Abstract
Alcohol use disorder (AUD) screening frequently involves questionnaires complemented by laboratory work to monitor alcohol use and/or evaluate AUD-associated complications. Here, we suggest that measuring aldehyde-induced DNA and protein adducts produced during alcohol metabolism may allow an earlier detection of AUD and AUD-associated complications compared with existing biomarkers. Using aldehyde-induced adducts to monitor AUD may also be important when considering that approximately 540 million people bear a genetic variant of aldehyde dehydrogenase 2 (ALDH2), predisposing this population to aldehyde-induced toxicity with alcohol use. We posit that measuring aldehyde-induced adducts may provide a means to improve precision medicine approaches, taking into account lifestyle choices and genetics to evaluate AUD and AUD-associated complications.
Keywords: reactive aldehydes, alcohol use disorder, acetaldehyde, alcohol, 4-hydroxynonenal, biomarker
Alcohol use disorder and risky alcohol use: a global problem
An estimated ~16 million individuals in the USA have an AUD [1]. The complications caused by alcohol use also produce an annual economic burden of ~250 billion and AUD constitutes the fourth leading cause of preventable death in the United States [2, 3]. Just as concerning is that a recent analysis of a national survey from the United States conducted in 2012–2013, compared to the results from this survey in 2001–2002, highlights that alcohol use, high risk drinking, and AUD are steadily increasing across all socioeconomic groups [4].
In East Asia, alcohol use is also on the rise and the Republic of Korea has the highest annual alcohol consumption per capita among all countries within the Asia Pacific region (12.3L) [5]. this is concerning as nearly 540 million people of East Asian descent are carriers of the aldehyde dehydrogenase 2 (ALDH2) genetic variant, ALDH2*2, and cannot metabolize acetaldehyde (the metabolite of alcohol) efficiently. The resultant accumulation of acetaldehyde leads to an increased risk for developing alcohol-induced complications such as head and neck cancers, including esophageal cancer, relative to individuals with the ALDH2*1 genetic variant [6].
Aldehydes, including acetaldehyde, possess an electrophilic carbon that reacts with nucleophilic groups in DNA or protein resulting in adduct formation [7]. Here, we propose that measuring aldehyde-induced adducts may be used to complement existing alcohol biomarkers for the earlier detection of AUD and AUD-related complications. This may allow for prompt intervention and could be exploited to achieve a precision medicine approach to diagnosing AUD at an early stage. In this article, we discuss the biochemistry of aldehyde-induced adduct formation and delineate recent studies measuring aldehyde-induced adducts to identify AUD and AUD-related complications.
Alcohol use disorder screening
AUD is determined by a DSM-V diagnosis, and is on a spectrum of alcohol consumption (that also includes risky use) known as unhealthy alcohol use. To receive a diagnosis of AUD, individuals must meet at least two of 11 outlined DSM-V criteria for AUD during a 12-month period [8].
To complement the AUD diagnosis, acute alcohol concentrations can be detected by breath or blood tests and are commonly used by law enforcement officials to monitor for acute alcohol intoxication. Further, monitoring for urine ethanol biomarkers including ethyl glucuronide (EtG) and ethyl sulfate (EtS) can detect alcohol consumption up to ~80 hours after use [9]. Although these biomarkers can detect ethanol use, they do not provide a means to identify AUD or the extent of AUD-associated complications. In turn, biomarkers such as circulating liver enzymes, red blood cell volume, and transferrin can gauge the damage caused by alcohol consumption (see Box 1). However, these biomarkers are only elevated after extensive cellular damage occurs and are not a means to formally identify AUD [75].
Box 1. Current Alcohol Use Disorder Biomarkers.
Acute consumption of alcohol can be detected by a blood or breath test. In addition, urine biomarkers including EtG and EtS can be used to detect whether alcohol was used within 80 hours. Further, blood work to monitor AUDs can involve the measurement of circulating liver enzymes (aspartate aminotransferase [AST], alanine aminotransferase [ALT], and γ-glutamyl transferase [GGT]), in addition to red blood cell mean corpuscular volume (MCV) and carbohydrate-deficient transferrin (CDT) [71]. Chronic alcohol abuse is suggested if blood work measures an AST:ALT ratio of at least 2:1 and an increased MCV [72]. Recent moderate alcohol consumption is indicated by an increased GGT and CDT. In turn, CDT levels are also used to monitor abstinence from alcohol [72] The sensitivity and specificity of these biomarkers are imperfect and frequently lower than required for diagnostic purposes [71]. The expression of these biomarkers may vary depending on the amount and pattern of alcohol consumption in addition to gender, age, weight, and the existence of other diseases [73].
The elevated concentration of these biomarkers is primarily caused by alcohol-induced organ injury [75]. The elevation of AST, ALT, and GGT can be secondary to damage of hepatocytes, or for GGT, biliary tract damage that occurs in alcoholic liver disease [75]. Increased MCV in alcohol consumption is caused by direct toxicity to the bone marrow in addition to folic acid deficiency or impaired B12 absorption associated with alcoholism [72]. CDT elevation following alcohol consumption are independent of liver disease severity and caused by transient changes in the glycosylation pattern of transferrin [74].
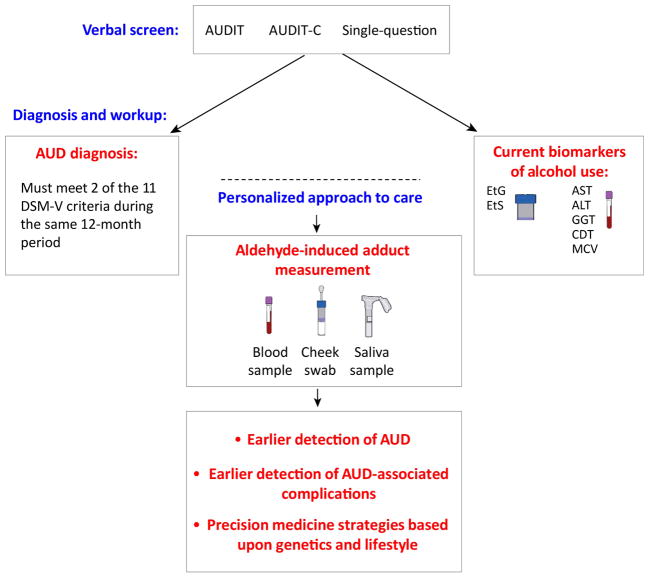
Since some AUD screening tools are based upon an evaluation of the number of standard drinks consumed (such as the AUDIT, shortened AUDIT-C, or single-question screen) [10–12], assessing aldehyde-induced adducts, particularly for those with a deficiency in aldehyde metabolism (which can lead to an accumulation of more acetaldehyde for each standard drink consumed), could provide valuable information in regards to the carcinogenic effects of acetaldehyde on the individual. This monitoring could potentially be used to fill in a void in identifying AUD and AUD-associated complications earlier in order to provide more timely interventions (Figure 1).
Figure 1. Proposed Scheme to incorporate the measurement of aldehyde-induced adducts in clinical practice.
