Abstract
Although type-2–induced (T2-induced) epithelial dysfunction is likely to profoundly alter epithelial differentiation and repair in asthma, the mechanisms for these effects are poorly understood. A role for specific mucins, heavily N-glycosylated epithelial glycoproteins, in orchestrating epithelial cell fate in response to T2 stimuli has not previously been investigated. Levels of a sialylated MUC4β isoform were found to be increased in airway specimens from asthmatic patients in association with T2 inflammation. We hypothesized that IL-13 would increase sialylation of MUC4β, thereby altering its function and that the β-galactoside α-2,6-sialyltransferase 1 (ST6GAL1) would regulate the sialylation. Using human biologic specimens and cultured primary human airway epithelial cells (HAECs),we demonstrated that IL-13 increases ST6GAL1-mediated sialylation of MUC4β and that both were increased in asthma, particularly in sputum supernatant and/or fresh isolated HAECs with elevated T2 biomarkers. ST6GAL1-induced sialylation of MUC4β altered its lectin binding and secretion. Both ST6GAL1 and MUC4β inhibited epithelial cell proliferation while promoting goblet cell differentiation. These in vivo and in vitro data provide strong evidence for a critical role for ST6GAL1-induced sialylation of MUC4β in epithelial dysfunction associated with T2-high asthma, thereby identifying specific sialylation pathways as potential targets in asthma.
Keywords: Inflammation, Pulmonology
Keywords: Asthma, Th2 response
β-galactoside α-2,6-sialyltranferase 1 (ST6GAL1)-induced sialylation of MUC4β is critical for airway epithelial differentiation and proliferation, and is enhanced in Type-2 inflammation.
Introduction
Asthma is a complex heterogeneous disease in which type-2 (T2) immune-associated goblet cell hyperplasia and hypersecretion of mucins are considered clinically important pathophysiologic features (1–3). Mucins are a large multifunctional family of heavily N- and O-glycosylated proteins whose primary functions are to protect and lubricate epithelial surfaces. However, changes in glycosylation of mucins could also affect their function, thereby affecting epithelium homeostasis and contributing to disease (4).
Glycosylation is an enzymatic cotranslational and posttranslational protein modification, which attaches glycans to proteins or lipids on specific sites in the rough ER (5). There are 5 major types of glycosylations; N- and O-linked are most common. N-linked glycans, in particular, are important for glycoprotein secretion, folding and stability, and cell-cell and cell-extracellular matrix attachments (6, 7) as well as receptor-ligand interactions of relevance to cell signaling (8). Some of N-glycan moieties are capped with sialic acids (a 9-carbon monosaccharide) at the terminal end (9).
Secreted mucins, including MUC5AC and MUC5B, normally associated with O-linked glycosylation, have been widely investigated in airway disease, but less is known about membrane-bound mucins or the role of N-glycosylation (10–13). MUC4, traditionally a tethered mucin reported to be upregulated in asthma (14–16), comprises 2 subunits: the 850-kDa MUC4α, with a high degree of O-glycosylation sites, but also a much smaller approximately 78- to 80-kDa MUC4β, which is suggested to have 18 potential N-glycosylation sites (17, 18). N-glycosylation of MUC4β is known to be induced by IL-4 and IL-9 in epithelial cell lines (19, 20), but the pattern, regulation, and functional effect are poorly studied.
Among many enzymes involved in the glycosylation and sialylation process, the β-galactoside α-2,6-sialyltransferase 1 (ST6GAL1) catalyzes terminal α 2,6-sialylation of N-glycans to add sialic acid residues (21). It is present in plasma and lungs of rats and humans, supporting its secretion (22–24). Previous gene expression profiling of freshly brushed human airway epithelial cells (HAECs) identified an association between ST6GAL1 expression and the T2 biomarker fractional exhaled nitric oxide (FeNO) (25), supporting a potential T2 immune relationship. However, its target proteins (and their N-glycosylation) remain largely unknown.
We hypothesized that ST6GAL1 would promote MUC4β N-glycosylation and sialylation, both of which would be upregulated in asthma, particularly in association with T2 inflammation. In vitro, IL-13–induced ST6GAL1 would promote terminal sialylation of MUC4β N-glycans in HAECs, which would affect their differentiation and proliferation. This was evaluated by identifying ST6GAL1, MUC4β, and related pathways in freshly isolated, as well as cultured, HAECs and sputum supernatants from healthy control subjects (HC) and a range of asthmatic patients to determine the expression, regulation, and functional effect of MUC4β N-glycosylation/sialylation in epithelial function and asthma.
Results
Demographics.
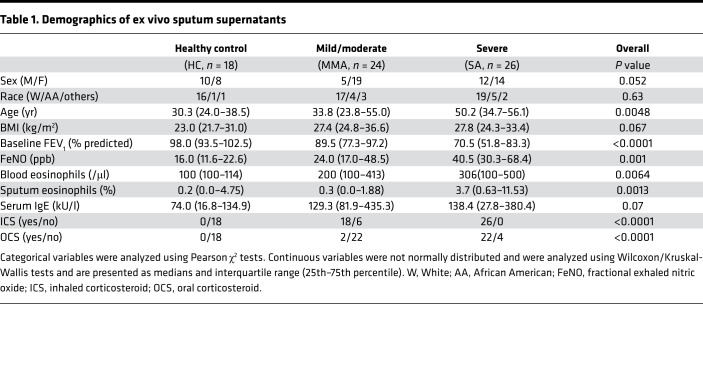
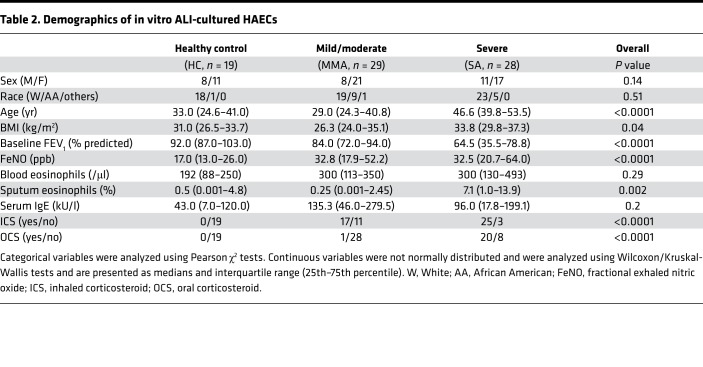
Ex vivo sputum supernatants were obtained from 18 HC, 24 mild-moderate (MMA) participants, and 26 severe asthma (SA) participants (Table 1). The groups did not differ by race, but SA patients were older, with the lowest forced expiratory volume in 1 second (FEV1) percentage predicted despite inhaled and oral corticosteroid use. SA patients also had the highest levels of FeNO and blood and sputum eosinophils. Microarray data (GEO accessionGSE63142) were further analyzed (25). Quantitative real-time PCR (qRT-PCR) validation was performed on a subset of 7 HC, 13 MMA participants, and 9 SA participants (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122475DS1), and Western blot was performed on separate freshly isolated HAECs (9 HC, 13 MMA, and 11 SA) (Supplemental Table 2). In vitro studies were performed on HAECs from 19 HC, 29 MMA participants, and 28 SA participants (Table 2).
Table 1. Demographics of ex vivo sputum supernatants.
Table 2. Demographics of in vitro ALI-cultured HAECs.
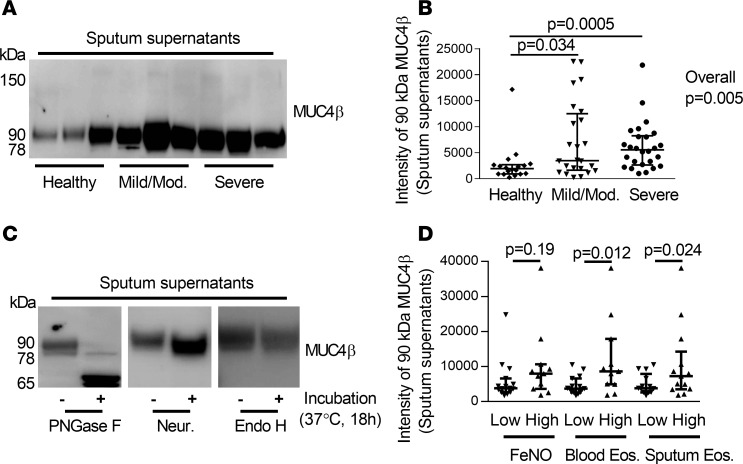
N-glycosylated MUC4βlevels are higher in sputum supernatants from asthmatic subjects compared with HC. Human sputum is an accumulation of extracellular secretions and cells from lower airways (26). MUC4β was present in sputum cell-free supernatants, and the 90-kDa isoform (by immunoblot) predominated. The intensity of the 90-kDa isoform was significantly higher in sputum supernatants from both MMA and SA participants compared with HC (Figure 1, A and B) and when controlling for age and sex. To determine whether the predominant 90-kDa isoform represented an N-glycosylated/sialylated isoform, sputum supernatants were treated with PNGase F to remove mature and immature N-glycans, neuraminidase to remove sialic acids from mature O- and N-linked glycans, or Endo H to remove only immature N-glycans. Following treatment, the 90-kDa band shifted primarily to a 65-kDa band when digested with PNGase F, consistent with the presence of approximately 8 N-glycans (3 kDa per N-glycan). Neuraminidase treatment shifted the 90-kDa isoform to 78 kDa, a potential desialylated isoform of MUC4β, while Endo H had no effect (Figure 1C). Thus, the 90-kDa band represents an N-glycosylated isoform of MUC4β that contains mature N-glycans with terminal sialic acid residues.
