Abstract
Lysophospholipids (LPLs) are important lipid-signaling molecules in plants, of which lysophosphatidylcholine (lysoPC) is one of the most well-characterized LPLs, having important roles in plant stress responses. It is broken down by lysophospholipases, but the molecular mechanism involved in lysoPC degradation is unclear. Recombinant Arabidopsis thaliana ACYL-CoA–BINDING PROTEIN2 (AtACBP2) has been reported to bind lysoPC via its acyl-CoA–binding domain and also LYSOPHOSPHOLIPASE 2 (AtLYSOPL2) via its ankyrin repeats in vitro. To investigate the interactions of AtACBP2 with AtLYSOPL2 and lysoPC in more detail, we conducted isothermal titration calorimetry with AtACBP270–354, an AtACBP2 derivative consisting of amino acids 70–354, containing both the acyl-CoA–binding domain and ankyrin repeats. We observed that the interactions of AtACBP270–354 with AtLYSOPL2 and lysoPC were both endothermic, favored by solvation entropy and opposed by enthalpy, with dissociation constants in the micromolar range. Of note, three AtLYSOPL2 catalytic triad mutant proteins (S147A, D268A, and H298A) bound lysoPC only weakly, with an exothermic burst and dissociation constants in the millimolar range. Furthermore, the binding affinity of lysoPC-premixed AtACBP270–354 to AtLYSOPL2 was 10-fold higher than that of AtACBP270–354 alone to AtLYSOPL2. We conclude that AtACBP2 may play a role in facilitating a direct interaction between AtLYSOPL2 and lysoPC. Our results suggest that AtACBP270–354 probably binds to lysoPC through a hydrophobic interface that enhances a hydrotropic interaction of AtACBP270–354 with AtLYSOPL2 and thereby facilitates AtLYSOPL2's lysophospholipase function.
Keywords: lysophospholipid, thermodynamics, isothermal titration calorimetry (ITC), enzyme mutation, structural model, molecular docking, ankyrin, lipid metabolism, lysophosphatidylcholine, protein–protein interaction
Introduction
Phospholipids are crucial components of many biological membranes (1). Lysophospholipids (LPLs)2 and free fatty acids are produced through the removal of an O-acyl chain (sn-1/sn-2) from phospholipids by phospholipase hydrolysis (2). LPLs are believed to be critical lipid-signaling molecules in cellular membranes (3). Plant LPLs normally exist in low amounts but can be elevated by environmental stimuli, such as low temperature and pathogen stress (4, 5). One of the most well-characterized LPLs is lysophosphatidylcholine (lysoPC) (6, 7). LysoPC has been reported to enhance pathogen susceptibility in tobacco plants via signaling pathways associated with the accumulation of reactive oxygen species and ethylene (8). As membrane-derived signaling molecules in eukaryotes, LPLs perform various biological functions through activating distinct signal transduction pathways (6, 9). LysoPC activates human T lymphocytes (10), up-regulates the expression of P-selectin in mammalian platelets and endothelial cells (11), and promotes nerve growth factor–induced signals in rat pheochromocytoma PC12 cells and neurotrophin-like activity in cerebellar granule neurons and PC12 cells (12). In mammalian cells, LPL signaling is supported by G protein–coupled plasma membrane receptors (13–15).
Lysophospholipases belong to a family of hydrolases that hydrolyze lysoPC by cleaving its carboxylic ester bond (16). In this enzymatic reaction, lysoPC and water are the substrates, and glycerophosphocholine and free fatty acid are the products (17). Based on sequence alignments, two classes of lysophospholipases have emerged in Arabidopsis thaliana, including LYSOPHOSPHOLIPASE1 (AtLYSOPL1; AT2G39400) and five AtLYSOPL1-like proteins (AT2G39400, AT2G39410, AT2G39420, AT3G55180, and AT3G55190), and LYSOPHOSPHOLIPASE2 (AtLYSOPL2; AT1G52760) (18). AtLYSOPL1, which functions in plant defense, was reported to be pathogen- and salicylic acid–inducible, but few studies have been conducted on the others (18, 19). AtLYSOPL2 can be grouped with monoacylglycerol lipases (MAGL), including 15 Arabidopsis homologues of the AtLYSOPL1 family, because they all showed high sequence homology in BLAST scores (≥75) to human monoglyceride lipase (MGL) (GB NP_009214), and all 16 Arabidopsis putative MAGLs (AtMAGLs) displayed 3D structures that are similar to a Homo sapiens MAGL (HsMAGL) (Protein Data Bank code 3HJU) (20, 21). In addition, AtLYSOPL2 (AT1G52760) was also identified as a caffeoyl shikimate esterase, an enzyme central to the lignin biosynthetic pathway (22).
The interaction between AtLYSOPL2 and Acyl-CoA–binding Protein2 (AtACBP2; AT4G27780) was first demonstrated by yeast two-hybrid analysis and co-immunoprecipitation assays (19). Their subcellular interaction was verified by the co-localization of autofluorescence-tagged AtACBP2 and AtLYSOPL2 to the plasma membrane by confocal microscopy of agroinfiltrated tobacco leaves (19). Both proteins have been reported to independently function in conferring tolerance to cadmium (Cd) and oxidative stresses in transgenic Arabidopsis (19, 23).
