Abstract
Numerous high-throughput omics studies have been conducted in schizophrenia, providing an accumulated catalog of susceptible variants and genes. The results from these studies, however, are highly heterogeneous. The variants and genes nominated by different omics studies often have limited overlap with each other. There is thus a pressing need for integrative analysis to unify the different types of data and provide a convergent view of schizophrenia candidate genes (SZgenes). In this study, we collected a comprehensive, multidimensional dataset, including 7819 brain-expressed genes. The data hosted genome-wide association evidence in genetics (eg, genotyping data, copy number variations, de novo mutations), epigenetics, transcriptomics, and literature mining. We developed a method named mega-analysis of odds ratio (MegaOR) to prioritize SZgenes. Application of MegaOR in the multidimensional data resulted in consensus sets of SZgenes (up to 530), each enriched with dense, multidimensional evidence. We proved that these SZgenes had highly tissue-specific expression in brain and nerve and had intensive interactions that were significantly stronger than chance expectation. Furthermore, we found these SZgenes were involved in human brain development by showing strong spatiotemporal expression patterns; these characteristics were replicated in independent brain expression datasets. Finally, we found the SZgenes were enriched in critical functional gene sets involved in neuronal activities, ligand gated ion signaling, and fragile X mental retardation protein targets. In summary, MegaOR analysis reported consensus sets of SZgenes with enriched association evidence to schizophrenia, providing insights into the pathophysiology underlying schizophrenia.
Keywords: schizophrenia, omics, tissue specificity, candidate genes, brain, spatiotemporal expression
Introduction
Schizophrenia is a chronic and severe brain disorder whose pathophysiology remained largely unknown.1,2 Over the past decade, numerous studies have been conducted to understand the genetic and pathophysiologic architecture of schizophrenia. These studies utilized the approaches in genetics,3–5 epigenetics,6 transcriptomics,7 proteomics, metabolomics,8,9 and functional genomics, among others. Genetic factors contribute significantly to risk for schizophrenia.10 So far, there have been hundreds of inherited genetic variants that were identified through genome-wide association studies (GWAS)2,11,12 and hundreds of rare variants and de novo mutations (DNMs) identified through next-generation sequencing (NGS) technologies.13 More than 10 studies reported DNMs in schizophrenic patients, though a recent study found only 1 gene, ie, SETD1A, had reached genome-wide significance.13 In addition to single nucleotide polymorphisms (SNPs), a stronger burden of rare copy number variations (CNVs) was observed in schizophrenic patients.14,15 In addition, more than 10 large-scale epigenetics studies reported differentially methylated CpG sites that showed significant association with schizophrenia.16–25 Because schizophrenia has long been considered a brain disorder, transcriptomic studies using postmortem human brain tissue are helpful for exploring molecular mechanisms of schizophrenia, such as genes and pathways abnormally expressed in patients.7,26,27 However, human brain development involves complex and rigorous regulation of transcriptional programs in a manner with temporal and spatial specificity,28 placing additional layers of complexity in the investigation of schizophrenia genes.
Several challenges exist in the integration of heterogeneous data to prioritize disease candidate genes.29 First, the correlations among the multidimensional data remain elusive. Specific properties of genes or mutations vary among original studies (eg, RNA-sequencing [RNA-seq] measures mRNA level of a gene whereas methylation measures the epigenetic level of a gene) and at different scales or resolution (eg, linkage studies report large genomic regions but NGS at single-base resolution). Some datasets presumably have high correlations (such as GWAS signals and linkage peaks) whereas others barely have good correlation (such as GWAS signals and DNMs). Although integration of GWAS signals with expression quantitative trait loci (eQTL) has revealed regulatory roles of schizophrenia-associated SNPs,30 integrative and cross-talk analysis among other types of association data has been lacked so far. Second, it is not known a priori which data types are more predictive than others for disease candidate genes. We previously developed a weighted-sum algorithm that estimated a relative weight for each dimension of data based on a set of gold-standard genes and assigned a weighted-sum score for each gene to prioritize promising candidates.31 However, as we will show in our results, such strategies are no longer suitable because historically important genes (HIGs), which were used as the gold-standard genes in previous works,31 were mainly detected using conventional technologies (eg, candidate gene association studies) and they have limited power in evaluating genes from newly explored data types (eg, methylation). Therefore, new approaches are needed to effectively integrate a variety of omics data for the discovery of disease candidate genes and can be easily expanded when new data are generated.
Here, we curated comprehensive, multidimensional omics data conducted for schizophrenia, involving 10 058 candidate genes (7819 expressed in brain).32 All datasets were constructed using schizophrenia cases and normal controls and all genes had at least one kind of evidence indicating their association with schizophrenia. We proposed a method called mega-analysis of odds ratio (MegaOR) to prioritize consensus sets of schizophrenia candidate genes (SZgenes). We demonstrated the robustness of the SZgenes by using independent omics data from normal brain samples. Such computational benchmarking revealed tissue specificity, temporal and spatial expression pattern, intensive connection, and functional enrichment of SZgenes in brain. To the best of our knowledge, this is the largest ever integrative analysis of schizophrenia genes, which leverages on nearly all types of variants and genes that had been reported in schizophrenia.
Materials and Methods
Multidimensional Datasets in Schizophrenia
We used the omics data collected in our SZGR 2.0 database32 and organized the data into 8 categories based on the types of evidence (tables S1 and S2). A ground rule for the data to be included in our analyses was that all variants and genes must be collected from studies using schizophrenic patients and normal control samples so that the association relationship could be calculated. We included GWAS “top hit” genes at genome-wide significance, CNV genes, GWAS Pascal genes (gene-based combined GWAS association information measured by PPascal33), Sherlock genes (gene-based combined GWAS and eQTL information measured by PSherlock34), genes with DNMs (gene-based enrichment of DNMs measured by Transmission And De novo Association (TADA)35P value), differentially expressed genes (DEGs), differentially methylated genes (DMGs), and genes nominated by the literature co-occurrence. A detailed description of the preprocessing steps is presented in the Supplementary material. All genes and their evidence are presented in table S1.
