Abstract
As anionic biopolymers, oligonucleotides can have biological functions independent from their roles as the medium for the storage and flow of genetic information. In this paper, we investigated the interaction between DNA and the pro-inflammatory cytokine tumor necrosis factor-α (TNFα). Although various forms of DNA bind to TNFα with low μm dissociation constants, the interaction stabilizes the trimeric form of TNFα and enhances its cytotoxic effect. Based on this mechanism, a photoswitchable TNFα (TNFα–2-nitroveratryloxycarbonyl) has been designed whose sensitivity to DNA-mediated up-regulation of TNFα activity can be tuned by light irradiation. The mechanism described in this study represents a general model to understand the involvement of nonspecific interactions among biomolecules in regulating their biological functions. Because the interaction is not DNA sequence–specific, the resulting effect should be considered for oligonucleotide-based therapeutics in general.
Keywords: tumor necrosis factor (TNF), DNA–protein interaction, circular dichroism (CD), allosteric regulation, protein–nucleic acid interaction, activity regulation, biolayer interferometry, homoquenching, nonspecific interaction, photo-switchable protein
Introduction
Biomolecular noncovalent interactions are involved in all biological processes, such as activation/inhibition of enzymes, or self-assembling and self-organization at different hierarchical levels. Although most studies aim to illustrate the structural basis for specific recognition, many biomolecules in high abundance (e.g. albumin and immunoglobulin in blood, actin and tubulin in cells, or collagen and glycosaminoglycan in extracellular matrix) could be involved in many different specific or promiscuous interactions relevant under pathophysiological conditions. Some important signaling pathways between two molecules of large concentration difference could be overlooked: only a very small fraction of the abundant molecule (e.g. molecule R of mm) is directly involved in the interaction with the molecule at low concentration (e.g. molecule A of pm). It is difficult to reveal the R–A complex and the regulatory effect of R on A. In principle, a unspecific interaction between biomolecules is more ubiquitous than a specific one. For example, positively charged sequences in extracellular matrix protein laminin have been shown to promiscuously bind to multiple growth factors (1). However, because it is very difficult to dissect them from each other, the biological functions of most unspecific interactions remain unknown.
DNA is ubiquitous, whereas extracellular DNA is secreted from various sources, including apoptotic cells, NETotic neutrophils, and bacterial biofilms. Fluctuations of DNA concentration in blood have been observed under many pathological conditions, including some autoimmune diseases (2, 3). DNA can have biological functions independent from their roles as the medium for the storage and flow of genetic information. For example, systemic lupus erythematosus, an autoimmune disease, is characterized by the expression of anti-DNA antibodies. These antibodies form immune complexes with DNA, stimulate inflammatory cytokine production, and incite inflammation and tissue damage (4). Given that DNA is an anionic polymer, its interaction with other biomolecules through electrostatic forces could provide a regulatory mechanism more direct than those associated with the immune complex or genetic information.
TNFα3 is a pro-inflammatory cytokine and is involved in most autoimmune diseases. It represents one of the most valuable therapeutic targets for various immune-mediated disorders (5, 6). It has been suggested that the soluble 51-kDa trimeric TNFα dissociates at concentration below the nanomolar range, thereby becoming inactive (7–9). TNFα in serum is in the low-to-high picomolar range depending on the pathophysiological state (10–12). Because the concentration affecting the ratio between monomeric and trimeric forms is too low for most biochemical methods, the correlation between the regulation of trimeric TNFα level and its pro-inflammatory effect remains elusive (13). To understand the regulation of TNFα activity beyond a simple function of its concentration could shed new light in its role in various autoimmune reactions (14, 15). Like many cytokines and morphogens, TNFα has a heparin-binding domain, because the positively charged residues can interact with the highly sulfated glycosaminoglycan (16). In this work, we investigated the interaction between TNFα and negatively charged biopolymers such as DNA and heparin, to illustrate the potential mechanism in regulating this cytokine activity.
Results
Binding of TNFα to anionic biopolymers
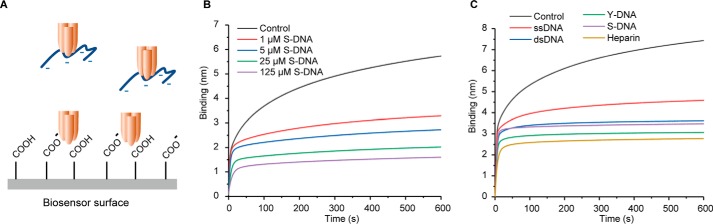
We first investigated whether TNFα could interact with negatively charged biopolymer using biolayer interferometry (BLI). As shown in Fig. 1, TNFα showed strong binding to a carboxylated surface. We then investigated whether DNA can inhibit the binding of TNFα to the negatively charged surface. We analyzed the effects of single-stranded DNA (ssDNA), dsDNA, ϒ-shaped DNA (ϒ-DNA), and salmon sperm DNA (S-DNA). Different forms of DNA (1 μm) reduced the binding of TNFα to the negatively charged surface. Heparin exhibited a similar effect. Interestingly, among the four different forms of DNA, ϒ-DNA has shown the strongest effect, although it is approximately three times smaller than the average size of S-DNA (Table S1). The TNFα concentration used in these experiments (1 μm) is much higher than the dissociation constant of the homotrimer (17). The competition experiment indicates that the trimeric TNFα is in slight favor of binding to ϒ-DNA, probably because their interaction is geometrically more favorable. More remarkable effects were observed when high concentrations of DNA were used (Fig. 1C).
Figure 1.