Following a positive verbal screen, we propose practitioners can formally diagnose AUD using both criteria outlined in the DSM-V (which includes meeting two of 11 DSM-V criteria) and measurement of aldehyde-induced adducts. Aldehyde-induced protein and DNA adducts can be collected by a blood sample, saliva sample, or a cheek swab, and the sample can be analyzed by mass spectroscopy or an immunoassay such as ELISA for quantification. Adhering to current patient guidelines, standard biomarkers of alcohol use, including AST, ALT, GGT, CDT, and red blood cell MCV may be tested concurrently. ALT = alanine aminotransferase, AST = aspartate aminotransferase, AUDIT = Alcohol Use Disorders Identification Test, CDT = carbohydrate deficient transferrin, ELISA = enzyme-linked immunosorbent assay, GGT = gamma-glutamyl transferase, MCV = mean corpuscular volume.
DNA and protein adduct formation during alcohol consumption
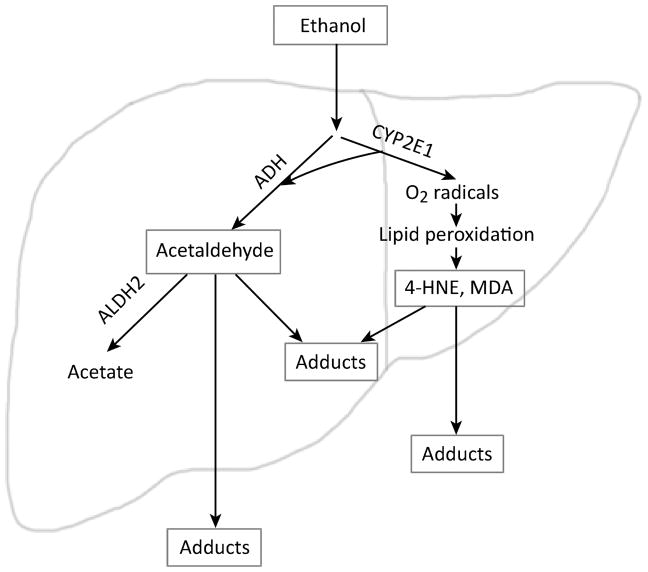
Alcohol metabolism in humans occurs through oxidation reactions in the liver primarily by alcohol dehydrogenase (ADH) and aldehyde dehydrogenase 2 (ALDH2) (Figure 2). Metabolism of ethanol to acetaldehyde also occurs through an alternative pathway by cytochrome P4502E1 (CYP2E1), which is activated either when ADH becomes saturated or by chronic alcohol consumption [13, 14]. During alcohol metabolism, CYP2E1 induction by ethanol is a predominant source of reactive oxygen species (ROS). The ROS produced causes lipid peroxidation of the cell membrane which forms toxic reactive aldehydes including malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) [15].
Figure 2. Production of aldehyde-induced adducts following alcohol consumption in humans.
The enzyme alcohol dehydrogenase converts ethanol to the highly reactive intermediate acetaldehyde. Acetaldehyde is then converted by aldehyde dehydrogenase 2 to the nontoxic molecule acetate. Alternatively, CYP2E1 metabolizes alcohol when ADH is saturated and is induced by chronic alcohol consumption. Acetaldehyde is highly reactive and can form complexes with protein or DNA known as adducts. CYP2E1 also generates acetaldehyde from ethanol, and its induction is a major source of oxygen radicals that can react with lipids in the cell to form the reactive aldehydes 4-hydroxynonenal (4-HNE) and malondialdehyde (MDA). 4-HNE and MDA also lead to DNA and protein adducts. Acetaldehyde, 4-HNE, and MDA can easily diffuse through cell membranes, forming aldehyde-induced adducts in the blood or other tissues.
The reaction forming an aldehyde-induced adduct occurs by either a Michael addition or a Schiff base. In the Michael addition, the β-carbon of the aldehyde reacts with the nucleophilic group to form a double bond. For a Schiff base, the carbonyl carbon of the aldehyde reacts with a DNA amine group or lysine residue [7]. Several aldehydes, including acetaldehyde and MDA, can also react with DNA or protein together to generate hybrid adducts; yielding a product such as an MDA-acetaldehyde (MAA) adduct [16].
Aldehydes preferentially form DNA adducts at the deoxyguanosine amino group, and less frequently at deoxyadenosine and deoxycytocine amino groups [76]. By causing DNA damage, aldehyde-induced DNA adducts can promote carcinogenesis through transversion. Under cellular stress, DNA can become oxidized and form 8-hydroxydeoxyguanosine (8-OH-dG) adducts which can occur with alcohol consumption. More specific to alcohol is the aldehyde-induced DNA adduct N2-ethylidene-deoxyguanosine (N2-ethylidene-dG). However, since N2-ethylidene-dG is highly unstable, this adduct is frequently measured by quantifying a stabilized and reduced form of N2-ethylidene-deoxyguanosine, N2-ethyl-2′-deoxyguanosine (N2-ethyl-dG) [77]. Acetaldehyde can also form crotonaldheyde by the condensation of two acetaldehyde molecules which can produce the DNA adduct -methyl-γ-OH-propano-deoxyguanosine; also known as crotonaldehyde-derived propano-deoxyguanosine (Cr-PdG) [78]. Additional adducts such as malondialdehyde-deoxyguanosine and 4-hydroxynonenal-deoxyguanosine are also formed.
For proteins, aldehydes form adducts primarily at lysine, histidine, and cysteine amino acids and protein function may be altered particularly when aldehydes bind to a protein at a critical location [18, 28, 79]. One role of aldehyde-induced adducts are to function as an auto-feedback mechanism to limit reactive aldehyde formation. This occurs when aldehyde-induced protein adducts form on cytochrome CYP2E1 and function as an auto-feedback mechanism to reduce aldehyde accumulation. This limits the CYP2E1-dependent metabolism of alcohol which can produce reactive aldehydes [17]. Conversely, reactive aldehydes can reduce the enzymatic activity of ALDH2 (for example, 50μM of 4-HNE in vitro) [18]. Although more detailed studies are needed, this auto-feedback mechanism inhibiting ALDH2 -- which likely increases acetaldehyde levels -- may cause a behavioral aversion to consuming additional alcohol. This seems likely given that the ALDH2 inhibitor disulfiram, an alcohol aversion therapy agent, leads to the avoidance of alcohol by causing nausea and vomiting with alcohol consumption.
Relevant to this discussion, several other aldehyde-induced protein adducts from alcohol consumption have been reported. [16, 17, 19–39] (table 1, Key Table). We discuss some of these examples in the following sections.
Table 1 Key Table.
