Abstract
Probiotics are live microorganisms conferring health benefits when administered in adequate amounts. However, the passage through the gastrointestinal tract represents a challenge due to pH variations, proteases, and bile salts. This study aimed to evaluate the proteomic response of Saccharomyces boulardii to simulated gastrointestinal digestion and the influence of encapsulation on yeast viability. Different pH values and time periods simulating the passage through different sections of the gastrointestinal tract were applied to unencapsulated and encapsulated yeasts. Encapsulation in 0.5% calcium alginate did not improve yeast survival or induce changes in protein patterns whereas protein extracts from control and digested yeasts showed remarkable differences when separated by SDS-PAGE. Protein bands were analyzed by tandem mass spectrometry. Protein identification revealed unique proteins that changed acutely in abundance after simulated digestion. Carbohydrate metabolism, protein processing, and oxide-reduction were the biological processes most affected by simulated gastrointestinal digestion in S. boulardii.
Electronic supplementary material
The online version of this article (10.1007/s10068-018-0508-9) contains supplementary material, which is available to authorized users.
Keywords: Electrophoresis, Probiotic yeast, Stress, Tandem mass spectrometry, Viability
Introduction
Currently, consumers seek functional or bioactive foods that provide additional health benefits (Kailasapathy, 2015). Probiotics are live microorganisms capable of providing these benefits when supplied viable and in adequate amounts [≥ 107 Colony-Forming Units (CFU)/g or mL of food] (Mortazavian et al., 2007). The main foods used as a vehicle for probiotics are those of dairy origin. However, they can also be included in other types of food (Corona-Hernández et al., 2013).
Before being included in food products, probiotics must demonstrate to be innocuous, functional, genetically stable and fulfill important requirements for the industry (Saarela et al., 2000). In addition, it is essential that they can reach the intestine, adhere and colonize it (often transiently) to exert beneficial effects (Bustos et al., 2015).
Encapsulation is a technique by which the desired product is protected from detrimental external influences. Alginate is a common biomaterial used for encapsulation or immobilization of probiotics (Basu et al., 2018). It is food grade, low cost, there are automated methods to generate capsules, and has been used extensively in the concentration range of 0.5–5%, as reported in old and recent reports (Hansen et al., 2002; Mandal et al., 2006; Mathews, 2017).
Encapsulation help probiotics acclimated or not to industrial conditions to reach viable and in larger quantities to the intestine; however, even not encapsulated, a small proportion of probiotics can survive the passage through the Gastrointestinal Tract (GIT), which leads us to question how the probiotic yeast S. boulardii faces this type of stress. Identification of changes in protein accumulation by proteomic tools have been used successfully for identification of stress adaptation markers, as well as in the prediction of the behavior of probiotics in the food matrix and its ability to survive the passage through the GIT, as well as to understand the molecular mechanisms behind the functional properties and the tolerance to adverse conditions in probiotics (Ruiz et al., 2016).
Although genetically close to Saccharomyces cerevisiae, the behavior of Saccharomyces boulardii is very different in growth yield and resistance to conditions faced by probiotic microorganisms such as acid and temperature stress (Fietto et al., 2004). Furthermore, it is widely used as a treatment or prophylactic for several diseases related to a bacterial infection and bowel inflammation (Wu et al., 2014). The aim of this work was to evaluate the viability of Saccharomyces boulardii and to identify changes in the abundance in total soluble proteins in unencapsulated cells and those encapsulated in 0.5% calcium alginate when both were subjected to simulated gastrointestinal digestion (SGID) involving the passage of cells through 5 different buffer pH/enzyme conditions corresponding to each section of the GIT over a total of 10.25 h, using a gel-based proteomic approach.
Materials and methods
Probiotic strain isolation and 18S rRNA sequencing
Saccharomyces cerevisiae subs. boulardii was purchased from a commercial lyophilizate used as a food supplement (Florastor®, Biocodex Inc. Redwood City, CA, USA). Lyophilized yeast contained in a capsule (250 mg), was growth in 100 mL of liquid Yeast Extract Peptone Dextrose (YPD) medium (1% Yeast extract, 2% Bacteriological peptone and 2% Glucose) by incubation at 30 °C, for 48 h, under aerobic conditions. Then, cells were collected by centrifugation at 10,000×g for 15 min at 4 °C (Sorvall Lynx 4000, Thermo Scientific, San Jose, CA, USA). The supernatant was discarded, and two additional washes were made with sterile water. This sample was stored as a stock at − 70 °C. Colony PCR was performed from a culture on solid medium. The ITS1-5.8S-IT2 region was amplified by PCR and sequenced. Identification corresponded to a Saccharomyces ‘boulardii’ isolate.
Inoculum standardization and yeast encapsulation
A freezer stock sample was incubated overnight in YPD medium at 30 °C, yeast culture absorbance was recorded, and cells were diluted in 0.9% sodium chloride and adjusted to an absorbance of 1.0 ± 0.05 at 600 nm (≈ 1.3 × 107 CFU/mL). The encapsulation mixture consisted of the adjusted cell suspension, brought to a volume of 10 mL (1.49 × 1010 CFU) and 40 mL of sodium alginate that were mixed by agitation until obtaining a homogeneous mixture, with a final concentration of 0.5% of alginate. Capsules were generated by the extrusion method, using a new and sterile 20 mL syringe, with a 27G gauge needle (0.40 mm diameter), loaded with the aforementioned mixture that was dropped into a 0.4 M calcium chloride solution which was kept under constant stirring and the drops were allowed to gel for 20 min (Ghorbani-Choboghlo et al., 2015). The resulting capsules were washed with Milli-Q water and stored at 4 °C for 24 h.
Encapsulation efficiency
The efficiency of encapsulation was estimated by the method used by Mokarram et al. (2009) with some modifications. Ten grams of the encapsulated probiotic were taken to a volume of 50 mL with sterile 1% sodium citrate solution. The mixture was kept in constant agitation until full capsules breakdown. Encapsulation efficiency was determined by serial dilutions and plate count by triplicate in YPD medium. Efficiency was estimated from the yeasts trapped in the alginate network, and that survived the encapsulation process.
