Abstract
Multiple myeloma (MM) is an incurable plasma cell malignancy with frequent treatment failures and relapses, suggesting the existence of pathogenic myeloma stem/progenitor populations. However, the identity of MM stem cells remains elusive. We used a murine model of MM with transgenic overexpression of the unfolded protein response sensor X-box binding protein 1 (XBP1s) in the B cell compartment to define MM stem cells. We herein report that a post–germinal center, pre–plasma cell population significantly expands as MM develops. This population has the following characteristics: (a) cell surface phenotype of B220+CD19+IgM–IgD–CD138–CD80+sIgG–AA4.1+FSChi; (b) high expression levels of Pax5 and Bcl6 with intermediate levels of Blimp1 and XBP1s; (c) increased expression of aldehyde dehydrogenase, Notch1, and c-Kit; and (d) ability to efficiently reconstitute antibody-producing capacity in B cell–deficient mice in vivo. We thus have defined a plasma cell progenitor population that resembles myeloma stem cells in mice. These results provide potentially novel insights into MM stem cell biology and may contribute to the development of novel stem cell–targeted therapies for the eradication of MM.
Keywords: Hematology, Immunology
Keywords: B cells, Immunoglobulins, Mouse stem cells
A post–germinal center, pre–plasma cell population significantly expands as multiple myeloma develops in a murine model with transgenic overexpression of the unfolded protein response sensor XBP1s in B cells.
Introduction
Multiple myeloma (MM), an incurable blood malignancy that occurs in 20,000 new patients and causes 10,000 deaths yearly in the US alone, is characterized by extensive proliferation of plasma cells (PCs) in the bone marrow (BM), leading to hematopoietic failure, bone lytic lesions, and kidney damage (1). Though various treatments for MM have been developed, a high relapse rate in patients has led to speculation that a myeloma cancer stem cell (CSC) (2–4) induces recurrence of the disease, a concept established in a number of other cancer types (5–8). Therapeutic resistance has been attributed to a mechanism known as cell adhesion–mediated drug resistance that requires complex cellular stroma signaling, which is used by normal stem/progenitor populations and established in many stroma-mediated diseases (9–12). In many studies of clonal myeloma cells, circulating B cell populations in myeloma patients express surface monoclonal protein and harbor chromosomal abnormalities similar to PCs (13–17). Circulating B cells also exhibit extensive somatic hypermutation without intraclonal heterogeneity, consistent with a post–germinal center memory B cell population (18–20). Furthermore, these B cells lack the plasma surface antigen CD138/syndecan-1 and could serially engraft NOD/SCID mice in transplantation studies (21, 22). In vitro clonogenic assays further established the memory-like CD138– B cell population as a myelomagenic population (21, 23, 24). Although these studies suggest that a clonal population in the B cell lineage induces myelomagenesis, the exact phenotype of the MM CSC population is currently unknown (2).
To secrete high levels of antibodies, MM cells use the unfolded protein response (UPR), a highly conserved protein quality control system allowing cells to resist death induced by proteotoxic stress in the secretory pathway. UPR is initiated as a result of endoplasmic reticulum (ER) stress and consists of 3 pathways: activating transcription factor 6 (ATF6) (25), dsRNA-activated protein kinase-like ER kinase (PERK) (26), and IRE1 (27), resulting in generation of the unconventional spliced form of X-box binding protein 1 (XBP1s) (28, 29). XBP1s is involved in the differentiation of B cells into PCs and in the stabilization and secretion of antibodies from PCs (30–32). Ploegh and colleagues also uncovered a number of unique functions of XBP1s that do not seem to associate with UPR, including promoting IgM synthesis and secretion, but are indispensable for degradation of glycoproteins in primary B cells (33), for BCR signaling, for expression of IRF4 and Blimp1 transcription factors, and for homing of plasmablasts to BM as a result of the regulating response to CXCL2 (34). Clinically, the expression of XBP1s varies across the stages of myeloma and thus provides a potential biomarker for diagnosing disease progression from premalignant to monoclonal gammopathy of undetermined significance (MGUS) to MM (35, 36). Moreover, increased expression of XBP1s in the BM microenvironment has also been shown to promote myeloma cell growth (37). A murine model of MM using Tg expression of XBP1s in B cells (XBP1s-Tg) demonstrated critical roles of XBP1s in myelomagenesis by inducing PC proliferation, lytic lesions in bone, and increased monoclonal antibody accumulation in the serum and kidney, which promotes kidney dysfunction (38). This model closely resembles the clinical development of MM during the transition from premalignant MGUS to MM. However, no studies have examined the CSC biology of MM in this murine model.
In this study, we demonstrate an age-dependent increase in B cell lymphopoiesis in the XBP1s-Tg mouse that correlates with myeloma progression. We also identified a B cell population that increases with aging and disease progression in the mice and exhibits phenotypes of a stem/progenitor-like population. This population displays surface markers associated with poor survival and enhanced disease progression in MM patients. More importantly, we demonstrated that this MM stem cell–like population was able to differentiate into PCs in vitro and converted to antibody-generating cells in vivo in a manner that is more efficient than PC conversion into antibody-generating cells.
Results
Tg expression of XBP1s in B cells promotes lymphoproliferative disease.
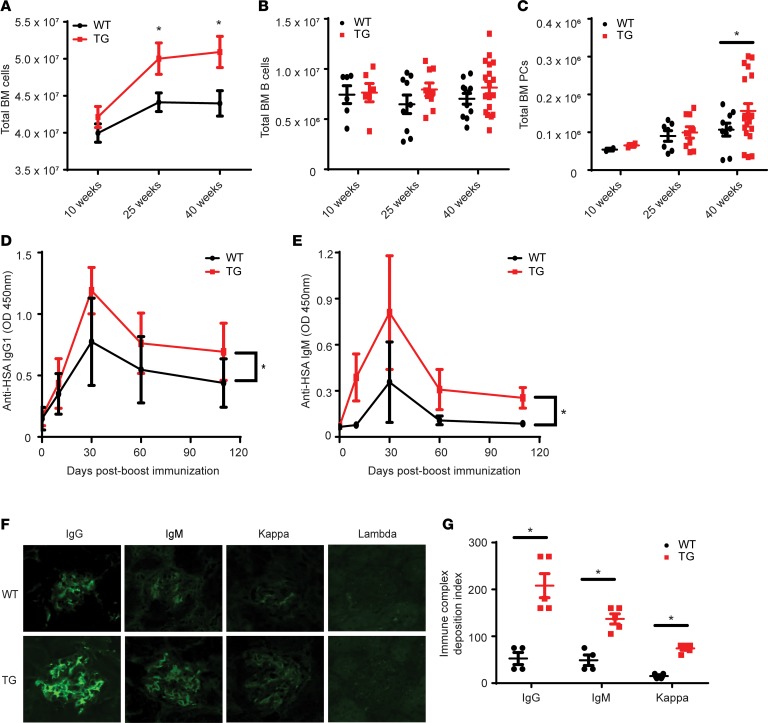
It was previously reported that development of B cells in the BM and spleen was not immediately altered by Tg expression of XBP1s (38). However, detailed analysis of the B cell compartment was not performed. To this end, we characterized the hematopoietic system of Tg mice kinetically. We found an age-dependent increase in total BM cell number in Tg mice (Figure 1A) although the B cell lineage (B220+CD19+) did not expand significantly during the studied time frame (Figure 1B). However, at 40 weeks, a significant increase in BM PCs (B220–CD138+) in Tg mice was seen, suggesting that XBP1s overexpression induces a slow, collective accumulation of PCs (Figure 1C). We also examined for mature BM B cells (B220+CD19+IgM+IgD+) and splenic total cells, B cells, and PCs, finding no significant difference in these populations between WT and Tg mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124698DS1). A complete blood count (CBC) and differential on the peripheral blood (PB) revealed a higher frequency of lymphocytes with a corresponding decrease in granulocytes and monocytes in Tg mice compared with WT at 40 weeks of age (data not shown). Thus XBP1s-Tg mice developed a mild lymphoproliferative disease at the advanced age, which corresponds with the onset of myeloma as reported (38).
