Abstract
Manganese (Mn) is an essential element, but in humans, chronic and/or acute exposure to this metal can lead to neurotoxicity and neurodegenerative disorders including Parkinsonism and Parkinson’s Disease by unclear mechanisms. To better understand the effects that exposure to Mn2+ exert on eukaryotic cell biology, we exposed a non-essential deletion library of the yeast Saccharomyces cerevisiae to a sub-inhibitory concentration of Mn2+ followed by targeted functional analyses of the positive hits. This screen produced a set of 43 sensitive deletion mutants that were enriched for genes associated with protein biosynthesis. Our follow-up investigations demonstrated that Mn reduced total rRNA levels in a dose-dependent manner and decreased expression of a β-galactosidase reporter gene. This was subsequently supported by analysis of ribosome profiles that suggested Mn-induced toxicity was associated with a reduction in formation of active ribosomes on the mRNAs. Altogether, these findings contribute to the current understanding of the mechanism of Mn-triggered cytotoxicity. Lastly, using the Comparative Toxicogenomic Database, we revealed that Mn shared certain similarities in toxicological mechanisms with neurodegenerative disorders including amyotrophic lateral sclerosis, Alzheimer’s, Parkinson’s and Huntington’s diseases.
Subject terms: Bioinorganic chemistry, Neurodegeneration
Introduction
All trace elements play an important role in the balance of life on our planet. The ability of cells to effectively utilize these elements depends strongly on their concentration, chemical speciation and fractionation1. Manganese (Mn) is the twelfth most abundant element in the earth’s crust. Natural levels of Mn range from 1–200 μg/L in fresh water and 410–6700 mg/kg (dry weight) in sediment2. In aquatic environments, Mn2+ is the most dominant and stable water-soluble species when the pH and redox potential are kept low2. The Mn3+ ion is soluble only in complex form and Mn4+ has very limited solubility3. Different Mn species can interconvert via oxidative or reductive processes depending on the redox environment1,3. Additionally, it is known that Mn is an vital trace mineral in nutrition4. Locally, levels of Mn can rise significantly in certain areas due to geogenic factors or anthropogenic activities such as mining5–8. Higher levels of Mn can have negative consequences for environmental health.
Epidemiological and toxicological studies suggest that Mn can be detrimental to specific biological processes and beneficial to the others in a concentration-dependent manner. This is also influenced by developmental stage, disease state9,10 cell type11, and/or the organism itself. Cell models from various organs including the liver, kidney and brain, suggest that neurotoxicity is the principal effect of this metal11. However, the underlying mechanism(s) are unclear and remain the subject of current studies12. Some investigations provide strong evidence that Mn can disturb the vital flow of genetic information from DNA to RNA to protein13–16. Recently; an interesting in vitro study demonstrated that Mn2+ had similar effects that Fe2+ and Mg2+ on rRNA folding and it can replace Mg2+ as the dominant divalent cation during translation of mRNA to functional protein17.
The addition of MnCI2 (5 mM) to highly-purified membrane rat-liver fractions caused a 30% increase in the polysome-binding capacity of stripped rough endoplasmic reticulum (ER) membranes, while four- to five-fold increases were observed with smooth ER membranes15. Previous studies in yeast suggest that the Mn2+ inhibits protein synthesis, disrupts nuclear DNA replication, and demonstrates mutagenic activity when under selective pressure13,14. Overwhelming evidence indicates that when stressed or undergoing environmental adaptation, cells accumulate non-synonymous mutations18. Studies investigating Mn-trafficking in humans suggest that Mn-induced Parkinsonism can result from mutations in SLC30A10, ATP13A2 or ZnT1019,20. Also, a His → Asn reversion mutant in ZnT10 conferred Mn transport activity and loss of zinc transport activity20. ZnT10 codes for a protein that is localized to the plasma membrane and is involved in zinc subcellular homeostasis20, while SLC30A10 codes for a surface-localized Mn efflux transporter that reduces cellular intake of Mn and protects against Mn-induced toxicity in neurons and worms19. The synthesis of these proteins is controlled by ribosomal activity in connection with the ER21. The ER is a large, continuous membrane-bound organelle with distinct domains and numerous contact sites with the plasma membrane, Golgi, mitochondria, and other cellular components including the SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors) complex that participates in the ER formation, fusion and function22. Protein synthesis is a crucial process for all living cells. Due to its central importance to cell survival and high energy requirements, protein synthesis is firmly regulated and strongly connected to other cellular processes, including the cell cycle and metabolic pathways23,24. Also, the mechanisms that govern the protein synthesis are highly-conserved through the course of evolution from higher to lower eukaryotes, as well as prokaryotes. In line with this, several aggregation-prone yeast proteins have human homologues that are implicated in protein misfolding associated diseases, suggesting that similar mechanisms may apply in both organisms and that yeast can serve as a good model organism to study such processes25. Studies using ribosome profiling and/or polysome profiling and classic gene expression analyses have provided new insights into the identification of novel genes that can affect this process as well as the mechanism of protein synthesis itself, which is often considered the endpoint of gene expression26,27.
Compound genome-wide toxicity is best studied using a systems biology approach, which can decipher the role(s) of individual components of complex biological systems under certain conditions by examining interactions on a global scale. To this end, large-scale chemical-genomic studies using yeast have been employed to identify individual chemical-genetic interactions (CGIs) and generate interaction profiles to infer mechanism(s) of action28,29. A category of these interactions occurs when the deletion of a single gene causes significant sensitivity or resistance to a target compound. Such interactions can suggest a functional relationship between the deleted gene and cell’s responses to the target compound. By screening for chemical-genetic interactions across the genome, significant insights into genotoxicity pathways can be drawn. In a similar context, protein-protein interaction (PPI) networks also provide a useful resource to better understand the mecheanism of toxicity. PPIs underlay nearly all biological processes, including cell-to-cell interactions, and metabolic and developmental controls. Depending on structural properties and functional characteristics, PPIs can range from transient interactions that generally participate in signalling pathways tothemore permanent interactions required to form stable protein complexes. It has been revealed that over 80% of proteins do not operate alone but in complexes30,31. PPIs can be studied in vitro, in vivo, and in silico31.
Connections between environmental toxin exposure and several human diseases have stimulated increased investigation into the toxicity of environmental contaminants using different model organisms32,33. Despite the evolutionary distance between yeast and humans, the underlying molecular players of numerous important pathways including programmed cell death, cell cycle progression and gene expression are conserved between the two species, allowing for the study of neurotoxins using highly-developed omics approaches in yeast34. Mn is one such compound that is applicable for large-scale screening in yeast. However, the range of intra-cellular Mn concentrations with physiological relevance or toxicity is quite large. Particularly, various studies done in yeast suggest that concentrations can range from between 2–100 nmol of Mn/(10 × 109 cells), or 0.04–2.0 mM Mn (assuming a single yeast cell has a volume of 50 femtoliters), without any impact on cell growth35. However, at levels below or above this, Mn induces toxicity stimulating cellular responses, including upregulating or downregulating cell surface and intra-cellular transport systems35. Consequently, yeast has been used to study events associated with Mn homeostasis, neurotoxic cell death, and neurodegeneration34–37. Generally, these studies have used Mn concentrations above of 2 mM, but have hardly explored the large network of physiological pathways that involve Mn, and hence the role of Mn in these processes remains unclear. In the current study, we provide evidence to connect Mn toxicity to the gene expression pathway in yeast. The impairment of protein biosynthesis by Mn2+ revealed in this study improves our current understanding of Mn-induced neurotoxicity and neurodegenerative disorders such as including amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD), Parkinson’s disease (PD) and Huntington’s disease (HD).
Results
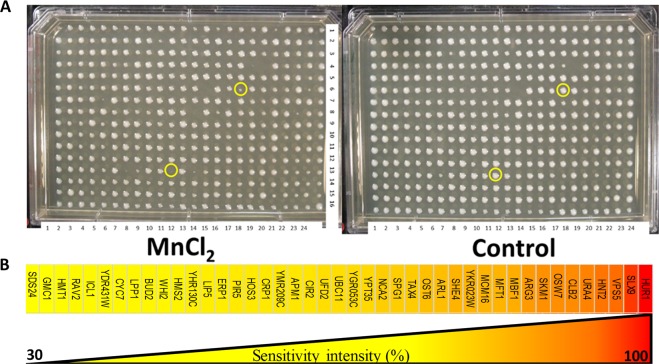
To identify pathways that are influenced by Mn exposure, we screened for gene deletion strains that demonstrate increased sensitivity to Mn2+ using the yeast non-essential gene deletion array (yGDA), Fig. 1A. These types of screens can provide a CGI profile for a target toxin and contribute to our knowledge of the cell’s global stress responses to that toxin. To this end, we performed sensitivity analysis by screening approximately 4700 gene deletion strains, under two conditions (presence and absence of MnCl2), for a total of approximately 28,000 individual analyses. Sensitivity was investigated by determining the relative colony growth size in the presence/absence of the target compound. In this way, we identified 68 gene deletion mutants with significantly altered growth profiles (Supplementary Material, Table SM1), of which 43 were confirmed to display high sensitivity to a sub-inhibitory concentration (1.35 mM) of Mn (a high concentration of a bioactive/toxic compound where growth of a wildtype (WT) strain is not completely inhibited), Fig. 1B. These genes represent a CGI profile for Mn sensitivity. They often represent “double hits” where the gene deletion and Mn treatment target compensating pathways generating an aggravated effect. The hits identified here were then subjected to further analysis.