Simulated gastrointestinal conditions
SGID was carried out as reported by Gbassi et al. (2011). Such conditions are commonly used in studies in which viability of probiotics is evaluated under SGID. Briefly: Phosphate buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl) was used as the gastrointestinal fluid and was maintained at a constant temperature of 37 °C under agitation at 100 rpm, with a shaker (New Brunswick Scientific Co., INC., Enfield, CT, USA). Table 1 shows the different pH values and time periods of exposure of the samples to the SGID. In each experimental run, 4 conditions were evaluated, unencapsulated and encapsulated S. boulardii, submitted to SGID, each with a control that was maintained at a constant pH of 6.5 during the 10.25 h of the experiment (Gbassi et al., 2011).
Table 1.
Simulated gastrointestinal digestion
| Parameter | Compartments | ||||
|---|---|---|---|---|---|
| Stomach | Duodenum | Jejunum | Ileum | Caecum | |
| pH value | 2.5 | 5.5 | 6.5 | 7.5 | 7.5 |
| Incubation time (h) | 2 | 0.25 | 3 | 4 | 1 |
| Pepsin 3 g/L | Pancreatin 10 g/L; bile salts 3 g/L | ||||
Simulated gastrointestinal digestion for unencapsulated and encapsulated yeasts consisted of five compartments with a specific pH value and incubation time for each compartment (for a total of 10.25 h). Pepsin was added in the stomach whereas pancreatin and bile salts were added when simulating the duodenum. Whereas unencapsulated and encapsulated control yeasts were maintained at constant pH 6.5 for 10.25 h (Gbassi et al., 2011)
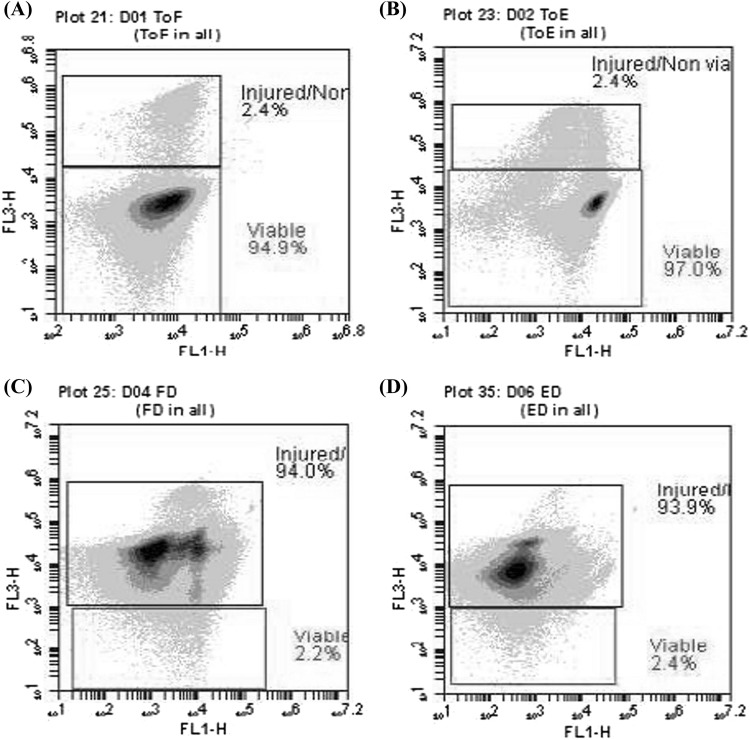
After 10.25 h of simulation, 0.1 mL was taken individually from every sample by triplicate (unencapsulated and encapsulated yeasts subjected to control conditions and SGID) and serial dilutions were made in sterile saline (0.9% NaCl) and inoculated in Petri dishes containing YPD medium for plate count in order to estimate yeast survival. The viability of unencapsulated and encapsulated yeast cells was also evaluated with BD Cell Viability Kit (BD Biosciences, San Jose CA, USA). Yeast samples (500 μL) were stained with thiazole orange, propidium iodide, and fluorescent counting beads to calculate absolute counts. Samples were incubated 15 min at room temperature. A volume of 30 μL was analyzed on an Accuri C6 flow Cytometer (BD Biosciences, San Jose CA, USA) and BD CSampler software used to process samples. The remaining sample was centrifuged at 13,000×g for 15 min at 4 °C. The supernatant was discarded and the pellet was washed twice with saline under the same conditions and the final pellet was stored at − 70 °C. Three independent experiments (biological replicates) were carried out.
Protein extraction and SDS-PAGE
Frozen pellets were placed in 15 mL tubes, and protein extraction was performed by the method reported by Faurobert et al. (2007). Total soluble proteins were suspended in 1 mL of rehydration buffer [8 M Urea, 2% CHAPS, 0.5% IPG buffer (pH 3–10), 20 mM dithiothreitol]. Samples were cleaned by second precipitation with 0.15 M ammonium acetate. Protein pellets resulting from the cleaning procedure were suspended in 550 μL of rehydration buffer. Protein was determined by the Bradford method using bovine serum albumin as standard. Proteins (30 μg for every sample) were separated by SDS-PAGE under denaturing and reducing conditions in a 13% polyacrylamide gel, and 2 μL of wide range molecular weight was used as a reference (Bio-Rad, Hercules, CA, USA).
In-gel digestion and tandem mass spectrometry analysis (LC–MS/MS)
Protein band sections were excised from gels, reduced with 10 mM DTT in 25 mM ammonium bicarbonate followed by protein alkylation with 55 mM iodoacetamide. Protein digestion was carried out overnight at 37 °C with sequencing grade trypsin (Promega, Madison, WI, USA).
LC separation of tryptic peptides was performed by ultra-performance liquid chromatography using the 1290 Infinity LC System (Agilent Technologies, Santa Clara, CA, USA) equipped with an analytical column ZORBAX 300SB-C8 (5 µm × 2.1 mm × 150 mm, Agilent Technologies, Santa Clara, CA, USA) equilibrated with 1% acetonitrile (ACN) and 0.1% formic acid (FA) that was maintained at 35 °C. Peptides were separated by using a flow of 400 μL/min and the following chromatographic conditions: 1 min with 99% A (H2O with 0.1% FA) and 1% B (ACN with 0.1% FA); followed by a linear gradient until reaching 70% of B in 15 min and a final gradient of 1 min until reaching 1% of B; with an equilibrium period of 3 min (1% B) between each run. Peptides eluted from the column were ionized by electrospray with a Dual AJS ESI ionization source (Agilent Technologies, Santa Clara, CA, USA) applying 3.5 kV and analyzed by tandem mass spectrometry through a data-dependent analysis in the 6530 Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) LC/MS system (Agilent Technologies, Santa Clara, CA, USA) operated in positive mode. Data was acquired in auto MS/MS mode, the mass range in MS mode was 400–2000 m/z (3 spectra/s) and in MS/MS mode from 50 to 2000 m/z (1 spectrum/s), with a maximum of 5 precursors per cycle.
