Abstract
Gcn5 and sirtuins are highly conserved histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes that were first characterized as regulators of gene expression. Although histone tails are important substrates of these enzymes, they also target many nonhistone proteins that function in diverse biological processes. However, the mechanisms used by these enzymes to choose their nonhistone substrates are unknown. Previously, we used SILAC-based MS to identify novel nonhistone substrates of Gcn5 and sirtuins in yeast and found a shared target consensus sequence. Here, we use a synthetic biology approach to demonstrate that this consensus sequence can direct acetylation and deacetylation targeting by these enzymes in vivo. Remarkably, fusion of the sequence to a nonsubstrate confers de novo acetylation that is regulated by both Gcn5 and sirtuins. We exploit this synthetic fusion substrate as a tool to define subunits of the Gcn5-containing SAGA and ADA complexes required for nonhistone protein acetylation. In particular, we find a key role for the Ada2 and Ada3 subunits in regulating acetylations on our fusion substrate. In contrast, other subunits tested were largely dispensable, including those required for SAGA stability. In an extended analysis, defects in proteome-wide acetylation observed in ada3Δ mutants mirror those in ada2Δ mutants. Altogether, our work argues that nonhistone protein acetylation by Gcn5 is determined in part by specific amino acids surrounding target lysines but that even optimal sequences require both Ada2 and Ada3 for robust acetylation. The synthetic fusion substrate we describe can serve as a tool to further dissect the regulation of both Gcn5 and sirtuin activities in vivo.
Keywords: acetylation, yeast, sirtuin, fusion protein, transcription coactivator, Gcn5, SAGA, synthetic biology, yeast
Introduction
The Gcn5 histone acetyltransferase (HAT)2 is a member of the GNAT (GCN5-related N-acetyltransferases) family of acetyltransferase enzymes. It functions in the context of a highly conserved protein complex called SAGA that contains at least 19 unique subunits. These subunits can be grouped into functional submodules that together regulate important aspects of eukaryotic gene transcription (1–4). Besides Gcn5, the HAT submodule contains Ada2, Ada3, and Sgf29 (1). The proteins of the HAT submodule also function in a distinct complex termed ADA that includes Ahc1 and Ahc2 (1, 5). SAGA deubiquitylation (DUB) and TATA-binding protein (TBP) regulatory modules (consisting of Spt3 and Spt8) mediate deubiquitylation of H2B Lys-123 and recruitment of TBP to gene promoters, respectively (6–10). A core structural module that includes subunits shared with general transcription factor TFIID serves as a scaffold for the SAGA complex (1, 4). A variant of the SAGA complex called SLIK has partially overlapping functions in the regulation of gene expression. The SLIK complex contains the retrograde response protein Rtg2 but lacks Spt8 and has a truncated version of Spt7 (11, 12).
Ada2 and Ada3 play important roles in promoting Gcn5 activity toward histone substrates, particularly in the context of nucleosomes (13–15). Although Sgf29 is largely dispensable for Gcn5 activity in vitro, it plays a critical role in global histone acetylation in vivo as sgf29Δ cells show decreased acetylation of histone H3 Lys-9, Lys-14, and Lys-18, paralleling what is observed for gcn5Δ and ada3Δ mutants (16). This function of Sgf29 is likely due to the ability of its Tudor domain to bind to methylated H3 Lys-4 (16). SAGA integrity is also required for histone acetylation in vivo as deletion of genes encoding scaffold elements Spt20 or Spt7 results in decreased global H3 acetylation (17).
Like HATs, histone deacetylase (HDAC) enzymes are grouped into families based on common structural and biochemical characteristics. The NAD+-dependent family of sirtuin HDACs, consisting of Sir2 and Hst1–Hst4, are conserved enzymes that can be inhibited with a by-product of their reactions called nicotinamide (18, 19). Sirtuins Hst3 and Hst4 deacetylate H3 Lys-56 (20, 21), which is important for DNA repair and the maintenance of genome integrity (22, 23). Hst4 also localizes to the mitochondria where it regulates protein deacetylation in response to biotin starvation (24). Sir2 and Hst1 function in gene silencing and transcriptional control at select genomic loci (25–28). Finally, Hst2 is the only cytoplasmic sirtuin (29, 30), and its function remains poorly characterized.
Although acetylation was originally characterized as a histone modification and regulator of gene transcription, thousands of nonhistone substrates have been described using high-throughput approaches in organisms from bacteria to humans (31–34). In yeast, at least one-third of all proteins are acetylated (35). Although regulation of histone acetylation and deacetylation activities is mediated by temporal and spatial changes in HAT and HDAC recruitment to specific chromatin loci, the factors governing selection of nonhistone substrates are less clear.
In previous work, we used SILAC labeling of yeast cells coupled with affinity enrichment of acetylated peptides and MS to uncover candidate substrates of the Gcn5 and Esa1 HATs and the sirtuin family of HDACs (34). Analysis of high-confidence candidate targets uncovered preferred amino acid motifs surrounding regulated acetylated lysines (34). Intriguingly, there were similarities between these “consensus” target sequences for Gcn5 and sirtuin enzymes with SXK(ac)(K/R)P being preferred for both enzymes. This shared sequence was distinct from that previously identified for Gcn5 (36) and from that of Esa1, which bore significant resemblance to the glycine-rich H4 tail (34).
Here, we used a synthetic biology approach to demonstrate that this shared sequence can direct Gcn5- and sirtuin-regulated acetylation in vivo. A protein construct containing GFP fused to variants of the consensus sequence, in conjunction with an antibody directed against that acetylated consensus, serves as a toolkit to probe sirtuin and Gcn5 functions in vivo. Our work with this toolkit points to a model where Gcn5 activity toward lysine residues within preferred sequence contexts depends on association with Ada2 and Ada3 but is largely independent of other SAGA proteins.
Results
A shared consensus sequence predicts Spt2 as a novel target of Gcn5 and sirtuins
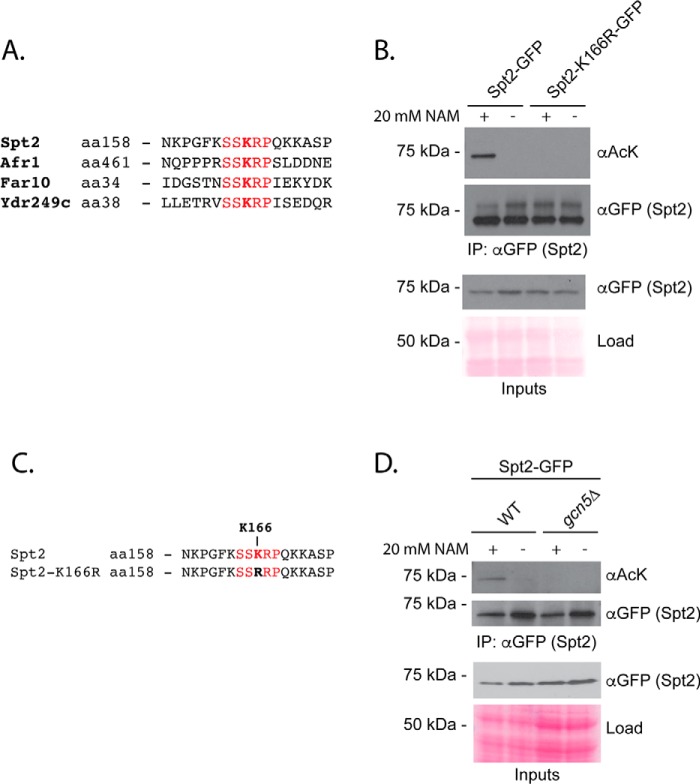
We previously identified a shared consensus sequence of SXK(ac)(K/R)P for Gcn5 and sirtuin enzymes by carrying out SILAC-based acetylome analyses for gcn5Δ and hst1Δ hst2Δ sir2Δ triple mutant cells (34). We first wondered whether this sequence could be used to predict new sites regulated by these enzymes. We focused on the SSK(ac)RP sequence, which represents the most frequently observed amino acids surrounding Gcn5-depdendent acetylations, corrected for relative amino acid frequencies in yeast (34). Four proteins contain an exact match: Spt2, Far10, Afr1, and Ydr249c (Fig. 1A). We were able to generate GFP-tagged versions of Spt2, Far10, and Ydr249c. Spt2 is a transcriptional regulator that physically interacts with the SWI–SNF chromatin remodeling complex (37). Far10 is a member of the conserved STRIPAK (striatin-interacting phosphatase and kinase) complex that mediates pheromone and TORC2-dependent signaling pathways in yeast (38, 39). Ydr249c is a largely uncharacterized protein (40). We immunopurified these GFP fusion proteins and tested for reactivity with monoclonal antibodies recognizing acetylated lysine in the context of defining features of the SXK(ac)(K/R)P sequence (Fig. S1, A and B; see “Experimental procedures”). In this experiment, FAR10-GFP and YDR249C-GFP were expressed from the inducible GAL promoter (41) to allow recovery of a sufficient level of protein, whereas SPT2-GFP was expressed at high enough levels under its endogenous promoter. We observed no evidence of Far10-GFP and Ydr249c-GFP acetylation, although in the case of Far10-GFP this could be related to the low level of protein recovery (Fig. S1C). In contrast, Spt2-GFP showed reactivity with monoclonal anti-acetyllysine antibody following recovery from cells treated with the sirtuin inhibitor nicotinamide (Fig. 1B), and expression of GFP-tagged Spt2 mutated for the lysine residue (Lys-166) within its SSKRP consensus sequence completely eliminated the signal (Fig. 1, B and C). Finally, as predicted from the consensus sequence, acetylation depended on GCN5 (Fig. 1D). Altogether, these data are consistent with Gcn5-regulated acetylation of Spt2-GFP Lys-166 and the reversal of this modification by sirtuin enzymes. The data highlight that acetylation consensus sequences derived from high-throughput MS data could be used to identify novel targets for HAT and HDAC enzymes. Notably, in contrast to our sequence-specific monoclonal antibodies, a commonly used pan-acetyllysine antibody (Cell Signaling Technology, 9441) did not detect regulated acetylations on Spt2-GFP (data not shown). This is the first description of regulation for the Lys-166 acetylation site. Although we previously detected sirtuin (but not Gcn5)-regulated sites on Spt2 using MS, Lys-166 was not one of these and was not identified in this previous work (34).