Figure 1. Hematopoietic and Ig analysis of the XBP1s-Tg mouse during aging.
(A) Total BM nucleated cell count in WT and Tg mice at indicated ages (n = 8). WT (black line) versus Tg (red line). (B) Total BM B cell count (n = 8). WT versus Tg. (C) Total BM PC (B220–CD138+) count defined by flow cytometry (n = 8). WT versus Tg. (D and E) WT (black line) or Tg (red line) mice were boosted (tertiary, 52 weeks old) with HSA, and the serum HSA-specific IgG1 (D) and IgM (E) were examined (n = 8). One-way ANOVA was used to calculate significance. *P < 0.05. (F) Immunofluorescence for glomerular deposition of IgG, IgM, κ chains, and λ chains in the kidneys of WT and Tg mice. (G) Immune complex deposition index of Ig in kidneys of WT and Tg mice. n = 3. In A, C, and G, *P < 0.05 was calculated using Student’s t test and error bars denote ± SEM.
Constitutive expression of XBP1s in B cells leads to increased antibody production.
To test whether T cell–dependent responses were altered in XBP1s-Tg mice, we immunized WT and Tg mice with human serum albumin (HSA) absorbed on alum. There were only slight differences in Ig levels in the sera of immunized WT and Tg mice even upon primary (3 weeks after) and secondary (12 weeks after) immunizations. However, a tertiary boost 6 months after the secondary immunization led to significantly more serum IgG1 (Figure 1D) and IgM (Figure 1E) in Tg mice than in WT mice. Additionally, consistent with the development of MM, we found that Tg but not WT mice over 40 weeks of age had significant deposition of IgG, IgM, and κ chain in the glomeruli (Figure 1, F and G).
A post–germinal center, pre–plasma B cell population increases with myeloma disease progression.
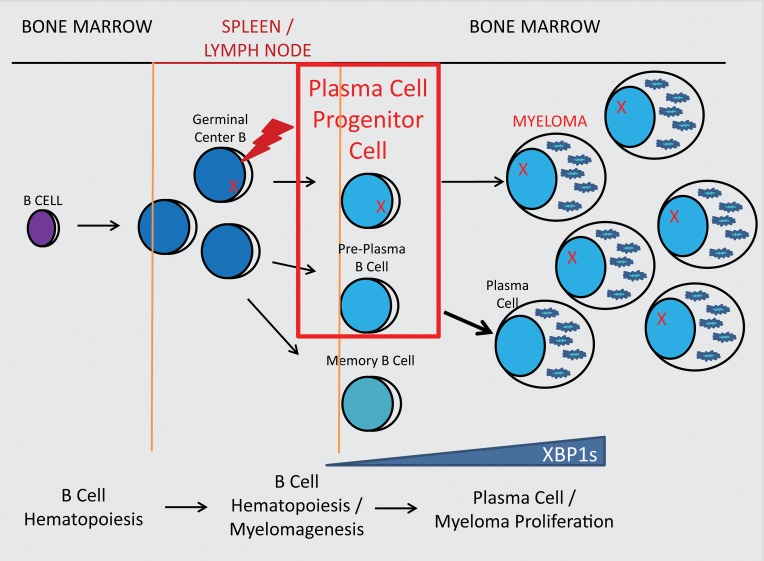
Given the clinical inability to eradicate MM, multiple studies have suggested that a clonal population derived from the B cell lineage survives therapy and drives disease relapse (13, 14, 19, 24). This population most likely arises from post–germinal center, class-switched B cells that are CD19+B220+IgM–IgD–. Furthermore, IgM–IgD– B cells have been shown to express CD80 (39, 40), particularly on transitional pre-plasmablasts in the BM (41). Finally, as a pre-PC, the PC progenitors likely would not express PC surface antigen CD138/syndecan-1. We thus reasoned that the multiple myeloma plasma progenitors (MMPPs) in mice reside within the cellular compartment with the cell surface phenotype of B220+CD19+IgM–IgD–CD138–CD80+.
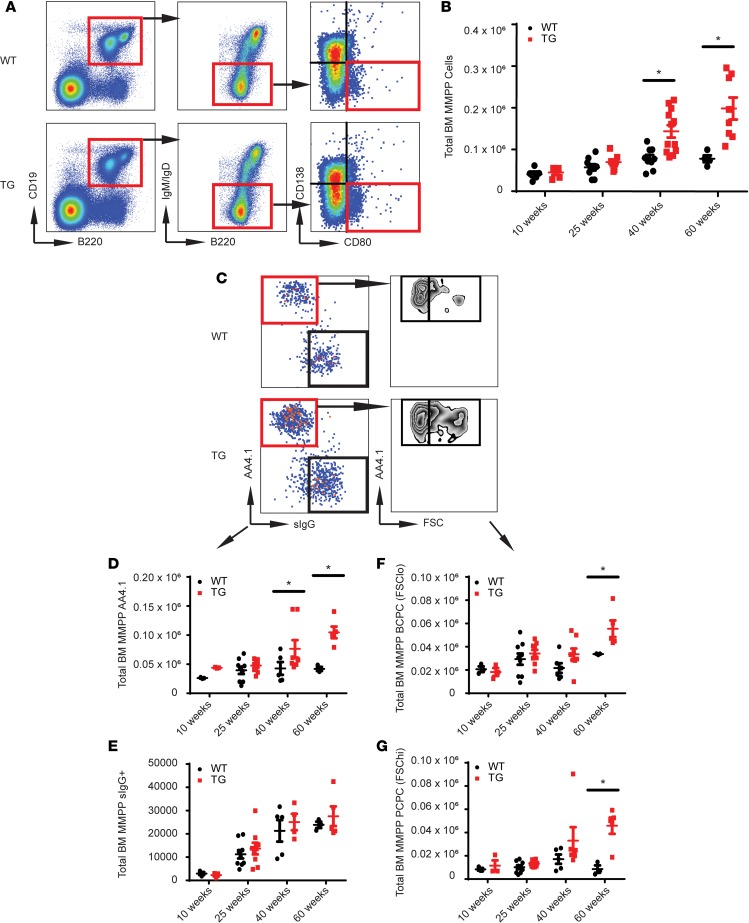
We found that the MMPP population was significantly increased in Tg mice by 40 and 60 weeks of age, whereas the stabilizing trend in WT mice suggests possible homeostasis of memory B cell and PC populations over time in nonpathological settings (Figure 2, A and B). However, elevated total numbers and flow scatter heterogeneity prompted us to further characterize this population using surface IgG (sIg), which identifies memory-like B cells, and AA4.1, which identifies early B/pro-B cells. Flow cytometry allowed us to segregate MMPP cells into 2 unique populations, AA4.1+sIgG– and AA4.1–sIgG+ (Figure 2C). At 40 and 60 weeks the MMPP AA4.1+ population in the Tg mice was significantly increased, while the memory-like MMPP IgG+ population was only slightly increased (Figure 2, D and E). Because immature, developing B cells are smaller in size, whereas mature BM B cells and post–germinal center B cells are larger, we used FSC to segregate the low- and high-scatter cell populations of the AA4.1+sIgG– population. Both the FSClo and FSChi fractions of the B220+CD19+IgM–IgD–CD138–CD80+sIgG–AA4.1+ population were significantly increased in Tg mice compared with WT mice (Figure 2, F and G). Chevrier et al. recently detected AA4.1/CD93 expression on BM B cells that was downregulated in the spleen until the development of pre-PC and PC phenotypes (42). Overall, these results suggested that the Tg overexpression of XBP1s in the B cell lineage promoted survival or proliferation of both early B cells and a post–germinal center, pre-PC population that might contain MM CSCs. We named the B220+CD19+IgM–IgD–CD138–CD80+sIgG–AA4.1+FSChi population the plasma cell progenitor cells (PCPCs) and the B220+CD19+IgM–IgD–CD138–CD80+sIgG–AA4.1+FSClo population the B cell progenitor cells (BCPCs) because the latter phenotypically resembles an early developing B cell. Uniquely, we did not detect these populations accumulating in the spleens of either the WT or Tg mice, confirming these phenotypes defined a BM population (Supplemental Figure 2).