Protein identification and functional classification
MS/MS raw data (.d files) were processed in the Spectrum Mill MS Proteomics Workbench server (Agilent Technologies, Santa Clara, CA, USA) to obtain .mzXML files which were transformed into .mgf files in the MSConvert program (available at http://proteowizard.sourceforge.net/). Proteins were then identified using the .mgf files and the MASCOT search engine (http://www.matrixscience.com). Searches were conducted against fungi subset of the SwissProt protein database (32,838 sequences, October 2017). Trypsin was used as the specific protease, and one missed cleavage was allowed. The mass tolerance for precursor and fragment ions was set to 20 ppm and 0.1 Da, respectively. Carbamidomethyl cysteine was set as fixed modification and oxidation of methionine was specified as variable modification. Protein identifications were considered successful when at least two peptides and significant MASCOT scores (> 39) were obtained, indicating the identity or extensive homology at p < 0.01. Unique proteins identified only under control conditions and only under SGID were classified based on the biological process in which they participate according to the Gene Ontology annotations using Blast2GO (available at www.blast2go.com).
Results and discussion
Encapsulation efficiency
Plate count results for the initial inoculum and the yeasts released from alginate capsules were calculated on the basis of three different experiments (biological replications). Results showed an average concentration of 1.47 × 108 CFU/mL for free yeasts, whereas after encapsulation and release there was a survival of 2.99 × 107 CFU/mL (51.1% encapsulation efficiency using 0.5% alginate). This result is similar to previous reports (Hugues-Ayala, 2013), where encapsulation rates of 45 and 66% were obtained, for Lactobacillus rhamnosus GG using alginate at 1 and 1.5% respectively. Encapsulation efficiency depends on conditions used to generate the capsules such as alginate concentration, encapsulation method and whether or not an automated device was used (Anal and Singh, 2007). Our results indicate that the parameters and conditions used to encapsulate S. boulardii by means of the extrusion method, allowed reaching the recommended quantities of probiotics to be subjected to SGID.
Survival of Saccharomyces boulardii to SGID
Unencapsulated and encapsulated cells were subjected SGID, in which unencapsulated cells and those protected with 0.5% alginate were exposed to changes in the pH of the medium and the presence of proteases (pepsin and pancreatin) and bile salts. SGID lasted 10.25 h. In the end, plate count was performed by serial dilutions to estimate the survival of probiotic yeast (Table 2). Such low survival proportion was also observed when viability was evaluated by flow cytometry analysis (Fig. 1). If it is considered the encapsulation efficiency, a lower number of encapsulated cells (almost half) were subjected to SGID. Indicating that encapsulation with alginate 0.5% promoted the survival of probiotics in a tiny proportion. However, this effect must be considered not significant for practical purposes.
Table 2.
Survival of S. boulardii subjected unencapsulated or encapsulated to simulated gastrointestinal digestion
| Free yeasts | Encapsulated yeasts | |||
|---|---|---|---|---|
| CFU (mL) | Survival (%) | CFU (mL) | Survival (%) | |
| Initial amount | 1.95 × 108 | – | 9.96 × 107 | – |
| Controls (pH 6.5 for 10.25 h) | 7.2 × 107 | 37.0 | 3.3 × 107 | 33.4 |
| Simulated digestion (10.25 h)a | 4.3 × 106 | 2.2 | 3.3 × 106 | 3.4 |
Probiotic yeasts were stored for 12 h at 4 °C prior to simulated gastrointestinal digestion
aSimulated gastrointestinal digestion consisted of five compartments with a specific pH value and incubation time for each compartment. Pepsin, pancreatin and bile salts were added as indicated in Table 1
Fig. 1.
Viability of S. boulardii before being subjected unencapsulated (A) or encapsulated (B) to simulated gastrointestinal digestion. Viability of unencapsulated (C) and encapsulated (D) probiotic yeasts after simulated gastrointestinal digestion
Probiotics should be supplied in quantities of 1 × 108–1 × 109 CFU/mL in food and that after the SGID should prevail between 1 × 106 and 1 × 107 CFU/mL to be able to exert the benefits attributed to probiotics (Shori, 2017). Probiotic yeast amounts used at the beginning and those reached at the end of SGID for both unencapsulated and encapsulated yeasts used in our experiment coincide with that recommended elsewhere in the literature.
Proteomic changes in free and encapsulated probiotic yeast subjected to simulated digestion
Total soluble proteins were extracted from S. boulardii subjected to four experimental conditions: (1) unencapsulated yeasts subjected to control conditions (UC); (2) unencapsulated yeasts subjected to SGID (UD); (3) encapsulated yeasts subjected to control conditions (EC); and (4) encapsulated yeasts subjected to SGID (ED), which, after quantification by the Bradford method, were separated by 13% SDS-PAGE as shown in Fig. 2.
Fig. 2.
