Abstract
Purpose
With growing evidence that rare single gene disorders present in the neonatal period, there is a need for rapid, systematic, and comprehensive genomic diagnoses in ICUs to assist acute and long-term clinical decisions. This study aimed to identify genetic conditions in neonatal (NICU) and paediatric (PICU) intensive care populations.
Methods
We performed trio whole genome sequence (WGS) analysis on a prospective cohort of families recruited in NICU and PICU at a single site in the UK. We developed a research pipeline in collaboration with the National Health Service to deliver validated pertinent pathogenic findings within 2–3 weeks of recruitment.
Results
A total of 195 families had whole genome analysis performed (567 samples) and 21% received a molecular diagnosis for the underlying genetic condition in the child. The phenotypic description of the child was a poor predictor of the gene identified in 90% of cases, arguing for gene agnostic testing in NICU/PICU. The diagnosis affected clinical management in more than 65% of cases (83% in neonates) including modification of treatments and care pathways and/or informing palliative care decisions. A 2–3 week turnaround was sufficient to impact most clinical decision-making.
Conclusions
The use of WGS in intensively ill children is acceptable and trio analysis facilitates diagnoses. A gene agnostic approach was effective in identifying an underlying genetic condition, with phenotypes and symptomatology being primarily used for data interpretation rather than gene selection. WGS analysis has the potential to be a first-line diagnostic tool for a subset of intensively ill children.
Electronic supplementary material
The online version of this article (10.1007/s00134-019-05552-x) contains supplementary material, which is available to authorized users.
Keywords: Whole genome sequencing, Genetics, Genomics, Critically ill children, NICU, PICU
Take-home message
| Children in intensive care frequently have a rare underlying genetic condition. Rapid genetic testing reveals a diagnosis in ~20% of case, but the clinical presentation is often atypical. Greater than 65% of diagnoses resulted in modification of treatments or care pathways and a 2–3 week diagnostic pipeline was sufficient to impact most clinical decision making. |
Introduction
Given the life-threatening circumstances of children entering intensive care, effective management relies on comprehensive monitoring, data acquisition and rapid clinical responses. In 2016 in the UK, there were 636,401 deliveries in National Health Service (NHS) hospitals and 95,000 babies (15%) were admitted to neonatal intensive care post-delivery, remaining there for 4–93 days depending on gestational age [1, 2]. With growing evidence that rare diseases (defined as < 1 in 2000 individuals) are collectively relatively common (6% of the population) and that rare genetic variants underlie many congenital presentations, whole exome and genome sequencing is increasingly being used in this clinical setting [3–9].
Studies have shown that early molecular diagnoses improve outcomes and reduce healthcare costs [10–12]. Provisioning of a rapid but affordable genomic testing strategy within a national healthcare service in order to deliver equity of access is nevertheless challenging [13]. As with all intensive care procedures, balancing the extreme stress of families with the complexities of an informed consent requires skill and sensitivity while incomplete understanding of new technology creates uncertainty for care providers [14]. Lack of consensus on reporting uncertainty and additional findings [15, 16] requires careful consideration of genome-wide sequence analysis in the intensive care setting.
The objectives of this study were (1) to establish a whole genome sequencing (WGS) analysis pipeline in the intensive care context and deliver clinically relevant results in a timely manner, (2) determine the prevalence of underlying genetic conditions in NICU and PICU populations and whether diagnosis influences clinical management, (3) to investigate the correlation of clinical features and genotype in very young children and (4) to create a recallable resource of children with rare disease for long-term follow-up and research.
Methods
Recruitment and consent
Participants for this study were recruited through NHS Cambridge University Hospitals Foundation Trust under Cambridge South Research Ethics Committee approval 13/EE/0325. The neonatal and paediatric intensive care units have 40 and 13 beds respectively and have on average 896 and 639 admissions per year. The recruitment criteria were broad and inclusive for any cases with a possible single gene disorder (Supp. Table 1). In NICU, exclusion criteria were children admitted for short stay post-delivery surveillance, prematurity without additional features, babies with a clear antenatal or delivery history suggestive of a non-genetic cause and those babies where a genetic diagnosis was already made. Babies with congenital anomalies, neurological symptom including seizures, suspected metabolic disease, surgical necrotizing enterocolitis, extreme intrauterine growth retardation and unexplained critical illness of likely genetic etiology were all eligible for the study. In PICU, inclusion criteria were the same as for NICU but cases of clear trauma, malignancy and bronchiolitis or respiratory tract infections in an otherwise well child were ineligible.
For all consented children, the detailed clinical history and clinical features were extracted from the electronic medical records. These were converted into standardized terms for comparison using a human phenotype ontology term (HPO [17]) and a pedigree was completed with the parents at the time of consenting.
Whole genome sequence analysis
A minimum of 500 µL of fresh blood in EDTA was requested to yield a minimum of 1 µg of DNA for WGS. Extracted DNA samples were shipped to an external WGS sequencing facility (Illumina, UK). WGS (30–40× coverage for the nuclear genome and 800–1000× for the mitochondrial genome), quality control, read alignment to the human reference genome build 37, and variant calling were performed by Illumina as described [18]. Genome data has been deposited at the European Genome-phenome Archive (EGA) under accession number EGAD00001004357. Variant annotation and filtering were performed in house (see Supplementary Methods) [19–22]. No gene list was used if both parents were available but a filter for 3809 genes with potential early-onset presentation was used for singletons.