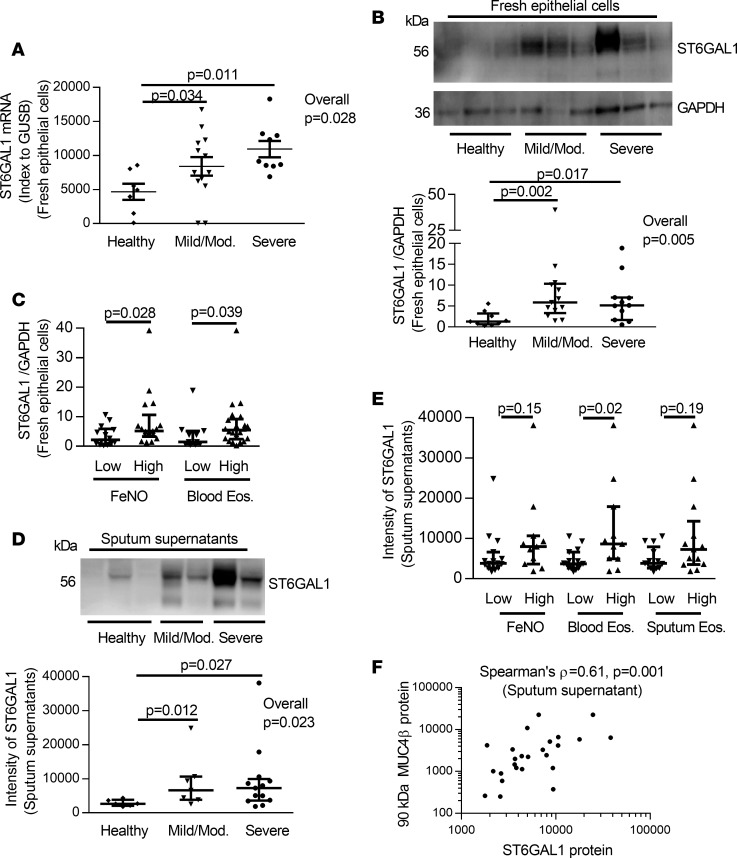
Figure 1. N-glycosylated 90-kDa MUC4β was higher in asthmatic sputum supernatants and associated with a T2 signature.
(A and B) Western blot and densitometry of MUC4β (18 HC, 24 MMA, 26 SA). Since there were no loading controls for sputum supernatant, the same amount of 40 μg total proteins were loaded for each sample, and 1 internal control sample was loaded for each gel. (C) Deglycosylation with PNGase F, neuraminidase (Neur.), and Endo H (n = 3). Untreated samples were added reaction buffer without above enzymes. (D) The 90-kDa MUC4β isoform was associated with the T2 inflammation markers blood and sputum eosinophils but not FeNO. The data are presented as median with interquantile range. Wilcoxon tests identified the overall differences among the groups. When overall P ≤ 0.05, intergroup comparisons were further assessed using Wilcoxon signed rank. One-way analysis Wilcoxon test was used for 2 independent group comparison.
T2 immune pathways are associated with N-glycosylated MUC4βlevels ex vivo, and IL-13 enhances its N-glycosylation in vitro. To begin to determine mechanisms driving the predominant 90-kDa MUC4β levels in asthmatic sputum, MUC4β was compared with levels of T2 biomarkers (FeNO and blood and sputum eosinophils). The subjects were defined as “high” T2 on the basis of a FeNO at ≥25 ppb (27, 28), blood eosinophils at ≥300 eosinophils/μl, or sputum eosinophils at ≥2% (29–32). The intensity of the 90-kDa MUC4β isoform was significantly higher in the T2/blood and sputum eosinophil–high group but not higher in FeNO-high group as compared with the low groups (Figure 1D).
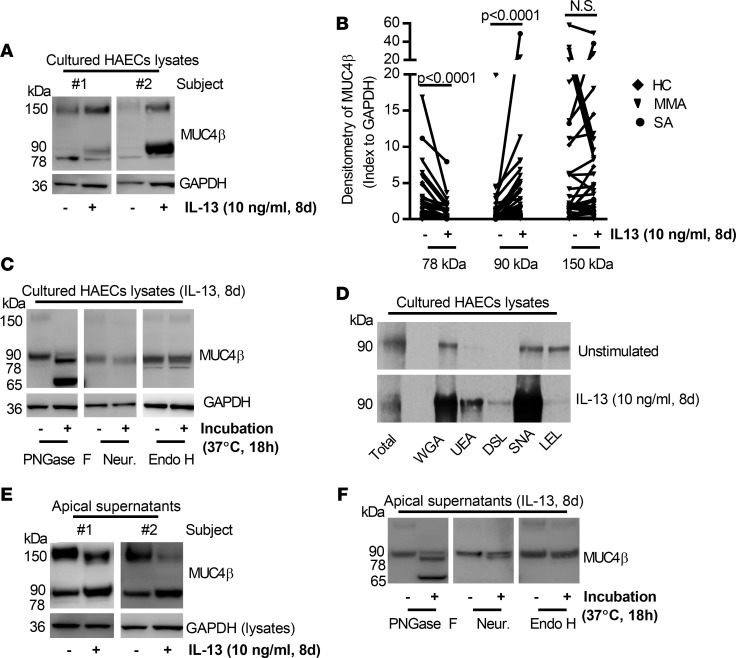
To determine whether the higher sputum N-glycosylated MUC4β in the presence of T2 biomarkers could be due to induction by T2 cytokines, air-liquid interface–cultured (ALI-cultured) HAECs were treated with IL-13 (10 ng/ml) for 8 days, and apical supernatants and cell lysates were harvested for MUC4β expression. IL-13 decreased the level of 78-kDa protein and increased the 90-kDa isoform levels but had inconsistent effects on a 150-kDa isoform (Figure 2, A and B). In contrast, IL-13 modestly but significantly decreased MUC4β mRNA (~20%) (Supplemental Figure 1). To confirm that the MUC4β-associated bands in vitro were N-glycosylated/sialylated isoforms, HAEC lysates were treated with PNGase F, neuraminidase, or Endo H. Similar to sputum, PNGase F shifted the 90-kDa band to a 65-kDa isoform. However, contrasting with sputum, neither neuraminidase nor Endo H had any effect (Figure 2C). Thus, IL-13 affects the levels of MUC4β isoforms in a posttranslational manner, while having a modest negative effect on mRNA.
Figure 2. Effects of IL-13 on MUC4β N-glycosylation/secretion and lectin-binding capacity in 8-day ALI-cultured HAECs.
(A and B) Representative Western blot of 78-, 90-, 150-kDa MUC4β isoforms influenced by IL-13 (indexed to GAPDH) (n = 37). (C) Deglycosylation with PNGase F, neuraminidase (Neur.), and Endo H in IL-13–stimulated HAECs (n = 3). (D) Representative Western blot of lectin (WGA, UEA, DSL, SNA, and LEL) binding of 90-kDa MUC4β in the absence and presence of IL-13 for 8 days (n = 3). (E) Representative Western blot of apical supernatant MUC4β isoforms in ALI-cultured HAECs. (F) Deglycosylation with PNGase F, neuraminidase, and Endo H in IL-13–stimulated apical supernatant (n = 3). Nonparametric paired t test identified the difference between IL-13 and unstimulated condition.
N-glycosylation/sialylation functionally alters MUC4β, affecting its lectin binding and apical secretion. To determine the functional effect of N-glycosylation of MUC4β, the binding of immunopurified MUC4β to lectins, wheat germ agglutinin (WGA), Ulex europaeus agglutinin (UEA), Datura stramonium lectin (DSL), Sambucus nigra lectin (SNA), and Lycopersicon esculentum (tomato) lectin (LEL/LEA), was assessed by pull-down assays with agarose-coupled lectins (33). IL-13 treatment for 8 days enhanced the fraction of immunopurified MUC4β binding to WGA (which binds to N-glycan chitobiose core structures as well as sialic acid), UEA (which binds terminal α-linked fucose residues) and SNA (which binds to sialic acid attached to terminal galactose in an α-2,6 configuration), while reducing binding to LEL (which binds polylactosamine present on the terminus of either O-linked glycans or mature N-linked glycans) (Figure 2D). These data support both sialylation and fucosylation of MUC4β by IL-13 and reduction in terminal polylactosamine.
To determine whether N-glycosylation/sialylation potentiated the secretion of MUC4β (similar to that observed in sputum sol phase), apically secreted MUC4β was measured after 8 days in IL-13–treated HAECs. IL-13 consistently increased apical secretion of the 90-kDa MUC4β, while having no consistent effect on the 150-kDa isoform (Figure 2E). To confirm that the secreted MUC4β-associated bands were N-glycosylated/sialylated isoforms, HAEC apical supernatants were treated with PNGase F, neuraminidase, or Endo H. Similar to sputum, PNGase F shifted the 90-kDa band to a 65-kDa isoform, while neuraminidase shifted the apical 90-kDa band to a 78 kDa isoform and Endo H had no effect (Figure 2F).
To confirm the effect of N-glycans on MUC4β secretion, HAECs cultured for 8 days with IL-13 were metabolically pulsed with [35S] Met/Cys for 30 minutes and chased for 0, 3, and 24 hours before recovery of cells and apical medium for MUC4β immunoprecipitation and analysis by SDS-PAGE. Radiolabeled proteins on SDS-PAGE were transferred to nitrocellulose for subsequent Western blotting. MUC4β was immunoprecipitated with the polyclonal anti-ASGP-2 (rat MUC4β) Ab. Radioactive imaging of IL-13–stimulated HAECs showed the presence of both the 78-kDa MUC4β and 90-kDa N-glycosylated/sialylated isoforms after 30 minutes of [35S]Met/Cys pulse (t = 0). As the chase period continued, the 78-kDa isoform decreased while the 90-kDa isoform increased, suggesting a shift to the more heavily sialylated isoform. The 90-kDa sialylated MUC4β isoform was measurable in the apical supernatant as early as 3 hours after stimulation, while very little 78-kDa MUC4β was measurable. Overall, cell-associated 78-kDa MUC4β decreased over the chase period, while the 90-kDa MUC4β isoform increased in the apical supernatants both by 35S imager and immunoblot (Supplemental Figure 2). The 150-kDa isoform was apparent in the cells and also disappeared with time but, interestingly, did not appear in the apical supernatant. Thus, N-glycosylation/sialylation of MUC4β appears to be associated with increased apical secretion.