Mega-analysis of Odds Ratio
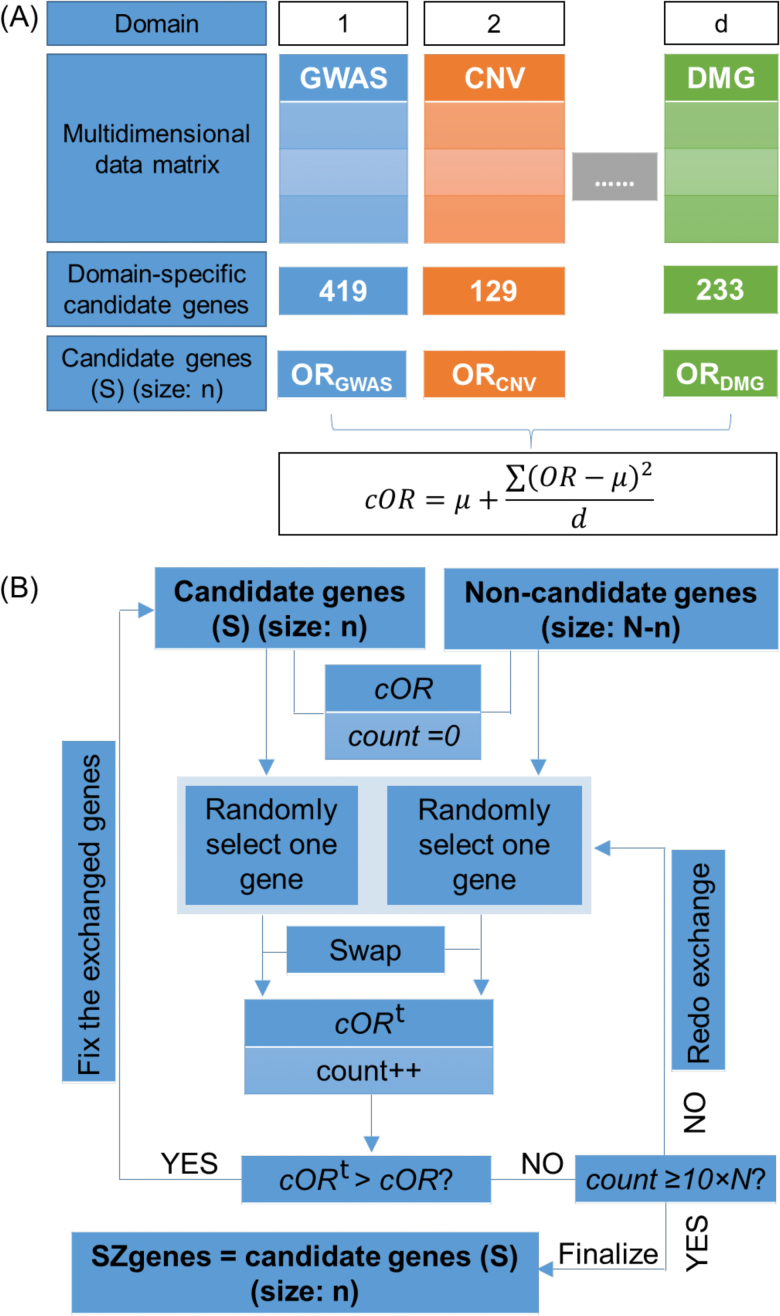
We aim to identify a set of candidate genes that collectively have the most intensive load of evidence for their association with schizophrenia. We propose MegaOR, an unsupervised approach that does not rely on a predefined gold-standard gene set (ie, a training set) to identify such consensus gene sets (figure 1). MegaOR implements an iterative “try-and-fix” procedure and requires 2 inputs: a multidimensional data matrix, with a value of 1 indicating a gene was a positive candidate based on the corresponding evidence and 0 otherwise, and a predefined set size of candidate genes (n). MegaOR calculates an odds ratio (OR) for each single dimension and a combined OR (cOR) across multidimensional data for a given set of candidate genes (S, with size n): , where OR is defined for each dimension, μ is the average value of all ORs, and d = 8 indicates the number of dimensions. The part was included as penalty to control deviation of any dimensional OR from the average OR. cOR is iteratively optimized by exchanging genes in the temporary candidate gene list S and genes not in S (ie, the trying process) and fixing the change if cOR improves (ie, the fixing process). Such “try-and-fix” steps will continue until cOR reaches a stable value. The number of candidate genes, n, depends on the particular polygenic burden of the disease model. Although it is far from accurate understanding in schizophrenia, previous studies have suggested n to be hundreds to a few thousands.29 Thus, we tested multiple representative parameters, ie, n = 200, 300, 400, 500, 600, 700, 800, 900, and 1000. A detailed description is available in the Supplementary material.
Fig. 1.
A schematic pipeline of the mega-analysis of odds ratio (MegaOR) method. (A) Definition of odds ratio (OR) in each dimension and the combined OR (cOR). (B) Illustration of the MegaOR pipeline.