(References cited in Table: 84–90)
| Adducted Protein | Reactive aldehyde used or studied to determine adduct formation | Protein Source | Proposed Pathophysiologic implications |
|---|---|---|---|
| α-tubulin | [14C]Acetaldehyde | Rodent Liver | Impaired microtubule formation [28, 84] |
| 4-HNE | Bovine brain | ||
| Collagen type I and type III | Acetaldehyde (using an antibody) | Human liver biopsies of alcoholic and non-alcoholic subjects | Inflammation; fibrosis [20, 29] |
| [14C]Acetaldehyde | Calf skin | ||
| ALDH2 | 4-HNE | Human recombinant ALDH2 | Possible negative feedback mechanism to facilitate alcohol aversion [26] |
| AMPK | 4-HNE | Recombinant AMPK, HepG2 cells, and liver from rodents fed an alcohol or non-alcohol diet | Regulation of β-oxidation[85] |
| Calmodulin | [14C] Acetaldehyde | Bovine brain | May Impair calcium homeostasis [30] |
| Cytochrome C oxidase | 4-HNE | Liver from rodents fed an alcohol or non-alcohol diet | Modulate electron transport chain [27, 86] |
| MDA | Liver from rodents fed an alcohol or non-alcohol diet | ||
| CYP2E1 | Acetaldehyde | Liver from rodents fed an alcohol or non-alcohol diet | May limit aldehyde accumulation [17] |
| Electron transfer flavoprotein α | 4-HNE (using an antibody) | Liver from rodents fed an alcohol or non-alcohol diet | Altered metabolism of fatty acyl-CoA[67] |
| ERK1/2 | 4-HNE | Rodent primary hepatocyte culture, liver from rodents fed an alcohol or non-alcohol diet | Inhibit prosurvival and proliferation pathways [87] |
| Glutathione S-transferase | Acetaldehyde | Rodent Liver | Limit capacity to handle cellular stress [31] |
| GRP78 | 4-HNE | Liver from rodents fed an alcohol or non-alcohol diet, human recombinant GRP78 | Limit capacity to handle cellular stress[88] |
| Ketosteroid reductase | Acetaldehyde (using an antibody) | Liver from rodents fed an alcohol or non-alcohol diet | Possible abnormal bile synthesis [21] |
| Surfactant Protein D | MAA (using an antibody) | Rodent Lung | May Interfere with surfactant production or function; inflammation [32] |
| Albumin | Acetaldehyde (using an antibody) | Human serum from subjects with alcohol versus non-alcohol history | Possible Immune-mediated pathology [19, 89] |
| [14C] Acetaldehyde | Bovine Serum | ||
| Carbonic anhydrase II | Acetaldehyde | Human recombinant protein, Human blood samples | Possible link to osteoporosis or renal tubular acidosis [33] |
| Coagulation factors I, II, IIa, VII, X, Xa | Acetaldehyde | Reconstituted factors | Increased clotting time [34–36, 90] |
| Coagulation factor IX | Acetaldehyde | Reconstituted factor | Decreased clotting time [34] |
| Hemoglobin | Acetaldehyde (using an antibody) | Human blood collected from subjects with alcohol versus non-alcohol history | May decrease oxygen binding; RBC vulnerability to hemolysis [23, 37, 38] |
| Acetaldehyde | Human blood | ||
| VLDL and LDL | Acetaldehyde (using an antibody) | Human plasma collected from subjects with alcohol versus non-alcohol history | Possible accelerated clearance of VLDL and LDL, decreased conversion of VLDL to LDL [22] |
Described are the types of aldehyde-induced protein adducts that form following alcohol consumption or exposure to ethanol-derived reactive aldehydes, the reactive aldehydes causing an adduct, the source of the studied adduct, and documented associated pathophysiology based on the aldehyde-induced biomarkers. LDL, low-density lipoprotein; RBCs, red blood cells; VLDL, very-low-density lipoprotein.
Monitoring aldehyde-induced adducts and influence of the ALDH2*2 genetic variant
Individuals with an ALDH2*2 genetic variant tend to limit alcohol consumption due to the unpleasant side effects of acetaldehyde accumulation, including elevated heart rate and facial flushing [40]. Although the ALDH2*2 genotype is considered to curb alcohol consumption, there are concerning trends that the number of heterozygotes for the ALDH2*2 genetic variant with AUD are steadily rising in East Asian countries [41, 42].
Heterozygotes for the ALDH2*2 variant who consume alcohol are associated with an increased risk for developing head and neck related cancers, including esophageal cancer, compared to individuals with the ALDH2*1 variant [41]. To support this association of aldehyde-induced adducts as a possible mechanism for developing esophageal cancer, N2-ethylidene-dG concentrations were measured in the esophagus of Aldh2 knockout mice, and were linked to ~100-fold greater level of DNA damage in the esophagus following 8 weeks of alcohol consumption relative to wild type controls under the same alcohol regimen [43]. Consistent with these data, in human esophageal keratinocytes, a ~15 fold increase in N2-ethylidene-dG concentrations were observed upon siRNA knockdown of ALDH2 relative to untreated human keratinocytes in vitro [43].
To support feasibility measuring N2-ethylidene-dG in humans, after 5 male and 5 female participants without AUD consumed alcohol (to achieve a target blood alcohol level of 0.03%), the DNA adduct N2-ethylidene-dG was measured in cells collected from the upper digestive tract with a saline wash and from cell types collected from blood [44, 45]. The highest level of N2-ethylidene-dG following alcohol consumption was present in the upper digestive track and levels of N2-ethylidene-dG decreased for samples collected further away from the upper digestive track [46]. In addition, heterozygote ALDH2*2 individuals with AUD could exhibit significant elevations of N2-ethyl-dG in DNA isolated from blood samples compared to ALDH2*1 individuals with AUD [47]. Consumption of a standard drink also resulted in higher concentrations of acetaldehyde-induced hemoglobin adduct formation in ALDH2*2 heterozygotes compared with ALDH2*1 individuals [48]. Together, these data suggested that in humans, it is possible to survey for alcohol-induced DNA damage and specifically monitor individual cell types within the body for the presence of aldehyde-induced protein adducts.
With alcohol abstinence, recent evidence would also imply that the DNA damage caused by alcohol consumption might be reversible. Individuals with AUD who abstained from alcohol reduced the amount of alcohol-induced cellular dysplasia identified by esophagoduodenoscopy [49]. Taken together, these data suggest developing strategies to measure acetaldehyde-induced DNA adducts at or near the esophagus or within the blood; this may prove to be a valuable approach for detecting potential risks of developing esophageal cancer and determining how to prevent advanced stages of development, although extensive testing will be needed.
Individuals with an ALDH2*2 variant might receive lower scores on an AUDIT or AUDIT-C questionnaire since, as discussed, smaller quantities of alcohol produce intoxicating effects in individuals heterozygotic for ALDH2*2 compared to ALDH2*1. Therefore, surveys based upon the number of standard drinks consumed may not accurately screen for AUD in these individuals. We propose that it might be more informative to combine written screening tools with aldehyde-induced adduct quantification in order to determine a threshold of alcohol consumption that is harmful. Although this remains to be tested, we hypothesize that alcohol consumption levels might be evaluated to assess the genetic predisposition of individuals to metabolize acetaldehyde at a personalized level. This approach might be potentially useful to set the stage for establishing a precision medicine platform to diagnose and treat individuals with AUD.
Aldehyde-induced adduct biomarkers to detect specific AUD related complications
Several additional examples exist of how aldehyde-induced biomarkers might be useful to detect and monitor specific AUD complications such as alcohol-induced cardiomyopathy, malignancy, and liver disease [50, 52, 53, 80]. The mechanistic role of aldehyde production and aldehyde metabolism in alcoholic cardiomyopathy has been recently reviewed [50]. New mechanistic insights on alcoholic cardiomyopathy may encourage the development of tools that might specifically detect alcohol-induced cardiomyopathy to correlate with disease progression. Below we briefly discuss the implications that such putative biomarkers may have for the detection of alcohol-induced liver disease and/or various malignancies.