Figure 1.
Representative illustration of Mn-induced disruption in yeast gene deletion array after exposure to MnCl2 (1.35 mM) for 24 hours. Mutants that showed a relative reduction in growth (sensitivity) of 30% or more were selected as hits (p < 0.05). Examples of hit stains are indicated using yellow circles.
Functional proteomic and Gene Ontology (GO) analysis of sensitive mutants identifies multiple pathways including protein synthesis
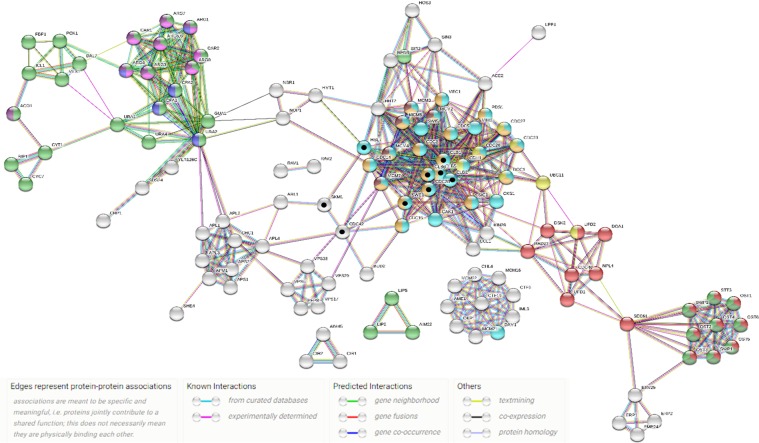
To have a comprehensive coverage of the hits identified in our sensitivity screen, the String database was used to expand the obtained CGI profile for Mn on the basis of PPI data38. String uses physical interactions and functional associations to study a defined set of proteins and expand it by including associated proteins. In this way, the network of functional interactors for Mn was increased to approximately 600 edges (p-value < 1.0e-16), of which more than 85% are known interactions. For example, approximately 87% of the interactions have been experimentally verified and almost 98% are from curated databases. A schematic representation of these interactors is shown in Fig. 2.
Figure 2.
The inferred and enriched PPI network from 43 genes that sensitize yeast to Mn when deleted. Analysis performed using the String database. Network properties are as follows: The minimum required interaction score, to be included at the predicted network, was accepted with a threshold on the high confidence equal 0.7; Number of nodes: 143; Number of edges: 594; Expected number of edges: 287; Average (avg) node degree: 8.31; avg. local clustering coefficient: 0.652; PPI enrichment p-value: <1.0e-16. Nodes and edges represent proteins and PPIs, respectively. Red nodes (protein processing in endoplasmic reticulum); blue nodes (metabolic pathways); dark green nodes (N-Glycan biosynthesis); cyan nodes (cell cycle); yellow nodes (ubiquitin-mediated proteolysis); orange nodes (meiosis); maroon nodes (DNA replication); purple nodes (amino acid biosynthesis); magenta nodes (arginine and proline metabolism); lime green nodes (alanine, aspartate and glutamate metabolism). The protein with black points in the center represent the MAPK signaling pathway. Proteins that are not connected to at least one partner are not shown.
Enrichment of cellular pathways represented by the expanded list of proteins is shown in Table 1. As expected, proteins associated with cellular development and protein metabolism were highly enriched39. Particularly, the ER associated activities have been highly connected to Mn toxicity21,40–43. However, a direct connection between protein synthesis and Mn toxicity has not been previously reported. This led us to further investigate the influence of Mn toxicity on protein biosynthesis.
Table 1.
Cellular pathways enriched in the Mn-induced interaction network.
| Cellular Processes | Pathway description | Number of observed genes | FDR |
|---|---|---|---|
| Cell cycle | Cell cycle | 29 | 8.46E-21 |
| Meiosis | 21 | 1.26E-11 | |
| DNA replication | 5 | 4.39E-03 | |
| Biosynthesis and Metabolism of Proteins | Protein processing in endoplasmic reticulum | 17 | 1.78E-11 |
| N-Glycan biosynthesis | 9 | 2.09E-07 | |
| Arginine and proline metabolism | 8 | 2.84E-06 | |
| Ubiquitin mediated proteolysis | 7 | 6.42E-04 | |
| Alanine aspartate and glutamate metabolism | 5 | 4.12E-03 | |
| Biosynthesis of amino acids | 8 | 3.47E-02 | |
| Metabolism | Metabolic pathways | 36 | 2.83E-06 |
This table was produced by performing GO analysis on the PPI network associated with genes influenced by Mn exposure (Fig. 1). This network was generated using the String database.
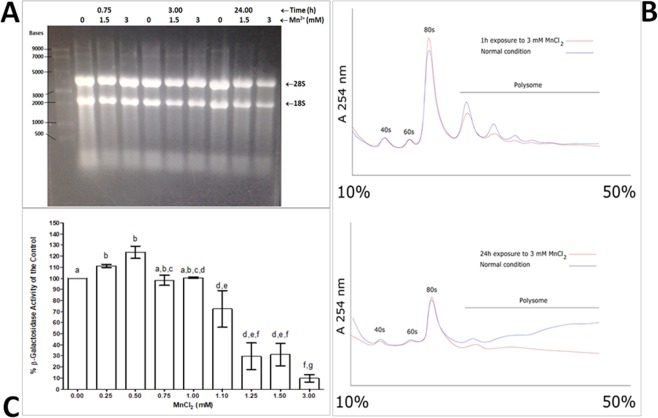
To study if Mn may also affect gene expression at the translation level, total RNA levels were analyzed. In response to the presence of Mn, we observed decreased levels of rRNA molecules (Fig. 3A). After treating the cells with 1.5 mM and 3.0 mM Mn for 45 minutes, total rRNA levels are reduced in a dose-dependent manner. We repeated this experiment by increasing the duration of Mn treatment to 3 and 24 hours. We observed similar results indicating that total levels of rRNA molecules seem to be reduced in response to Mn. Next, we investigated ribosome profiles of cells in response to Mn treatment (Fig. 3B). We observed a reduction in the pool of polysomes in response to treatment with 3 mM Mn for 1 hour, in addition to an increase in the pool of 80S ribosomes. Reduction in polysomes is interpreted as a decrease in the number of ribosomes that are active and engaged in synthesizing proteins. An increase in 80S monosomes is generally regarded as stalled initiation of translation. Treatment of the cells with Mn for 24 hours, resulted in additional reduction in polysomes in comparison to control conditions.
Figure 3.
Influence of Mn on protein biosynthesis. (A) rRNA levels are reduced in response to Mn (1.5 and 3 mM), for 0.75, 3 and 24 hours. Total rRNA (1 µg) run on a 1.2% agarose gel. (B) Ribosome profile analysis suggests that the number of active polysomes are reduced in response to Mn 3 mM, for 1 and 24 hours. (C) The relative expression of β-galactosidase is reduced in response to the presence on increasing concentrations of Mn. Bars represent the mean value of at least 3 independent experiments and error bars represent (mean ± SEM). Differences were stipulated by ANOVA one-way, followed by a Bonferroni post-test. Letters indicate statistically significant differences among treatments (p < 0.05).
Lastly, using an expression vector we investigated the expression of β-galactosidase, used as a reporter, in response to Mn. In a dose-dependent manner, the presence of Mn2+ reduced the expression of β-galactosidase (Fig. 3C). Importantly, this trend differs significantly when other divalent ions such as Ca2+, Mg2+ and Zn2+ are used suggesting that the decreased rate of translation is unique to Mn2+ stress and not a general byproduct (Supplementary Material, Fig. SM2). Altogether, these follow-up investigations connect Mn toxicity to the process of protein biosynthesis, which were identified as an enriched cellular process in our GDA analysis.
Manganese-induced disturbance of processes that converge to protein biosynthesis in yeast, which mimics molecular pathways associated with neurodegeneration
It has been postulated that the ER has various active domains and membrane contact sites that are required for multiple cellular processes including protein and lipid biosynthesis, calcium regulation, and the exchange of macromolecules22. In this study, multiple approaches including GDA and PPI analysis (Figs 1 and 2), GO ontology enrichment (Table 1), total rRNA analysis, ribosome profiling and a β-galactosidase reporter assay (Fig. 3), together suggest that the Mn induces a significant perturbation of protein biosynthesis and associated pathways.
