Abstract
Contactin-associated protein 1 (CASPR1 or CNTNAP1) was recently reported to be expressed in brain microvascular endothelial cells (BMECs), the major component of the blood–brain barrier. To investigate CASPR1's physiological role in BMECs, here we used CASPR1 as a bait in a yeast two-hybrid screen to identify CASPR1-interacting proteins and identified the β3 subunit of Na+/K+-ATPase (ATP1B3) as a CASPR1-binding protein. Using recombinant and purified CASPR1, RNAi, GST-pulldown, immunofluorescence, immunoprecipitation, and Na+/K+-ATPase activity assays, we found that ATP1B3's core proteins, but not its glycosylated forms, interact with CASPR1, which was primarily located in the endoplasmic reticulum of BMECs. CASPR1 knockdown reduced ATP1B3 glycosylation and prevented its plasma membrane localization, phenotypes that were reversed by expression of full-length CASPR1. We also found that the CASPR1 knockdown reduces the plasma membrane distribution of the α1 subunit of Na+/K+-ATPase, which is the major component assembled with ATP1B3 in the complete Na+/K+-ATPase complex. The binding of CASPR1 with ATP1B3, but not the α1 subunit, indicated that CASPR1 binds with ATP1B3 to facilitate the assembly of Na+/K+-ATPase. Furthermore, the activity of Na+/K+-ATPase was reduced in CASPR1-silenced BMECs. Interestingly, shRNA-mediated CASPR1 silencing reduced glutamate efflux through the BMECs. These results demonstrate that CASPR1 binds with ATP1B3 and thereby contributes to the regulation of Na+/K+-ATPase maturation and trafficking to the plasma membrane in BMECs. We conclude that CASPR1-mediated regulation of Na+/K+-ATPase activity is important for glutamate transport across the blood–brain barrier.
Keywords: Na+/K+-ATPase, brain, endothelial cell, endoplasmic reticulum (ER), glutamate, ATP1B3, blood-brain barrier, contactin-associated protein 1 (CASPR1), CNS homeostasis, CNTNAP1
Introduction
The blood–brain barrier (BBB)4 is a physical barrier that is essential in regulating the flux of ions and molecules into and out of brain parenchyma and protecting central nervous system homeostasis (1, 2). The BBB is composed of brain microvascular endothelial cells (BMECs), basal lamina, end-feet of astrocytes, and pericytes. The BMECs, distinct from endothelial cells in other tissues, are unique as they have continuous intercellular tight junctions and undergo extremely low rates of transcytosis, which greatly limits both the paracellular and transcellular transport of molecules through the BBB (3–6). The BMECs contain multiple substrate-specific transport systems that control transport of ions, metabolites, and specific molecules. For example, the sodium pump, localized on the abluminal membrane of BMECs, regulates Na+ influx into the brain interstitial fluid in exchange for K+ (7). The sodium-dependent transporters for the amino acids exist at the abluminal membrane, transfer amino acids from the brain to endothelial cells, and then transfer amino acids from endothelial cells into the circulation (4, 8). Large proteins, such as transferrin and low-density lipoproteins, utilize receptor-mediated transport systems to cross the BBB (9, 10).
CASPR1 (contactin-associated protein 1, also known as CNTNAP1) is a 190-kDa transmembrane protein highly concentrated at paranodes, which was originally identified as an adhesion molecule that interacted with contactin in neurons (11, 12). CASPR1 forms a complex with contactin and neurofascin-155 and acts as a barrier between the nodes of Ranvier and internodes to ensure the propagation of action potentials in myelinated axons (13–15). Decreased expression of CASPR1 was associated with the axonal demyelination in multiple sclerosis (16, 17). Recently, we identified the expression of CASPR1 in the BMEC in vivo and in vitro (18). We found that CASPR1 acts as a host receptor for bacterial virulence factor to trigger the penetration of pathogenic Escherichia coli through the BBB in the condition of bacterial meningitis (18). However, the physiological function of CASPR1 in brain endothelial cells remains unknown.
In this study, we found that CASPR1 directly interacted with ATP1B3, the β3 subunit of Na+/K+-ATPase. The Na+/K+-ATPase, also known as the sodium pump, transports three Na+ out of the cell and two K+ into the cell and plays a crucial role in maintaining the low concentrations of intracellular Na+ ions and high concentrations of intracellular K+ ions (19). The Na+/K+-ATPase belongs to the P-type ATPase family and consists of two subunits, α and β (20). The α subunit of Na+/K+-ATPase, containing ATP and ligand-binding sites, is considered as the catalytic subunit, whereas the β subunit is essential for the membrane targeting and full function of the Na+/K+-ATPase (20, 21). Here, we demonstrated that CASPR1 interacts with ATP1B3, and this interaction is required for the effective trafficking of ATP1B3 to the plasma membrane. Functionally, we found that CASPR1 interacted with ATP1B3 to regulate the activity of Na+/K+-ATPase, which is involved in the efflux of glutamate, considered as the major excitatory neurotransmitter in the brain, across the BBB formed by brain endothelial cells.
Results
CASPR1 interacts with ATP1B3 in brain endothelial cells
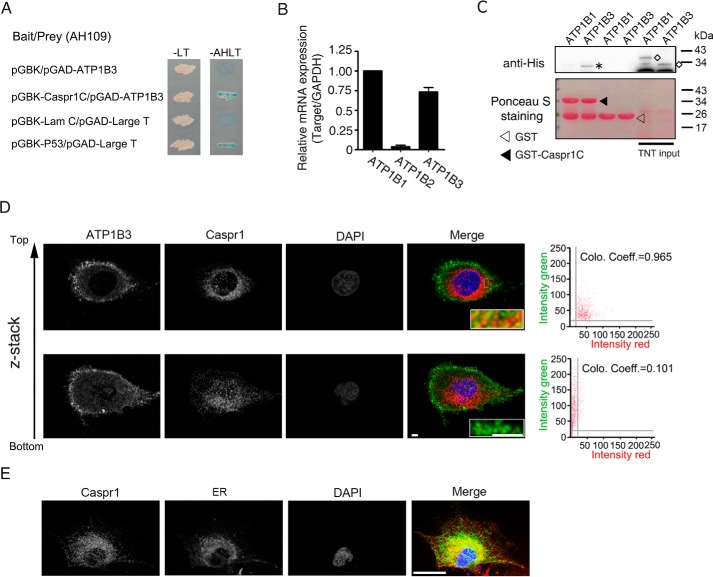
To investigate the biological function of CASPR1, we performed yeast two-hybrid analysis to identify the binding partner of CASPR1. Human CASPR1 protein contains a large extracellular domain (aa 1–1283), a single transmembrane domain (aa 1284–1304), and a short intracellular domain (aa 1305–1384). Here, to screen the intracellular binding protein of CASPR1, the intracellular domain of CASPR1 was used as a bait to screen the human fetal brain cDNA library. From the results of yeast two-hybrid analysis, we obtained several positive clones encoding the β3 subunit of Na+/K+-ATPase (ATP1B3). Yeast cells co-transformed with the bait vector (pGBK) containing the CASPR1 intracellular domain and the prey vector (pGAD) containing ATP1B3 were able to grow and form blue colonies on the selection plates, suggesting the interaction of the cytoplasmic domain of CASPR1 with ATP1B3 (Fig. 1A). Previous reports showed that the β subunits of Na+/K+-ATPase include three isoforms, ATP1B1, ATP1B2, and ATP1B3 (22–24). Our real-time RT-PCR results revealed that ATP1B1 and ATP1B3 are expressed at relatively high levels in human brain microvascular endothelial cells (HBMECs) (Fig. 1B). Then we performed GST pulldown assays to assess the binding of Na+/K+-ATPase β subunits with the cytoplasmic domain of CASPR1 (CASPR1-C). The constructs containing the full-length cDNA of ATP1B1 and ATP1B3 with a His tag were translated with an in vitro transcription/translation system, and the products were incubated with GSH-Sepharose 4B beads prebound with the cytoplasmic domain of CASPR1 tagged with GST (GST-CASPR1-C), with GST serving as control. The following Western blotting results showed the robust binding of GST-CASPR1-C with ATP1B3, whereas GST-CASPR1-C could not bind with ATP1B1 (Fig. 1C), suggesting the specific binding of the cytoplasmic domain of CASPR1 with ATP1B3. Further results from immunoprecipitation revealed the interaction of endogenous CASPR1 protein with ATP1B3 in HBMECs (see Fig. 2C for details). We also used immunofluorescence to analyze the co-localization of ATP1B3 with CASPR1 in HBMECs. We found that ATP1B3 was expressed at the plasma membrane, with positive intracellular staining at the perinuclear region (Fig. 1D). Furthermore, we performed Z-stack scanning, and the results showed that ATP1B3 was co-localized with CASPR1 mainly at the perinuclear region (Fig. 1D). Immunofluorescence results showed that CASPR1 was mostly present in the endoplasmic reticulum (ER) of HBMECs, co-localized with ER marker (Fig. 1E). These data demonstrated that CASPR1 interacted with ATP1B3 in brain endothelial cells.