Figure 1.
Consensus target sequences predict Spt2 as a candidate Gcn5 and sirtuin target. A, alignment of yeast proteins containing an exact match to the SSKRP consensus sequence. B, WT or Spt2-GFP K166R was immunoprecipitated in the presence or absence of NAM treatment (20 mm; two generations). Eluates were separated via SDS-PAGE, transferred to PVDF membrane, and probed with αGFP or mAb developed to recognize the acetylated SSKRP sequence. C, alignment of Spt2 target sequences showing the Lys-to-Arg mutation used in B. D, acetylation of Spt2-GFP was measured in a gcn5Δ strain in the presence and absence of nicotinamide. aa, amino acids; AcK, acetyllysine.
Of Spt2, Far10, and Ydr249c, only Spt2 showed regulated acetylation of its SSKRP consensus sequence via Gcn5 and sirtuins (Fig. 1, B and D, and Fig. S1C). As such, the presence of the consensus sequence alone is not sufficient to confer regulated acetylation. Of the three candidate targets, only Spt2 has demonstrated localization to the nucleus (42). Thus, it is possible that nuclear localization is required for acetylation by Gcn5. Sequence accessibility is also likely to be an important regulatory mechanism.
A synthetic nonhistone substrate is acetylated in vivo
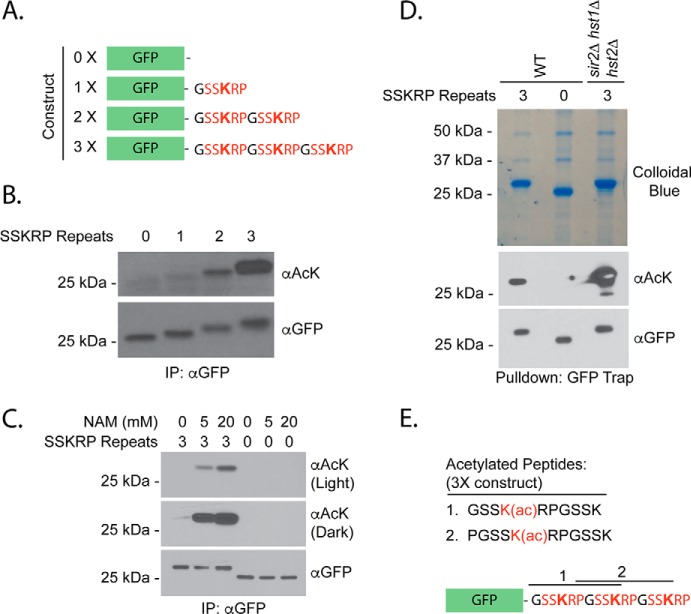
To further probe the contribution of the shared Gcn5/sirtuin sequence to protein acetylation, we asked whether the addition of this sequence to a nonsubstrate would confer its acetylation in vivo. To test this idea, we fused increasing numbers of SSKRP consensus sequence to GFP (0X–3X; Fig. 2A). We chose GFP because it does not react with anti-acetyllysine antibodies in IP–Western experiments (see below) and can localize throughout the cell (43, 44). We expressed these fusion constructs or GFP alone from a constitutive ADH1 promoter and used an IP–Western strategy to recover and compare their acetylation using our SSK(ac)RP-reactive monoclonal antibodies. We detected acetylation on our fusion constructs but not GFP alone (Fig. 2B). Moreover, the acetylation signal increased with the number of consensus repeats (Fig. 2B; see Figs. S3 and S4 for all input analyses).
Figure 2.
A synthetic acetylation substrate is acetylated in vivo. A, synthetic fusion constructs used for acetylation analyses. B, GFP fusions were purified from strains expressing the constructs in A using an antibody against GFP. Eluates were analyzed via SDS-PAGE and probed either with anti-acetyllysine (αAcK) that recognizes the consensus or αGFP following Western blotting. Diluted forms () of immunoprecipitated protein samples were loaded for GFP detection. C, the indicated constructs were purified from strains treated with 0, 5, or 20 mm NAM for 30 min and analyzed as in B. D, indicated constructs were purified from the strains shown using the GFP Trap reagent prior to NuPAGE analysis, staining by colloidal Coomassie, and analysis of excised bands by MS. E, acetylated peptides detected by MS analysis. Acetylations on these sites were detected in two of three independent purifications. Also see Fig. S2.
Regulation of the synthetic substrate by sirtuins
We predicted that our synthetic substrate would be regulated by enzymes used to derive the consensus sequence, namely sirtuin HDACs and the Gcn5 HAT. To test whether our substrate was regulated by sirtuin enzymes, we measured the acetylation on our synthetic substrate with three consensus repeats (3X) following its purification from yeast strains treated with the sirtuin inhibitor nicotinamide. The acetylation observed on the 3X substrate increased with nicotinamide treatment, and this effect was concentration-dependent (Fig. 2C). In contrast, nicotinamide had no impact on acetylation of GFP alone (Fig. 2C). To determine the sirtuins that contribute to this effect, we analyzed the acetylation of the 3X construct in hst1Δ, hst2Δ, or sir2Δ deletion mutants. We observed an increase in acetylation only in hst2Δ mutants (Fig. S2A). Hst2 also deacetylated the purified substrate in vitro (Fig. S2B), consistent with the possibility of direct deacetylation. We next tested acetylation of the 3X substrate in an hst1Δ hst2Δ sir2Δ triple mutant, used previously to generate the consensus sequences investigated in this work. We observed a dramatic increase in acetylation of our substrate in this mutant background beyond that observed in hst2Δ strains (Fig. S2A). Because no single mutant recapitulated the effect of the sirtuin triple mutant, we suggest that sirtuins act redundantly to deacetylate the synthetic substrate.
