Abstract
Game meat potentially harbors a number of parasitic and bacterial pathogens that cause foodborne disease. It is thus important to monitor the prevalence of such pathogens in game meats before retail and consumption to ensure consumer safety. In particular, Sarcocystis spp. and Shiga toxin-producing Escherichia coli (STEC) have been reported to be causative agents of food poisoning associated with deer meat consumption. To examine the prevalence of these microbiological agents on-site at a slaughterhouse, the rapid, simple and sensitive detection method known as the “DNA strip” has been developed, a novel tool combining loop-mediated isothermal amplification and a lateral flow strip. This assay has achieved higher sensitivity and faster than conventional PCR and is suitable for on-site inspection.
Keywords: deer meat, DNA strip, loop-mediated isothermal amplification, Sarcocystis spp., STEC
Due to increasing numbers of sika deer (Cervus nippon) in Japan, the government recommends hunting these animals and consuming their meats as game meat in local restaurants and retail meat shops [12]. However, since Japanese abattoir law does not apply to the slaughter of game animals, the hunters must dissect and inspect the game by themselves. To guarantee the meat’s sanitation, a easy and quick assay that can be used during the slaughter of game animals to determine the presence of foodborne agents is needed. Many rapidly assay methods have been developed for foodborne bacteria, viruses and parasites, including immunochemical assays, conventional polymerase chain reaction (PCR), real time PCR and the loop-mediated isothermal amplification (LAMP) assay [18, 21, 28]. However, few of these methods have been validated using deer meat.
Outbreaks of food poisoning have been caused by parasites, bacteria and viruses associated with the consumption of deer meat in Japan [1, 15, 24]. We recently reported that Sarcocystis spp. was one of such causative agents of food poisoning [14]. Kabeya et al. [13] also reported that Shiga toxin-producing Escherichia coli (STEC) detected in the feces of sika deer possessed potential human pathogenicity. Due to the high contamination frequency in meat or feces in Japan, Sarcocystis spp. and STEC [9, 13] were chosen as the subjects in this study.
Given this background, we developed an easy, rapid and sensitive assay for the detection of Sarcocystis spp. and STEC in deer meat. The assay is called the “DNA strip” [23] and may be useful for on-site inspection in slaughterhouses.
The “DNA strip” enables the detection of the amplification products by LAMP on a lateral flow DNA strip and has been validated by Takasaki et al. [23]. Specific primers for the detection of Sarcocystis spp. and STEC were designed by attaching unique oligonucleotides as a tag modification at the 5-terminal of the forward inner primer, while the 5′-terminal of the backward inner primer was biotin-modified. In this way, successfully amplified target DNA will have unique oligonucleotide tags attached and bind to immobilized complementary tags on the DNA strip. The biotin will bond to streptavidin provided in the DNA strip buffer, generating blue color enhancement.
First, to confirm the performance of the “DNA strip” for the detection of 18S rRNA of Sarcocystis spp. (SarcoR) as well as the Shiga toxin genes (stx1 and stx2) of STEC, a LAMP assay was performed using the Sarcocyctis spp. and STEC isolates in PBS solution. Cyst of Sarcocystis spp. was collected from deer meat obtained from Yamanashi Prefecture, Japan. The STEC O157:H7 strain NIHS0106, which possesses both the stx1 and stx2 genes, was used as a positive control. These DNA were extracted using a QIAamp DNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The DNA concentration was measured by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, U.S.A.), and the extracted DNA was prepared at 10-fold dilution to be amplified with the reaction mixture. The reaction mixture contained of 1.5x Isothermal Master Mix (OptiGene Ltd., West Sussex, U.K.), tagged specific primer for the detection of Sarcocyctis spp. and STEC, respectively, and 10 × template DNA. DNA amplification was performed for 60 min at 65°C. The tubes were put on ice for a minimum of 5 min to avoid contamination by completely stopping the reaction and performing evaporation at the end of the amplification. The amplification product (1 µl) was diluted 40-fold with distilled water and then mixed with 20 µl of development solution containing Latex beads (TBA Co., Ltd., Sendai, Japan). The DNA strip was dipped into the solution for 15 min, and the results were determined visually. The specific primer of SarcoR in Sarcocystis spp. and stx1 and stx2 in STEC for the LAMP assay are shown in Table 1.