TNFα binding to anionic biopolymers DNA and heparin. A, inhibition of the binding of TNFα to a negatively charged biosensor surface by DNA and heparin. B and C, binding kinetics of the mixture of TNFα (1 μm) with various concentration of S-DNA (B) and several DNA structures or heparin at 1 μm (C) to a negatively charged sensor.
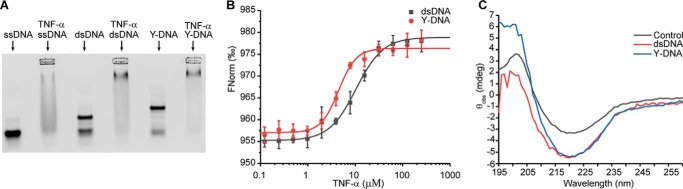
To directly probe the interaction between DNA and TNFα, we performed band-shift assays by mixing fluorescently labeled ssDNA, dsDNA, or Y-DNA with TNFα. At a high concentration of TNFα (50 μm), the bands of DNA shifted to smears (Fig. 2A). Moreover, residue amounts of DNA could be observed in the wells in a concentration-dependent manner, which was caused by noncovalent assembly between the trimeric protein and DNA. Then we titrated DNA with different concentrations of TNFα. Although the titration experiments indicated that the Kd values between TNFα and various forms of DNA are in the low micromolar range, as expected, the ϒ-DNA showed higher affinity to TNFα than ssDNA and dsDNA (Fig. S3). Band-shift assays using DNA staining gave similar results. Interestingly, fluorescently labeled TNFα was not able to affect the mobility of DNA (Fig. S4). Labeling TNFα using an amine-reactive dye reduced the protein surface charge, thus affecting its interaction with negatively charged DNA. This principle will be explored later to generate a modified TNFα, whose responsiveness to DNA can be switched by light. Finally, we used microscale thermophoresis (MTS) to determine the binding constants between TNFα and different forms of DNA. As shown in Fig. 2B, TNFα binds to dsDNA and Y-DNA with Kd values of 10.7 ± 2.0 and 4.9 ± 1.1 μm, respectively.
Figure 2.
A, band-shift assay of 1 μm Cy5–DNA incubated with 50 μm TNFα. B, DNA binding to TNFα measured by MTS, where the concentrations of Cy5–dsDNA and Cy5–Y-DNA are 10 nm, and TNFα concentration ranged from 122.5 nm to 250 μm. C, CD spectra of TNFα (10 μm) in the presence or absence of DNA (10 μm) after subtracting the DNA spectra.
The interaction between DNA and TNFα is not stoichiometric 1:1, because both the trimeric protein and various forms of DNA can form multiple interactions, e.g. to assemble into aggregates (Fig. 2A). Nevertheless, all these binding assays have indicated their binding constants in the low micromolar range, coincident with the range of fluctuation of DNA in blood (18–20).
Conformational change of TNFα upon DNA binding
We applied CD spectroscopy to study whether the interaction with DNA could cause structural change in TNFα. The CD spectra of TNFα is stable over time (Fig. S5A). As shown on Fig. 2C, incubation of dsDNA or Y-DNA with TNFα caused a remarkable increase of ellipticity at 217 nm. It is important to note that the DNAs have very weak signal at 217 nm (Fig. S5B). Therefore, the changes could be assigned to the conformational switch in the protein, indicating an increased content of β-sheet structure.
DNA enhances TNFα activity
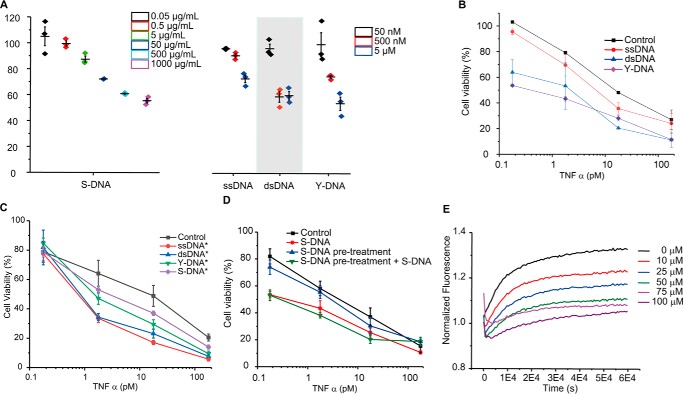
We then investigated whether binding to DNA could affect TNFα activity. Treating mouse fibrosarcoma L929 cells with TNFα and a low dose of cytotoxic agent represents a well-established model to test the biological activity of TNFα (21). Co-treatment of cells using TNFα with cycloheximide (CHX, 8 μm) led to concentration-dependent decrease of cell viability (Fig. S7A). The presence of DNA increased the sensitivity to TNFα treatment in a concentration-dependent manner (Fig. 3A). The TNFα concentration-dependent cytotoxic effects were measured in the presence of 5 μm of DNA of different forms (Fig. 3B). The effect is not caused by the interaction between DNA and Toll-like receptors, because randomly generated sequences as well as designed sequences excluding any potential CpG motif can up-regulate the TNFα activity (Fig. 3C). To exclude the possibility that the enhanced cytotoxicity is caused by an TNFα-independent effect of DNA, the cells were pretreated with DNA followed by removing the DNA-containing supernatant. TNFα treatment of cells after removing DNA did not exhibit the effect of TNFα and DNA co-treatment (Fig. 3D). To exclude the possibility that the effect is caused by the interaction between DNA and the protein His tag, we performed the experiment with an untagged TNFα. The untagged and His-tagged TNFα have shown similar response to 5 μm dsDNA (Fig. S6). Treating cells using CHX and DNA or heparin also did not affect cell viability (Fig. S7B). Thus, the interaction between TNFα and DNA is important for the enhanced cytotoxic effect. Like CHX, a low dose of actinomycin D (ACT-D, 1.6 μm) also induced TNFα-associated cytotoxicity (Fig. S7A). However, it is known that DNA interacts with ACT-D, thus diminishing the cytotoxicity of the drug (22). As shown in Fig. S7B, adding DNA to ACT-D and TNFα co-treated cells did not reduce cell viability. Instead, it rescued the cells. Because ACT-D is a guanine-specific DNA intercalator and does not interact with heparin, a remarkably enhanced cytotoxic effect was observed when heparin is used. All these results indicate that the up-regulation of TNFα effect is caused by the interaction between the cytokine and a negatively charged polymer.