Contribution of individual sites to acetylation of tandem consensus sequences
To confirm that acetylation was occurring on the consensus sequence, we purified our fusion protein and mapped acetylation sites following separation by NuPAGE, trypsin digestion, and analysis by Orbitrap MS. We observed acetylations on the first and second lysine residues when the substrate was purified from sirtuin mutant cells, confirming acetylation of the target sequence in vivo (Fig. 2, D and E, and Fig. S2, C and D). To test whether individual lysine residues were equally important, we focused on the 2X substrate. We generated variants of the 2X consensus where the first (R1), second (R2), or both (DM) lysine residues were mutated to arginine, which maintains the charge of a lysine residue but cannot be acetylated (Fig. S2E). The mutation of only the first lysine residue (R1) resulted in decreased acetylation as measured by IP–Western blot analysis (Fig. S2F). In contrast, the mutation of the second lysine residue (R2) had little effect (Fig. S2F). As expected, mutation of both lysine residues (DM) prevented acetylation altogether (Fig. S2F). Because its conversion to arginine resulted in the greatest loss of signal, it appears that the lysine within the first consensus repeat is normally more heavily acetylated than the lysine in the second repeat. It is possible that acetyltransferases have difficulty in acetylating residues very close to the C terminus of protein sequences. Decreased acetylation observed for the R1 mutant, but not the double mutant, was rescued by mutation of sirtuin enzymes (Fig. S2F), consistent with our results using the 3X substrate (Fig. S2A). Therefore, a second possibility is that sirtuins prefer to target the distal site. In the sirtuin triple mutant we were also able to detect acetylation on the 1X substrate, which was not readily apparent in the WT background (Fig. S2F).
In vivo regulation of the consensus sequence by Gcn5
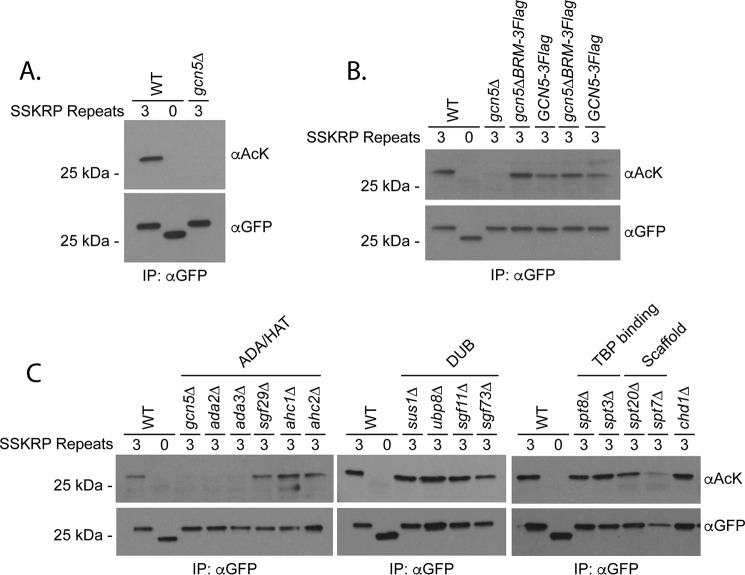
We next tested the contribution of Gcn5 to the acetylation of the 3X synthetic substrate. Interestingly, expression of the construct was decreased in gcn5Δ mutants relative to WT controls (Fig. S3C). Nevertheless, our optimized IP protocol recovered similar levels of protein in gcn5Δ strains, allowing us to make direct comparisons regarding overall acetylation. Acetylation was eliminated in cells lacking the Gcn5 HAT, confirming dependence on this enzyme in vivo (Fig. 3A). The regulation of our synthetic substrate by the opposing activities of the Gcn5 HAT and sirtuin HDACs validates the consensus sequences for these enzymes and suggests that target sequences are an important determinant of acetylation. To our knowledge, this is the first demonstration of a portable HAT consensus sequence that directs acetylation in vivo.
Figure 3.
Acetylation of a synthetic substrate depends on Gcn5. A, the indicated constructs were purified from the strains shown using an antibody against GFP prior to analysis by SDS-PAGE, Western blotting, and detection with anti-acetyllysine (αAcK) or αGFP. Diluted forms () of immunoprecipitated protein samples were loaded for GFP detection. B, the role of the Gcn5 bromodomain in substrate acetylation was tested as in A using the strains shown. C, the indicated constructs were analyzed in SAGA mutants shown as described in A.
Previous work suggested that Gcn5's bromodomain plays a role in regulating acetylation of histone tails (45). Whether this is a general property of Gcn5 function that is also applicable to nonhistone substrates is unknown. To test this, we assayed the acetylation of the 3X substrate recovered from strains where Gcn5 was mutated for its bromodomain (Gcn5ΔBRM). Unexpectedly, the substrate showed increased acetylation in Gcn5ΔBRM strains relative to matched controls (Fig. 3B). The increase in acetylation may stem from a moderate increase in Gcn5 levels that was observed in the absence of the bromodomain (Fig. S3D). Although these data suggest that the Gcn5 bromodomain does not contribute to the overall acetylation of our construct, we cannot exclude the possibility that it regulates the relative distribution of acetylation marks among individual lysines.
Regulation of nonhistone protein acetylation by key subunits of SAGA complex
We next used our synthetic substrate as a tool to test the contribution of individual SAGA subunits to nonhistone protein acetylation in vivo. As was the case with gcn5Δ, the expression level of the construct varied in SAGA mutants considerably (Fig. S3E), but we were able to compare acetylations on equal amounts of recovered protein following our IP protocol. We found that acetylation was largely unaffected by deletion of genes encoding the DUB, TBP-binding, and structural proteins as well as the potential SAGA-binding protein Chd1 (46) (Fig. 3C). Although we recovered less substrate from spt7Δ mutants, the protein that we did recover was acetylated at near WT levels (Fig. 3C). The lack of effect in spt7Δ and spt20Δ mutants is particularly intriguing because Spt7 and Spt20 are required for SAGA complex formation (1, 47). The impact of the HAT subcomplex varied depending on the subunit in question. A striking defect in acetylation of our substrate was observed in the absence of HAT submodule proteins Ada2 and Ada3 (Fig. 3C). In contrast, Sgf29 and ADA subcomplex–specific components Ahc1 and Ahc2 were largely dispensable. The dependence of acetylation on substrate on Ada2 and Ada3 is consistent with the known role of these binding partners in acetylation of histone tails (13).
Ada3 is a global regulator of acetylation
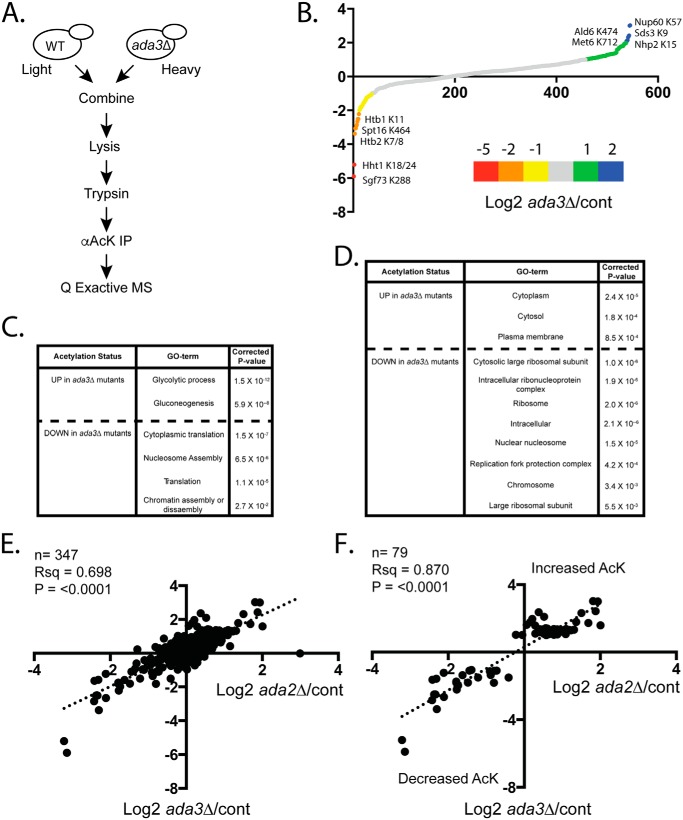
Having identified Ada3 as a potential regulator of nonhistone protein acetylation using our synthetic substrate, we carried out acetylome profiling for cells mutated for ada3Δ to validate our results and test for effects on acetylations proteome-wide (Fig. 4A). Our rationale was that any mutant showing defects in Gcn5-dependent acetylation of a substrate with an optimized target sequence is likely to impact other Gcn5 targets. We obtained SILAC ratios for 548 acetylated peptides, with 38 showing ≥2-fold down-regulation relative to WT (Fig. 4B and Table S3). GO-term analysis revealed that regulated proteins function predominantly in translation and chromatin-related processes (Fig. 4, C and D). These functional categories are reminiscent of what we observed previously for Gcn5 targets (34). Included in this group were previously identified Gcn5 targets such as Sgf73 Lys-288 (34) and novel targets, including Spt16 Lys-464 (Fig. 4B). These data confirm a global role of Ada3 in the regulation of protein acetylation. Perhaps unexpectedly, we also uncovered 88 acetylated peptides that were up-regulated ≥2-fold in ada3Δ relative to WT cells (Fig. 4B and Table S3). GO-term analysis showed enrichment for cytosolic proteins involved in glycolysis and gluconeogenesis (Fig. 4, C and D). Up-regulated protein acetylations could be the result of indirect effects on other HAT and HDAC enzymes.
