL-22–producing Th cells and gp130-mediated trans-signaling pathways for AD pathogenesis (36). CD4+ T cells from Ets1ΔdLck mice under AD-like skin inflammation highly produced pathogenic Th cell response–related cytokines, including IL-17A and IL-22 (Figure 3, F and K). An increased Il6st (gp130) level might be the key step in this procedure (Figure 5, A and B, and Supplemental Figure 9A). gp130 has been known as the key upstream component of Th17 cell signaling through functional enhancement of STAT3 activity (Figure 5E) (37), and it may serve as a key mediator of pathogenic Th17 cell response–related gene expression (50) in Ets1ΔdLck mice under AD-like skin inflammation conditions. Ets1 may also contribute to the progression of autoimmunity by controlling the expression of gp130 in IL-6–mediated signaling (36, 51). In addition, Ets1 may also regulate Th17 differentiation by orchestrating inhibitory (Il2 expression through Aiolos) (52) and facilitative (IL-6 signaling sensitivity through its receptor expression) mechanisms. Indeed, Ets1 deficiency in CD4+ T cells enhanced the expression of Ikzf3 (Aiolos), a known repressor for IL-2 expression in T cells (52) (Supplemental Figure 9, B–D), through direct binding in the Ikzf3 locus (Supplemental Figure 9E), thereby reducing IL-2 expression (Supplemental Figure 9F). In addition to CD4+ T cells, Ets1 may also regulate the expression levels of molecules related with skin barrier integrity. Compared with LMC mice, exaggerated skin inflammation was observed in Ets1ΔdLck mice (Supplemental Figure 6B). Skin lesions from Ets1ΔdLck mice showed significantly altered gene expression levels, including Flg2, Cldn1, and Lor and antimicrobial proteins S100A8/A9, a known target of IL-17A and IL-22 (53, 54). Taking one more step forward from these cellular mechanisms, we endeavored to uncover the molecular functions of Ets1 in CD4+ T cells. It has been shown that Ets family transcription factors mainly act as coactivators (55), but our data also suggest a dominant role of Ets1 as the transcriptional corepressor (Figure 4B; Figure 5, A and B; and Supplemental Figure 9, B and C). Our unbiased interactome analysis of Ets1 by IP/liquid chromatography/tandem mass spectrometry revealed that Ets1 may form a corepressor complex through interaction with HDAC1, mSin3a, HCFC1, and Suds3 (56–58) (Supplemental Figure 10). Moreover, we confirmed increased Il6st in Ets1-deficient CD4+ T cells accompanied by epigenetic modification (Figure 5D). Ets1 may act as a molecular switch by cooperating with the Sin3A repressor complex to prevent pathogenic Th cell differentiation and AD pathogenesis in CD4+ T cells.
While our study substantially advances the current understanding of AD pathogenesis, several questions remain. Correlation analysis between ETS1 levels and disease, upon analyzing ETS1 levels in CLA+ memory CD4+ T cells in AD patients, may help in substantiating a direct link between ETS1 expression and AD pathogenesis. It will be also interesting to define whether Ets1 also plays an important role in regulation of Th17-mediated inflammatory disorders, such as an infection model and skin psoriasis (59). Moreover, it will be interesting to test whether treatment with IL-17 or IL-17 receptor blocking antibody could ameliorate AD-like pathogenesis in Ets1ΔdLck mice and AD patients.
Our study suggests the role of Ets1 as a key regulator of pathogenic Th cell–mediated AD development. Dysregulation of Th17 cell response and gp130 in Ets1 deficiency may be causative to pathogenesis of various autoimmune diseases. IL-17A– and IL-22–producing Th cells are major pathogenic regulators not only in skin inflammatory disease, but also in various autoimmune diseases. In this regard, modulation of Ets1 activity or its target genes could be considered as potentially novel therapeutic targets for inflammatory immune disorders, including AD and autoimmune diseases.
Methods
Mice.