Figure 3.
A, TNFα activity was enhanced by the addition of DNA in a concentration-dependent manner. Cell viability was measured in the presence of TNFα (0.1 ng/ml) and S-DNA, ssDNA, dsDNA, or Y-DNA of different concentrations. B and C, cell viability was measured in the presence or absence of DNA (5 μm) and TNFα of different concentrations. The sequences of DNA are either random (B) or without potential CpG motif (C). D, TNFα was incubated with 5 μm S-DNA for 24 h before adding to the cells (labeled as DNA), or TNFα was added to the cells after a 2-h preincubation with and removal of 5 μm S-DNA (as DNA pretreatment), or TNFα and S-DNA were added to the cells after a 2-h preincubation with and removal of 5 μm S-DNA (as DNA pretreatment + S-DNA). E, homoquenched fluorescence release upon adding 2 μm TNFα to 300 nm TNFα-fluorescein in the presence of different concentrations of S-DNA.
DNA binding provides a mechanism to fine-tune its activity. We found that changing DNA concentration from high nanomolar to low micromolar can regulate cytokine activity, which is, interestingly, coincident with their binding constants. As discussed later, as well as in a simulation experiment, the Kd value limits that range of DNA concentration fluctuation where the cytokine activity can be regulated.
DNA binding affects TNFα homotrimer dissociation
To investigate whether the binding of DNA can influence the dynamics of TNFα homotrimer and thus affect its dissociation, we performed a homoquenching assay (Fig. 3E). The fluorescently labeled TNFα possesses low fluorescence signal because of the closely assembled homotrimeric structure, whereas dissociated monomer can be incorporated into unlabeled TNFα, resulting in enhanced fluorescent signal (23, 24). To avoid affecting the electrostatic interaction with DNA through positively charged residues, TNFα containing free cysteine residues was used for labeling. In the absence of unlabeled TNFα, the time course of labeled TNFα is stable over time. When unlabeled TNFα was added in large excess, a gradual increase of fluorescent signal was observed. Interestingly, adding S-DNA decelerated the process (Fig. 3E), indicating that binding to DNA stabilizes the homotrimer. S-DNA inhibited the dissociation of TNFα in a concentration-dependent manner. Heparin showed a similar effect stabilizing the TNFα trimer (Fig. S8). Without adding unlabeled TNFα in large excess, neither S-DNA nor heparin affects the signal of labeled TNFα. It is important to note that the effective concentration to inhibit TNFα dissociation is in the same range as the Kd value determined previously by MTS and band-shift assays.
Light switch of DNA-regulated TNFα activity
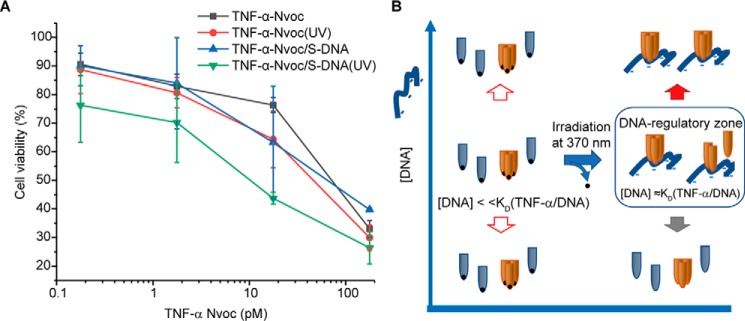
To design a light-switchable system, which can be used to regulate TNFα activity based on its interaction with negatively charged polymer such as DNA, will not only provide further evidence for the regulatory mechanism, but also help develop therapeutics that can be fine-tuned at disease sites by light. We modified TNFα with a photocleavable 2-nitroveratryloxycarbonyl (Nvoc) group. Although neutralizing the positively charged amino acids could reduce its binding to DNA, photolysis of the Nvoc group is expected to resume the interaction. Like TNFα, in combination with a low dose of CHX, Nvoc modified light-switchable TNFα (TNFα–Nvoc) and its photolysis product have shown similar concentration-dependent cytotoxic effects (Fig. 4A). Adding 5 μm S-DNA to TNFα–Nvoc caused a minor decrease of cell viability. In contrast, a short irradiation of the sample using light of 370 nm (near UVA1) enhanced the TNFα cytotoxicity remarkably, whereas irradiation itself showed no effect on the cell viability. Photolysis of Nvoc switches TNFα–Nvoc from the DNA insensitive state (Kd ≫ [DNA]) to the DNA-sensitive state (Kd ≈ [DNA]).
Figure 4.
A, influence on cell viability of TNFα–Nvoc with and without UV exposure in the presence or absence of 5 μm S-DNA. B, photolysis of Nvoc switches TNFα–Nvoc from the DNA-insensitive state to the DNA-sensitive state.