Figure 2. A post–germinal center B cell increases in Tg mice with age.
(A) Representative flow diagram depicting the PC stem/progenitor population phenotype in the BM of WT and Tg mice (n = 6). (B) Total number of MMPP cells in the BM of WT and Tg mice over advancing ages (n = 6). (C) Representative flow plot using AA4.1, sIg, and forward scatter (FSC) to identify B cell populations in the BM (n = 6). (D) Total number of AA4.1+ MMPP cells (n = 6) in WT versus Tg mice. (E) Total number of surface IgG+ MMPP cells (n = 6) in WT versus Tg mice. (F) Total number of AA4.1+FSClo PCPCs (n = 6). (G) Total number of AA4.1+FSChi PCPCs (n z= 6). In all panels, Student’s t test was used to calculate *P < 0.05, and error bars denote ± SEM.
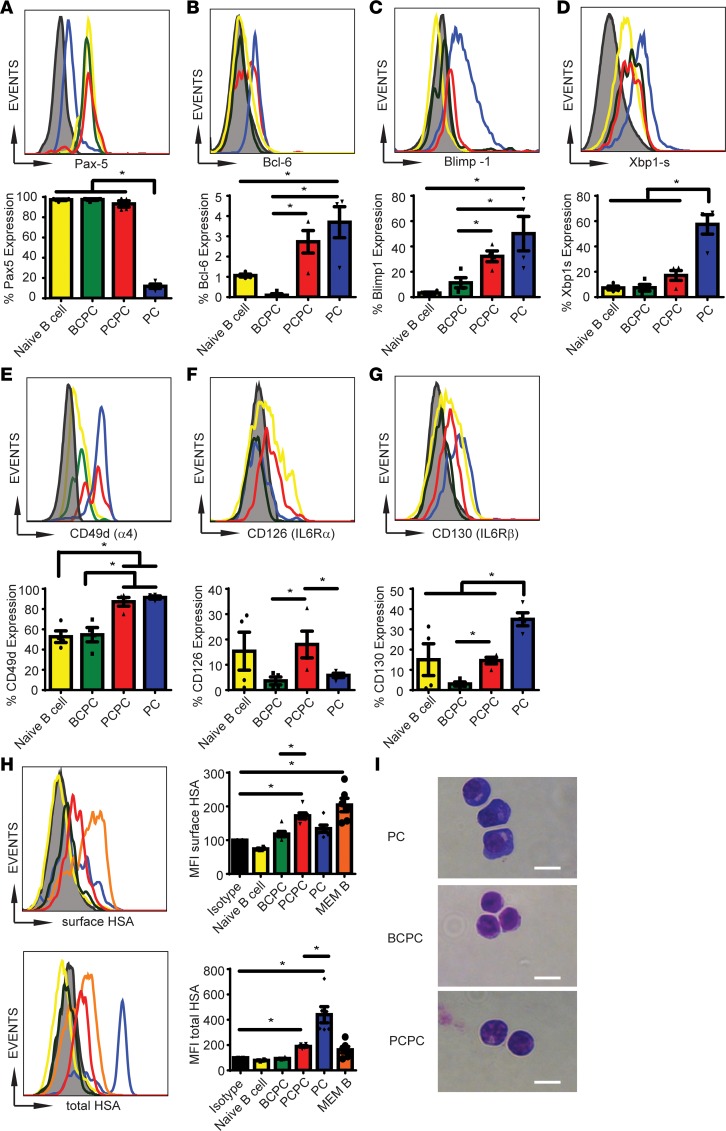
We next performed a transcription factor analysis on the BCPCs, PCPCs, and PCs to better characterize each population (Figure 3). We used Tg mice for this analysis because of expansion of the B cell compartment, though importantly, all the studied populations were present in normal mice. Among them, Pax5 is an important factor for maintaining B cell phenotypes and distinguishes the B cell lineage from the PC population by inhibiting PC formation (43). Bcl6 is expressed in post–germinal center cells, such as memory B cells, and has been shown to induce self-renewal in B cells (44). Blimp1 is required for PC generation and induces activation of endogenous XBP1s, which maintains PC function (30, 32, 45). Naive B cells (B220+CD19+IgM+CD138–AA4.1+), BCPCs, and PCPCs had significantly higher expression of Pax5 while the PC population expressed minimal Pax5 (Figure 3A). Expression of Bcl6 was negligible in BCPCs (0.2%), with minimal expression in naive B cells (1%), and we found more significant expression in both PCPCs and PCs (Figure 3B). We detected high levels of Blimp1 in the PCs, little expression in the BCPCs and naive B cells, and intermediate levels in the PCPCs when compared with the BCPCs, suggesting that this population may undergo transition from a B cell to a PC phenotype (Figure 3C). However, XBP1s expression was significantly lower among all populations compared with the PCs (Figure 3D).
Figure 3. The PCPC population demonstrates a late-B, early PC-like cell phenotype.
Naive B cells (yellow line), BCPCs (green line), PCPCs (red line), and PCs (blue line) were examined for total percentage of the expression of Pax5 (A), Bcl6 (B), Blimp1 (C), Xbp1s (D), CD49d (E), CD126 (F), and CD130 (G). Black line indicates isotype control. (A–G) Top panels are representative flow histograms, and bottom panels are bar graphs denoting percentage of expression in each population (n = 4, *P < 0.05). (H) Mice were immunized with HSA. Various cellular populations in the BM were examined 10 weeks later for binding to HSA-FITC to the cell surface (top row) or total cells (surface and intracellular, bottom row). Orange bar represents memory B cells (B220+CD19+IgM–IgD–sIgG+). Bar graphs denote MFI expression (n = 6, *P < 0.05). (I) Hema 3 stain of sorted cell populations, followed by microscopic examination. Scale bar: 10 μm. Error bars denote ± SEM. In A–H, 1-way ANOVA and Tukey’s multiple-comparisons test was used to calculate *P < 0.05.
Because the MM CSC is thought to require niche or stromal interactions for survival and evasion of therapy, we next analyzed the expression of molecules involved in cell adhesion and interaction with the BM microenvironment, including CD49d (α4 integrin) (9, 10, 12, 46, 47). We found that both PCPCs and PCs expressed a high level of CD49d (Figure 3E). We also found that PCPCs had increased expression of CD126 (IL-6Rα) and CD130 (gp130/IL-6Rβ) (Figure 3, F and G), both of which are regulated by the BM niche and have been implicated in MM pathogenesis (48, 49). No detectable expression of CD126 or CD130 in the BCPCs was observed, while PCs had increased expression of CD130 only. Naive B cells expressed high levels of CD126 and CD130, which is consistent with a population targeted for maturation and mobilization to the spleen and lymph nodes for antigen exposure.