C57BL/6, CD45.1, distal-Lck (dLck)-cre, and Rag1–/– mice were originally obtained from The Jackson Laboratory. BALB/c mice were purchased from Orient Bio Inc. Ets1–/– mice carrying a germline deletion of Ets1 were previously described (25) and provided by I-Cheng Ho (Harvard Medical School, Boston, Massachusetts, USA). Ets1fl/fl mice were generated by Toolgen and Macrogen (Seoul, Korea). In brief, RNA-guided endonucleases (RGENs) mediating targeted genome modification was performed against the seventh exon of Ets1, which contains part of the DNA binding domain (28). T cell–specific conditional Ets1-KO mice (Ets1ΔdLck) were generated by breeding Ets1fl/fl mice to dLck-Cre mice. Mice were maintained in the animal facility of POSTECH Biotech Center in SPF conditions. Mixtures of male and female mice were used, matched between groups. Mice were sacrificed between the ages of 8–10 weeks.
Study subjects.
Skin biopsy specimens were obtained after informed consent from all patients. All the patients (n = 22) were from outpatient clinics of Chonnam National University Hospital (Gwangju, South Korea). Each subject was classified into 3 groups based on their clinical symptoms. A severity score was recorded for each of the 4 regions of the body. The severity score is the sum of the intensity scores for 4 signs. The 4 signs are redness, thickness, scratching, and lichenification. The average intensity of each sign in each body region is assessed as none, mild, moderate, and severe.
Induction of AD-like skin inflammation.
Induction of experimental AD-like skin inflammation was performed by following a protocol described previously (27). Briefly, surfaces of both ear lobes were stripped 5 times with surgical tape (Medi-Korea). After stripping, 20 μl of 2% 2,4-dinitrochlorobenzene (DNCB, MilliporeSigma) dissolved in acetone/olive oil (1:3) solution was painted on each ear. After 4 days, 20μl of 10 mg/ml HDM extract (dermatophagoides farinae, GREER) dissolved in PBS containing 0.5% Tween 20 was repainted. The DNCB/HDM treatment was repeated weekly for 4 weeks. SC144 treatment was performed via i.p. route at a concentration of 7 mg/kg/day. The treatment started the week after the first DNCB application and was repeated daily for another 4 days. After a 2-day pause, this 5 days–on and 2 days–off SC144 treatment protocol was repeated. Ear thickness was measured 24 hours after DNCB or HDM application by using a dial thickness gauge (Kori Seiki MFG Co.). The animals were sacrificed on day 31. Blood samples were collected by eye bleeding. Plasma was prepared from the blood samples and stored at –70°C for further analysis. After blood collection, the ears were removed and subjected to histopathological analysis. For AD-like skin inflammation induction in Ets1-/- mice, naive CD4+ T cells (5 × 105 per mouse) from LMC and Ets1ΔdLck mice sorted by flow cytometry (MoFlo XDP and MoFlo Asterios, Beckman-Coulter) were i.v. transferred into Rag1−/− mice, together with WT CD45.1+ CD19+ B cells (2 × 106 per mouse). One day later, the recipient mice were induced with experimental AD-like skin inflammation as described above. The AD-like disease incidence rate of experimental AD induction with Ets1–/–, Ets1ΔdLck, and Rag1–/– adoptive transfer with Ets1-deficient CD4+ T cells showed more than 90%.
Histological analysis.
Skin biopsies from the patients and ear tissues from the mice were prepared, fixed in 4% paraformaldehyde solution in PBS, embedded in paraffin, and sectioned for staining with H&E. Histopathology were examined based on the severity of tissues destruction, eosinophil infiltration, and lymphocytic infiltration (Weak, no tissue destruction and rare eosinophil and lymphocyte infiltration; moderate, no tissue destruction and severe lymphocytes and eosinophil infiltration in dermis; severe, destruction of skin formation and severe lymphocytic and eosinophilic infiltration in dermis and epidermis). For IHC with patient samples, collected inflamed skin regions were stained with anti-ETS1 (Santa Cruz Biotechnology Inc., sc-350) and appropriate peroxidase-conjugated secondary antibodies (Abcam, catalog ab6721). Stained ETS1 protein was visualized with EnVision FLEX+, Mouse, High pH (Agilent, Dako) according to manufacturer protocol. For each case, lymphocytes were counted in three 400× fields with the aid of a grid and a mechanical counting device. Numbers of ETS1+ cells in the same fields were counted and expressed as a ratio of total lymphocytes.