Figure 1.
ATP1B3 is identified as the binding protein of the CASPR1 C terminus. A, the bait plasmids containing cytoplasmic domain of CASPR1 (pGBK-CASPR1-C) and prey plasmids containing ATP1B3 (pGAD-ATP1B3) were co-transformed into yeast AH109 cells, and then the cells were grown on SD/−Leu−Trp (−LT) selection medium and SD/−Ade−His−Leu−Trp (−AHLT) medium containing X-α-Gal. Co-transformation of lamin C and large T served as a negative control, whereas p53 and large T were a positive control. B, the total RNA of HBMECs was extracted and reverse-transcribed with reverse transcriptase. Then real-time PCR was performed to detect the mRNA expression levels of ATP1B family members, including ATP1B1, ATP1B2, and ATP1B3, in HBMECs, with GAPDH used as an internal control. Data are normalized to the expression level of ATP1B1, which was defined as 1 (n = 3). C, the full-length ATP1B1 and ATP1B3 protein with a His tag were obtained by in vitro transcription and translation, respectively, and then incubated with GST-tagged CASPR1 intracellular domain (GST-CASPR1-C), with GST as a negative control. Precipitates were analyzed with anti-His antibody. An asterisk represents the precipitated ATP1B3, whereas the rhombi indicate the input proteins (top). Images are representative of three independent experiments. The blotting membrane was stained with Ponceau S as a loading control (bottom). D, HBMECs were fixed, and immunofluorescence was performed with antibodies recognizing CASPR1 (ab34151, red) and ATP1B3 (ab137055, green). DAPI (blue) was used for counterstaining. Continuous z-axis images were obtained using the confocal laser-scanning microscopy. The colocalization of CASPR1 and ATP1B3 was analyzed by the scatter plots and the colocalization coefficient. Scale bar, 4 μm. E, HBMECs were fixed, and immunofluorescence was performed with CASPR1 antibody (green) and ER tracker (red) to stain the endoplasmic reticulum. DAPI (blue) was used for counterstaining. Scale bar, 20 μm. Error bars, S.D.
Figure 2.
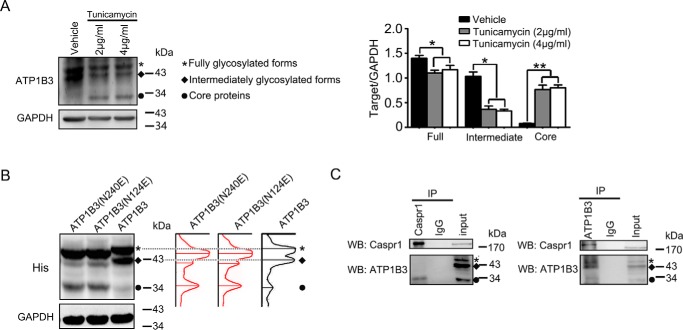
The core protein of ATP1B3 interacts with CASPR1 in HBMECs. A, HBMECs were pretreated with the indicated concentrations of tunicamycin for 24 h, and then the cell lysates were subjected to Western blotting with ATP1B3 antibody (ab67409). GAPDH was served as loading control. An asterisk represents fully glycosylated forms of ATP1B3, and a rhombus indicates the intermediately glycosylated forms of ATP1B3. A circle indicates the core proteins of ATP1B3. The labels in B and C are the same as in A. The changes of the different forms of ATP1B3 were obtained by calculating the band densitometry and normalized to the band intensity of GAPDH (right). Values are mean ± S.D. (error bars) from three independent experiments. *, p < 0.05. **, p < 0.01 (one-way ANOVA). B, HEK293T cells were transiently transfected with WT ATP1B3 and ATP1B3 mutant (N124E/N240E) with His tag for 48 h, respectively. Then the cell lysates were analyzed using anti-His antibody, with GAPDH as loading control. C, HBMECs lysates were immunoprecipitated (IP) with CASPR1 (ab34151) and ATP1B3 (H00000483-D01) antibody, respectively, and then the precipitated proteins were analyzed by Western blotting (WB) using antibodies against ATP1B3 (ab67409) and CASPR1 (ab133634).
CASPR1 binds with the core proteins of ATP1B3 in brain endothelial cells
Previous studies showed that ATP1B3 is a glycosylated protein, and there are fully and intermediately glycosylated forms of ATP1B3 in mammalian cells (25, 26). To evaluate the glycosylation of ATP1B3 in HBMECs, the cell were pretreated with tunicamycin, an inhibitor of glycosylation, and the cell lysates were subjected to Western blotting. We found that the fully and intermediately glycosylated forms of ATP1B3 were reduced after tunicamycin treatment compared with the control, whereas the core proteins of ATP1B3 were increased (Fig. 2A). It was reported that ATP1B3 mutants (N124E and N240E), containing the mutations at residues 124 and 240 with N-linked glycosylation, could affect ATP1B3 maturation in polarized hepatic cells (26). Here, we transfected the constructs containing His-tagged WT ATP1B3 or ATP1B3 mutants (N124E and N240E) into HEK293T cells, respectively, and the cell lysates were subjected to Western blot analysis with anti-His antibody. The results showed that compared with WT ATP1B3, the levels of core protein of mutated ATP1B3 (N124E and N240E) were boosted (Fig. 2B). Furthermore, the migration rate of the bands of the glycosylated forms of mutated ATP1B3 were increased (right panel in Fig. 2B), suggesting alleviated glycosylation caused by the N124E and N240E mutation in ATP1B3. These results revealed the presence of different glycosylated forms of ATP1B3 in HBMECs. To determine which forms of ATP1B3 in brain endothelial cells could bind with CASPR1, a reciprocal immunoprecipitation assay was performed with HBMECs lysates. The results showed that CASPR1 interacted with the core proteins, but not the glycosylated forms, of ATP1B3 in HBMECs (Fig. 2C). These data illustrated that CASPR1 was physically associated with the core proteins of ATP1B3, but not its glycosylated forms, in brain endothelial cells.