We next examined these populations in an antigen-specific system after HSA immunization (Figure 3H), using B220+CD19+IgM–IgD–sIgG+ memory B cells as a positive control. Mice were immunized with HSA as shown in Figure 1, D and E, and BM cells were examined for each population for cell surface binding to HSA. Indeed, HSA-specific memory B cells captured high levels of HSA-FITC. The PC population exhibited high total but little surface binding to HSA, indicating downregulated sIg, as expected. The PCPCs had a significant increase in surface binding to HSA-FITC while no HSA surface reactivity was seen with BCPCs. Naive B cells did not express either surface or intracellular HSA, which is consistent with a naive population absent of prior antigen exposure. This experiment definitely demonstrated that the PCPC population expresses B cell receptor and may represent an intermediate population residing in the lineage between memory B cells and PCs.
Morphological characterization of each population revealed a unique appearance of the PCPC (Figure 3I). Although BCPCs were smaller and had a large nucleus/cytoplasm ratio, the PCs were larger cells with large basophilic cytoplasm with pale zones that would indicate the abundance of the Golgi apparatus and ER organelles required for antibody secretion. The PCPCs were intermediate in size between the BCPCs and PCs and had increasingly larger cytoplasmic regions with developing pale zones, resembling a B cell transitioning to a PC phenotype.
PCPCs exhibit stem/progenitor-like phenotypes.
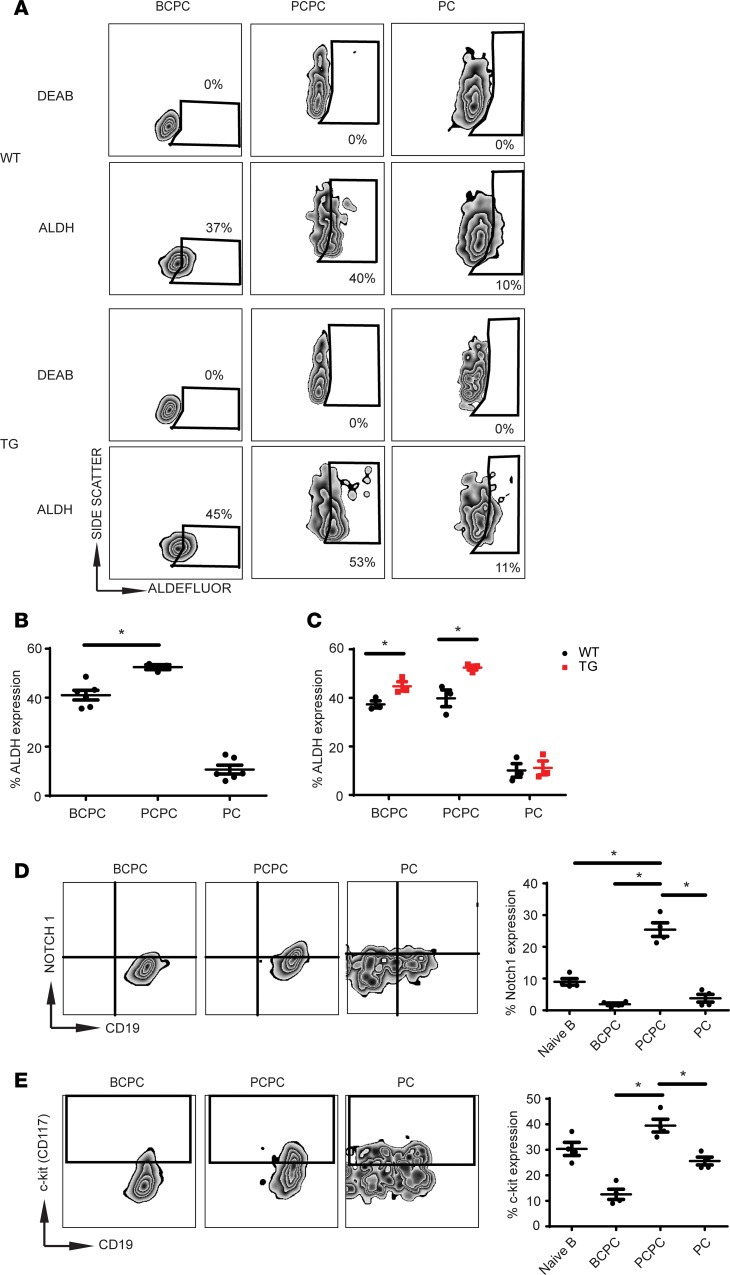
Recent reports have demonstrated high expression of aldehyde dehydrogenase (ALDH) in primitive hematopoietic stem cells (50). In addition, ALDH is upregulated in CSCs isolated from breast, liver, brain, pancreatic, and hematopoietic cancers (51–54). Additional studies have suggested that MM CSCs might exhibit ALDH activity (24, 55, 56). Using the ALDH enzymatic inhibitor DEAB to validate our staining protocol, we found that the PCPC population expressed significantly higher levels of ALDH compared with BCPCs (Figure 4, A and B). In contrast, the PC population expressed negligible levels of ALDH. The significantly higher levels of ALDH in Tg cells compared with WT suggest a possible role for XBP1s in driving the PCPC phenotype (Figure 4C).
Figure 4. The PCPC population exhibits a “stem cell–like” phenotype.
(A) Representative flow dot plots of various cellular populations from the BM of WT and Tg mice. ALDH expression was determined by Aldefluor staining. The ALDH inhibitor DEAB was used to set the gate for negative staining (n = 3). (B) Total percentage of ALDH expression in each population (n = 6). (C) Total percentage of ALDH expression of indicated populations from WT and Tg mice (n = 3). (D) Expression of Notch1 determined by intracellular stain. Representative flow plots are depicted in the left and middle. Bar graph (far right) is an average of 4 mice. (E) Expression of c-Kit (CD117) in indicated cell populations. Representative flow plots are depicted in the left and middle. Bar graph (far right) is the average of 4 mice. In all panels, error bars denote ± SEM. Student’s t test was used to calculate significance, and *P < 0.05.
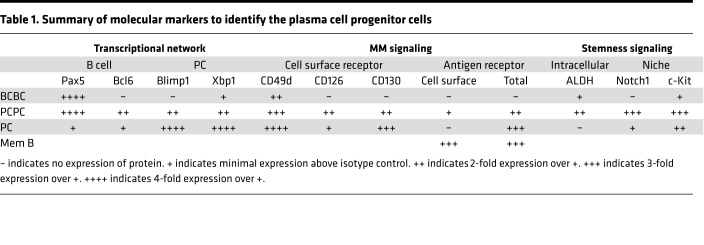
We next examined whether the PCPCs expressed Notch1 and c-Kit, both of which are important receptors for maintaining stem/progenitor cell populations at the BM niche and are expressed in myeloma clonal cell populations (55). The PCPCs exhibited a marked increase in Notch1 and c-Kit expression compared with the BCPCs or PCs (Figure 4, D and E, and Table 1), consistent with the stem-like qualities of this population. To determine whether the PCPC population represents a stem-like/progenitor population, we used a classical BrdU assay to determine the proliferative/cell cycle capacity of the populations (Supplemental Figure 3). Although total BrdU was unchanged in the BM and spleens of WT and Tg mice over 14 days (Supplemental Figure 3A), as was seen in total B cells (B220+CD19+) (Supplemental Figure 3B), the PCPCs did exhibit higher levels of BrdU at day 14 compared with naive B cells or the BCPCs but similar in expression of a more static population in PCs (Supplemental Figure 3C). This further suggests that the PCPC population is actively cycling and thus may represent a progenitor-like population.