The sialyltransferase ST6GAL1 is higher in lung samples from asthmatics in association with T2 inflammation, and the results are mirrored by IL-13 stimulation in vitro.
Lectin binding and neuraminidase treatments suggest MUC4β is an N-glycosylated mucin with terminal sialic acids modifications induced by T2 stimulation. Global epithelial gene profiling identified a strong correlation of ST6GAL1 with FeNO (ρ = 0.508, P = 0.000024) (25). Further analysis showed that ST6GAL1 mRNA (microarray data) was significantly higher in asthmatics compared with HC and that severity and inhaled corticosteroid (ICS) use had only a minimal effect on differences (Supplemental Figure 3A). However, it was higher in all subjects with elevations in both T2 biomarkers, as previously defined (Supplemental Figure 3, B and C). These differences in ST6GAL1 by microarray were validated by qRT-PCR on freshly isolated epithelial cells, 80% of which overlapped with cells from subjects analyzed by microarray (Figure 3A). ST6GAL1 protein from freshly isolated HAECs was also higher in asthmatic patients compared with HC (overall P = 0.005; MMA vs. HC, P = 0.002; SA vs. HC, P = 0.017) (Figure 3B). Additionally, higher ST6GAL1 protein was associated with high FeNO (P = 0.028) and blood eosinophils (P = 0.039) (Figure 3C).
Figure 3. The levels of ST6GAL1 mRNA/protein and association with T2 inflammation in freshly isolated epithelial cells and sputum supernatant among HC, MMA, and SA.
(A) ST6GAL1 mRNA by qRT-PCR in freshly isolated epithelial cells (7 HC, 13 MMA, 9 SA). (B) Representative Western blot and densitometry of ST6GAL1 protein (9 HC, 13 MMA, and 11 SA) in freshly isolated epithelial cells. (C) Presence of T2 inflammation (FeNO and blood eosinophils) associated with increased epithelial cell ST6GAL1 protein. (D) Representative Western blot and densitometry of sputum supernatant ST6GAL1 protein (6 HC, 7 MMA, 13 SA). (E) Sputum supernatant ST6GAL1 protein was associated with blood eosinophils but not sputum eosinophils and FeNO. (F) ST6GAL1 protein positively correlated with 90-kDa MUC4β protein in sputum supernatant. The data are presented as median with interquantile range. Wilcoxon tests identified the overall differences for multiple comparisons, followed by Wilcoxon each pair comparison when overall P ≤ 0.05. One-way analysis Wilcoxon test was used for independent comparison between 2 groups. Spearman’s ρ was applied for nonparametric correlation analysis.
As ST6GAL1 has also been reported in plasma (34), ST6GAL1 protein was measured in sputum supernatants by immunoblotting and similarly was higher in asthmatic subjects as compared with HC (overall P = 0.023; MMA vs. HC, P = 0.012; SA vs. HC, P = 0.027), without differences in groups at different levels of severity (Figure 3D). Importantly, higher ST6GAL1 protein in sputum was associated with high blood eosinophils (P = 0.02) but not with high sputum eosinophils (P = 0.19) and FeNO (P = 0.15) (Figure 3E). Not surprisingly, secreted ST6GAL1 protein in the sputum sol phase strongly correlated with the 90-kDa MUC4β isoform (Spearman’s ρ = 0.61, P = 0.001) (Figure 3F), suggesting that the relationship of ST6GAL1 to sialylated MUC4β is due to the enzymatic activity of ST6GAL1.
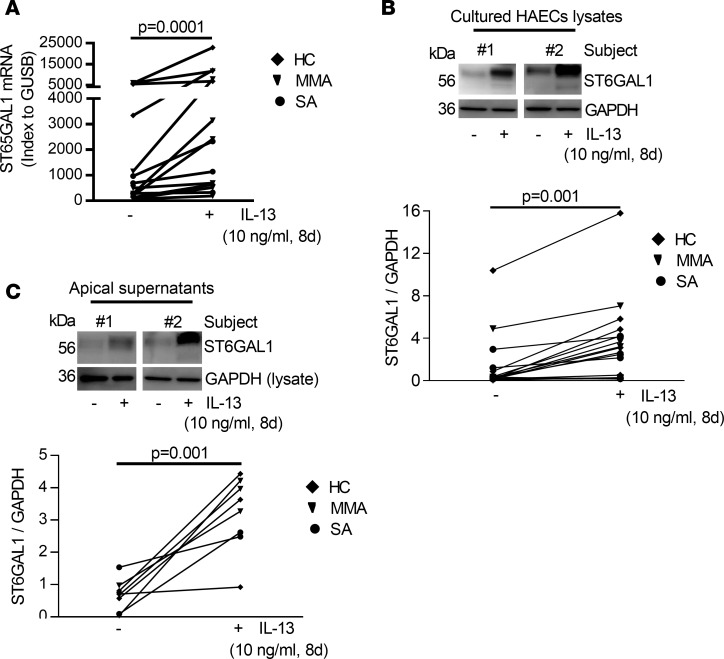
To determine whether IL-13 stimulation of HAECs would recapitulate the ex vivo findings, ST6GAL1 expression was measured. IL-13 increased expression of ST6GAL1 mRNA (P = 0.0001) and protein (P = 0.001) in whole cell lysates (Figure 4, A and B) as well as in apical supernatants (P = 0.001) (Figure 4C), mirroring the ex vivo findings. To determine the specificity of IL-13 for ST6GAL1 expression, dose-response studies were performed with other immune/inflammatory factors of relevance to asthma, including TNF-α, IL-1β, IL-33, TSLP, and IFN-γ. Unlike the strong consistent effects of IL-13, these cytokines did not consistently increase ST6GAL1 protein. However, higher doses of TSLP generally induced ST6GAL1 protein expression, although to a lesser degree than IL-13. In contrast, the type 1 cytokine IFN-γ consistently decreased expression of ST6GAL1 protein with concomitant IL-13 stimulation (Supplemental Figure 4). Thus, only IL-13 and the T2-associated cytokine TSLP affected ST6GAL1 protein expression.
Figure 4. IL-13–induced ST6GAL1 mRNA and protein expression in intracellular and apical supernatants in ALI-cultured HAECs.
(A) IL-13 increased ST6GAL1 mRNA expression. (B) Representative Western blot and densitometry of ST6GAL1 in IL-13–stimulated HAECs (indexed to GAPDH). (C) Representative Western blot and densitometry of apical supernatant ST6GAL1 (indexed to whole cell lysates GAPDH; since there were no loading controls for apical supernatant and the cells directly controlled secretion). Nonparametric paired t test identified the difference between IL-13 and unstimulated condition.
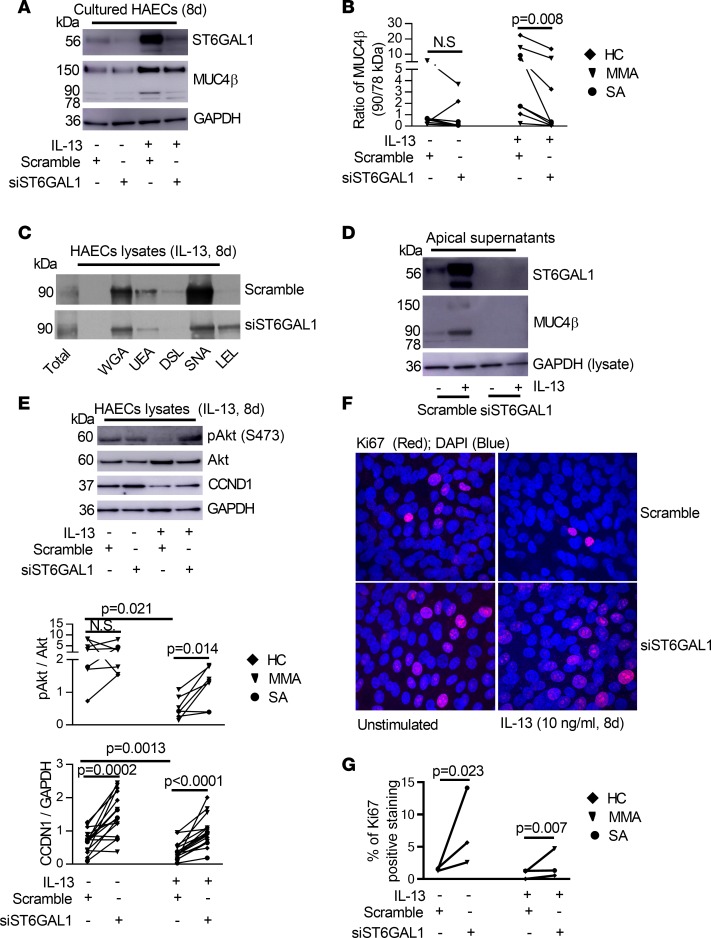
ST6GAL1 sialylates MUC4βunder T2 conditions and alters its lectin binding. The strong correlation between sputum ST6GAL1 and MUC4β suggested an enzyme-substrate association. To confirm the effect of ST6GAL1 on MUC4β sialylation, ST6GAL1 silencing was performed with Dicer siRNA transfection in the absence and presence of IL-13. ST6GAL1 knockdown decreased IL-13–induced ST6GAL1 mRNA (P = 0.001) and protein (P = 0.0005) (Supplemental Figure 5, A and B, and Figure 5A). Importantly, ST6GAL1 knockdown markedly decreased the ratio of the 90-kDa isoform to 78-kDa MUC4β in the presence of IL-13 (P = 0.008) (Figure 5, A and B) as well as the ratio of the 150-kDa isoform to 78-kDa MUC4β (P = 0.016) (Supplemental Figure 5C) in whole cell lysates. In addition, ST6GAL1 knockdown reduced the binding of immunopurified MUC4β to WGA, UEA, and SNA, while it recovered the LEL-binding capacity (Figure 5C), essentially reversing the effects of IL-13 reported in Figure 2D. As expected, ST6GAL1 knockdown inhibited secretion of apical ST6GAL1 and 90-kDa MUC4β in the presence of IL-13, supporting the necessity of N-glycosylation for secretion (Figure 5D and Supplemental Figure 5D). Thus, IL-13 increases MUC4β sialylation through ST6GAL1 enzymatic activity, which substantially alters its structure, function, and location.