Multidisciplinary team assessment, clinical reporting and counselling
Consistent with NHS clinical genetics pathways, candidate variants were reviewed at a multidisciplinary team meeting (MDT), which included research bioinformatics analysts, clinical scientists, clinical geneticists, neonatal and paediatric intensivists, and neurologists as required. Additional genetic findings not related to the presenting complaint [16] were not assessed or reported as per the consent and genetic variants of uncertain significance (VUS) were not reported; however, VUS were curated for research and re-analysis. Pathogenic/likely pathogenic variants were confirmed by Sanger sequence analysis and a clinical report was issued according to the American College of Medical Genetics (ACMG) variant classification scheme. Results were communicated to the family by a geneticist or appropriate specialist and intensivists referred families to Clinical Genetics for further evaluation of all abnormal results.
Phenotypic comparison analysis
Pairwise similarity scores between clinical features using HPO term profiles (either proband-proband or proband-gene) were calculated using the R analysis package ontologyX and HPO release 2018-10-09 [17, 23]. The proband-proband scores were used to hierarchically cluster and categorise probands into phenotypic groups. Enrichment analysis of HPO terms was performed with Fisher’s exact test, FDR < 0.1. See Supplementary Methods for details.
Results
Recruitment of cohort
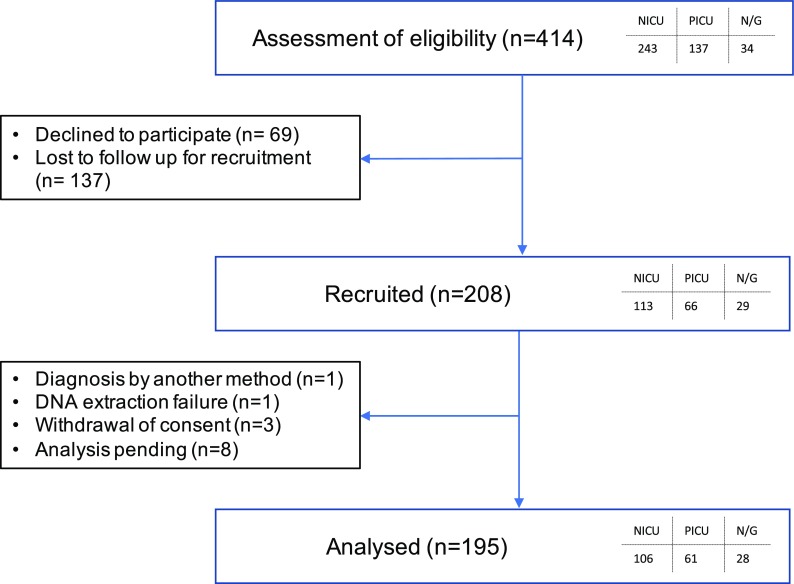
A total of 414 families were approached and offered WGS analysis between the period of December 2016 and September 2018 (Fig. 1), 380 were families with a child in the NICU or PICU and the rest were referred from other departments. In NICU, 113 families consented to join the study (47% of eligible cases, ~ 10% of all babies in the unit over the recruitment period). In PICU, 66 families consented to join the study (48% of eligible cases, ~ 8% of all children in the unit). Of the recruited families, 13 were not sequenced or were pending (see Fig. 1).
Fig. 1.
Recruitment summary of cohort. Families were recruited from the neonatal intensive care unit (NICU), paediatric intensive care unit (PICU) and paediatric neurology or clinical genetics department (N/G)
Recruitment criteria were broad and inclusive for any case with a possible undiagnosed single gene disorder (Supp. Table 1). In total, 208 families were recruited (50% of total eligible cases) and 69 families actively declined to take part (17%). The remainder were either lost to follow-up or remained undecided about participation by the time of discharge or repatriation to a more local hospital.
Of the 195 probands and their families that were analysed (567 genomes total), both parents were included in 90% of cases (Table 1). Most were recruited from the NICU (54%) and PICU (31%) (Table 1, Supp. Table 2). Twenty-eight probands (15%) were referred from paediatric neurology or clinical genetics. In the NICU, the ages of the probands at consent ranged from 1 day postnatal to 6 months old and the median age was 12 days postnatal (equivalent to a median corrected gestational age of + 4 days because 50% of the neonates were premature) (Supp. Fig. 1). For the others, the age at consent ranged from 8 days to 16 years (median 24 months, plus an outlier of 23 years). Only 50% of the probands had routine comparative genomic hybridization (aCGH) microarray testing, which is the first-line test for children with multiple congenital abnormalities. Overall, recruitment rates were much higher than expected given that blood samples were requested from both parents.
Table 1.