Measurement of IgG and IgE.
Serum IgG and IgE level was measured by ELISA kit (BD Biosciences) following manufacture protocol. Three wells were prepared for each antigen and serum, and the concentration and OD value indicated the average of each well and serum. Mean absorbance of an antigen wells minus mean absorbance of a nonantigen well was used as the OD value of the antigen.
Isolation of CD4+ T cells and total ear residual cells.
Naive CD4+ T cells were purified from the dLNs of the mice by using a CD4+ negative selection method (Stem Cell Technologies) or flow cytometry–based cell sorting for CD4+CD25–CD44loCD62Lhi cells. Ears were removed from the mice, cut into 3 pieces, and washed with PBS. The ear segments were gently stirred in flasks with solution (PBS containing 25 ml 10 mM EDTA [Tech&Innovation], 3% FBS [HyClone Laboratories], 20 mM HEPES [Tech&Innovation], and 1 mM sodium pyruvate [Welgene Inc.]) for 20 minutes at 37°C and then washed 3 times with PBS. The ear pieces were minced and incubated 5 ml RPMI 1640 containing 1 mg/ml collagenase type V (MilliporeSigma) for 45 minutes in 37°C incubator (Hanyang Scientific Equiment Co., Ltd) on the magnetic stirrer . Finally, the soup containing total ear cells was centrifuged and washed with ice-cold PBS and cultured in T cell media.
In vitro Th cell differentiation.
Naive CD4+ T cells were stimulated with plate-bound anti-CD3 (1 μg/ml) (BioXcell, clone 145-2C11) and anti-CD28 (2 μg/ml) (BioXcell, clone 37.51) in medium supplemented as follows: for Th1 differentiation, IL-12 (10 ng/ml) and anti–IL-4 (10 μg/ml) (BioXcell, clone 11B11); for Th2 differentiation, IL-4 (10 ng/ml) and anti–IFN-γ (10 μg/ml); for conventional Th17 differentiation, IL-6 (20 ng/ml), TGF-β1 (2 ng/ml), anti–IFN-γ (10 μg/ml) (BioXcell, clone XMG1.2), and anti–IL-4 (10 μg/ml); for pathogenic Th17 differentiation, IL-1β (20 ng/ml), IL-6 (20 ng/ml), IL-23 (20 ng/ml), anti–IFN-γ (10 μg/ml), and anti–IL-4 (10 μg/ml) (32). All antibodies are purchased from the BioXcell, and recombinant cytokines are purchased from the Miltenyi Biotec.
Flow cytometric analysis.
To detect tissues infiltrated population, anti–CD4-Alexa488, anti–CD8a-PE, and anti–CD19-BV421 (BioLegend; catalogs 100529, 100707, 115549, respectively) were used. For checking T cell differentiation into various Th cells and ex vivo cells, cells were stained with anti–Tbet-Alexa647 (BD Bioscience, 561267), anti–GATA3-PE, anti–RORγt-APC, anti–AHR-Alexa488 (eBioscience; catalogs 12-9966-42, 17-6981-82, 53-5925-82, respectively), and anti–Aiolos-PE (BioLegend, 653204). To analyze intracellular IL-4, IL-13, IL-10, IFN-γ, IL-17A, and IL-22, cells were permeabilized with IC fixation buffer (eBioscience), washed and stained with anti–IL4-APC, anti–IL13-PE, anti–IFN-γ eFluor450, anti–IL22-PE (eBioscience; catalogs 17-7041-81, 12-7133-81, 48-7311-82, 12-7221-80, respectively), anti–IL10-BV421, and anti–IL17A-APC (BioLegend; catalogs 505021, 506916, respectively). Antibodies for anti–Foxp3-PE, anti–Ki-67-PerCP-eFluor710 (eBioscience; catalogs 12-5773-82, 46-5698-82, respectively), anti–Helios-Alexa488, anti–CD44-APC, anti–CD62L-BV421, anti–IL6Rα-APC (BioLegend; catalogs 137223, 103012, 104436, 115812, respectively), and anti–gp130-PE (R&D systems, FAB4681P) were used in some experiments. The samples were analyzed using the LSRFortessa flow cytometer, and data were analyzed using FlowJo software.