Figure 5. ST6GAL1 knockdown decreased sialylated MUC4β but enhanced proliferative signaling in ALI-cultured HAECs.
(A) Representative Western blot of ST6GAL1 and MUC4β after ST6GAL1 knockdown in HAECs. (B) The ratio of 90-kDa/78-kDa MUC4β was decreased by ST6GAL1 knockdown in IL-13 condition. (C) Representative Western blot of MUC4β lectin pulldown after ST6GAL1 knockdown in IL-13–stimulated HAECs for 8 days (n = 3). (D) Representative Western blot of apical secreted ST6GAL1 and MUC4β in the absence and presence of IL-13 (n = 5). (E) Representative Western blot and densitometry of pAkt/Akt and CCND1/GAPDH by ST6GAL1 knockdown in the absence and presence of IL-13 for 8 days. (F and G) Representative Ki67 immunofluorescent staining and quantification by ST6GAL1 knockdown in the absence and presence of IL-13 for 8 days in ALI-cultured HAECs (n = 3). Original magnification, ×600. Nonparametric paired t test identified the difference between IL-13 and unstimulated condition and siST6GAL1 vs. scramble.
Antiproliferative effects of T2 inflammation may be mediated by ST6GAL1 in vivo and in vitro.
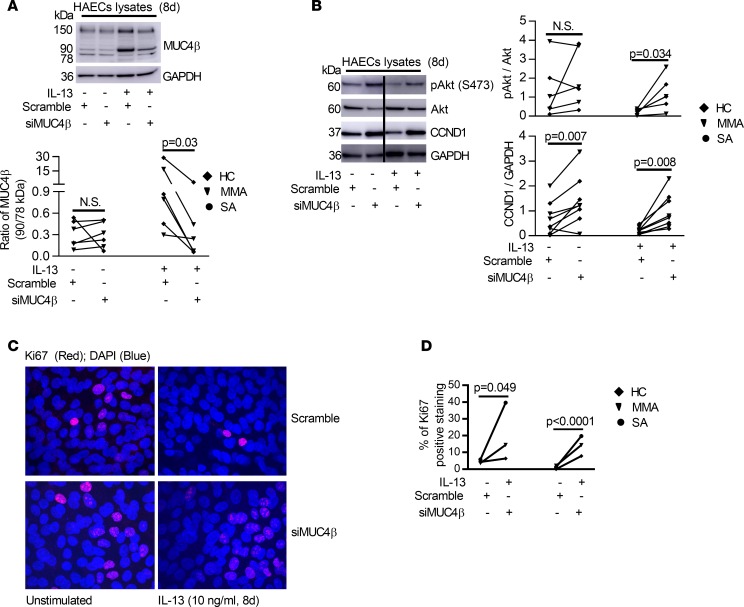
Evidence exists for ongoing epithelial damage and repair in asthma (35, 36). ST6GAL1 mRNA negatively correlated with the cell cycle protein cyclin D1 (CCND1) in the epithelial microarrays (r = –0.39, P < 0.0001), potentially indicating an effect on cell proliferation. To confirm the hypothesis that ST6GAL1 has antiproliferative effects on HAECs, the effect of ST6GAL1 knockdown on pAkt/Akt, CCND1, and the proliferation marker Ki67 was assessed (37, 38). PI3K/Akt is one of the most important signaling pathways for cell proliferation, cell cycle, and apoptosis (39). In order to verify whether IL-13 or IL-13–induced ST6GAL1 was associated with PI3K/Akt signaling or cell cycle, ST6GAL1 knockdown was performed and proliferation markers were analyzed.
IL-13 has been reported to increase Akt phosphorylation following acute (2-hour) exposure (40). Acute IL-13 stimulation (from 0.25–24 hours) similarly increased pAkt and CCND1 (Supplemental Figure 6). In sharp contrast, chronic IL-13 stimulation for 8 days markedly decreased pAkt (relative to total Akt) (P = 0.021, IL-13 vs. unstimulated) and CCND1 (relative to GAPDH) (P = 0.0013, IL-13 vs. unstimulated) (Figure 5E). Interestingly, ST6GAL1 knockdown reversed IL-13–attenuated relative pAkt and CCND1 protein (pAkt/Akt, P = 0.014; CCND1/GAPDH, P < 0.0001, siST6GAL1 vs. scramble). ST6GAL1 knockdown also increased CCND1 in the absence of IL-13 (P = 0.0002, siST6GAL1 vs. scramble) (Figure 5E), suggesting that the effect did not depend on IL-13. The functional effect of the higher pAkt and CCND1 levels induced by ST6GAL1 knockdown was confirmed by the parallel restoration of Ki67 immunofluorescent staining (unstimulated, P = 0.023; IL-13, P = 0.007, siST6GAL1 vs. scramble) (Figure 5, F and G).
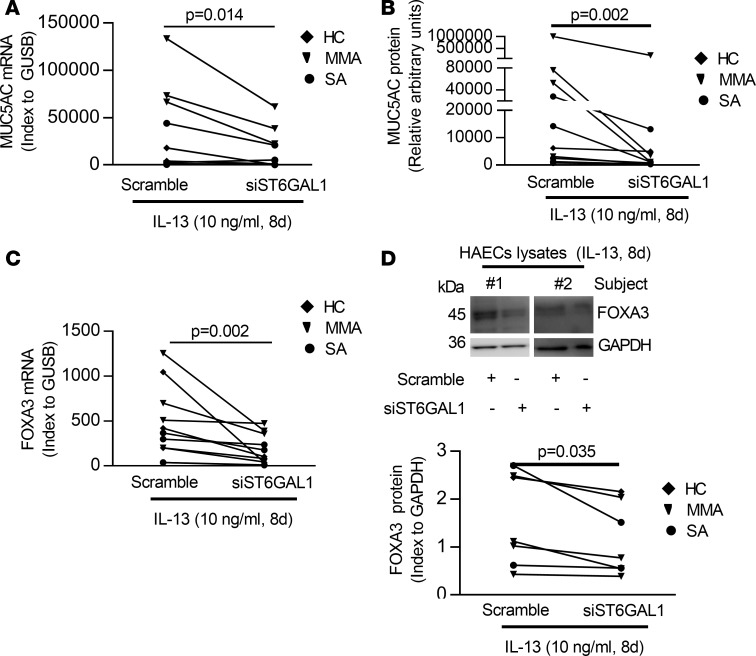
Knockdown of ST6GAL1 decreases MUC5AC and the goblet cell–associated transcription factor FOXA3.
FOXA3 induces MUC5AC and goblet cell metaplasia in response to infection or T2 stimulation (41, 42). To determine whether ST6GAL1 affected goblet cell differentiation, the effect of ST6GAL1 knockdown on expression of MUC5AC and FOXA3 mRNA/protein under IL-13 conditions was evaluated. ST6GAL1 knockdown reduced mRNA/protein expression of MUC5AC and FOXA3 (MUC5AC mRNA, P = 0.014; secreted MUC5AC protein, P = 0.002; FOXA3 mRNA, P = 0.002; FOXA3 protein, P = 0.035) (Figure 6), supporting a critical role for ST6GAL1 in goblet cell differentiation via FOXA3 and MUC5AC.
Figure 6. ST6GAL1 knockdown attenuated T2-caused goblet cell differentiation in ALI-cultured HAECs.
(A and B) Knockdown of ST6GAL1 decreased the MUC5AC mRNA by qRT-PCR and protein by ELISA. (C) Knockdown of ST6GAL1 decreased FOXA3 mRNA by qRT-PCR. (D) Representative Western blot of FOXA3 and densitometry (index to GAPDH). Nonparametric paired t test identified the difference between siST6GAL1 and scramble.
ST6GAL1-induced MUC4βN-glycosylation is necessary for IL-13–induced effects on HAEC proliferation and differentiation. To confirm whether the effects of ST6GAL1 were through MUC4β sialylation, MUC4β knockdown was performed in the presence and absence of IL-13. Silencing MUC4β preferentially decreased the ratio of the 90-kDa sialylated isoform to 78-kDa MUC4β (P = 0.03, siMUC4β vs. scramble) and the ratio of the 150-kDa isoform to the 78-kDa MUC4β (P = 0.03, siMUC4β vs. scramble) in the presence of IL-13 (Figure 7A and Supplemental Figure 7), suggesting a preferential effect on the most highly expressed (N-glycosylated) forms, including the 90-kDa isoform. Similar to the effects of ST6GAL1 knockdown, silencing MUC4β increased pAkt (relative to Akt) (IL-13 condition, P = 0.034), CCND1 (relative to GAPDH) (P = 0.007 for unstimulated and P = 0.008 for IL-13 stimulation) (Figure 7B), and Ki67+ immunofluorescent staining in the absence and presence of IL-13 (unstimulated, P = 0.049; IL-13, P < 0.0001, siMUC4β vs. scramble) (Figure 7, C and D). Due to limited primary HAEC numbers, knockdown experiments were set up using a single culture well treated with the same scramble control. Cells from this well were utilized as the control condition for both siST6GAL1 and siMUC4β experiments (Figure 5F and Figure 7C).
Figure 7. Knockdown of MUC4β primarily decreased its N-glycosylated/sialylated isoform but increased proliferative signaling in 8-day ALI-cultured HAECs in the absence or presence of IL-13.
(A) Representative Western blot of MUC4β and densitometry of MUC4β ratios (90 kDa/78 kDa) after MUC4β knockdown. (B) Representative Western blot and densitometry of pAkt (indexed to total Akt) and CCND1 (indexed to GAPDH) by MUC4β knockdown. (C and D) Representative Ki67 immunofluorescent staining and quantification by MUC4β knockdown in the absence and presence of IL-13–stimulated HAECs (n = 3). For transfection experiments, the same scramble siRNA was utilized as a negative control well/condition for both siST6GAL1 and siMUC4β. Therefore, the Ki67 immunofluorescent images for scramble siRNA are identical to the images in Figure 5F. Original magnification, ×600. Nonparametric paired t test identified the difference between siMUC4β and scramble.