CASPR1 is required for the plasma membrane localization of ATP1B3 in brain endothelial cells
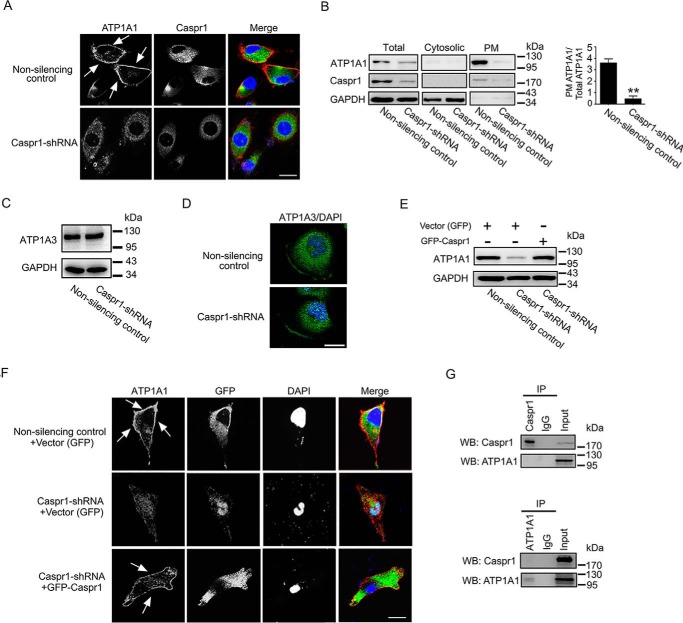
To investigate the relationship between CASPR1 and ATP1B3, the stable CASPR1-silenced HBMECs cell line was established. The construct containing shRNA targeting CASPR1 was transfected into HBMECs followed by G418 selection until the formation of cell clones. The single clone–derived cells were cultured, and the silencing effect was examined by Western blotting (Fig. 3A). The results showed that the CASPR1 was effectively suppressed in HBMECs compared with the nonsilencing shRNA control (Fig. 3A, left and right). Then we analyzed the expression of ATP1B3 in CASPR1-silenced HBMECs. The Western blotting results showed that the glycosylated forms of ATP1B3 were decreased in CASPR1-silenced HBMECs compared with the nonsilencing control (Fig. 3A, left and right). Because the mRNA level of ATP1B3 was not altered in CASPR1 knockdown HBMECs (Fig. S1A), we considered that CASPR1 was critical for the post-translational glycosylation of ATP1B3. Consistently, in HBMECs transiently transfected with CASPR1 siRNA, we observed similar reduced glycosylation of ATP1B3 (Fig. S1, B and C). Our immunostaining results revealed the localization of ATP1B3 at the plasma membrane in HBMECs, which is consistent with its function as a component of Na+/K+-ATPase at the plasma membrane (Fig. 1D and Fig. 3B (top panels)). Interestingly, we found ATP1B3 was remarkably aggregated in the cytoplasm of HBMECs with CASPR1 knockdown (Fig. 3B (bottom)). In contrast, the glycosylation of ATP1B1, another β subunit of Na+/K+-ATPase in HBMECs, was not affected by CASPR1 knockdown (Fig. S1D). Further immunofluorescence results showed that the plasma membrane localization of ATP1B1 remained unchanged in CASPR1-silenced HBMECs (Fig. S1E). The following immunoprecipitation confirmed that the biotin-labeled ATP1B3 on the plasma membrane was decreased in CASPR1-silenced HBMECs compared with the control (Fig. 3C). Further results showed that the full-length CASPR1 can effectively rescue the decreased glycosylated ATP1B3 (Fig. 3D) and the reduced plasma membrane localization of ATP1B3 (Fig. 3E). These results indicated that CASPR1 was required for the glycosylation and plasma membrane localization of ATP1B3, but not ATP1B1, in brain endothelial cells.
Figure 3.
Knockdown of CASPR1 reduced the glycosylation and plasma membrane localization of ATP1B3 in HBMECs. A, CASPR1-specific shRNA was stably transfected to HBMECs. Then the cell lysates were analyzed by Western blotting using CASPR1 and ATP1B3 (ab67409) antibody, with GAPDH as loading control. An asterisk represents the fully glycosylated ATP1B3, a rhombus indicates the intermediately forms of ATP1B3, and a circle indicates the core proteins of ATP1B3 (left). The knockdown effect was calculated as the band density of CASPR1 divided by that of GAPDH (right). The change of the fully glycosylated, intermediately glycosylated, and core protein of ATP1B3 was calculated as the band density of three forms of ATP1B3 divided by that of GAPDH (right). Values are mean ± S.D. (error bars) from three independent experiments. **, p < 0.01 (Student's t test). B, HBMECs with CASPR1 knockdown were immunostained with antibodies recognizing ATP1B3 (ab137055, red) and CASPR1 (ab34151, green). DAPI (blue) was used for counterstaining. Arrows, plasma membrane localization of ATP1B3. Images are representative of three independent experiments. Scale bar, 20 μm. C, HBMECs with CASPR1 knockdown were incubated with medium containing Sulf-NHS-LC-biotin for 30 min to label the plasma membrane protein, and then the cell lysates were immunoprecipitated (IP) with ATP1B3 antibody (H00000483-D01). The precipitated proteins were analyzed by Western blotting (WB) with ATP1B3 antibody (ab67409) as well as horseradish peroxidase–conjugated streptavidin to recognize the biotin-labeled proteins. The total cell lysates (TCL) were analyzed by Western blotting with ATP1B3 antibody (ab67409), with GAPDH as loading control (left). The percentage of plasma membrane ATP1B3 was calculated as the band density of biotin-labeled ATP1B3 divided by that of precipitated ATP1B3 (right). Values are mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). D and E, for rescue experiments, the HBMECs with CASPR1 knockdown were transfected with constructs encoding GFP-tagged CASPR1, with the empty vector (GFP alone) serving as control. The transfected cells were lysed and analyzed by Western blotting with ATP1B3, with GAPDH as loading control (D). As indicated, the transfected cells were analyzed by immunofluorescence with ATP1B3 antibody (ab137055, red) and GFP (green). DAPI (blue) was used for counterstaining (E). Images are representative of three independent experiments. Scale bar, 20 μm.
Plasma membrane trafficking of ATP1A1 depends on CASPR1
Previous studies showed that the newly synthesized β subunit of Na+/K+-ATPase is folded in the ER. The properly folded Na+/K+-ATPase β subunits can assemble with the α subunit forming α/β heterodimers, which will be trafficked to the plasma membrane (27, 28). The results above showed that CASPR1 knockdown reduced ATP1B3 glycosylation; thus, we hypothesized that CASPR1 may participate in the folding of ATP1B3 in ER, which will affect the trafficking of Na+/K+-ATPase, formed by α and β subunits, to the plasma membrane. To verify this, we analyzed the effect of CASPR1 knockdown on the distribution of the α1 subunit of Na+/K+-ATPase (ATP1A1), which was expressed at high levels in HBMECs (Fig. S2A). Immunofluorescence results revealed that the localization of ATP1A1 at the plasma membrane was reduced in CASPR1-silenced HBMECs (Fig. 4A, bottom). In contrast, ATP1A1 was primarily localized at the plasma membrane in the control cells (Fig. 4A, top). Consistently, biochemical subfractionation analysis revealed that the levels of ATP1A1 in the fraction of plasma membrane were significantly reduced in CASPR1-knockdown HBMECs compared with the control (Fig. 4B). The expression level and plasma membrane localization of ATP1A3, another member of the Na+/K+-ATPase α subunit, was not affected in CASPR1-silenced HBMECs (Fig. 4, C and D). Furthermore, we found that the full-length CASPR1 (GFP-CASPR1) effectively rescued the reduced ATP1A1 expression in the plasma membrane fraction (Fig. 4E) as well as the redistribution of cellular ATP1A1 in CASPR1 knockdown HBMECs (Fig. 4F), whereas the vector alone (GFP) had no effect. These data supported the idea that CASPR1 is involved in the folding of ATP1B3 in ER before its formation with the α subunit of Na+/K+-ATPase, which is essential for their proper trafficking to the plasma membrane. To exclude the possibility that CASPR1 may directly associate with ATP1A1, contributing to its membrane trafficking, we performed an immunoprecipitation assay to test their interaction. The negative binding of CASPR1 with ATP1A1 in HBMECs indicated by immunoprecipitation (Fig. 4G) demonstrated that CASPR1 specifically bound with the β subunit (ATP1B3) but not the α subunit (ATP1A1) of Na+/K+-ATPase. Thus, the redistribution of cellular ATP1A1 was caused by the attenuated interaction of CASPR1 with ATP1B3 in CASPR1-silenced HBMECs.
Figure 4.