To augment this finding and more-closely study individual participants, we selected several genes linked to processes that converge on protein biosynthesis and then analyzed their relative transcription levels using qPCR (Fig. 4). Indeed, the presence of Mn induced alterations in the expression of these genes. For example, we observed decreased expressionof key translation initiation factor eIF4A (TIF1) and upregulation of the essential translation elongation factor eIF-5A (HYP2). Additionally, several other genes associated with translation and/or ribosome biogenesis had significantly altered levels of transcription including the downregulation of NSR1, NOP1 and up-regulation of the gene RPS15. However, protein biosynthesis is a complex process that involve other pathways. For instance, we observed perturbation in the expression of genes such as UFD1, UFD2, STT3, DSK2. Additionally, OST2 and OST6, involved in post-translational processing in the ER (PPER), were significantly downregulated and upregulated, respectively. Similar alterations are inferred for the N-glycan biosynthesis pathway, which is partially regulated by OST2 and OST6 activity. We also observed decreased-expression of ARG3 involved in amino acid biosynthesis, including arginine, proline, alanine, aspartate and glutamate metabolism and increased expression of URA2.
Figure 4.
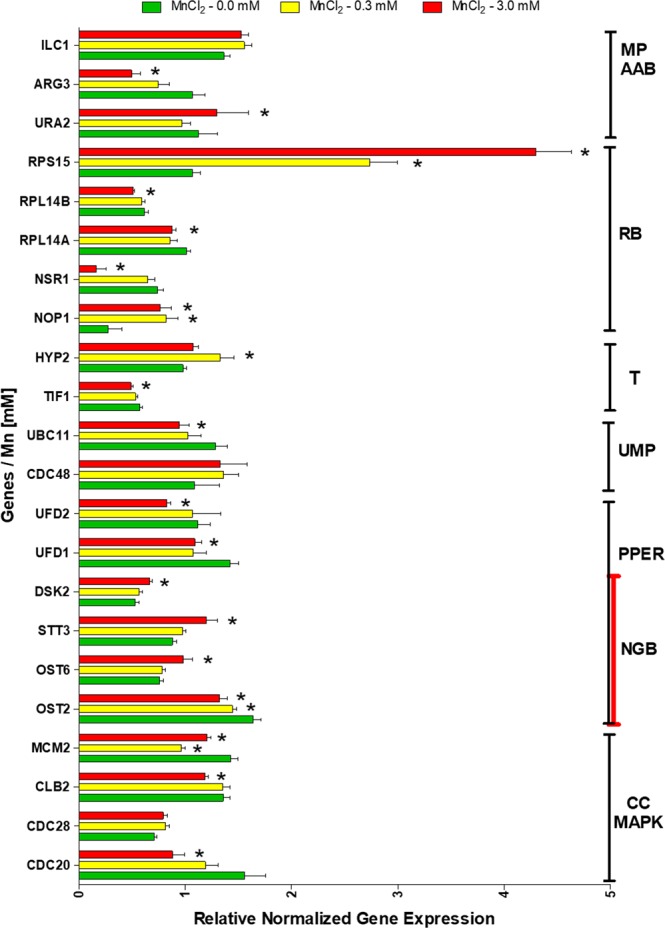
Relative gene-transcription of preselected hits from yeast stressed with Mn, after analysis of protein-protein interaction network (figure 2), β-galactosidase expression assay and ribosome profile, which are an evidence of perturbation of protein biosynthesis and other associated pathways (protein processing in endoplasmic reticulum, metabolic pathways, N-Glycan biosynthesis, cell cycle, ubiquitin mediated proteolysis, amino acid biosynthesis, MAPK signaling pathway and translation control analysis) was performed by qPCR. Bars represent the mean value of at least 3 independent experiments and error bars represent (mean ± SEM). Preliminarily, we verified some trends to be different between Mn treatment and the control using t-test (+p < 0.05). Then, we confirmed several significant differences by ANOVA two-way, followed of Bonferroni post-test (*p < 0.05).
Also of interest, genes such as CDC20 and UBC11, which are related to ubiquitin proteolysis were significantly disrupted. Protein synthesis underpins much of cell growth and multiplication44. Coincidently, impairment of CDC20 suggests a direct relationship among dysregulation of protein biosynthesis and alteration of MAPK signaling pathways, cell cycle and DNA replication respectively.
Mn-induced molecular impairment in yeast mimics pathways associated with neurodegeneration
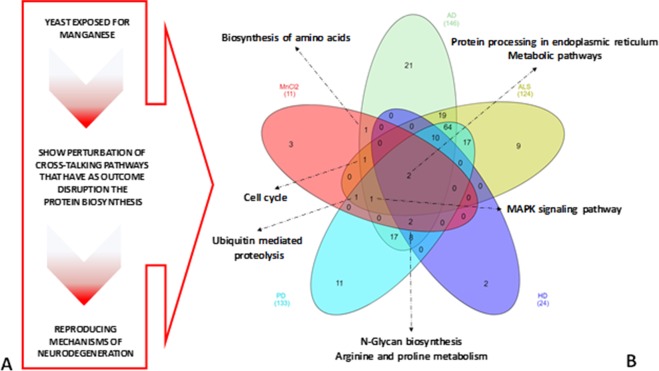
We identified alterations in various pathways that lead to impairment of protein biosynthesis, which is a conflicting topic in neurodegeneration research. Some reports have viewed this as a therapeutic target, while others suggest that it provokes the onset of certain neurodegenerative disorders45. Due to the conservation of the key cellular processes and genes, yeast has been used as a model organism to study human neurodegenative diseases34. In this sense, we conducted an additional analysis of the pathways affected by Mn using both the String database and the Comparative Toxicogenomics Database – CTD46, which permits the development of novel hypotheses about the relationships between chemicals and diseases47. The results are shown in Fig. 5.
Figure 5.
Mn-induced disruption pathways in yeast (A) share similarities with certain pathways linked to neurotoxicity and neurodegeneration (B). Analysis performed with the Comparative Toxicogenomic Databasewhich containscurated data regarding the mechanisms of action for neurodegenetative disorders. The results these analyses indicate overlapping pathways related to Mn Toxicity, AD (Alzheimer’s Disease), Amyotrophic Lateral Sclerosis (ALS), HD (Huntington’s Disease), and PD (Parkinson’s Disease).
We verified that approximately 31% (44 proteins/genes, Supplementary Material - Fig. SM 3) of the inferred network for hits (genes) affected by Mn (Fig. 2) have homologues in human, of which approximately 73% (32 proteins/genes, Supplementary Material – Fig. SM 3) are potentially linked to neurodegeneration, according to the CTD46. The genes affected by Mn suggest that this cation-induced toxicity in yeast involves disruption of several pathways which together lead to impairment of protein biosynthesis (Fig. 5A). These alterations shared characteristics with pathways involved in neurodegenerative diseases (Fig. 5B). For example, we identified that the MnCl2 affects the CDC20 involved in the metabolism of proteins46,the cell cycle and MAPK signaling pathways and is potentially involved in the development of neurodegenerative disorders such as AD, ALS, HD and PD46. At the same time, UFD1 is associated with protein processing in endoplasmic reticulum and potentially linked to the evolution of AD, ALS and PD46. Altogether; our findings and subsequent inferences suggest that the developmental impairment induced by Mn, according to cell cycle disruption, is mainly influenced by collective perturbation of pathways that converge to disturbance of protein biosynthesis. We demonstrated a decrease in total RNA, polysome and β-galactosidase activity as well as potential alterations in the expression of genes directly associated with translation (HPY2 and TIF1) and ribosome biogeneses (RPS15, NSR1 and NOP1). Together findings suggest a plausible hypothesis for Mn-induced neurotoxicity and neurodegeneration (Fig. 5B). Further analysis in higher-order animal models is needed to confirm this theory.
Discussion
The role of Mn in toxicity, particularly in relation to neurotoxicity and neurodegeneration disorders, remains unclear with several proposed hypotheses12. The CTDdescribes Mn as an essential trace element, with possible connections to approximately 570 biological processes and/or pathways48. In this work, using a functional genomics and systems biology approach, we observe a connection between Mn and cellular processes such as cell cycle progression, cell signaling, and protein metabolism. Agreeably, previous studies have suggested the possibility that Mn may disturb cellular development processes49,50 as well as the flow of genetic information that could influence protein synthesis14–16, including ER stress21,40–43.