RNA isolation, cDNA synthesis, and quantitative PCR.
Total RNA was extracted from cells with TRIzol reagent (Molecular Research Center) according to the manufacturer’s protocol. For reverse transcription, up to 1 μg total RNA was used per sample, and cDNA was generated using an oligo (dT) primer (Promega) and ImProm-II reverse transcriptase (Promega) in a total volume of 20 μl. The mRNA level was determined using 1 μl of diluted cDNA by real-time PCR (Rotor-gene Q, Qiagen) with SYBR Green (Takara), using a protocol provided by the manufacturer. Mouse hypoxanthine phosphoribosyltransferase (HPRT) primer was used for quantitative PCR (qPCR) to normalize the amount of cDNA used for each condition. The following primers, in-house designed or obtained from other references and PrimerBank (https://pga.mgh.harvard.edu/primerbank/) (60), were used in the study. Hprt: forward, 5′-TCAGTCAACGGGGGACATAAA-3′ and reverse, 5′-GGGGCTGTACTGCTTAACCAG-3′ (PrimerBank ID 7305155a1); Ets1: forward, 5′-TTTCTGACCCAGATGAGGTGG-3′ and reverse, 5′-GCATCCGGCTTTACATCCAG-3′; Flg2: forward, 5′-GAGCAAGGATGAGCTAAAGGAAC-3′ and reverse, 5′-GCCACGCCTATGCTTCTTTGAC-3′; Cldn1: forward, 5′- ATCGCAATCTTTGTGTCCACCATTG-3′ and reverse, 5′-ATTCTGTTTCCATACCATGCTGTGG-3′ (61); Lor: forward, 5′-CACATCAGCATCACCTCCTTCC-3′ and reverse, 5′-CCTCCTCCACCAGAGGTCTTTC-3′ (62); S100A8: forward, 5′-CTTCAAGACATCGTTTGAAAGG-3′ and reverse, 5′-ATATTCTGCACAAACTGAGGAC-3′; S100A9: forward, 5′-TAGCCTTGAGCAAGAAGATGG-3′ and reverse, 5′-CTGATTGTCCTGGTTTGTGTCC-3′; Il6st: forward, 5′-CCGTGTGGTTACATCTACCCT-3′ and reverse, 5′-CGTGGTTCTGTTGATGACAGTG-3′ (PrimerBank ID 6754338a1); Il6ra: forward, 5′-CCTGAGACTCAAGCAGAAATGG-3′ and reverse, 5′-AGAAGGAAGGTCGGCTTCAGT-3′ (PrimerBank ID 52693a1); Ikzf3: forward, 5′-CTGAATGACTACAGCTTGCCC-3′ and reverse, 5′-GCTCCGGCTTCATAATGTTCT-3′ (PrimerBank ID 2150044a1); Il2: forward, 5′-TGAGCAGGATGGAGAATTACAGG-3′ and reverse, 5′-GTCCAAGTTCATCTTCTAGGCAC-3′ (PrimerBank ID 1504135a1).
Immunoblotting (IB), IP/LC-MS/MS analysis and co-IP.
Total cell protein extracts were prepared in RIPA buffer and subjected to immunoblotting assay as described previously (63). IB was performed using standard procedures with anti-Ets1 (Santa Cruz Biotechnology Inc.), anti–phospho-STAT3, anti-STAT3 (Cell Signaling Technology), and anti-Actin (Abcam) and was then visualized using ImageQuant LAS 4000 (GE Healthcare). Band intensity was quantified using ImageJ software (NIH). For IP/LC-MS/MS analysis, 20 mg of cell lysate from stimulated primary CD4+ T cells was immunoprecipitated with 40 μg of anti-Ets1 overnight at 4°C, and 100 μl of protein A magnetic bead (Invitrogen) was added and further incubated for 2 hours at 4°C. Each immunoprecipitate was washed with cell lysis buffer 3 times, washed with 1× PBS 3 times. Precipitates were eluted in 8 M Urea for further LC-MS/MS analysis. For co-IP, 1 mg of cell lysate from ex vivo CD4+ T cells was immunoprecipitated with 4 μg of anti-Ets1 overnight at 4°C. Protein A magnetic bead (20 μl) was added and further incubated for 2 hours at 4°C. Each immunoprecipitate was washed with cell lysis buffer 3 times, followed by 1× PBS 3 times, separated by SDS-PAGE, and then transferred onto a supported nitrocellulose membrane (Bio-Rad). Immunoblotting was performed using standard procedures with anti-HDAC1 (Abcam) and anti-mSin3A (Santa Cruz Biotechnology Inc.).