CASPR1 is required for plasma membrane trafficking of ATP1A1 in HBMECs. A, HBMECs with CASPR1 knockdown were fixed and stained with antibodies recognizing ATP1A1 (ab7671, red), and CASPR1 (green). DAPI (blue) was used for counterstaining. Images are representative of three independent experiments. Scale bar, 20 μm. B, the cytosolic, plasma membrane (PM), and total proteins extracted from HBMECs with CASPR1 knockdown were analyzed by Western blotting with ATP1A1 (ab7671) and CASPR1 antibody. GAPDH served as loading control (left). The percentage of plasma membrane ATP1A1 was calculated as the band density of plasma membrane ATP1A1 divided by total ATP1A1 (right). Values are mean ± S.D. (error bars) from three independent experiments. **, p < 0.01 (Student's t test). C, HBMECs with CASPR1 knockdown were were lysed, and the cell lysates were subjected to Western blotting using ATP1A3 antibody, with nonsilencing siRNA as a control. D, HBMECs with CASPR1 knockdown were immunostained with antibodies recognizing ATP1A3 (green). DAPI (blue) was used for counterstaining. Scale bar, 20 μm. E and F, HBMECs with CASPR1 knockdown were transfected with constructs encoding GFP-tagged CASPR1 for rescue experiments, with the empty GFP vector as control. The cell lysates of transfected cells were analyzed by Western blotting with ATP1A1 antibody (ab7671), with GAPDH as loading control (E). The transfected cells were fixed, and immunofluorescence was performed with ATP1A1 antibody (red) and GFP (green) (F). DAPI (blue) was used for counterstaining. Images are representative of three independent experiments. Scale bar, 20 μm. G, HBMECs lysates were immunoprecipitated (IP) with CASPR1 and ATP1A1 (ab2872) antibody, respectively, and then the precipitated proteins were analyzed by Western blotting (WB) using antibodies against ATP1A1 (ab7671) and CASPR1 (ab133634).
CASPR1 regulates the activity of Na+/K+-ATPase in brain endothelial cells
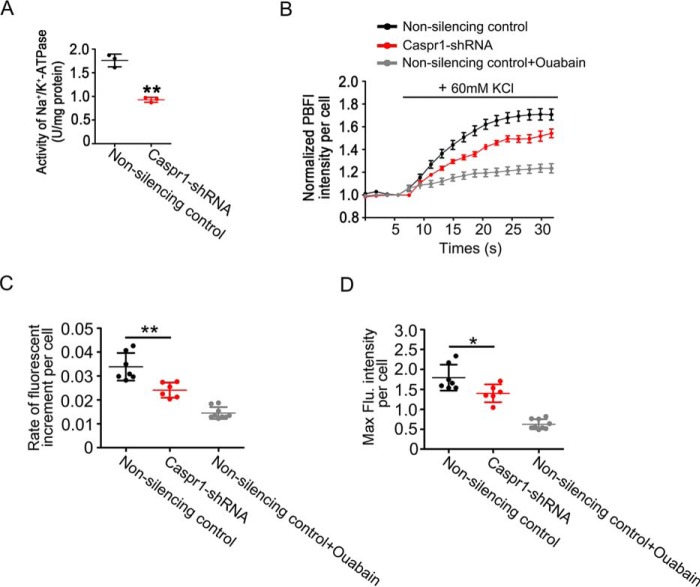
To identify whether CASPR1 could affect the activity of Na+/K+-ATPase in brain endothelial cells, we first measured the activity of Na+/K+-ATPase, determined as the amount of Pi released from ATP hydrolysis by Na+/K+-ATPase, in HBMECs with CASPR1 knockdown. As shown in Fig. 5A, the activity of Na+/K+-ATPase was significantly reduced in CASPR1-silenced HBMECs (0.93 ± 0.05 units/mg protein) compared with that in nonsilencing control (1.76 ± 0.13 units/mg protein).
Figure 5.
CASPR1 regulates the activity of Na+/K+-ATPase in HBMECs. A, the activity of Na+/K+-ATPase in HBMECs with CASPR1 knockdown was analyzed using an Na+/K+-ATPase assay kit as described under “Materials and methods.” Data are mean ± S.D. (error bars) from three independent experiments. **, p < 0.01 (Student's t test). B, HBMECs with CASPR1 knockdown were incubated with culture medium containing a 10 μm concentration of the K+-sensitive fluorophore PBFI at 37 °C for 30 min. Then the cells were mounted in an imaging chamber in the presence of K+ channel inhibitor TEA (20 mm), followed by perfusion with KCl (60 mm) solution. Measurement of PBFI fluorescence in living cells was performed using a two-photon microscope. When indicated, the cells were pretreated with the Na+/K+-ATPase inhibitor ouabain (positive control). Data are normalized to the fluorescence intensity of PBFI before the perfusion with KCl, which was defined as 1 (n = 3). C, the increase rate of PBFI intensity, indicating the rate of K+ uptake into cells, was calculated as the increment of PBFI intensity divided by the times (18.64 s) after administration of 60 mm KCl. Data are mean ± S.D. from three independent experiments. **, p < 0.01 (Student's t test). D, the peak fluorescence intensities of PBFI after perfusion with 60 mm KCl were quantified. Data are mean ± S.D. from three independent experiments. *, p < 0.05 (Student's t test).
By hydrolysis of ATP, the Na+/K+-ATPase can export three Na+ ions and import two K+ ions through the plasma membrane of the cells (29). Studies showed that K+ absorption under high concentration of extracellular K+ was predominantly dependent on K+ channels and Na+/K+-ATPase activity in astrocytes (30, 31). Thus, in the presence of K+ channel blocker (tetraethylammonium chloride, TEA), the alterations of intracellular K+ in response to the elevation of extracellular K+ could reflect the activity of the Na+/K+-ATPase. Then the intracellular K+ ions of HBMECs upon KCl (60 mm) perfusion in the presence of TEA were detected by the K+ indicator fluorophore potassium-binding benzofuran isophthalate acetoxymethyl ester (PBFI) by live-cell imaging under a two-photon microscope. We found that the increased rate of intracellular K+ in CASPR1 knockdown HBMECs was significantly reduced compared with nonsilencing control cells (Fig. 5, B and C). As a positive control, the application of inhibitor of Na+/K+-ATPase, ouabain, remarkably attenuated the increase rate of intracellular K+ (Fig. 5, B and C). Also, the maximum intensity of PBFI, indicating the highest intracellular concentration of K+ in response to the elevated extracellular K+, was lower in CASPR1-silenced HBMECs than that of controls (Fig. 5D). These results demonstrated that CASPR1 knockdown significantly attenuated the activity of Na+/K+-ATPase in brain endothelial cells.
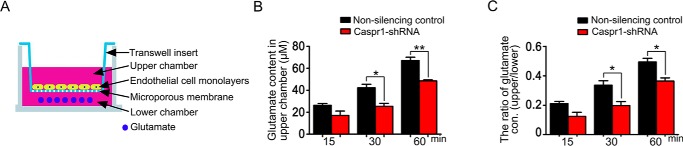
Knockdown of CASPR1 reduces the glutamate efflux through brain endothelial cell monolayers
Glutamate, the primary excitatory neurotransmitter in the brain, could be transported into astrocytes and neurons by glutamate transporters. Glutamate transporters are sodium-dependent proteins that depend on the sodium and potassium gradients generated principally by Na+/K+-ATPase (32, 33). Recently, the role of brain endothelial cells of the blood–brain barrier in the brain-to-blood glutamate efflux was reported. Our findings that CASPR1 regulates the activity of Na+/K+-ATPase prompt us to test whether CASPR1 is involved in the glutamate transport in brain endothelial cells. To establish the in vitro glutamate efflux model of BBB, the HBMECs were cultured on the Transwell inset until the formation of confluent monolayers. Then the glutamate was added to the lower chamber of Transwell, and the glutamate concentrations in the upper chamber were measured at the indicated times (Fig. 6A). The results showed that the glutamate concentrations in the upper chamber were in the range of 26.3–67.1 μm (Fig. 6B), which is remarkably lower than the glutamate concentration (200 μm) in the lower chamber. This indicated that the HBMEC monolayers formed at the Transwell insert were not permeable to the glutamate molecules; thus, the glutamate concentrations in the upper chamber represented the glutamate transportation from the lower chamber through the HBMEC monolayers. Interestingly, we found that the glutamate concentrations in the upper chamber were reduced by CASPR1 knockdown, showing statistical significance at 30 min after the addition of glutamate compared with that in nonsilencing controls (Fig. 6B), indicating that CASPR1 down-regulation reduced the glutamate transport through the HBMEC monolayers. When the concentration of glutamate in the lower chamber was reduced to 50 μm, we observed similar attenuated glutamate transportation through the in vitro BBB model caused by CASPR1 knockdown (Fig. S3). Consistently, the ratio of glutamate concentrations in the upper chamber divided by glutamate concentrations in the lower chamber were decreased by CASPR1 knockdown (Fig. 6C). These results demonstrated that CASPR1 is involved in the efflux of glutamate in brain endothelial cells of the blood–brain barrier.