Discussion
Biomolecular interaction is fundamental to all biological processes. Although potent and specific molecular recognition has been considered as the biochemical basis for myriads of well-orchestrated intra-cellular and intercellular events, they have also been the subject of most structural biology studies. In recent years, weak and specific interactions have also been found to underlie numerous cellular processes, including the positive selection in the thymus through the weak interaction between the T-cell receptors and self-peptide and major histocompatibility complex proteins (25–27). In contrast, weak and promiscuous binding is thought to be less relevant, because they seem not to be able to affect a biochemical process in a specific manner. In this paper, we suggest that specificity can rise through another type of mechanism, although the molecular recognition is weak and nonspecific.
Interaction between two molecules is dependent not only on the dissociation constant, but also on their concentrations. The concentrations of various biomolecules in blood cover a broad range of multiple amplitudes, from high millimolar to low concentrations (e.g. low femtomolar) at the limit of detection. Considering the following scenario: A is a molecule whose activity must be subtly regulated under physiological conditions, e.g. a cytokine of low concentration. R is a highly abundant molecule that can interact with A and affect its activity. Thus, the activity of A could be influenced by its own concentration ([A]), as well as the concentration of R ([R]) (Figs. 5, A–C). If the [R] is constantly much higher than the dissociation constant between R and A (Kd), the presence of R becomes a pseudo-intrinsic feature of the biological activity of A. If the [R] is constantly much lower than Kd, it will have no effect on the activity of A. However, if [R] is in the range of the Kd, fluctuation in [R] at certain pathophysiological condition can cause a change in the biological function of A.
| (Eq. 1) |
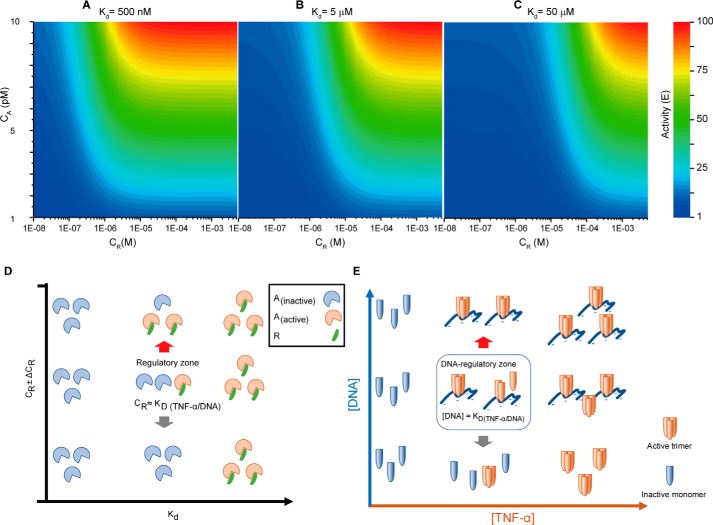
where [R] and [A] are the concentrations of R and A, respectively; [R] ≫ [A]; Kd is the dissociation constant between R and A; and γ is a value ≥ 0. When binding of R to A has an activation effect, γ > 1. When binding of R to A has an inhibitory effect, γ < 1. E is the biological effect of A. The regulation of biological effect of A by varying the concentrations of A and R is simulated with three different Kd values of 500 nm, 5 μm, and 50 μm, assuming binding of R to A having an activation effect with γ equal to 10. As shown in Fig. 5, dependent on the Kd value, fluctuation of R concentration within a certain range can influence the biological effect of A.
Figure 5.
Regulation of TNFα activity by DNA. A–C, the regulation of biological effect of A by varying the concentrations of A (CA) and R (CR) is simulated with three different Kd values of 500 nm (A), 5 μm (B), and 50 μm (C), assuming binding of R to A having an activation effect with γ equal to 10. D, dependent on the Kd value, fluctuation of R concentration within a certain range can influence the biological effect of A. E, fluctuation of DNA concentration can influence the biological effect of TNFα through stabilizing the trimer.
The potential to regulate cytokine A by altering the concentration of R is not related to the binding specificity, but the relationship between Kd and [R] (Fig. 5D). We demonstrated this type of regulatory mechanism using the nonspecific interaction between TNFα and DNA (Fig. 5E). Because circulating DNA level in blood can alter drastically in the range of low ng/ml to μg/ml and is related to various pathophysiological conditions (12, 18–20), it could have profound influence on the biological effects of TNFα. More generally, any TNFα-binding anionic biopolymer can cause similar effect, if its concentration fluctuates in the Kd range. Even more generally, this could be applied to any soluble factors of low concentration that interact with anionic biopolymers, whereas many of them have been classified as heparin-binding morphogens. The cytotoxic effect of TNFα involves necrosis, whereas apoptosis has been suggested as a secondary mechanism in addition to necrosis (21, 28, 29). Upon necrosis, DNA fragments released by cells will interact with TNFα and cause an additional effect (as positive feedback). The maximal amount of DNA released by cells in our experiment (30,000 cells/well) is ∼1 μg/ml. This is in the concentration range at which genomic DNA starts to exhibit an effect (Fig. 3A), whereas a stronger effect was observed at much higher concentrations (e.g. 500 and 1500 μg/ml). To which extent the positive feedback associated with necrosis contributes to TNFα activity under physiological conditions remains to be investigated in the future.