Our global chemical-genetic sensitivity screen, followed by GO term enrichment of interaction network of participants, suggest that Mn disturbed anabolic metabolism pathways In line with this, we provide evidence to suggest that disruptions in the biosynthesis of amino acids through decreased expression of ARG3 which is involved in the biosynthesis of arginine from ornithine carbamoyltransferase51. Previous works have suggested that ornithine deficiency causes hyperammonemia and neurotoxicity in humans52. Specifically, alteration of arginine and proline metabolism has been associated with development of ALS53. The impairment of amino acid biosynthesis can directly disrupt translation efficiency54, a process that is energetically very costly23,24. Furthermore, these events appear to be associated with inactivation of MAPK pathways that can lead to translation repression23,55, antiapoptotic activities56 and/or cell cycle arrest23,44,56. Interestingly, we identified and inferred significant impairment of genes involved in the cell cycle and MAPK pathways (Figs 1, 2 and 4) such as CDC20 and CLB2, which correlated with our qPCR results. Cell cycle disruption has been associated with AD, PD and ALS57. At the same time, aberrations from strictly controlled of MAPK signaling pathway have also been implicated in the development of different human diseases including AD, PD and ALS56,58.
According to the results discussed above, Mn-induced toxicity in yeast appears to be associated with essential pathways linked to protein metabolism. In the current study we inferred that Mn may induce ER stress (Fig. 2), which was demonstrated through qPCR analysis showing up-regulation of OST6 and downregulation of OST2 (Fig. 4). This is in agreement with previous in vitro and yeast studies that suggested that the ATPase activity of ER gene SPF1 is compromised under exposure to Mn resulting in severe ER stress43. Other works have suggested that Mn-induced ER stress can be mediated through iron depletion, increased phosphorylation of the eukaryotic translation initiation factor 2α (phospho-eIF2α)59, activation of PERK and IRE1 signaling pathways41,42 and ER tumefaction60. An RNA-Seq approach in Caenorhabditis elegans revealed that Mn induced both up and down-regulation of ER-related protein families (FKB and ABU) which are both implicated in ER stress40. ER stress can trigger a signaling reaction known as the unfolded protein response (UPR), which induces adaptive programs that improve protein folding. In certain neurodegenerative diseases such as AD, ALS, HD and PD, when the cell damage is irreversible, UPR can also activate apoptosis61,62.
Moreover, we found that Mn could potentially influence glycosylation throughOST2 and OST663,64. Aminoglycoside antibiotics have been proposed to introduce errors in post-translational modifications such as glycosylation and protein misfolding that can lead to destabilized membranes and chronic stress65. Other studies suggest that alterations of SLC39A8 links Mn deficiency to inherited glycosylation disorders, specifically impairment of Mn-dependent enzymes activity, most notably the Golgi enzyme β-1,4-galactosyltransferase, which is essential for biosynthesis of the carbohydrates in glycoproteins63. Moreover, Golgi glycosylation defects may also be the result of Gdt1p/TMEM165 deficiencies that stem from Golgi Mn homeostasis defects64. Collectively, this evidence suggest that ER stress in yeast treated with Mn, may be associated with the impairment of N-glycan biosynthesis66, which could consequently lead to arrest the protein biosynthesis.
Additionally, ER stress can be exacerbated by the impairment of endosome-to-Golgi retrograde trafficking67. Since the retromer complex, comprised of vacuolar protein sorting, is essential to the bidirectional transport between the trans-Golgi network and endosomes. It is one of the key vesicular trafficking pathways in the cell68, particularly the transport of protein to endoplasmic reticulum67,69,70.
Vacuole protein sorting appears disrupted in the presence of Mn. VPS5 mutants are hypersensitive to Mn (Fig. 1B), and the PPI network analysis implicatedprotein/genes with similar function such as Vps35, Vps29, Vps17 and PEP8 (Fig. 2). Interestingly, previous studies have reported that Mn is linked to yeast VPS1, VPS53 and PEP871. Vesicle transport is considered to play an important role in yeast and mammalian models of ALS as well72. Furthermore, other studies have verified Mn down-regulated the expression of SNAP-25 and up-regulated the expression of VAMP-2, which interacted with Synaptophysin73. Using the FM1-43 dye (N-(3-Triethylammoniumpropyl)-4-(4-(Dibutylamino) Styryl) Pyridinium Dibromide), an excellent reagent both for identifying actively firing neurons and for investigating the mechanisms of activity-dependent vesicle cycling74, various authors verified that FM1-43-labeled synaptic vesicles treated with Mn resulted in an initial increase followed by a decrease in the number of vesicles73,75. Other studies linked Mn neurotoxicity to the disruption of genes with transport functions including SLC30A10, ATP13A2 and ZnT1019,20.
Alternatively, Gitler et al.76 identified that YPK9 overexpression significantly rescued the ability of proteins to leave the ER and traffic to the Golgi, which reduced the toxic effects of α–syn intracellular accumulation and Mn toxicity, suggesting a close connection between genetic and environmental causes of neurodegeneration. Ypk9 is a yeast orthologue of human PARK9/ATP13A2, whose expression in animal models of PD is capable of rescuing neurodegeneration76. At the same time, Golgi dysfunction can lead to the rapid repression of rRNA and ribosomal proteins23, affecting protein biosynthesis. Indeed, we observed that translation arrest in yeast was notably increased after 24 hours of exposure to Mn (decreasing of β-galactosidase activity), suggesting that long or chronic exposure of Mn2+ appears more effective in yeast than short or acute exposure which is similar to observations made by others11,77. We conducted the same analyses using other divalent cations such as Ca2+, Mg2+ and Zn2+ at equivalent or higher concentrations than used with Mn to determine if this was a general effect of metal stress and did not observe decreased rates of expression like the dose-dependent response to Mn seen in Fig. 3C. In addition, our ribosome profile analysis revealed a reduction in heavy polysomes fractions in response to Mn suggesting that Mn reduces efficiency of translation (Fig. 3B). This may be in agreement with a recent study in human SH-SY5Y cells that identified Mn-induced ER stress associated with increased phosphorylation of translation initiation factor eIF2α59. Similar profiles have been reported when using anti-translation drugs including pactamycin and harringtonine78.
Furthermore, we verified direct impairment of protein biosynthesis, including disruption of ribosome biogenesis due to down-regulation of the genes NOP1, NSR1; although this can be partially composed through up-regulation of the gene RPS15. A review at the CTD48 revealed that RPS15 is a marker of Disease Progression, including memory impairment in transgenic mice modelingAD treated with copper79 as well as RPL14, potentially affected by Mn, is a marker of PD), which have been observed in case of residential exposure to maneb,. It is very interesting because, while paraquat is a derived of bipyridine, maneb is a polymeric complex of Mn. Unpublished studies from our group have identified maneb-induced impairment of protein biosynthesis in cerebellar granule neurons.
Final Considerations
Literature in this field has consolidated robust hypotheses regarding Mn induced-neurotoxicity and neurodegeneration that include mitochondrial dysfunction, energy impairment, oxidative stress, disruption of neurotransmitters, ER stress, neuroinflammation, DNA damage and epigenetic alterations, apoptosis, autophagy, and many others9,10,80,81. However, occasionally, the accuracy of these hypotheses is challenged in different models. All processes cited above either occur after the process of protein synthesis is completed and/or are directly linked to it. In this study we identified protein synthesis as a key target of Mn-induced toxicity in S. cerevisiae. Defects in protein synthesis have been documented in different neurodegenerative disorders in humans. Since Mn-induced toxicity has been linked to human neurodegenerative disorders, the data presented in the current study may provide a connection between Mn-induced toxicity and human neurodegenerative disorders through the process of protein synthesis. Altogether, our findings provide strong evidence that Mn-toxicity can occur at multiple levels simultaneously, which appear to be associated with disruption of orchestrated essential pathways including metabolism and protein biosynthesis. In this way the presented study adds to our current understanding of the Mn-induced mode of toxicity. Additional experiments with mammalian models must be conducted to validate if these findings apply to other systems.
Experimental
Gene expression analysis
Manganese sensitivity/resistance screening using yeast gene deletion array
Approximately 4700 MATa haploid yeast, S. cerevisiae strains (BY4741, MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) from the non-essential Gene Deletion Array (yGDA) were manually arrayed onto agar plates as previously described by Alamgir, et al.82 in the presence or absence of sub-inhibitory concentration (a high concentration of a bioactive/toxic compound where growth of a WT strain is not completely inhibited) of MnCl2 (1.35 mM). Plates were incubated at 30 °C overnight. Finally, digital images of plates were used to analyze the growth of individual colonies, by automatized visual density comparison between control and their respective Mn treatment through of the available in-house and online software “SGAtools” from the University of Toronto83. The experiment was repeated between three and five times (Supplementary Material, Table SM 1). Colonies that showed 30% reduction or more in at least three repeats were considered hits84–86.
Transcriptomics experiment using q-PCR
The quantitative PCR (q-PCR) assay has been used to study the effects of a gene deletion on expression under specified conditionsgene deletion in previous works87,88. Primers of selected genes were synthetized (Table 2). Total RNA was isolated from each strain, using a Qiagen RNA isolation kit; followed by cDNA construction, using an iScript cDNA synthesis kit and finally used SYBR green supermix (Bio-Rad) for the multiplex real time PCR assay; according to the instructions of the manufacturer. The quantification of mRNA was performed by q-PCR on a Rotor-Gene RG-300 from Corbett research, according to Samanfar et al.85.