Cd, highly toxic to plants, accumulates in food chains leading to adverse effects on human and animal health (24). Plants absorb Cd through their roots and accumulate it in shoots via zinc (Zn)/Cd-transporting ATPases (25, 26) and phytochelatin transporters (26, 27). Cd, “resembling common metal cofactors” such as Zn and calcium, inhibits protein function by binding to cysteine residues (28) and disrupts enzyme activity and signal transduction (29, 30). Cd accesses plant cells via calcium (Ca), iron (Fe), and Zn transporters/channels (24, 31) and causes deleterious effects via nitric oxide and reactive oxygen species (ROS) that can result in cell death (32). The roots of Arabidopsis mutants lacking SNF1-RELATED PROTEIN KINASE TYPE 2 displayed lower Cd-induced ROS accumulation, suggesting that these kinases regulate Cd-induced ROS (33). Transgenic Arabidopsis overexpressing AtLYSOPL2 and those overexpressing AtACBP2 were more tolerant to Cd than the WT (19). It has been suggested that AtLYSOPL2 overexpressors were more tolerant to H2O2 and Cd than the wildtype (WT) because AtLYSOPL2 enhances phospholipid repair following lipid peroxidation (19). Both AtLYSOPL2 and AtACBP2 mRNAs were elevated by Cd treatment; Zn and hydrogen peroxide (H2O2), but not lead (Pb), Cd, or copper (Cu), induced AtLYSOPL2 expression in shoots, whereas only H2O2 up-regulated AtLYSOPL2 expression in roots (19). Microarray data from the Arabidopsis Electronic Fluorescent Pictograph Browser revealed that AtLYSOPL2 was inducible by biotic stresses caused by salicylic acid, bacterial-derived elicitor Flg22, and Pseudomonas syringae, besides various abiotic stresses, including cold, drought, genotoxicity (bleomycin plus mitomycin), oxidative treatment (methyl viologen), UV-B light, wounding, heat, and selenium (34). Western blot analysis using AtLYSOPL2 antibodies indicated that Cd and Zn treatments resulted in AtLYSOPL2 accumulation in shoots and roots, although Cd treatment down-regulated AtLYSOPL2 expression in Northern blotting (19). Northern blotting analyses revealed a higher AtLYSOPL2 expression in stems, flowers, and roots than in siliques and leaves (19).
To better understand the role of AtACBP2 and AtLYSOPL2 in stress tolerance, the in vitro energetics of their interactions were investigated. The thermodynamic analysis reported here provides new insights on AtACBP2, AtLYSOPL2, and lysoPC interactions.
Results
AtACBP270–354 binds both lysoPC (C16:0) and palmitoyl-CoA thioester
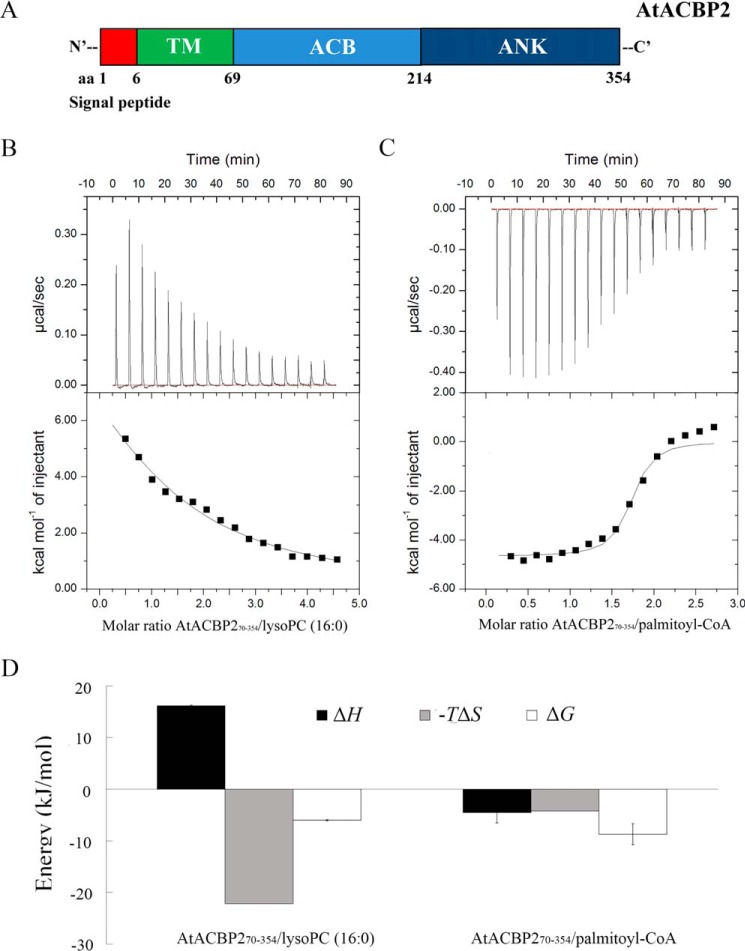
AtACBP2 (AT4G27780) consists of amino acids (aa) 1–354 comprising a signal peptide (aa 1–6), transmembrane domain (aa 7–69), the acyl-CoA–binding domain (aa 70–214) and the ankyrin repeats (aa 215–354) (Fig. 1A). The transmembrane domain (aa 7–69) was deleted in AtACBP270–354, because the truncated version (AtACBP270–354) enhanced AtACBP2 expression and solubility. The AtACBP270–354 interaction with lipid ligands and AtLYSOPL2 was assessed by isothermal titration calorimetry (ITC), which quantifies the binding equilibrium directly by measuring the heat change resulting from the association of a ligand with its binding partner (35). ITC thermograms showed that AtACBP270–354, an AtACBP2 derivative consisting of aa 70–354 inclusive of the acyl-CoA–binding domain and the ankyrin repeats, binds to both lysoPC (C16:0) and palmitoyl-CoA (C16:0–CoA) thioesters (Fig. 1, B and C). The primary heat change of AtACBP270–354–lysoPC (Fig. 1B) and AtACBP270–354–C16:0–CoA (Fig. 1C) interactions was fitted using a simple one-site binding model to yield the thermodynamic parameters (Table 1). AtACBP270–354 showed a higher binding affinity to C16:0–CoA than lysoPC, with KD values of 0.64 and 39.2 μm, respectively (Table 1).