Table 1. The sequences and modification of the LAMP and PCR primers used in this study.
| Method | Assay | Primar name | Sequence | Ref. |
|---|---|---|---|---|
| DNA strip | 18S rRNA of Sarcocystis spp. (SarcoR) | F3 | TGAAAACCTTGGCCGATAGG | This study |
| B3 | CCTTGTTACGACTTCTCCTTCCTC | |||
| FIP | [tag]-GTAATCGGCGCAAGCTGCTGACGGTAATCTTTTGAGTATGCATCG | |||
| BIP | [bi]-TGTACACACCGCCCGTCGCTCTTTCCATTCCGCACACGTTG | |||
| LF | CTACTAGGCATTCCTCGTTGAAGAT | |||
| LB | CTACCGATTGAGTGTTCCGGTGA | |||
| stx1 | F3 | GCGATTTATCTGCATCCCCGTATGTCTGGTGACAGTAGCTAT | [4] | |
| B3 | GGAACCTCACTGACGCAGTCCTTCAGCTGTCACAGTAACA | |||
| FIP | ACTGATCCCTGCAACACG | |||
| BIP | TGTGGCAAGAGCGATGTT | |||
| LF | ACAACAGCGGTTACATTGT | |||
| LB | GATCATCCAGTGTTGTACGAA | |||
| stx2 | F3 | GGCGTCATCGTATACACAGGAGCGCTTCAGGCAGATACAG | ||
| B3 | AGACGTGGACCTCACTCTGAAACTCTGACACCATCCTCTC | |||
| FIP | CAGACAGTGCCTGACGAA | |||
| BIP | GGCGAATCAGCAATGTGC | |||
| LF | GCATCCAGAGCAGTTCTG | |||
| LB | CAGTATAACGGCCACAGTC | |||
| Conventional PCR | 18S rRNA of Sarcocystis spp. (SarcoR) | F | GGATAACCGTGGTAATTCTATG | [18] |
| R | TCCTATGTCTGGACCTGGTGAG | |||
| stx1 | mStx1_F | GGATAATTTGTTTGCAGTTGATGTC | [17] | |
| stx1 | mStx1_R | CAAATCCTGTCACATATAAATTATTTCGT | ||
| stx2 | mStx2_F | GGGCAGTTATTTTGCTGTGGA | ||
| stx2 | mStx2_R | GAAAGTATTTGTTGCCGTATTAACG | ||
According to a previous paper, some species of Sarcocystis have been parasitic to Japanese sika deer [9], such as S. wapiti, S. sybillensis, and S. hofmanni. However, except for S. wapiti, the DNA sequences of these species have never deposited in databases, so the sequences of the predominant Sarcocystis species in deer meat obtained from Yamanashi Prefecture, Japan, were determined in order to construct LAMP primers for SarcoR.
The 18S rRNA region was PCR-amplified from DNA extracted from deer meat using universal 18S rRNA region primers, as described by Pritt et al. [20], that were able to detect the 18S rRNA region of Sarcocystis spp. universally (Table 1). We then followed by cloning into pCRTM 2.1-TOPO® in E. coli TOP10 obtained from the TOPO® TA Cloning Kit for Sequencing (Thermo Fisher Scientific Inc.). The successful transformants were then subjected to a cycle sequencing reaction using the BigDye terminator in an ABI 3730×l system to determine the target gene sequences. Finally, six sequences were obtained (LC405946-LC405951), and the LAMP primers were designed based on the obtained consensus sequences using a LAMP Designer (OptiGene, Ltd.). We used the oligonucleotide primers for the stx1 and stx2 genes reported elsewhere [6]. As shown in Fig. 1, we first evaluated the detection performance of the three target genes by LAMP-DNA strip testing. The results indicated that these genes could be discriminated on the DNA strip based on the attached tag sequences.
Fig. 1.