Table 2.
Genes selected for q-PCR analysis. Primer sequences and associated parameters are included.
| # | Gene Symbol |
Sequence (5′->3′) | Template strand | Length | Start | Stop | Tm | GC (%) | Self complementarity | Self 3′ complementarity | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | OST2 | Forward primer | TCCCGCTAAAAACGCATTGA | Plus | 20 | 516602 | 516621 | 58.48 | 45 | 4 | 2 |
| Reverse primer | CAGCACGTCATCTGCAGTCT | Minus | 20 | 516803 | 516784 | 60.39 | 55 | 6 | 3 | ||
| Product length | 202 | ||||||||||
| 2 | OST6 | Forward primer | CGCTGACAACTACCCACTGT | Plus | 20 | 233594 | 233613 | 59.97 | 55 | 3 | 3 |
| Reverse primer | TGGCAACTCATGCCGTTACT | Minus | 20 | 233692 | 233673 | 59.96 | 50 | 4 | 2 | ||
| Product length | 99 | ||||||||||
| 3 | STT3 | Forward primer | TTCGGTGACTTCGTGAAGGG | Plus | 20 | 453244 | 453263 | 59.97 | 55 | 6 | 2 |
| Reverse primer | TCAAGGCAGAAAGTCCGACC | Minus | 20 | 453352 | 453333 | 59.97 | 55 | 5 | 3 | ||
| Product length | 109 | ||||||||||
| 4 | DSK2 | Forward primer | GGACCCTAATGCCGGTATGG | Plus | 20 | 819486 | 819505 | 59.96 | 60 | 6 | 3 |
| Reverse primer | TTCGTGTTGGAGCCTTCCTC | Minus | 20 | 819578 | 819559 | 59.97 | 55 | 3 | 1 | ||
| Product length | 93 | ||||||||||
| 5 | ARG3 | Forward primer | GTTGCTGAGAGAAACGGTGC | Plus | 20 | 269462 | 269481 | 59.76 | 55 | 5 | 2 |
| Reverse primer | GCTTGGCCTGTTTCGCAAAT | Minus | 20 | 269588 | 269569 | 60.32 | 50 | 4 | 2 | ||
| Product length | 127 | ||||||||||
| 6 | ICL1 | Forward primer | ACCCAGCCTTTGGATGAAGG | Plus | 20 | 285532 | 285551 | 59.96 | 55 | 4 | 2 |
| Reverse primer | GTTACAGAGGTGGGACGCAA | Minus | 20 | 285764 | 285745 | 59.97 | 55 | 3 | 1 | ||
| Product length | 233 | ||||||||||
| 7 | CLB2 | Forward primer | CAGAGACAGACGGTGCATGT | Plus | 20 | 772641 | 772660 | 60.04 | 55 | 4 | 3 |
| Reverse primer | CAGCTGCTGCACACAATGAG | Minus | 20 | 772871 | 772852 | 60.11 | 55 | 7 | 3 | ||
| Product length | 231 | ||||||||||
| 8 | CDC28 | Forward primer | GCCAAGCTTTCCTCAATGGC | Plus | 20 | 560815 | 560834 | 60.11 | 55 | 6 | 2 |
| Reverse primer | GGGTCATACGCGAGGAGTTT | Minus | 20 | 560916 | 560897 | 59.82 | 55 | 4 | 1 | ||
| Product length | 102 | ||||||||||
| 9 | rpl14A | Forward primer | TTGGCCGCTATCGTCGAAAT | Plus | 20 | 431999 | 432018 | 60.18 | 50 | 6 | 3 |
| Reverse primer | TTGCCAGCACTTTTTCGTAGC | Minus | 21 | 432120 | 432100 | 60 | 47.62 | 4 | 2 | ||
| Product length | 122 | ||||||||||
| 10 | rpl14B | Forward primer | ACCTAAAACCCACCGTGGAC | Plus | 20 | 104415 | 104434 | 59.89 | 55 | 7 | 3 |
| Reverse primer | GTTGGCGGTCCCTGAACATA | Minus | 20 | 104492 | 104473 | 60.04 | 55 | 3 | 2 | ||
| Product length | 78 | ||||||||||
| 11 | TIF1 | Forward primer | ACTGGTAAGACCGGTACCTTTT | Plus | 22 | 555194 | 555215 | 59.03 | 45.45 | 6 | 2 |
| Reverse primer | GCTTGAGGAGCCTTGACAGA | Minus | 20 | 555264 | 555245 | 59.68 | 55 | 4 | 1 | ||
| Product length | 71 | ||||||||||
| 12 | HYP2 | Forward primer | TGAACATGGACGGTGACACT | Plus | 20 | 85983 | 86002 | 59.24 | 50 | 5 | 3 |
| Reverse primer | GCTTCTTCACCCATAGCGGA | Minus | 20 | 86112 | 86093 | 59.82 | 55 | 3 | 2 | ||
| Product length | 130 | ||||||||||
| 13 | UFD1 | Forward primer | GCGGCGGAAATGGTTTTGTA | Plus | 20 | 589851 | 589870 | 59.76 | 50 | 3 | 2 |
| Reverse primer | ATTTTCCCGCCGAAGTTTGC | Minus | 20 | 589968 | 589949 | 60.04 | 50 | 3 | 2 | ||
| Product length | 118 | ||||||||||
| 14 | UFD2 | Forward primer | CGGCGAAAGCAATCGTTCAA | Plus | 20 | 121396 | 121415 | 60.11 | 50 | 4 | 3 |
| Reverse primer | GTGCCTCAAGGCTCAACTCT | Minus | 20 | 121511 | 121492 | 59.96 | 55 | 6 | 1 | ||
| Product length | 116 | ||||||||||
| 15 | CDC48 | Forward primer | CCAGTACCAGGGGGACCATA | Plus | 20 | 237886 | 237905 | 60.03 | 60 | 4 | 2 |
| Reverse primer | CAGTGAGGAAAGGCGACCAT | Minus | 20 | 238201 | 238182 | 60.04 | 55 | 3 | 2 | ||
| Product length | 316 | ||||||||||
| 16 | UBC11 | Forward primer | ATCGTCTACGGGAAACGCAC | Plus | 20 | 958728 | 958747 | 60.46 | 55 | 5 | 1 |
| Reverse primer | AGTAGAAGAGGGTGGTTGCG | Minus | 20 | 958827 | 958808 | 59.39 | 55 | 2 | 2 | ||
| Product length | 100 | ||||||||||
| 17 | CDC20 | Forward primer | TTGCGTCCCCAACAAAGCTA | Plus | 20 | 289873 | 289892 | 60.18 | 50 | 4 | 2 |
| Reverse primer | ATTAACGGTGGTGCCCCAAT | Minus | 20 | 290059 | 290040 | 59.96 | 50 | 6 | 2 | ||
| Product length | 187 | ||||||||||
| 18 | MCM2 | Forward primer | GTGGCCAATCTTTCGTCTGC | Plus | 20 | 175437 | 175456 | 59.83 | 55 | 6 | 2 |
| Reverse primer | CGCGCTGCTCAATTATTGCT | Minus | 20 | 175602 | 175583 | 59.97 | 50 | 7 | 3 | ||
| Product length | 166 | ||||||||||
| 19 | NSR1 | Forward primer | CTAGAGCCTTCTTGGCGTCC | Plus | 20 | 806681 | 806700 | 60.18 | 60 | 4 | 2 |
| Reverse primer | CCGTCCGTATCCCAACACAT | Minus | 20 | 806773 | 806754 | 59.82 | 55 | 2 | 2 | ||
| Product length | 93 | ||||||||||
| 20 | NOP1 | Forward primer | ATTGCCCCAGGCAAGAAAGT | Plus | 20 | 427853 | 427872 | 60.18 | 50 | 7 | 1 |
| Reverse primer | TCTCTGCCTGGTCTGTGAGA | Minus | 20 | 427980 | 427961 | 59.89 | 55 | 4 | 3 | ||
| Product length | 128 | ||||||||||
| 21 | URA2 | Forward primer | CAAATTCTGGATGGCGCCTG | Plus | 20 | 166665 | 166684 | 59.9 | 55 | 6 | 2 |
| Reverse primer | ACGGAAGAAGCAATCGCTGA | Minus | 20 | 166793 | 166774 | 60.04 | 50 | 6 | 2 | ||
| Product length | 129 | ||||||||||
| 22 | MCM2 | Forward primer | GTGGCCAATCTTTCGTCTGC | Plus | 20 | 175437 | 175456 | 59.83 | 55 | 6 | 2 |
| Reverse primer | CGCGCTGCTCAATTATTGCT | Minus | 20 | 175602 | 175583 | 59.97 | 50 | 7 | 3 | ||
| Product length | 166 | ||||||||||
Protein synthesis analysis
Total rRNA analysis
The yeast wild type strains were preincubated and grown overnight, then grown on YPD media at 30 °C to an OD600 of 0.8–1.0, in either the absence or presence of Mn (1.5 mM or 3 mM), for 0.75, 1 and 24 hours respectively. Total RNA was isolated from each strain using a Qiagen RNA isolation kit (RNeasy mini kit). RNA electrophoresis was carried out in 1X MOPS running buffer diluted from 10X MOPS buffer [40.8 g 3-(N-morpholino) propanesulfonic acid (MOPS); 6.8 g sodium acetate; 3.8 g ethylenediamine tetraacetic acid (EDTA)]. Volume was completed up to 1000 ml by the addition of ultrapure water treated with diethyl pyrocarbonate (DEPC), and the pH was adjusted to 7.0 using sodium hydroxide (NaOH)). The concentration of agarose used in the RNA gels was 1.2% (w/v). Samples were prepared in 80% v/v deionized formamide, heated at 65 °C for 5 minutes, then immediately cooled on ice. Before loading the samples on the gel, 1/10th of sample volume 10X RNA loading dye (0.0125 g Bromophenol Blue; 10 µl 0.5 M EDTA; 2.5 ml 100% glycerol; 2.5 ml DEPC-treated water; mixed by vortexing and autoclaved) was added to the samples for a final concentration of 1 × (1 µg).
