Abstract
Background
The clinical and pathological features of primary melanoma are not sufficiently sensitive to accurately predict which patients are at a greater risk of relapse. Recently, a 31‐gene expression profile (DecisionDx‐Melanoma) test has shown promising results.
Objectives
To evaluate the early prognostic performance of a genetic signature in a multicentre prospectively evaluated cohort.
Methods
Inclusion of patients with AJCC stages IB and II conducted between April 2015 and December 2016. All patients were followed up prospectively to assess their risk of relapse. Prognostic performance of this test was evaluated individually and later combined with the AJCC staging system. Prognostic accuracy of disease‐free survival was determined using Kaplan–Meier curves and Cox regression analysis. Results of the gene expression profile test were designated as Class 1 (low risk) and Class 2 (high risk).
Results
Median follow‐up time was 26 months (IQR 22–30). The gene expression profile test was performed with 86 patients; seven had developed metastasis (8.1%) and all of them were in the Class 2 group, representing 21.2% of this group. Gene expression profile was an independent prognostic factor for relapse as indicated by multivariate Cox regression analysis, adjusted for AJCC stages and age.
Conclusions
This prospective multicentre cohort study, performed in a Spanish Caucasian cohort, shows that this 31‐gene expression profile test could correctly identify patients at early AJCC stages who are at greater risk of relapse. We believe that gene expression profile in combination with the AJCC staging system could well improve the detection of patients who need intensive surveillance and optimize follow‐up strategies.
Introduction
The incidence of melanoma is rapidly increasing and resulting mortality has risen significantly over the past 30 years.1 An increased incidence of invasive melanoma has been reported worldwide together with a corresponding rise in intermediate‐risk patients.2, 3, 4 After the diagnosis of a primary melanoma, patients are enrolled in follow‐up schedules depending on their risk of relapse, but due to its increasing prevalence, the workload and costs of surveillance programs have increased markedly.5, 6, 7, 8 Many surveillance strategies have been proposed, mainly based on initial AJCC clinical–pathological staging;9, 10, 11 nevertheless, there are still many ‘low‐risk’ melanoma patients who suffer relapses, resulting in high mortality.9 In addition, approximately two thirds of patients whose melanoma will metastasize leading to death were initially sentinel lymph node biopsy (SLNB) negative (American Joint Committee on Cancer [AJCC] stages I and II).12, 13
Due to this problem, many biomarkers have been developed to improve the identification of patients who will relapse, but with no real clinical impact until now.14 Recently, new prognostic tests based on genetic profiling signatures have been developed to better identify those patients who are at a higher risk of developing metastasis leading to death.15, 16, 17, 18, 19, 20 A gene expression profile (GEP) test (DecisionDx‐Melanoma, Castle Biosciences, Inc., Friendswood, TX, USA) evaluates 31 genes of primary cutaneous melanoma tumours and based on the expression of these genes (genetic signature) it classifies patients into two groups of risk of relapse, low (class 1) or high (class 2). This test has been validated in several historical cohorts showing that its prognostic ability is independent from the clinical and pathological features of the tumour.15, 16, 17, 18, 19, 20, 21, 22, 23, 24 However, this test has never before been evaluated in a prospective multicentre cohort.
Here we present a multicentre prospectively evaluated cohort of 86 patients where a 31‐gene expression profile (GEP) test was used to assess their risk of relapse.
Material and methods
Study design
We conducted a prospective multicentre cohort study with patients from five tertiary melanoma referral centres in Spain included between April 2015 and December 2016, who were followed up until December 2018. Patients with resected pathologic American Committee on Cancer (AJCC, 7th edition, 2009) stages IB and II primary cutaneous melanoma were invited to participate in the study (Fig. 1). Furthermore, exclusion criteria included no evidence of disease within 3 months of primary surgery.
Figure 1.