Alcohol-induced liver disease
Aldehyde-induced adducts contribute to the initial development and later stages of alcohol-induced liver disease [51–53]. For instance, in rats receiving a diet containing ethanol, acetaldehyde, 4-HNE, and MDA adducts co-localize with perivenous lipid deposits, indicative of the earliest lesion observed in alcohol-induced liver disease, known as steatosis [51, 52]. A similar co-localization of acetaldehyde-induced adducts and steatosis has been reported in alcoholic individuals with no clinical signs or laboratory tests suggesting the presence of liver disease [53]. Moreover, in non-alcoholics, acetaldehyde-induced adducts have not been identified in liver biopsies [60].
In humans with advanced stages of liver disease, aldehyde-induced adducts are localized in hepatic stellate cells associated with fibrosis and cirrhosis, as well as in myofibroblasts in liver regions with fibrotic bridging [54]. Aldehyde hybrid adducts can additionally stimulate the secretion/production of fibronectin in vitro by hepatic stellate cells, as well as inflammatory cytokines and adhesion molecules from endothelial cells [55]. These findings are relevant because hybrid adducts can cause increased development of scar tissue and fibrosis compared to individual adducts [56]. In addition, aldehyde-induced adducts can directly activate immune cells, Kupffer cells, and endothelial cells to produce profibrogenic mediators [81]. Consequently, the ability to monitor specific aldehyde-induced protein cellular adducts such as these may provide a basis to specifically monitor the progression of liver disease.
Autoimmunity to aldehyde-induced adducts might also be implicated in the pathogenesis of alcoholic liver disease. To support this notion, IgA, IgM, and IgG antibodies directed against acetaldehyde-induced protein adducts have been documented in the serum of chronicalcoholics compared with non-alcoholics [57]. In patients with alcoholic liver disease, antiadduct IgA titers significantly correlated with the combined clinical and laboratory index of liver disease severity [57]. IgG antibodies reacting with MDA-, 4HNE-, and MAA-induced adducts were also significantly increased in alcoholic patients with alcohol-induced hepatitis or cirrhosis compared with alcoholics without liver damage, patients with nonalcoholic liver disease, and healthy controls. The titers measured in alcoholic patients with alcohol-induced hepatitis or cirrhosis also correlated with the severity of liver damage [58, 59]. Although further studies are warranted, these studies taken together suggest that antibodies against aldehyde-induced protein adducts might have potential as putative biomarkers to stratify liver disease severity in AUD.
Alcohol-induced malignancies
AUD has been associated with an increased risk for developing cancers of the head and neck, gastrointestinal tract, breast, and liver [49, 82, 83] with aldehyde-induced adducts contributing to DNA damage that can promote carcinogenesis [15, 60, 61]. Alcohol consumption associated with developing hepatocellular carcinoma is caused by transversion of p53 at codon 249 (by a G to T transversion). 4-HNE is known to form DNA adducts on deoxyguanosine, and when 4-HNE is directly applied to wild type p53 TK-6 lymphoblastoid cells, 4-HNE causes an increased transversion of p53 at codon 249 [62, 63]. Therefore, aldehyde-induced formation of DNA adducts can potentially lead to a transversion that leads to a higher risk for carcinogenesis. Specifically quantifying acetaldehyde-induced adducts may be a powerful method for earlier detection and cancer screening.
Recent Promising techniques for detecting acetaldehyde and aldehyde-induced adducts
Although techniques to measure aldehyde-induced DNA and protein adducts have existed for several decades, new technology for assessing aldehyde-induced adducts can now be exploited to improve upon existing biomarkers, in addition to developing novel candidate biomarkers to detect AUD and AUD-associated complications. Here, we discuss these novel methods including probes to quantify aldehydes using live cell imaging techniques, the advances made in detecting DNA-induced aldehyde adducts, and the use of mass spectrometry to further identify proteins modified by aldehyde-induced adducts.
Live cell detection of aldehydes
One challenge in quantifying reactive aldehydes is that with the techniques available, the sample requires processing to evaluate aldehyde-induced adducts. However, quite recently a reporter probe was developed by using dark hydrazone fluorescence labeling which can quantify alkyl aldehydes (such as acetaldehyde and 4-HNE) in live cells. This study illustrated the ability to measure dose-dependent changes in alkyl aldehyde levels in HeLa cells by both fluorescence imaging and flow cytometry [64]. Additionally, a hydrazinyl naphthalimide fluorescent probe was recently developed that can monitor aldehyde load within lung epithelial cells exposed to ethanol [65]. Overall, the recent reports of fluorescent dyes to monitor aldehydes in live cells, not presently available until now, provide new valuable research tools to potentially study AUD and AUD-associated complications in cellular systems.
DNA adduct detection
Recently, an effective method of quantifying N2-ethyl-dG was developed using liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) with hydrophobic interaction chromatography (HILIC) in order to improve the ionization efficiency of detecting N2-ethyl-dG. Although further corroborating evidence is needed, using HILIC increased the mass spectrometry signal intensity 97-fold when compared to using reversed phase chromatography [66]. This is encouraging since a LC-ESI-MS/MS system may allow for quantification of N2-ethyl-dG without requiring additional tools such as a nano-electrospray interface in order to detect N2-ethyl-dG; potentially making N2-ethyl-dG quantification easier and more feasible [66].
Protein adduct detection
Since a number of proteins that form aldehyde-induced protein adducts were identified over a decade ago, detecting new and novel targets with existing technology for AUD and AUD-associated complications may be possible. Potentially, antibodies which detect aldehyde-induced protein adducts, such as MDA and 4-HNE, can be used in combination with mass spectrometry to understand specifically how aldehyde-induced adducts of the proteome are altered when exposed to alcohol. This approach recently identified in rat liver mitochondria fractions, several proteins harboring 4-HNE-induced adducts [67]. In particular, electron transfer flavoprotein alpha was identified to exhibit significant enhancement of 4-HNE-induced adducts in rats fed a Lieber-DeCarli ethanol diet for 5 weeks when compared to rats not receiving alcohol [67]. Further application of this and similar methods may uncover a subset of the proteome and/or novel putative protein targets previously unrecognized to being modified by aldheyde-induced adducts. Presumably, these might also be used as biomarkers for AUD and AUD-associated complications.
Limitations to Quantifying Aldehyde-induced Adducts
One potential challenge in implementing aldehyde-induced adducts to monitor AUD and AUD-associated complications is that other endogenous and exogenous sources of acetaldehyde other than alcohol upon consumption may exist. Although acetaldehyde is produced by bacteria within the gastrointestinal tract, the levels of alcohol-induced acetaldehyde adducts -- particularly DNA adducts -- are present at sufficient concentrations to be detected above the natural biological processes of bacteria; this is based on studies measuring DNA adducts within epithelial cells and in blood samples [45, 48]. However, if additional biomarkers are developed, the utility of these could be potentially limited if changes in aldehyde-induced adducts caused by alcohol consumption are subtle when compared to natural biological processes producing aldehydes.
