The pineal body is a small endocrine gland in the midline of the brain that secretes melatonin to modulate circadian rhythms. A group of primary tumors which arise from the pineal gland termed pineal parenchymal tumors are classified as pineocytoma (grade I), pineal parenchymal tumor of intermediate differentiation (PPTID; grade II or III), or pineoblastoma (grade IV). Most pineoblastomas arise in children, whereas pineocytomas and PPTIDs typically occur later in life. Pineocytomas are associated with favorable prognosis, with 5-year survival exceeding 90% following gross total resection. In contrast, pineoblastomas are embryonal tumors with a propensity for cerebrospinal dissemination and poor outcome despite resection, craniospinal radiation, and systemic chemotherapy [3]. PPTIDs are morphologically heterogeneous with intermediate histologic features and variable clinical outcomes [3, 5]. Recently, pineoblastomas were identified to harbor frequent mutations of the DICER1 gene or homozygous deletion of the DROSHA gene that both encode microRNA processing enzymes [2, 6, 7]. However, the genetic alterations responsible for driving PPTID and pineocytoma are unknown.
To investigate the molecular pathogenesis of pineal parenchymal tumors, we performed whole-exome sequencing on genomic DNA extracted from tumor and matched normal tissue from eight patients (Figure 1a) as described in the Supplementary Methods (Online Resource 1). The clinical features and outcomes of this patient cohort are presented in Supplementary Table 1 (Online Resource 2). Imaging features are shown in Supplementary Figure 1 (Online Resource 1). The four pineoblastomas were primitive small round blue cell tumors with frequent mitoses, karyorrhectic debris, and foci of necrosis (Supplementary Figure 2 [Online Resource 1]). In contrast, the three PPTID were histologically characterized by sheets of tumor cells with uniform round nuclei containing delicate chromatin and contained only occasional mitotic figures (Supplementary Figure 3 [Online Resource 1]). The one pineocytoma contained numerous pineocytomatous rosettes and lacked appreciable mitotic activity (Supplementary Figure 4 [Online Resource 1]).
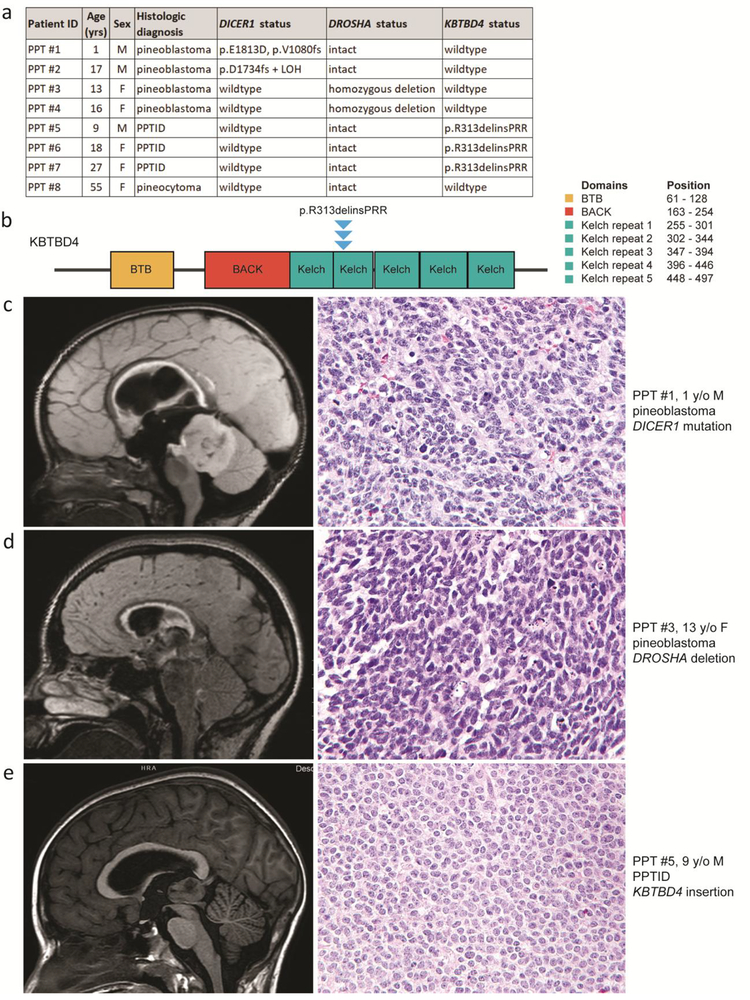
Fig. 1.