Multiple Tissue and Brain Expression Data
We utilized 3 sets of gene expression data with different aspects to explore the tissue specificity and spatiotemporal expression pattern of the SZgenes. Of note, all validation data were collected from normal samples, which had no overlap with the samples used in SZgene prioritization. First, the GTEx data (version 6) were included to investigate the tissue-specific expression patterns of each gene.36 A total of 27 tissues were considered, each with ≥ 30 samples. For each gene, we defined a z score to m easure its tissue specificity: , where is the average gene expression of the gene in the i th tissue, E represents the collection of its average gene expression in all tissues, and indicates the standard deviation of E. A higher z score indicates the gene to be more specifically expressed in the investigated tissue. Second, the BrainSpan dataset measured the developmental transcriptome of normal brain from fetus to adulthood and was included to explore the spatiotemporal expression patterns of genes.37 The transcriptome of multiple brain regions was investigated using RNA-seq. Third, the BrainCloud dataset was included to explore the temporal expression pattern of genes,28 including 269 human postmortem dorsolateral prefrontal cortex samples. Following the original work, we calculated a β value for each gene to measure the expression changes before birth (n = 38 fetal samples) and after birth (n = 231 samples aging up to 80 years). Specifically, we fit a regression model for each gene as , where y is the vector of expression for the gene, Xfetal is an indicator variable with xi = 0 for fetal samples and xi = 1 otherwise, and represents surrogate variables as suggested by the original work to control for potential biases.28,38
Results
Multidimensional Association Evidence for Schizophrenia
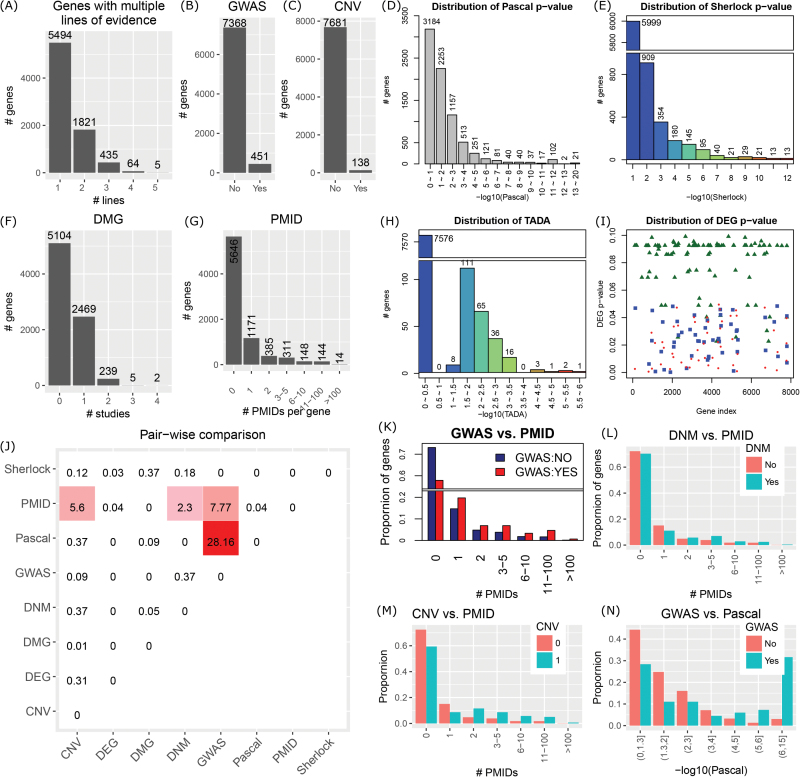
We obtained a total of 7819 brain-expressed genes, each with at least one type of evidence supporting its association with schizophrenia: 451 GWAS genes, 138 CNV genes, 4146 genes with PPascal < .05, 707 genes with PSherlock < .05, 243 genes with PTADA < .05, 149 DEGs, 246 (2715) DMGs supported by ≥2 (≥1) studies, and 617 (2173) genes co-mentioned with schizophrenia key words in ≥3 (≥1) PubMed records (figure 2B–I). Among them, 5494 (70.26%) genes had only 1 line of evidence and no gene was supported by ≥6 lines of evidence (figure 2A). Some example genes included CACNA1C (a GWAS gene, PPascal = 1.00 × 10−12, PSherlock = 2.98 × 10−3, a DMG, and # PMIDs [PubMed identifier]) = 77), GRIN2A (a GWAS gene, PPascal = 2.30 × 10−5, a DMG, PTADA = .024, and # PMIDs = 25), and GABRB3 (located in the duplication region on chromosome 15,39PPascal = 5.53 × 10−4, and # PMIDs = 9).
Fig. 2.
Overview of the multidimensional data covering 7819 genes with at least 1 line of evidence. (A) Distribution of genes with multiple lines of evidence. Genome-wide association studies (GWAS) genes (B) and copy number variation (CNV) genes (C) were binary definition. The gene-based P values by Pascal (D), by Sherlock (E), and by TADA were continuous variables (H). (F) Distribution of differentially methylated genes (DMGs) reported by the number of studies. (G) Distribution of PubMed identifier (PMID) genes co-occurring with schizophrenia keywords. (I) Distribution of adjusted P values for differentially expressed genes (DEGs) from 2 datasets. Triangles indicate DEGs defined in Zhao et al.,7 whereas rectangles indicate DEGs using HBB_BA9 and dots indicate DEGs using CC_BA10 from Maycox et al.27 (J) Overall representation of the pairwise comparison. Genes were categorized as candidates if they had PPascal < .05, PSherlock < .05, or PTADA < .05, or if they were GWAS genes, CNV genes, DEGs, DMGs with ≥2 studies, or PMID genes co-occurring with schizophrenia keywords in ≥3 publications. The values in each cell are—log10(P) from 1-sided χ2 test. (K–N) Demonstration of pairwise comparison. The comparison between PSherlock and PPascal was not shown.
We evaluated HIGs using our data but did not find overrepresentation of evidence. HIGs referred to those that were implied in the widely recognized hypotheses in schizophrenia (neurodevelopment, glutamate, dopamine, immune, and mood disorder)40,41 and have been extensively studied, such as BDNF, DISC1,42 and DTNBP1.43 HIGs had been frequently used as gold-standard genes in many studies to evaluate and predict novel candidate genes for schizophrenia.31 Here, we collected 45 HIGs from our previous work (n = 38)31 and others (n = 25)40 and re-evaluated the evidence for these HIGs. Eight genes were removed due to low expression in brain. Among the remaining 37 HIGs, none had ≥4 lines of evidence (table S4). Within each data dimension, there were <50% HIGs with evidence: 8 GWAS genes, 3 CNV genes, 14 genes with PPascal < .05, no gene with PSherlock < .05, 2 genes with PTADA < .05, no DEG, and 8 DMGs. This obvious lack of support, which is consistent with a recent study,44 made HIGs short of power to evaluate other genes with the multidimensional evidence data, and thus, HIGs might not be appropriate to serve as the gold standard to prioritize candidate genes in omics-based studies.