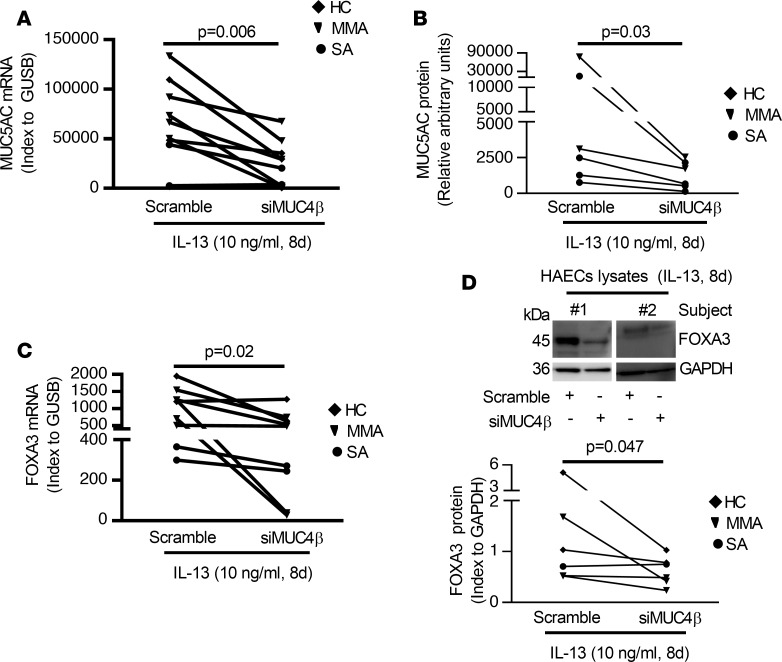
Similar to silencing ST6GAL1, knockdown of MUC4β decreased mRNA/protein expression of MUC5AC and FOXA3 (MUC5AC mRNA, P = 0.006; secreted MUC5AC protein, P = 0.03; FOXA3 mRNA, P = 0.02; FOXA3 protein, P = 0.047) (Figure 8). Thus, MUC4β knockdown recapitulates the differentiation and proliferation effects of ST6GAL1 knockdown, suggesting that posttranslationally modified MUC4β plays an intermediary role in these functional changes.
Figure 8. Knockdown of MUC4β decreased T2-induced goblet cell differentiation in 8-day IL-13–stimulated ALI-cultured HAECs.
(A and B) Knockdown of MUC4β decreased the MUC5AC mRNA by qRT-PCR and protein by ELISA. (C) Knockdown of MUC4β decreased FOXA3 mRNA by qRT-PCR. (D) Representative Western blot and densitometry of FOXA3 (index to GAPDH) in MUC4β-silenced HAECs under IL-13 conditions. Nonparametric paired t test identified the difference between siMUC4β and scramble.
Discussion
It has become increasingly apparent that posttranslational modifications, such as N- glycosylation and sialylation, are essential for regulating biologic responses. Results from this study confirm a critical role for the sialyltransferase ST6GAL1 to promote the terminal sialylation of MUC4β, resulting in subsequent goblet cell differentiation and antiproliferative responses in airway epithelium. Sialylated MUC4β is increased ex vivo in sputum supernatants, generally appears at higher levels in T2-high participants, and is concordant with high levels of ST6GAL1 in both sputum supernatant and freshly isolated HAECs. These ex vivo results correspond with the ability of IL-13–induced ST6GAL1 to increase MUC4β sialylation in ALI-cultured HAECs. Importantly, N-glycosylated/sialylated MUC4β enhances specific lectin (WGA and SNA) binding, while decreasing LEL binding, thus altering its function and secretion. ST6GAL1 knockdown reverses these effects, restoring HAEC proliferation and inhibiting goblet cell differentiation. These results uniquely define a central role for N-glycosylation/sialylation in epithelial cell differentiation, with implications for asthma and more broadly in other epithelial processes, including cancer.
Asthma has long been characterized by mucin abnormalities (43). Mucins are large family of high MW, heavily glycosylated proteins produced by epithelial cells (14). MUC4β is the N-glycosylated transmembrane subunit of MUC4, which has been suggested to be both membrane bound and secreted (44, 45). MUC4β has multiple potential N-glycosylation sites (46), consistent with the multiple MW isoforms reported here (78, 90, and 150 kDa). Only an approximately 140-kDa isoform has previously been reported, in response to IL-4 and IL-9 in human bronchoalveolar carcinoma cell lines (19, 20), with limited data to suggest an indirect role for neutrophil elastase in MUC4β N-glycosylation (47). However, no specific N-glycosylating enzymes have been identified and the functional effect has not been outlined in HAECs.
In the current study, IL-13 most consistently induced a 90-kDa MUC4β isoform in primary HAECs, with the 150 kDa isoform inconsistently altered. Importantly, this 90-kDa isoform was selectively secreted both into cultured HAEC apical supernatants and asthmatic subject sputum, with higher levels preferentially associated with elevations in T2 biomarkers. PNGase F decreased the MW of both the secreted and intracellular glycoproteins, while neuraminidase only reduced the MW of the secreted form, suggesting that sialylation was functionally contributing to its secretion. To confirm the presence of sialic acid residues on the 90-kDa MUC4β isoform, lectin pulldown studies were performed. The highly specific lectin binding to SNA demonstrated the characteristic presence α 2, 6 sialic acid residues on the 90-kDa isoform and was consistent with the neuraminidase studies.
However, in addition to affecting secretion, sialylation is likely to have other effects. Sialylation can mask biological recognition sites or represent specific recognition epitopes as ligands, all widely changing immune responses (48, 49). Previous reports showed that sialylation of N-glycans was responsible for apical delivery of a model mucin-like protein (endolyn) to the surface of polarized MDCK cells — the sialylation allowed interaction with galectin-9 that crosslinks the protein and retains it on the cell surface (50). This suggests that N-glycan sialylation can have substantial effects on protein expression and stability (51, 52). Sialylation of MUC1, for instance, influences cell-cell and cell-matrix adhesion, cell motility, and host cell–pathogen interactions (53, 54). Thus, the effect of sialylation of MUC4β on epithelial differentiation is in-line with that shown in previous studies. Importantly, IL-13 appears to contribute to the addition of alternative N-glycosylation effects as well, as it enhances UEA binding of MUC4β, consistent with fucosylation, while decreasing terminal polylactosamine processing of N-glycans, as demonstrated by reduced LEL binding. Further studies of the precise glycoprotein interactions driving these additional functional changes to MUC4β and their functional effect will be needed.
Given the evidence for the addition of α 2,6-sialic acid residues to MUC4β, it is perhaps not surprising that ST6GAL1 was found to contribute to this process. ST6GAL1, highly expressed in airway epithelial cells (55), was identified on our previous microarrays as one of several N-glycosylating enzymes strongly correlated with the T2 biomarker FeNO in human airways (25). ST6GAL1 broadly distributes in different tissues and is necessary for the addition of almost all α 2,6-sialylated N-glycans (56). The current results add to these by reporting higher ST6GAL1 mRNA and protein in asthmatic HAECs and sputum in association with elevated T2 biomarkers. The ex vivo relationship to T2 inflammation was then mechanistically confirmed in vitro, as IL-13 strongly induced intracellular and secreted ST6GAL1 protein. The modestly variable relationships of ST6GAL1 or MUC4β with T2 biomarkers (FeNO, blood/sputum eosinophils) suggest other stimuli may contribute to their expression. In fact, other inflammatory factors (TNF-α, IL-1β, IL-33, and IFN-γ) had less or no stimulatory effect on ST6GAL1, suggesting that T2 cytokines are likely the most important inducer for ST6GAL1 protein. A potential stimulatory role for TSLP in this expression requires further study. While in vitro and ex vivo data support a stimulatory role for T2 and related cytokines in the upregulation of ST6GAL1, the actual cytokine or mixture of cytokines driving the increase in ST6GAL1 expression in individual patients cannot be determined from these studies.
Altered glycosylation is known to be a common feature of epithelial-derived cancer cells, where aberrant glycosylation interferes with regulation of cell adhesion, migration, and proliferation and links to tumor initiation, progression, and metastasis (48). The effect of specific N-glycosylation events varies, with some promoting tumor progression, while others limit progression/metastases. Elevated expression of ST6GAL1 has previously been observed in multiple types of adenocarcinomas and linked to both worsened and improved outcomes (57, 58). These contradictory observations may relate to differences in cancer mucin expression and/or effect on PI3K/Akt-related proliferation pathways (59, 60). In primary differentiated noncancerous HAECs, ST6GAL1 inhibited PI3K/Akt pathways and associated proliferation signaling in the presence of IL-13, while ST6GAL1 knockdown restored a more proliferative/progenitor-like cell type. These results contrast with cancerous cells, which also express high levels of ST6GAL1. Cancerous cells can have similarities with human induced pluripotent stem cells, which continuously divide until a stimulus is received to exit the division cycle and cell differentiation occurs (61). High ST6GAL1 expression was recently associated with human induced pluripotent stem cells, they were even required for their perpetuation (62), such that ST6GAL1 could play distinctly different roles, depending on the differentiation state of the cell.
The evolving data on ST6GAL1 in asthma yield conflicting views. Overexpression of ST6GAL1 was reported to promote TGF-β–induced epithelial-mesenchymal transition via glycosylation of multiple proteins (60). While epithelial-mesenchymal transition of HAECs is controversial, IL-13 increases TGF-β2 expression in HAECs, although a role for ST6GAL1 remains to be determined (63). The data presented here show ST6GAL1 upstream of FOXA3 in orchestrating goblet cell differentiation, widely observed in asthma, and at least partially dependent on MUC4β sialylation (10, 42). Similarly, increased expression of ST6GAL1 has been reported on T2 lymphocytes (64). However, a global ST6GAL1-knockout mouse study demonstrated worsened eosinophilic inflammation to allergen challenge (65). Effects on airway hyperresponsiveness were not reported in that study, and further targeted deletion studies will be required.