Table 1. Summary of molecular markers to identify the plasma cell progenitor cells.
PCPC population differentiates into PCs.
We next performed an experiment to compare the ability of naive B cells, PCPCs, and PCs to give rise to antibodies in vivo. Naive B cells (B220+CD19+IgM+CD138–AA4.1–), PCPCs, and PCs were sorted using the gating strategy described (Figure 5A), with purity confirmed after sorting for each population (Figure 5B). Very little cross-contamination of various subpopulations was seen. Sorted cells were adoptively transferred into B cell–deficient B6.129S2-Ighmtm1Cgm/J (μMT) mice, where the only possible source of antibody in the recipient mice was either transferred cells or their derivatives. The mice were kept in specific pathogen-free conditions without immunizations. As expected, we did not detect any total IgM or IgG in the sera of naive B cell–transplanted mice over 6 months (4-month data are shown). However, significant levels of IgG1 and IgM antibodies were detected in the PCPC-transplanted group. Strikingly, adoptive transfer of the terminal differentiated PC population did not reconstitute Ig-producing capacity in the B cell–deficient mice as much as PCPCs did (Figure 5C). Because the PCPC is clearly not a memory B cell (sIgG+) or PC (B220–CD138+), this study strongly suggests that it is indeed part of a precursor population that is capable of efficiently generating antibody-producing PCs. Finally, we sorted BCPCs, PCPCs, memory B cells, and PCs from XBP1-Tg mice and cultured them in vitro in a PC maintenance condition for 7 days, without antigen stimulation. PCs retained high-scatter properties with expression of CD138+ after 7 days. The memory B cells and the PCPC population began to blast to a size similar to that of the PCs with subsequent upregulation of CD138 expression. The BCPC population did not acquire a PC phenotype. When examining total PCs generated in culture, we found a significant increase in PC generation among the PCPCs compared with the memory B cells (Figure 5D). The PCs in this in vitro culture were blasting in size as expected from a PC differentiation culture (Supplemental Figure 4). Our data from both in vivo and in vitro experiment therefore suggested strongly that PCPCs are precursors of PCs (Figure 6).
Figure 5. The PCPC population differentiates into PCs.
(A–C) Naive B cells (B220+CD19+IgM+CD138–AA4.1–), PCPCs, and PCs (B220–CD138+) were sorted from XBP1-Tg mice and adoptively transferred into μMT mice (n = 4 mice/group). (A) FACS staining profile of nucleated cells before sorting. (B) Characterization of naive, PCPC, and PC populations after sorting but before the adoptive transfer. The percentages of each population in the final sorted cellular products are indicated. (C) Total IgG1 and IgM concentrations in the sera of recipient mice were measured by ELISA 4 months after the adoptive transfer. *P < 0.05 and error bars denote ± SEM. (D) PCs (B220–CD138+), BCPCs, PCPCs, and memory-like B cells (PCPC sIgG+) were sorted to over 90% purity and placed into PC-culturing conditions for 7 days. The total numbers of PCs generated from 300,000 total cells are shown (n = 4). In C and D, 1-way ANOVA with Tukey’s multiple-comparisons test was used to calculate *P < 0.05. All error bars denote ± SEM.
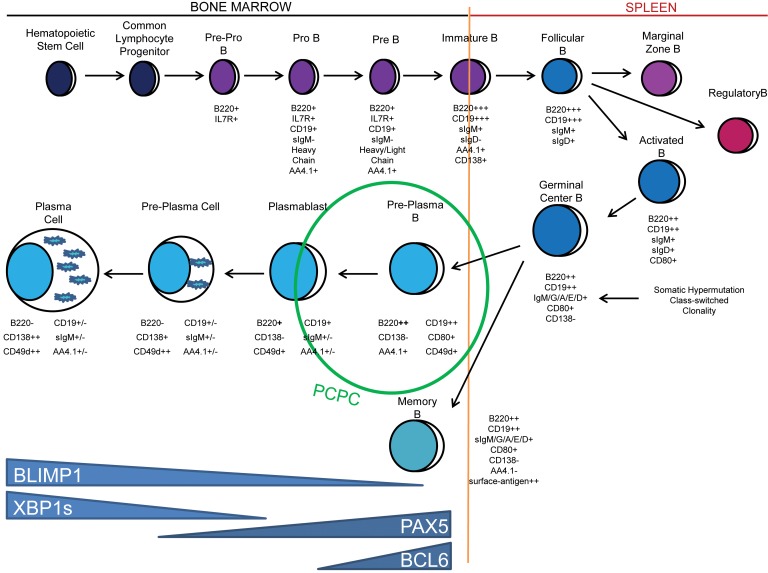
Figure 6. Proposed lineage location and the molecular characteristics of the PCPC population.
The diagram depicts schematically the B cell lineage progression from the BM to the spleen and the return of antigen-experienced, mature B cell and PC populations back to the BM. The developmental location of PCPCs, their phenotype, and their expression pattern of key B cell differentiation factors are illustrated.
Discussion
Despite the advancements in both diagnosis and treatment of MM, it remains incurable, with relapse being the major cause of morbidity and mortality. The concept of an MM CSC has been hypothesized based on the clonal origin of the recurrent disease and progression similar to other clonal tumors (57, 58). However, the inability to identify the progenitor-like population driving MM progression has presented a challenge. In this study, we aimed to identify an MM progenitor–like population by using a murine model of MM that demonstrated transitional and progressive MM disease with similar characteristics to human diseases. We defined the mouse PCPC as B220+CD19+IgM–IgD–CD138–CD80+sIgG–AA4.1+FSChi (summarized in Figure 6). To our knowledge, this is the first study identifying a progenitor population of myeloma cells with the ability to drive antibody production in vivo in a syngeneic system.
XBP1s is upregulated in myeloma cells and is involved in the maintenance of PCs (35, 37). Tg overexpression of XBP1s in B cells induced a lymphoproliferative disease and myeloma with increased serum Ig, kidney Ig depositions, and bone lytic lesions, mimicking the clinical observation in MM patients (38, 59). The significant finding in this study was the accumulation of post–germinal center B cell and PC populations that increased with disease progression, consistent with studies that have established the impact of XBP1s expression in MM pathogenesis (35, 60, 61).
The late–B cell and the pre-PC population is traditionally assumed to contain the elusive MM stem cell population (21). The stem cell–like characteristics of ALDH activity along with longer retention of BrdU and the ability to reconstitute antibody production in B cell–deficient mice also suggest the PCPCs we defined in this study are capable of long-term maintenance in vivo (54, 55, 62). Recent evidence has supported that the BM microenvironment protects MM by inducing oncogene activity, upregulating adhesion properties or decreasing the cell cycle, thus promoting survival and self-renewal (63, 64). Additionally, the loss of immunosurveillance mechanisms has been proposed to contribute to the persistence of CSCs (65, 66). The various stem cell–like qualities and transcriptional profile we have uncovered support the PCPC as a potential MM CSC in the XBP1s-Tg model. Indeed, a link between Bcl6 and the subsequent survival of CSCs is becoming more established (67, 68). Bcl6 transcriptional signaling pathways have also been implicated in lymphomagenesis (69, 70). Further, our study is consistent with a study suggesting that XBP1s-negative pre-plasmablast B cells are responsible for mediating clinical therapeutic resistance (71).