Summary of recruitment demographics, family structure, and types of mutation reported including mode of inheritance
| Number of probands sequenced | 195 |
|---|---|
| Family structure | Singleton: 3 (1%), Parent + child: 18 (9%), Trio: 169 (87%), Othera: 5 (3%) |
| Gender | Male: 99 (51%), Female: 96 (49%) |
| Recruitment ward | NICUb: 106 (54%), PICU: 61 (31%), Paediatric Neurology: 23 (12%), Clinical Genetics: 5 (3%) |
| Age at recruitment | NICU: 1 day–6 months (median 12 days) Other: 8 days–16.8 yearsc (median 24 months) |
| Genetic diagnosis via WGS | 40 (21%) |
|---|---|
| ACMG classification | Pathogenic: 21, Likely pathogenic: 17, VUSd: 2 |
| Contribution to phenotype | Fully explained: 38, Partially explained: 2 |
| Inheritance pattern | De novo: 27 (autosomal dominant: 24, X-linked dominant: 2, X-linked recessive: 1) Inherited: 13 (homozygous: 4, compound heterozygous: 6, X-linked recessive: 2, X-linked dominant: 1) |
| Variant type | Loss of function: 18 (frameshift: 8, stop gain: 3, splicing: 4, structural variant: 3), missense: 21, in-frame deletion: 1 |
aIncludes trio + sibling (4) and trio + grandparents (1)
bIncludes two cases from other postnatal wards
cIn addition there is one 23-year-old
dOne case (44) was a duo and likely pathogenic pending confirmation of de novo; one case (138) was a missense VUS in trans with a pathogenic loss of function variant
Rapid diagnoses
Forty cases received a diagnosis via WGS analysis (21% diagnostic rate), which either fully (95%) or partially (5%) explained the phenotype. The time from recruitment to preliminary findings reduced over the course of the project and reached steady state at about 3 weeks (Supp. Fig. 2). From January 2018 onwards, the median time from recruitment to accredited report was less than 5 weeks with the fastest turnaround being 21 days. Much of the variability stemmed from difficulty in collecting parental samples in a timely manner (Supp. Fig. 3a). Sequencing and analysis took a median of 16 days (Supp. Fig. 3b–d) while MDT meetings, confirmation, and clinical reporting took 11 days (Supp. Fig. 3e–f). Iterative redesign of processes allowed continuous improvement of the pipeline as shown by the reduced median turnaround time to diagnosis over the recruitment period. Two diagnoses were made after subsequent re-analysis as these disease-causing genes were newly reported in the literature.
The diagnostic rate in the NICU was 13% and in the PICU it was 25%. The referrals from the paediatric neurology and/or clinical genetics departments had a 39% diagnostic rate (Supp. Table 3). Two-thirds of the diagnoses were de novo variants; familial X-linked, compound heterozygous and homozygous variants were also observed (Table 1). Parental sequence data were crucial for calling pathogenic bi-allelic compound heterozygous variants in trans. Thus, accelerated WGS testing is feasible in an NHS-compliant genetics pipeline with an overall diagnostic yield of 21%.
Impact on clinical care
The 40 diagnoses reported included pathogenic or likely pathogenic variants in a broad range of rare disorders including encephalopathies, myopathies, skeletal dysplasia, and various syndromes (Supp. Tables 4 and 5). The age at recruitment for the diagnosed probands ranged from 1 day to 15 years. The most common clinical impact of the diagnoses was to improve ongoing management by informing established specialist care pathways for older children (35%) or initiating new specialist care pathways (48%), especially for neonates and infants (Fig. 2). In a few cases (13%), the diagnosis affected acute clinical management decisions and modified treatment: change in epilepsy medication due to an SCN1A mutation, Dravet syndrome; prophylactic antibiotic use for a primary ciliary dyskinesia (DNAH11); and consideration of a ketogenic diet for children with pyruvate dehydrogenase deficiency (PDHA1), Dravet syndrome (SCN1A), and severe West syndrome with a CDKL5 mutation.
Fig. 2.
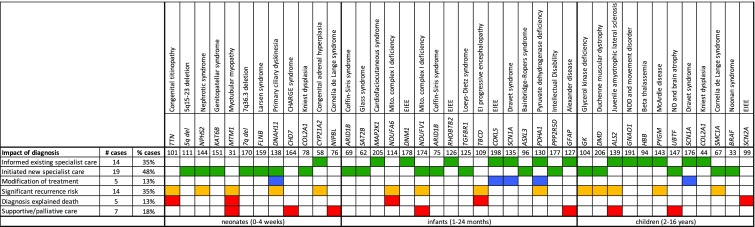
Impact of diagnosis on each case. Cases are ordered by increasing age. Green, specialist care. Blue, modification of treatment. Yellow, recurrence risk. Red, deceased and/or lethal condition. EIEE early infantile epileptic encephalopathy, Mito. mitochondrial, EI early infantile, NDD neurodevelopmental disorder, ND neurodegeneration
To date, for at least seven cases, distinguishing between inherited and de novo variants informed subsequent reproductive decisions. For 14 cases, the pathogenic variant(s) were inherited, conferring a significant recurrence risk for subsequent pregnancies. For example, in the case of family 114, the proband died from a novel mitochondrial disorder [24] due to inherited compound heterozygous variants in a mitochondrial complex I deficiency gene, NDUFA6 and prenatal testing was offered in a subsequent pregnancy (Fig. 2).