Pineoblastoma is characterized by mutually exclusive DICER1 mutation and DROSHA homozygous deletion and absence of KBTBD4 mutation, whereas pineal parenchymal tumor of intermediate differentiation (PPTID) is characterized by a recurrent KBTBD4 small in-frame insertion and absence of DICER1 mutation or DROSHA deletion. a, Clinicopathologic features and identified genetic alterations in the eight pineal parenchymal tumors. b, Diagram of human KBTBD4 protein with the location of the recurrent p.R313delinsPRR small in-frame insertion identified in the three PPTID. UniProt Q9NVX7, RefSeq NM_018095. c, Pre-operative magnetic resonance imaging and tumor histology for patient PPT #1 with pineoblastoma harboring DICER1 mutation. d, Pre-operative magnetic resonance imaging and tumor histology for patient PPT #3 with pineoblastoma harboring DROSHA homozygous deletion. e, Pre-operative magnetic resonance imaging and tumor histology for patient PPT #5 with PPTID harboring KBTBD4 small in-frame insertion.
Among the four pineoblastomas, two harbored somatic mutations in DICER1 and the other two harbored focal homozygous deletions of DROSHA on chromosome 5p13 (Figure 1c, Figure 1d, and Supplementary Figures 5–7 [Online Resource 1]). Pineoblastoma PPT #1 contained a somatic DICER1 frameshift mutation (p.V1080fs) on one allele and a hotspot missense mutation within the C-terminal Ribonuclease III domain on the other allele (p.E1813D) that has been recurrently found in pleuropulmonary blastomas, cystic nephromas, Sertoli-Leydig cell tumors, and other neoplasms known to be driven by DICER1 mutation (Catalog Of Somatic Mutations In Cancer database). Pineoblastoma PPT #2 contained a somatic DICER1 frameshift mutation (p.D1734fs) with elimination of the remaining wildtype allele due to loss of chromosome 14q. The DICER1 mutations were somatic in both PPT #1 and PPT #2, with no evidence of constitutional mosaicism in either patient (variant allele frequency in normal samples of 0%). Other than DICER1, no other genes harbored recurrent somatic nonsynonymous mutations among the four pineoblastomas (Supplementary Tables 2 and 3 [Online Resource 2]).
In contrast, none of the three PPTID harbored DICER1 mutation or DROHSA deletion. Instead, the three PPTID were all found to harbor the identical somatic small in-frame insertion (p.R313delinsPRR) in the KBTBD4 gene (Figure 1b, Figure 1e, and Supplementary Figure 8 [Online Resource 1]), which encodes Kelch repeat- and BTB domain-containing protein 4 that is reported to regulate the Cullin3-based E3 ubiquitin ligase complex [1]. Similar small in-frame insertions in KBTBD4 at codons 308–313 within a Kelch repeat domain were recently identified in a subset of Group 3 and Group 4 medulloblastomas that lacked other known genetic drivers such as MYC or MYCN amplification, GFI1B rearrangement, or SNCAIP duplication/PRDM6 rearrangement [4]. Other than medulloblastoma, no other human tumor types have been identified to harbor recurrent KBTBD4 mutations, and the functional mechanism by which these KBTBD4 mutations drive tumorigenesis in PPTID and medulloblastomas is uncertain at present. Other than KBTBD4, no other genes harbored recurrent somatic nonsynonymous mutations among the three PPTID (Supplementary Tables 2 and 3 [Online Resource 2]).
The single case of pineocytoma lacked deletion of DROSHA and mutation of DICER1 and KBTBD4. A small number of somatic nonsynonymous mutations were identified of uncertain significance (Supplementary Tables 2 and 3 [Online Resource 2]), but identification of a recurrent genetic driver in pineocytoma awaits assessment of additional tumor samples.