Importantly, we have demonstrated that the PCPC population can be differentiated into PCs in vitro and antibody-producing cells in vivo, a finding that is critical to establishing this population as a PC precursor. Though in vitro differentiation occurred, it is unclear if these PCs were rapidly secreting antibodies. Unfortunately, in vitro culturing of PCs has not been well documented, and most studies of in vitro PC differentiation use human cells cocultured with stromal cell lines for additional growth factor and cytokine support (72–74). Future studies may identify specific factors of PC differentiation and maintenance in vitro that allow them to develop into PCs from germinal center B cells to memory B cells or plasmablasts. Nevertheless, the adoptive transfer of PCPCs into mice lacking B cells and the subsequent generation of antibodies definitively showed the progenitor status of the PCPCs. The ability to detect secreted antibody in mice after transferring only 5 × 104 total cells suggests proliferation of the PCPC and plasmablast populations that may contribute to progression and maintenance of MM, although we were not able to track these cells in the animal because of the low number of cells being transferred. Nonetheless, the ability of PCPCs to reconstitute antibody pools in B cell–deficient mice was impressive compared with studies of B cells in which adoptive transfer typically used 10- to 100-fold greater numbers of B cells (75–77).
This study supports the role of stem/progenitor cell signaling in MM and the role of XBP1s in development and maintenance of MM. Although some aspects of the PCPC phenotype have been demonstrated in other studies, the lack of more specific markers on these populations prevents efficient targeting of these cells (15, 21). Additionally, these studies used human cells, requiring xenogenic transplantation and potentially limiting the ability to determine “stemness” of a clonal population in generating and progressing myeloma in vivo (22). Murine models of myeloma allow for studies of myeloma progenitor cells in vivo with an intact host immune system. Using the XBP1s-Tg model, we have previously reported that genetic deletion of gp96, a critical ER chaperone mediating the UPR in PCs, can attenuate MM, demonstrating the usefulness of a syngeneic model where genetic manipulation can be done to alter specific pathways and study disease progression (59). Though one caveat of the XBP1s-Tg model is the length of time required to accumulate the PCPC population in mice, the ability to transition through all stages of MM as it is observed clinically provides us a unique capability to study accrual of potential stem/progenitor populations of MM. Newer or more improved murine models of myeloma need to be developed that accelerate studies of the stem-like population to induce MM tumorigenesis (78). Recent adoptive transfer models have also shown promise in generating repeatable syngeneic transplantation results that promote MM (79, 80). Additionally, the phenotype identified for the PCPC population needs to be translated into the human setting and phenotyped in patients specifically with relapsing/refractory myeloma, which may exhibit larger numbers of this population. Future experiments involving genetic profiling or proteomics may introduce a novel marker that might specifically identify the PCPC/myeloma progenitor to other B cells or plasma cells, thus providing a means for specific targeting through pharmacological or immunotherapeutic approaches.
Taken together, we have identified a potentially novel progenitor population in a murine model of MM that is phenotypically and transcriptionally different from naive B cells, memory B cells, and PCs but has the robust ability to differentiate into PCs in vitro and in vivo. Although limited to murine studies and a mostly cellular approach, our study has provided an important step forward in identifying and validating the “stem” cell population in MM, which may lead to the potential development of novel stem cell–targeted therapies for the eradication of MM.
Methods
Mice.
XBP1s-Tg mice were obtained from Ronald DePinho (MD Anderson Cancer Center, Houston, Texas, USA). Blood was collected by tail bleed, and differential counts of the CBC were performed on an abc blood counter (scil Vet). B6.129S2-Ighmtm1Cgm/J (μMT) mice were obtained from The Jackson Laboratory (catalog 002288). Mice were bred and maintained in a specific pathogen-free facility, in accordance with the established guidelines and a protocol approved by the Medical University of South Carolina Institutional Animal Care and Use Committee.
Reagents.
Antibodies used for flow cytometry were obtained from eBioscience, BD Biosciences, and BioLegend (Supplemental Table 1). All other chemicals were obtained from Sigma-Aldrich and Thermo Fisher Scientific. IGF-1, IL-6, IL-11, and IL-21 were purchased from PeproTech. IFN-α was provided by Beichu Guo (Medical University of South Carolina, Charleston, South Carolina, USA).
Immunization and ELISA.
We immunized 12-week-old mice with 100 μg HSA coabsorbed on 2 mg alum per mouse, with primary and secondary boosts at 3 and 16 weeks, respectively. Tertiary immunization was performed 6 months after the secondary one using 100 μg HSA/mouse coabsorbed with alum. PB was drawn by tail bleed. An ELISA plate was coated with 10 μg/ml HSA, and a 1:5,000 dilution of sera was used (81). Ig levels were determined using a kit from SouthernBiotech.
Immunofluorescence and immune complex deposition.
We fixed 5-μm-thick cryosections of kidneys with cold acetone for 10 minutes and then blocked them with normal rabbit serum for 2 hours at room temperature. After washing, the sections were stained with goat anti–mouse IgG, IgM, κ, and λ antibodies (SouthernBiotech) for 30 minutes, followed by staining with FITC-conjugated anti–goat IgG (Abcam) for 30 minutes at room temperature. Images of sections were taken under a fluorescent microscope (×200) (Zeiss) and analyzed by AxioVision 4.4 software (Carl Zeiss MicroImaging). The percentage of glomerulus involvement was determined after examining at least 60 nonoverlapping glomeruli per kidney section, as described previously (82, 83). The immune complex (IC) deposition was quantitatively graded based on an intensity scale from 1+ to 3+. The IC deposition index was determined by multiplying the percentage of involvement with the average glomerular IC deposition grade.
Flow cytometry protocols.
Transcription factors were stained using BD Transcription Factor Analysis Kit according to the manufacturer’s protocol (BD Biosciences).
For identification of HSA-specific B cells, HSA was conjugated to FITC using FluoroTag FITC Conjugation Kit (Sigma-Aldrich). HSA-specific B cells were detected by staining with HSA-FITC. Cells with intracellular HSA-specific antibody were identified by first staining the cell surface markers, followed by fixation and permeabilization with BD Transcription Factor Analysis Kit, then by staining with HSA-FITC.
Cell sorting.
After staining BM cells with various antibodies, BCPC (B220+CD19+IgM–IgD–CD138–CD80+AA4.1+sIgG–FSClo), PCPC (B220+CD19+IgM–IgD–CD138–CD80+AA4.1+sIgG–FSChi), and PC (B220–CD138+) populations were sorted using a MoFlow Astrios (Beckman Coulter) or a BD FACSAria IIu cytometer. The purity of the desired populations was routinely confirmed after sorting.
All flow procedures followed manufacturer’s protocols and recommendations. Cell acquisition was performed on an LSR Fortessa (Becton Dickinson) followed by analysis using FlowJo software (Tree Star Inc.).
Cytospin and morphology stain.
Cells were spun onto microscope slides using a Cytospin (Cytospin 4, Thermo Fisher Scientific) and fixed and stained with Fisher HealthCare Protocol Hema 3 (Thermo Fisher Scientific) following the manufacturer’s protocol. Images were taken with a Zeiss AX10 microscope (Zeiss).
PC in vitro culture.
After FACS, memory B cells, PCs, BCPCs, and PCPCs were placed into IMDM supplemented with Glutamax and 10% FBS (Invitrogen). Cultures were supplemented with CD Hybridoma (11 μl/ml, Invitrogen), 1× Lipid Mixture 1 (200×, Sigma-Aldrich), and 1× MEM Amino Acids solution (50×, Sigma-Aldrich). All cells were plated and cultured in PC maintenance conditions as described (74), including IL-6 (50 ng/ml), IL-21 (50 ng/ml) and IFN-α (500 U/ml), without antigen stimulation. After 7 days, cells were analyzed for the presence of CD138+FSChi PCs.