Figure 1.
ITC analysis of AtACBP270–354 interactions with lysoPC (C16:0) and C16:0–CoA. A, schematic representation of the domains in Arabidopsis AtACBP2. The signal peptide (aa 1–6), transmembrane (TM) domain (aa 7–69), acyl-CoA binding (ACB) domain (aa 70 to 214), and ankyrin repeats (ANK) (aa 215–354) of AtACBP2 are shown in red, green, light blue, and dark blue, respectively. AtACBP270–354 consists of a derivative lacking the transmembrane domain, with the ACB and ANK domains intact. AtACBP2215–354 consists of the ANK domain. B, AtACBP270–354 and lysoPC (C16:0) binding measured by titrating 30–40 μm AtACBP270–354 in the chamber with 600–800 μm lysoPC (C16:0) in the syringe. C, AtACBP270–354 and C16:0–CoA binding measured by titrating 30–40 μm AtACBP270–354 in the chamber with 600–800 μm C16:0–CoA in the syringe. Top panel, raw heating power over time; bottom panel, fit of the integrated energy values normalized for injected protein. D, binding signature (ΔH, −TΔS, and ΔG) plotted for AtACBP270–354–lysoPC (C16:0) and AtACBP270–354–C16:0–CoA interactions. Binding enthalpy, entropy, and free energy are shown in black, gray, and white, respectively. The values originate from Table 1. Error bars denote S.E., n = 2.
Table 1.
ITC binding constants and thermodynamic parameters for AtACBP270–354 interactions with palmitoyl-CoA and lysoPC
The values are plotted in Fig. 1. Experiments were carried out at 25 °C, and each value is the mean of at least two independent titrations. n is number of binding sites (n = ligand/receptor); KD is dissociation constant; ΔH is enthalpy change; ΔS is entropy change; ΔG, Gibbs free energy. The binding entropy and enthalpy determine ligand binding. Positive is unfavorable, and negative is favorable (61, 62).
| Combinations | n | KD | ΔH | −TΔS | ΔG |
|---|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| AtACBP270–354/palmitoyl-CoA | 1.73 ± 0.03 | 0.64 ± 0.09 | −4.51 ± 0.13 | −3.93 | −8.44 ± 0.13 |
| AtACBP270–354/lysoPC | 1.22 ± 0.42 | 39.2 ± 8.4 | 16.17 ± 6.8 | −22.17 | −6.05 ± 6.8 |
Interestingly, the thermodynamic characteristics of interactions between AtACBP270–354–lysoPC (Fig. 1B) and AtACBP270–354–C16:0–CoA (Fig. 1C) were fundamentally different. AtACBP270–354–lysoPC interaction represented an unusual endothermic association with favorable entropy (−TΔS −22.17 kcal mol−1) and opposed by enthalpy (ΔH 16.17 kcal mol−1) (Table 1), which suggests that the binding of AtACBP270–354 to lysoPC is supported by hydrophobic interactions (Fig. 1D). For lysoPC, the stoichiometric value (n) was 1.22, indicating that one molecule of AtACBP270–354 binds to only one lysoPC ligand (Table 1).
The AtACBP270–354–C16:0–CoA interaction was exothermic, favored by both enthalpy (ΔH −4.51 kcal mol−1) and entropy (−TΔS −3.93 kcal mol−1), suggesting that the binding of AtACBP270–354 to C16:0–CoA is composed of hydrogen bonding and hydrophobic interactions (Fig. 1D). In addition, the interaction between AtACBP270–354 and C16:0–CoA showed an n value of 1.73 (Table 1), indicating that one molecule of AtACBP270–354 likely binds to two C16:0–CoA ligands. The stoichiometry value observed here resembles that (n = 1.7) previously reported for the C2B and C2G domains of dysferlin, which was presumed to bind to two Ca2+ ions (36).
AtACBP270–354 binds to WT AtLYSOPL2 and the WT AtLYSOPL2–lysoPC complex
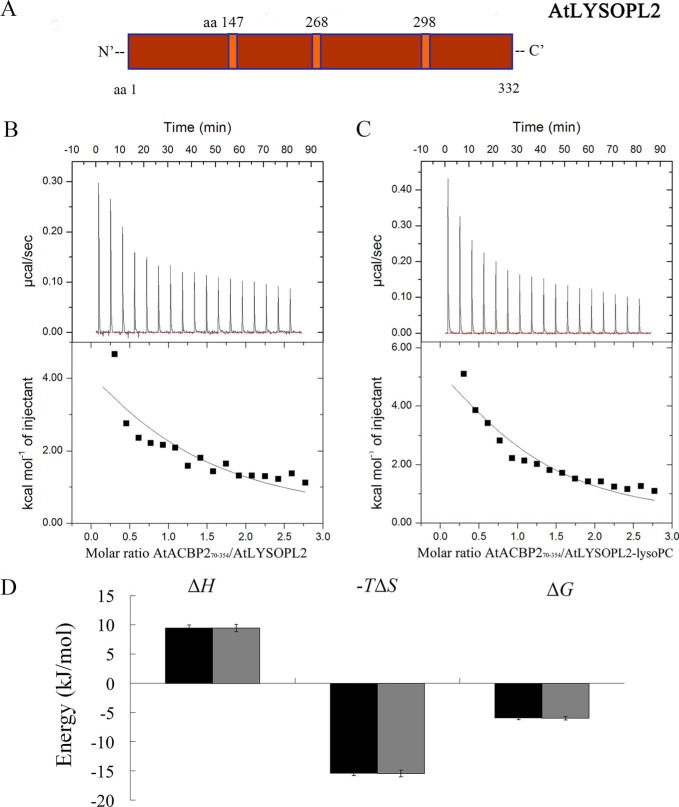
The effect of lysoPC on the AtLYSOPL2–AtACBP270–354 (Fig. 2A) interaction was determined by ITC. Theoretical fits to the experimental data were achieved using a single binding site model with the thermodynamic parameters listed in Table 2, including dissociation constant (KD), stoichiometry of the ligand-to-protein binding (n), enthalpy change (ΔH), the entropy change (ΔS), and free energy change (ΔG). ITC thermograms revealed that binding of AtLYSOPL2 (Fig. 2B) or the AtLYSOPL2–lysoPC complex (Fig. 2C) to AtACBP270–354 was highly endothermic. The interactions were driven by favorable entropy −TΔS of −15.38 and −15.29 kcal mol−1 and opposed by enthalpy changes ΔH of 9.55 and 9.65 kcal mol−1, respectively (Table 2). The binding mechanism of AtLYSOPL2–AtACBP270–354 is similar to AtACBP270–354–lysoPC and is assumed to be based on hydrophobic interactions following a conformational change (Fig. 2D).