In four cases of perinatal death, WGS provided a valuable etiological explanation. For family 101, the diagnosis of congenital titinopathy [25] was made in the second affected neonate. In four neonatal/infant cases, the molecular diagnosis informed discussion with the family about palliative care. Three older children were diagnosed with neurodegenerative conditions due to mutations in GFAP (Alexander disease), ALS2 (juvenile amyotrophic lateral sclerosis) and UBTF, a recently described syndrome of neurodegeneration and brain atrophy (families 127, 139 and 147). For these families, supportive care and the resolution of diagnostic uncertainty was achieved but also brought painful realization of severe and currently incurable conditions being diagnosed. These findings indicate a variety of ways that WGS impacts clinical management of intensively ill children and their families.
Phenotype as predictor of genotype in young children
All probands were extensively phenotyped using Human Phenotype Ontology (HPO) terms extracted from the electronic health record. Analysis excluded maternal phenotypes and those relating to birth. The median number of non-redundant HPO terms per proband was 11, with 90% having at least five terms (Supp. Fig. 4). For most probands, the phenotypes fell into more than one major subclass of the root HPO term ‘Phenotypic abnormality’ and 80% have phenotypes from at least five subclasses. The most common phenotypes were abnormalities of the nervous (70% of probands), respiratory (68%), digestive (67%), and cardiovascular (61%) systems (Supp. Fig. 5).
In order to investigate the phenotypic composition of our cohort and to test the predictive value of specific phenotypes, we clustered the probands by phenotypic profile similarity and determined which phenotypes were enriched in each subgroup (see Supplementary Methods for details).
Clustering of the 106 patients recruited from the NICU (Supp. Fig. 6) revealed a few small groups with substantially increased diagnostic rates (> 30% compared to 13% for the NICU cohort overall). The cases with dysmorphic phenotypes such as hypertelorism, cleft palate, and micrognathia and those with abnormal renal function had the highest diagnostic yield (groups 1–4, Table 2, Supp. Table 6). Cases with congenital heart disease, decreased body weight or sepsis were the next most likely cases to have a diagnosis (groups 5–10). The groups with no diagnoses to date (groups 11–15) were enriched for suspected hypoxic ischaemic encephalopathy (HIE) and those with a general physiological disturbance.
Table 2.
Probands grouped by phenotype similarity
| Ward | Group(s) | Diagnostic rate | Number of probands | Enriched phenotypesa | Diagnosed genes |
|---|---|---|---|---|---|
| NICU | 1 | 100% | 2 | Hypertelorism, Talipes | COL2A1, FLNB |
| 2 | 50% | 4 | None significant. Top hitsb: Abnormal renal physiology, Abnormal urine output | CYP21A2, NPHS2 | |
| 3 | 50% | 2 | Abnormal heart valve morphology | KAT6B | |
| 4 | 33% | 3 | Cleft palate, Micrognathia | 16 Mb deletion | |
| 5–10 | 10–25% (avg. 17%) | 4–15 (avg. 8) | 5: Abnormal vascular physiologyb; 6: Abnormal morphology of the great vesselsb; 7: Abnormality of the nervous system; 8: Tachypnoea; 9: Decreased body weight; 10: Sepsis | TTN; CHD7, NIPBL, 24 Mb deletion; DNAH11; MAP2K1; MTM1; SATB2 | |
| 11–15 | 0% | 4–14 (avg. 9) | 11: Abnormality of nervous system physiology (HIE); 12: Hyperglycaemiab; 13: Abnormality of the amniotic fluid; 14: Meconium-stained amniotic fluid; 15: Ventriculomegaly | – | |
| PICU | 1 | 50% | 4 | None significant. Top hitb: Abnormality of the amniotic fluid | BRAF, RHOBTB2 |
| 2 | 50% | 2 | None significant. Top hitb: Lactic acidosis | NDUFA6 | |
| 3 | 40% | 5 | Cerebral palsy | SCN2A , COL2A1 c | |
| 4 | 36% | 11 | Abnormality of body height, Abnormality of skull size, Short stature | SMC1A, ARID1B, PPP2R5D, HBB c | |
| 5 | 33% | 3 | Ventilator dependence with inability to wean | PYGM | |
| 6–8 | 13–25% (avg. 20%) | 4–15 (avg. 9) | 6: Congenital malformation of the great arteries; 7: Feeding difficultiesb; 8: Abnormality of immune system physiology, Sepsis, Respiratory tract infection |
TGFBR1; ASXL3, ARID1B; DMD, GK | |
| 9–11 | 0% | 2–4 (avg. 3) | 9: Functional respiratory abnormalityb; 10: Joint hypermobility; 11: Prolonged neonatal jaundice | – |
aPhenotypes significantly enriched in group over cohort from ward (Fisher’s exact test, FDR < 0.1) and found in at least half of the probands in the group. Simplified for ontology redundancy. For rows representing multiple groups, the most descriptive terms are listed for each
bMost enriched term in a group with no significantly enriched terms
cGene only partially explains phenotype
In the 61 PICU cases, the phenotypic groups with the highest diagnostic rate were children with lactic acidosis and those with cerebral palsy and/or epilepsy (group1–3) (Table 2, Supp. Table 7, Supp. Fig. 7). Children with a distinct skeletal dysplasia and dysmorphology or long-term ventilator dependency were the next highest yield (group 4 and 5). Children with infection, sepsis and immune or respiratory dysfunction were the least likely to receive a molecular genetic diagnosis (groups 6–11).