Figure 6.
CASPR1 affects the glutamate efflux through brain endothelial cells. A, schematic representation of in vitro glutamate efflux assay with HBMECs. The HBMECs were cultured on the Transwell insert placed on a 24-well plate for 5 days until formation of endothelial barrier with tight junctions. Then the glutamate (200 μm) was added to the lower chamber of the Transwell, and the glutamate in the upper chamber of Transwell was measured at the indicated time points. B and C, the HBMECs with CASPR1 knockdown were cultured on the Transwell insert, and the in vitro glutamate efflux assay was performed, with nonsilencing shRNA as control. The glutamate concentrations in the upper chamber of the Transwell were measured at the indicated time points (B). The glutamate concentrations in the lower chamber of Transwell were measured, and the ratio of glutamate concentrations in the upper chamber divided by glutamate concentrations in the lower chamber was calculated (C). Data are mean ± S.D. (error bars) from three independent experiments. *, p < 0.05. **, p < 0.01 (one-way ANOVA, Student's t test).
Discussion
Our recent study identified the expression of CASPR1 in the brain endothelial cells (18). CASPR1, acting as a host receptor, is required for the penetration of pathogenic E. coli through the BBB during the development of meningitis (18). The function of CASPR1 in brain endothelial cells under physiological conditions remains unknown. In this study, we found that CASPR1 interacts with the core protein of β3 subunit of Na+/K+-ATPase, ATP1B3, to facilitate the glycosylation and maturation of ATP1B3 in brain endothelial cells. CASPR1-mediated ATP1B3 maturation is necessary for the trafficking of Na+/K+-ATPase to the plasma membrane. For the first time, we found CASPR1 is associated with the transportation of neurotransmitter glutamate in brain endothelial cell by regulating the activity of Na+/K+-ATPase.
Previous studies showed that the folding of the β subunits of Na+/K+-ATPase is completed in the ER, predominantly dependent on its interaction with the ER chaperones (27, 34). ER chaperones contained several family members including heat shock proteins (Hsp), calreticulin, calnexin, etc. The β1 subunit of Na+/K+-ATPase is associated with the ER chaperone calnexin for its glycosylation and maturation, whereas β2 subunit binds with the chaperone Bip, a member of the Hsp70 family (28, 35). So far, the ER chaperone for β3 subunit (ATP1B3) maturation remains unclear. Here, we found that only the core proteins of ATP1B3 could interact with CASPR1 primarily present in ER in HBMECs. Depletion of CASPR1 significantly reduced the glycosylation of ATP1B3. These findings suggested that CASPR1 participates in the maturation process of ATP1B3 in ER. In other words, the glycosylation of ATP1B3 occurred in the ER before its assembly with the α subunit of Na+/K+-ATPase. Our further results showed that CASPR1 did not interact with ATP1A1, the α subunit of Na+/K+-ATPase (Fig. 4G). Thus, the assembly of ATP1B3 with the α subunit ATP1A1 may take place after the dissociation of ATP1B3 from CASPR1. However, the lack of chaperone-like structural domain in CASPR1 excluded the role of CASPR1 as chaperone. We have performed MS to identify the proteins precipitated with CASPR1, and the results revealed the binding of CASPR1 with HSPA8 (Hsp70 member 8) (Table S1). Interaction of Hsp70 with Na+/K+-ATPase was reported in renal epithelial cells (36). Thus, we think that CASPR1 may act as a co-chaperone or adaptor of HSPA8 to facilitate the folding of nascent β3 subunits of Na+/K+-ATPase in ER. It will be interesting to characterize the formation of chaperone machinery complex constituted by CASPR1, HSPA8, and ATP1B3 in ER during the maturation of Na+/K+-ATPase in future studies.
We noticed that the core proteins of ATP1B3 were reduced when CASPR1 was knocked down in HBMECs (Fig. 3C). The mRNA level of ATP1B3 was not altered in CASPR1 knockdown HBMECs (Fig. S1A), suggesting that the reduced core proteins of ATP1B3 was not caused by transcriptional regulation. Previous studies showed that the properly folded Na+/K+-ATPase β subunits could assemble with the α subunit, which allows the export of α/β heterodimers from the ER to the plasma membrane (27, 28). The β subunit of Na+/K+-ATPase that is not correctly assembled with α subunit is retained in the ER and degraded rapidly (37). Thus, the reduced core proteins of ATP1B3 induced by CASPR1 knockdown were likely caused by the degradation of incompletely folded ATP1B3 due to less availability of CASPR1. Regarding the residual function of Na+/K+-ATPase following CASPR1 silencing (Fig. 5B), we think it could be due to the presence of remaining Na+/K+-ATPase composed of other α and β subunits rather than ATP1B3 and ATP1A1. As we described above, the glycosylation and membrane localization of ATP1B1 remained unchanged by CASPR1 knockdown. Furthermore, we found that the expression level and plasma membrane localization of ATP1A3, another member of the Na+/K+-ATPase α subunit, was not affected in CASPR1-silenced HBMECs (Fig. 4, C and D). Also, the expression of FXYD family members, which are thought to be key regulators of Na+/K+-ATPase (38), was also analyzed in HBMECs with CASPR1 knockdown. The results demonstrated that the FXYD5 mRNAs are expressed at relatively high levels in HBMECs, and CASPR1 silencing did not affect the expression and plasma membrane localization of FXYD5 in HBMECs (Fig. S2, B–D). Thus, the residual function of Na+/K+-ATPase following CASPR1 silencing may reflect the remaining function of Na+/K+-ATPase composed by ATP1B1 and ATP1A3.
Glutamate is the most important excitatory neurotransmitter in the brain. However, abnormally elevated glutamate concentration in the brain may cause excitotoxicity in certain disease states, such as epilepsy, ischemic stroke, multiple sclerosis, and Alzheimer's disease (39, 40). Glutamate transporters present in the neurons and astrocytes contribute to the maintenance of the concentration of glutamate at relatively low levels in the brain (41, 42). An alternative, equally important pathway for glutamate transport is the efflux of glutamate from the brain to the peripheral blood, during which brain endothelial cells play a vital role (43, 44). By establishment of an in vitro model to measure glutamate efflux of BBB, we found that down-regulation of CASPR1 reduced the efflux of glutamate through the BBB. In contrast, CASPR1 knockdown did not affect the glutamate metabolism in brain endothelial cells (Fig. S4). These findings revealed the novel function of CASPR1 in glutamate transport through the blood–brain barrier. Given the well-known expression of CASPR1 in neurons, whether CASPR1 is involved in the glutamate transport at synaptic termini remains an interesting issue to be explored.
In summary, our results demonstrated that CASPR1 interacts with the core proteins of ATP1B3 to promote maturation of ATP1B3 in brain endothelial cells. CASPR1-mediated ATP1B3 maturation is essential for the assembly of full Na+/K+-ATPase and its trafficking to the plasma membrane. CASPR1 can regulate the activity of Na+/K+-ATPase in brain endothelial cells to facilitate glutamate efflux in the blood–brain barrier.
Materials and methods
Cell culture
HBMECs were a generous gift from Dr. K. S. Kim (Johns Hopkins University, Baltimore, MD). The HBMECs were cultured in complete RPMI 1640 medium supplemented with 10% Nu-serum (BD Biosciences), 10% fetal bovine serum (Hyclone), 1 mm sodium pyruvate, 2 mm glutamine, 1× nonessential amino acid, and 1× minimum Eagle's medium vitamin. Human embryonic kidney 293T (HEK293T) cells were cultured in high glucose Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum.