This study does not define the specifics of the structural changes to MUC4β. While ST6GAL1 is a specific sialyltransferase that alters the glycostructure and subsequent biology of MUC4β, most likely through addition of sialic acid residues to N-glycans, it is conceivable that other glycosylating enzymes, and even alterations in O-glycosylation, may also have a functional effect, and they await further study. This is further emphasized by the lectin-binding studies reported here, which suggest MUC4β is also fucosylated. The enzyme controlling that reaction, the actual structural change or functional effect, is not yet known, although a reduction in UEA binding with ST6GAL1 knockdown suggests ST6GAL1 itself may be involved.
Limitations of this study include the possible off-target effects of the ST6GAL1 or MUC4β DsiRNA. However, the results from ST6GAL1 and MUC4β knockdown appear to be highly complementary. The pathway by which MUC4β affects PI3K/Akt signaling and proliferation also remains unclear. Whether there are differences in the target proteins for the surface-bound versus secreted forms of these proteins is also not clear but is certainly likely. For example, secreted ST6GAL1 could alter airway luminal immunoglobulin glycosylation status, substantially changing its function (66).
Taken together, these results identify ST6GAL1 as the primary driver for the terminal sialylation of MUC4β N-glycans, and the apical secretion of the glycoprotein, both in vitro and in vivo, predominantly in T2-high asthma. Most importantly, ST6GAL1 and MUC4β orchestrate IL-13–induced goblet cell differentiation, cell cycling, and proliferation upstream of FOXA3, identifying this axis as an epithelial target for asthma.
Methods
Subjects.
Participants were 18–65 years old and enrolled in the Severe Asthma Research Program (SARP) and the Electrophilic Fatty Acid Derivatives and Immune Epithelial Interactions in Severe Asthma studies. Participants were nonsmokers or had a smoking history of less than 5 packs year, with no smoking in last the year. HC had normal lung function, without a history of any acute or chronic respiratory illness. SA was defined using the 2014 European Respiratory Society-American Thoracic Society guidelines (67). Patients with MMA had an FEV1 equal or greater than 75% predicted and were on no ICS or a low/medium dose, with or without a second controller (68). All participants were extensively characterized, including spirometry, FeNO, and blood/sputum eosinophils (68, 69). T2-high inflammation was identified as FeNO levels above 25 ppb (T2/FeNO high), blood eosinophils above 300 eosinophils/μl (T2/Eos. high), or sputum eosinophils above 2% (29, 31, 32, 69). T2-low inflammation was defined as levels below all of these cut-off points.
Bronchoscopy and sample processing.
Bronchoscopy with epithelial brushings was performed following the SARP Manual of Procedures as previously described (68, 70). Induced sputum was processed according to SARP protocol, using a slightly modified method of Fahy et al. (71).
Primary HAEC culture under ALI and DsiRNA transfection.
Bronchoscopically obtained HAECs placed in submerged culture were proliferated and then passaged onto Transwell plates and cultured under ALI for 8 days in the absence and presence of IL-13 (R&D Systems, catalog 213-ILB-025, 10 ng/ml) as previously described (63, 72). Apical supernatants were collected after incubating with fresh medium at 37°C for 1 hour, while cells were harvested for mRNA and protein analysis. Meanwhile, similar to the initial experiments with IL-13, other immune/inflammatory cytokines (R&D Systems) (TNF-α, catalog 210-TA-010; IL-1β, catalog 201-LB-005; IL-33, catalog 3625-IL-010; and TSLP, catalog 1398-TS-010) in different doses were applied to measure ST6GAL1 expression in ALI-cultured HAECs. For IFN-γ (catalog 285-IF-100), the HAECs were pretreated with IL-13 (1 ng/ml) for 6 days and then further stimulated with either IL-13 or different doses of IFN-γ (1, 10, and 100 ng/ml) for 2 days before collecting the cells.
Transfection of ST6GAL1 or MUC4β DsiRNA was performed as previously described with slight modification (73). In general, scrambled (negative control) or ST6GAL1/MUC4β DsiRNA (siST6GAL1/siMUC4β) was mixed with Lipofectamine RNAiMAX (catalog 13778150, Invitrogen) in Opti-MEM I (catalog 31-985-070, Gibco) incubated at room temperature for 20 minutes and added to full media, including 200,000 cells per well. Following gentle mixing, the siRNA-RNAiMAX-cell mixture was added to the Transwell insert (final siRNA concentration 50 nM) and incubated at 37°C in a 5% CO2 incubator. After reaching confluency at 24 hours, IL-13 was added to the lower chamber every 48 hours with each media change. At days 8, apical supernatant was harvested to measure apical ST6GAL1 and MUC4β protein and cells harvested for mRNA/protein analysis.
Microarray mRNA data analysis.
Our published microarray profile from freshly brushed HAECs reported that ST6GAL1 positively correlated with FeNO (25). The previously reported gene expression data (National Center for Biotechnology Information’s GEO database; accession GSE63142) were utilized to better establish the relationship between ST6GAL1 and FeNO and blood eosinophils. Gene expression of ST6GAL1 (probe: A24_P397043) was also compared among HC, MMA, and SA. ST6GAL1 mRNA/protein expression was validated by qRT-PCR and Western blot using freshly isolated HAECs.
qRT-PCR.
mRNA expression was determined by qRT-PCR performed on the ABI prism 7900 sequence detection system (Applied Biosystems). Primers and probes were purchased from Applied Biosystems (Assays on Demand; ST6GAL1: Hs00949382_m1; MUC4β: Hs00366414_m1; CCND1: Hs00765553_m1; MUC5AC: Hs01365616_m1; FOXA3: Hs00270130_m1). The mRNA levels were determined relative to β glucuronidase (GUSB: Hs99999908_m1) using the ΔCt method.
SDS-PAGE and Western blotting.
The expression of ST6GAL1 (1:1000, catalog AF5924, R&D Systems), MUC4β (1:1000, catalog NBP1-52193, Novus), CCND1 (1:500, catalog ab134175, Abcam), phosphorylated Akt (1:1000, catalog 9271S, Cell Signaling) and total Akt (1:1000, catalog 9272S, Cell Signaling), and FOXA3 (1:100, catalog sc-5361, Santa Cruz) was measured by Western blotting on 4%–12% SDS-PAGE (Invitrogen). GAPDH (1:1000, catalog NB 300-320, Novus) or β-Actin (1:2000, catalog A5441, MilliporeSigma) served as a loading control. Amersham imager 600 (GE Healthcare Life Sciences) with SuperSignal West Femto Maximum Sensitivity Substrate (catalog 34096, Thermo Fisher) was utilized to develop the blots. Sputum supernatant ST6GAL1 or MUC4β levels were normalized by loading 40 μg of total protein per sample. As no loading control was available for sputum supernatants, a single HC sputum sample was run on all the gels, and ST6GAL1 or MUC4β levels were indexed to it. The apical supernatant ST6GAL1 or MUC4β was indexed using the loading control from whole cell lysates.
Glycosidase digestion reactions.
PNGase F (catalog P0704S), neuraminidase (catalog P0720S), and Endo H (catalog P07025) were purchased from New England Biolabs. Glycosidase digestion reactions were performed on sputum supernatant, ALI-cultured HAECs lysates, and apical supernatant based on the products’ protocol. The samples were denatured, digested by glycosidases, and resolved on 4%–12% SDS-PAGE and blotted with anti-MUC4β Ab. For control reactions buffer was substituted for the enzyme. The untreated and enzyme-treated samples were split from the same sample, so no loading control was needed for the supernatant or apical supernatants.
Pulse-chase protein labeling.
Following 8 days of ALI culture, cells in medium (with or without IL-13) lacking methionine (Met) or cysteine (Cys), were then metabolically pulsed labeled for 30 minutes in 0.5 ml of the same medium containing [35S] Met/Cys (catalog NET027X250UC, Perkin Elmer); then chased for 0, 3, and 24 hours with nonradioactive medium; and washed. Apical supernatants and whole cell lysates were harvested for immunoprecipitation using anti-MUC4β Ab (Novus). SDS-PAGE was performed, proteins were transferred to nitrocellulose, and radiolabeled bands were identified with a Bio-Rad Imager and then blotted with anti-MUC4β Ab. Radiolabeling was essentially carried out as previously described (74).
Lectin-binding assays.
Lectin-binding assays were performed on detergent extracts from cells grown with and without IL-13 or siST6GAL1 for 8 days. After immunoprecipitation using anti-rat ASGP-2 Ab (provided by Kermit L. Carraway, University of Miami, Coral Gables, Florida, USA), samples were eluted in 1% SDS, diluted with buffer (0.5% Triton X-100 in 10 mM HEPES, pH 7.4, 150 mM NaCl), and incubated with 50 μl of the indicated lectin-agarose beads (WGA, UEA, DSL, SNA, and LEL) by end-over-end mixing at 4°C overnight, washed with buffer, analyzed by SDS-PAGE, and blotted with anti-MUC4β Ab. WGA, DSL, SNA and LEL were from Vector Laboratories, and UEA was from EY Laboratories. Lectin-binding studies were carried out as previously described (33).
Immunofluorescence staining for Ki67 and quantification.
ALI-cultured HAECs were transit transfected with siST6GAL1 or siMUC4β in the absence or presence of IL-13 for 8 days and then fixed on 2% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked by blocking serum, and then incubated with anti-Ki67 Ab (1:1000, catalog ab15580, Abcam), followed by incubation with Alexa Fluor 594 donkey anti-rabbit IgG (1:400, catalog A21207, Invitrogen). DAPI (1:1000) was applied for nuclear staining. No primary Ab control was performed as negative control. The sections were observed under Nikon A1 confocal microscope. To quantify Ki67 in immunofluorescent staining, the percentage of positive Ki67 cells was calculated based on the total numbers of nuclei stained with DAPI.