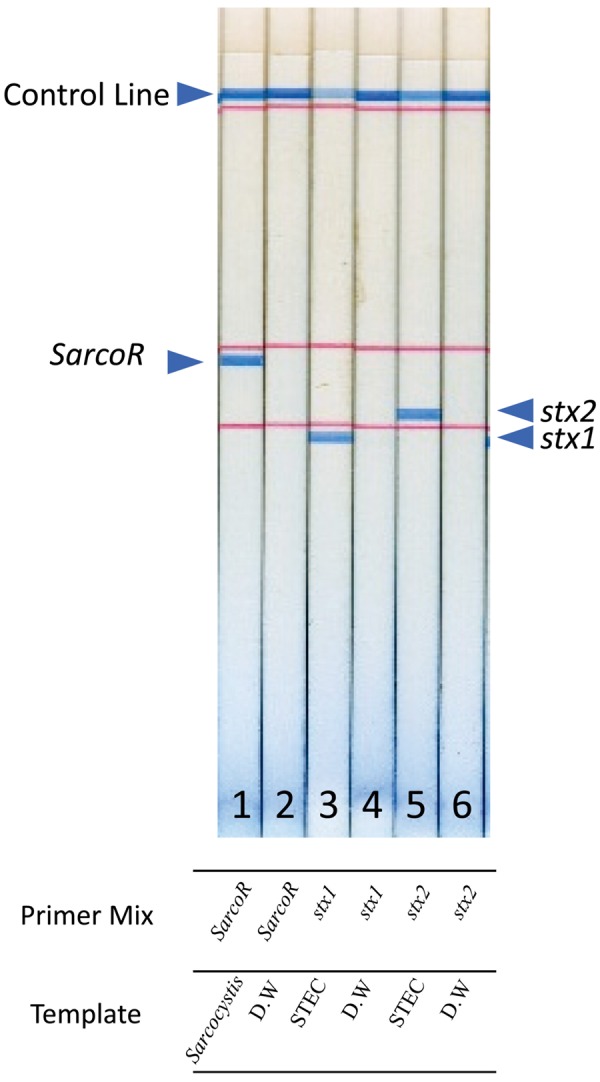
Performance of the DNA strip for the detection of the Sarcocystis spp. 18S rRNA gene (SarcoR) and the stx1 and stx2 genes. The template DNA was prepared from a cyst of Sarcocystis spp. or STEC O157:H7 strain NIHS0106. The primer mix:1,2: Sarco R, 3,4: stx 1, 5,6: stx 2 and 1,3 and 4 was with template. The arrow indicates each LAMP product.
Next, to determine the limit of detection (LOD) of this assay for both targets, LODs of conventional PCR for these targets were compared using various concentrations of bradizoids and the STEC strain. Conventional PCR for the rRNA of Sarcocystis spp. and for the stx1 and stx2 genes was performed according to the method of Pritt et al. [20] and Nielsen et al. [19], respectively. The bradizoids were prepared from Sarcocystis spp. according to the previous paper [9]. The Sarcocystis spp. (3.6 × 105 bradizoids/ml) or STEC strain (3.4 × 108 cfu/ml) was added to PBS containing 10% deer meat extract that had been confirmed to be free from Sarcocystis spp. as well as STEC and the solutions were diluted serially. The DNA in these dilutions was extracted using a QIAamp DNA mini kit (QIAGEN). In STEC, all serial dilutions of extracted DNA were used for this assay and conventional PCR. In Sarcocystis spp., dilutions from 3.6 × 105 to 3.6 × 101 bradizoids/ml were used for both assays. As a negative control, 10% deer meat extract alone in PBS was used. As a positive control for conventional PCR, 3.4 × 108 cfu/ml in PBS for STEC or a cyst of Sarcocystis spp. with 3.6 × 106 bradizoids/ml (data not shown) in PBS was used.
As shown in Fig. 2, the visual LOD of bradizoids in this assay was determined to be 3.6 × 103 bradizoids/ml (Fig. 2A), which was 100 times more efficient than that of conventional PCR (Fig. 2B). The negative control showed no visual signals. The difference in the sensitivity seemed to be due to the amplification efficiency and detection procedure.
Fig. 2.