Correlation Among Multidimensional Data
Pairwise comparisons of all 8 data types revealed only 4 pairs of cross talk with positive correlations (figure 2J). Genes co-occurring with schizophrenia key words (# PMIDs ≥3) had 3 correlated data types: GWAS genes (P = 1.71 × 10−8, 1-sided Fisher’s exact test [FET]; figure 2K), CNV genes (P = 2.53 × 10−6; figure 2M), and DNM genes (P = 5.03 × 10−3; figure 2L). GWAS genes tended to have small Pascal P values (P = 6.99 × 10−29, 1-sided t test; figure 2N). The remaining pairs showed no favorable overlaps or correlations. For example, DEGs, DMGs, and DNM genes were not correlated with any other data types; Pascal genes showed no difference between CNV and non-CNV genes, DNM and non-DNM genes, DEGs and non-DEGs, or DMGs and non-DMGs. Surprisingly, no significant enrichment was found between Pascal genes and Sherlock genes, even though both were derived from the Psychiatric Genomics Consortium (PGC)11 GWAS data. This lack of correlation suggested that there might be unique information in each of them. Collectively, these results implied that the 8 categories of evidence data were highly heterogeneous and diversely correlated, motivating an effort to develop an efficient method to integrate them, as shown in the following sections.
SZgenes by MegaOR
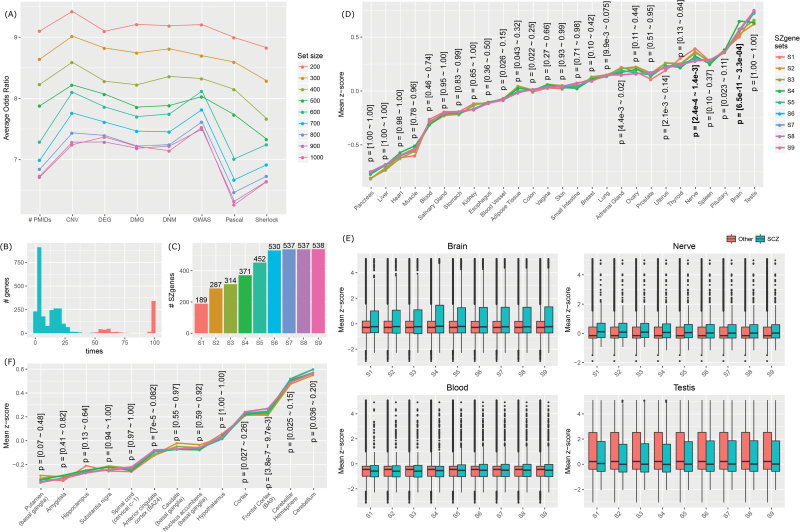
Considering that the candidate genes supported by each single category were rather sparse, we applied MegaOR with the ultimate goal for identifying a subset of candidate genes that collectively have the most intensive load of evidence to support their association with schizophrenia. To ensure high confidence of candidate genes, we utilized the following criteria: GWAS and CNV genes as originally mapped, PPascal < .001, PSherlock < .05, DEGs with adjusted P < .05 in the original studies, PTADA < .05, DMGs reported by ≥2 studies, and PMID genes co-occurring with schizophrenia key words in ≥3 publications. We tested 9 set sizes (n = 200, 300, 400, 500, 600, 700, 800, 900, and 1000) and for each set size, we conducted MegaOR for 100 times, resulting in 100 stable sets, each with n genes. Figure 3A shows the average OR values of the 100 stable sets at each sizes. When set size increased, the OR values decreased. For each set size, the difference of ORs from each dimension did not exceed by 1.5, implying that none of the 8 types of evidence dominated the resultant genes, mainly due to the penalty we included in the calculation of cOR (see the section “Materials and Methods”). We chose the genes that were retained in the stable sets for more than 50% times (figure 3B). In total, we obtained 9 sets of SZgenes. We referred them to S1 for set size n = 200 (final SZgenes: 189), S2 for n = 300 (287), S3 for n = 400 (314), S4 for n = 500 (371), S5 for n = 600 (452), S6 for n = 700 (530), S7 for n = 800 (537), S8 for n = 900 (537), and S9 for n = 1000 (538). SZgenes obtained using large set sizes could cover nearly all the SZgenes obtained using lower set sizes. For example, S2 had 287 genes, including all the 189 genes in S1. At size n = 700, SZgenes converged to a stable status—no substantial extension of SZgenes even when the set size increased to 1000 (figure 3C). Thus, we suggested that the 530 SZgenes in S6, all of which were included in S7–S9, were close to a consensus set of SZgenes that could reach the global maximum load of evidence. Importantly, all 530 SZgenes were supported by at least 2 lines of evidence (figure S1). We also conducted a systematic evaluation of the evidence (Supplementary material), particularly for GWAS, Pascal, DEG, and PMID (figures S2, S3, and S4). Our results indicated that each of them contributed a unique part of genes in the consensus SZgene sets. Thus, we chose to keep all 8 lines in our analyses.
Fig. 3.
Application of mega-analysis of odds ratio (MegaOR) to identify schizophrenia candidate genes (SZgenes). (A) Odds ratio (OR) distribution in each dimension for each set size: n = 200, 300, 400, 500, 600, 700, 800, 900, and 1000. Each dot line in the same color indicates the average ORs in each dimension for 100 stable sets resulted from MegaOR (see main text). (B) The frequency of genes covered by 100 stable sets at an example size n = 700. Genes on the left part of the plot were less frequently covered, ie, <50% times. Genes on the right part of the plot were selected as the final SZgenes for the corresponding set size. (C) Distribution of SZgenes at each set size. (D) Tissue-specific expression of 9 SZgene sets (S1–S9) in 27 tissues from GTEx. The y-axis is the average z scores of SZgenes in each tissue. The x-axis was ordered by the average z scores in S1. P values for each tissue were obtained using 1-sided t test comparing z scores of SZgenes and non-SZgenes. For each tissue, the range of P values from 9 SZgene sets were labeled. (E) Illustration of z scores in tissues where SZgenes had significantly higher z scores than other genes (brain and nerves) and in tissues where SZgenes showed no difference (blood) or showed decreased z scores (testis) compared to non-SZgenes. (F) Brain region-specific expression of 9 SZgene sets (S1–S9) using GTEx data. The y-axis is the average z scores of SZgenes in each brain region. The x-axis was ordered by the average z scores in S1.