ChIP.
ChIP assays were performed as described previously (63). In brief, cells were cross-linked with formaldehyde at a final concentration 1%, lysed, and sonicated to shear DNA. After IP with anti-H3K27ac (Abcam, ab4729) or rabbit IgG (Vector Laboratories, I-1000) at 4°C overnight, Ab/DNA complexes were eluted, and cross-linking was reversed by boiling. After reversal of cross-links, the presence of selected DNA sequences was assessed by real-time PCR using the following primers. Il6st Enh1: forward, 5′-CTCAAAGCTCAAGTACTACAGCA-3′ and reverse, 5′-AATACTCCCAGGAGCTCATACAC-3′; Il6st Enh2: forward, 5′-ACTTAAGATCAGAACTCTGGAGTC-3′ and reverse, 5′-CTCTGAGCATGCAAAGTGTG-3′; Il6st Prom: forward, 5′-CGATCTAGTCTAGGAAAGGCGA-3′ and reverse, 5′-CTCATTGGCTCTGGTCAGTC-3′. As a loading control, the qPCR was done directly on input DNA purified from chromatin before IP. Data are presented as the amount of DNA recovered relative to the input control. Rpl30 and Mest (Cell Signaling Technology, 7015 and 12928) were used as positive and negative control, respectively.
RNA-seq, ChIP-seq, and analysis.
dLNs near sites of inflammation under AD-like skin inflammation were excised from LMC and Ets1ΔdLck mice and prepared into single cell suspensions. CD4+ T cells were purified by CD4+ negative selection method (Mouse CD4+ T cell negative selection kit; Stem Cell Technologies). Three replicates of each group were prepared. RNA were isolated using RNeasy Mini Kit (Qiagen) according to manufacturer’s instruction. Libraries were prepared using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina), and sequencing was done on an Illumina NextSeq 500, 75 paired-end cycles. The quality of the sequenced reads was determined using the FastQC tool, and contaminated adaptor sequences were trimmed using the Cutadapt tool. The trimmed sequence reads were aligned to the mouse reference genome mm9 using Bowtie2. The abundance of transcripts was estimated using the Tuxedo protocol (Tophat and Cufflinks). Fragments per kilobase of transcript per million mapped reads (FPKM), which is normalized expression level, was generated with Cuffnorm, and differentially expressed genes (DEGs) were identified using Cuffdiff. DEGs were defined as the genes that were differentially expressed in CD4+ T cells from LMC and Ets1ΔdLck mice, showing less than a FDR-adjusted P value cutoff of 0.05. We further discarded genes showing less than 1.5-fold changes of up- or downregulated genes between conditions. To identify gene sets significantly involved in known pathways, gene set enrichment analysis (GSEA) was performed with the identified up- or downregulated genes. RNA-seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE122795) (https://www.ncbi.nlm.nih.gov/geo/). Mouse Ets1 ChIP-seq from T conventional cells and CD4+Foxp3– T cells were downloaded from GEO with the following accession numbers, respectively: NCBI SRA accession number SRX2680314 and GEO database accession number GSM999187. Reads from the ChIP-seq data were aligned to the mouse reference genome mm9 using Bowtie2. Genome-wide Ets1 binding sites were identified using HOMER (64) with default parameters. Integrative genomics viewer (IGV) (65) was used to examine loci of interest associated with Ets1 binding.