Figure 4.
Ada3 regulates acetylations proteome-wide. A, schematic of SILAC-based MS protocol used for acetylome analysis. B, average log2 -fold change for ada3Δ/WT for all peptides detected in SILAC experiments. The graph includes combined results of forward and reverse label experiments (total biological replicates = 4). C, GO-term analysis for “biological process” was done using DAVID 6.8. Regulated peptides are ≥2-fold changed in the indicated direction. D, GO-term analysis for “cellular component” using DAVID 6.8. Regulated peptides are ≥2-fold changed in the indicated direction. E, comparison of ada3Δ/control versus ada2Δ/control ratios for acetylated peptides in Downey et al. (34). F, as in E but just peptides found to be ≥2-fold changed in ada3Δ/control experiments in either direction. AcK, acetyllysine.
We compared the results of our ada3Δ experiments with those of similar experiments performed in ada2Δ mutants (34) and found significant correlation of the data sets (Fig. 4E). This correlation persisted when unregulated peptides were excluded from the analysis (Fig. 4F). Together, the analysis suggests that Ada2 and Ada3 work together to regulate protein acetylation of nonhistone substrates by Gcn5.
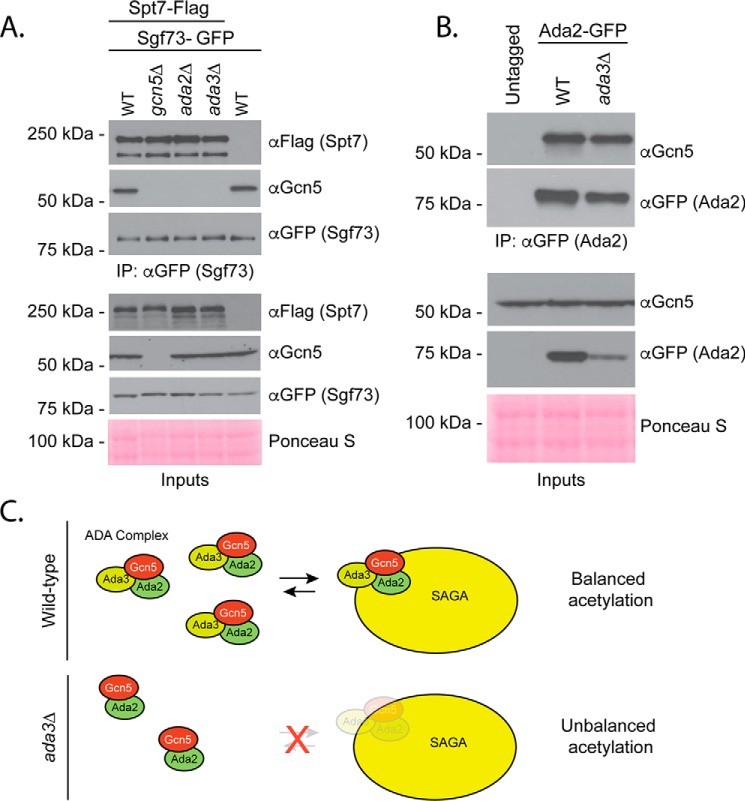
To investigate the mechanism by which Ada3 impacts protein acetylation, we used a coimmunoprecipitation strategy to compare Gcn5-binding partners in ADA3 and ada3Δ cells. Consistent with our previous findings in ada2Δ mutants, Gcn5 failed to bind to SAGA protein Spt7 in the absence of ada3Δ (Fig. 5A). However, this is unlikely to explain the lack of acetylation on our synthetic substrate as critical SAGA subunits were not required for its acetylation (Fig. 3C). Interestingly, Ada2 is able to retain interaction with Gcn5 in the absence of Ada3, although analysis of input material reveals less Ada2 protein overall (Fig. 5B). This observation is consistent with a previous report that found LexA-Ada2 fusions were poorly expressed in the absence of Ada3 (48). Altogether, our data support a model where Ada2 and Ada3 cooperate with Gcn5 to regulate the acetylation of nonhistone substrates. In the absence of Ada3, Ada2 can still bind to Gcn5, but this subcomplex is less abundant and incapable of maintaining balanced levels of nonhistone protein acetylation (Fig. 5C).
Figure 5.
ADA3 mutation prevents Gcn5 binding to SAGA but is permissive for Ada2–Gcn5 interaction. A, SAGA subunit Sgf73 was immunoprecipitated via the GFP tag in the indicated strains, and Spt7–3Flag or Gcn5 was detected with the antibodies shown following SDS-PAGE and Western blotting. B, Ada2 was immunoprecipitated with a GFP tag in the indicated strains, and Gcn5 was detected with an αGcn5 antibody following SDS-PAGE and Western blotting. C, model for Ada3's role in Gcn5-dependent acetylation of nonhistone targets.
Discussion
Consensus sequences as regulators of acetylation
Despite the identification of thousands of lysine acetylation sites in high-throughput studies (33–35), there has been little effort to identify and evaluate common target sequences that direct activities of HAT and HDAC enzymes toward target lysines. Here, we focused on a consensus sequence shared by Gcn5 and sirtuins that was developed from acetylome profiling data in yeast (34). The critical finding of our work is that fusion of this consensus sequence to a nonsubstrate confers regulated acetylation of that substrate in vivo. Our work validates the utility of deriving consensus sequences from high-throughput acetylome profiling data and, to our knowledge, is the first to demonstrate the portability of an experimentally determined acetylation target sequence in vivo. The SSK(ac)RP sequence employed in our work is derived from dozens of high-confidence regulated acetylation sites (34). As such, it is likely to represent an optimal target sequence for both Gcn5 and sirtuins. Of the three native yeast proteins that contain this sequence that we tested, only one, Spt2, showed evidence of Gcn5 and sirtuin regulation in vivo. Thus, we cannot claim that an optimized consensus sequence is necessarily sufficient to confer acetylation in vivo. Other factors such as subcellular localization are also likely to be important for substrate targeting. Importantly, the quality of the acetylation signal observed in our Western blots using the combination of our synthetic substrate and mAb was more consistent and robust than that observed previously for other nonhistone substrates using other commercially available reagents. This made the substrate an ideal tool to study nonhistone acetylation in vivo.
Regulation of acetylation by Gcn5 and SAGA
Acetylation of our synthetic substrate depends on Gcn5, and although we cannot exclude that Gcn5 indirectly regulates the activity of other HATs that target the consensus sequence, we favor a model where Gcn5 acetylates the SSKRP sequence directly. Prior work demonstrated that Gcn5's acetyllysine-binding bromodomain promoted total acetylation of H3 and defined the pattern of acetylation across lysines in the N-terminal tail (45, 50). Notably, our assay does not permit us to determine whether the pattern of acetylation among the three target lysines in our fusion substrate is impacted by the absence of the Gcn5 bromodomain. However, total acetylation of our 3X construct was marginally increased rather than decreased in strains where the bromodomain was deleted. The role of the Gcn5 bromodomain in substrate acetylation may depend on the individual amino acids surrounding target lysines. Effects of the bromodomain may also require access to additional substrate molecules in trans as is the case for adjacent nucleosomes (50).