ALDH enzymatic assay.
Cells were cultured and stained for ALDH with Aldefluor following the manufacturer’s protocol (Stem Cell Technologies).
Adoptive transfer.
BM cells from 5 XBP1s-Tg mice were collected in a sterile manner and pooled. Naive B cell (B220+CD19+IgM+CD138–AA4.1–), PCPC (B220+CD19+IgM/IgD–CD138–CD80+AA4.1+FSChi), and PC (B220–CD138+) populations were sorted using a BD Aria IIu cytometer. We transferred 5 × 104 cells from each population into μMT mice via tail vein injection (n = 4/group). Serum was collected from mice via tail bleed at monthly intervals, and the concentrations of Ig were measured by ELISA.
Statistics.
Error bars depict ± SEM. Either an unpaired 2-tailed Student’s t test or 1-way ANOVA with Tukey’s multiple-comparisons test was used for statistical analysis. Each applied statistical test is indicated in figure legends. P values less than 0.05 were considered statistically significant. All statistical testing was done on GraphPad Prism 6. Values are expressed as mean ± SEM unless stated otherwise.
Study approval.
All animal experiments adhered to laws and regulations of the US Department of Agriculture and were performed according to guidelines from the NIH (Guide for the Care and Use of Laboratory Animals, National Academies Press, 2011). Experiments were approved by the Institutional Animal Care and Use Committee of the Medical University of South Carolina.
Author contributions
JK, ZL, and BL conceived ideas. JK, CWF, and BL performed the experiments. All authors contributed to the data analysis and writing of the manuscript.
Supplementary Material
Acknowledgments
The work was supported by grants from the NIH P01CA186866, R01CA188419, and R01AI077283 (to ZL) and R01CA193939 (to BL) and by the Flow Cytometry and Cell Sorting Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313). ZL is the Abney Chair Remembering Sally Abney Rose in Stem Cell Biology & Therapy and was supported by the SmartState Endowed Chair Program of South Carolina.
Version 1. 04/04/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(7):e124698. https://doi.org/10.1172/jci.insight.124698.
Contributor Information
Caroline Wallace, Email: wallacc@musc.edu.
Bei Liu, Email: liube@musc.edu.
Zihai Li, Email: zihai@musc.edu.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Kellner J, Liu B, Kang Y, Li Z. Fact or fiction — identifying the elusive multiple myeloma stem cell. J Hematol Oncol. 2013;6:91. doi: 10.1186/1756-8722-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaccoby S. Two states of myeloma stem cells. Clin Lymphoma Myeloma Leuk. 2018;18(1):38–43. doi: 10.1016/j.clml.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Johnsen HE, et al. The myeloma stem cell concept, revisited: from phenomenology to operational terms. Haematologica. 2016;101(12):1451–1459. doi: 10.3324/haematol.2015.138826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 9.Damiano JS, Cress AE, Hazlehurst LA, Shtil AA, Dalton WS. Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood. 1999;93(5):1658–1667. [PMC free article] [PubMed] [Google Scholar]
- 10.Li ZW, Dalton WS. Tumor microenvironment and drug resistance in hematologic malignancies. Blood Rev. 2006;20(6):333–342. doi: 10.1016/j.blre.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Hazlehurst LA, Damiano JS, Buyuksal I, Pledger WJ, Dalton WS. Adhesion to fibronectin via beta1 integrins regulates p27kip1 levels and contributes to cell adhesion mediated drug resistance (CAM-DR) Oncogene. 2000;19(38):4319–4327. doi: 10.1038/sj.onc.1203782. [DOI] [PubMed] [Google Scholar]
- 12.Noborio-Hatano K, et al. Bortezomib overcomes cell-adhesion-mediated drug resistance through downregulation of VLA-4 expression in multiple myeloma. Oncogene. 2009;28(2):231–242. doi: 10.1038/onc.2008.385. [DOI] [PubMed] [Google Scholar]
- 13.Bakkus MH, Van Riet I, Van Camp B, Thielemans K. Evidence that the clonogenic cell in multiple myeloma originates from a pre-switched but somatically mutated B cell. Br J Haematol. 1994;87(1):68–74. doi: 10.1111/j.1365-2141.1994.tb04872.x. [DOI] [PubMed] [Google Scholar]
- 14.Bergsagel PL, Smith AM, Szczepek A, Mant MJ, Belch AR, Pilarski LM. In multiple myeloma, clonotypic B lymphocytes are detectable among CD19+ peripheral blood cells expressing CD38, CD56, and monotypic Ig light chain. Blood. 1995;85(2):436–447. [PubMed] [Google Scholar]
- 15.Billadeau D, Ahmann G, Greipp P, Van Ness B. The bone marrow of multiple myeloma patients contains B cell populations at different stages of differentiation that are clonally related to the malignant plasma cell. J Exp Med. 1993;178(3):1023–1031. doi: 10.1084/jem.178.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilarski LM, Giannakopoulos NV, Szczepek AJ, Masellis AM, Mant MJ, Belch AR. In multiple myeloma, circulating hyperdiploid B cells have clonotypic immunoglobulin heavy chain rearrangements and may mediate spread of disease. Clin Cancer Res. 2000;6(2):585–596. [PubMed] [Google Scholar]
- 17.Szczepek AJ, Seeberger K, Wizniak J, Mant MJ, Belch AR, Pilarski LM. A high frequency of circulating B cells share clonotypic Ig heavy-chain VDJ rearrangements with autologous bone marrow plasma cells in multiple myeloma, as measured by single-cell and in situ reverse transcriptase-polymerase chain reaction. Blood. 1998;92(8):2844–2855. [PubMed] [Google Scholar]
- 18.Bakkus MH, Heirman C, Van Riet I, Van Camp B, Thielemans K. Evidence that multiple myeloma Ig heavy chain VDJ genes contain somatic mutations but show no intraclonal variation. Blood. 1992;80(9):2326–2335. [PubMed] [Google Scholar]
- 19.Rasmussen T, Lodahl M, Hancke S, Johnsen HE. In multiple myeloma clonotypic CD38–/CD19+/CD27+ memory B cells recirculate through bone marrow, peripheral blood and lymph nodes. Leuk Lymphoma. 2004;45(7):1413–1417. doi: 10.1080/10428190410001655157. [DOI] [PubMed] [Google Scholar]
- 20.Vescio RA, et al. Myeloma Ig heavy chain V region sequences reveal prior antigenic selection and marked somatic mutation but no intraclonal diversity. J Immunol. 1995;155(5):2487–2497. [PubMed] [Google Scholar]
- 21.Matsui W, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103(6):2332–2336. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilarski LM, et al. Leukemic B cells clonally identical to myeloma plasma cells are myelomagenic in NOD/SCID mice. Exp Hematol. 2002;30(3):221–228. doi: 10.1016/S0301-472X(01)00788-3. [DOI] [PubMed] [Google Scholar]
- 23.Agematsu K, Hokibara S, Nagumo H, Komiyama A. CD27: a memory B-cell marker. Immunol Today. 2000;21(5):204–206. doi: 10.1016/S0167-5699(00)01605-4. [DOI] [PubMed] [Google Scholar]
- 24.Matsui W, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68(1):190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 27.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 28.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 29.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5(3):a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 32.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4(4):321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 33.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J Exp Med. 2005;202(4):505–516. doi: 10.1084/jem.20050575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 2009;28(11):1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagratuni T, et al. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood. 2010;116(2):250–253. doi: 10.1182/blood-2010-01-263236. [DOI] [PubMed] [Google Scholar]
- 36.Davies FE, et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003;102(13):4504–4511. doi: 10.1182/blood-2003-01-0016. [DOI] [PubMed] [Google Scholar]