Figure 2.
ITC analysis of AtLYSOPL2, AtACBP270–354, and lysoPC interactions. A, schematic representation of the structure of Arabidopsis AtLYSOPL2. The catalytic triad of AtLYSOPL2 is shown in orange. B, interactions between AtLYSOPL2 and AtACBP270–354. Binding was tested in ITC by titrating 30–40 μm AtLYSOPL2 in the chamber with 400–500 μm AtACBP270–354 in the syringe. C, interactions between the AtLYSOPL2–lysoPC (molar ratio 1:1) complex and AtACBP270–354. Binding was tested in ITC by titrating 30 μm AtLYSOPL2–lysoPC complex in the chamber with 400–500 μm AtACBP270–354 in the syringe. Top panel, raw heating power over time; bottom panel, fit of the integrated energy values normalized for injected protein. D, energetics of AtLYSOPL2 interaction with AtACBP270–354. Plots of the binding signature (ΔH, −TΔS, and ΔG) with interactions of AtLYSOPL2 and AtACBP270–354 in the absence of lysoPC shown in black, and AtLYSOPL2 and AtACBP270–354 in the presence of lysoPC in gray. The values were obtained from data presented in Table 2. Error bars denote S.E., n = 3. The values originate from Table 2.
Table 2.
ITC binding constants and thermodynamic parameters for interactions of AtLYSOPL2 wildtype or mutant proteins with AtACBP270–354 with/without lysoPC
The values are plotted in Figs. 2 and 5. Experiments were carried out at 25 °C, and each value is the mean of three independent titrations. n is number of binding sites (n = ligand/receptor); KD is dissociation constant; ΔH is enthalpy change; ΔS is entropy change; ΔG is Gibbs free energy. The binding entropy and enthalpy determine ligand binding. Positive is unfavorable, and negative is favorable (61, 62).
| Combinations | n | KD | ΔH | −TΔS | ΔG |
|---|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| AtACBP270–354/wildtype AtLYSOPL2 | 1.05 ± 0.01 | 28.98 ± 3.36 | 9.55 ± 0.26 | −15.38 ± 0.13 | −5.93 ± 0.12 |
| AtACBP270–354/wildtype AtLYSOPL2-lysoPC | 1.17 ± 0.5 | 29.99 ± 3.86 | 9.65 ± 4 | −15.29 ± 3.87 | −5.63 ± 2.87 |
| AtACBP270–354/AtLYSOPL2 S147A | 0.56 ± 0.2 | 20.7 ± 1.03 | 10.71 ± 3.19 | −15.7 ± 3.21 | −4.99 ± 2.2 |
| AtACBP270–354/AtLYSOPL2 S147A-lysoPC | 0.36 ± 0.09 | 15.02 ± 1.63 | 18.1 ± 2.17 | −24.47 ± 1.77 | −6.37 ± 1.9 |
| AtACBP270–354/AtLYSOPL2 D268A | 0.37 ± 0.08 | 12.39 ± 0.24 | 9.27 ± 1.16 | −15.94 ± 1.18 | −6.67 ± 1.4 |
| AtACBP270–354/AtLYSOPL2 D268A-lysoPC | 0.31 ± 0.12 | 10.17 ± 4.32 | 9.63 ± 2.46 | −16.42 ± 1.79 | −6.79 ± 1.88 |
| AtACBP270–354/AtLYSOPL2 H298A | 0.84 ± 0.03 | 25.64 ± 2.49 | 6.42 ± 1.2 | −12.67 ± 1.45 | −5.27 ± 1.84 |
| AtACBP270–354/AtLYSOPL2 H298A-lysoPC | 0.67 ± 0.66 | 23.04 ± 2.14 | 7.63 ± 0.7 | −14.23 ± 1.12 | −6.62 ± 1.11 |
The KD value for the AtLYSOPL2 and AtACBP270–354 interaction was 28.98 μm (Table 2). A similar affinity as reflected by a KD value of 29.99 μm when AtACBP270–354 was titrated to the preformed AtLYSOPL2–lysoPC complex (molar ratio 1:1) demonstrating that lysoPC, when pre-bound to AtLYSOPL2, did not promote the AtLYSOPL2 and AtACBP270–354 interaction (Table 2). Only one binding site was predicted for the AtLYSOPL2 and AtACBP270–354 interaction, because ITC data showed an n value of 1.05 (Table 2).