Ada3 was required for the acetylation of our synthetic substrate and for a broad range of intracellular targets also impacted by Gcn5 and Ada2. The requirement for Ada3 is consistent with our inability to observe acetylation of the substrate by bacterially purified Gcn5 in vitro (Fig. S5). Although Ada3 was required for Gcn5 interaction with SAGA protein Spt7, it is unclear whether it is also required for interaction with other members of the ADA complex besides Ada2 such as Sgf29, Ahc1, and Ahc2. Regardless, as was the case for SAGA proteins, these ADA components did not make dramatic contributions to acetylation of our synthetic substrate in vivo. We favor a model wherein Ada2 and Ada3 make distinct and relatively direct contributions to Gcn5's activity even toward ideal target sequences such as that present in our 3X consensus substrate. This interpretation is consistent with recent structural work that provides clear evidence for Ada2's role in acetyl-CoA binding (51). We also find that overall levels of Ada2-GFP were reduced in ada3Δ cells, which may suggest a role for Ada3 in promoting Ada2 stability and explain at least part of the defects observed in ada3Δ mutant cells. Notably, it has been shown previously that Ada2 is not required for all Gcn5-regulated acetylations, although the reasons for this are unclear (34, 52).
Still, Gcn5 may target our substrate as a member of one or more multisubunit complexes. Although many SAGA proteins are not required for the acetylation of our synthetic substrate, this observation does not necessarily imply that the population of Gcn5 enzyme targeting our substrate is not SAGA-bound under most circumstances. Moreover, we do not discount the possibility that SAGA, ADA, or SLIK proteins play important roles in directing Gcn5 activity toward specific nonhistone substrates. Indeed, we showed previously that Gcn5's acetylation of the ribosomal protein transcription factor Ifh1 depends on SPT7, which is required for SAGA stability (11, 47, 53). In this context, the dependence of Ifh1 acetylation on SPT7 is consistent with the observation that this acetylation event occurs at ribosomal protein promoters (53) where SAGA is recruited (54, 55). In contrast, we suggest that our synthetic substrate functions as a “generic” target whose acetylation, or lack thereof, is predictive of intimate effects on Gcn5 or its nearest neighbors (i.e. Ada2 and Ada3). This would make our synthetic construct an ideal method to evaluate direct regulators of Gcn5 activity. As we demonstrated for ada3Δ, mutations or treatments that impact our substrate are likely to have broad consequences across the entire acetylome.
Regulation of acetylation by sirtuins
Acetylation of our synthetic substrate was negatively regulated by multiple sirtuins. Of hst1Δ, hst2Δ, and sir2Δ, only hst2Δ impacted acetylation as a single mutant. Bacterially purified Hst2 also deacetylated our synthetic substrate in vitro, consistent with a model where Hst2 is acting directly. Hst2 is cytoplasmic, and therefore our synthetic substrate may be deacetylated at least partially in the cytoplasm. However, this remains to be formally tested as Hst2 may also shuttle between the cytoplasm and nucleus under some circumstances (29, 30). Increased acetylation of the substrate in the sirtuin triple mutant (hst1Δ hst2Δ sir2Δ) suggests additional contributions from Sir2 and/or Hst1. Sir2 may cooperate with Hst1 to deacetylate the fusion substrate in the nucleus. This model is reminiscent of the proposed cooperative sirtuin-dependent regulation of Ifh1 and Sgf73, which we and others suggested previously (34, 53, 56). Sirtuin HDACs may function redundantly on the same acetylation sites, target different acetylation sites on the same protein, or act on unique protein populations that reside in distinct cellular compartments. Alternatively, Sir2 and Hst1 may act indirectly to promote Gcn5-dependent acetylation. Notably, although we demonstrated the utility of our synthetic acetylation tool by examining the contribution of SAGA subunits to nonhistone protein acetylation, it could also be used to study the impact of Sir2 complexes (e.g. RENT (regulator of nucleolar silencing and telophase exit) or SIR (silent information regulator)) (57); Hst1 binders Sum1 and Rfm1 (58, 59); or Crm1, which regulates the nuclear export of Hst2 and may promote its accumulation in the cytoplasm (29).
Gcn5 and sirtuins appear to regulate many of the same acetylation sites, and this is exemplified by our synthetic substrate whose shared consensus sequence is oppositely regulated by both enzymes in vivo. However, the mechanisms that facilitate this coordinated activity are still poorly understood. Recently, Sir2 has been reported to physically interact with SAGA via the DUB module (60). This interaction may promote Sir2 reversal of SAGA-regulated acetylations. Whether the ADA subcomplex also interacts physically with Sir2 or other sirtuins is unclear. Future work will use the synthetic substrate described in this work to address this important question. The extension of our synthetic biology approach to other HAT and HDAC enzymes across model systems will generate a toolkit to compare and contrast mechanisms of regulation in vivo.
Experimental procedures
Yeast strains
Yeast strains are in the S288C background and are described in Table S1. All strains were generated using standard procedures (41, 61) and verified using a combination of PCR analysis of colony-purified transformants and Western blotting where appropriate as described previously (62). Primer sequences used to confirm strains are available upon request. Plasmids, described below, were introduced into yeast using high-efficiency lithium acetate transformation followed by selection on synthetic complete medium lacking uracil.
Plasmids
To construct the entry vector for GFP-consensus constructs, the multicloning site from pRS406-ADH1/CYC1 (a gift from Nicolas Buchler and Fred Cross; Addgene plasmid number 15974) was cloned into pRS316 using the restriction enzymes KpnI (New England Biolabs, R3142) and SacI (New England Biolabs, R3156S). GFP or GFP-consensus constructs were generated by amplifying GFP from plasmid pFA6-GFP-His3MX using PAGE-purified oligonucleotides (Eurofins; sequences available upon request). The 5′ oligo included an EcoRI restriction site. The 3′ oligo included consensus sequences followed by a HindIII site. Constructs and vectors were digested with HindIII (New England Biolabs, R0104S) and EcoRI (New England Biolabs, R0101S) for 90 min at 37 °C. Agarose gel–purified fragments were ligated using T4 DNA ligase (New England Biolabs, MO202) prior to transformation into chemically competent DH5α cells (Thermo Fisher, 18263012) and recovery via plasmid miniprep (Biobasics, BS614). Construct sequences were verified via Sanger sequencing (McGill University and Génome Québec Innovation Centre) using primers within the ADH1 promoter, GFP coding sequence, and the CYC1 terminator. For SPT2-GFP plasmids, pRS316 was first digested using HindIII-HF (New England Biolabs, R3104S) and EcoRI-HF (New England Biolabs, R3101T) for 15 min at 37 °C. SPT2-GFP cassette was amplified using primers providing homology with pRS316. Agarose gel–purified fragments were combined in Gibson Assembly Mix (New England Biolabs, E5510S) and incubated for 15 min at 50 °C. Product was then transformed into chemically competent cells (New England Biolabs, E5510S) and recovered via plasmid miniprep followed by Sanger sequencing verification. spt2-K166R-GFP mutant plasmid was created by amplifying sequencing primers with overlapping primers to introduce two separate nucleotide changes that add an XbaI restriction site (noncoding) in addition to a mutation conferring the desired lysine-to-arginine change. Reaction mixture was digested with DpnI, transformed into chemically competent cells, and then recovered via plasmid miniprep. Plasmids were first confirmed by digestion using XbaI (New England Biolabs, R0145) followed by verification by Sanger sequencing. Plasmids are now available through Addgene.
Whole-cell extract generation and immunoprecipitation
40–80 A600 equivalents of log-phase cells were collected and lysed using acid-washed glass beads in 750 μl of chilled IP lysis buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 150 mm NaCl, 0.5% Nonidet P-40) with inhibitors (10 mm glycerol 2-phosphate, 5 mm NaF (Sigma, 201154), 1 mm DTT (BioBasic, DB0058), 1.75 mm phenylmethylsulfonyl fluoride (Sigma, P7626), Complete protease inhibitor tablet (without EDTA; Roche Applied Science, 4693132001), 10 mm sodium butyrate (Sigma, 303410), and 10 mm nicotinamide (NAM) (Sigma, N3376)). Lysis was carried out in screw-cap tubes with eight timed pulses of 1.5 min on a BioSpec Mini Beadbeater with incubation on ice in between bursts. Tubes were punctured with an 18-gauge needle, and supernatant was collected in 75-mm tubes (Falcon, reference number 352054) via centrifugation, transferred to microcentrifuge tubes, and spun for 4 min at 17,000 × g. Supernatants were transferred and spun again for 4 min at 17,000 × g before transferring again to a clean microcentrifuge tube. 20–50 μl of cell extract was saved for inputs. Remaining supernatants were incubated at 4 °C for 2 h with 0.5 μl of anti-GFP antibody (Abcam, ab290) and then another hour with 20 μl of washed magnetic beads coupled to Protein A (Bio-Rad, 161-4013). Beads, antibody, and bound proteins were recovered on the magnetic Dynarack and washed three times in IP lysis buffer followed by elution in 1–2× SDS sample buffer (with DTT at a final concentration of 100 mm) at 65 °C for 10 min. Eluates were transferred to new tubes prior to heating at 100 °C for 5 min and analysis via SDS-PAGE.