The long chain of DNA or heparin can interact with more than one TNFα monomer through electrostatic interactions. The multivalent interaction can thus further stabilize the trimeric protein. Our group has previously established a method called DNA-encoded dynamic combinatorial chemical libraries. In the DNA-encoded dynamic combinatorial chemical libraries setup, two DNA strands interact with each other through only 6 or 8 bp; thus, the resulting duplex is not stable and undergoes constant reshuffling. When both DNA strands are modified with chemical groups and both can bind to a protein, the multivalent interaction can stabilize the duplex and reduce the reshuffling rate of this dynamic system (Fig. S9). In this work, we have described a similar system, in which the dynamic component is the trimeric protein TNFα. A long anionic polymer chain can interact with TNFα monomers and stabilize the trimer through multivalent interaction.
DNA is the medium to encode genetic information, whereas recent evidence has suggested that other forms of DNA, e.g. the I-motif structures (30), can also be present in the cells and associated with key regulatory roles in the genome. Moreover, DNA has found many new applications, such as aptamer technology (31), encoding chemical structures of combinatorial libraries (32), self-assembled nanostructures (33), computation (34), and data storage (35). However, as an anionic biopolymer, DNA could also have a biological role independent from the nucleobases. The enhanced biological activity of TNFα through interacting DNA can be caused by different mechanisms. We have shown that binding to DNA can induce a conformational change of TNFα, whereas its direct impact on TNFα activity remains to be investigated. Another possible mechanism is that DNA binding can affect the dynamics of the TNFα homotrimer. The Kd of TNFα homotrimer is ∼100 pm, whereas its serum concentration is also in the picomolar range (17, 36). Thus, the concentration is too low for directly measuring the dissociation in solution. By using a homoquenching assay, we demonstrated that the presence of DNA decelerates the dissociation of TNFα, indicating that DNA can stabilize the homotrimer, enhancing the ratio of homotrimer to the inactive monomer. The photoswitching experiment shows that the biochemical interaction can be used to design a tuneable cytokine responsive to external stimuli (Fig. 4A). Some severe autoimmune diseases, such as systemic lupus erythematosus, are associated with subtle immune system defects, causing chronic inflammatory responses. In the future, DNA-mediated up-regulation of TNFα activity will be investigated in more biomedically relevant contexts, especially in autoimmune diseases. Moreover, to identify inhibitors to block the interaction would present a novel avenue for immunosuppressive treatment. Oligonucleotide-based therapeutics have become increasingly interesting, including gene-silencing (37), RNA/DNA aptamer (38), and DNA origami (39). Because the interaction with TNFα described in this study is not DNA sequence-specific, the resulting effect should be considered for oligonucleotide-based therapeutics in general. Because oligonucleotides in various forms possess very different half-lives in blood, it is not easy to draw a simple conclusion to set a dosage limit. It remains to be studied in biomedically relevant models.
Experimental procedures
Cell culture
The L929 cell line from mouse (C3H/An connective tissue) was kindly provided by the Garbe group (Center for Regenerative Therapies, Dresden, Germany). L929 were cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Biochrom) and 2 mm l-glutamine (Life Technologies) in polystyrene culture flasks (Greiner Bio-One). For subculturing or prior counting, L929 cells were treated with trypsin-EDTA (Life Technologies) to detach from the surface. L929 cells in suspension were spun down at 1000 rpm for 3 min at room temperature. The pellet was resuspended and transferred into new culture flasks. For the experiments, the cells were seeded in 96-well plates at 30,000 cell/well, in 100 μl of completed medium for 24 h before the incubation with TNFα.
TNFα expression and purification
Human recombinant His-tagged TNFα was expressed in BL21 (DE3). Single colonies of Escherichia coli were picked and inoculated into 20 ml of LB medium containing kanamycin (50 μg/ml). The cultures were grown overnight at 37 °C with vigorous shaking. Subsequently, 1 liter of prewarmed medium (2 × YT, 50 μg/ml kanamycin) was inoculated with 15 ml of overnight cultures and again incubated at 37 °C, with vigorous shaking until A600 was 0.8–1. Expression was induced by isopropyl β-d-thiogalactopyranoside (final concentration, 300 μm) and again incubating overnight at 25 °C, with 200 rpm shaking. The cells were harvested by centrifugation at 8000 rpm for 20 min, and the supernatant was discarded.
Protein purification: cells were resuspended in 50 ml of buffer A (35 mm HEPES, 500 mm NaCl, 40 mm imidazole, pH 7.8) with 1 mm DTT and 1 mm phenylmethylsulfonyl fluoride. The cells were lysated by passing three times through the French press (EmulsiFlex-C3, AVESTIN) at 4 °C under 1000 p.s.i. The lysate obtained was centrifuged at 45,000 rpm at 4 °C for 1 h (Beckmann LE-80K ultracentrifuge; Beckmann, Palo Alto, CA; rotor SW 45Ti). The supernatant was collected and loaded on a Histrap HP column at 4 ml/min, followed by linear gradient change up to 100% Buffer B (35 mm HEPES, 500 mm NaCl, 500 mm imidazole, pH 7.8) for 30 min. The eluted fractions were characterized by MS. High purity ones were pooled, concentrated, and stored at −80 °C. The full sequence is on Table S2, the mass spectrum on Fig. S1, and the UV spectrum on Fig. S2.