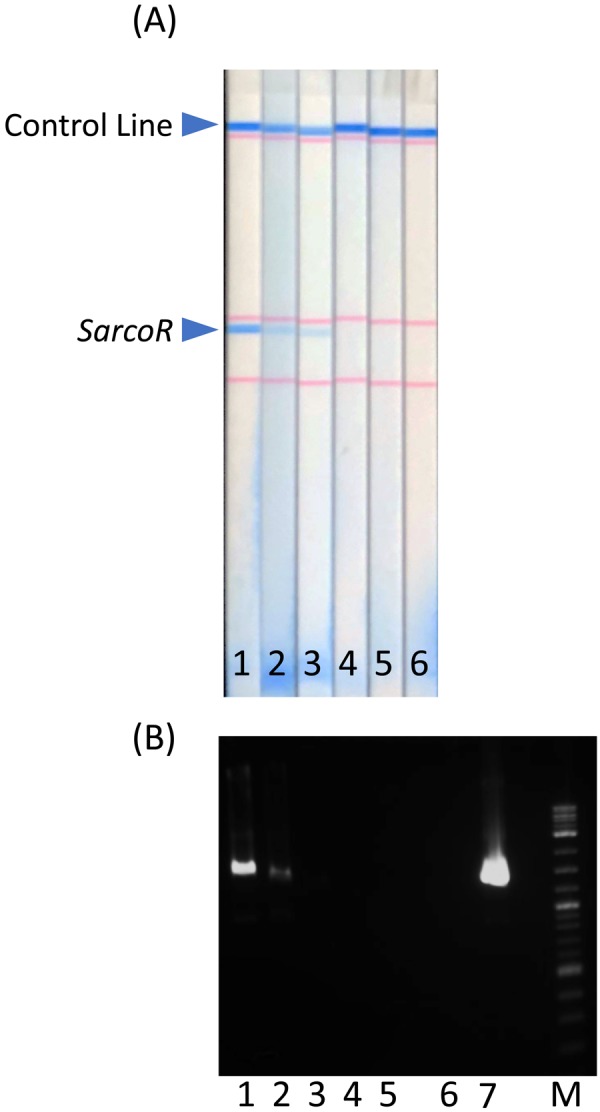
Sensitivity of (A) the DNA strip and (B) conventional PCR forthe detection of the 18S rRNA gene of Sarcocystis spp. 1: 3.6 × 105 bradizoids/ml in 10% deer meat extract, 2: 3.6 × 104 bradizoids/ml in 10% deer meat extract, 3: 3.6 × 103 bradizoids/ml in 10% deer meat extract, 4: 3.6 × 102 bradizoids/ml in 10% deer meat extract, 5: 3.6 × 101 bradizoids/ml in 10% deer meat extract, 6: negative sample (10% deer meat extract only), 7: positive sample (Sarcocyst) in PBS. M: molecular marker, SarcoR: Sarcocystis spp.18S rRNA gene.
The LODs of the stx1 and stx2 genes with the DNA strip were 3.4 × 104 and 3.4 × 103 cfu/ml of STEC, respectively (Fig. 3A). In contrast, the LODs of these genes with conventional PCR were 3.4 × 106 cfu/ml of STEC for both targets (Fig. 3B). The results revealed that the DNA strip showed 100- and 1000-fold greater sensitivity for the stx1 and stx2 genes, respectively, than conventional PCR. Our previous study revealed that the LAMP assay showed increased sensitivity for the detection of the stx1 and stx2 genes from STEC O157 compared with real-time PCR [26]. The difference in sensitivity is likely related to the high performance of the LAMP procedure in the DNA strip.
Fig. 3.
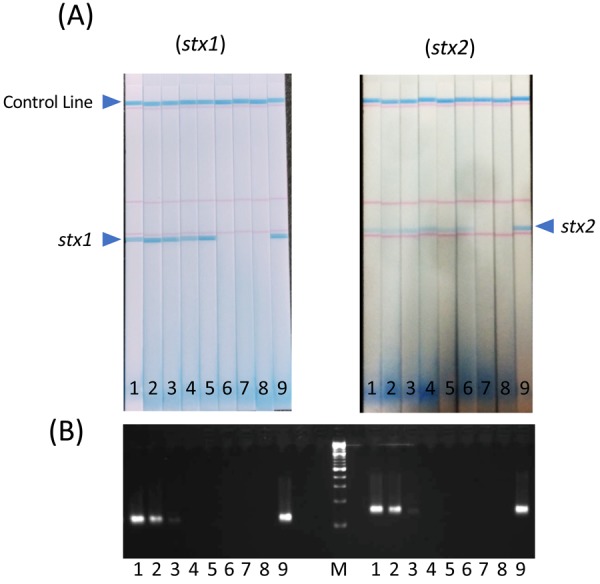
Sensitivity of (A) the DNA strip and (B) conventional PCR for the detection of stx1 and stx2 genes. 1: 3.4 × 108 CFU/ml in 10% deer meat extract, 2: 3.4 × 107 CFU/ml in 10% deer meat extract, 3: 3.4 × 106 CFU/ml in 10% deer meat extract, 4: 3.4 × 105 CFU/ml in 10% deer meat extract, 5: 3.4 × 104 CFU/ml in 10% deer meat extract, 6: 3.4 × 103 CFU/ml in 10% deer meat extract, 7: 3.4 × 102 CFU/ml in 10% deer meat extract, 8: negative sample (10% deer meat extract only), 9: positive sample (3.4 × 108/ml in PBS), M: molecular marker.