Immunoblotting
Unless indicated otherwise, gels were 10% SDS-polyacrylamide with 37.5:1 acrylamide:bisacrylamide (Bio-Rad, 1610158). Gels were transferred to PVDF membrane (Bio-Rad, 162-0177) at 75 V. All membranes were blocked in 5% milk or BSA in TBS with 0.1% Tween (TBS-T). Primary antibody mixtures were made at a 1:2000 dilution unless otherwise mentioned in either 5% milk or BSA in TBS-T with 0.01% sodium azide. Membranes were incubated at 4 °C overnight, washed three times for 10 min each with TBS-T before probing with HRP-coupled secondary antibodies (also made in 5% milk or BSA at 1:10,000 dilution) for 30–50 min. Blots were then washed an additional three times in TBS-T for 10 min each prior to application of ECL reagent (Millipore) and exposure to autoradiography film (Progene). Product numbers and concentrations of antibodies used are summarized in Table S2. Representative Western blots of IP-Western experiments are shown (minimum n = 2 biological replicates).
KDAC assay
KDAC assays were carried out similarly to those described previously (34, 53). Purified GFP-3X substrate from a sirtuin triple mutant was used in a final volume of 25 μl with 5 μl of 5× HDAC reaction buffer (250 mm Tris-HCl, pH 8.0, 2.5 mm DTT, one Roche Applied Science protease inhibitor tablet without EDTA/10 ml), 2.5 μl of GST-Hst2 (∼1 μg total), and 125 μm NAD+. Nicotinamide (Sigma, N3376) was used at a final concentration of 10 mm. Reactions were incubated for 1 h at 30 °C. Reactions were stopped with the addition of SDS-PAGE sample buffer with 0.1 m DTT and boiled to remove GFP from beads. GST and HST2-GST plasmids used for expression in BL21 cells were gifts from Adam Rudner. Protein constructs were purified using GSH-Sepharose (Thermo Fisher, 16101) and stored at −80 °C in glycerol until use.
HAT assay
In vitro HAT assays were performed as described previously (53). Recombinant His6-TRX-Gcn5 WT and E173Q mutant constructs were combined with purified GFP control and GFP-3X substrate (isolated from gcn5Δ cells) in a final volume of 50 μl made with 25 μl of HAT reaction buffer (10% glycerol, 200 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.2 mm EDTA, 2 mm sodium butyrate, 2 mm DTT) supplemented with 800 μm acetyl-CoA. Reactions were incubated for 1 h at 30 °C and then stopped by addition of 3× SDS-PAGE sample buffer (0.1 m DTT) followed by boiling. Control reactions were carried out using 0.5 μg of human H3.3 (New England Biolabs, N25075).
Mass spectrometry
Determination of acetylation sites on GFP-consensus constructs
Indicated constructs were immunoprecipitated via GFP Trap (Chromotek). Bound proteins were eluted with SDS-PAGE sample buffer and analyzed on a NuPAGE Novex Bis-Tris (4–12%) gel (Thermo Fisher, NP0336BOX) run at 200 V for 50 min according to the manufacturer's instructions and as described elsewhere (63). Staining was with Invitrogen Colloidal Blue staining kit (LC6025) following the manufacturer's directions for Novex Bis-Tris gels. Preparation of excised gel slices was carried out as described (64).
ELITE LC-MS/MS was completed as described previously with minor modifications (65). Briefly, our analysis employed an Ekspert nanoLC 400 (Eksigent, Dublin, CA) and an Orbitrap ELITE MS (Thermo Fisher Scientific, San Jose, CA). The MS was operated in the positive ion mode. Peptides were resuspended in 30 μl of 0.5% formic acid prior to injection into an analytical column of 75-μm internal diameter and packed with 1.9-μm C18 resin (Dr. Maisch, GmbH, Ammerbuch, Germany). Elution was with a flow rate of 300 nl/min. A 120-min gradient of 5–30% (v/v) acetonitrile with 0.1% (v/v) formic acid was used. The heating capillary was set at 300 °C. The spray voltage was fixed at 2.2 kV. The MS scan used ranged from 350 to 1750 m/z. The MS/MS scan was conducted on the 20 most intense ions. Exclusion duration was 90 s with one repeat count and a 30-s repeat duration.
Acetylome analysis
SILAC labeling for paired WT and ada3Δ mutant cells, cell lysis, chemical treatments, trypsin digestion (Thermo Fisher, 90058), anti-acetyllysine IP (ImmuneChem, ICP0380), elution, and peptide purification prior to MS analysis were as described previously (34). HPLC-electrospray ionization-tandem MS (MS/MS) for yeast acetylome analyses was completed using a Q Exactive mass spectrometer (Thermo Fisher Scientific). Conditions used were similar to those described elsewhere (66). Briefly, the Q Exactive instrument was operated in positive ion mode. Peptides immunoprecipitated with anti-acetyllysine antibody were first resuspended in 0.5% (v/v) formic acid and injected onto a 75-μm-internal-diameter analytical column packed with 1.9-μm C18 resin (Dr. Maisch, GmbH). Peptides were eluted using a 200 nl/min flow rate. A 120-min gradient was used with increasing acetonitrile concentration (5–30% (v/v), 0.1% (v/v) formic acid). The MS scan employed ranged from 300 to 1800 m/z with subsequent selection of the 12 most intense ions for data-dependent MS/MS scan. A dynamic exclusion repeat count of 2 and repeat exclusion duration of 30 s were used.
Database searches
Xcalibur software (Thermo Fisher Scientific) was used to acquire data. Following acquisition, a search was performed using MaxQuant software version 1.5.3.30 (67) against a Saccharomyces cerevisiae database (downloaded from UniProt February, 9, 2017). Parameters used were: multiplicity of 2 (heavy label, Lys8); trypsin digest, a maximum of two missed cleavages; fixed modification of cysteine carbamidomethylation; variable modifications of methionine oxidation, acetyllysine, and N-terminal acetylation; minimum peptide length of seven amino acids; 0.5 Da for ion mass tolerance; peptide and protein false discovery rate fixed at 1%. For in-gel analysis, the GFP-3X consensus fusion sequence was added to the database.
Bioinformatics analyses
GO-term enrichments were determined using DAVID version 6.8 (https://david.ncifcrf.gov) with S. cerevisiae as the background and with default settings (68, 69).
Antibody generation
Antibody protocols were developed with the intention to generate a reagent that recognizes the critical features of the acetylated consensus without being specific to an exact amino acid sequence. Hybridoma clone A1504705 was developed by immunizing four BALB/c mice with 25 μg of KLH-conjugated peptide AAASAK(ac)RPAAA prepared in complete Freund's adjuvant (Sigma-Aldrich, catalog number F5881). Two weeks later, each mouse was boosted with 12.5 μg of KLH-conjugated peptide GAPANK(ac)RPRRG prepared in incomplete Freund's adjuvant (Sigma-Aldrich, catalog number F5506) followed by another boost with 12.5 μg of KLH-conjugated peptide SSVSYK(ac)RVCGG prepared in incomplete Freund's adjuvant 2 weeks apart. Three days after the boosting, sera of immunized mice were collected and tested against BSA-conjugated peptides AAASAK(ac)RPAAA, GAPANK(ac)RPRRG, SSVSYK(ac)RVCGG, and AAASAKRPAAA in ELISA. The mouse with the best response to the first three peptides was subsequently boosted with 10 μg each of KLH-conjugated peptides AAASAK(ac)RPAAA, GAPANK(ac)RPRRG, and SSVSYK(ac)RVCGG. Lymphocytes from the mouse that received the final antigen boost were harvested 3 days later and fused with myeloma cells sp2/0 using a GenomOne kit (Cosmo Bio, catalog number ISK-CF-001-EX). Clone A1504705 was selected based on its reactivity to BSA-conjugated peptides GAPANK(ac)RPRRG, SSVSYK(ac)RVCGG, and AAASAK(ac)RPAAA but not to BSA-conjugated peptide AAASAKRPAAA in ELISA. Hybridoma cell culture supernatants were collected, and the antibody was purified via a Protein G column (Sigma-Aldrich, catalog number GE17-0618-01). After supernatant binding, the resin was washed with PBS (pH 7.2) and antibodies were eluted with 50 mm diethanolamine (pH 11.0) (Sigma-Aldrich, catalog number 31589). Subsequently, eluted antibodies were neutralized by 1 m Tris (pH 8.0). The antibody was prepared by dialyzing against PBS (pH 7.2) with 0.09% of sodium azide (Sigma-Aldrich, catalog number 71289).