Exogenous sources of aldehyde exposure, such as tobacco cigarettes, could also be difficult to differentiate from aldehyde-induced adducts produced by alcohol l[32, 68]. However, it is important to recognize that acetaldehyde levels measured in the saliva are 7 fold higher when smoking tobacco cigarettes with alcohol when compared to alcohol consumption alone [68]. This interplay between lifestyle choices and genetics is also eloquently supported by a small clinical study reporting the odds ratio of developing esophageal cancer as the highest among people drinking alcohol, smoking tobacco cigarettes, and presenting the ALDH2*2 genotype [69].
Overall, although measuring acetaldehyde-induced adducts may not be specific to a process which only occurs with alcohol consumption, these measurements may provide a valuable tool to defining a level of risky alcohol use that may be relevant to developing potential AUD-associated complications when factoring in additional lifestyle choices, such as cigarette smoking.
Although such methods of aldehyde-induced adduct detection may not be easily applied within communities of varied socio-economic status, developing such biomarker tools may allow for stratifications of patient populations based on lifestyle and genetics. These in turn, might provide general recommendations for patients which cannot receive testing to assess aldehyde-induced adducts. Therefore, although biomarkers of aldehyde-induced adducts are not intended to replace existing verbal screening tools and urine EtG and EtS quantification, they might potentially provide an added dimension to evaluating AUD and AUD-associated complications.
Concluding Remarks
We posit that aldehyde-induced adducts may prove to be promising biomarkers for AUD and AUD-associated complications. Additional research is still required to validate these findings and to fully develop these candidate biomarkers for possible clinical use (see “Outstanding Questions and Box 2). Since adduct quantification can correlate with the amount of alcohol consumed both acutely and chronically, aldehyde-induced adducts might be exploited to better characterize the degree of unhealthy alcohol use (risky use or AUD), and presumably, the severity of AUD. Additionally, the concept of DNA and protein adduct detection might also be extended as a possible useful tool to monitor other substance abuse disorders, such as cocaine [70].
Outstanding Questions Box.
How does the aldehyde-induced DNA adduct, N2-ethylidene-2′-dG, specifically contribute to human diseases caused by alcohol consumption?
Following alcohol consumption, what is the time course of adduct formation in biospecimens and which may involve different proteins? How does that compare to the time course of DNA adducts isolated from different cell lines?
How will blood test, saliva sample, or cheek swab results differ in the quantification of aldehyde-induced adducts? Is one test best suited to detect the concentration of alcohol that can lead to AUD or the risk for developing specific alcohol-related diseases?
Box 2. Clinician’s Corner.
Alcohol consumption, especially 12-month alcohol use, high-risk drinking, and alcohol use disorder (AUD), are on the rise in the United States when compared to a decade ago.
Following alcohol consumption, reactive aldehydes are produced which modify DNA and protein. These modifications are called aldehyde-induced adducts and can lead to cellular damage that can potentially result in alcohol-induced complications such as cancer or cardiomyopathy.
People of East Asian descent carry a genetic variant in the enzyme aldehyde dehydrogenase 2 (ALDH2) known as ALDH2*2. The ALDH2*2 variant severely limits aldehyde metabolism after alcohol consumption and results in facial flushing and tachycardia. Frequent alcohol consumption by people heterozygous for the ALDH2*2 variant is associated with an increased risk of developing head and neck cancers, including esophageal cancer. Monitoring aldehyde-induced adducts may be a means to develop more precise care for this particular patient population.
In the future, monitoring the presence of aldehyde-induced adducts in patient biospecimens may complement questionnaires and biomarkers for AUD and AUD-associated complications. This could ultimately improve management strategies, stratify risk for alcohol-related diseases, and allow for timely interventions for AUD and AUD-associated complications.
In the era of precision medicine, aldehyde-induced adducts combined with written questionnaires might provide more detailed tools to help evaluate AUD and AUD-associated complications. in the future, It will be exciting to see if quantification of aldehyde-induced adducts by a blood test, saliva sample, or a cheek swab might allow for an earlier detection and possible intervention for AUD and AUD-associated complications.
HIghlights.
During alcohol consumption, reactive aldehydes are formed which can damage DNA and proteins. Recent evidence suggests aldehyde-induced DNA adducts formed following human alcohol consumption are detectable in epithelial cells from the upper digestive tract and from DNA in blood samples.
Biomarker tools to monitor aldehyde-induced adducts are particularly relevant since~560 million people worldwide cannot efficiently metabolize reactive aldehydes. These individuals are more susceptible to AUD-associated complications even though they may consume less alcohol.
Measuring aldehyde-induced adducts may serve as a promising biomarker to advance precision medicine for AUD by allowing for earlier detection and more precise management strategies for AUD and AUD-associated complications.
Acknowledgments
This work was supported by NIH GM119522 (ERG), a California TRDRP high impact pilot research award (ERG), and a FAER medical student anesthesia research fellowship (HH).
Glossary
- The Alcohol Use Disorders Identification Test (AUDIT)
A 10-item clinician-administered or self-reported screening tool that utilizes the concept of a standard drink to screen for alcohol consumption, drinking behavior, and alcohol-related problems.
- The Alcohol Use Disorders Identification Test (AUDIT-C)
A modified 3-item version of the 10-item AUDIT.
- Adduct
A complex that forms when a chemical reacts with a cellular macromolecule such DNA or protein.
- Alcohol-induced liver disease
Alcohol abuse leads to liver disease pathology that progresses from steatosis to steatohepatitis, fibrosis, cirrhosis which leads to end-stage liver disease.
- Aldehyde dehydrogenase 2 (ALDH2)
A mitochondrial enzyme encoded on chromosome 12q24 that detoxifies and removes acetaldehyde and other reactive aldehydes.
- ALDH2*1
The wild type allele coding for ALDH2 which metabolizes acetaldehyde.
- ALDH2*2
The East Asian variant coding for ALDH2, caused by a single point mutation of guanine to adenine which decreases the ability to metabolize reactive aldehydes by 60–90% when compared to those carriers of the ALDH2*1 gene.
- Cirrhosis
A late stage of progressive liver fibrosis, generally considered irreversible.
- Cytochrome P4502E1 (CYP2E1)
enzyme responsible for metabolizing several molecules including ethanol to acetaldehyde when ALDH2 is saturated. CYP2E1 activity generates ROS that cause aldehyde formation through lipid peroxidation.
- DSM-V
The diagnostic and Statistical Manual of Mental Disorders (DSM, fifth edition) published by the American Psychiatric Association is an outline of the standard criteria for the classification of mental disorders.
- Fibrosis
The formation of excess fibrous connective tissue, representing (in this case) the liver’s response to injury.
- Kupffer cells
group of resident macrophages accounting for 20% of non-parenchymal cells in the liver. These cells are responsible for clearing toxins, microorganisms, and cell debris.
- Hepatic Stellate Cells
Liver pericytes involved in regulating the turnover of extracellular matrix. In response to cytokines produced in chronic injury, stellate cells differentiate into myofibroblasts.
- Hybrid Adducts
Several aldehydes, in particular acetaldehyde and malondialdehyde, can react together with DNA or protein to form these mixed compounds.