The specificity of the DNA strip for each hazard was examined. To validate the specificity for stx1 and stx2 genes in STEC, DNA extracts obtained from genera non-Shiga toxin-producing E. coli and other pathogenic bacteria were used. To investigate the specificity of Sarcocystis spp. in Japanese sika deer, DNA extracts obtained from the closely related genera (Toxoplasma gondii, Theileria parva) and Sarcocystis fayeri were used. As shown in Table 2, none of the bacterial DNA extracts used in this experiment showed cross reactions, suggesting that the DNA strip had high sensitivity for the stx1 and stx2 genes in STEC. However, the DNA strip showed cross-reactions for T. gondii and S. fayeri, but not T. parva, demonstrating that our primer reacted with the 18S rRNA of T. gondii and S. fayeri because the 18S rRNA of these parasites were similar. And T. gondii has been reported to infect in Japanese sika deer [4] and cause food poisoning [16]. The results also suggested that this DNA strip might be applicable to the detection of Sarcocystis spp. in other animals, including wild deer and T. gondii.
Table 2. The specificity of DNA strip for detection of Sarcocystis spp. and STEC in Japanese sika deer.
| Subject of DNA strip | Strain | Species | Cross reaction |
|---|---|---|---|
| STEC | PE7 | EAECa) | Negative |
| HP1001 | EPECb) | Negative | |
| WHO1 | ETECc) | Negative | |
| NIHS_00214 | STEC O157: H7 stx (−) | Negative | |
| NIHS_00069 | Salmonella Enteritidis | Negative | |
| ATCC8739 | Commensal E. coli | Negative | |
| ATCC43864 | Citrobacter freundii | Negative | |
| ATCC10145 | Pseudomonas aeruginosa | Negative | |
| Lm0132 | Listeria monocytogenes | Negative | |
| Sarcocystis spp. | AFSS-0002 | Sarcocystis fayeri | Positive |
| RH strain (NIHS) | Toxoplasma gondii | Positive | |
| AV-Tp001 | Theileria parva | Negative | |
a) EAEC: enteroaggregative Escherichia coli, b) EPEC: enteropathogenic Escherichia coli, c) ETEC: enterotoxigenic Escherichia coli.
Finally, we assessed the prevalence of Sarcocystis spp. and STEC in a total of 47 samples of meat from deer that had been slaughtered and processed in Yamanashi Prefecture, Japan using the DNA strip in combination with the conventional PCR approach. Toxoplasma gondii in serum was examined for 36 samples out of 47 samples using commercial ELISA kit (ID Screen® Toxoplasmosis Indirect Multi-species, ID.Vet, Grabels, France). As shown in Table 3, 32 samples were positive for conventional PCR while 37 samples were positive for the DNA strip, and T. gondii was negative in all samples assayed. Therefore it is considered that the positive samples in this study were contaminated with Sarcocystis spp. predominantly. Since the negative samples detected by the DNA strip were also negative on conventional PCR, we concluded that the DNA strip was able to detect Sarcocystis spp. from deer meat more efficiently than conventional PCR. Parallel experiments revealed that no STEC were detected in either assay. We previously reported that STEC OUT:H25 was isolated from only 1 out of 120 venison samples [2]. This suggests that the prevalence rates of STEC in deer meat might be less than roughly 1–2%.