β-Galactosidase expression assay
The efficiency of translation was quantified using an inducible β-galactosidase reporter gene in the p416 plasmid82,89. β-galactosidase is a model of intracellular protein synthesis that can provide a profile of aberrancy in the rate of protein synthesis and an estimate of gene expression. Thus, yeast cells transformed with p416 plasmid were preincubated for 1 hour and then exposed to a crescent toxicological curve of Mn (0.25–3 mM) and other divalent ions (Ca2+, Mg2+ and Zn2+ at 0.5, 1.5, 5 mM and a higher 15 mM concentration for Mg2+) for 3 hours at 30 °C, followed of spectrophotometric determination of β-galactosidase activity82. Metal ion concentrations were suggested by previous works90–93.
Ribosome profile analysis
Ribosome profiling94, allows for the monitoring of translation dynamics in vivo. Yeast wild type strains were preincubated 1 hr and then grown on YPD media at 30 °C to an OD600 of 0.8–1.0, in the absence or presence of 3 mM Mn, for 1 hr and 24 hrs respectively at 30 °C. Immediately before harvest, cycloheximide was added to all samples, to a final concentration of 100 μg/ml, and the culture was incubated again at 30 °C for 15 minutes, followed by a cold snap in an ice water bath. Cells were harvested, washed with a cycloheximide/water solution (100 μg/ml) and centrifuged at 4000 rpm for 4 min at 4 °C using a Sorvall SLA-1500 rotor to separate the supernatant. Cell pellets were resuspended in 10 ml of ice-cold lysis buffer A (YA buffer: 10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 30 mM MgCl2, cycloheximide 50 μg/ml, heparin 200 μg/ml) and centrifuged at 4000 rpm for 4 min at 4 °C (Sorvall SS34 rotor) twice. Pellets were resuspended in 750 µl of YA buffer, lysed by vortexing with glass beads, transferred to microtubes, and centrifuged at 13000 rpm for 10 minutes at 4 °C. The supernatant was preserved for the quantitative determination of total RNA, followed by fractionation on 10–50% sucrose gradients containing 50 mM Tris-acetate [pH 7.0], 50 mM NH4Cl, 12 mM MgCl2, and 1 mM dithiothreitol. The extract was centrifuged for 2 h at 40,000 rpm using a SW40-Ti rotor in a Beckman LE-80 K at 4 °C. The polysome profiles were analyzed via a Biocomp gradient station and the absorbance was recorded at 254 nm using a spectrophotometer (Bio-Rad Econo UV monitor) coupled with the Biocomp station. In this method, free mRNAs from the top fractions were separated from polysome-associated mRNAs from the bottom fractions95.
Protein-protein interaction (PPI) prediction and gene ontology (GO) analysis
A PPI network can be described as a heterogeneous network of proteins joined by interactions as edges. Protein network and GO enrichment analysis were based on the data from the current project and analyzed using the STRING database (http://string-db.org)38. Additional GO analysis was conducted at the Comparative Toxicogenomic Database – CTD (http://ctdbase.org/)48 to test the hypothesis of a conserved mode of action of Mn between yeast and humans. Both STRING and the CTD database were accessed on November 18th, 2018.
Data analysis
The results were expressed as mean ± sem of at least three independent experiments. To detect statistically significant differences, ANOVA (analysis of variance) followed by Bonferroni’s tests was be used; preceded of single t-test analysis between pairs of treatments. Fitting and statistical analyses were performed using GraphPad Prism (GraphPad 4.0 Software Inc, San Diego, CA, USA).
Supplementary information
Acknowledgements
FAPESP (15-24207-9, 16/00371-7, 16/50483-6), NSERC (Discovery Grant 2013–2018).
Author Contributions
Professors Hernández and Golshani are responsible for conceptual development of this study, co-executor of all experiments, and coordinators of the grants that supported this work. H. Moteshareie, a PhD candidate, contributed to translation experiments as well as data analysis. D. Burnside, a PhD candidate, contributed to the bioinformatic approaches and data analysis. Professor McKay contibuted to the ribosome profile analysis. All authors contributed to the writing of the manuscript.
Data Availability
All data generated and/or analyzed during this study are included in this published article and/or its Supplementary Material Files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Raúl Bonne Hernández, Houman Moteshareie and Ashkan Golshani contributed equally.
Contributor Information
Raúl Bonne Hernández, Email: rbhernandez@unifesp.br.
Ashkan Golshani, Email: ashkan_golshani@carleton.ca.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42907-2.
References
- 1.Templeton, D. M. et al. Guidelines for terms related to chemical speciation and fractionation of elements. Definitions, structural aspects, and methodological approaches (IUPAC Recommendations 2000). Pure Appl. Chem. 72 (2000).
- 2.Howe, M. P. D., Malcolm, H. M. & Dobson, D. S. Manganese and Its Compounds: Environmental Aspects. World Health Organization (2004).
- 3.Kenneth Klewicki J, Morgan JJ. Kinetic behavior of Mn(III) complexes of pyrophosphate, EDTA, and citrate. Environ. Sci. Technol. 1998;32:2916–2922. doi: 10.1021/es980308e. [DOI] [Google Scholar]
- 4.Luo XG, et al. Gene expression of manganese-containing superoxide dismutase as a biomarker of manganese bioavailability for manganese sources in broilers. Poult. Sci. 2007;86:888–894. doi: 10.1093/ps/86.5.888. [DOI] [PubMed] [Google Scholar]
- 5.Hafeman D, Factor-Litvak P, Cheng Z, van Geen A, Ahsan H. Association between manganese exposure through drinking water and infant mortality in Bangladesh. Environ. Health Perspect. 2007;115:1107–1112. doi: 10.1289/ehp.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ljung K, Vahter M. Time to re-evaluate the guideline value for manganese in drinking water? Environ. Health Perspect. 2007;115:1533–1538. doi: 10.1289/ehp.10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordão CP, Pereira JL, Jham GN, Bellato CR. Distribution of Heavy Metals in Environmental Samples Near Smelters and Mining Areas in Brazil. Environ. Technol. 1999;20:489–498. doi: 10.1080/09593332008616844. [DOI] [Google Scholar]
- 8.Bonne Hernández R, Oliveira E, Espósito BP. Distribution and behavior of manganese in the Alto do Paranapanema Basin. J. Environ. Monit. 2009;11:1236–43. doi: 10.1039/b822579f. [DOI] [PubMed] [Google Scholar]
- 9.Peres, T. V. et al. Manganese-induced neurotoxicity: A review of its behavioral consequences and neuroprotective strategies. BMC Pharmacol. Toxicol. 17 (2016). [DOI] [PMC free article] [PubMed]
- 10.Pfalzer AC, Bowman AB. Relationships Between Essential Manganese Biology and Manganese Toxicity in Neurological Disease. Curr. Environ. Heal. reports. 2017;4:223–228. doi: 10.1007/s40572-017-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández RB, et al. Mechanisms of manganese-induced neurotoxicity in primary neuronal cultures: The role of manganese speciation and cell type. Toxicol. Sci. 2011;124:414–423. doi: 10.1093/toxsci/kfr234. [DOI] [PubMed] [Google Scholar]
- 12.RB, H. Current Challenges about Understanding of Manganese-Induced Neurotoxicity. Toxicol. Open Access01, 58201 (2015).