Validation of SZgenes: Tissue Specificity
By examining the tissue-specific gene expression patterns, we found SZgenes tended to be significantly expressed in brain and nerve (figure 3D). We utilized expression data for 27 tissues from GTEx36 and performed a 1-sided t test for each tissue to compare the z scores of SZgenes and non-SZgenes. As shown in figures 3D and S5, SZgenes showed statistically significant difference in several tissues, including brain (P = [6.5 × 10−11 – 3.3 × 10−4] for 9 SZgene sets) and nerve (P = [2.4 × 10−4 – 1.4 × 10−3]). Importantly, although blood samples were widely used in schizophrenia research, SZgenes showed no difference in blood compared with non-SZgenes, highlighting the importance of tissue-specific expression profile in studying disease genes. Similarly, although SZgenes had high z scores in testis, our significance test eliminated testis from the tissue list in which SZgenes showed significant specificity, as SZgenes did not have significantly higher z scores than those of non-SZgenes (figure 3E).
As our genes were prefiltered as “brain-expressed genes,” we conducted additional analyses to ensure that the prefiltering criteria did not confound the tissue-specific patterns we observed. Specifically, we performed the t test in following 4 ways: (1) compare SZgenes with other genes in the transcriptome; (2) compare SZgenes with other genes in the evidence data matrix; (3) compare SZgenes with other genes in the transcriptome, while requiring each gene to have an average Reads Per Kilobase of transcript per Million mapped reads (RPKM) > 1 in the tissue of investigation (a similar criterion as we used to define brain-expressed genes); and (4) compare SZgenes with other genes in the evidence data matrix, while requiring each gene to have an average RPKM > 1 in the tissue of investigation. In addition, for each test, we conducted a randomization test by randomly selecting the same number of genes from the evidence data matrix. An empirical P value was calculated as the number of random sets that had a P value lower than the actual P value. As shown in table S5, in all the tests, brain is the only tissue in which our SZgenes showed significant tissue-specific expression.
We further explored the brain region specificity of SZgenes (figure 3F). The nine SZgene sets showed increased specificity in gene expression data from brain regions (cerebellum, cerebellar hemisphere, frontal cortex (BA9), and cortex datasets), which had been implied in schizophrenia previously. However, the difference between SZgenes and non-SZgenes was not statistically significant in most regions, implying that the region specificity might not be prominent (see the section “Lack of Regional Variability”).
We performed a cell-type specific expression analysis45 of the 530 consensus SZgenes. We found these SZgenes were significantly enriched in Ntsr+ neurons of cortex (P = 6.24 × 10−4, adjusted PBH = .007), Drd1+ medium spiny neurons of striatum (P = 2.47 × 10−4, PBH = .004), and Drd2+ medium spiny neurons of striatum (P = 5.20 × 10−5, PBH = .002) (figure S10).
Validation of SZgenes: Protein–Protein Interaction
Examination of protein–protein interaction (PPI) data revealed that SZgenes interacted with each other more frequently than by chance. We and others have previously shown that schizophrenia genes, eg, those harboring DNMs, tended to interact with each other more closely than with other genes in human PPI networks.46 For each SZgene set, we recorded the number of interactions among SZgenes and resampled 10 000 random gene sets, each matching the SZgenes in size. The number of random gene sets that had interactions exceeding the actual number of interactions was used to calculate an empirical P value. We performed this analysis using 3 independent human PPI networks: a combined network of STRING and HPRD,47 HumanNet v.1,48 and PathwayCommons.49 These networks have been utilized in the analysis of GWAS data or other common diseases,48 each with different focuses (table S6). Strikingly, SZgenes showed significantly more PPIs than those from random gene sets in all 3 networks (table 1).
Table 1.
SZgenes Interact With Each Other More Often Than by Chance
| Reference network | Total | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 |
|---|---|---|---|---|---|---|---|---|---|---|
| STRING and HPRD |
V = 10 349 E = 52 663 |
V = 133 E = 9 P = .3488 |
V = 209 E = 38 P = .0253 |
V = 229 E = 57 P = .0053 |
V = 280 E = 115 P < 1 × 10−4 |
V = 329 E = 127 P = 2 × 10−4 |
V = 375 E = 167 P < 1 × 10−4 |
V = 380 E = 171 P = 1 × 10−4 |
V = 380 E = 171 P < 1 × 10−4 |
V = 381 E = 172 P = 1 × 10−4 |
| Pathway Common |
V = 16 305 E = 369 884 |
V = 180 E = 113 P = .0028 |
V = 276 E = 325 P < 1 × 10−4 |
V = 301 E = 404 P < 1 × 10−4 |
V = 356 E = 662 P < 1 × 10−4 |
V = 430 E = 835 P < 1 × 10−4 |
V = 500 E = 1081 P < 1 × 10−4 |
V = 506 E = 1101 P < 1 × 10−4 |
V = 506 E = 1101 P < 1 × 10−4 |
V = 507 E = 1103 P < 1 × 10−4 |
| HumanNet bench |
V = 5236 E = 269 410 |
V = 75 E = 74 P = .0973 |
V = 114 E = 211 P = .0037 |
V = 126 E = 237 P = .0099 |
V = 163 E = 419 P = .0015 |
V = 188 E = 549 P = .0011 |
V = 209 E = 739 P = 1 × 10−4 |
V = 212 E = 739 P = 2 × 10−4 |
V = 212 E = 739 P < 1 × 10−4 |
V = 212 E = 739 P = 1 × 10−4 |
| HumanNet joint |
V = 16 117 E = 474 714 |
V = 185 E = 108 P = .0084 |
V = 280 E = 272 P = 3 × 10−4 |
V = 305 E = 357 P < 1 × 10−4 |
V = 360 E = 561 P < 1 × 10−4 |
V = 431 E = 726 P < 1 × 10−4 |
V = 500 E = 1003 P < 1 × 10−4 |
V = 506 E = 1025 P < 1 × 10−4 |
V = 506 E = 1025 P < 1 × 10−4 |
V = 506 E = 1025 P < 1 × 10−4 |
SZgenes = schizophrenia candidate genes.