TNFα modification
TNFα-fluorescein was obtained by labeling His-tagged TNFα with maleimide-fluorescein in 1× PBS, pH 7.5, overnight at 4 °C. The final product was then purified using a PD10 desalting column (GE Healthcare). TNFα–Nvoc was made by reacting each His-tagged TNFα monomer with 5 or 25 eq of Nvoc-Cl in 1× PBS, pH 7.5, in a glass vial with a stirrer, for 2 h on ice, protected from light. Then the reaction was quenched with 0.5 mm Tris, pH 9, for 20 min, and the product was purified using a PD10 desalting column. The concentration of Nvoc that reacted to TNFα was 190.8 μm (determined via A350, using an extinction coefficient of 5485 m−1 cm−1) (40). Nvoc also has absorbance at 280 nm (A280), and the value is 0.52 times A350. The extinction coefficient of TNFα monomer is 20,820 m−1 cm−1. The protein monomer molar concentration of TNFα–Nvoc was 23.9 μm [(A280 −0.52 × A350)/20,820 m]. For each monomer, eight Nvoc groups reacted. For the cell assay, the TNFα–Nvoc control was kept away from light exposure, and the photoswitching was performed by irradiating TNFα–Nvoc for 30 min with a 366-nm UV lamp.
Oligonucleotides
The oligonucleotide sequences were based on a previously published Y-structure (41) or were designed in house (longer dsDNA). The oligonucleotides were diluted in an annealing buffer solution (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA) and heated for 5 min at 95 °C. Then they were slowly cooled to room temperature.
Binding assay by BLI
For the binding assay, in a BLI platform (Octet Red 384; ForteBio), COOH-functionalized (amine reactive second generation) sensors were used as: baseline for 50 s, loading for 600 s, dissociation for 50 s, and regeneration for 300 s. The tested regeneration buffers were 150 mm NaOH, and 10 mm glycine with 2 m NaCl, pH 1.5. The final working buffer was 1× PBS with 0.05% Tween 20. Two sensors were treated as controls, incubated only with TNFα, and two other sensors were treated as test sensors, incubated with TNFα plus DNA or heparin. The result was shown in Fig. 1.
DNA band-shift assays
The DNA constructs were incubated with TNFα for 1 h, at 4 °C on a shaker and in Tris buffer (100 mm NaCl, 20 mm Tris-HCl, pH 7.6, 2 mm MgCl2, 5 mm KCl, 1 mm CaCl2, 0.02% Tween 20). The incubation product was resolved with an electrophoresis run on 4% agarose gels, which were run for 2.5 h at 80 V and 4 °C and with cold 1× TBE buffer. The Cy5 or fluorescein signals were monitored on a Stella 3200, Xstella 1.00 imaging, and its analysis system (Raytest). The results are shown in Fig. 2A and Figs. S3 and S4.
Binding assay by MST
After a short incubation of 122.5 nm to 250 μm TNFα with 10 nm Cy5–DNA in 1× PBS with 0.05% Tween 20, the samples were loaded onto MST standard glass capillaries. Then the analysis was done on the red channel, with MST power 20%, and excitation power 1%, in a Monolith NT. Automated (Nanotemper). The result was shown in Fig. 2B.
CD spectroscopy
CD spectroscopy measurements were performed in a Chirascan plus (Applied Photophysics) at 25 °C, after incubation of DNA with TNFα in 10 mm sodium phosphate buffer, pH 7.4, for 60 min. The data were collected in the interval from 190 to 260 nm, and the final spectra were averages of five scans. The DNA spectra were collected alone. Then a highly concentrated TNFα solution was added directly to the 1-mm cuvette, followed by vigorous shaking. TNFα and DNA interaction was monitored at 0, 30, and 60 min. The results are shown in Fig. 2B and Fig. S5.
TNFα cytotoxicity
L929 cells were seeded in 96-wells plates at 30,000 cell/well, in 100 μl of complete medium for 24 h before the incubation with TNFα. Varying concentrations of TNFα were preincubated for 30 min at room temperature with several testing structures (e.g. DNA, heparin) in media containing 8 μm CHX or 1.6 μm ACT-D. After, all media were removed, and 90 μl of each test condition was distributed through the wells. Then the cells were incubated for 24 h at 37 °C and 5% CO2. On the next day, a solution of MTT in 1× PBS was added to each well to a final concentration of 0.5 mg/ml and incubated for another 90 min at 37 °C, 5% CO2. Finally, all medium was removed, and 1 V DMSO was used to solubilize the formazan crystals. Their absorbance was read at 540 nm. Cell viability (CV) was calculated as CV (%) = [(X − B)/(C − B)] × 100, where B is the blank (wells only with medium), C is the control (wells with medium plus CHX), and X is the absorbance from the test wells. The results were shown in Figs. 3 and 4 and Fig. S7.
Homoquenched fluorescence assay
The incubation of 2 μm TNFα with 300 nm TNFα-fluorescein and varying concentrations of S-DNA was done during 16 h in a 384-well plate with a clear bottom, nonbinding, and black walls (Greiner Bio-One). Each well had 210 μl of each condition tested, and the working buffer was PBS Superblock (Thermo Fisher Scientific). The fluorescein signal was monitored and registered as an average of three replicates on a Paradigm plate reader (Beckman Coulter). The results are shown in Fig. 3E and Fig. S8.
Statistical analysis
The number of biological and/or technical replicates for each experiment is stated in the figure legends.
Author contributions
H. A., W. L., and Y. Z. conceptualization; H. A., W. L., and Y. Z. data curation; H. A., W. L., and Y. Z. formal analysis; H. A., W. L., and Y. Z. validation; H. A., W. L., and Y. Z. investigation; H. A., W. L., and Y. Z. methodology; H. A. and Y. Z. writing-original draft; H. A. and Y. Z. project administration; H. A., W. L., and Y. Z. writing-review and editing; W. L. and Y. Z. software; W. L. and Y. Z. supervision; W. L. and Y. Z. visualization; Y. Z. resources; Y. Z. funding acquisition.
Supplementary Material
Acknowledgment
The L929 cell line from mouse (C3H/An connective tissue) was kindly provided by the Garbe group (Center for Regenerative Therapies, Dresden, Germany).