Given that dysmorphism was a predictor of a molecular diagnosis in both NICU and PICU, we calculated the diagnostic yield in children who did not have documented dysmorphology. In NICU the yield was 9% compared to 13% overall and in PICU 18% compared to 25%.
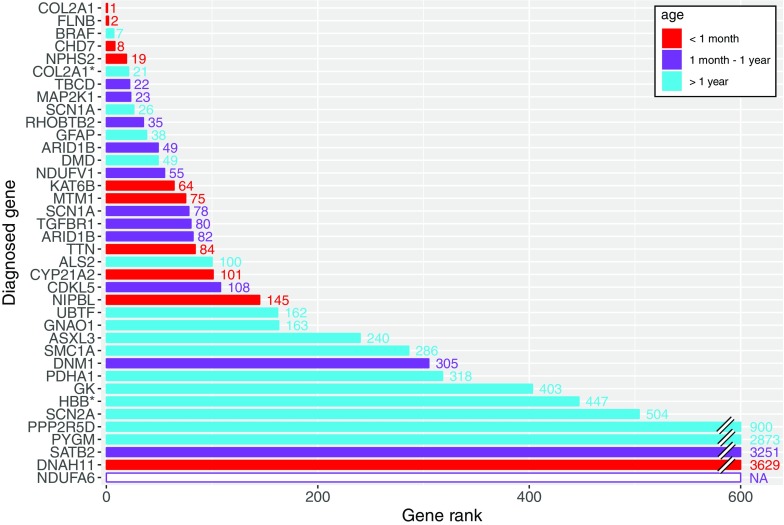
We then compared the phenotypes of the diagnosed probands with the known phenotypes reported to be associated with the identified disease-causing genes. To do this, we calculated the HPO term profile similarity score between each proband and every published gene in OMIM (Online Mendelian Inheritance in Man, www.omim.org) that has been annotated with HPO terms. Only in 10% of cases did the phenotype of the child predict accurately the most likely gene to cause disease. For the remaining 90% of cases the gene that caused disease could not be easily predicted (Fig. 3). In one case, the diagnosed gene (NDUFA6) was so recently published that it was not yet included in the OMIM-to-HPO build [24]. Importantly, more extensive phenotyping in the proband did not correlate with improved similarity ranking of the gene (Supp. Fig. 8a) and there was no difference in the number of HPO terms per proband between the diagnosed and non-diagnosed sets (Supp. Fig. 9). However, the number of HPO terms associated with a specific gene can improve specificity. Genes with a large number of phenotypes previously reported to be associated with the gene tend to rank higher, although for some genes an unusual combination of only a few HPO terms was sufficient to rank highly (Supp. Fig. 8b).
Fig. 3.
Bar chart showing where the diagnosed gene ranked in phenotype similarity score to the proband, compared to all 3926 HPO-typed genes in OMIM. The white bar indicates that the diagnosed gene was only recently reported and not yet HPO-typed. An asterisk (*) indicates that the gene only partially explained the phenotype. Families 111 and 170 are not included because both are large deletions where a specific gene has not been implicated. Red, under 1 month old at recruitment. Purple, 1 month–1 year old at recruitment. Blue, over 1 year old at recruitment
Discussion
Whole genome sequence analysis is increasingly being used to diagnose rare diseases in order to stratify patients for anti-seizure control, cancer treatments and clinical trials including gene therapy [26, 27]. Because the genome encodes information relevant to both acute and distant health risks, maximum benefit could be achieved by applying this technique early in life. Whilst the use of WGS in newborn screening is still under discussion, there is broad consensus that WGS has the potential to improve acute treatment of seriously ill babies [28].
WGS shows that underlying genetic conditions are prevalent in approximately 20% of intensively ill neonates and children selected for testing
We have created a timely-turnaround WGS trio analysis process for diagnosing severely ill children within the NHS infrastructure for clinical practice implementation. The study sought to identify a genetic cause of disease in children admitted to PICU and NICU and the recruitment criteria required there to be phenotypic features predicting a rare genetic cause of disease and did not include all admissions. The selection of this cohort is therefore likely to be enriched for a diagnosis and would be lower if all admissions were tested. The opportunity for WGS analysis was well received in the NICU and PICU by staff and parents. Parental motivations for consenting to the study included finding a cause for their child’s illness, the ability to potentially rule out a genetic condition, and an altruistic component to support improvement in healthcare for families in similar circumstances. Nevertheless, half of families who were eligible declined or did not respond to the offer of recruitment. Reasons given by families when declining participation can be broadly categorised as not being ready for a genetic diagnosis in the neonatal period, being too overwhelmed during the child’s critical illness, believing their child’s symptoms were not due to a genetic cause, or concerns over a genetic ‘label’. We noted that timing and appropriateness of the approach were crucial to full and informed consent and required a dedicated and trained person. Future studies are needed to focus on further understanding family perceptions surrounding these issues and ascertainment bias of the consented cohort.