S1–S9 represents the SZgene sets. V: number of nodes. E: number of edges. P values were obtained through 10 000 randomization sets selected from the background network with the same number of each SZgenes set (S1–S9).
Developmental Gene Expression Patterns of SZgenes
To explore the expression patterns of SZgenes during brain development, we used 2 datasets of normal brain tissue from BrainSpan (multiple regions)37 and BrainCloud (prefrontal cortex).28 We conducted 2-stage (before and after birth) and 3-stage (before birth, infancy, and childhood to adulthood) analyses.
2-Stage Expression Patterns.
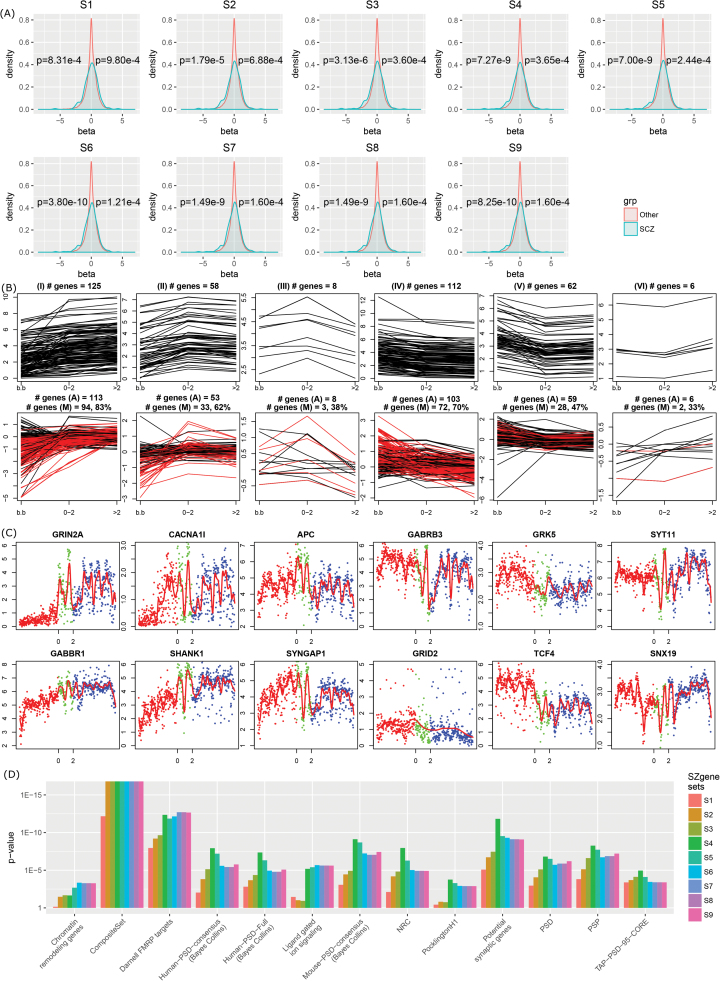
SZgenes showed overrepresentation of genes with more dramatic changes before and after birth (figure 4A). To quantitatively assess the pattern, we used Wilcoxon rank-sum test (1 sided) to compare SZgenes and other genes respectively in 2 scenarios: genes that were overexpressed after birth (β > 0) and genes that were overexpressed before birth (β < 0). In either case, we tested if SZgenes had β values further apart from 0 (ie, large |β| values). As shown in figure 4A, using BrainSpan data, we observed an increased proportion of genes with large |β| values on both sides, represented by the increased shoulders on both sides. This is in line with previous studies that reported genes with prenatal transcript abundance in several psychiatric diseases, including intellectual disability and autism disorder.38 The same pattern was validated using the BrainCloud data (figure S6). Because a larger |β| value reflects stronger change, these results implied that these SZgenes would act on their roles through expression during brain development.
Fig. 4.
Expression and functional characterization of schizophrenia candidate genes (SZgenes). (A) Density plot of 2-stage gene expression pattern for SZgenes using the BrainSpan dataset. β value (x-axis) was obtained by comparison of expression before and after birth. A positive β value indicates the gene has higher expression after birth than before birth. One-sided t test was performed for genes with positive β values and genes with negative β values. (B) Six classes of expression patterns (I–VI) of SZgenes. x-axis: developmental stages: before birth (b.b), infancy (age ≤ 2), and childhood to adulthood (age > 2). y-axis: expression intensity. The top panel showed the expression pattern using the BrainSpan data (RNA-sequencing), with each line indicating 1 gene. The bottom panel showed the expression pattern using the BrainCloud data (microarray data), with each line indicating 1 probe. In the bottom panel, #genes (A) indicates the number of genes from the corresponding cluster in the BrainSpan data (top panel) that were also available in the BrainCloud data, and #genes (M) indicates the number of genes showing the matched expression pattern in the BrainCloud data (highlighted as red lines). (C) Example genes for each class using the BrainSpan data. Each dot represents a gene in a sample; dot color indicates stages. (D) Functional gene set enrichment analysis. PSD: postsynaptic densities. NRC: N-methyl-d-aspartate receptor complex. PSP: postsynaptic proteome. Details of the gene sets are provided in table S3. Only gene sets with significant enrichment in any of the 9 SZgene sets (P < .05, Bonferroni method) were shown.
3-Stage Expression Patterns.