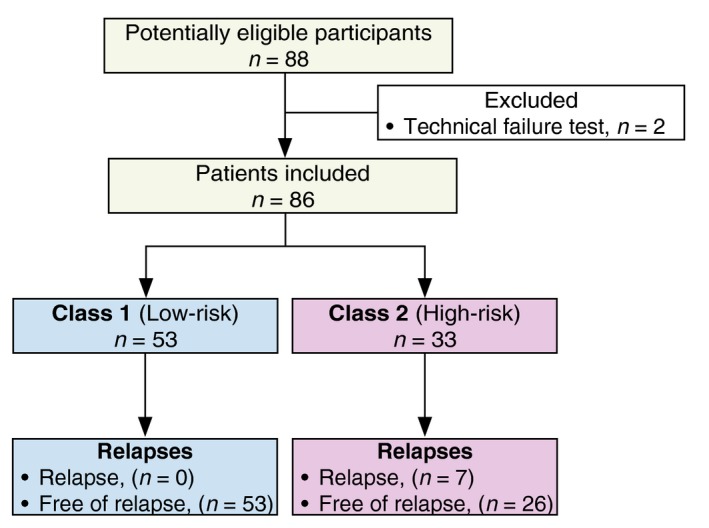
Flow chart of the cohort.
Patients were treated with conventional surgery followed by wide excision depending on Breslow thickness of the tumour. Patients with tumours T1b and above were staged with sentinel lymph node biopsy (SLNB) following each institution protocol. For the purpose of this study, patients with positive SLNB were excluded. The study was approved by the ethics committees of all Hospitals.
In all patients, tumour specimens were obtained from the hospital biobank. The formalin‐fixed paraffin‐embedded primary cutaneous melanoma tissue was analysed with the 31‐GEP test (DecisionDx‐Melanoma, Castle Biosciences, Inc).15, 25 Moreover, the RT‐PCR‐based test classifies patients into a low‐risk (Class 1) or high‐risk (Class 2) category for recurrence, as previously reported and validated.15
Disease‐free survival (DFS) was defined as the period of time in months from the date of diagnosis to the date of relapse. Patients that were free of disease at the time of the last follow‐up or died during the study period, by any other causes, were treated as censored cases for evaluation purposes. All patients were followed up prospectively according to each institution protocol to identify relapses.
According to marked differences in 5‐year survival curves for AJCC stages IIA and IIB patients (AJCC 79% vs. 68% respectively), all included subjects were further classified into two risk groups. Patients with AJCC stages IB and IIA were considered as low‐risk and IIB and IIC as high‐risk.18, 26
For better reporting of the evaluation conducted, the REMARK checklist recommendations have been applied.
Statistical analysis
Pearson's chi‐squared and Student's t‐tests were used to compare categorical and continuous variables, respectively. Mann–Whitney–Wilcoxon was used to compare samples not distributed normally. Primary survival end‐points (disease‐free survival) were evaluated using Kaplan–Meier curves and univariate and multivariate Cox regression analyses. Due to the substantial censoring of survival times and several highly predictive covariates, we applied Firth's Correction to our Cox regression model.27 The starting point for all cases was the diagnosis of primary melanoma. P‐values <0.05 were considered significant. Statistical analyses were performed using SPSS v.25.0 (IBM Corp., Armonk, NY, USA), and Cox regression analysis was performed using R studio (R Studio Team (2015). RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, USA).
Results
After applying the inclusion/exclusion criteria, 88 patients were included in the study, but two were later excluded because of a technical failure in the test. Of the remaining 86 patients, 40 patients (46.5%) were male and 46 female (53.5%), with a median age at diagnosis of 59.2 years (interquartile range [IQR] 47–72). Characteristics of this cohort are summarized in Table 1. Patients were followed up for 2206 person‐months, with a metastasis incidence rate of 3.17 cases per 1000 person‐months. Overall median follow‐up time was 26 months (IQR 22–30).
Table 1.