Diagnosis has clinical utility
The ability to find a diagnosis in a timely manner for neonates and children was well received by professionals caring for these children. The availability of a result to inform discussions with families around palliative care was welcomed where a genetic diagnosis was associated with a known and poor prognosis. For some cases, early diagnosis allowed for interventions such as gene/disorder-specific medications (e.g. for epilepsy) and for surveillance and monitoring that can reduce harm in the long term (Fig. 2). A molecular diagnosis can obviate the need for muscle biopsy for mitochondrial and muscle diseases or further MRI scanning. For parents, a diagnosis and the removing of uncertainty for the family were valued; however, where the diagnosis was associated with a poor prognosis, it brought grief and loss. For 14 families the diagnosis gave reproductive choice not previously available (Fig. 2) and a number of couples had subsequent unaffected pregnancies.
Evidence that WGS should be offered to NICU/PICU patients
Whilst there were some phenotypic groups with increased likelihood of a genetic diagnosis (e.g. dysmorphology, neurological features), restricting recruitment to these specific phenotypes would have missed many important diagnoses. We found that the genotype is sufficient to drive the diagnosis with phenotypes assisting with variant interpretation rather than gene selection, especially since de novo pathogenic mutations were common (68%) in this cohort. In-depth phenotyping is critical when interrogating multiple different potentially pathogenic variants (common for singleton cases). However, we observed that even extensive phenotyping did not always strongly imply a particular gene or genes, indicating that predicting the disease gene based on phenotype will often be inaccurate. In neonates and infants, it is difficult to distinguish between transient and constitutive symptoms. Thus, the ability to perform a comprehensive gene agnostic trio analysis with WGS on 90% of our samples was advantageous over any single gene or panel tests as it did not require a phenotype-based hypothesis. Additionally, a negative result was more informative as almost all known disease-causing genes were assessed, significantly decreasing the likelihood of a genetic cause. In a cohort of intensively ill neonates and young children, our findings support the use of gene agnostic trio WGS for cases where a single gene disorder is suspected.
Advantages of a WGS pipeline and caveats
Trio WGS analysis has many advantages over singleton whole exome sequencing (WES) including improved copy number calling, mitochondrial and inter-genetic mutations, and increased analysis speed and robustness, but it does carry a higher up-front cost. We note that 30× WGS (and WES) short read analysis lacks the capacity to identify all known diagnostic regions of the genome routinely needed in neonatal care such as repetitive sequences, homologous genes, and epigenetic modifications. These include homozygous exon 7 and 8 deletions in SMN1 causing spinal muscular atrophy type 1 (one case in this cohort) and methylation defects (two cases in this cohort: Kagami–Ogata and Beckwith–Wiedemann syndrome). In addition, very low levels of mosaicism in the child will not be reliably detected.
Variants of uncertain significance (VUS) and additional findings were not reported to families as per first tier consent, which included only the resolution of diagnostic uncertainty. This cautious approach to data feedback was adopted because the use of systematic WGS in the neonatal setting in the UK is novel [28]. However, there is still uncertainty regarding the interpretation of some variants due to the paucity of reports of congenital presentations of these disorders. This can render the WGS data analysis complex because of the blurred boundaries between diagnostic and predictive findings. Although by default we did not include autosomal dominant inherited variants in order to avoid identifying additional findings, some diseases with variable penetrance or expressivity have incidence in the childhood period. To mitigate this, variants in specific genes were examined on request when suspected by clinicians, though no such findings have been made to date.
An enduring research database resource
All families were additionally consented to the NIHR BioResource, permitting ongoing research with the data. Establishing this as a research platform enabled iterative re-evaluation of phenotypes and genotypes over time and the continuing influence on care of patients and families. WGS allows for discovery of novel disease-causing elements, including genes and regulatory elements [18, 29]. Re-analysis of WGS with improved variant calling and new gene discovery can substantially increase the diagnostic yield [30]. Indeed, in this cohort, three of the diagnoses were for genes first published since August 2017, including one case that contributed to the reporting of a novel mitochondrial disorder [24]. Improvements to genomic analysis pipelines are ongoing, such as assessing the analytical validity of an algorithm developed to detect abnormalities associated with expansion disorders relevant to the neonatal period (ExpansionHunter) [31].
Conclusion
In summary, this study shows that rapid whole genome sequencing for diagnostic purposes is feasible at scale within the existing infrastructure of the NHS. WGS analysis of trios in NICU and PICU identified the underlying cause of disease in 13–25% of individuals who were selected for testing. A genotype-driven approach ensured that all genes were considered equally as the phenotype of the child in PICU and NICU was a poor predictor of the specific gene identified. Finally, our findings suggest that WGS in neonatal and paediatric intensive care provides a unique opportunity to build a research resource of children with early detection of genetic diseases, which are eligible for clinical trials of rare diseases at a potentially more therapeutically responsive stage in the disease. Adoption of these procedures could alter acute management and life course outcomes for children with chronic diseases using stratified therapeutics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The study was supported by the Rosetrees Trust, Newton Trust, National Institute for Health Research (NIHR) for the Cambridge Biomedical Research Centre and NIHR BioResource (Grant Number RG65966).