Mixed BM chimera generation.
BM cells from CD45.2 LMC or CD45.2 Ets1ΔdLck mice were mixed with BM cells from congenic CD45.1 WT mice at a 1:1 ratio. These BM mixtures (5 × 106) were injected into irradiated Rag1−/− hosts (300 cGy). At 6–8 weeks after BM cell transfer, cells from LN and spleen were analyzed by flow cytometry.
Statistics.
Statistical analyses were performed using Prism (GraphPad Software) by the unpaired, 2-tailed Student’s t test, 1-way ANOVA, or 2-way ANOVA. P values below 0.05 were considered significant in the following manner: *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; ****P ≤ 0.0001.
Study approval.
All human studies were reviewed and approved by the Cheonnam National University Hospital and IRB (IRB approval number CNUH-2014-053). Written informed consent was obtained from all subjects. All animal experiments were in accordance with protocols approved by the POSTECH IACUC.
Author contributions
CGL took the lead throughout the entire course of a project. CGL and HKK designed the studies, performed experiments, and wrote the manuscript. CGL and HK designed and performed the in vivo experiments. CGL, HKK, and HK participated in data acquisition and analyzed the data. YK, JN, and YW provided skin biopsies and performed the experiments related to it. SP contributed to the performance of the in vivo experiments. DR and CDJ provided intellectual suggestions during the course of the study and edited the manuscript, along with SHI. TK and KK contributed to the bioinformatics analysis of RNA-seq and ChIP-seq. ZP designed and guided proteomic experiment. SHI supervised the entire work and edited and approved the manuscript.
Supplementary Material
Acknowledgments
We thank the flow cytometry facility at AIM for technical assistance and cell sorting, as well as members of the Immune Regulation & Tolerance (IRT) laboratory for technical support and assistance. This work was supported by project IBS-R005 of the IBS, Korean Ministry of Science, Information/Communication Technology, and Future Planning.
Version 1. 03/07/2019
Electronic publication
Funding Statement
This work was supported by project IBS-R005 of the Institute for Basic Science, Korean Ministry of Science, Information/Communication Technology and Future Planning
Footnotes
TK’s present address is: Department of Biological Sciences, Korea Advanced Institute of Science and Technology, Daejeon, South Korea.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(5):e124202. https://doi.org/10.1172/jci.insight.124202.
Contributor Information
Choong-Gu Lee, Email: cglee0708@kist.re.kr.
Ho-Keun Kwon, Email: aspirationpassion@gmail.com.
Hyeji Kang, Email: Cindyjee14@gmail.com.
Young Kim, Email: youngkim2017@jnu.ac.kr.
Jong Hee Nam, Email: jhnam@chonnam.ac.kr.
Young Ho Won, Email: yhwon@jnu.ac.kr.
Sunhee Park, Email: ddony369@ibs.re.kr.
Taemook Kim, Email: taemook14@gmail.com.
Keunsoo Kang, Email: kangk1204@dankook.ac.kr.
Dipayan Rudra, Email: rudrad@ibs.re.kr.
Chang-Duk Jun, Email: cdjun@gist.ac.kr.
Zee Yong Park, Email: zeeyong@gist.ac.kr.