Role of the conserved catalytic triad in AtLYSOPL2
Structural sequence alignments of AtLYSOPL2, AtLYSOPL1, and human MGL were analyzed using the Jalview version 1.6 software (http://www.jalview.org/)3 (Fig. S1) (63). The Fig. S1 identified three conserved catalytic residues (Ser-147, Asp-268, and His-298) in AtLYSOPL2, corresponding to the catalytic triad (Ser-132, Asp-249, and His-279) of MGL (21). AtLYSOPL2 showed 28% aa sequence homology to MGL and 34% homology to AtLYSOPL1, whereas AtLYSOPL1 showed 31% homology to MGL. Phylogenetic analysis indicated that AtLYSOPL2 stands by itself and displayed low aa sequence identities with other homologues in A. thaliana (Fig. S2).
To construct a 3D structural model for AtLYSOPL2, the X-ray crystal structure of human MGL, an orthologue of AtLYSOPL2, was used as a structural template for analysis with the MODELLER program (Fig. 3A). When the positions for lysoPC binding were predicted using the AutoDock program (http://autodock.scripps.edu/),3 lysoPC (C16:0) was found to lie in a pocket-like binding site on AtLYSOPL2 (Fig. 3, B and C). The conserved AtLYSOPL2 catalytic triad, Ser-147, Asp-268, and His-298, was observed located adjacent to lysoPC, suggesting its potential significance in an enzymatic reaction (Fig. 3D).
Figure 3.
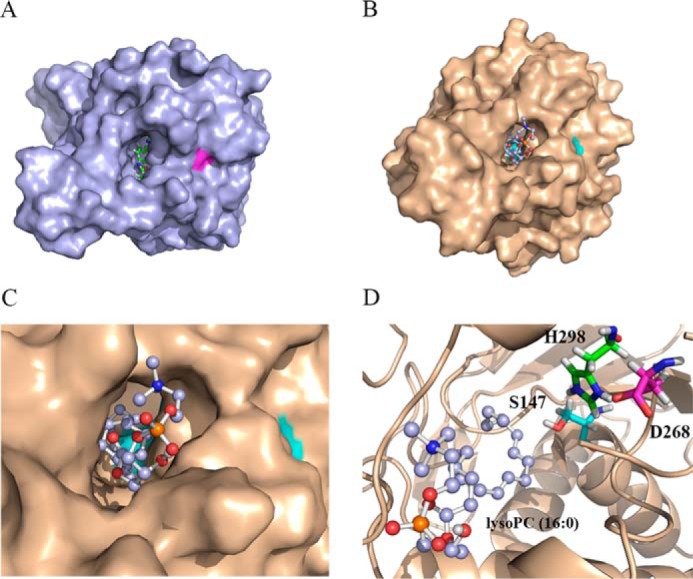
Homology modeling and dock simulation of lysoPC (C16:0) binding to AtLYSOPL2. A, structure of human MGL with its inhibitor JZL184 (Protein Data Bank code 3JWE (21). B and C, 3D structure model of AtLYSOPL2 with lysoPC (C16:0). The bottom catalytic residues in the pocket-like cleavage site are shown in cyan. D, putative scissor aa residues Ser-147, Asp-268, and His-298 in the cleavage site of AtLYSOPL2. LysoPC (C16:0) is shown as ball and stick.
To validate these docking analyses, single point mutations (i.e. S147A, D268A, and H298A) were generated by site-directed mutagenesis and proteins purified (Fig. S3). Results from GC-MS analysis, using lysoPC (C16:0) as a substrate in enzymatic assays, demonstrated that hexadecanoic acid was produced, verifying AtLYSOPL2 function as a typical lysophospholipase in vitro. In contrast, AtLYSOPL2 mutant proteins S147A, D268A, and H298A lack lysophospholipase activity, and little to no hexadecanoic acid was produced (Fig. 4, A and B). The relative lysophospholipase activities of the AtLYSOPL2 mutant proteins S147A, D268A, and H298A were 0.10, 0.08, and 0.12, respectively, and compared with the WT AtLYSOPL2 protein value of 1 (Fig. 4A), indicating that all three conserved catalytic aa residues are essential for lysophospholipase catalysis.
Figure 4.
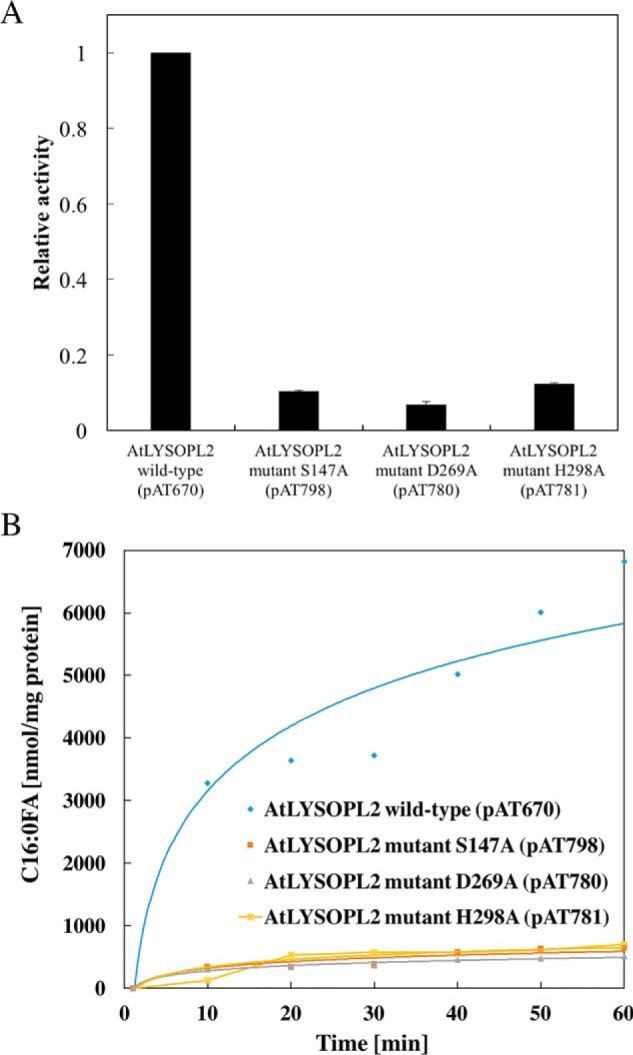
Lysophospholipase activity assays on AtLYSOPL2 and its mutant derivatives. A, comparison of lysophospholipase activity among WT AtLYSOPL2 and its mutant proteins (S147A, D268A, and H298A). The values are shown as the relative activity to WT AtLYSOPL2 (defined value of 1). B, amount of C16:0-FA detected at different time intervals.