Semiquantitative MUC5AC ELISA.
MUC5AC protein was measured from apical supernatant in ALI-cultured HAECs using a semiquantitative sandwich ELISA. All results are given in relative arbitrary units/ml (42).
Statistics.
Statistical analysis was performed with JMP SAS software (SAS Institute). Data were analyzed for normality. Most data were not normally distributed and are presented as medians, with 25%–75% interquartile range. Wilcoxon tests identified the overall significant differences among the groups or conditions. When a significant difference was observed, intergroup comparisons were further assessed using Wilcoxon signed rank and a Bonferroni correction for multiple comparisons. For in vitro experiments, nonparametric signed-rank tests compared mRNA or protein levels for paired conditions of ST6GAL1, MUC4β, pAkt, CCND1, MUC5AC, and FOXA3. Pearson correlation was used for normally distributed data, and Spearman for correlations was used for nonparametric data. Quantification of immunoblots was performed using ImageJ software (NIH). P < 0.05 was considered statistically significant. Student’s 2-tailed t test was used.
Study approval.
The study was approved by the individual IRBs located at the University of Wisconsin, the Cleveland Clinic, Wake Forest University, as well as the University of Pittsburgh. All participants provided written informed consent prior to inclusion in the study and were identified by different numbers, not by names.
Author contributions
XZ and SEW designed research. XZ, CLK, RPH, MJ, and HI performed research. EE and JBT performed HAEC culture and sputum processing. BDM, NK, ERB, DAM, NNJ, and FH performed studies with SARP samples and/or microarray data. XZ, MJ, and SEW analyzed the data. XZ and SEW wrote the manuscript. AR and SEW supervised the conception and overall supervision of the project and main revision. All authors agreed to all of the content of the submitted manuscript.
Supplementary Material
Acknowledgments
This study was supported by the NIH (R01 HL069174); the National Heart, Lung, and Blood Institute (R01 HL064937, R01 HL069116, P01 HL103453, R01 HL69167, U01 HL109086, U10 HL109152, R21 AI122071); the National Institute of Allergy and Infectious Diseases (P01 AI106684); and a donation from the Dellenback family. We thank Kermit Carraway for providing ASGP-2 Ab and Callen Wallace, Claudette St. Croix, and Simon C. Watkins at Center for Biologic Imaging for their wonderful technical support on immunofluorescent staining and quantification. This study was partially supported by Nikon A1 (NIH 1S10OD019973-01). We also thank Serpil C. Erzurum for providing samples and John R. Tedrow and Jadranka Milosevic for microarray analysis.
Version 1. 02/07/2019
In-Press Preview
Version 2. 03/07/2019
Electronic publication
Footnotes
Conflict of interest: ERB has served as a paid consultant for AstraZeneca/MedImmune, GSK, Novartis, Regeneron, and Sanofi Genzyme. SEW has been site and study PI on studies sponsored by AstraZeneca, GSK, and Sanofi-Aventis.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(5):e122475. https://doi.org/10.1172/jci.insight.122475.
Contributor Information
Xiuxia Zhou, Email: xiz48@pitt.edu.
Carol L. Kinlough, Email: clk9@pitt.edu.
Rebecca P. Hughey, Email: hughey@dom.pitt.edu.
Mingzhu Jin, Email: jinmz@pitt.edu.
Hideki Inoue, Email: hiyumi2001@gmail.com.
Emily Etling, Email: emilyetling@gmail.com.
Brian D. Modena, Email: modena.brian@gmail.com.
Naftali Kaminski, Email: naftali.kaminski@yale.edu.
Eugene R. Bleecker, Email: erbleecker@email.arizona.edu.
Deborah A. Meyers, Email: dameyers@email.arizona.edu.
Nizar N. Jarjour, Email: nnj@medicine.wisc.edu.
John B. Trudeau, Email: JBT8@pitt.edu.
Fernando Holguin, Email: Fernando.holguin@UCDENVER.EDU.
Anuradha Ray, Email: raya@pitt.edu.
Sally E. Wenzel, Email: swenzel@pitt.edu.
References
- 1.Wawrzyniak P, et al. Regulation of bronchial epithelial barrier integrity by type 2 cytokines and histone deacetylases in asthmatic patients. J Allergy Clin Immunol. 2017;139(1):93–103. doi: 10.1016/j.jaci.2016.03.050. [DOI] [PubMed] [Google Scholar]
- 2.Morcillo EJ, Cortijo J. Mucus and MUC in asthma. Curr Opin Pulm Med. 2006;12(1):1–6. doi: 10.1097/01.mcp.0000198064.27586.37. [DOI] [PubMed] [Google Scholar]
- 3.Fahy JV. Goblet cell and mucin gene abnormalities in asthma. Chest. 2002;122(6 Suppl):320S–326S. doi: 10.1378/chest.122.6_suppl.320s. [DOI] [PubMed] [Google Scholar]
- 4.Dorscheid DR, Wojcik KR, Yule K, White SR. Role of cell surface glycosylation in mediating repair of human airway epithelial cell monolayers. Am J Physiol Lung Cell Mol Physiol. 2001;281(4):L982–L992. doi: 10.1152/ajplung.2001.281.4.L982. [DOI] [PubMed] [Google Scholar]
- 5.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4):43R–56R. doi: 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- 6.Li JH, et al. N-linked glycosylation at Asn152 on CD147 affects protein folding and stability: promoting tumour metastasis in hepatocellular carcinoma. Sci Rep. 2016;6:35210. doi: 10.1038/srep35210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki O, Abe M, Hashimoto Y. Sialylation by β-galactoside α-2,6-sialyltransferase and N-glycans regulate cell adhesion and invasion in human anaplastic large cell lymphoma. Int J Oncol. 2015;46(3):973–980. doi: 10.3892/ijo.2015.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hang Q, Isaji T, Hou S, Im S, Fukuda T, Gu J. Integrin α5 suppresses the phosphorylation of epidermal growth factor receptor and its cellular signaling of cell proliferation via N-glycosylation. J Biol Chem. 2015;290(49):29345–29360. doi: 10.1074/jbc.M115.682229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baycin-Hizal D, et al. Physiologic and pathophysiologic consequences of altered sialylation and glycosylation on ion channel function. Biochem Biophys Res Commun. 2014;453(2):243–253. doi: 10.1016/j.bbrc.2014.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, et al. Interleukin-13-induced MUC5AC is regulated by 15-lipoxygenase 1 pathway in human bronchial epithelial cells. Am J Respir Crit Care Med. 2009;179(9):782–790. doi: 10.1164/rccm.200811-1744OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alevy YG, et al. IL-13-induced airway mucus production is attenuated by MAPK13 inhibition. J Clin Invest. 2012;122(12):4555–4568. doi: 10.1172/JCI64896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra D, Coleman JM, Herazo-Maya JD. MUC5B and pulmonary fibrosis, omalizumab for severe allergic asthma, and interstitial lung abnormalities in smokers with emphysema. Am J Respir Crit Care Med. 2012;185(9):1021–1022. doi: 10.1164/rccm.201110-1846RR. [DOI] [PubMed] [Google Scholar]
- 13.Kraft M, et al. Mycoplasma pneumoniae induces airway epithelial cell expression of MUC5AC in asthma. Eur Respir J. 2008;31(1):43–46. doi: 10.1183/09031936.00103307. [DOI] [PubMed] [Google Scholar]
- 14.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29(20):2893–2904. doi: 10.1038/onc.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia P, et al. Cell membrane-anchored MUC4 promotes tumorigenicity in epithelial carcinomas. Oncotarget. 2017;8(8):14147–14157. doi: 10.18632/oncotarget.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanaoka J, et al. Analysis of MUC4 mucin expression in lung carcinoma cells and its immunogenicity. Cancer. 2001;92(8):2148–2157. doi: 10.1002/1097-0142(20011015)92:8<2148::AID-CNCR1557>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Moniaux N, Nollet S, Porchet N, Degand P, Laine A, Aubert JP. Complete sequence of the human mucin MUC4: a putative cell membrane-associated mucin. Biochem J. 1999;338(Pt 2):325–333. [PMC free article] [PubMed] [Google Scholar]
- 18.Chaturvedi P, Singh AP, Batra SK. Structure, evolution, and biology of the MUC4 mucin. FASEB J. 2008;22(4):966–981. doi: 10.1096/fj.07-9673rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damera G, Xia B, Ancha HR, Sachdev GP. IL-9 modulated MUC4 gene and glycoprotein expression in airway epithelial cells. Biosci Rep. 2006;26(1):55–67. doi: 10.1007/s10540-006-9000-5. [DOI] [PubMed] [Google Scholar]
- 20.Damera G, Xia B, Sachdev GP. IL-4 induced MUC4 enhancement in respiratory epithelial cells in vitro is mediated through JAK-3 selective signaling. Respir Res. 2006;7:39. doi: 10.1186/1465-9921-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedlund M, Ng E, Varki A, Varki NM. alpha 2-6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 2008;68(2):388–394. doi: 10.1158/0008-5472.CAN-07-1340. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan HA, Woloski BM, Hellman M, Jamieson JC. Studies on the effect of inflammation on rat liver and serum sialyltransferase. Evidence that inflammation causes release of Gal beta 1 leads to 4GlcNAc alpha 2 leads to 6 sialyltransferase from liver. J Biol Chem. 1983;258(19):11505–11509. [PubMed] [Google Scholar]
- 23.Paulson JC, Weinstein J, Schauer A. Tissue-specific expression of sialyltransferases. J Biol Chem. 1989;264(19):10931–10934. [PubMed] [Google Scholar]
- 24.Kitagawa H, Paulson JC. Differential expression of five sialyltransferase genes in human tissues. J Biol Chem. 1994;269(27):17872–17878. [PubMed] [Google Scholar]
- 25.Modena BD, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190(12):1363–1372. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chanez P, Godard P. [Analysis of induced sputum in the management of asthma: a valid technique, a service rendered by community pneumonologists] Rev Mal Respir. 2002;19(6):689–691. [PubMed] [Google Scholar]