Basal clinicopathological characteristics of the cohort and relapse data
| Overall | Class 1 Low risk | Class 2 High risk | P value | |
|---|---|---|---|---|
| n | 86 | 53 | 33 | – |
| Follow‐up time median (IQR) | 26 (22–30) | 27 (23–32) | 24 (20–29) | 0.066 |
| Sex | ||||
| Male | 40 (46.5%) | 21 (40%) | 19 (58%) | 0.105 |
| Female | 46 (53.5%) | 32 (60%) | 14 (42%) | |
| Age | ||||
| Median (IQR) | 59.2 (47–72) | 57 (46–68) | 68 (55–74) | 0.025 |
| Localization | ||||
| Acral | 5 (6%) | 2 (4%) | 3 (9%) | 0.738 |
| Head and neck | 11 (13%) | 7 (13%) | 4 (12%) | |
| Legs | 21 (24%) | 15 (28%) | 6 (18%) | |
| Arms | 12 (14%) | 7 (13%) | 5 (15%) | |
| Trunk | 37 (43%) | 22 (41%) | 15 (45%) | |
| Breslow | ||||
| Mean (SD) | 2.5 (2.3) | 1.7 (1.4) | 3.7 (2.9) | <0.001 |
| ≤1.00 mm | 18 (21%) | 17 (32%) | 1 (3%) | <0.001 |
| 1.00–2.00 mm | 33 (38%) | 23 (43%) | 10 (30%) | |
| 2.01–4.00 mm | 22 (26%) | 10 (19%) | 12 (36%) | |
| >4.00 mm | 13 (15%) | 3 (6%) | 10 (30%) | |
| Mitotic rate (mm2) | ||||
| <1 mm2 | 9 (11%) | 6 (11%) | 3 (9%) | 0.684 |
| ≥1 mm2 | 76 (88%) | 46 (87%) | 30 (91%) | |
| N.A. | 1 (1%) | 1 (2%) | 0 | |
| Ulceration | ||||
| Absent | 60 (70%) | 51 (96%) | 9 (27%) | <0.001 |
| Present | 26 (30%) | 2 (4%) | 24 (73%) | |
| AJCC stage | ||||
| Low‐risk (IB‐IIA) | 62 (72%) | 48 (91%) | 14 (42% | <0.001 |
| High‐risk (IIB‐IIC) | 24 (28%) | 5 (9%) | 19 (58%) | |
| Relapse | ||||
| No | 79 (91%) | 53 (100%) | 26 (79%) | <0.001 |
| Yes | 7 (9%) | 0 | 7 (21%) | |
| Relapse site (n = 7) | ||||
| Skin | – | 0 | 2 (29%) | – |
| Lymph node | – | 0 | 2 (29%) | |
| Visceral | – | 0 | 3 (43%) | |
AJCC, American Joint Committee on Cancer; IQR, Interquartile range; N.A., Not Available; SD, Standard Deviation.
P‐values are bold where they are less than or equal to the significance level cut‐off of 0.05.
Class 1 and Class 2 patients showed a mean Breslow of 1.7 mm (standard deviation [SD] 1.4) and 3.7 mm (SD 2.9), respectively, which was statistically significant (P < 0.001). Moreover, Class 2 melanomas were ulcerated more often and presented significantly higher AJCC staging (Table 1).
Relapses were identified in seven patients (8.1%), all corresponding to Class 2 (high‐risk) by the GEP test (P < 0.001). Furthermore, the GEP risk score identified 19 patients (22.1%) with a risk score different from that predicted by AJCC classification. Five (5.8%) patients with high‐risk AJCC stage were rated as Class 1 (low risk), and 14 subjects (16.3%) with low‐risk AJCC stage were identified as Class 2 (high risk) by the GEP test. Five patients (5.8%) presented relapses with a high‐risk GEP test score and AJCC high‐risk at the same time, while two subjects (2.3%) patient were identified as Class 2 (high‐risk) by the GEP test although belonging to AJCC low‐risk.
Kaplan–Meier survival curves were analysed to evaluate DFS for the two 31‐GEP risk classes and showed statistically significant differences between the two groups (log rank P < 0.001). When combining the GEP test with the AJCC (log rank P = 0.001) the significance was maintained (Fig. 2).
Figure 2.
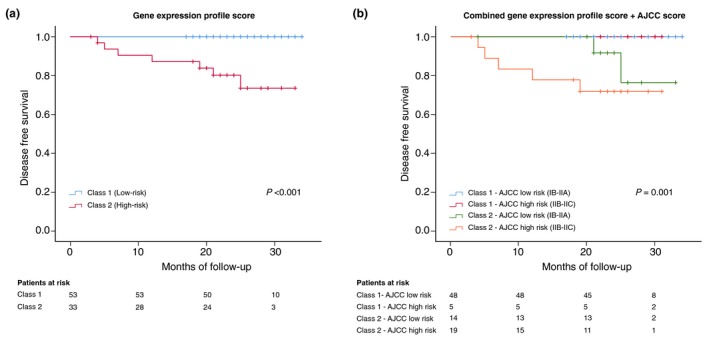
Kaplan–Meier estimates of disease‐free survival. (a) Kaplan–Meier curve showing disease‐free survival for the two GEP cohorts. (b) displays four subgroups of patients according to the outcomes predicted by the AJCC staging system in combination with gene GEP testing. Survival table is shown at the bottom of each figure. AJCC, American Joint Committee on Cancer.