Table 3. Comparison of conventional PCR and DNA strip for detection of Sarcocyctis spp., Toxoplasma gondii and STEC in Japanese sika deer.
| Sample No. |
Sex | Sarcocystis spp. | Toxoplasma gondii | STEC | ||
|---|---|---|---|---|---|---|
| Conventional PCR | DNA strip | ELISA | Conventional PCR | DNA strip | ||
| 1 | M | - | - | NT | - | - |
| 2 | F | - | - | NT | - | - |
| 3 | M | - | - | NT | - | - |
| 4 | M | - | - | NT | - | - |
| 5 | F | - | - | NT | - | - |
| 6 | M | - | + | NT | - | - |
| 7 | M | - | - | NT | - | - |
| 8 | F | + | + | NT | - | - |
| 9 | F | + | + | - | - | - |
| 10 | F | + | + | - | - | - |
| 11 | F | - | - | - | - | - |
| 12 | M | - | - | - | - | - |
| 13 | F | + | + | - | - | - |
| 14 | F | + | + | NT | - | - |
| 15 | F | + | + | - | - | - |
| 16 | M | + | + | - | - | - |
| 17 | F | + | + | - | - | - |
| 18 | F | + | + | - | - | - |
| 19 | M | + | + | - | - | - |
| 20 | F | + | + | - | - | - |
| 21 | M | + | + | - | - | - |
| 22 | M | + | + | - | - | - |
| 23 | M | + | + | - | - | - |
| 24 | M | - | + | - | - | - |
| 25 | M | - | + | - | - | - |
| 26 | F | + | + | - | - | - |
| 27 | F | + | + | - | - | - |
| 28 | F | + | + | - | - | - |
| 29 | F | + | + | - | - | - |
| 30 | F | + | + | - | - | - |
| 31 | F | - | + | - | - | - |
| 32 | F | + | + | - | - | - |
| 33 | F | - | - | - | - | - |
| 34 | F | + | + | - | - | - |
| 35 | F | + | + | - | - | - |
| 36 | M | + | + | - | - | - |
| 37 | F | + | + | - | - | - |
| 38 | F | + | + | - | - | - |
| 39 | M | + | + | - | - | - |
| 40 | F | - | + | - | - | - |
| 41 | unknown | + | + | - | - | - |
| 42 | M | + | + | - | - | - |
| 43 | F | - | - | - | - | - |
| 44 | M | + | + | - | - | - |
| 45 | unknown | + | + | - | - | - |
| 46 | unknown | + | + | NT | - | - |
| 47 | unknown | + | + | NT | - | - |
| Total number of positive sample | 32 | 37 | 0 | 0 | 0 | |
NT, not tested; +, positive; -, negative.
In slaughterhouses for game meat in Japan, self-sanitation systems are needed to ensure excellent food hygiene, as laws concerning slaughter are not applied to these meats. Therefore on-site sanitary check systems should be required. Since the DNA strip method is rapid (within 60 min after DNA extraction), simple, and requires no special equipment, it will prove a promising method for on-site investigations in slaughterhouses.
Game meat, including deer meat, carries many different risk factors for food poisoning. Sarcocystis spp. is one such novel intrinsic risk factor for food poisoning [10]. We previously reported the presence of at least four species of Sarcocystis spp. in Japanese sika deer meat samples obtained from Yamanashi Prefecture, Japan [9], and designed our primers based on information in the 18S rRNA region of the predominant Sarcocystis spp., among the 21 reported species. The results of cross-reaction experiment suggested that DNA strip for Sarcocystis spp. could be applied to other species of deer [8, 22] and T. gondii. However, whether or not the stx1 and stx2 gene primers used in this study collectively detect a series of stx variants is unclear. As deer-originating STEC can show variations in their stx genotypes [3, 5, 7, 11, 13], further studies will be required to clarify the above issues and expand the application of this detection method.
Despite these issues, the DNA strip approach developed in this study has several advantages, including its rapidity, multiplicity, and no need for special equipment, over other detection methods, such as conventional PCR, immunochromatography, and real-time PCR. Of further note, LAMP-lateral flow combined assays have also been recently developed for the detection of several other pathogenic microbes, such as Toxoplasma [17] and Staphylococcus aureus [25, 27].
In conclusion, our study showed that the “DNA strip” was able to detect Sarcocystis spp. and STEC in deer meat with high sensitivity. However, the improvement for the specificity of DNA strips is required to achieve specific reaction of the pathogens. Nevertheless, the application of this system will enable the quick and simple on-site inspection of food poisoning factors in deer meat.