- 27.Dweik RA, et al. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med. 2011;184(5):602–615. doi: 10.1164/rccm.9120-11ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malinovschi A, Van Muylem A, Michiels S, Michils A. FeNO as a predictor of asthma control improvement after starting inhaled steroid treatment. Nitric Oxide. 2014;40:110–116. doi: 10.1016/j.niox.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Fajt ML, Wenzel SE. Asthma phenotypes and the use of biologic medications in asthma and allergic disease: the next steps toward personalized care. J Allergy Clin Immunol. 2015;135(2):299–310. doi: 10.1016/j.jaci.2014.12.1871. [DOI] [PubMed] [Google Scholar]
- 30.Belda J, Leigh R, Parameswaran K, O’Byrne PM, Sears MR, Hargreave FE. Induced sputum cell counts in healthy adults. Am J Respir Crit Care Med. 2000;161(2 Pt 1):475–478. doi: 10.1164/ajrccm.161.2.9903097. [DOI] [PubMed] [Google Scholar]
- 31.Spanevello A, et al. Induced sputum cellularity. Reference values and distribution in normal volunteers. Am J Respir Crit Care Med. 2000;162(3 Pt 1):1172–1174. doi: 10.1164/ajrccm.162.3.9908057. [DOI] [PubMed] [Google Scholar]
- 32.Szefler SJ, et al. Asthma outcomes: biomarkers. J Allergy Clin Immunol. 2012;129(3 Suppl):S9–23. doi: 10.1016/j.jaci.2011.12.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinlough CL, et al. Core-glycosylated mucin-like repeats from MUC1 are an apical targeting signal. J Biol Chem. 2011;286(45):39072–39081. doi: 10.1074/jbc.M111.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catera M, et al. Identification of novel plasma glycosylation-associated markers of aging. Oncotarget. 2016;7(7):7455–7468. doi: 10.18632/oncotarget.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modena BD, Wenzel SE. Consistency of T2 gene signatures in severe asthma. Key to effective treatments or merely the tip of the iceberg? Am J Respir Crit Care Med. 2017;195(4):411–412. doi: 10.1164/rccm.201609-1854ED. [DOI] [PubMed] [Google Scholar]
- 36.Brightling CE, Gupta S, Gonem S, Siddiqui S. Lung damage and airway remodelling in severe asthma. Clin Exp Allergy. 2012;42(5):638–649. doi: 10.1111/j.1365-2222.2011.03917.x. [DOI] [PubMed] [Google Scholar]
- 37.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 38.Bruno S, Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 1992;25(1):31–40. doi: 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]
- 39.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer Drugs. 2005;16(8):797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q, et al. IL-13 dampens human airway epithelial innate immunity through induction of IL-1 receptor-associated kinase M. J Allergy Clin Immunol. 2012;129(3):825–833.e2. doi: 10.1016/j.jaci.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen G, et al. Foxa3 induces goblet cell metaplasia and inhibits innate antiviral immunity. Am J Respir Crit Care Med. 2014;189(3):301–313. doi: 10.1164/rccm.201306-1181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao J, et al. Preferential generation of 15-HETE-PE induced by IL-13 regulates goblet cell differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol. 2017;57(6):692–701. doi: 10.1165/rcmb.2017-0031OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonser LR, Erle DJ. Airway mucus and asthma: the role of MUC5AC and MUC5B. J Clin Med. 2017;6(12):112. doi: 10.3390/jcm6120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carraway KL, Funes M, Workman HC, Sweeney C. Contribution of membrane mucins to tumor progression through modulation of cellular growth signaling pathways. Curr Top Dev Biol. 2007;78:1–22. doi: 10.1016/S0070-2153(06)78001-2. [DOI] [PubMed] [Google Scholar]
- 45.Ordoñez CL, et al. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am J Respir Crit Care Med. 2001;163(2):517–523. doi: 10.1164/ajrccm.163.2.2004039. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi T, et al. N-Glycosylation affects the stability and barrier function of the MUC16 mucin. J Biol Chem. 2017;292(26):11079–11090. doi: 10.1074/jbc.M116.770123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer BM, et al. Neutrophil elastase increases MUC4 expression in normal human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;284(4):L671–L679. doi: 10.1152/ajplung.00220.2002. [DOI] [PubMed] [Google Scholar]
- 48.Vajaria BN, Patel KR, Begum R, Patel PS. Sialylation: an avenue to target cancer cells. Pathol Oncol Res. 2016;22(3):443–447. doi: 10.1007/s12253-015-0033-6. [DOI] [PubMed] [Google Scholar]
- 49.Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol. 2009;19(5):507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mo D, et al. Sialylation of N-linked glycans mediates apical delivery of endolyn in MDCK cells via a galectin-9-dependent mechanism. Mol Biol Cell. 2012;23(18):3636–3646. doi: 10.1091/mbc.e12-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Partridge EA, et al. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306(5693):120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 52.Ohtsubo K, Takamatsu S, Minowa MT, Yoshida A, Takeuchi M, Marth JD. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123(7):1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 53.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 54.Lillehoj EP, et al. NEU1 sialidase expressed in human airway epithelia regulates epidermal growth factor receptor (EGFR) and MUC1 protein signaling. J Biol Chem. 2012;287(11):8214–8231. doi: 10.1074/jbc.M111.292888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swindall AF, Londoño-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 2013;73(7):2368–2378. doi: 10.1158/0008-5472.CAN-12-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones MB. IgG and leukocytes: Targets of immunomodulatory α2,6 sialic acids. Cell Immunol. 2018;333:58–64. doi: 10.1016/j.cellimm.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, et al. Differential expression of ST6GAL1 in the tumor progression of colorectal cancer. Biochem Biophys Res Commun. 2017;486(4):1090–1096. doi: 10.1016/j.bbrc.2017.03.167. [DOI] [PubMed] [Google Scholar]
- 58.Jung YR, et al. Silencing of ST6Gal I enhances colorectal cancer metastasis by down-regulating KAI1 via exosome-mediated exportation and thereby rescues integrin signaling. Carcinogenesis. 2016;37(11):1089–1097. doi: 10.1093/carcin/bgw091. [DOI] [PubMed] [Google Scholar]
- 59.Park JJ, et al. Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem Pharmacol. 2012;83(7):849–857. doi: 10.1016/j.bcp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Lu J, et al. β-Galactoside α2,6-sialyltranferase 1 promotes transforming growth factor-β-mediated epithelial-mesenchymal transition. J Biol Chem. 2014;289(50):34627–34641. doi: 10.1074/jbc.M114.593392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruijtenberg S, van den Heuvel S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15(2):196–212. doi: 10.1080/15384101.2015.1120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang YC, et al. Glycosyltransferase ST6GAL1 contributes to the regulation of pluripotency in human pluripotent stem cells. Sci Rep. 2015;5:13317. doi: 10.1038/srep13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu HW, et al. Transforming growth factor-beta2 induces bronchial epithelial mucin expression in asthma. Am J Pathol. 2004;165(4):1097–1106. doi: 10.1016/S0002-9440(10)63371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basu SK, Whisler RL, Yates AJ. Effects of lectin activation on sialyltransferase activities in human lymphocytes. Biochemistry. 1986;25(9):2577–2581. doi: 10.1021/bi00357a044. [DOI] [PubMed] [Google Scholar]
- 65.Nasirikenari M, et al. Altered eosinophil profile in mice with ST6Gal-1 deficiency: an additional role for ST6Gal-1 generated by the P1 promoter in regulating allergic inflammation. J Leukoc Biol. 2010;87(3):457–466. doi: 10.1189/jlb.1108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones MB, Oswald DM, Joshi S, Whiteheart SW, Orlando R, Cobb BA. B-cell-independent sialylation of IgG. Proc Natl Acad Sci USA. 2016;113(26):7207–7212. doi: 10.1073/pnas.1523968113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung KF, Wenzel S, European Respiratory Society/American Thoracic Society Severe Asthma International Guidelines Task Force From the authors: International European Respiratory Society/American Thoracic Society guidelines on severe asthma. Eur Respir J. 2014;44(5):1378–1379. doi: 10.1183/09031936.00120714. [DOI] [PubMed] [Google Scholar]
- 68.Moore WC, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405–413. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fajt ML, et al. Prostaglandin D2 pathway upregulation: relation to asthma severity, control, and TH2 inflammation. J Allergy Clin Immunol. 2013;131(6):1504–1512. doi: 10.1016/j.jaci.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie M, et al. IL-27 and type 2 immunity in asthmatic patients: association with severity, CXCL9, and signal transducer and activator of transcription signaling. J Allergy Clin Immunol. 2015;135(2):386–394. doi: 10.1016/j.jaci.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fahy JV, Liu J, Wong H, Boushey HA. Cellular and biochemical analysis of induced sputum from asthmatic and from healthy subjects. Am Rev Respir Dis. 1993;147(5):1126–1131. doi: 10.1164/ajrccm/147.5.1126. [DOI] [PubMed] [Google Scholar]
- 72.Trudeau J, Hu H, Chibana K, Chu HW, Westcott JY, Wenzel SE. Selective downregulation of prostaglandin E2-related pathways by the Th2 cytokine IL-13. J Allergy Clin Immunol. 2006;117(6):1446–1454. doi: 10.1016/j.jaci.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 73.Ramachandran S, et al. Efficient delivery of RNA interference oligonucleotides to polarized airway epithelia in vitro. Am J Physiol Lung Cell Mol Physiol. 2013;305(1):L23–L32. doi: 10.1152/ajplung.00426.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanisch FG, Kinlough CL, Staubach S, Hughey RP. MUC1 membrane trafficking: protocols for assessing biosynthetic delivery, endocytosis, recycling, and release through exosomes. Methods Mol Biol. 2012;842:123–140. doi: 10.1007/978-1-61779-513-8_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.