The development of human brain starts in fetal life and continues several years after birth. Through hierarchical cluster analysis of all samples from the BrainSpan dataset, we defined 3 developmental stages: fetal development (stage 1, age < 0), infancy (stage 2, age ≤ 2), and childhood to adulthood (stage 3, age > 2) (figure S7). We compared the median gene expression of each SZgene in each of the 3 stages, resulting in 6 expression clusters (figure 4B). In clusters I, II, and III, gene expression increased from stage 1 to stage 2, and then increased (cluster I), decreased (cluster II), or further decreased (cluster III) from stage 2 to stage 3. In contrast, in clusters IV, V, and VI, gene expression decreased from stage 1 to stage 2, and then decreased (cluster IV), increased (cluster V), or further increased (VI) from stage 2 to stage 3. The temporal expression pattern was largely validated using the independent BrainCloud dataset (figure 4B, bottom panel). Among the 6 clusters, the majority of genes (≥62%) in 3 clusters (I, II, IV) were found with at least 1 probe with the same pattern whereas genes in the other 3 clusters had reproducible patterns in 47% (cluster V), 33% (cluster VI), and 38% (cluster III) genes. For each cluster, we chose 2 example SZgenes to demonstrate the expression changes over development stages (figure 4C).
Lack of Regional Variability
The BrainSpan data represents 4 major brain regions: subcortical regions (SC), sensorimotor regions (SM), frontal cortex (FC), and temporal–parietal cortex (TP). In our examination of the developmental expression pattern in each of the 4 regions, we found different brain regions displayed similar trends (figure S8). Those genes in the 6 clusters showed no significant change of expression in these brain regions except marginal significance in FC (using all genes: PFC = .11, PSC = .34, PSM = .63, and PTP = .16; using genes with variance > 0.05: PFC = .27, PSC = .30, PSM = .66, and PTP = .48; χ2 test). Two clusters had the strongest change: cluster I, where genes showed continuous increase during all 3 stages of lifespan, and cluster IV, where genes showed continuous decrease during all 3 stages. These results were consistent with our analysis using GTEx data (figure 3F), where SZgenes did not show statistical significance among brain regions, although region-specific trend was observed.
Gene Set Enrichment Analysis
Enrichment analysis using customized functional gene sets (table S3, Supplementary material) revealed that SZgenes were consistently enriched in G-protein-coupled receptor (GPCR) signaling, postsynaptic density (PSD) gene groups, N-methyl-d-aspartate receptor complex, ligand-gated ion signaling, potential synaptic genes,50,51 and fragile X mental retardation protein (FMRP) targets52,53 (figure 4D). In particular, SZgenes were enriched in a set of FMRP targets (n = 780 genes) generated using mouse brain52 but not in an independent set of FMRP targets from cultured human embryonic kidney cells (HEK293, n = 899).53 Another group of interest was the composite set, which comprised 1796 genes that were implied in schizophrenia in previous studies.29 On the contrary, single dimensional data did not show many enrichment results (table S7), with a few functional gene sets associated with ≥2 types of omics data, such as various PSD complex, synaptosome, and FMRP genes.
During the revision of this work, a transcriptome-wide association study (TWAS) was published that integrated the analysis of GWAS, gene expression, splicing, and chromatin variation to identify genes whose expression is genetically correlated with schizophrenia.54 We compared SZgenes with the 157 TWAS-significant genes and found that 35.7% (56 of 157) of the TWAS genes were included in our SZgenes, indicating a high recovery rate (P = 4.12 × 10−30, FET; figure S9).
Discussion
The past decade has overseen an incredible growth of genomic data for schizophrenia and other psychiatric disorders. Yet an effective integration and cross talk has been lacking for the heterogeneous omics data. We presented a systematic catalog of schizophrenia-associated variants and genes and a comprehensive multidimensional analysis to prioritize SZgenes, which leveraged on association evidence ranging in genetics, epigenetics, transcriptomics, and functional annotations. The resultant SZgenes had intensive association evidence. Their complex expression patterns and other characteristics provided insights for our understanding of the pathological architecture underlying schizophrenia. The proposed method, MegaOR, combines multidimensional omics data in an unbiased fashion and is applicable to many other complex diseases with heterogeneous data.
Among the SZgenes, we observed many well-studied candidates for schizophrenia, such as Gamma Amino Butyric Acid (GABA) receptors (GABBR1, GABBR2, GABRA2, GABRA5, GABRB3, and GABRG2), G-protein receptors (GNAO1, GRK5, and GRM3), genes in the major histocompatibility complex (MHC) region, and neuron-related genes (NRG1, NRG3, NRGN, NRXN1, NT5C2, and NTRK3). GABA is the main inhibitory neurotransmitter in the mammalian central nervous system. GABA receptor genes and the GABAergic system have long been considered as being involved in schizophrenia, autism, and other psychiatric disorders.55 Six GABA receptor genes in our SZgenes all had PPascal < .05 and are located in 5 different chromosomes, indicating that these genes represented independent genetic association signals. In addition, novel candidate genes were also observed that have not been studied in schizophrenia before. For example, NREP (neuronal regeneration-related protein) was implied by GWAS hit and eQTL support (PSherlock = 1.55 × 10−5); GPM6B (glycoprotein M6B) was a DEG and had eQTL support (PSherlock = 1.53 × 10−3).
Our characterization of the SZgenes provided insights into the convergent molecular processes in schizophrenia. SZgenes were found to undergo dramatic changes during brain development, interact with each other more frequently than by chance, and converge on functional gene sets that are of high interest in schizophrenia. The expression pattern of SZgenes reinforced the importance of studying disease genes in the context of disease-relevant tissues with temporal and spatial information. Although schizophrenia mainly occurs in adults, the enrichment of SZgenes with increased load of disturbance before and after birth implies that the risk factors may function in the early stages of patients’ life, an observation that was similarly reported in other psychiatric disorders.56 One limitation of our work is that the multi-omics data we used are still limited. For example, we only used DEGs from 2 datasets whereas more data could be included.26 In addition, further strategies can be developed to better validate the SZgenes.