Following curve comparison by Kaplan–Meier method, we compared the prognostic accuracy of the GEP test to the AJCC and age using Cox regression analysis. Both univariate and multivariate analysis showed the GEP test to be an independent predictor of metastasis. Hazard ratios for the GEP test were 28.37 (95% CI 3.46–3682.91; P < 0.01) for the univariable analysis and 18.82 (95% CI 1.81–2549.76; P = 0.01) for the multivariable analysis (Table 2).
Table 2.
Univariable and multivariate Cox regression analysis of prognostic factors in 86 melanoma patients for relapse free survival
| Factor (high‐risk variable) | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| GEP (Class 2) | 28.37 (3.46–3682.91) | <0.01 | 18.82 (1.81–2,549.76) | 0.01 |
| AJCC risk group (IIB‐IIC) | 6.33 (1.52–35.28) | 0.01 | 1.52 (0.36–8.77) | 0.58 |
| Age of onset (>50) | 7.26 (0.88–942.67) | 0.07 | 3.7 (0.43–486.05) | 0.29 |
AJCC, American Joint Committee on Cancer; CI, Confidence interval; GEP, gene expression profile.
Discussion
This paper describes the results of a 31‐GEP test performed to better categorize patients with low to intermediate‐risk melanoma according to the AJCC staging system. We present the data from a multicentre cohort study in which patients with malignant melanoma were followed up prospectively after the 31‐GEP test. We observed that seven patients (8.1%) presented relapses within a median of 12 months (IQR 5–21), all seven belonging to the Class 2 group (overall, 21.2% of the Class 2 group). Kaplan–Meier survival curves were statistically different between both groups with an increased risk of recurrence in the Class 2 group with a hazard ratio of 28.37 and 18.82, for univariate and multivariate Cox regression analysis, respectively. Similarly, a previous validation study of the 31‐GEP test showed that the metastatic risk was predicted with high accuracy in the Class 2 cohort of primary cutaneous melanomas, with a receiver operating characteristic (ROC) curve value of 0.91–0.93.15
Subsequent studies have analysed the use of the 31‐GEP test in combination with the AJCC stage system and found that this approach improves the identification of patients at risk of relapse.18, 19 In this report, we also combined GEP with the AJCC score confirming that patients with higher AJCC (IIB‐IIC) staging and Class 2 GEP are at a greater risk of relapse. Nevertheless, two patients with a low‐risk AJCC stage and a Class 2 GEP metastasized. Based solely on AJCC staging, many patients classified at low‐risk will relapse as seen in the study by Ferris et al. where 43% of the cases classified as Class 2 GEP and AJCC low risk, relapsed. This has a profound impact on high‐risk patients based on the GEP test, but at lower AJCC stages who could opt for intensive follow‐up,9 but are at early stages of the AJCC. Moreover in the new era of adjuvant therapies, patients at greater risk of relapse could be selected for these treatment schemes and improve overall survival.
Moreover, GEP tests have been used in combination with SLNB status resulting in different outcomes of disease‐free survival, distant metastasis free survival, and overall survival curves.16 The authors have observed that patients with negative SLNB and class 2 GEP have a worse prognosis than patients with positive SLNB and class 1 GEP status.16, 21 In the present study, we only included SLN negative patients, to evaluate the performance of this test in identifying those at a high risk of melanoma relapsing during early stages. It may be that the use of genetic signatures in conjunction with the classic clinical, pathological criteria of AJCC and the SLNB status will significantly improve the detection of patients at risk of relapse. Accordingly, we believe that validating a gene assay test will be an important step towards the goal of optimizing follow‐up protocols and we propose personalized adjuvant therapy in low to intermediate‐risk melanoma patients.
As a limitation of this study, we observed that patients who tested as Class 2 GEP had a tendency towards thicker melanomas, higher frequency of ulceration and higher AJCC stages. However, univariate and multivariate Cox regression analyses showed that the GEP test was an independent prognostic factor. Moreover, we had a low incidence of events, so the external validity should be interpreted with caution. This could be explained by the follow‐up time being relatively short, although most of the metastases developed within the first two years of follow‐up. The number of relapses is then, in accordance with that published in the literature.8, 13 Moreover, due to the multicentre and prospective design of this study and the assessment of this test in ‘real daily practice’, the GEP test gives useful information about what to expect during the first two years of follow‐up, as we observed that 21% of the patients in the class 2 group developed metastasis.