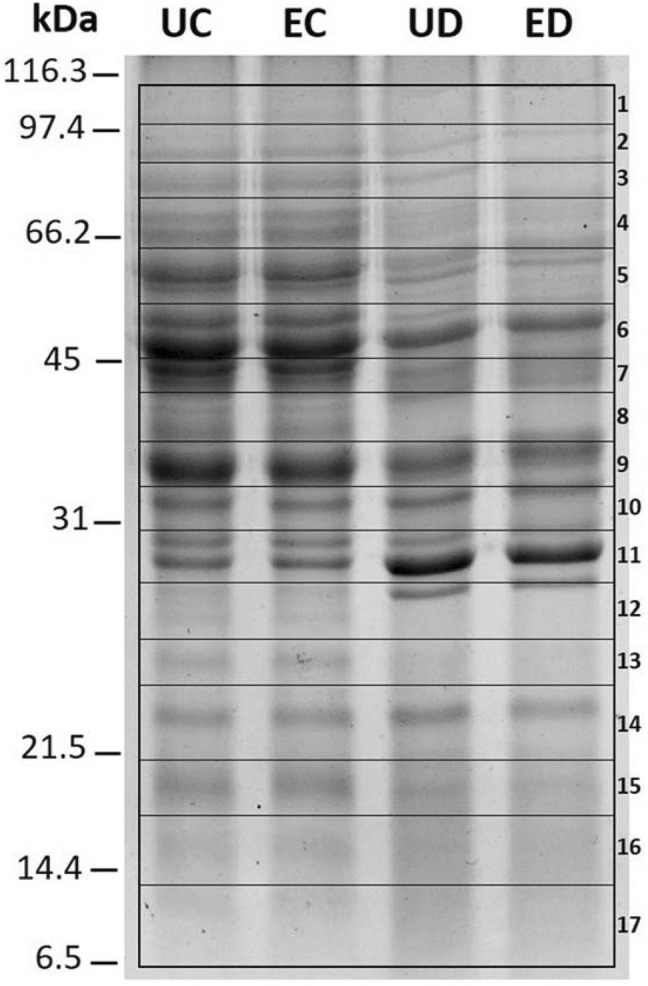
SDS-PAGE (13%) of total soluble proteins of Saccharomyces boulardii extracted from unencapsulated (U) or encapsulated (E) probiotic, subjected to control conditions (C) or after simulated gastrointestinal digestion (D). Numbers indicate gel sections used for protein identification based on tandem mass spectrometry
Electrophoretic patterns from yeast proteins subjected to SGID were dramatically different from those of yeasts subjected to control conditions, independently of the encapsulation process. Whereas the electrophoretic patterns between unencapsulated and encapsulated samples but exposed to the same treatment (control or SGID) showed no noticeable changes in the band pattern (Fig. 2), indicating that encapsulation did not affect protein accumulation patterns.
Seventeen gel sections with molecular masses from 6.4 to 112 kDa (As indicated with numbers in Fig. 2) for each of the 4 experimental conditions (UC, UD, EC, ED), were digested (in-gel) with trypsin and analyzed by LC–MS/MS. MS data for each of the 68 samples were analyzed with the Mascot search engine (www.matrixscience.com) for identification against the fungi subset of the SwissProt protein database.
Interestingly, 29 proteins were identified only under control conditions regardless of whether yeasts were unencapsulated or encapsulated, whereas 10 unique proteins were identified only in yeasts subjected to SGID, regardless of whether yeast was encapsulated or not. Only two proteins were identified only in unencapsulated yeasts, whereas one protein was identified only in encapsulated yeasts (Table 3, Supplementary Table 1).
Table 3.
Identification of differentially accumulated proteins in the probiotic yeast S. boulardii
| Banda | Protein | Identifierb | Exp. Mwc | Theor. Mw/pId | Mascot score | PM/SCe |
|---|---|---|---|---|---|---|
| Control conditions | ||||||
| 17 | 10 kDa heat shock protein, mitochondrial | CH10_YEAST | 6.4–13.0 | 11.4/8.96 | 79 | 2/25% |
| 15 | 40S ribosomal protein S12 | RS12_YEAST | 18.4–20.9 | 15.5/4.68 | 66 | 2/14% |
| 11 | 40S ribosomal protein S5 | RS5_YEAST | 28.1–30.7 | 25.1/8.63 | 409 | 3/17% |
| 13 | 40S ribosomal protein S7-A | RS7A_YEAST | 24.2–26.2 | 21.6/9.83 | 55 | 2/13% |
| 9 | 60S acidic ribosomal protein P0 | RLA0_YEAST | 34.4–38.2 | 33.7/4.75 | 89 | 4/14% |
| 15 | 60S ribosomal protein L14-A | RL14A_YEAST | 18.4–20.9 | 15.2/10.94 | 118 | 3/26% |
| 13 | 60S ribosomal protein L18-A | RL18A_YEAST | 24.2–26.2 | 20.6/11.70 | 72 | 2/11% |
| 15 | 60S ribosomal protein L25 | RL25_YEAST | 18.4–20.9 | 15.7/10.11 | 198 | 2/18% |
| 17 | 60S ribosomal protein L30 | RL30_YEAST | 6.4–13.0 | 11.4/9.80 | 74 | 3/45% |
| 16 | 60S ribosomal protein L31-A | RL31A_YEAST | 13.0–18.4 | 12.9/9.99 | 47 | 2/23% |
| 16 | 60S ribosomal protein L36-A | RL36A_YEAST | 13.0–18.4 | 11.1/11.60 | 48 | 2/21% |
| 6 | 6-Phosphogluconate dehydrogenase, decarboxylating 1 | 6PGD1_YEAST | 45.5–54.2 | 53.9/6.19 | 164 | 4/11% |
| 3 | Aconitate hydratase, mitochondrial | ACON_YEAST | 79.3–88.1 | 85.7/8.17 | 197 | 8/11% |
| 2 | Aminopeptidase 2, mitochondrial | APE2_YEAST | 88.1–98.1 | 108.1/8.11 | 47 | 4/5% |
| 11 | Heat shock protein 26 | HSP26_YEAST | 28.1–30.7 | 23.9/5.31 | 60 | 2/13% |
| 4 | Heat shock protein SSA2 | HSP72_YEAST | 64.4–79.3 | 69.6/4.95 | 240 | 11/25% |
| 4 | Heat shock protein SSC1, mitochondrial | HSP77_YEAST | 64.4–79.3 | 70.6/5.48 | 121 | 8/16% |