AtACBP270–354 binds to AtLYSOPL2 catalytic triad mutant proteins and the AtLYSOPL2 catalytic triad mutant—lysoPC complexes
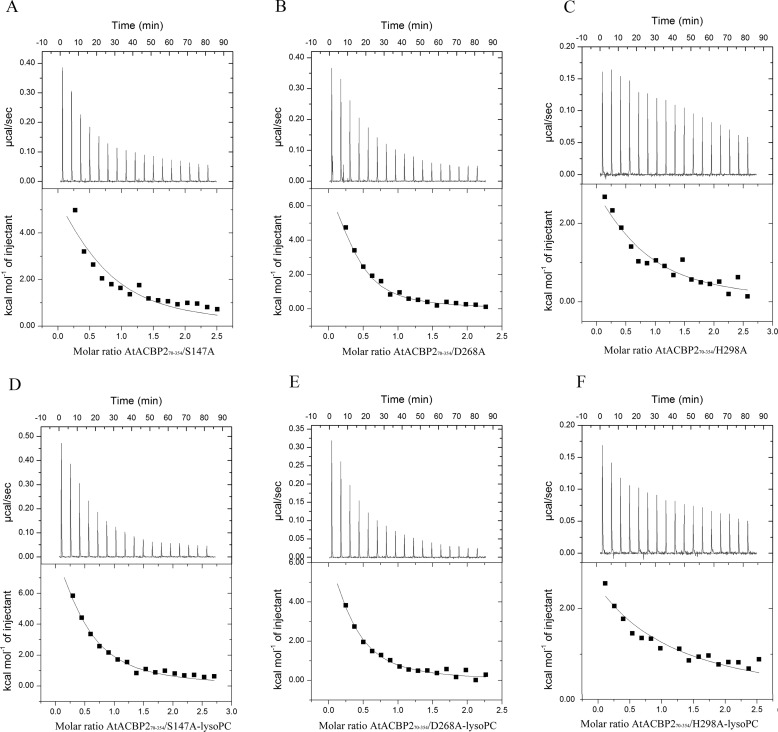
To analyze whether the AtLYSOPL2 catalytic triad influences the interaction between AtLYSOPL2 and AtACBP2, ITC experiments were performed using AtLYSOPL2 WT and mutant (S147A, D268A, and H298A) proteins in the presence and absence of lysoPC and AtACBP270–354. The ITC results were again fitted using a single binding site model. Thermodynamic properties of interactions between WT and mutant AtLYSOPL2 to AtACBP270–354 were similar. All appeared to be endothermic, and the interactions were favored by a change of entropy (−TΔS), but opposed by an unfavorable enthalpy (ΔH) change (Fig. 5 and Table 2).
Figure 5.
ITC analysis of interactions between AtACBP270–354 and AtLYSOPL2 mutant proteins in the absence or presence of lysoPC. A–C, AtACBP270–354interactions with AtLYSOPL2 mutants S147A (A), D268A (B), and H298A (C) in the absence of lysoPC. Interactions were measured by titrating 20–30 μm AtLYSOPL2 (S147A, D268A, and H298A) in the chamber with 300–400 μm AtACBP270–354 in the syringe. D–F, AtACBP270–354 interactions with AtLYSOPL2 mutants S147A (D), D268A (E), H298A (F) in the presence of lysoPC. Interactions were measured by titrating 30 μm AtLYSOPL2 (S147A, D268A, and H298A)–lysoPC complex (molar ratio 1:1) in the chamber with 400–500 μm AtACBP270–354 in the syringe. Top panel, raw heating power over time; bottom panel, fit of the integrated energy values normalized for the injected protein. The values originate from Table 2.
The binding affinities of S147A, D268A, and H298A to AtACBP270–354 were similar to WT AtLYSOPL2 and AtACBP270–354 (KD = 28.98 μm). In the absence of lysoPC, interactions between AtACBP270–354 and S147A, D268A, or H298A yielded KD values of 20.7, 12.39, and 25.64 μm, respectively (Fig. 5, A–C, and Table 2), which were similar to interactions between AtACBP270–354 and S147A, D268A, and H298A in the presence of lysoPC (KD = 15.02, 10.17, and 23.04 μm, respectively) (Fig. 5, D–F, and Table 2). These results demonstrated that the presence of lysoPC did not enhance interactions between AtACBP270–354 and S147A, D268A, and H298A, when premixed with the mutant protein at a 1:1 molar ratio.