Antibodies and inhibitors
Anti-CASPR1 (ab34151 and ab133634), anti-ATP1A1 (ab2872 and ab7671), anti-FXYD5 (ab190957), and anti-ATP1B3 (ab67409 and ab137055) were purchased from Abcam. Anti-ATP1B3 (H00000483-D01) was from Abnova. Anti-ATP1B1 (15192-1-AP), anti-His (60001-1-Ig), and anti-GFP (50430-2-AP) were from Protein Tech Group, Inc. Anti-GAPDH and anti-ATP1A3 (sc-365744) were from Santa Cruz Biotechnology, Inc. DAPI was from Roche Applied Science. Secondary antibodies used for immunofluorescence and Western blotting were from Jackson ImmunoResearch Laboratories. The glycosylation inhibitor tunicamycin (catalog no. 3516), voltage-gated K+ channels inhibitor tetraethylammonium (catalog no. 3068), and Na+/K+-ATPase inhibitor Ouabain (catalog no. 1076) were purchased from Tocris.
Yeast two-hybrid screening
The nucleotide sequence of CASPR1 mRNA (NM_003632) corresponding to CASPR1 cytoplasmic domain (1305–1384 aa) was cloned into the PGBKT7 as bait. The strain Y187 (Clontech) tansformed with bait plasmid was mated with yeast AH109 containing human fetal cDNA library cloned into pGAD. The yeast diploid was selected on SD/−Ade−His−Leu−Trp (−AHLT) medium containing 5-Bromo-4-chloro-3-indoxyl-alpha-d-galactopyranoside (X-α-Gal). The positive clones were obtained and identified by DNA sequencing. AH109 co-transformed with SV40 large T antigen and p53 was used as a positive control. AH109 co-transformed with SV40 large T antigen and lamin C served as a negative control.
Plasmid construction and transfection
The full-length cDNAs of human ATP1B1 and ATP1B2 obtained by RT-PCR from HBMECs were cloned into pcDNA3.1 myc/his B vector, respectively, for the TNT coupled transcription/translation system. The mutant ATP1B3 cDNA N124E and N240E was cloned into pcDNA3.1 myc/his B vector, respectively, for transfection of HEK293T. The nucleotide sequence corresponding to the cytoplasmic domain of CASPR1 (aa 1305–1384) amplified with PCR was inserted into pGEX4T-3 for fusion protein purification. The primers used for amplification and the cloning vectors are listed in Table S2. The constructs confirmed by DNA sequencing were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
RNA isolation and real-time quantitative RT-PCR analysis
The total RNA isolated with TRIzol reagent (Invitrogen) was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Promega). Real-time PCR was performed on an ABI 7500 real-time PCR system with a SYBR Selected Master Mix (Applied BioSystems) according to the manufacturer's instructions. The primers for ATP1B family members are listed in Table S3. The amplification conditions were as follows: 95 °C for 30 s and 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The comparative cycle threshold method was used to calculate the relative gene expression level, with GAPDH as the internal control. Real-time PCR products were verified by DNA sequencing.
Recombinant protein purification
The cytoplasmic domain of CASPR1 (listed in Table S2) was cloned into pGEX4T-3 and transformed into E. coli BL21 (DE3). The bacteria were grown to exponential phase (A260 = 0.5), and then isopropyl 1-thio-β-d-galactopyranoside was added to 0.5 mm for an additional 3 h. The cells were pelleted by centrifugation and resuspended in ice-cold PBS containing lysozyme (1 mg/ml). Cells were disrupted by sonication, and Triton X-100 was added to 1%, followed by incubation at 4 °C for 30 min. Insoluble materials were removed by centrifugation, and the supernatant was purified by affinity chromatography with GSH-Sepharose 4B.
GST pulldown
Equal amounts of GST and cytoplasmic domain of CASPR1 (aa 1305–1384) coupled with GST fusion proteins (GST-CASPR1-C) fusion proteins were immobilized with GSH-Sepharose 4B. Then ATP1B1 or ATP1B3 protein obtained from the TNT coupled transcription/translation system (Promega) was co-incubated with GSH-Sepharose 4B prebound with GST-IbeA overnight at 4 °C, with GST as control. The bound proteins were washed with binding buffer (20 mm Tris, 50 mm NaCl, 10% glycerol, 1% Nonidet P-40), and the samples were analyzed by Western blotting. Each experiment was repeated at least three times.
RNAi
Three siRNA sequences targeting human CASPR1 corresponding to the coding region, siRNA1 (TGAGCATGATGGACGCTGCTA, nucleotides 1647–1667), siRNA2 (CAGTTCCTTTGTTCGTGACTA, nucleotides 3300–3320), and siRNA3 (ACGGCTATGTGCAGCGCTTTA, nucleotides 849–869), were obtained from Shanghai Genepharma Corp. The nonsilencing siRNA sequence (TTCTCCGAACGTGTCACGT) was used as a control. The siRNA sequences were transfected into HBMECs using Lipofectamine 2000, and the expression of CASPR1 was examined by Western blot analysis. The siRNA effectively knocking down CASPR1 expression was synthesized and cloned into pRNA-U6.1/Neo (GenScript). The recombinant constructs with shRNA were transfected into HBMECs by Lipofectamine 2000. The stable HBMEC cell line with CASPR1 knocked down was selected with G418.
Western blotting
The experimental procedure was performed as described previously (18). Briefly, The cells were lysed with radioimmune precipitation assay buffer (Beyotime, Nantong, China) containing protease inhibitor mixture. The protein samples were separated by SDS-PAGE and then transferred to polyvinylidene difluoride membrane. The polyvinylidene difluoride membrane was blocked with 5% nonfat milk and incubated with the primary antibody (1:1000 dilution). Then the blots were incubated with a horseradish peroxidase–conjugated secondary antibody. Immunoreactive bands were visualized by Super Signal West Pico chemiluminescent substrate using an LAS-3000 mini imaging system. For quantification, the protein band intensities of the Western blotting images were quantified with ImageJ software. Data are represented as mean intensity of bands from three independent experiments.
Immunofluorescence
HBMECs cultured on coverslips were fixed with 4% paraformaldehyde and then permeabilized with 0.02% Triton X-100. Then the cells were blocked with 5% donkey serum and stained with the antibody CASPR1, ATP1B3, or ATP1A1 diluted at 1:100. Following incubation with secondary antibody conjugated with Alexa Fluor 488 and Alexa Fluor 594 (1:200 dilution; Invitrogen) and DAPI staining, the coverslips were mounted and analyzed under confocal laser-scanning microscopy (Zeiss LSM880).
Immunoprecipitation
Cells were washed with ice-cold PBS and lysed with Nonidet P-40 lysis buffer (Beyotime) containing protease inhibitors. The cell lysates were centrifuged, and the supernatant was collected. A total of 1 mg of protein was incubated with appropriate antibody (3 μg/ml) overnight at 4 °C and then incubated with protein A/G–agarose (Santa Cruz Biotechnology). The proteins precipitated from immune complexes were eluted in SDS sample buffer for Western blotting. Each experiment was repeated at least three times.
Plasma membrane protein assay
Cells were cultured until ∼80–90% confluence and then starved with RPMI 1640 medium overnight. Then cells were incubated with RPMI 1640 medium containing Sulfo-NHS-LC-biotin (0.5 mg/ml) for 30 min to label the cell plasma membrane proteins. The biotin-labeled cells were lysed with Nonidet P-40 lysis buffer containing protease inhibitors, and then the cell lysates were incubated with anti-ATP1B3 antibody. Immunoblotting was performed to analyze the total precipitated ATP1B3 with ATP1B3 antibody, and the plasma membrane ATP1B3 was analyzed using horseradish peroxidase–conjugated streptavidin.
Isolation of plasma membrane protein
107 cells were prepared to isolate plasma membrane proteins and cytosolic proteins according to the Plasma Membrane Protein Isolation Kit user manual (Invent Biotechnologies, Eden Prairie, MN). The total protein of cells was extracted following the Total Protein Extraction Kit user manual (Invent Biotechnologies).