- 13.Putrament A, Baranowska H, Ejchart A, Prazmo W. Manganese Mutagenesis in Yeast. A Practical Application of Manganese for the Induction of Mitochondrial Antibiotic-resistant Mutations. J. Gen. Microbiol. 1975;90:265–270. doi: 10.1099/00221287-90-2-265. [DOI] [PubMed] [Google Scholar]
- 14.Putrament A, Baranowska H, Ejchart A, Jachymczyk W. Manganese mutagenesis in yeast - VI. Mn2+ uptake, mitDNA replication and ER induction. Comparison with other divalent cations. MGG Mol. Gen. Genet. 1977;151:69–76. doi: 10.1007/BF00446914. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson SG, Fox OF, Kishore GS, Carubelli R. Effect of manganese ions on the interaction between ribosomes and endoplasmic reticulum membranes isolated from rat liver. Biosci. Rep. 1981;1:727 LP–731. doi: 10.1007/BF01116471. [DOI] [PubMed] [Google Scholar]
- 16.Dambach M, et al. The Ubiquitous yybP-ykoY Riboswitch Is a Manganese-Responsive Regulatory Element. Mol. Cell. 2015;57:1099–1109. doi: 10.1016/j.molcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bray MS, et al. Multiple prebiotic metals mediate translation. Proc. Natl. Acad. Sci. 2018;115:12164 LP–12169. doi: 10.1073/pnas.1803636115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan T. Adaptive Translation as a Mechanism of Stress Response and Adaptation. Annu. Rev. Genet. 2013;47:121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leyva-Illades D, et al. SLC30A10 Is a Cell Surface-Localized Manganese Efflux Transporter, and Parkinsonism-Causing Mutations Block Its Intracellular Trafficking and Efflux Activity. J. Neurosci. 2014;34:14079–14095. doi: 10.1523/JNEUROSCI.2329-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishito Y, et al. Direct comparison of manganese detoxification/efflux proteins and molecular characterization of ZnT10 protein as a manganese transporter. J. Biol. Chem. 2016;291:14773–14787. doi: 10.1074/jbc.M116.728014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F, Canadeo LA, Huibregtse JM. Ubiquitination of newly synthesized proteins at the ribosome. Biochimie. 2015;114:127–133. doi: 10.1016/j.biochi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.English A. R., Voeltz G. K. Endoplasmic Reticulum Structure and Interconnections with Other Organelles. Cold Spring Harbor Perspectives in Biology. 2013;5(4):a013227–a013227. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warner JR. The economics of ribosome biosynthesis in yeast. Trends in Biochemical Sciences. 1999;24:437–440. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 24.Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013;126:4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 25.Ibstedt S, Sideri TC, Grant CM, Tamas MJ. Global analysis of protein aggregation in yeast during physiological conditions and arsenite stress. Biol. Open. 2014;3:913–923. doi: 10.1242/bio.20148938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia NT. Ribosome profiling: New views of translation, from single codons to genome scale. Nature Reviews Genetics. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 27.Chassé H, Boulben S, Costache V, Cormier P, Morales J. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017;45:e15. doi: 10.1093/nar/gkw1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnside Daniel, Moteshareie Houman, Marquez Imelda G., Hooshyar Mohsen, Samanfar Bahram, Shostak Kristina, Omidi Katayoun, Peery Harry E., Smith Myron L., Golshani Ashkan. Bioactive Natural Products. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2015. Use of Chemical Genomics to Investigate the Mechanism of Action for Inhibitory Bioactive Natural Compounds; pp. 9–32. [Google Scholar]
- 29.Galván Márquez I, et al. Disruption of protein synthesis as antifungal mode of action by chitosan. Int. J. Food Microbiol. 2013;164:108–112. doi: 10.1016/j.ijfoodmicro.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Bessarabova M, Ishkin A, JeBailey L, Nikolskaya T, Nikolsky Y. Knowledge-based analysis of proteomics data. BMC Bioinformatics. 2012;13(Suppl 1):S13. doi: 10.1186/1471-2105-13-S16-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao VS, Srinivas K, Sujini GN, Kumar GNS. Protein-Protein Interaction Detection: Methods and Analysis. Int. J. Proteomics. 2014;2014:1–12. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McHale CM, et al. Assessing health risks from multiple environmental stressors: Moving from G × E to I × E. Mutation Research - Reviews in Mutation Research. 2018;775:11–20. doi: 10.1016/j.mrrev.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Escher BI, et al. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ. Int. 2017;99:97–106. doi: 10.1016/j.envint.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun RJ, Büttner S, Ring J, Kroemer G, Madeo F. Nervous yeast: modeling neurotoxic cell death. Trends in Biochemical Sciences. 2010;35:135–144. doi: 10.1016/j.tibs.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Reddi AR, Jensen LT, Culotta VC. Manganese homeostasis in saccharomyces cerevisiae. Chem. Rev. 2009;109:4722–4732. doi: 10.1021/cr900031u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason RP, Giorgini F. Modeling huntington disease in yeast: Perspectives and future directions. Prion. 2011;5:269–276. doi: 10.4161/pri.18005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Büttner S, et al. Endonuclease G mediates α-synuclein cytotoxicity during Parkinson’s disease. EMBO J. 2013;32:3041–3054. doi: 10.1038/emboj.2013.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ash David E. Manganese in Metabolism and Enzyme Function. 1986. METHODS OF Mn(II) DETERMINATION11Studies of Mn(II) levels in rat hepatocytes were supported by NIH grants AM-25551 to Dr. V.L. Schramm, Temple University and AM-17517 to Dr. G.H. Reed, University of Pennsylvania. Studies of Mn(II) uptake by Chlamydomonas were supported by Biomedical Research Support Grant S07 RR05417 to Dr. D.E. Ash and NSF grant PCM-8402330 to Dr. J.H. Hoober; pp. 327–356. [Google Scholar]
- 40.Rudgalvyte M, Peltonen J, Lakso M, Nass R, Wong G. RNA-Seq Reveals Acute Manganese Exposure Increases Endoplasmic Reticulum Related and Lipocalin mRNAs in Caenorhabditis elegans. J. Biochem. Mol. Toxicol. 2016;30:97–105. doi: 10.1002/jbt.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu B, et al. Endoplasmic reticulum stress signaling involvement in manganese-induced nerve cell damage in organotypic brain slice cultures. Toxicol. Lett. 2013;222:239–246. doi: 10.1016/j.toxlet.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Xu B, et al. Alpha-synuclein is involved in manganese-induced ER stress via PERK signal pathway in organotypic brain slice cultures. Molecular Neurobiology. 2014;49:399–412. doi: 10.1007/s12035-013-8527-2. [DOI] [PubMed] [Google Scholar]
- 43.Cohen Yifat, Megyeri Márton, Chen Oscar C. W., Condomitti Giuseppe, Riezman Isabelle, Loizides-Mangold Ursula, Abdul-Sada Alaa, Rimon Nitzan, Riezman Howard, Platt Frances M., Futerman Anthony H., Schuldiner Maya. The Yeast P5 Type ATPase, Spf1, Regulates Manganese Transport into the Endoplasmic Reticulum. PLoS ONE. 2013;8(12):e85519. doi: 10.1371/journal.pone.0085519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polymenis M, Aramayo R. Microb. Cell. 2015. Translate to divide: сontrol of the cell cycle by protein synthesis; pp. 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dasuri K, Zhang L, Keller JN. Oxidative stress, neurodegeneration, and the balance of protein degradation and protein synthesis. Free Radical Biology and Medicine. 2013;62:170–185. doi: 10.1016/j.freeradbiomed.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Negga R, et al. Exposure to Mn/Zn ethylene-bis-dithiocarbamate and glyphosate pesticides leads to neurodegeneration in Caenorhabditis elegans. Neurotoxicology. 2011;32:331–341. doi: 10.1016/j.neuro.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King, B. L., Davis, A. P., Rosenstein, M. C., Wiegers, T. C. & Mattingly, C. J. Ranking Transitive Chemical-Disease Inferences Using Local Network Topology in the Comparative Toxicogenomics Database. PLoS One7 (2012). [DOI] [PMC free article] [PubMed]
- 48.Davis AP, et al. The Comparative Toxicogenomics Database: Update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar KK, et al. Cellular manganese content is developmentally regulated in human dopaminergic neurons. Sci. Rep. 2014;4:6801. doi: 10.1038/srep06801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuormaa, T. E. The Adverse Effects of Manganese Deficiency on Reproduction and Health: A Literature Review. 27–29 (1996).