| 5 | Hexokinase-1 | HXKA_YEAST | 54.2–64.4 | 53.9/5.28 | 142 | 6/18% |
| 16 | Histone H4 | H4_YEAST | 13.0–18.4 | 11.36/11.3 | 61 | 3/29% |
| 6 | Homocysteine/cysteine synthase | CYSD_YEAST | 45.5–54.2 | 48.7/5.97 | 167 | 3/7% |
| 4 | Isocitrate lyase | ACEA_YEAST | 64.4–79.3 | 62.7/5.97 | 115 | 5/10% |
| 7 | Malate dehydrogenase, cytoplasmic | MDHC_YEAST | 41.2–45.5 | 41.0/6.41 | 150 | 3/16% |
| 7 | Mannose-1-phosphate guanylyltransferase | MPG1_YEAST | 41.2–45.5 | 39.7/5.95 | 154 | 3/12% |
| 8 | Putative pyridoxal reductase | PLR1_YEAST | 38.2–41.2 | 38.9/5.66 | 104 | 3/11% |
| 16 | Restriction of telomere capping protein 3 | SDO1L_YEAST | 13.0–18.4 | 12.0/5.05 | 108 | 3/44% |
| 8 | Ribonucleoside-diphosphate reductase small chain 2 | RIR4_YEAST | 38.2–41.2 | 40.1/5.11 | 81 | 4/19% |
| 7 | Sphingolipid long chain base-responsive protein PIL1 | PIL1_YEAST | 41.2–45.5 | 38.3/4.54 | 63 | 4/17% |
| 8 | Transaldolase | TAL1_YEAST | 38.2–41.2 | 37.1/6.09 | 119 | 4/15% |
| 17 | Ubiquitin-60S ribosomal protein L40 | RL40A_YEAST | 6.4–13.0 | 14.8/9.87 | 153 | 3/26% |
| Simulated gastrointestinal digestion | ||||||
| 7 | Dihydrolipoyl dehydrogenase, mitochondrial | DLDH_YEAST | 41.2–45.5 | 54.3/8.07 | 160 | 4/12% |
| 10 | Glyceraldehyde-3-phosphate dehydrogenase 1 | G3P1_YEAST | 30.7–34.4 | 35.8/8.29 | 138 | 5/23% |
| 2 | Invertase 2 | INV2_YEAST | 88.1–98.1 | 60.7/4.61 | 84 | 6/11% |
| 8 | Ribonucleoside-diphosphate reductase small chain 1 | RIR2_YEAST | 38.2–41.2 | 46.2/5.14 | 115 | 3/7% |
| 6 | S-(hydroxymethyl)glutathione dehydrogenase | FADH_YEAST | 45.5–54.2 | 41.9/6.33 | 58 | 3/8% |
| 7 | Saccharopepsin | CARP_YEAST | 41.2–45.5 | 44.7/4.70 | 224 | 7/20% |
| 6 | Serine hydroxymethyltransferase, cytosolic | GLYC_YEAST | 45.5–54.2 | 52.5/6.98 | 87 | 4/10% |
| 14 | Superoxide dismutase [Cu–Zn] | SODC_YEAST | 20.9–24.2 | 16.0/5.62 | 207 | 9/88% |
| 5 | Threonine synthase | THRC_YEAST | 54.2–64.4 | 57.6/5.46 | 190 | 5/12% |
| 5 | Vacuolar aminopeptidase 1 | AMPL_YEAST | 54.2–64.4 | 57.3/5.55 | 181 | 4/12% |
| Encapsulated cells | ||||||
| 12 | 40S ribosomal protein S8-A | RS8A_YEAST | 26.7–28.1 | 22.6/10.67 | 105 | 3/20% |
| Unencapsulated cells | ||||||
| 3 | Acetyl-coenzyme A synthetase 1 | ACS1_YEAST | 79.3–88.1 | 74.5/6.15 | 58 | 2/3% |
| 10 | Succinate-CoA ligase [ADP-forming] subunit alpha, mitochondrial | SUCA_YEAST | 30.7–34.4 | 35.2/8.59 | 81 | 3/11% |
aBand numbers correspond to Fig. 2
bMnemonic identifier according to SwissProt
cExperimental molecular mass
dTheoretical molecular mass/isoelectric point
ePeptides matched/sequence coverage
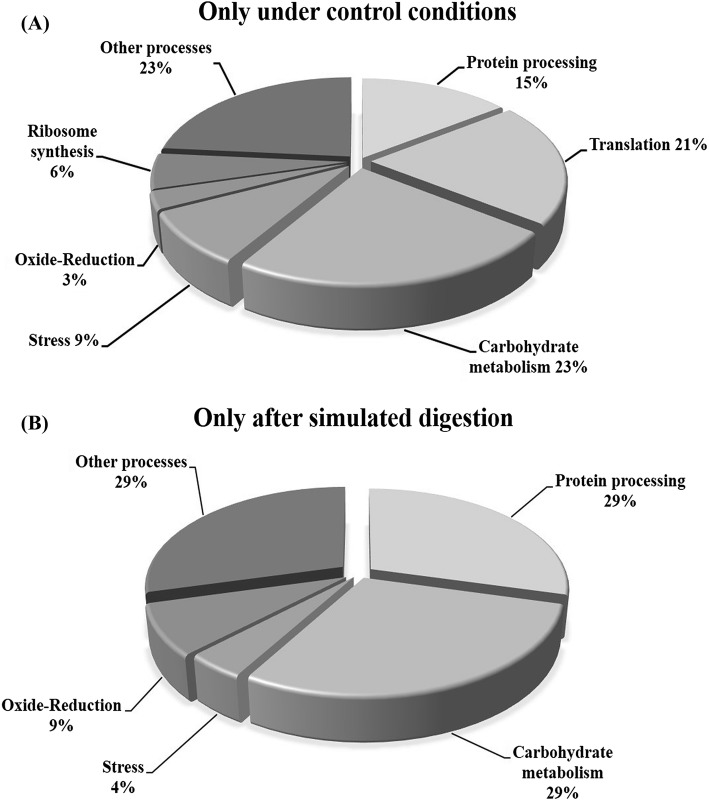
Proteins increased or decreased in the abundance in response to SGID could provide more information about the metabolic changes occurred during the digestion, in particular during the final phase of the process. Protein patterns obtained from yeasts subjected to SGID correspond to a snapshot of the final stage of the process. The study conducted does not provide information on the changes in abundance of proteins that occurred during the different stages of the process. However, it is at this final stage of the process that probiotics must frequently cope with pathogens and adhere, transiently, to the intestine in order to exert probiotic effects. In response to SGID, carbohydrate metabolism, protein processing, and oxide-reduction were the biological processes most affected in S. boulardii, as illustrated in Fig. 3.
Fig. 3.