Binding of WT AtLYSOPL2 or AtLYSOPL2 catalytic triad mutant proteins to AtACBP270–354–lysoPC complex
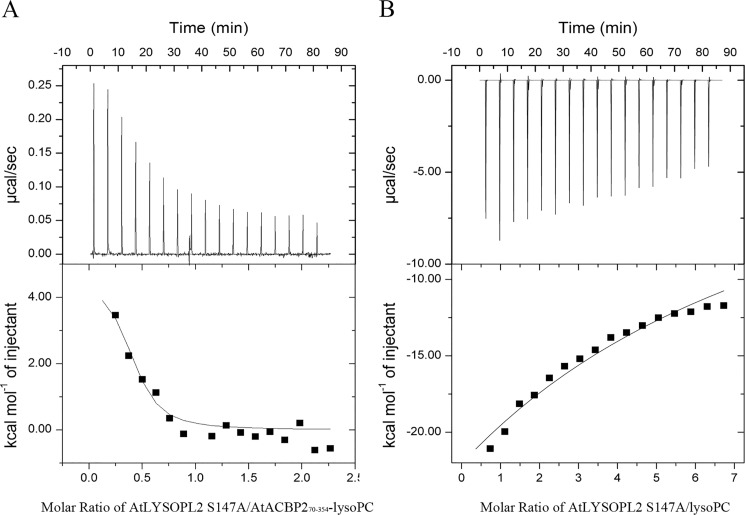
To identify the effect of the preformed AtACBP270–354–lysoPC complex (molar ratio 1:1) on interaction with WT AtLYSOPL2 or AtLYSOPL2 catalytic triad mutant proteins (S147A, D268A, and H298A), thermodynamic parameters were again determined by ITC and fitted through a simple one-site binding model. The AtACBP270–354–lysoPC (molar ratio 1:1) complex was titrated to the WT AtLYSOPL2 or AtLYSOPL2 mutant proteins (S147A, D268A, and H298A). The results revealed that the interactions of the AtACBP270–354–lysoPC complex with WT AtLYSOPL2 and AtLYSOPL2 catalytic triad mutant proteins were endothermic, favored entirely by entropy (−TΔS) and opposed by enthalpy (ΔH) (Fig. 6A and Table 3). However, the dissociation constant (KD = 1.69 μm) (Table 3) declined 12-fold compared with AtACBP270–354 binding to AtLYSOPL2 S147A (KD = 20.7 μm) (Table 3).
Figure 6.
ITC analysis of AtLYSOPL2 catalytic triad mutant S147A interactions with the AtACBP270–354–lysoPC complex and lysoPC. A, binding of AtLYSOPL2 S147A and AtACBP270–354–lysoPC complex was measured by titrating 20–30 μm AtLYSOPL2 S147A in the chamber with 400–500 μm of the AtACBP270–354–lysoPC (molar ratio 1:1) complex in the syringe. B, AtLYSOPL2 S147A and lysoPC interaction was measured by titrating 20–30 μm AtLYSOPL2 S147A in the chamber with 600–800 μm lysoPC in the syringe. Top panel, raw heating power over time; bottom panel, fit of the integrated energy values normalized for the injected protein. The values originate from Tables 3 and 4.
Table 3.
ITC binding constants and thermodynamic parameters for interactions of AtLYSOPL2 wildtype, or mutant proteins with truncated AtACBP2 proteins with/without lysoPC
The values are plotted in Fig. 6 and Fig. S4. AtACBP270–214 and AtACBP2215–354 represent AtACBP2 lacking ankyrin repeats and AtACBP2 lacking the acyl-CoA–binding domain, respectively. Experiments were carried out at 25 °C, and each value is the mean of at least three independent titrations. n is number of binding sites (n = ligand/receptor); KD is dissociation constant; ΔH is enthalpy change; ΔS is entropy change; ΔG is Gibbs free energy. The binding entropy and enthalpy determine ligand binding. Positive is unfavorable, and negative is favorable (61, 62). ND indicates not determinable values due to lack of binding.
| Combinations | n | KD | ΔH | −TΔS | ΔG |
|---|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| Wildtype AtLYSOPL2 | |||||
| with AtACBP270–354-lysoPC | 0.89 ± 0.27 | 33.48 ± 0.08 | 9.38 ± 0.20 | −15.14 ± 0.65 | −5.76 ± 0.68 |
| with AtACBP270–354 | 1.05 ± 0.01 | 28.98 ± 3.36 | 9.55 ± 0.26 | −15.38 ± 0.13 | −5.93 ± 0.12 |
| with AtACBP270–214-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP270–214 | ND | ND | ND | ND | ND |
| with AtACBP2215–354-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP2215–354 | ND | ND | ND | ND | ND |
| AtLYSOPL2 S147A | |||||
| with AtACBP270–354-lysoPC | 0.38 ± 0.08 | 1.69 ± 0.11 | 4.85 ± 1.5 | −12.46 ± 0.12 | −7.88 ± 1.5 |
| with AtACBP270–354 | 0.56 ± 0.17 | 20.7 ± 1.03 | 10.71 ± 3.19 | −15.7 ± 3.21 | −4.99 ± 2.2 |
| with AtACBP270–214-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP270–214 | ND | ND | ND | ND | ND |
| with AtACBP2215–354-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP2215–354 | ND | ND | ND | ND | ND |
| AtLYSOPL2 D268A | |||||
| with AtACBP270–354-lysoPC | 0.53 ± 0.02 | 1.38 ± 0.42 | 5.10 ± 0.16 | −13.68 ± 0.20 | −8.58 ± 0.26 |
| with AtACBP270–354 | 0.37 ± 0.08 | 12.39 ± 0.24 | 9.27 ± 1.16 | −15.94 ± 1.18 | −6.67 ± 1.4 |
| with AtACBP270–214-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP270–214 | ND | ND | ND | ND | ND |
| with AtACBP2215–354-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP2215–354 | ND | ND | ND | ND | ND |
| AtLYSOPL2 H298A | |||||
| with AtACBP270–354-lysoPC | 0.61 ± 0.06 | 0.89 ± 0.05 | 4.98 ± 0.06 | −13.69 ± 1.35 | −8.71 ± 1.35 |
| with AtACBP270–354 | 0.84 ± 0.03 | 25.64 ± 0.17 | 6.42 ± 1.2 | −12.67 ± 1.45 | −5.27 ± 1.84 |
| with AtACBP270–214-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP270–214 | ND | ND | ND | ND | ND |
| with AtACBP2215–354-lysoPC | ND | ND | ND | ND | ND |
| with AtACBP2215–354 | ND | ND | ND | ND | ND |
The binding affinities of AtLYSOPL2 mutant proteins (S147A, D268A, and H298A) to AtACBP270–354 were similar to that of WT AtLYSOPL2 with AtACBP270–354 (KD = 28.98 μm). In the absence of lysoPC, the interactions between AtACBP270–354 and AtLYSOPL2 mutant proteins (S147A, D268A, and H298A) yielded KD values of 20.7, 12.39, and 25.64 μm, respectively (Table 3). However, the dissociation constant declined in interactions between the AtACBP270–354–lysoPC complex and AtLYSOPL2 mutant proteins (S147A, D268A, and H298A), which had KD values of 1.69, 1.38, and 0.89 μm, respectively (Table 3). These data demonstrated that lysoPC enhanced the interactions between AtACBP70–354 and AtLYSOPL2 mutant proteins (S147A, D268A, and H298A) when it was premixed with AtACBP270–354 at a 1:1 molar ratio.