References
- 1.Leung DY. Atopic dermatitis: new insights and opportunities for therapeutic intervention. J Allergy Clin Immunol. 2000;105(5):860–876. doi: 10.1067/mai.2000.106484. [DOI] [PubMed] [Google Scholar]
- 2.Eyerich K, Eyerich S, Biedermann T. The Multi-Modal Immune Pathogenesis of Atopic Eczema. Trends Immunol. 2015;36(12):788–801. doi: 10.1016/j.it.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113(5):651–657. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biedermann T, Skabytska Y, Kaesler S, Volz T. Regulation of T Cell Immunity in Atopic Dermatitis by Microbes: The Yin and Yang of Cutaneous Inflammation. Front Immunol. 2015;6:353. doi: 10.3389/fimmu.2015.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homey B, Steinhoff M, Ruzicka T, Leung DY. Cytokines and chemokines orchestrate atopic skin inflammation. J Allergy Clin Immunol. 2006;118(1):178–189. doi: 10.1016/j.jaci.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 6.Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol. 2008;128(11):2625–2630. doi: 10.1038/jid.2008.111. [DOI] [PubMed] [Google Scholar]
- 7.Guilloteau K, et al. Skin Inflammation Induced by the Synergistic Action of IL-17A, IL-22, Oncostatin M, IL-1{alpha}, and TNF-{alpha} Recapitulates Some Features of Psoriasis. J Immunol. 2010;184(9):5263–5270. doi: 10.4049/jimmunol.0902464. [DOI] [PubMed] [Google Scholar]
- 8.Ma HL, et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest. 2008;118(2):597–607. doi: 10.1172/JCI33263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung DY, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol. 2014;134(4):769–779. doi: 10.1016/j.jaci.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werfel T, et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J Allergy Clin Immunol. 2016;138(2):336–349. doi: 10.1016/j.jaci.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Czarnowicki T, Krueger JG, Guttman-Yassky E. Novel concepts of prevention and treatment of atopic dermatitis through barrier and immune manipulations with implications for the atopic march. J Allergy Clin Immunol. 2017;139(6):1723–1734. doi: 10.1016/j.jaci.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Morar N, Willis-Owen SA, Moffatt MF, Cookson WO. The genetics of atopic dermatitis. J Allergy Clin Immunol. 2006;118(1):24–34. doi: 10.1016/j.jaci.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Paternoster L, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47(12):1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinds DA, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet. 2013;45(8):907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kola I, et al. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc Natl Acad Sci USA. 1993;90(16):7588–7592. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9(6):1551–1565. [PubMed] [Google Scholar]
- 18.Barton K, et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9(4):555–563. doi: 10.1016/S1074-7613(00)80638-X. [DOI] [PubMed] [Google Scholar]
- 19.Walunas TL, Wang B, Wang CR, Leiden JM. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J Immunol. 2000;164(6):2857–2860. doi: 10.4049/jimmunol.164.6.2857. [DOI] [PubMed] [Google Scholar]
- 20.Mouly E, et al. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207(10):2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez K, et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36(6):921–932. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eyquem S, Chemin K, Fasseu M, Bories JC. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc Natl Acad Sci USA. 2004;101(44):15712–15717. doi: 10.1073/pnas.0405546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clements JL, John SA, Garrett-Sinha LA. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J Immunol. 2006;177(2):905–912. doi: 10.4049/jimmunol.177.2.905. [DOI] [PubMed] [Google Scholar]
- 24.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5(12):e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med. 2005;201(4):615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204(12):2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon HK, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA. 2010;107(5):2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CJ, et al. The Transcription Factor Ets1 Suppresses T Follicular Helper Type 2 Cell Differentiation to Halt the Onset of Systemic Lupus Erythematosus. Immunity. 2018;49(6):1034–1048.e8. doi: 10.1016/j.immuni.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Zhang DJ, et al. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol. 2005;174(11):6725–6731. doi: 10.4049/jimmunol.174.11.6725. [DOI] [PubMed] [Google Scholar]
- 30.Sebastian M, Lopez-Ocasio M, Metidji A, Rieder SA, Shevach EM, Thornton AM. Helios Controls a Limited Subset of Regulatory T Cell Functions. J Immunol. 2016;196(1):144–155. doi: 10.4049/jimmunol.1501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyara M, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30(6):899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 2010;467(7318):967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18(2):173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA. 2010;107(7):2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8(9):1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishihara M, et al. IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int Immunol. 2007;19(6):695–702. doi: 10.1093/intimm/dxm045. [DOI] [PubMed] [Google Scholar]