Measurement of cytoplasmic K+ concentration
The cultured HBMECs were washed with salt solution (4.5 mm KCl, 125 mm NaCl, 5 mm CaCl2, 1 mm MgCl2) and then cultured with culture medium (10 mm glucose, 10 mm Hepes, pH 7.3) containing a 10 μm concentration of the K+-sensitive fluorophore PBFI and 0.05% Pluronic F-127 (Invitrogen) at 37 °C for 30 min. Then cells were washed and mounted in FCS3 chamber with 4.5 mm KCl salt solution in the presence of voltage-gated K+ channel inhibitor TEA (20 mm), followed by 60 mm KCl solution perfusion. Measurements of cytoplasmic K+ concentration with PBFI from regions of interest were performed using confocal laser-scanning microscopy (Zeiss LSM880) throughout the course of an experiment. When indicated, the cells were pretreated with the Na+/K+-ATPase inhibitor ouabain (1 mm).
Na+/K+-ATPase activity assay
107 HBMECs were suspended with physiological saline and lysed by the ultrasonic wave following the Na+/K+-ATPase assay kit instructions. Briefly, the protein concentrations of cell lysates were determined by the BCA method (Pierce). Then Na+/K+-ATP enzyme activity was analyzed using a SpectraMax M5 reader (Molecular Devices, Sunnyvale, CA).
Glutamate efflux assay
105 HBMECs were cultured on Transwell membrane for 5 days. The cells were replaced with HBSS and incubated at 37 °C for 10 min. Then 600 μl of fresh HBSS containing 50 μm l-glutamate were added to the lower chamber, and 100 μl of HBSS were added to the upper chamber. The l-glutamate concentration in the upper chamber was analyzed at the indicated time using a glutamate assay kit (ab138883) according to the user manual.
Statistical analysis
All values were analyzed by GraphPad Prism software. The quantitative variables are presented as mean ± S.D. Statistical significance between two groups was analyzed by Student's t test. One-way analysis of variance (ANOVA) was used to compare multiple groups. A p value of <0.05 was considered significant.
Author contributions
S.-H. Z. and D.-X. L performed most of the experiments. L. W., Y.-H. L., Y.-H. W., H. Z., Z.-K. S., W.-G. F., X.-X. Q., D.-S. S., B. L., and X.-N. H. performed some of the experiments. S.-H. Z. and D.-X. L. drafted the manuscript. Y.-H. C. and W.-D. Z. conceived the project and finalized the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Drs. Monique Stins and Kwang Sik Kim (Department of Pediatrics, John Hopkins University School of Medicine) for providing HBMECs.
This work was supported by National Natural Science Foundation of China Grants 31771258, 31171291, 31571057, 81171537, 81201255, 31670845, and 81671168; the National 1000 Plan Program; Innovation Team Program Foundation of the Ministry of Education of China Grants IRT13101 and IRT_17R107; the Program of Distinguished Professor of Liaoning Province; Foundation of Liaoning Province Grants LT2011011, LR2012025, LS201609, and LQNK201731; and Intramural Program of China Medical University Grant JQ20160002. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1–S3.
- BBB
- blood–brain barrier
- BMEC
- brain microvascular endothelial cell
- HBMEC
- human BMEC
- aa
- amino acids
- ER
- endoplasmic reticulum
- TEA
- tetraethylammonium chloride
- PBFI
- benzofuran isophthalate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DAPI
- 4′,6-diamidino-2-phenylindole
- HBSS
- Hank's balanced salt solution
- X-α-Gal
- 5-bromo-4-chloro-3-indoxyl-α-d-galactopyranoside.
References
- 1. Keaney J., and Campbell M. (2015) The dynamic blood-brain barrier. FEBS J. 282, 4067–4079 10.1111/febs.13412 [DOI] [PubMed] [Google Scholar]
- 2. Abbott N. J., Patabendige A. A., Dolman D. E., Yusof S. R., and Begley D. J. (2010) Structure and function of the blood-brain barrier. Neurobiol. Dis. 37, 13–25 10.1016/j.nbd.2009.07.030 [DOI] [PubMed] [Google Scholar]
- 3. Ben-Zvi A., Lacoste B., Kur E., Andreone B. J., Mayshar Y., Yan H., and Gu C. (2014) Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509, 507–511 10.1038/nature13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zlokovic B. V. (2008) The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 57, 178–201 10.1016/j.neuron.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 5. Rubin L. L., and Staddon J. M. (1999) The cell biology of the blood-brain barrier. Annu. Rev. Neurosci. 22, 11–28 10.1146/annurev.neuro.22.1.11 [DOI] [PubMed] [Google Scholar]
- 6. Obermeier B., Daneman R., and Ransohoff R. M. (2013) Development, maintenance and disruption of the blood-brain barrier. Nat. Med. 19, 1584–1596 10.1038/nm.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vorbrodt A. W. (1988) Ultrastructural cytochemistry of blood-brain barrier endothelia. Prog. Histochem. Cytochem. 18, 1–99 [DOI] [PubMed] [Google Scholar]
- 8. Hawkins R. A., O'Kane R. L., Simpson I. A., and Viña J. R. (2006) Structure of the blood-brain barrier and its role in the transport of amino acids. J. Nutr. 136, 218S–226S 10.1093/jn/136.1.218S [DOI] [PubMed] [Google Scholar]
- 9. Jefferies W. A., Brandon M. R., Hunt S. V., Williams A. F., Gatter K. C., and Mason D. Y. (1984) Transferrin receptor on endothelium of brain capillaries. Nature 312, 162–163 10.1038/312162a0 [DOI] [PubMed] [Google Scholar]
- 10. Méresse S., Delbart C., Fruchart J. C., and Cecchelli R. (1989) Low-density lipoprotein receptor on endothelium of brain capillaries. J. Neurochem. 53, 340–345 10.1111/j.1471-4159.1989.tb07340.x [DOI] [PubMed] [Google Scholar]
- 11. Einheber S., Zanazzi G., Ching W., Scherer S., Milner T. A., Peles E., and Salzer J. L. (1997) The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination. J. Cell Biol. 139, 1495–1506 10.1083/jcb.139.6.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peles E., Nativ M., Lustig M., Grumet M., Schilling J., Martinez R., Plowman G. D., and Schlessinger J. (1997) Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 16, 978–988 10.1093/emboj/16.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhat M. A., Rios J. C., Lu Y., Garcia-Fresco G. P., Ching W., St. Martin M., Li J., Einheber S., Chesler M., Rosenbluth J., Salzer J. L., and Bellen H. J. (2001) Axon-glia interactions and the domain organization of myelinated axons requires neurexin IV/Caspr/Paranodin. Neuron 30, 369–383 10.1016/S0896-6273(01)00294-X [DOI] [PubMed] [Google Scholar]
- 14. Rios J. C., Melendez-Vasquez C. V., Einheber S., Lustig M., Grumet M., Hemperly J., Peles E., and Salzer J. L. (2000) Contactin-associated protein (Caspr) and contactin form a complex that is targeted to the paranodal junctions during myelination. J. Neurosci. 20, 8354–8364 10.1523/JNEUROSCI.20-22-08354.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sherman D. L., Tait S., Melrose S., Johnson R., Zonta B., Court F. A., Macklin W. B., Meek S., Smith A. J., Cottrell D. F., and Brophy P. J. (2005) Neurofascins are required to establish axonal domains for saltatory conduction. Neuron 48, 737–742 10.1016/j.neuron.2005.10.019 [DOI] [PubMed] [Google Scholar]
- 16. Coman I., Aigrot M. S., Seilhean D., Reynolds R., Girault J. A., Zalc B., and Lubetzki C. (2006) Nodal, paranodal and juxtaparanodal axonal proteins during demyelination and remyelination in multiple sclerosis. Brain 129, 3186–3195 10.1093/brain/awl144 [DOI] [PubMed] [Google Scholar]