Acknowledgments
This study was financially supported in part by grants from the Ministry of Health, Labour and Welfare, Japan (H27-shokuhin-ippan-011, H30-shokuhin-ippan-004). We thank Drs. Yasuyuki Morishima, Kisaburo Nagamune, Hiromu Sugiyama (National Institute of Infectious Disease), and Fujiko Sunaga (Azabu University) for providing the DNA extracts of the parasites.
REFERENCES
- 1.Aoki K., Ishikawa K., Hayashi K., Saito M., Sugita-Konishi Y., Watanabe M., Kamata Y.2013. An outbreak of suspected food poisoning related to deer meat containing Sarcocystis cysts. Jpn. J. Food Microbiol. 30: 28–32. [Google Scholar]
- 2.Asakura H., Kawase J., Ikeda T., Honda M., Sasaki Y., Uema M., Kabeya H., Sugiyama H., Igimi S., Takai S.2017. Microbiological quality assessment of game meats at retail in Japan. J. Food Prot. 80: 2119–2126. [DOI] [PubMed] [Google Scholar]
- 3.Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K.1998. Detection and genetical characterization of Shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 42: 815–822 . [DOI] [PubMed] [Google Scholar]
- 4.Cong W., Qin S. Y., Meng Q. F., Zou F. C., Qian A. D., Zhu X. Q.2016. Molecular detection and genetic characterization of Toxoplasma gondii infection in sika deer (Cervus nippon) in China. Infect. Genet. Evol. 39: 9–11 . [DOI] [PubMed] [Google Scholar]
- 5.Díaz-Sánchez S., Sánchez S., Herrera-León S., Porrero C., Blanco J., Dahbi G., Blanco J. E., Mora A., Mateo R., Hanning I., Vidal D.2013. Prevalence of Shiga toxin-producing Escherichia coli, Salmonella spp. and Campylobacter spp. in large game animals intended for consumption: relationship with management practices and livestock influence. Vet. Microbiol. 163: 274–281 . [DOI] [PubMed] [Google Scholar]
- 6.Dong H. J., Cho A. R., Hahn T. W., Cho S.2014. Development of a multiplex loop-mediated isothermal amplification assay to detect shiga toxin-producing Escherichia coli in cattle. J. Vet. Sci. 15: 317–325 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggert M., Stüber E., Heurich M., Fredriksson-Ahomaa M., Burgos Y., Beutin L., Märtlbauer E.2013. Detection and characterization of Shiga toxin-producing Escherichia coli in faeces and lymphatic tissue of free-ranging deer. Epidemiol. Infect. 141: 251–259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gjerde B., Vikøren T., Hamnes I. S.2017. Molecular identification of Sarcocystis halieti n. sp., Sarcocystis lari and Sarcocystis truncata in the intestine of a white-tailed sea eagle (Haliaeetus albicilla) in Norway. Int. J. Parasitol. Parasites Wildl. 7: 1–11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda M., Sawaya M., Taira K., Yamazaki A., Kamata Y., Shimizu H., Kobayashi N., Sakata R., Asakura H., Sugita-Konishi Y.2018. Effects of temperature, pH and curing on the viability of Sarcocystis, a Japanese sika deer (Cervus Nippon centralis) parasite, and the inactivation of their diarrheal toxin. J. Vet. Med. Sci. 80: 1337–1344 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irikura D., Saito M., Sugita-Konishi Y., Ohnishi T., Sugiyama K. I., Watanabe M., Yamazaki A., Izumiyama S., Sato H., Kimura Y., Doi R., Kamata Y.2017. Characterization of Sarcocystis fayeri’s actin-depolymerizing factor as a toxin that causes diarrhea. Genes Cells 22: 825–835 . [DOI] [PubMed] [Google Scholar]
- 11.Ishii S., Meyer K. P., Sadowsky M. J.2007. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl. Environ. Microbiol. 73: 5703–5710 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Japan Ministry of Environments 2014. Protection and control of wild birds and mammals and hunting management act. http://elaws.e-gov.go.jp/search/elawsSearch/elaws_search/lsg0500/detail?lawId=414AC0000000088&openerCode=1 [accessed on July 1, 2018].