- Lipid Peroxidation
Initiated by ROS, the oxidative degradation of fatty acids in the cell plasma membrane results in a series of autocatalytic reactions. During the process, a variety of small molecules are produced including reactive aldehydes.
- Myofibroblasts
Derived from stellate cells, myofibroblasts have contractive, proinflammatory, and fibrogenetic properties, and are a key mediators of liver fibrosis.
- Precision medicine
Medical care that takes into account individual variability in genes, environment, and lifestyle.
- Sarcolemma
The membrane surrounding each striated muscle fiber cell.
- Single-question screen
one-question screen for alcohol use which asks how many times in the past year an individual has had ≥X drinks in a day (X=4 for women, 5 for men).
- Standard Drink
constitutes approximately 14 grams of alcohol, equivalent to 12 ounces of beer, 5 ounces of wine, or 1.5 ounces of 80 proof spirits.
- Steatosis
The abnormal infiltration of liver cells with fat. Also known as fatty liver, steatosis comprises the earliest stage of alcoholic liver disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.SubstanceAbuseandMentalHealthServicesAdministration Key Substance Use and Mental Health Indicators in the United States: Results from the 2016 National Survey on Drug Use and Health (HHS Publication No.SMA17-5044, NSDUH SeriesH52), Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration. 2017 https://www.samhsa.gov/data/
- 2.Centers for Disease Control and Prevention (CDC) Alcohol and Public Health: Alcohol-Related Disease Impact (ARDI) Average for United States 2006–2010 – Alcohol-Attributable Deaths Due to Excessive Alcohol Use, CDC.
- 3.Sacks JJ, et al. 2010 National and State Costs of Excessive Alcohol Consumption. Am J Prev Med. 2015;49(5):e73–9. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 4.Grant BF, et al. Prevalence of 12-Month Alcohol Use, High-Risk Drinking, and DSM-IV Alcohol Use Disorder in the United States, 2001–2002 to 2012–2013: Results From the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry. 2017;74(9):911–923. doi: 10.1001/jamapsychiatry.2017.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monzavi SM, et al. Alcohol Related Disorders in Asia Pacific Region: Prevalence, Health Consequences and Impacts on the Nations. Asia Pacific Journal of Medical Toxicology. 2015;4:1–8. [Google Scholar]
- 6.Brooks PJ, et al. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6(3):e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esterbauer H, et al. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 8.APA . Diagnostic and Statistical Manual of Mental Disorders. 5th edition. American Psychiatric Association; 2013. [Google Scholar]
- 9.Helander A, et al. Detection times for urinary ethyl glucuronide and ethyl sulfate in heavy drinkers during alcohol detoxification. Alcohol Alcohol. 2009;44:55–61. doi: 10.1093/alcalc/agn084. [DOI] [PubMed] [Google Scholar]
- 10.Babor TF, et al. The Alcohol Use Disorders Identification Test: Guidelines for Use in Primary Care. World Health Organization; 2010. [Google Scholar]
- 11.Bush K, et al. The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med. 1998;158:1789–1795. doi: 10.1001/archinte.158.16.1789. [DOI] [PubMed] [Google Scholar]
- 12.Smith PC, et al. Primary care validation of a singlequestion alcohol screening test. J Gen Intern Med. 2009;24:783–788. doi: 10.1007/s11606-009-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieber CS, DeCarli LM, et al. Ethanol oxidation by hepatic microsomes: adaptive increase after ethanol feeding. Science. 1968;162:917–918. doi: 10.1126/science.162.3856.917. [DOI] [PubMed] [Google Scholar]
- 14.Lieber CS, DeCarli, et al. Hepatic microsomal ethanol-oxidizing system. In vitro characteristics and adaptive properties in vivo. J Biol Chem. 1970;245:2505–2512. [PubMed] [Google Scholar]
- 15.Wang Y, et al. Ethanol-induced cytochrome P4502E1 causes carcinogenic etheno-DNA lesions in alcoholic liver disease. Hepatology. 2009;50:453–461. doi: 10.1002/hep.22978. [DOI] [PubMed] [Google Scholar]
- 16.Tuma DJ, et al. Acetaldehyde and malondialdehyde react together to generate distinct protein adducts in the liver during long-term ethanol administration. Hepatology. 1996;23:872–880. doi: 10.1002/hep.510230431. [DOI] [PubMed] [Google Scholar]
- 17.Behrens UJ, et al. Formation of acetaldehyde adducts with ethanol-inducible P450IIE1 in vivo. Biochem Biophys Res Commun. 1998;154:584–590. doi: 10.1016/0006-291x(88)90180-5. [DOI] [PubMed] [Google Scholar]
- 18.Doorn JA, Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem Biol Interact. 2003;143–144:93–100. doi: 10.1016/s0009-2797(02)00178-3. [DOI] [PubMed] [Google Scholar]
- 19.Romanazzi V, et al. Immune response to acetaldehydehuman serum albumin adduct among healthy subjects related to alcohol intake. Environ Toxicol Pharmacol. 2013;36:378–383. doi: 10.1016/j.etap.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Svegliati-Baroni G, et al. Collagen-acetaldehyde adducts in alcoholic and nonalcoholic liver diseases. Hepatology. 1994;20:111–118. doi: 10.1016/0270-9139(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. Identification of the 37-kd rat liver protein that forms an acetaldehyde adduct in vivo as delta 4-3-ketosteroid 5 beta-reductase. Hepatology. 1996;23:115–122. doi: 10.1002/hep.510230116. [DOI] [PubMed] [Google Scholar]
- 22.Wehr H, et al. Acetaldehyde adducts and autoantibodies against VLDL and LDL in alcoholics. J Lipid Res. 1993;34:1237–1244. [PubMed] [Google Scholar]
- 23.Koivisto H, et al. Long-term ethanol consumption and macrocytosis: diagnostic and pathogenic implications. J Lab Clin Med. 2006;147:191–196. doi: 10.1016/j.lab.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Yoon Y, et al. Ethanol-induced alterations of the microtubule cytoskeleton in hepatocytes. Am J Physiol. 1998;274:G757–G766. doi: 10.1152/ajpgi.1998.274.4.G757. [DOI] [PubMed] [Google Scholar]
- 25.Garro AJ, et al. The effects of chronic ethanol consumption on carcinogen metabolism and on O6-methylguanine transferase-mediated repair of alkylated DNA. Alcohol Clin Exp Res. 1986;10:73S–77S. doi: 10.1111/j.1530-0277.1986.tb05184.x. [DOI] [PubMed] [Google Scholar]
- 26.Doorn JA, et al. Inhibition of human mitochondrial aldehyde dehydrogenase by 4-hydroxynon-2-enal and 4-oxonon-2-enal. Chem Res Toxicol. 2006;19:102–110. doi: 10.1021/tx0501839. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, et al. Formation of malondialdehyde adducts in livers of rats exposed to ethanol: role in ethanol-mediated inhibition of cytochrome c oxidase. Alcohol Clin Exp Res. 2000;24:544–552. [PubMed] [Google Scholar]