NIHR BioResource—Rare Disease Consortium
Salih Tuna1, Prof Timothy J Aitman2, Sofie Ashford1, Willian J Astle1, David L Bennet3, Marta Bleda1, Keren J Carss1, Prof Patrick F Chinnery1,9, Sri V V Deevi1, Debra Fletcher1, Daniel P Gale4, Stefan F Gräf1, Fengyuan Hu1, Roger James1, Mary A Kasanicki1,9, Nathalie Kingston1, Ania B Koziell5, Hana Lango Allen1, Prof Eamonn R Maher1,9, Prof Hugh S Markus1,9, Stuart Meacham1, Prof Nicholas W Morrell1,9, Christopher J Penkett1, Prof Irene Roberts6, Alba Sanchis-Juan1, Prof Kenneth G C Smith1,9, Hannah Stark1, Kathleen E Stirrups1, Ernest Turro1, Prof Hugh Watkins7, Prof Catherine Williamson8, Timothy Young1, Prof John R Bradley1,9, Prof Willem H Ouwehand1, Prof F Lucy Raymond1,9 on behalf of the NIHR BioResource
1. School of Clinical Medicine, University of Cambridge, Cambridge Biomedical Campus, Cambridge, CB2 0SP, UK. 2. Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK. 3. The Nuffield Department of Clinical Neurosciences, University of Oxford, John Radcliffe Hospital, Oxford, UK. 4. UCL Centre for Nephrology, University College London, London, UK. 5. Department of Experimental Immunobiology, King’s College London, London, UK. 6. MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, University of Oxford, Oxford, UK. 7. Department of Cardiovascular Medicine, Radcliffe Department of Medicine, University of Oxford, Oxford, UK. 8. Division of Women’s Health, King’s College London, London, UK. 9. Cambridge University Hospitals NHS Foundation Trust, Cambridge Biomedical Campus, Hills Road, Cambridge, CB2 0QQ, UK.
Next Generation Children Project
Shruti Agrawal2, Ruth Armstrong2, Kathryn Beardsall2, Gusztav Belteki2, Marion Bohatschek2, Susan Broster2, Rosalie Campbell2, Rajiv Chaudhary2, Cristine Costa2, Angela D’Amore2, Annie Fitzsimmons2, Jennifer Hague2, Joanne Harley2, Shazia Hoodbhoy2, Riaz Kayani2, Wilf Kelsall2, Sarju G Mehta2, Roddy O’Donnell2, Samantha O’Hare2, Amanda Ogilvy-Stuart2, Stergios Papakostas2, Soo-Mi Park2, Alasdair Parker2, Nazima Pathan2, Matina Prapa2, Audrienne Sammut2, Richard Sandford1,2, Katherine Schon2, Yogen Singh2, Kelly Spike2, Ana Lisa Taylor Tavares2, Doris Wari-Pepple2, Hilary S Wong1,2, Prof C Geoff Woods1,2
1. School of Clinical Medicine, University of Cambridge, Cambridge Biomedical Campus, Cambridge, CB2 0SP, UK. 2. Cambridge University Hospitals NHS Foundation Trust, Cambridge Biomedical Campus, Hills Road, Cambridge, CB2 0QQ, UK.
Conflicts of interest
The author(s) declare that they have no conflicts of interest.
Ethical approval
Participants for this study were recruited through NHS Cambridge University Hospitals Foundation Trust under Cambridge South Research Ethics Committee approval 13/EE/0325.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