Functional classification of (A) proteins identified only under control conditions; and (B) proteins identified only after the simulated gastrointestinal digestion; in both cases regardless of whether probiotic yeasts were encapsulated or not
Proteins identified under control conditions in probiotic yeast
Among the proteins identified only under control conditions and whose nonidentification after SGID may indicate that they decreased in abundance, six proteins related to carbohydrate metabolism were identified: 6-phosphogluconate dehydrogenase decarboxylating 1, aconitate hydratase, isocitrate lyase, malate dehydrogenase, mannose-1-phosphate guanylyltransferase, transaldolase and hexokinase-1. Kolkman et al. (2005), exposed S. cerevisiae to low glucose concentrations and identified differentially accumulated proteins by 2-DE and mass spectrometry and showed that most glycolytic enzymes, with the exception of hexokinase 1, were significantly up accumulated under glucose-limited conditions. Bruckmann et al. (2009), reported the increase of up to 4.7 times in hexokinase under anaerobiosis when analyzed the proteomic changes in S. cerevisiae under aerobic and anaerobic growth. This agrees with our results because under SGID probiotic yeast was subjected to anaerobiosis in addition to low glucose conditions.
Under control conditions representing nutrient limitation for probiotic yeast, the homocysteine/cysteine synthase, enzyme involved in the synthesis of the amino acids methionine and cysteine, was also identified, agreeing with that reported for fungi subjected to low amino acid availability, where an increase in proteins related to amino acid biosynthesis, in addition to carbon metabolism and a general stress response was reported (Kroll et al., 2014). Ubiquitin-60S ribosomal protein L40 was also identified only under control conditions; this is essential for translation of a subset of cellular transcripts. It is also known that polyubiquitin chains, when linked to a target protein, have different functions depending on the ubiquitin Lys residue linked. Commonly this label directs proteins to degradation. Increased ubiquitination in S. cerevisiae has been reported in response to several types of stress, such as high temperatures and starvation (Wang et al., 2015).
Seventeen ribosomal proteins were identified, nine corresponding to the small 40S subunit (S12, S13, S18-A, S19-A, S1-A, S22-A, S3, S5, and S7-A) and 8 corresponding to the large 60S subunit (P0, L11-A, L14-A, L18-A, L25, L30, L31-A and L36-A), these proteins are responsible for translation of messenger RNA to proteins (Lu et al., 2015). Alternate translational functions have been attributed to some ribosomal proteins, such as the ribosomal S3 protein involved in DNA repair, through endonuclease activity (Seong et al., 2012). Identification of these ribosomal proteins suggests a high synthesis of proteins, thus reflecting the efforts of the probiotic yeast to counteract the stress caused by the lack of nutrients.
Heat shock protein (HSP) 10 kDa (mitochondrial), HSP 26, HSP SSA2 and HSP SSC1 proteins were also identified. HSPs facilitate cell survival under stress conditions, mainly involving post-translational processes such as folding, stability, transport and protein degradation (Verghese et al., 2012). HSP 10 kDa (HSP10) and HSP 26 are both involved in protein folding, HSP 10 is also essential for mitochondrial protein biogenesis, and together with CPN60 contributes to the stabilization of the catalytic subunit of DNA polymerase-α (Ricke and Bielinsky, 2006), whereas HSP 26 is known to be repressed under optimal growth conditions, but it increases in abundance under stress conditions (Lytras et al., 2017). On the other hand, HSP SSA2 and SSC1 belong to the family of HSP 70, the members of this family assist in the folding of newly synthesized proteins, facilitate the translocation of proteins through the membranes and protect the cell from protein denaturing stress, all possess an N-terminal ATPase domain and a C-terminal peptide-binding domain that allow them to interact with other proteins and peptides (Shaner et al., 2004; Sun et al., 2017).
Proteins identified under SGID in probiotic yeast
In this study, GAPDH1 was identified under all conditions, whereas GAPDH3 was identified only after SGID. This agrees with that reported for different probiotics, for example, when exposing Lactobacillus strains to different pH conditions, differentially accumulated GAPDH isoforms were identified (Chen et al., 2017; Koponen et al., 2012). In addition to being a glycolytic enzyme, GADPH has been recognized as moonlight protein, which besides to its main function in glycolysis has additional functions in the cell (Giménez et al., 2014). Extracellular exportation of GAPDH has been linked with adhesion to mucin, whereas intestinal colonization and exposure to low pHs and/or bile salts have been related as triggering conditions of adhesion mechanisms in probiotics (Siciliano and Mazzeo, 2012). Invertase 1 (EC 3.2.1.26), was also identified only after simulated digestion, this enzyme catalyzes the hydrolysis of non-reducing ends of fructofuranosides, being sucrose one of the main substrates, resulting in one molecule of glucose and another of fructose, both recognized as main sources of carbon and energy (Kulshrestha et al., 2013). Identification of these proteins reveals that yeasts exposed to simulated digestion try to find alternate sources of energy, coming from carbohydrates other than glucose.
Four proteins related to amino acid catabolism were identified only after simulated digestion: Dihydrolipoyl dehydrogenase, S-(hydroxymethyl)glutathione dehydrogenase, Saccharopepsin, Threonine synthase and Vacuolar aminopeptidase 1. The first two are involved in the catabolism of amino acids, such as glycine, leucine, isoleucine, and valine. While saccharopepsin (EC 3.4.23.25) and vacuolar aminopeptidase 1 (EC 3.4.11.22) are catabolic enzymes involved in cellular autophagy, a process in which the cell digests part of its own cytoplasm and even organelles, often for the purpose of recycling macromolecules in response to starvation (Huang and Klionsky, 2002). Saccharopepsin has shown to be essential for vacuolar proteolysis under nutritional stress in S. cerevisiae and has been implicated with the activity of other hydrolases such as aminopeptidase 1, enzyme important for the formation of vesicles for vacuolar transport (Morales-Quinones et al., 2012; Parr et al., 2007). Taken together, these findings reveal a great activity in amino acid metabolism and indicate a critical condition in terms of nutrient and energy availability, reflecting the harsh environments that probiotics must cope when crossing the GIT regardless of whether they are encapsulated or not.