- 28.Stewart BJ, et al. Residue-specific adduction of tubulin by 4-hydroxynonenal and 4-oxononenal causes cross-linking and inhibits polymerization. Chem Res Toxicol. 2007;20:1111–1119. doi: 10.1021/tx700106v. [DOI] [PubMed] [Google Scholar]
- 29.Jukkola A, Niemela O. Covalent binding of acetaldehyde to type III collagen. Biochem Biophys Res Commun. 1989;159:163–169. doi: 10.1016/0006-291x(89)92418-2. [DOI] [PubMed] [Google Scholar]
- 30.Jennett RB, et al. Increased covalent binding of acetaldehyde to calmodulin in the presence of calcium. Life Sci. 1989;45:1461–1466. doi: 10.1016/0024-3205(89)90036-2. [DOI] [PubMed] [Google Scholar]
- 31.Sultana R, et al. Formation of acetaldehyde adducts of glutathione S-transferase A3 in the liver of rats administered alcohol chronically. Alcohol. 2005;35:57–66. doi: 10.1016/j.alcohol.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 32.McCaskill ML, et al. Hybrid malondialdehyde and acetaldehyde protein adducts form in the lungs of mice exposed to alcohol and cigarette smoke. Alcohol Clin Exp Res. 2011;35:1106–1113. doi: 10.1111/j.1530-0277.2011.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bootorabi F, et al. Modification of carbonic anhydrase II with acetaldehyde, the first metabolite of ethanol, leads to decreased enzyme activity. BMC Biochem. 2008;9:32. doi: 10.1186/1471-2091-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabol DA, et al. Coagulation protein function VII: diametric effects of acetaldehyde on factor VII and factor IX function. Dig Dis Sci. 1999;44:2564–2567. doi: 10.1023/a:1026615928562. [DOI] [PubMed] [Google Scholar]
- 35.Basista MH, et al. Acetaldehyde alters coagulation protein function. Dig Dis Sci. 1994;39:2421–2425. doi: 10.1007/BF02087660. [DOI] [PubMed] [Google Scholar]
- 36.Brecher AS, et al. Coagulation protein function. IV. Effect of acetaldehyde upon factor X and factor Xa, the proteins at the gateway to the common coagulation pathway. Alcohol. 1996;13:539–545. doi: 10.1016/s0741-8329(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 37.De Benedetto GE, Fanigliulo M. A new CE-ESI-MS method for the detection of stable hemoglobin acetaldehyde adducts, potential biomarkers of alcohol abuse. Electrophoresis. 2009;30:1798–1807. doi: 10.1002/elps.200800379. [DOI] [PubMed] [Google Scholar]
- 38.Tsuboi KK, et al. Acetaldehyde-dependent changes in hemoglobin and oxygen affinity of human erythrocytes. Hemoglobin. 1981;5:241–250. doi: 10.3109/03630268108997548. [DOI] [PubMed] [Google Scholar]
- 39.Hill GE, et al. Association of malondialdehyde-acetaldehyde (MAA) adducted proteins with atherosclerotic-induced vascular inflammatory injury. Atherosclerosis. 1998;141:107–116. doi: 10.1016/s0021-9150(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 40.Gross ER, et al. A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu Rev Pharmacol Toxicol. 2015;55:107–127. doi: 10.1146/annurev-pharmtox-010814-124915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoyama A, et al. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383–1387. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 42.Chang JS, et al. ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. J Biomed Sci. 2017;24:19. doi: 10.1186/s12929-017-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amanuma Y, et al. Protective role of ALDH2 against acetaldehyde-derived DNA damage in oesophageal squamous epithelium. Sci Rep. 2015;5:14142. doi: 10.1038/srep14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balbo S, et al. Time course of DNA adduct formation in peripheral blood granulocytes and lymphocytes after drinking alcohol. Mutagenesis. 2012;27:485–490. doi: 10.1093/mutage/ges008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balbo S, et al. Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol Biomarkers Prev. 2012;21:601–608. doi: 10.1158/1055-9965.EPI-11-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balbo S, et al. Increased levels of the acetaldehyde-derived DNA adduct N2-ethyldeoxyguanosine in oral mucosa DNA from rhesus monkeys exposed to alcohol. Mutagenesis. 2016;31:553–558. doi: 10.1093/mutage/gew016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuda T, et al. Increased DNA damage in ALDH2-deficient alcoholics. Chem Res Toxicol. 2006;19:1374–1378. doi: 10.1021/tx060113h. [DOI] [PubMed] [Google Scholar]
- 48.Takeshita T, Morimoto K. Accumulation of hemoglobin-associated acetaldehyde with habitual alcohol drinking in the atypical ALDH2 genotype. Alcohol Clin Exp Res. 2000;24:1–7. [PubMed] [Google Scholar]
- 49.Katada C, et al. Alcohol consumption and multiple dysplastic lesions increase risk of squamous cell carcinoma in the esophagus, head, and neck. Gastroenterology. 2016;151:860–869. e7. doi: 10.1053/j.gastro.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Ren J. ALDH2 in alcoholic heart diseases: molecular mechanism and clinical implications. Pharmacol Ther. 2011;132:86–95. doi: 10.1016/j.pharmthera.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sampey BP, et al. Immunohistochemical characterization of hepatic malondialdehyde and 4-hydroxynonenal modified proteins during early stages of ethanol-induced liver injury. Alcohol Clin Exp Res. 2003;27:1015–1022. doi: 10.1097/01.ALC.0000071928.16732.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li CJ, et al. Acetaldehyde-modified and 4-hydroxynonenal-modified proteins in the livers of rats with alcoholic liver disease. Hepatology. 1997;26:650–657. doi: 10.1002/hep.510260317. [DOI] [PubMed] [Google Scholar]
- 53.Holstege A, et al. Acetaldehyde-modified epitopes in liver biopsy specimens of alcoholic and nonalcoholic patients: localization and association with progression of liver fibrosis. Hepatology. 1994;19:367–374. [PubMed] [Google Scholar]
- 54.Paradis V, et al. Cellular and subcellular localization of acetaldehyde-protein adducts in liver biopsies from alcoholic patients. J Histochem Cytochem. 1996;44:1051–1057. doi: 10.1177/44.9.8773571. [DOI] [PubMed] [Google Scholar]
- 55.Thiele GM, et al. Rat sinusoidal liver endothelial cells (SECs) produce pro-fibrotic factors in response to adducts formed from the metabolites of ethanol. Biochem Pharmacol. 2005;70:1593–1600. doi: 10.1016/j.bcp.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Tuma DJ. Role of malondialdehyde-acetaldehyde adducts in liver injury. Free Radic Biol Med. 2002;32:303–308. doi: 10.1016/s0891-5849(01)00742-0. [DOI] [PubMed] [Google Scholar]
- 57.Viitala K, et al. Serum IgA, IgG, and IgM antibodies directed against acetaldehyde-derived epitopes: relationship to liver disease severity and alcohol consumption. Hepatology. 1997;25:1418–1424. doi: 10.1002/hep.510250619. [DOI] [PubMed] [Google Scholar]
- 58.Rolla R, et al. Detection of circulating antibodies against malondialdehyde-acetaldehyde adducts in patients with alcohol-induced liver disease. Hepatology. 2000;31:878–884. doi: 10.1053/he.2000.5373. [DOI] [PubMed] [Google Scholar]