- 38.Xu S, Grande F, Garofalo A, Neamati N. Discovery of a novel orally active small-molecule gp130 inhibitor for the treatment of ovarian cancer. Mol Cancer Ther. 2013;12(6):937–949. doi: 10.1158/1535-7163.MCT-12-1082. [DOI] [PubMed] [Google Scholar]
- 39.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 40.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. Altered sequence of the ETS1 transcription factor may predispose to rheumatoid arthritis susceptibility. Scand J Rheumatol. 2013;42(1):11–14. doi: 10.3109/03009742.2012.711367. [DOI] [PubMed] [Google Scholar]
- 42.Shan S, et al. ETS1 variants confer susceptibility to ankylosing spondylitis in Han Chinese. Arthritis Res Ther. 2014;16(2):R87. doi: 10.1186/ar4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lill CM, et al. Genome-wide significant association with seven novel multiple sclerosis risk loci. J Med Genet. 2015;52(12):848–855. doi: 10.1136/jmedgenet-2015-103442. [DOI] [PubMed] [Google Scholar]
- 44.Bruhn S, et al. Increased expression of IRF4 and ETS1 in CD4+ cells from patients with intermittent allergic rhinitis. Allergy. 2012;67(1):33–40. doi: 10.1111/j.1398-9995.2011.02707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Na SY, Park MJ, Park S, Lee ES. MicroRNA-155 regulates the Th17 immune response by targeting Ets-1 in Behçet’s disease. Clin Exp Rheumatol. 2016;34(6 Suppl 102):S56–S63. [PubMed] [Google Scholar]
- 46.Xiang N, et al. Expression of Ets-1 and FOXP3 mRNA in CD4(+)CD25 (+) T regulatory cells from patients with systemic lupus erythematosus. Clin Exp Med. 2014;14(4):375–381. doi: 10.1007/s10238-013-0263-4. [DOI] [PubMed] [Google Scholar]
- 47.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377(6550):639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 48.Nagarajan P, Chin SS, Wang D, Liu S, Sinha S, Garrett-Sinha LA. Ets1 blocks terminal differentiation of keratinocytes and induces expression of matrix metalloproteases and innate immune mediators. J Cell Sci. 2010;123(Pt 20):3566–3575. doi: 10.1242/jcs.062240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zook EC, et al. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J Exp Med. 2016;213(5):687–696. doi: 10.1084/jem.20150851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13(10):991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silver JS, Hunter CA. gp130 at the nexus of inflammation, autoimmunity, and cancer. J Leukoc Biol. 2010;88(6):1145–1156. doi: 10.1189/jlb.0410217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quintana FJ, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13(8):770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McAleer MA, Irvine AD. The multifunctional role of filaggrin in allergic skin disease. J Allergy Clin Immunol. 2013;131(2):280–291. doi: 10.1016/j.jaci.2012.12.668. [DOI] [PubMed] [Google Scholar]
- 54.Brunner PM, Guttman-Yassky E, Leung DY. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol. 2017;139(4S):S65–S76. doi: 10.1016/j.jaci.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson ML, et al. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc Natl Acad Sci USA. 2010;107(22):10026–10031. doi: 10.1073/pnas.0915137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadamb R, Mittal S, Bansal N, Batra H, Saluja D. Sin3: insight into its transcription regulatory functions. Eur J Cell Biol. 2013;92(8-9):237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Alland L, et al. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol Cell Biol. 2002;22(8):2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17(7):896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizzo HL, Kagami S, Phillips KG, Kurtz SE, Jacques SL, Blauvelt A. IL-23-mediated psoriasis-like epidermal hyperplasia is dependent on IL-17A. J Immunol. 2011;186(3):1495–1502. doi: 10.4049/jimmunol.1001001. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40(Database issue):D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lou H, et al. Expression of IL-22 in the Skin Causes Th2-Biased Immunity, Epidermal Barrier Dysfunction, and Pruritus via Stimulating Epithelial Th2 Cytokines and the GRP Pathway. J Immunol. 2017;198(7):2543–2555. doi: 10.4049/jimmunol.1600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grine L, Dejager L, Libert C, Vandenbroucke RE. Dual Inhibition of TNFR1 and IFNAR1 in Imiquimod-Induced Psoriasiform Skin Inflammation in Mice. J Immunol. 2015;194(11):5094–5102. doi: 10.4049/jimmunol.1403015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee CG, et al. Interaction of Ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J Immunol. 2012;188(5):2244–2253. doi: 10.4049/jimmunol.1101614. [DOI] [PubMed] [Google Scholar]
- 64.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.