A Superoxide dismutase [Mn], mitochondrial was identified under all conditions, whereas the Superoxide dismutase [Cu–Zn] was identified only after SGID. Superoxide dismutases (SODs, EC 1.15.1.1) maintain Reactive Oxygen Species (ROS) balance by converting O2 into hydrogen peroxide (H2O2); SODs are classified essentially by the metal cofactors in three known types: manganese (MnSOD), copper/zinc (Cu–ZnSOD),) and iron (FeSOD) which are localized in different cellular compartments. In eukaryotic cells, MnSOD is found in the mitochondria and peroxisomes, whereas Cu/ZnSOD isoenzymes are found in cytosolic space and in chloroplasts of higher plants, while FeSOD isoenzymes are usually associated to chloroplast when present in plants (Montero-Morán et al., 2015). It is widely known that oxidative stress results from other stressing conditions such as starvation, lack of nitrogen or glucose, and hypoxia (Kroll et al., 2014). Identification of Cu–Zn SOS only in yeasts exposed to SGID shows that probiotics underwent intense oxidative stress, as a result of the exposure to the different stressing conditions during the SGID such as acid pH, exposure to bile salts and lack of nutrients.
In conclusion, encapsulation in 0.5% calcium alginate showed no significant effect on the survival of S. boulardii exposed to SGID. As well as in the electrophoretic patterns of S. boulardii soluble proteins. Conversely, SGID induced dramatic changes in the electrophoretic patterns of soluble proteins evaluated by SDS-PAGE and 2-DE (Data not shown). Mass spectrometry and homology database search allowed identification of the main differentially accumulated proteins in response to the different conditions evaluated; showing that SGID mainly affects the accumulation proteins related to carbohydrate metabolism, oxide reduction processes, and protein processing. This leads us to ask: Once met the gastrointestinal challenge, are probiotics released from alginate as efficient as those supplied free/not encapsulated to adhere to the intestine? Will the encapsulation have any effect on the yeast surface proteins and molecules responsible for adherence capacity? Will the encapsulation have any effect on the proteins and molecules responsible for such adherence capacity? Without question, more experiments are required to understand better how S. boulardii survive the passage through the gastrointestinal tract and how, after that, can to colonize the intestine to exert its probiotic effects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Thanks to CONACYT-Mexico for awarding us Grant-251744-Infrastructure. Thanks to Plataforma Analítica Institucional-Centro de Investigación en Alimentación y Desarrollo, A.C. (Project PAI-10363). We also thank CONACYT-Mexico Project CB-169358. Martha Beatriz Morales-Amparano thanks CONACYT-Mexico for her MSc fellowship (713677).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Anal A, Singh H. Recent advances in microencapsulation of probiotics for industrial applications and targeted delivery. Trends Food Sci. Technol. 2007;18:240–251. doi: 10.1016/j.tifs.2007.01.004. [DOI] [Google Scholar]
- Basu S, Banerjee D, Chowdhury R, Bhattacharya P. Controlled release of microencapsulated probiotics in food matrix. J. Food Eng. 2018;238:61–69. doi: 10.1016/j.jfoodeng.2018.06.005. [DOI] [Google Scholar]
- Bruckmann A, Hensbergen PJ, Balog CI, Deelder AM, Brandt R, Snoek II, van Heusden GPH. Proteome analysis of aerobically and anaerobically grown Saccharomyces cerevisiae cells. J. Proteomics. 2009;71:662–669. doi: 10.1016/j.jprot.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Bustos A, Font G, Raya R, Martinho A, Fadda S, Taranto M. Proteomic analysis of the probiotic Lactobacillus reuteri CRL1098 reveals novel tolerance biomarkers to bile acid-induced stress. Food Res. Int. 2015;77:599–607. doi: 10.1016/j.foodres.2015.10.001. [DOI] [Google Scholar]
- Chen M, Tang H, Chiang M. Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol. 2017;66:20–27. doi: 10.1016/j.fm.2017.03.020. [DOI] [PubMed] [Google Scholar]
- Corona-Hernández R, Álvarez-Parrilla E, Lizardi-Mendoza J, Islas-Rubio A, Rosa L, Wall-Medrano A. Structural stability and viability of microencapsulated probiotic bacteria: a review. Compr. Rev. Food Sci. Food Saf. 2013;12:614–628. doi: 10.1111/1541-4337.12030. [DOI] [PubMed] [Google Scholar]
- Faurobert M, Pelpoir E, Chaïb J. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. In: Thiellement H, Zivy M, Damerval C, Méchin V, editors. Methods in Molecular Biology, Plant Proteomics: Methods and Protocols. Totowa, NJ, USA: Humana Press Inc.; 2007. pp. 9–14. [DOI] [PubMed] [Google Scholar]
- Fietto J, Araújo R, Valadão F, Fietto L, Brandão R, Neves M, Gomes F, Nicolo J, Castro I. Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can. J. Microbiol. 2004;50:615–621. doi: 10.1139/w04-050. [DOI] [PubMed] [Google Scholar]
- Gbassi G, Vandamme T, Yolou F, Marchioni E. In vitro effects of pH, bile salts and enzymes on the release and viability of encapsulated Lactobacillus plantarum strains in a gastrointestinal tract model. Int. Dairy J. 2011;21:97–102. doi: 10.1016/j.idairyj.2010.09.006. [DOI] [Google Scholar]
- Ghorbani-Choboghlo H, Zahraei-Salehi T, Ashrafi-Helan J, Yahyaraeyat R, Pourjafar H, Nikaein D, Balal A, Khosravi AR. Microencapsulation of Saccharomyces cerevisiae and its evaluation to protect in simulated gastric conditions. Iran. J. Microbiol. 2015;7:338–342. [PMC free article] [PubMed] [Google Scholar]
- Giménez R, Aguilera L, Ferreira E, Aguilar J, Baldomà L, Badia J. Glyceraldehyde-3-phosphate dehydrogenase as a moonlighting protein in bacteria. Recent Adv. Pharm. Sci. 2014;4:165–180. [Google Scholar]
- Hansen LT, Allan-Wojtas PM, Jin YL, Paulson AT. Survival of Ca-alginate microencapsulated Bifidobacterium spp. in milk and simulated gastrointestinal conditions. Food Microbiol. 2002;19:35–45. doi: 10.1006/fmic.2001.0452. [DOI] [Google Scholar]