- 13.Kabeya H., Sato S., Oda S., Kawamura M., Nagasaka M., Kuranaga M., Yokoyama E., Hirai S., Iguchi A., Ishihara T., Kuroki T., Morita-Ishihara T., Iyoda S., Terajima J., Ohnishi M., Maruyama S.2017. Characterization of Shiga toxin-producing Escherichia coli from feces of sika deer (Cervus nippon) in Japan using PCR binary typing analysis to evaluate their potential human pathogenicity. J. Vet. Med. Sci. 79: 834–841 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamata Y., Saito M., Irikura D., Yahata Y., Ohnishi T., Bessho T., Inui T., Watanabe M., Sugita-Konishi Y.2014. A toxin isolated from Sarcocystis fayeri in raw horsemeat may be responsible for food poisoning. J. Food Prot. 77: 814–819 . [DOI] [PubMed] [Google Scholar]
- 15.Keene W. E., Sazie E., Kok J., Rice D. H., Hancock D. D., Balan V. K., Zhao T., Doyle M. P.1997. An outbreak of Escherichia coli O157:H7 infections traced to jerky made from deer meat. JAMA 277: 1229–1231 . [DOI] [PubMed] [Google Scholar]
- 16.Kim C. S., Kim D. S., Jung H. R.2019. Toxoplasma lymphadenitis caused by ingestion of raw blood and meat of deer in a 10-year-old boy. Pediatr. Neonatol. 60: 112–113 . [DOI] [PubMed] [Google Scholar]
- 17.Lalle M., Possenti A., Dubey J. P., Pozio E.2018. Loop-mediated isothermal amplification-lateral-flow dipstick (LAMP-LFD) to detect toxoplasma gondii oocyst in ready-to-eat salad. Food Microbiol. 70: 137–142 . [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Fan P., Zhou S., Zhang L.2017. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 107: 54–61 . [DOI] [PubMed] [Google Scholar]
- 19.Nielsen E. M., Andersen M. T.2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41: 2884–2893 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pritt B., Trainer T., Simmons-Arnold L., Evans M., Dunams D., Rosenthal B. M.2008. Detection of sarcocystis parasites in retail beef: a regional survey combining histological and genetic detection methods. J. Food Prot. 71: 2144–2147 . [DOI] [PubMed] [Google Scholar]
- 21.Priyanka B., Patil R. K., Dwarakanath S.2016. A review on detection methods used for foodborne pathogens. Indian J. Med. Res. 144: 327–338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudaitytė-Lukošienė E., Prakas P., Butkauskas D., Kutkienė L., Vepštaitė-Monstavičė I., Servienė E.2018. Morphological and molecular identification of Sarcocystis spp. from the sika deer (Cervus nippon), including two new species Sarcocystis frondea and Sarcocystis nipponi. Parasitol. Res. 117: 1305–1315 . [DOI] [PubMed] [Google Scholar]
- 23.Takasaki K., Yamakoshi Y., Futo S.2018. Single-laboratory validation of rapid and easy DNA strip for porcine DNA detection in beef meatballs. J. AOAC Int. 101: 1653–1656 . [DOI] [PubMed] [Google Scholar]
- 24.Tei S., Kitajima N., Takahashi K., Mishiro S.2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362: 371–373 . [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Li H., Wang Y., Zhang L., Xu J., Ye C.2017. Loop-mediated isothermal amplification label-based gold nanoparticles lateral flow biosensor for detection of Enterococcus faecalis and Staphylococcus aureus. Front. Microbiol. 8: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamazaki A., Honda M., Kobayashi N., Ishizaki N., Asakura H., Sugita-Konishi Y.2018. The sensitivity of commercial kits in detecting the genes of pathogenic bacteria in venison. J. Vet. Med. Sci. 80: 706–709 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin H. Y., Fang T. J., Wen H. W.2016. Combined multiplex loop-mediated isothermal amplification with lateral flow assay to detect sea and seb genes of enterotoxic Staphylococcus aureus. Lett. Appl. Microbiol. 63: 16–24 . [DOI] [PubMed] [Google Scholar]
- 28.Zhao X., Lin C. W., Wang J., Oh D. H.2014. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 24: 297–312 . [DOI] [PubMed] [Google Scholar]