In summary, we have identified that pineoblastoma is characterized by mutually exclusive DICER1 mutation or DROSHA homozygous deletion and absence of KBTBD4 mutations. In contrast, PPTID is characterized by recurrent KBTBD4 small in-frame insertions and absence of DICER1 mutation or DROSHA homozygous deletion. Further studies are warranted to confirm these findings in a larger cohort of pineal parenchymal tumors. Nonetheless, these findings indicate a fundamental role for deregulation of microRNA processing in the pathogenesis of pineoblastoma, whereas PPTID appear to arise independently of disruption of microRNA processing genes. The recurrent KBTBD4 mutation in PPTID identified here will likely be useful in helping to diagnostically distinguish this tumor entity from pineoblastoma. Future studies will determine the potential prognostic significance of DICER1 mutation versus DROSHA homozygous deletion in pineoblastoma, and also the oncogenic mechanism by which KBTBD4 mutation promotes tumorigenesis of the pineal gland. The KBTBD4 mutations identified in PPTID in this study, as well as those found in medulloblastomas [4], are all heterozygous mutations that cluster at a hotspot within one of the Kelch repeat domains. This genetic pattern of heterozygous variants that cluster at a mutational hotspot within a functional domain is strongly suggestive that these are activating, gain-of-function mutations, as opposed to inactivating, loss-of-function events. Thus, KBTBD4 is likely to function as an oncogene, rather than a tumor suppressor gene, in PPTID and medulloblastoma.
Supplementary Material
The online version of this article (https://doi.org/#######) contains supplementary material, which is available to authorized users:
Online Resource 1 – Supplementary Methods and Supplementary Figures 1–8
Online Resource 2 – Supplementary Tables 1–3
Acknowledgements
This study was supported by the NIH Director’s Early Independence Award (DP5 OD021403) and a Career Development Award from the UCSF Brain Tumor SPORE grant (NIH P50 CA097257) to D.A.S. We thank the UCSF Brain Tumor Tissue Bank (supported by NIH P50 CA097257) for supplying key samples.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Data availability
Scanned image files of the H&E stained slides from the eight pineal parenchymal tumors from which representative images are presented are available for downloading and viewing at the following link: https://figshare.com/projects/Pineal_parenchymal_tumors/59621. Sequencing data files are available from the authors upon request.
Compliance with ethical standards
This study was approved by the Committee on Human Research of the University of California, San Francisco, with a waiver of patient consent.
Conflict of interest
The authors declare that they have no competing interests related to this study.
References
- 1.Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A et al. (2013) Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem 288:7803–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Kock L, Sabbaghian N, Druker H, Weber E, Hamel N, Miller S et al. (2014) Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol 128:583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauchon F, Jouvet A, Paquis P, Saint-Pierre G, Mottolese C, Ben Hassel M et al. (2000) Parenchymal pineal tumors: a clinicopathological study of 76 cases. Int J Radiat Oncol Biol Phys 46:959–968 [DOI] [PubMed] [Google Scholar]
- 4.Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T et al. (2017) The whole-genome landscape of medulloblastoma subtypes. Nature 547:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raleigh DR, Solomon DA, Lloyd SA, Lazar A, Garcia MA, Sneed PK et al. (2017) Histopathologic review of pineal parenchymal tumors identifies novel morphologic subtypes and prognostic factors for outcome. Neuro Oncol 19:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabbaghian N, Hamel N, Srivastava A, Albrecht S, Priest JR, Foulkes WD (2012) Germline DICER1 mutation and associated loss of heterozygosity in a pineoblastoma. J Med Genet 49:417–419 [DOI] [PubMed] [Google Scholar]
- 7.Snuderl M, Kannan K, Pfaff E, Wang S, Stafford JM, Serrano J et al. (2018) Recurrent homozygous deletion of DROSHA and microduplication of PDE4DIP in pineoblastoma. Nat Commun 9:2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version of this article (https://doi.org/#######) contains supplementary material, which is available to authorized users:
Online Resource 1 – Supplementary Methods and Supplementary Figures 1–8
Online Resource 2 – Supplementary Tables 1–3