- Huang W, Klionsky D. Autophagy in yeast: a review of the molecular machinery. Cell. Struct. Funct. 2002;27:409–420. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- Hugues-Ayala A. Microencapsulation of Lactobacillus rhamnosus GG in a matrix of calcium alginate coated with buttermilk proteins. MS thesis, Research Center for Food and Development. Hermosillo, Sonora, Mexico (2013)
- Kailasapathy K. Encapsulation and controlled release techniques for administration and delivery of bioactive components in the health food sector. In: Boye JI, editor. Nutraceutical and Functional Food Processing Technology. Hoboken, NJ, USA: John Wiley & Sons, Ltd.; 2015. pp. 307–346. [Google Scholar]
- Kolkman A, Olsthoorn M, Heeremans C, Heck A, Slijper M. Comparative proteome analysis of Saccharomyces cerevisiae grown in chemostat cultures limited for glucose or ethanol. Mol. Cell. Proteomics. 2005;4:1–11. doi: 10.1074/mcp.M400087-MCP200. [DOI] [PubMed] [Google Scholar]
- Koponen J, Laakso K, Koskenniemi K, Kankainen M, Savijoki K, Nyman T, Varmanen P. Effect of acid stress on protein expression and phosphorylation in Lactobacillus rhamnosus GG. J. Proteomics. 2012;75:1357–1374. doi: 10.1016/j.jprot.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Kroll K, Pähtz V, Kniemeyer O. Elucidating the fungal stress response by proteomics. J. Proteomics. 2014;97:151–163. doi: 10.1016/j.jprot.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Kulshrestha S, Tyagi P, Sindhi V, Yadavilli K. Invertase and its applications—a brief review. J. Pharm. Res. 2013;7:792–797. [Google Scholar]
- Lu H, Zhu Y, Xiong J, Wang R, Jia Z. Potential extra-ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae. Microbiol. Res. 2015;177:28–33. doi: 10.1016/j.micres.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Lytras G, Zacharioudakis I, Tzamarias D. Asymmetric inheritance of the yeast chaperone Hsp26p and its functional consequences. Biochem. Biophys. Res. Commun. 2017;491:1055–1061. doi: 10.1016/j.bbrc.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Mandal S, Puniya AK, Singh K. Effect of alginate concentrations on survival of microencapsulated Lactobacillus casei NCDC-298. Int. Dairy J. 2006;16:1190–1195. doi: 10.1016/j.idairyj.2005.10.005. [DOI] [Google Scholar]
- Mathews S. Microencapsulation of probiotics by calcium alginate and gelatin and evaluation of its survival in simulated human gastro-intestinal condition. Int. J. Curr. Microbiol. Appl. Sci. 2017;6:2080–2087. doi: 10.20546/ijcmas.2017.604.245. [DOI] [Google Scholar]
- Mokarram R, Mortazavi S, Habibi-Najafi M, Shahidi F. The influence of multi stage alginate coating on survivability of potential probiotic bacteria in simulated gastric and intestinal juice. Food Res. Int. 2009;42:1040–1045. doi: 10.1016/j.foodres.2009.04.023. [DOI] [Google Scholar]
- Montero-Morán GM, Sampedro J, Saab-Rincón G, Cervantes-González MA, Huerta-Ocampo JA, De León-Rodríguez A, Barba de la Rosa AP. Biochemical and molecular characterization of a novel Cu/Zn superoxide dismutase from Amaranthus hypochondriacus L.: an intrinsically disordered protein. Appl. Biochem. Biotechnol. 2015;176:2328–2345. doi: 10.1007/s12010-015-1721-0. [DOI] [PubMed] [Google Scholar]
- Morales-Quinones M, Winston J, Stromhaug P. Propeptide of Aminopeptidase 1 mediates aggregation and vesicle formation in the Cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 2012;287:10121–10133. doi: 10.1074/jbc.M111.311696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavian A, Razavi S, Ehsani M, Sohrabvandi S. Principles and methods of microencapsulation of probiotic microorganisms. Iran. J. Biotechnol. 2007;5:1–18. [Google Scholar]
- Parr C, Keates R, Bryksa B, Ogawa M, Yada R. The structure and function of Saccharomyces cerevisiae proteinase A. Yeast. 2007;24:467–480. doi: 10.1002/yea.1485. [DOI] [PubMed] [Google Scholar]
- Ricke R, Bielinsky A. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-α in budding yeast. J. Biol. Chem. 2006;281:18414–18425. doi: 10.1074/jbc.M513551200. [DOI] [PubMed] [Google Scholar]
- Ruiz L, Hidalgo C, Blanco-Míguez A, Lourenço A, Sánchez B, Margolles A. Tackling probiotic and gut microbiota functionality through proteomics. J. Proteomics. 2016;147:28–39. doi: 10.1016/j.jprot.2016.03.023. [DOI] [PubMed] [Google Scholar]
- Saarela M, Mogensen G, Fonden R, Mättö J, Mattila-Sandholm T. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 2000;84:197–215. doi: 10.1016/S0168-1656(00)00375-8. [DOI] [PubMed] [Google Scholar]
- Seong K, Jung S, Kim H, Kim H, Jung Y, Choi S, Kim J. Yeast ribosomal protein S3 possesses a β-lyase activity on damaged DNA. FEBS Lett. 2012;586:356–361. doi: 10.1016/j.febslet.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Shaner L, Trott A, Goeckeler J, Brodsky J, Morano K. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- Shori A. Microencapsulation improved probiotics survival during gastric transit. HAYATI J. Biosci. 2017;24:1–5. doi: 10.1016/j.hjb.2016.12.008. [DOI] [Google Scholar]
- Siciliano R, Mazzeo M. Molecular mechanisms of probiotic action: a proteomic perspective. Curr. Opin. Microbiol. 2012;15:390–396. doi: 10.1016/j.mib.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Sun S, Gong Y, Li T, Yu C. Candida albicans heat shock proteins and Hsps-associated signaling pathways as potential antifungal targets. Front. Cell. Infect. Microbiol. 2017;7:520. doi: 10.3389/fcimb.2017.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano K. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Canadeo L, Huibregtse J. Ubiquitination of newly synthesized proteins at the ribosome. Biochimie. 2015;114:127–133. doi: 10.1016/j.biochi.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Teng D, Wang X, Dai C, Wang J. Saccharomyces boulardii prevention of the hepatic injury induced by Salmonella Enteritidis infection. Can. J. Microbiol. 2014;60:681–686. doi: 10.1139/cjm-2014-0259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.