ELISA
The ELISA was carried out at BioLegend. Briefly, peptides were conjugated to BSA and coated to 96-well plates overnight at 4 °C. Plates were washed with PBS, and antibody was added at the indicated concentrations in 10% BSA for 45 min at room temperature. Plates were washed with PBS and incubated with 100 μl of HRP-coupled goat anti-mouse IgG in PBS with 10% BSA (1:2,500; BioLegend) for 45 min at room temperature. Plates were again washed and incubated with 50 μl/well tetramethylbenzidine for 2 min. Reactions were stopped with the addition of 50 ml of 0.2 n H2SO4 and read at 450 nm. Graphs were prepared using GraphPad Prism.
Data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (49) partner repository with the dataset identifier PXD012608.
Author contributions
A. R. and M. D. conceptualization; A. R. and M. D. formal analysis; A. R., A. D., M.-S. L., and M. D. investigation; A. R. and M. D. writing-original draft; A. R. and M. D. writing-review and editing; A. D. and M.-S. L. resources; M. D. supervision; M. D. funding acquisition; M. D. project administration.
Supplementary Material
Acknowledgments
We thank members of the Downey laboratory for critical reading of the manuscript. We thank members of the Figeys laboratory for assistance with protein purification and MS. We thank Adam Rudner for plasmids.
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Grant RGPIN-2016-05015 (to M. D.). Mong-Shang Lin is an employee of BioLegend, and the monoclonal antibody generation was carried out in BioLegend's facility in San Diego, CA.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012608.
This article contains Figs. S1–S5 and Tables S1–S3.
- HAT
- histone acetyltransferase
- DUB
- deubiquitylation
- TBP
- TATA-binding protein
- HDAC
- histone deacetylase
- SILAC
- stable isotope labeling with amino acids in cell culture
- IP
- immunoprecipitation
- GO
- gene ontology
- NAM
- nicotinamide
- PVDF
- polyvinylidene difluoride
- TBS
- Tris-buffered saline
- HRP
- horseradish peroxidase
- KDAC
- lysine deacetylase
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- KLH
- keyhole limpet hemocyanin
- SAGA
- Spt-Ada-Gcn5-acetyltransferase
- SLIK
- SAGA-like.
References
- 1. Lee K. K., Sardiu M. E., Swanson S. K., Gilmore J. M., Torok M., Grant P. A., Florens L., Workman J. L., and Washburn M. P. (2011) Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7, 503 10.1038/msb.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Helmlinger D., and Tora L. (2017) Sharing the SAGA. Trends Biochem. Sci. 42, 850–861 10.1016/j.tibs.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spedale G., Timmers H. T., and Pijnappel W. W. (2012) ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527–541 10.1101/gad.184705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han Y., Luo J., Ranish J., and Hahn S. (2014) Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33, 2534–2546 10.15252/embj.201488638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eberharter A., Sterner D. E., Schieltz D., Hassan A., Yates J. R. 3rd, Berger S. L., and Workman J. L. (1999) The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6621–6631 10.1128/MCB.19.10.6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ingvarsdottir K., Krogan N. J., Emre N. C., Wyce A., Thompson N. J., Emili A., Hughes T. R., Greenblatt J. F., and Berger S. L. (2005) H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25, 1162–1172 10.1128/MCB.25.3.1162-1172.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warfield L., Ranish J. A., and Hahn S. (2004) Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 18, 1022–1034 10.1101/gad.1192204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sermwittayawong D., and Tan S. (2006) SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25, 3791–3800 10.1038/sj.emboj.7601265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laprade L., Rose D., and Winston F. (2007) Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics 177, 2007–2017 10.1534/genetics.107.081976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee K. K., Florens L., Swanson S. K., Washburn M. P., and Workman J. L. (2005) The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25, 1173–1182 10.1128/MCB.25.3.1173-1182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sterner D. E., Belotserkovskaya R., and Berger S. L. (2002) SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11622–11627 10.1073/pnas.182021199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pray-Grant M. G., Schieltz D., McMahon S. J., Wood J. M., Kennedy E. L., Cook R. G., Workman J. L., Yates J. R. 3rd, Grant P. A. (2002) The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22, 8774–8786 10.1128/MCB.22.24.8774-8786.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balasubramanian R., Pray-Grant M. G., Selleck W., Grant P. A., and Tan S. (2002) Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277, 7989–7995 10.1074/jbc.M110849200 [DOI] [PubMed] [Google Scholar]
- 14. Sterner D. E., Wang X., Bloom M. H., Simon G. M., and Berger S. L. (2002) The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277, 8178–8186 10.1074/jbc.M108601200 [DOI] [PubMed] [Google Scholar]
- 15. Marcus G. A., Silverman N., Berger S. L., Horiuchi J., and Guarente L. (1994) Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13, 4807–4815 10.1002/j.1460-2075.1994.tb06806.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bian C., Xu C., Ruan J., Lee K. K., Burke T. L., Tempel W., Barsyte D., Li J., Wu M., Zhou B. O., Fleharty B. E., Paulson A., Allali-Hassani A., Zhou J. Q., Mer G., et al. (2011) Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842 10.1038/emboj.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng W., Togawa C., Zhang K., and Kurdistani S. K. (2008) Regulators of cellular levels of histone acetylation in Saccharomyces cerevisiae. Genetics 179, 277–289 10.1534/genetics.107.085068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brachmann C. B., Sherman J. M., Devine S. E., Cameron E. E., Pillus L., and Boeke J. D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9, 2888–2902 10.1101/gad.9.23.2888 [DOI] [PubMed] [Google Scholar]
- 19. Wierman M. B., and Smith J. S. (2014) Yeast sirtuins and the regulation of aging. FEMS Yeast Res. 14, 73–88 10.1111/1567-1364.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maas N. L., Miller K. M., DeFazio L. G., and Toczyski D. P. (2006) Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23, 109–119 10.1016/j.molcel.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 21. Celic I., Masumoto H., Griffith W. P., Meluh P., Cotter R. J., Boeke J. D., and Verreault A. (2006) The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr. Biol. 16, 1280–1289 10.1016/j.cub.2006.06.023 [DOI] [PubMed] [Google Scholar]
- 22. Che J., Smith S., Kim Y. J., Shim E. Y., Myung K., and Lee S. E. (2015) Hyper-acetylation of histone H3K56 limits break-induced replication by inhibiting extensive repair synthesis. PLoS Genet. 11, e1004990 10.1371/journal.pgen.1004990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Celic I., Verreault A., and Boeke J. D. (2008) Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics 179, 1769–1784 10.1534/genetics.108.088914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madsen C. T., Sylvestersen K. B., Young C., Larsen S. C., Poulsen J. W., Andersen M. A., Palmqvist E. A., Hey-Mogensen M., Jensen P. B., Treebak J. T., Lisby M., and Nielsen M. L. (2015) Biotin starvation causes mitochondrial protein hyperacetylation and partial rescue by the SIRT3-like deacetylase Hst4p. Nat. Commun. 6, 7726 10.1038/ncomms8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mead J., McCord R., Youngster L., Sharma M., Gartenberg M. R., and Vershon A. K. (2007) Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases. Mol. Cell. Biol. 27, 2466–2475 10.1128/MCB.01641-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Froyd C. A., and Rusche L. N. (2011) The duplicated deacetylases Sir2 and Hst1 subfunctionalized by acquiring complementary inactivating mutations. Mol. Cell. Biol. 31, 3351–3365 10.1128/MCB.05175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imai S., Armstrong C. M., Kaeberlein M., and Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 10.1038/35001622 [DOI] [PubMed] [Google Scholar]