This work was supported by Bundesministerium für Bildung und Forschung Grants 03Z2EN12 and 03Z2E511. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2 and Figs. S1–S9.
- TNF
- tumor necrosis factor
- ACT-D
- actinomycin D
- BLI
- biolayer interferometry
- CHX
- cycloheximide
- MTS
- microscale thermophoresis
- Nvoc
- 2-nitroveratryloxycarbonyl
- S-DNA
- salmon sperm DNA
- ssDNA
- single-stranded DNA
- Y-DNA
- ϒ-shaped DNA.
References
- 1. Ishihara J., Ishihara A., Fukunaga K., Sasaki K., White M. J. V., Briquez P. S., and Hubbell J. A. (2018) Laminin heparin-binding peptides bind to several growth factors and enhance diabetic wound healing. Nat. Commun. 9, 2163 10.1038/s41467-018-04525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagata S., Hanayama R., and Kawane K. (2010) Autoimmunity and the clearance of dead cells. Cell 140, 619–630 10.1016/j.cell.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 3. Haugbro K., Nossent J. C., Winkler T., Figenschau Y., and Rekvig O. P. (2004) Anti-dsDNA antibodies and disease classification in antinuclear antibody positive patients: the role of analytical diversity. Ann. Rheum. Dis. 63, 386–394 10.1136/ard.2003.016303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su K.-Y., and Pisetsky D. S. (2009) The role of extracellular DNA in autoimmunity in SLE. Scand. J. Immunol. 70, 175–183 10.1111/j.1365-3083.2009.02300.x [DOI] [PubMed] [Google Scholar]
- 5. Li P., Zheng Y., and Chen X. (2017) Drugs for autoimmune inflammatory diseases: from small molecule compounds to anti-TNF biologics. Front. Pharmacol. 8, 460 10.3389/fphar.2017.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palladino M. A., Bahjat F. R., Theodorakis E. A., and Moldawer L. L. (2003) Anti-TNF-α therapies: the next generation. Nat. Rev. Drug Discov. 2, 736–746 10.1038/nrd1175 [DOI] [PubMed] [Google Scholar]
- 7. Idriss H. T., and Naismith J. H. (2000) TNF-α and the TNF receptor superfamily: structure–function relationship(s). Microsc. Res. Tech. 50, 184–195 [DOI] [PubMed] [Google Scholar]
- 8. Eck M. J., and Sprang S. R. (1989) The structure of tumor necrosis factor-α at 2.6 Å resolution: implications for receptor binding. J. Biol. Chem. 264, 17595–17605 [DOI] [PubMed] [Google Scholar]
- 9. Smith R. A., and Baglioni C. (1987) The active form of tumor necrosis factor is a trimer. J. Biol. Chem. 262, 6951–6954 [PubMed] [Google Scholar]
- 10. Intiso D., Zarrelli M. M., Lagioia G., Di Rienzo F., Checchia De Ambrosio C., Simone P., Tonali P., and Cioffi Dagger R. P. (2004) Tumor necrosis factor α serum levels and inflammatory response in acute ischemic stroke. Neurol. Sci. 24, 390–396 10.1007/s10072-003-0194-z [DOI] [PubMed] [Google Scholar]
- 11. Michalaki V., Syrigos K., Charles P., and Waxman J. (2004) Serum levels of IL-6 and TNF-α correlate with clinicopathological features and patient survival in patients with prostate cancer. Br. J. Cancer 90, 2312–2316 10.1038/sj.bjc.6601814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arican O., Aral M., Sasmaz S., and Ciragil P. (2005) Serum levels of TNF-α, IFN-γ, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005, 273–279 10.1155/MI.2005.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bazzoni F., and Beutler B. (1996) The tumor necrosis factor ligand and receptor families. N. Engl. J. Med. 334, 1717–1725 10.1056/NEJM199606273342607 [DOI] [PubMed] [Google Scholar]
- 14. Corti A., Fassina G., Marcucci F., Barbanti E., and Cassani G. (1992) Oligomeric tumour necrosis factor α slowly converts into inactive forms at bioactive levels. Biochem. J. 284, 905–910 10.1042/bj2840905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bradley J. R. (2008) The role of nuclear organization in cancer. J. Pathol. 214, 149–160 10.1002/path.2287 [DOI] [PubMed] [Google Scholar]
- 16. Kenig M., Gaberc-Porekar V., Fonda I., and Menart V. (2008) Identification of the heparin-binding domain of TNF-α and its use for efficient TNF-α purification by heparin-Sepharose affinity chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 867, 119–125 10.1016/j.jchromb.2008.03.023 [DOI] [PubMed] [Google Scholar]
- 17. Alzani R., Cozzi E., Corti A., Temponi M., Trizio D., Gigli M., and Rizzo V. (1995) Mechanism of suramin-induced deoligomerization of tumor necrosis factor α. Biochemistry 34, 6344–6350 10.1021/bi00019a012 [DOI] [PubMed] [Google Scholar]
- 18. Tamkovich S. N., Bryzgunova O. E., Rykova E. Y., Permyakova V. I., Vlassov V. V., and Laktionov P. P. (2005) Circulating nucleic acids in blood of healthy male and female donors. Clin. Chem. 51, 1317–1319 10.1373/clinchem.2004.045062 [DOI] [PubMed] [Google Scholar]
- 19. Cheng C., Omura-Minamisawa M., Kang Y., Hara T., Koike I., and Inoue T. (2009) Quantification of circulating cell-free DNA in the plasma of cancer patients during radiation therapy. Cancer Sci. 100, 303–309 10.1111/j.1349-7006.2008.01021.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szpechcinski A., Chorostowska-Wynimko J., Struniawski R., Kupis W., Rudzinski P., Langfort R., Puscinska E., Bielen P., Sliwinski P., and Orlowski T. (2015) Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br. J. Cancer 113, 476–483 10.1038/bjc.2015.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humphreys D. T., and Wilson M. R. (1999) Modes of L929 cell death induced by TNF-α and other cytotoxic agents. Cytokine 11, 773–782 10.1006/cyto.1998.0492 [DOI] [PubMed] [Google Scholar]