- 37.Xu G, et al. Expression of XBP1s in bone marrow stromal cells is critical for myeloma cell growth and osteoclast formation. Blood. 2012;119(18):4205–4214. doi: 10.1182/blood-2011-05-353300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrasco DR, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11(4):349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med. 2007;204(9):2103–2114. doi: 10.1084/jem.20062571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya D, et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179(10):6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connor BP, Cascalho M, Noelle RJ. Short-lived and long-lived bone marrow plasma cells are derived from a novel precursor population. J Exp Med. 2002;195(6):737–745. doi: 10.1084/jem.20011626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chevrier S, et al. CD93 is required for maintenance of antibody secretion and persistence of plasma cells in the bone marrow niche. Proc Natl Acad Sci U S A. 2009;106(10):3895–3900. doi: 10.1073/pnas.0809736106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes ML, Carotta S, Corcoran LM, Nutt SL. Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev. 2006;20(8):933–938. doi: 10.1101/gad.1396206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheeren FA, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6(3):303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 45.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/S1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 46.Sanz-Rodríguez F, Teixidó J. VLA-4-dependent myeloma cell adhesion. Leuk Lymphoma. 2001;41(3-4):239–245. doi: 10.3109/10428190109057979. [DOI] [PubMed] [Google Scholar]
- 47.Katz BZ. Adhesion molecules — the lifelines of multiple myeloma cells. Semin Cancer Biol. 2010;20(3):186–195. doi: 10.1016/j.semcancer.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Barillé S, et al. CD130 rather than CD126 expression is associated with disease activity in multiple myeloma. Br J Haematol. 1999;106(2):532–535. doi: 10.1046/j.1365-2141.1999.01563.x. [DOI] [PubMed] [Google Scholar]
- 49.Rawstron AC, et al. The interleukin-6 receptor alpha-chain (CD126) is expressed by neoplastic but not normal plasma cells. Blood. 2000;96(12):3880–3886. [PubMed] [Google Scholar]
- 50.Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 51.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma S, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6(7):1146–1153. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- 53.Feldmann G, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67(5):2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther. 2008;3(4):237–246. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 55.Boucher K, et al. Stemness of B-cell progenitors in multiple myeloma bone marrow. Clin Cancer Res. 2012;18(22):6155–6168. doi: 10.1158/1078-0432.CCR-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reghunathan R, et al. Clonogenic multiple myeloma cells have shared stemness signature associated with patient survival. Oncotarget. 2013;4(8):1230–1240. doi: 10.18632/oncotarget.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fialkow PJ. Clonal origin of human tumors. Biochim Biophys Acta. 1976;458(3):283–321. doi: 10.1016/0304-419x(76)90003-2. [DOI] [PubMed] [Google Scholar]
- 58.Nowell PC. Mechanisms of tumor progression. Cancer Res. 1986;46(5):2203–2207. [PubMed] [Google Scholar]
- 59.Hua Y, et al. Molecular chaperone gp96 is a novel therapeutic target of multiple myeloma. Clin Cancer Res. 2013;19(22):6242–6251. doi: 10.1158/1078-0432.CCR-13-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munshi NC, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103(5):1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura M, et al. Activation of the endoplasmic reticulum stress pathway is associated with survival of myeloma cells. Leuk Lymphoma. 2006;47(3):531–539. doi: 10.1080/10428190500312196. [DOI] [PubMed] [Google Scholar]
- 62.Matsui W, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68(1):190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuhler GM, et al. Bone marrow stromal cell interaction reduces syndecan-1 expression and induces kinomic changes in myeloma cells. Exp Cell Res. 2010;316(11):1816–1828. doi: 10.1016/j.yexcr.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Hideshima T, et al. A proto-oncogene BCL6 is up-regulated in the bone marrow microenvironment in multiple myeloma cells. Blood. 2010;115(18):3772–3775. doi: 10.1182/blood-2010-02-270082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saudemont A, Quesnel B. In a model of tumor dormancy, long-term persistent leukemic cells have increased B7-H1 and B7.1 expression and resist CTL-mediated lysis. Blood. 2004;104(7):2124–2133. doi: 10.1182/blood-2004-01-0064. [DOI] [PubMed] [Google Scholar]
- 66.Tirapu I, et al. Low surface expression of B7-1 (CD80) is an immunoescape mechanism of colon carcinoma. Cancer Res. 2006;66(4):2442–2450. doi: 10.1158/0008-5472.CAN-05-1681. [DOI] [PubMed] [Google Scholar]
- 67.Niu H, Cattoretti G, Dalla-Favera R. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. J Exp Med. 2003;198(2):211–221. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hurtz C, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J Exp Med. 2011;208(11):2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Basso K, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115(5):975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7(5):445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 71.Leung-Hagesteijn C, et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24(3):289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jourdan M, et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009;114(25):5173–5181. doi: 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jourdan M, et al. Characterization of a transitional preplasmablast population in the process of human B cell to plasma cell differentiation. J Immunol. 2011;187(8):3931–3941. doi: 10.4049/jimmunol.1101230. [DOI] [PubMed] [Google Scholar]
- 74.Cocco M, et al. In vitro generation of long-lived human plasma cells. J Immunol. 2012;189(12):5773–5785. doi: 10.4049/jimmunol.1103720. [DOI] [PubMed] [Google Scholar]
- 75.Li Q, et al. Adoptive transfer of tumor reactive B cells confers host T-cell immunity and tumor regression. Clin Cancer Res. 2011;17(15):4987–4995. doi: 10.1158/1078-0432.CCR-11-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ray A, Basu S, Williams CB, Salzman NH, Dittel BN. A novel IL-10-independent regulatory role for B cells in suppressing autoimmunity by maintenance of regulatory T cells via GITR ligand. J Immunol. 2012;188(7):3188–3198. doi: 10.4049/jimmunol.1103354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jang HR, Gandolfo MT, Ko GJ, Satpute SR, Racusen L, Rabb H. B cells limit repair after ischemic acute kidney injury. J Am Soc Nephrol. 2010;21(4):654–665. doi: 10.1681/ASN.2009020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chesi M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13(2):167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Y, et al. A novel rapid-onset high-penetrance plasmacytoma mouse model driven by deregulation of cMYC cooperating with KRAS12V in BALB/c mice. Blood Cancer J. 2013;3:e156. doi: 10.1038/bcj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dechow T, et al. GP130 activation induces myeloma and collaborates with MYC. J Clin Invest. 2014;124(12):5263–5274. doi: 10.1172/JCI69094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu B, Li Z. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood. 2008;112(4):1223–1230. doi: 10.1182/blood-2008-03-143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dai J, Liu B, Cua DJ, Li Z. Essential roles of IL-12 and dendritic cells but not IL-23 and macrophages in lupus-like diseases initiated by cell surface HSP gp96. Eur J Immunol. 2007;37(3):706–715. doi: 10.1002/eji.200636643. [DOI] [PubMed] [Google Scholar]
- 83.Liu B, et al. TLR4 up-regulation at protein or gene level is pathogenic for lupus-like autoimmune disease. J Immunol. 2006;177(10):6880–6888. doi: 10.4049/jimmunol.177.10.6880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.