F. Lucy Raymond, Phone: +44 1223 762609, Email: flr24@cam.ac.uk.
NIHR BioResource—Rare Disease:
Salih Tuna, Timothy J. Aitman, Sofie Ashford, Willian J. Astle, David L. Bennet, Marta Bleda, Keren J. Carss, Patrick F. Chinnery, Sri V. V. Deevi, Debra Fletcher, Daniel P. Gale, Stefan F. Gräf, Fengyuan Hu, Roger James, Mary A. Kasanicki, Nathalie Kingston, Ania B. Koziell, Hana Lango Allen, Eamonn R. Maher, Hugh S. Markus, Stuart Meacham, Nicholas W. Morrell, Christopher J. Penkett, Irene Roberts, Alba Sanchis-Juan, Kenneth G. C. Smith, Hannah Stark, Kathleen E. Stirrups, Ernest Turro, Hugh Watkins, Catherine Williamson, Timothy Young, John R. Bradley, Willem H. Ouwehand, and F. Lucy Raymond
Next Generation Children Project:
Shruti Agrawal, Ruth Armstrong, Kathryn Beardsall, Gusztav Belteki, Marion Bohatschek, Susan Broster, Rosalie Campbell, Rajiv Chaudhary, Cristine Costa, Angela D’Amore, Annie Fitzsimmons, Jennifer Hague, Joanne Harley, Shazia Hoodbhoy, Riaz Kayani, Wilf Kelsall, Sarju G. Mehta, Roddy O’Donnell, Samantha O’Hare, Amanda Ogilvy-Stuart, Stergios Papakostas, Soo-Mi Park, Alasdair Parker, Nazima Pathan, Matina Prapa, Audrienne Sammut, Richard Sandford, Katherine Schon, Yogen Singh, Kelly Spike, Ana Lisa Taylor Tavares, Doris Wari-Pepple, Hilary S. Wong, and C. Geoff Woods
References
- 1.NHS Maternity Statistics, England 2016–17. https://digital.nhs.uk/data-and-information/publications/statistical/nhs-maternity-statistics/2016-17. Accessed 1 Aug 2018
- 2.Neonatal Data Analysis Unit, Imperial College London. https://www.imperial.ac.uk/neonatal-data-analysis-unit. Accessed 1 Aug 2018
- 3.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units. JAMA Pediatr. 2017;171:e173438. doi: 10.1001/jamapediatrics.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiffault I, Farrow E, Zellmer L et al (2018) Clinical genome sequencing in an unbiased pediatric cohort. Genet Med. 10.1038/s41436-018-0075-8 [DOI] [PMC free article] [PubMed]
- 5.Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855. doi: 10.1001/jamapediatrics.2017.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powis Z, Farwell Hagman KD, Speare V et al (2018) Exome sequencing in neonates: diagnostic rates, characteristics, and time to diagnosis. Genet Med. 10.1038/gim.2018.11 [DOI] [PubMed]
- 7.Stark Z (2018) Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med. 10.1038/gim.2018.37 [DOI] [PubMed]
- 8.Farnaes L, Hildreth A, Sweeney NM, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genomic Med. 2018;3:10. doi: 10.1038/s41525-018-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mestek-Boukhibar L, Clement E, Jones WD, et al. Rapid paediatric sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children. J Med Genet. 2018 doi: 10.1136/jmedgenet-2018-105396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howell KB, Eggers S, Dalziel K, et al. A population-based cost-effectiveness study of early genetic testing in severe epilepsies of infancy. Epilepsia. 2018;59:1177–1187. doi: 10.1111/epi.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith HS, Swint JM, Lalani SR et al (2019) Clinical application of genome and exome sequencing as a diagnostic tool for pediatric patients: a scoping review of the literature. Genet Med 21:3–16. 10.1038/s41436-018-0024-6 [DOI] [PubMed]
- 12.Vissers LELM, Van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19:1055–1063. doi: 10.1038/gim.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg JS, Agrawal PB, Bailey DB, et al. Newborn sequencing in genomic medicine and public health. Pediatrics. 2017;139:e20162252. doi: 10.1542/peds.2016-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Char DS, Lee SS-J, Magnus D, Cho M (2018) Anticipating uncertainty and irrevocable decisions: provider perspectives on implementing whole-genome sequencing in critically ill children with heart disease. Genet Med. 10.1038/gim.2018.25 [DOI] [PMC free article] [PubMed]
- 15.Braverman G, Shapiro ZE, Bernstein JA. Ethical issues in contemporary clinical genetics. Mayo Clin Proc Innov Qual Outcomes. 2018;2:81–90. doi: 10.1016/j.mayocpiqo.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan N, Amendola LM, O’Daniel JM, et al. Is “incidental finding” the best term?: a study of patients’ preferences. Genet Med. 2017;19:176–181. doi: 10.1038/gim.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler S, Vasilevsky NA, Engelstad M, et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017;45:D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carss KJ, Arno G, Erwood M, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese C, Simone D, Diroma MA, et al. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics. 2014;30:3115–3117. doi: 10.1093/bioinformatics/btu483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene D, Richardson S, Turro E. OntologyX: a suite of R packages for working with ontological data. Bioinformatics. 2017;33:1104–1106. doi: 10.1093/bioinformatics/btw763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alston CL, Heidler J, Dibley MG et al (2018) Biallelic mutations in NDUFA6 establish its role in early-onset isolated mitochondrial complex I deficiency. Am J Hum Genet. 10.1016/J.AJHG.2018.08.013 [DOI] [PMC free article] [PubMed]
- 25.Oates EC, Jones KJ, Donkervoort S, et al. Congenital titinopathy: comprehensive characterisation and pathogenic insights. Ann Neurol. 2018;83:1105–1124. doi: 10.1002/ana.25241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrander BEP, Butterfield RJ, Pedersen BS et al (2018) Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy. NPJ Genomic Med 3:22. 10.1038/s41525-018-0061-8 [DOI] [PMC free article] [PubMed]
- 27.Turnbull C, Scott RH, Thomas E, et al. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ. 2018;361:k1687. doi: 10.1136/bmj.k1687. [DOI] [PubMed] [Google Scholar]
- 28.Whole genome sequencing of babies, Nuffield Council on Bioethics. http://nuffieldbioethics.org/wp-content/uploads/Nuffield-Council-on-Bioethics-briefing-note-whole-genome-sequencing-of-babies.pdf. Accessed 30 Jul 2018
- 29.Short PJ, McRae JF, Gallone G, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555:611–616. doi: 10.1038/nature25983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright CF, McRae JF, Clayton S et al (2018) Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med. 10.1038/gim.2017.246 [DOI] [PMC free article] [PubMed]
- 31.Dolzhenko E, van Vugt JJFA, Shaw RJ, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.