- 17. Wolswijk G., and Balesar R. (2003) Changes in the expression and localization of the paranodal protein Caspr on axons in chronic multiple sclerosis. Brain 126, 1638–1649 10.1093/brain/awg151 [DOI] [PubMed] [Google Scholar]
- 18. Zhao W. D., Liu D. X., Wei J. Y., Miao Z. W., Zhang K., Su Z. K., Zhang X. W., Li Q., Fang W. G., Qin X. X., Shang D. S., Li B., Li Q. C., Cao L., Kim K. S., and Chen Y. H. (2018) Caspr1 is a host receptor for meningitis-causing Escherichia coli. Nat. Commun. 9, 2296 10.1038/s41467-018-04637-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang L. N., Sun Y. J., Pan S., Li J. X., Qu Y. E., Li Y., Wang Y. L., and Gao Z. B. (2013) Na+-K+-ATPase, a potent neuroprotective modulator against Alzheimer disease. Fundam. Clin. Pharmacol. 27, 96–103 10.1111/fcp.12000 [DOI] [PubMed] [Google Scholar]
- 20. Blanco G., and Mercer R. W. (1998) Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 275, F633–F650 10.1152/ajprenal.1998.275.5.F633 [DOI] [PubMed] [Google Scholar]
- 21. Marks M. J., and Seeds N. W. (1978) A heterogeneous ouabain-ATPase interaction in mouse brain. Life Sci. 23, 2735–2744 10.1016/0024-3205(78)90654-9 [DOI] [PubMed] [Google Scholar]
- 22. Blanco G., DeTomaso A. W., Koster J., Xie Z. J., and Mercer R. W. (1994) The α-subunit of the Na,K-ATPase has catalytic activity independent of the β-subunit. J. Biol. Chem. 269, 23420–23425 [PubMed] [Google Scholar]
- 23. Kaplan J. H. (2002) Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 10.1146/annurev.biochem.71.102201.141218 [DOI] [PubMed] [Google Scholar]
- 24. Malik N., Canfield V. A., Beckers M. C., Gros P., and Levenson R. (1996) Identification of the mammalian Na,K-ATPase 3 subunit. J. Biol. Chem. 271, 22754–22758 10.1074/jbc.271.37.22754 [DOI] [PubMed] [Google Scholar]
- 25. Vagin O., Turdikulova S., and Sachs G. (2005) Recombinant addition of N-glycosylation sites to the basolateral Na,K-ATPase β1 subunit results in its clustering in caveolae and apical sorting in HGT-1 cells. J. Biol. Chem. 280, 43159–43167 10.1074/jbc.M508262200 [DOI] [PubMed] [Google Scholar]
- 26. Lian W. N., Wu T. W., Dao R. L., Chen Y. J., and Lin C. H. (2006) Deglycosylation of Na+/K+-ATPase causes the basolateral protein to undergo apical targeting in polarized hepatic cells. J. Cell Sci. 119, 11–22 10.1242/jcs.02706 [DOI] [PubMed] [Google Scholar]
- 27. Beggah A., Mathews P., Beguin P., and Geering K. (1996) Degradation and endoplasmic reticulum retention of unassembled α- and β-subunits of Na,K-ATPase correlate with interaction of BiP. J. Biol. Chem. 271, 20895–20902 10.1074/jbc.271.34.20895 [DOI] [PubMed] [Google Scholar]
- 28. Tokhtaeva E., Sachs G., and Vagin O. (2010) Diverse pathways for maturation of the Na,K-ATPase beta1 and β2 subunits in the endoplasmic reticulum of Madin-Darby canine kidney cells. J. Biol. Chem. 285, 39289–39302 10.1074/jbc.M110.172858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishitsuji H., Sugiyama R., Abe M., and Takaku H. (2016) ATP1B3 protein modulates the restriction of HIV-1 production and nuclear factor κ light chain enhancer of activated B cells (NF-κB) activation by BST-2. J. Biol. Chem. 291, 4754–4762 10.1074/jbc.M115.679357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chever O., Djukic B., McCarthy K. D., and Amzica F. (2010) Implication of Kir4.1 channel in excess potassium clearance: an in vivo study on anesthetized glial-conditional Kir4.1 knock-out mice. J. Neurosci. 30, 15769–15777 10.1523/JNEUROSCI.2078-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murakami S., and Kurachi Y. (2016) Mechanisms of astrocytic K+ clearance and swelling under high extracellular K+ concentrations. J. Physiol. Sci. 66, 127–142 10.1007/s12576-015-0404-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hutchison H. T., Eisenberg H. M., and Haber B. (1985) High-affinity transport of glutamate in rat brain microvessels. Exp. Neurol. 87, 260–269 10.1016/0014-4886(85)90216-X [DOI] [PubMed] [Google Scholar]
- 33. Rose E. M., Koo J. C., Antflick J. E., Ahmed S. M., Angers S., and Hampson D. R. (2009) Glutamate transporter coupling to Na,K-ATPase. J. Neurosci. 29, 8143–8155 10.1523/JNEUROSCI.1081-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beggah A. T., Jaunin P., and Geering K. (1997) Role of glycosylation and disulfide bond formation in the beta subunit in the folding and functional expression of Na,K-ATPase. J. Biol. Chem. 272, 10318–10326 10.1074/jbc.272.15.10318 [DOI] [PubMed] [Google Scholar]
- 35. Tokhtaeva E., Munson K., Sachs G., and Vagin O. (2010) N-glycan-dependent quality control of the Na,K-ATPase β(2) subunit. Biochemistry 49, 3116–3128 10.1021/bi100115a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Riordan M., Sreedharan R., Wang S., Thulin G., Mann A., Stankewich M., Van Why S., Kashgarian M., and Siegel N. J. (2005) HSP70 binding modulates detachment of Na-K-ATPase following energy deprivation in renal epithelial cells. Am. J. Physiol. Renal Physiol. 288, F1236–F1242 10.1152/ajprenal.00438.2004 [DOI] [PubMed] [Google Scholar]
- 37. Tokhtaeva E., Sachs G., and Vagin O. (2009) Assembly with the Na,K-ATPase α(1) subunit is required for export of β(1) and β(2) subunits from the endoplasmic reticulum. Biochemistry 48, 11421–11431 10.1021/bi901438z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Béguin P., Crambert G., Monnet-Tschudi F., Uldry M., Horisberger J. D., Garty H., and Geering K. (2002) FXYD7 is a brain-specific regulator of Na,K-ATPase α 1-β isozymes. EMBO J. 21, 3264–3273 10.1093/emboj/cdf330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gillessen T., Budd S. L., and Lipton S. A. (2002) Excitatory amino acid neurotoxicity. Adv. Exp. Med. Biol. 513, 3–40 [DOI] [PubMed] [Google Scholar]
- 40. Nakanishi S. (1992) Molecular diversity of glutamate receptors and implications for brain function. Science 258, 597–603 10.1126/science.1329206 [DOI] [PubMed] [Google Scholar]
- 41. Hunsberger H. C., Rudy C. C., Batten S. R., Gerhardt G. A., and Reed M. N. (2015) P301L tau expression affects glutamate release and clearance in the hippocampal trisynaptic pathway. J. Neurochem. 132, 169–182 10.1111/jnc.12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson J. G., O'Donnell J. C., Takano H., Coulter D. A., and Robinson M. B. (2014) Neuronal activity and glutamate uptake decrease mitochondrial mobility in astrocytes and position mitochondria near glutamate transporters. J. Neurosci. 34, 1613–1624 10.1523/JNEUROSCI.3510-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gottlieb M., Wang Y., and Teichberg V. I. (2003) Blood-mediated scavenging of cerebrospinal fluid glutamate. J. Neurochem. 87, 119–126 10.1046/j.1471-4159.2003.01972.x [DOI] [PubMed] [Google Scholar]
- 44. O'Kane R. L., Martínez-López I., DeJoseph M. R., Viña J. R., and Hawkins R. A. (1999) Na+-dependent glutamate transporters (EAAT1, EAAT2, and EAAT3) of the blood-brain barrier: a mechanism for glutamate removal. J. Biol. Chem. 274, 31891–31895 10.1074/jbc.274.45.31891 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.