- 51.Jauniaux JC, Urrestarazu LA, Wiame JM. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J. Bacteriol. 1978;133:1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao Y, Wang J-D, Hung D-Z, Deng J-F, Yang C-C. Hyperammonemia following glufosinate-containing herbicide poisoning: A potential marker of severe neurotoxicity. Clin. Toxicol. 2011;49:48–52. doi: 10.3109/15563650.2010.539184. [DOI] [PubMed] [Google Scholar]
- 53.Patin F, et al. Omics to Explore Amyotrophic Lateral Sclerosis Evolution: the Central Role of Arginine and Proline Metabolism. Mol Neurobiol. 2017;54:5361–5374. doi: 10.1007/s12035-016-0078-x. [DOI] [PubMed] [Google Scholar]
- 54.Hu XP, Yang Y, Ma BG. Amino Acid Flux from Metabolic Network Benefits Protein Translation: The Role of Resource Availability. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shahbazian D, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darling NJ, Cook SJ, Krebs J, Moreau M. Biochimica et Biophysica Acta The role of MAPK signalling pathways in the response to endoplasmic reticulum stress ☆. BBA - Mol. Cell Res. 2014;1843:2150–2163. doi: 10.1016/j.bbamcr.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Liu D-Z, Ander BP. Cell cycle inhibition without disruption of neurogenesis is a strategy for treatment of aberrant cell cycle diseases: an update. ScientificWorldJournal. 2012;2012:491737. doi: 10.1100/2012/491737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta - Mol. Basis Dis. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 59.Seo YA, Li Y, Wessling-Resnick M. Iron depletion increases manganese uptake and potentiates apoptosis through ER stress. Neurotoxicology. 2013;38:67–73. doi: 10.1016/j.neuro.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang T, et al. ER stress and ER stress-mediated apoptosis are involved in manganese-induced neurotoxicity in the rat striatum in vivo. Neurotoxicology. 2015;48:109–119. doi: 10.1016/j.neuro.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Hetz C, Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017;13:477–491. doi: 10.1038/nrneurol.2017.99. [DOI] [PubMed] [Google Scholar]
- 62.Fruhmann G, et al. Yeast buddies helping to unravel the complexity of neurodegenerative disorders. Mechanisms of Ageing and Development. 2017;161:288–305. doi: 10.1016/j.mad.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Park JH, et al. SLC39A8 Deficiency: A Disorder of Manganese Transport and Glycosylation. Am. J. Hum. Genet. 2015;97:894–903. doi: 10.1016/j.ajhg.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potelle S, et al. Glycosylation abnormalities in Gdt1p/TMEM165 deficient cells result from a defect in Golgi manganese homeostasis. Hum. Mol. Genet. 2016;25:1489–1500. doi: 10.1093/hmg/ddw026. [DOI] [PubMed] [Google Scholar]
- 65.Allan Drummond D, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nature Reviews Genetics. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruddock LW, Molinari M. N-glycan processing in ER quality control. J. Cell Sci. 2006;119:4373–4380. doi: 10.1242/jcs.03225. [DOI] [PubMed] [Google Scholar]
- 67.Johannes, L. & Popoff, V. Review Tracing the Retrograde Route in Protein Trafficking. 5, 1175–1187 (2008). [DOI] [PubMed]
- 68.Progida, C. & Bakke, O. Bidirectional traffic between the Golgi and the endosomes – machineries and regulation. 1–12, 10.1242/jcs.185702 (2016). [DOI] [PubMed]
- 69.Tsvetanova, N. G. The secretory pathway in control of endoplasmic reticulum homeostasis. 4, 28–33 (2013). [DOI] [PMC free article] [PubMed]
- 70.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006;7:568. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 71.Chesi, A., Kilaru, A., Fang, X., Cooper, A. A. & Gitler, A. D. The Role of the Parkinson’ s Disease Gene PARK9 in Essential Cellular Pathways and the Manganese Homeostasis Network in Yeast. 7 (2012). [DOI] [PMC free article] [PubMed]
- 72.Van Damme P, Robberecht W, Van Den Bosch L. Modelling amyotrophic lateral sclerosis: progress and possibilities. Dis. Model. Mech. 2017;10:537–549. doi: 10.1242/dmm.029058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C, et al. Manganese exposure disrupts SNARE protein complex-mediated vesicle fusion in primary cultured neurons. Environ. Toxicol. 2017;32:705–716. doi: 10.1002/tox.22272. [DOI] [PubMed] [Google Scholar]
- 74.Lee SC, Pappone PA. ATP can stimulate exocytosis in rat brown adipocytes without apparent increases in cytosolic Ca2+ or G protein activation. Biophys. J. 1999;76:2297–2306. doi: 10.1016/S0006-3495(99)77385-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, C. et al. Inhibition of Calpains Protects Mn-Induced Neurotransmitter release disorders in Synaptosomes from Mice: Involvement of SNARE Complex and Synaptic Vesicle Fusion. Sci. Rep.7 (2017). [DOI] [PMC free article] [PubMed]
- 76.Gitler AD, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hernández RB, Nishita MI, Espósito BP, Scholz S, Michalke B. The role of chemical speciation, chemical fractionation and calcium disruption in manganese-induced developmental toxicity in zebrafish (Danio rerio) embryos. J. Trace Elem. Med. Biol. 2015;32:209–217. doi: 10.1016/j.jtemb.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Al-Jubran K, et al. Visualization of the joining of ribosomal subunits reveals the presence of 80S ribosomes in the nucleus. Rna. 2013;19:1669–1683. doi: 10.1261/rna.038356.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu J, et al. Identification of the Key Molecules Involved in Chronic Copper Exposure-Aggravated Memory Impairment in Transgenic Mice of Alzheimer’s Disease Using Proteomic. Analysis. J. Alzheimer’s Dis. 2015;44:455–469. doi: 10.3233/JAD-141776. [DOI] [PubMed] [Google Scholar]
- 80.Tarale P, et al. Global DNA methylation profiling of manganese-exposed human neuroblastoma SH-SY5Y cells reveals epigenetic alterations in Parkinson’s disease-associated genes. Arch. Toxicol. 2017;91:2629–2641. doi: 10.1007/s00204-016-1899-0. [DOI] [PubMed] [Google Scholar]
- 81.Bevan R, Ashdown L, McGough D, Huici-Montagud A, Levy L. Setting evidence-based occupational exposure limits for manganese. Neurotoxicology. 2017;58:238–248. doi: 10.1016/j.neuro.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 82.Alamgir, M., Erukova, V., Jessulat, M., Azizi, A. & Golshani, A. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem. Biol. 10 (2010). [DOI] [PMC free article] [PubMed]
- 83.Wagih, O. et al. SGAtools: One-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 41 (2013). [DOI] [PMC free article] [PubMed]
- 84.Memarian N, et al. Colony size measurement of the yeast gene deletion strains for functional genomics. BMC Bioinformatics. 2007;8:117. doi: 10.1186/1471-2105-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samanfar, B. et al. The sensitivity of the yeast, Saccharomyces cerevisiae, to acetic acid is influenced by DOM34 and RPL36A. PeerJ2017 (2017). [DOI] [PMC free article] [PubMed]
- 86.Galván Márquez I, et al. Zinc oxide and silver nanoparticles toxicity in the baker’s yeast, Saccharomyces cerevisiae. PLoS One. 2018;13:e0193111–e0193111. doi: 10.1371/journal.pone.0193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costa C, et al. Quantitative Real-Time PCR Assay for Rapid Identification of Deletion Carriers in Hemophilia. Clin. Chem. 2004;50:1269 LP–1270. doi: 10.1373/clinchem.2004.031609. [DOI] [PubMed] [Google Scholar]
- 88.Traverso M, et al. Multiplex real-time PCR for detection of deletions and duplications in dystrophin gene. Biochem. Biophys. Res. Commun. 2006;339:145–150. doi: 10.1016/j.bbrc.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 89.Alamgir, M., Eroukova, V., Jessulat, M., Xu, J. & Golshani, A. Chemical-genetic profile analysis in yeast suggests that a previously uncharacterized open reading frame, YBR261C, affects protein synthesis. BMC Genomics9 (2008). [DOI] [PMC free article] [PubMed]
- 90.Loukin, S. & Kung, C. Manganese Effectively Supports Yeast Cell-Cycle Progression in Place of Calcium. 13 (1995). [DOI] [PMC free article] [PubMed]
- 91.Blackwell, K. J. & Tobin, J. M. Manganese toxicity towards Saccharomyces cerevisiae: Dependence on intracellular and extracellular magnesium concentrations. 751–757 (1998). [DOI] [PubMed]
- 92.Daly, M. J. Manganese Complexes: Diverse Metabolic Routes to Oxidative Stress Resistance in Prokaryotes and Yeast. 19 (2013). [DOI] [PMC free article] [PubMed]
- 93.Liang Q, Zhou B. Copper and Manganese Induce Yeast Apoptosis via Different. Pathways. 2007;18:4741–4749. doi: 10.1091/mbc.E07-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Esposito, A. M. et al. Eukaryotic Polyribosome Profile Analysis. J. Vis. Exp.,10.3791/1948 (2010). [DOI] [PMC free article] [PubMed]
- 95.Faye, M. D., Graber, T. E. & Holcik, M. Assessment of Selective mRNA Translation in Mammalian Cells by Polysome Profiling. J. Vis. Exp., 10.3791/52295 (2014). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and/or analyzed during this study are included in this published article and/or its Supplementary Material Files).