- 22. Stull R. A., Zon G., and Szoka F. C. (1993) Single-stranded phosphodiester and phosphorothioate oligonucleotides bind actinomycin D and interfere with tumor necrosis factor-induced lysis in the L929 cytotoxicity assay. Antisense Res. Dev. 3, 295–300 10.1089/ard.1993.3.295 [DOI] [PubMed] [Google Scholar]
- 23. Hoffmann A., Kovermann M., Lilie H., Fiedler M., Balbach J., Rudolph R., and Pfeifer S. (2012) New binding mode to TNF-α revealed by ubiquitin-based artificial binding protein. PLoS One 7, e31298 10.1371/journal.pone.0031298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He M. M., Smith A. S., Oslob J. D., Flanagan W. M., Braisted A. C., Whitty A., Cancilla M. T., Wang J., Lugovskoy A. A., Yoburn J. C., Fung A. D., Farrington G., Eldredge J. K., Day E. S., Cruz L. A., et al. (2005) Small-molecule inhibition of TNF-α. Science 310, 1022–1025 10.1126/science.1116304 [DOI] [PubMed] [Google Scholar]
- 25. Klein L., Kyewski B., Allen P. M., and Hogquist K. A. (2014) Positive and negative selection of the T cell repertoire: what thymocytes see and don't see. Nat. Rev. Immunol. 14, 377–391 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vrisekoop N., Monteiro J. P., Mandl J. N., and Germain R. N. (2014) Thymic positive selection and the mature T cell repertoire for antigen revisited. Immunity 41, 181–190 10.1016/j.immuni.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kosmrlj A., Jha A. K., Huseby E. S., Kardar M., and Chakraborty A. K. (2008) How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc. Natl. Acad. Sci. U.S.A. 105, 16671–16676 10.1073/pnas.0808081105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fulda S. (2014) Therapeutic exploitation of necroptosis for cancer therapy. Semin. Cell Dev. Biol. 35, 51–56 10.1016/j.semcdb.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 29. Vanlangenakker N., Bertrand M. J., Bogaert P., Vandenabeele P., and Vanden Berghe T. (2011) TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2, e230 10.1038/cddis.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeraati M., Langley D. B., Schofield P., Moye A. L., Rouet R., Hughes W. E., Bryan T. M., Dinger M. E., and Christ D. (2018) I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 10, 631–637 10.1038/s41557-018-0046-3 [DOI] [PubMed] [Google Scholar]
- 31. Kimoto M., Nakamura M., and Hirao I. (2016) Post-ExSELEX stabilization of an unnatural-base DNA aptamer targeting VEGF165 toward pharmaceutical applications. Nucleic Acids Res. 44, 7487–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mannocci L., Zhang Y., Scheuermann J., Leimbacher M., De Bellis G., Rizzi E., Dumelin C., Melkko S., and Neri D. (2008) High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc. Natl. Acad. Sci. U.S.A. 105, 17670–17675 10.1073/pnas.0805130105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rothemund P. W. (2006) Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 10.1038/nature04586 [DOI] [PubMed] [Google Scholar]
- 34. Seelig G., Soloveichik D., Zhang D. Y., and Winfree E. (2006) Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 10.1126/science.1132493 [DOI] [PubMed] [Google Scholar]
- 35. Organick L., Ang S. D., Chen Y.-J., Lopez R., Yekhanin S., Makarychev K., Racz M. Z., Kamath G., Gopalan P., Nguyen B., Takahashi C. N., Newman S., Parker H.-Y., Rashtchian C., Stewart K., et al. (2018) Random access in large-scale DNA data storage. Nat. Biotechnol. 36, 242–248 10.1038/nbt.4079 [DOI] [PubMed] [Google Scholar]
- 36. Alzani R., Corti A., Grazioli L., Cozzi E., Ghezzi P., and Marcucci F. (1993) Suramin induces deoligomierization of human tumor necrosis factor α. J. Biol. Chem. 268, 12526–12529 [PubMed] [Google Scholar]
- 37. Ledford H. (2018) Gene-silencing technology gets first drug approval after 20-year wait. Nature 560, 291–292 10.1038/d41586-018-05867-7 [DOI] [PubMed] [Google Scholar]
- 38. Zhou J., and Rossi J. (2017) Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 10.1038/nrd.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li S., Jiang Q., Liu S., Zhang Y., Tian Y., Song C., Wang J., Zou Y., Anderson G. J., Han J.-Y., Chang Y., Liu Y., Zhang C., Chen L., Zhou G., et al. (2018) A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 10.1038/nbt.4071 [DOI] [PubMed] [Google Scholar]
- 40. Measey T. J., and Gai F. (2012) Light-triggered disassembly of amyloid fibrils. Langmuir 28, 12588–12592 10.1021/la302626d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chatterjee S., Lee J. B., Valappil N. V., Luo D., and Menon V. M. (2012) Probing Y-shaped DNA structure with time-resolved FRET. Nanoscale 4, 1568–1571 10.1039/c2nr12039a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.