Conclusion
We observed that a 31‐GEP test allows the accurate prediction of patients at a high risk of relapse, despite having a low‐risk AJCC staging. We believe that GEP in combination with AJCC staging system could well improve the detection of patients who need intensive surveillance and optimize follow‐up strategies.
Role of the sponsors
The sponsors had no role in the design and conduct of the study, nor in the collection, analysis and interpretation of data, nor in the preparation, review and approval of the manuscript nor in the decision to submit the manuscript for publication.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Acknowledgements
Thanks to our patients and their families who are the main reason for our studies; to nurses from the Melanoma Unit of Hospital Clínic of Barcelona, Daniel Gabriel, Pablo Iglesias and Maria E Moliner for helping to collect patient data and to Paul Hetherington for helping with English editing and correction of the manuscript.
Conflicts of interest
The authors declare that they have no conflict of interest.
Funding source
Funding for this project was provided by Castle Biosciences, Inc. The research at the Melanoma Unit in Barcelona is partially funded by Spanish Fondo de Investigaciones Sanitarias grants 09/1393, 12/00840 and 15/00716; CIBER de Enfermedades Raras of the Instituto de Salud Carlos III, Spain, co‐financed by European Development Regional Fund “A way to achieve Europe” ERDF; European Commission under the 6th Framework Programme, Contract No. LSHC‐CT‐2006‐018702 (GenoMEL) and by the European Commission under the 7th Framework Programme, Diagnoptics; The National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115), a grant from “Fundació La Marató de TV3, 201331‐30”, Catalonia, Spain; CERCA Programme / Generalitat de Catalunya, and a grant from “Fundación Científica de la Asociación Española Contra el Cáncer”, Spain. Part of the work was carried out at the Esther Koplowitz Center, Barcelona.
References
- 1. Glazer AM, Winkelmann RR, Farberg AS, Rigel DS. Analysis of trends in US melanoma incidence and mortality. JAMA Dermatol 2016; 153(2): 225. [DOI] [PubMed] [Google Scholar]
- 2. Puig S, Marcoval J, Paradelo C et al Melanoma incidence increases in the elderly of catalonia but not in the younger population: effect of prevention or consequence of immigration? Acta Dermato Venereol 2015; 95: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ríos L, Nagore E, López J et al Melanoma characteristics at diagnosis from the Spanish National Cutaneous Melanoma Registry: 15 years of experience. Actas Dermo‐Sifiliográficas 2013; 104: 789–799. [DOI] [PubMed] [Google Scholar]
- 4. Clarke CA, McKinley M, Hurley S et al Continued increase in melanoma incidence across all socioeconomic status groups in California, 1998–2012. J Invest Dermatol 2017; 137: 2282–2290. [DOI] [PubMed] [Google Scholar]
- 5. Podlipnik S, Moreno‐Ramírez D, Carrera C et al Cost‐effectiveness analysis of imaging strategy for an intensive follow‐up of patients with American Joint Committee on Cancer stage IIB, IIC and III malignant melanoma. Br J Dermatol 2018. [DOI] [PubMed] [Google Scholar]
- 6. Rueth NM, Cromwell KD, Cormier JN. Long‐term follow‐up for melanoma patients: is there any evidence of a benefit? Surg Oncol Clin N Am 2015; 24: 359–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guy GP, Thomas CC, Thompson T et al Vital signs: melanoma incidence and mortality trends and projections ‐ United States, 1982‐2030. MMWR Morb Mortal Wkly Rep 2015; 64: 591–596. [PMC free article] [PubMed] [Google Scholar]
- 8. Ribero S, Podlipnik S, Osella‐Abate S et al Ultrasound‐based follow‐up does not increase survival in early‐stage melanoma patients: a comparative cohort study. Eur J Cancer 2017; 85: 59–66. [DOI] [PubMed] [Google Scholar]
- 9. Podlipnik S, Carrera C, Sánchez M et al Performance of diagnostic tests in an intensive follow‐up protocol for patients with American Joint Committee on Cancer (AJCC) stage IIB, IIC, and III localized primary melanoma: a prospective cohort study. J Am Acad Dermatol 2016; 75: 516–524. [DOI] [PubMed] [Google Scholar]