Data dependence remains a challenge in multi-omics integrative analysis. In our work, GWAS, Pascal, and Sherlock were not completely independent from each other. GWAS top hits were collected from a number of GWAS, including the PGC GWAS data, which were used for Pascal and Sherlock results. When we counted once for overlapping genes, we found that the results would miss many important genes (figure S9). In complex disease such as schizophrenia, independence of data sources would be very hard to achieve, leading to a substantial reduction of the data content.
In summary, we provide a comprehensive catalog of schizophrenia risk genes based on a novel convergent analysis of the currently available data from many sources. These genes further supported the polygenic and neurodevelopment models in schizophrenia, but they may act as early as in fetal development stages.
Funding
This work was supported by National Institutes of Health grants (R01LM011177 and R01LM012806).
Conflict of Interest
The authors have declared that there are no conflicts of interest in relation to the subject of this study.
Supplementary Material
Acknowledgments
The authors thank Drs. Xiaoming Liu, Timothy O’Brien and Ms. Xueying Zhang for insightful discussion.
References
- 1. Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. [DOI] [PubMed] [Google Scholar]
- 2. Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Purcell SM, Wray NR, Stone JL et al. ; International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi J, Levinson DF, Duan J et al. . Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stefansson H, Ophoff RA, Steinberg S et al. ; Genetic Risk and Outcome in Psychosis (GROUP) Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishioka M, Bundo M, Kasai K, Iwamoto K. DNA methylation in schizophrenia: progress and challenges of epigenetic studies. Genome Med. 2012;4:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Z, Xu J, Chen J et al. . Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2015;20:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou K, Yang Y, Gao L et al. . NMDA receptor hypofunction induces dysfunctions of energy metabolism and semaphorin signaling in rats: a synaptic proteome study. Schizophr Bull. 2012;38:579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang J, Chen T, Sun L et al. . Potential metabolite markers of schizophrenia. Mol Psychiatry. 2013;18:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. [DOI] [PubMed] [Google Scholar]
- 11. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manolio TA, Collins FS, Cox NJ et al. . Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh T, Kurki MI, Curtis D et al. ; Swedish Schizophrenia Study; INTERVAL Study; DDD Study; UK10 K Consortium Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szatkiewicz JP, O’Dushlaine C, Chen G et al. . Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jaffe AE, Gao Y, Deep-Soboslay A et al. . Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Montano C, Taub MA, Jaffe A et al. . Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry. 2016;73:506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wockner LF, Noble EP, Lawford BR et al. . Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishioka M, Bundo M, Koike S et al. . Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J Hum Genet. 2013;58:91–97. [DOI] [PubMed] [Google Scholar]
- 20. Dempster EL, Pidsley R, Schalkwyk LC et al. . Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kano S, Colantuoni C, Han F et al. . Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol Psychiatry. 2013;18:740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rukova B, Staneva R, Hadjidekova S, Stamenov G, Milanova 5th, Toncheva D. Genome-wide methylation profiling of schizophrenia. Balkan J Med Genet. 2014;17:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mill J, Tang T, Kaminsky Z et al. . Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aberg KA, McClay JL, Nerella S et al. . Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Eijk KR, de Jong S, Strengman E et al. . Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur J Hum Genet. 2015;23:1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fromer M, Roussos P, Sieberts SK et al. . Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maycox PR, Kelly F, Taylor A et al. . Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol Psychiatry. 2009;14:1083–1094. [DOI] [PubMed] [Google Scholar]
- 28. Colantuoni C, Lipska BK, Ye T et al. . Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purcell SM, Moran JL, Fromer M et al. . A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richards AL, Jones L, Moskvina V et al. ; Molecular Genetics of Schizophrenia Collaboration (MGS); International Schizophrenia Consortium (ISC) Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun J, Jia P, Fanous AH et al. . A multi-dimensional evidence-based candidate gene prioritization approach for complex diseases-schizophrenia as a case. Bioinformatics. 2009;25:2595–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jia P, Han G, Zhao J, Lu P, Zhao Z. SZGR 2.0: a one-stop shop of schizophrenia candidate genes. Nucleic Acids Res. 2017;45:D915–D924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12:e1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He X, Fuller CK, Song Y et al. . Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet. 2013;92:667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He X, Sanders SJ, Liu L et al. . Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Melé M, Ferreira PG, Reverter F et al. ; GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. BRAINSPAN: atlas of the developing human brain http://www.brainspan.org/static/home. Accessed May, 2016.
- 38. Birnbaum R, Jaffe AE, Hyde TM, Kleinman JE, Weinberger DR. Prenatal expression patterns of genes associated with neuropsychiatric disorders. Am J Psychiatry. 2014;171:758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rees E, Walters JT, Georgieva L et al. . Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry. 2014;204:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farrell MS, Werge T, Sklar P et al. . Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20:555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun J, Jia P, Fanous AH et al. . Schizophrenia gene networks and pathways and their applications for novel candidate gene selection. PLoS One. 2010;5:e11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. [DOI] [PubMed] [Google Scholar]
- 43. Guo AY, Sun J, Riley BP, Thiselton DL, Kendler KS, Zhao Z. The dystrobrevin-binding protein 1 gene: features and networks. Mol Psychiatry. 2009;14:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, Keller MC. No evidence that schizophrenia candidate genes are more associated with schizophrenia than noncandidate genes. Biol Psychiatry. 2017;82:702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34:1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gulsuner S, Walsh T, Watts AC et al. ; Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hormozdiari F, Penn O, Borenstein E, Eichler EE. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2015;25:142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cerami EG, Gross BE, Demir E et al. . Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirov G, Pocklington AJ, Holmans P et al. . De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the Genes to Cognition database. Nucleic Acids Res. 2009;37:D846–D851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Darnell JC, Van Driesche SJ, Zhang C et al. . FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ascano M Jr, Mukherjee N, Bandaru P et al. . FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gusev A, Mancuso N, Won H et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50:538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Charych EI, Liu F, Moss SJ, Brandon NJ. GABA(A) receptors and their associated proteins: implications in the etiology and treatment of schizophrenia and related disorders. Neuropharmacology. 2009;57:481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Willsey AJ, Sanders SJ, Li M et al. . Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.