- 28. Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., and Sternglanz R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811 10.1073/pnas.110148297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson J. M., Le V. Q., Zimmerman C., Marmorstein R., and Pillus L. (2006) Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep. 7, 1247–1251 10.1038/sj.embor.7400829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perrod S., Cockell M. M., Laroche T., Renauld H., Ducrest A. L., Bonnard C., and Gasser S. M. (2001) A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 20, 197–209 10.1093/emboj/20.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weinert B. T., Wagner S. A., Horn H., Henriksen P., Liu W. R., Olsen J. V., Jensen L. J., and Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signal. 4, ra48 10.1126/scisignal.2001902 [DOI] [PubMed] [Google Scholar]
- 32. Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., and Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–1522 10.1074/mcp.M112.017251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 34. Downey M., Johnson J. R., Davey N. E., Newton B. W., Johnson T. L., Galaang S., Seller C. A., Krogan N., and Toczyski D. P. (2015) Acetylome profiling reveals overlap in the regulation of diverse processes by sirtuins, gcn5, and esa1. Mol. Cell. Proteomics 14, 162–176 10.1074/mcp.M114.043141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Downey M., and Baetz K. (2016) Building a KATalogue of acetyllysine targeting and function. Brief. Funct. Genomics 15, 109–118 10.1093/bfgp/elv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rojas J. R., Trievel R. C., Zhou J., Mo Y., Li X., Berger S. L., Allis C. D., and Marmorstein R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 10.1038/43487 [DOI] [PubMed] [Google Scholar]
- 37. Pérez-Martin J., and Johnson A. D. (1998) The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast. Mol. Cell. Biol. 18, 4157–4164 10.1128/MCB.18.7.4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pracheil T., and Liu Z. (2013) Tiered assembly of the yeast Far3-7-8-9-10-11 complex at the endoplasmic reticulum. J. Biol. Chem. 288, 16986–16997 10.1074/jbc.M113.451674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bloemendal S., Bernhards Y., Bartho K., Dettmann A., Voigt O., Teichert I., Seiler S., Wolters D. A., Pöggeler S., and Kück U. (2012) A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84, 310–323 10.1111/j.1365-2958.2012.08024.x [DOI] [PubMed] [Google Scholar]
- 40. Cherry J. M., Hong E. L., Amundsen C., Balakrishnan R., Binkley G., Chan E. T., Christie K. R., Costanzo M. C., Dwight S. S., Engel S. R., Fisk D. G., Hirschman J. E., Hitz B. C., Karra K., Krieger C. J., et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 10.1093/nar/gkr1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 42. Kruger W., and Herskowitz I. (1991) A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol. Cell. Biol. 11, 4135–4146 10.1128/MCB.11.8.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Niedenthal R. K., Riles L., Johnston M., and Hegemann J. H. (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12, 773–786 [DOI] [PubMed] [Google Scholar]
- 44. Yen Y. M., Roberts P. M., and Johnson R. C. (2001) Nuclear localization of the Saccharomyces cerevisiae HMG protein NHP6A occurs by a Ran-independent nonclassical pathway. Traffic 2, 449–464 10.1034/j.1600-0854.2001.20703.x [DOI] [PubMed] [Google Scholar]
- 45. Cieniewicz A. M., Moreland L., Ringel A. E., Mackintosh S. G., Raman A., Gilbert T. M., Wolberger C., Tackett A. J., and Taverna S. D. (2014) The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3. Mol. Cell. Proteomics 13, 2896–2910 10.1074/mcp.M114.038174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pray-Grant M. G., Daniel J. A., Schieltz D., Yates J. R. 3rd, Grant P. A. (2005) Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433, 434–438 10.1038/nature03242 [DOI] [PubMed] [Google Scholar]
- 47. Wu P. Y., and Winston F. (2002) Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22, 5367–5379 10.1128/MCB.22.15.5367-5379.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Candau R., Moore P. A., Wang L., Barlev N., Ying C. Y., Rosen C. A., and Berger S. L. (1996) Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16, 593–602 10.1128/MCB.16.2.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li S., and Shogren-Knaak M. A. (2009) The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J. Biol. Chem. 284, 9411–9417 10.1074/jbc.M809617200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun J., Paduch M., Kim S. A., Kramer R. M., Barrios A. F., Lu V., Luke J., Usatyuk S., Kossiakoff A. A., and Tan S. (2018) Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2. Proc. Natl. Acad. Sci. U.S.A. 115, 10010–10015 10.1073/pnas.1805343115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi J. K., Grimes D. E., Rowe K. M., and Howe L. J. (2008) Acetylation of Rsc4p by Gcn5p is essential in the absence of histone H3 acetylation. Mol. Cell. Biol. 28, 6967–6972 10.1128/MCB.00570-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Downey M., Knight B., Vashisht A. A., Seller C. A., Wohlschlegel J. A., Shore D., and Toczyski D. P. (2013) Gcn5 and sirtuins regulate acetylation of the ribosomal protein transcription factor Ifh1. Curr. Biol. 23, 1638–1648 10.1016/j.cub.2013.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cai L., Sutter B. M., Li B., and Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 10.1016/j.molcel.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robert F., Pokholok D. K., Hannett N. M., Rinaldi N. J., Chandy M., Rolfe A., Workman J. L., Gifford D. K., and Young R. A. (2004) Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16, 199–209 10.1016/j.molcel.2004.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cai L., McCormick M. A., Kennedy B. K., and Tu B. P. (2013) Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep. 4, 1063–1071 10.1016/j.celrep.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tanny J. C., Kirkpatrick D. S., Gerber S. A., Gygi S. P., and Moazed D. (2004) Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 24, 6931–6946 10.1128/MCB.24.16.6931-6946.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xie J., Pierce M., Gailus-Durner V., Wagner M., Winter E., and Vershon A. K. (1999) Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18, 6448–6454 10.1093/emboj/18.22.6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCord R., Pierce M., Xie J., Wonkatal S., Mickel C., and Vershon A. K. (2003) Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23, 2009–2016 10.1128/MCB.23.6.2009-2016.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McCormick M. A., Mason A. G., Guyenet S. J., Dang W., Garza R. M., Ting M. K., Moller R. M., Berger S. L., Kaeberlein M., Pillus L., La Spada A. R., and Kennedy B. K. (2014) The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep. 8, 477–486 10.1016/j.celrep.2014.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Goldstein A. L., and McCusker J. H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 62. Rössl A., Bentley-DeSousa A., Tseng Y. C., Nwosu C., and Downey M. (2016) Nicotinamide suppresses the DNA damage sensitivity of Saccharomyces cerevisiae independently of sirtuin deacetylases. Genetics 204, 569–579 10.1534/genetics.116.193524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bentley-DeSousa A., Holinier C., Moteshareie H., Tseng Y. C., Kajjo S., Nwosu C., Amodeo G. F., Bondy-Chorney E., Sai Y., Rudner A., Golshani A., Davey N. E., and Downey M. (2018) A screen for candidate targets of lysine polyphosphorylation uncovers a conserved network implicated in ribosome biogenesis. Cell Rep. 22, 3427–3439 10.1016/j.celrep.2018.02.104 [DOI] [PubMed] [Google Scholar]
- 64. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 65. Starr A. E., Deeke S. A., Ning Z., Chiang C. K., Zhang X., Mottawea W., Singleton R., Benchimol E. I., Wen M., Mack D. R., Stintzi A., and Figeys D. (2017) Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn's disease from UC. Gut 66, 1573–1583 10.1136/gutjnl-2015-310705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang X., Li L., Mayne J., Ning Z., Stintzi A., and Figeys D. (2018) Assessing the impact of protein extraction methods for human gut metaproteomics. J. Proteomics 180, 120–127 10.1016/j.jprot.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 67. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., and Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 10.1038/nprot.2009.36 [DOI] [PubMed] [Google Scholar]
- 68. Huang D. W., Sherman B. T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M. W., Lane H. C., and Lempicki R. A. (2007) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169–W175 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.