- 59.Mottaran E, et al. Lipid peroxidation contributes to immune reactions associated with alcoholic liver disease. Free Radic Biol Med. 2002;32:38–45. doi: 10.1016/s0891-5849(01)00757-2. [DOI] [PubMed] [Google Scholar]
- 60.Brooks PJ, Theruvathu JA. DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol. 2005;35:187–193. doi: 10.1016/j.alcohol.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 61.Linhart K, et al. The role of reactive oxygen species (ROS) and cytochrome P-450 2E1 in the generation of carcinogenic etheno-DNA adducts. Redox Biol. 2014;3:56–62. doi: 10.1016/j.redox.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balbo S, Brooks PJ. Implications of acetaldehydederived DNA adducts for understanding alcohol-related carcinogenesis. Adv Exp Med Biol. 2015;815:71–88. doi: 10.1007/978-3-319-09614-8_5. [DOI] [PubMed] [Google Scholar]
- 63.Hussain SP, et al. Increased p53 mutation load in nontumorous human liver of Wilson disease and hemochromatosis: oxyradical overload diseases. Proc Natl Acad Sci U S A. 2000;97:12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuen LH, et al. Dark Hydrazone Fluorescence Labeling Agents Enable Imaging of Cellular Aldehydic Load. ACS Chem Biol. 2016;11:2312–2319. doi: 10.1021/acschembio.6b00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reeves AG, et al. Imaging Acetaldehyde Formation During Ethanol Metabolism in Living Cells using a Hydrazinyl Naphthalimide Fluorescent Probe. Anal Methods. 2017;9:3418–3421. doi: 10.1039/C7AY01238A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murakami H, et al. Progress in a selective method for the determination of the acetaldehyde-derived DNA adducts by using HILIC-ESI-MS/MS. Talanta. 2018;177:12–17. doi: 10.1016/j.talanta.2017.09.055. [DOI] [PubMed] [Google Scholar]
- 67.Andringa KK, et al. Proteomic analysis of 4-hydroxynonenal (4-HNE) modified proteins in liver mitochondria from chronic ethanol-fed rats. Redox Biol. 2014;2:1038–1047. doi: 10.1016/j.redox.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salaspuro V, Salaspuro M. Synergistic effect of alcohol drinking and smoking on in vivo acetaldehyde concentration in saliva. Int J Cancer. 2004;111:480–483. doi: 10.1002/ijc.20293. [DOI] [PubMed] [Google Scholar]
- 69.Cui R, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137:1768–1775. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 70.Ndikum-Moffor FM, Roberts SM. Cocaine-protein targets in mouse liver. Biochem Pharmacol. 2003;66:105–113. doi: 10.1016/s0006-2952(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 71.Litten RZ, et al. Alcohol biomarkers in applied settings: recent advances and future research opportunities. Alcohol Clin Exp Res. 2010;34:955–967. doi: 10.1111/j.1530-0277.2010.01170.x. [DOI] [PubMed] [Google Scholar]
- 72.Conigrave KM, et al. Traditional markers of excessive alcohol use. Addiction. 2003;98(Suppl 2):31–43. doi: 10.1046/j.1359-6357.2003.00581.x. [DOI] [PubMed] [Google Scholar]
- 73.Niemela O. Biomarker-based approaches for assessing alcohol use disorders. Int J Environ Res Public Health. 2016;13:166. doi: 10.3390/ijerph13020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramskogler K, et al. CDT values are not influenced by epithelial cell apoptosis in chronic alcoholic patients – preliminary results. Alcohol Clin Exp Res. 2004;28:1396–1398. doi: 10.1097/01.alc.0000139818.68555.8a. [DOI] [PubMed] [Google Scholar]
- 75.Tavakoli HR, et al. Review of current clinical biomarkers for the detection of alcohol dependence. Innov Clin Neurosci. 2011;8:26–33. [PMC free article] [PubMed] [Google Scholar]
- 76.Vaca CE, et al. Studies of the reaction of acetaldehyde with deoxynucleosides. Chem Biol Interact. 1995;98:51–67. doi: 10.1016/0009-2797(95)03632-v. [DOI] [PubMed] [Google Scholar]
- 77.Wang M, et al. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem Res Toxicol. 2006;19:319–324. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcia CC, et al. [13C2]-Acetaldehyde promotes unequivocal formation of 1,N2-propano-20-deoxyguanosine in human cells. J Am Chem Soc. 2011;133:9140–9143. doi: 10.1021/ja2004686. [DOI] [PubMed] [Google Scholar]
- 79.Mauch TJ, et al. Covalent binding of acetaldehyde selectively inhibits the catalytic activity of lysine-dependent enzymes. Hepatology. 1986;6:263–269. doi: 10.1002/hep.1840060218. [DOI] [PubMed] [Google Scholar]
- 80.Munnia A, et al. Exocyclic malondialdehyde and aromatic DNA adducts in larynx tissues. Free Radic Biol Med. 2004;37:850–858. doi: 10.1016/j.freeradbiomed.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 81.Setshedi M, et al. Acetaldehyde adducts in alcoholic liver disease. Oxid Med Cell Longev. 2010;3:178–185. doi: 10.4161/oxim.3.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morgan TR, et al. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 83.McDonald JA, et al. Alcohol intake and breast cancer risk: weighing the overall evidence. Curr Breast Cancer Rep. 2013;5 doi: 10.1007/s12609-013-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jennett RB, et al. Preferential covalent binding of acetaldehyde to the alpha-chain of purified rat liver tubulin. Hepatology. 1989;9:57–62. doi: 10.1002/hep.1840090109. [DOI] [PubMed] [Google Scholar]
- 85.Shearn CT, et al. Identification of 50 AMP-activated kinase as a target of reactive aldehydes during chronic ingestion of high concentrations of ethanol. J Biol Chem. 2014;289:15449–15462. doi: 10.1074/jbc.M113.543942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen J, et al. Formation of 4-hydroxynonenal adducts with cytochrome c oxidase in rats following short-term ethanol intake. Hepatology. 1999;29:1792–1798. doi: 10.1002/hep.510290611. [DOI] [PubMed] [Google Scholar]
- 87.Sampey BP, et al. Ethanol-induced modulation of hepatocellular extracellular signal-regulated kinase-1/2 activity via 4-hydroxynonenal. J Biol Chem. 2007;282:1925–1937. doi: 10.1074/jbc.M610602200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Galligan JJ, et al. Oxidative stress-mediated aldehyde adduction of GRP78 in a mouse model of alcoholic liver disease: functional independence of ATPase activity and chaperone function. Free Radic Biol Med. 2014;73:411–420. doi: 10.1016/j.freeradbiomed.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Donohue TM, Jr, et al. Acetaldehyde adducts with proteins: binding of [14C]acetaldehyde to serum albumin. Arch Biochem Biophys. 1983;220:239–246. doi: 10.1016/0003-9861(83)90406-x. [DOI] [PubMed] [Google Scholar]
- 90.Brecher A, et al. Coagulation protein function. III. Effect of acetaldehyde upon the activation of prothrombin. Alcohol. 1996;13:423–429. doi: 10.1016/0741-8329(96)00025-0. [DOI] [PubMed] [Google Scholar]