- 10. Mangas C, Paradelo C, Puig S et al Initial evaluation, diagnosis, staging, treatment, and follow‐up of patients with primary cutaneous malignant melanoma. Consensus statement of the Network of Catalan and Balearic Melanoma Centers. Actas Dermo‐Sifiliográficas 2010; 101: 129–142. [PubMed] [Google Scholar]
- 11. Fong Z, Tanabe K. Comparison of melanoma guidelines in the U.S.A., Canada, Europe, Australia and New Zealand: a critical appraisal and comprehensive review. Br J Dermatol 2014; 170: 20–30. [DOI] [PubMed] [Google Scholar]
- 12. Morton DL, Thompson JF, Cochran AJ et al Final trial report of sentinel‐node biopsy versus nodal observation in melanoma. N Engl J Med 2014; 370: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leiter U, Buettner PG, Eigentler TK et al Hazard rates for recurrent and secondary cutaneous melanoma: an analysis of 33,384 patients in the German Central Malignant Melanoma Registry. J Am Acad Dermatol 2012; 66: 37–45. [DOI] [PubMed] [Google Scholar]
- 14. Griewank KG. Biomarkers in melanoma. Scand J Clin Lab Invest 2016; 76: S104–S112. [DOI] [PubMed] [Google Scholar]
- 15. Gerami P, Cook RW, Wilkinson J et al Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res 2015; 21: 175–183. [DOI] [PubMed] [Google Scholar]
- 16. Gerami P, Cook RW, Russell MC et al Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J Am Acad Dermatol 2015; 72: 780–785.e3. [DOI] [PubMed] [Google Scholar]
- 17. Berger AC, Davidson RS, Poitras KJ et al Clinical impact of a 31‐gene expression profile test for cutaneous melanoma in 156 prospectively and consecutively tested patients. Curr Med Res Opin 2016; 32: 1–23. [DOI] [PubMed] [Google Scholar]
- 18. Ferris LK, Farberg AS, Middlebrook B et al Identification of high‐risk cutaneous melanoma tumors is improved when combining the online American Joint Committee on Cancer Individualized Melanoma Patient Outcome Prediction Tool with a 31‐gene expression profile–based classification. J Am Acad Dermatol 2017; 76: 818–825.e3. [DOI] [PubMed] [Google Scholar]
- 19. Dillon LD, Gadzia JE, Davidson RS et al Prospective, multicenter clinical impact evaluation of a 31‐gene expression profile test for management of melanoma patients. Ski J Cutan Med 2018; 2: 111–121. [Google Scholar]
- 20. Zager JS, Gastman BR, Leachman S et al Performance of a prognostic 31‐gene expression profile in an independent cohort of 523 cutaneous melanoma patients. BMC Cancer 2018; 18: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lawson DH, Cook RW, Johnson C, et al Continued evaluation of a 31‐gene expression profile test (GEP) for prediction of distant metastasis (DM) in cutaneous melanoma (CM). J Clin Oncol 2015; 33: 9066. [Google Scholar]
- 22. Lawson DH, Russell MC, Wilkinson J, et al Gene expression profile test (GEP) prediction of metastasis‐free (MFS) and overall survival (OS) in a cohort of cutaneous melanoma (CM) patients undergoing sentinel lymph node biopsy (SLNB). J Clin Oncol 2014; 32: 9022. [Google Scholar]
- 23. Farberg AS, Glazer AM, White R, Rigel DS. Impact of a 31‐gene expression profiling test for cutaneous melanoma on dermatologists’ clinical management decisions. J Drugs Dermatol 2017; 16: 428–431. [PubMed] [Google Scholar]
- 24. Hsueh EC, DeBloom JR, Lee J et al Interim analysis of survival in a prospective, multi‐center registry cohort of cutaneous melanoma tested with a prognostic 31‐gene expression profile test. J Hematol Oncol 2017; 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cook RW, Middlebrook B, Wilkinson J et al Analytic validity of DecisionDx‐Melanoma, a gene expression profile test for determining metastatic risk in melanoma patients. Diagn Pathol 2018; 13: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Balch CM, Gershenwald JE, Soong S et al Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 2009; 27: 6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heinze G, Dunkler D. Avoiding infinite estimates of time‐dependent effects in small‐sample survival studies. Stat Med 2008; 27: 6455–6469. [DOI] [PubMed] [Google Scholar]