To address whether the ankyrin repeats or the acyl-CoA–binding domain of AtACBP2 influences its interaction with AtLYSOPL2, ITC experiments were performed by titrating AtACBP270–214 (lacking the ankyrin repeats) or AtACBP2215–354 (lacking the acyl-CoA–binding domain), in the presence or absence of lysoPC, into WT AtLYSOPL2 or AtLYSOPL2 catalytic triad mutant (S147A, D268A, and H298A) proteins. No interaction was detected in all combinations involving AtACBP270–214 or AtACBP2215–354 (Fig. S4 and Table 3). This demonstrated that the presence of both the ankyrin repeats and the acyl-CoA–binding domain of AtACBP2 was crucial for its binding to AtLYSOPL2.
Binding of WT AtLYSOPL2 or AtLYSOPL2 catalytic triad mutant proteins to lysoPC
Arising from its lysophospholipase activity (Fig. 4), WT AtLYSOPL2 could not be used in ITC experiments to determine the thermodynamic parameters for its binding to lysoPC (Fig. S5 and Table 4). Therefore, the AtLYSOPL2 catalytic triad mutant proteins (S147A, D268A, and H298A) were used to measure the thermodynamic parameters for lysoPC binding. The weak interactions between the AtLYSOPL2 catalytic triad mutant proteins (S147A, D268A, and H298A) and lysoPC were reflected in an exothermic heat burst with KD values of 101.72, 99.67, and 137.37 μm, respectively (Table 4). Unlike the AtACBP2–AtLYSOPL2 interaction, the thermodynamic forces from the AtLYSOPL2 catalytic triad mutant protein–lysoPC interactions were driven by a change in both enthalpy (ΔH) and entropy (−TΔS) (Fig. 6B), indicating that hydrogen bonding and hydrophobic forces contribute to the interaction of AtLYSOPL2 with lysoPC.
Table 4.
ITC binding constants and thermodynamic parameters for interactions of AtLYSOPL2 wildtype or mutant proteins with lysoPC
The values are plotted in Fig. 6 and Fig. S5. Experiments were carried out at 25 °C, and each value is the mean of at least three independent titrations. n is number of binding sites (n = ligand/receptor); KD is dissociation constant; ΔH is enthalpy change; ΔS is entropy change; ΔG is Gibbs free energy. The binding entropy and enthalpy determine ligand binding. Positive is unfavorable, and negative is favorable (61, 62). ND indicates not determinable values due to lack of binding.
| Combinations | n | KD | ΔH | −TΔS | ΔG |
|---|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| Wildtype AtLYSOPL2/lysoPC | ND | ND | ND | ND | ND |
| AtLYSOPL2 S147A/lysoPC | 1.08 ± 0.03 | 101.72 ± 0.19 | −1.67 ± 0.05 | −13.44 ± 0.40 | −15.11 ± 0.41 |
| AtLYSOPL2 D268A/lysoPC | 0.92 ± 0.03 | 99.67 ± 0.05 | −1.66 ± 0.01 | −9.56 ± 0.20 | −11.21 ± 0.20 |
| AtLYSOPL2 H298A/lysoPC | 1.02 ± 0.01 | 137.37 ± 0.11 | −1.68 ± 0.01 | −10.85 ± 0.38 | −12.52 ± 0.38 |
Discussion
Studies on lysophospholipases in bacteria, yeasts, and mammals suggest that these enzymes play a major role in LPL degradation (37, 38). In contrast, very few reports are available on plant lysophospholipases. Structural alignments on lysophospholipases indicate that they share a very conserved hydrolase tertiary structure, arising from convergent evolution, and hydrolase action is conserved among various species. AtLYSOPL2 belongs to the serine hydrolase superfamily (19), and several crystal structures of its homologues, including Serratia marcescens lipase lipA, Ophiostoma piceae sterol esterase, and human MGL, have been reported (21, 39, 40). As homology models are useful in examining ligand binding in many proteins, this strategy was used in the structural prediction of AtLYSOPL2 (Fig. 3).
Although the lysoPC-hydrolyzing activity (Fig. 4) of AtLYSOPL2 (19) has been confirmed by an independent group (41), this protein has also been identified as a caffeoyl shikimate esterase (22), suggesting that AT1G52760 is a multifunctional enzyme. In fact, the Ser–Asp–His catalytic triad, which is critical for the lysophospholipase (19, 20) and esterase activities (22) of AT1G52760, is conserved to catalyze lysophospholipase and esterase activities of other multifunctional enzymes, such as the Escherichia coli thioesterase I, also known as protease I and phospholipase L1 (EC 3.1.2.2) (42). Vijayaraj et al. (41) confirmed that AT1G52760 is a bifunctional enzyme with broad substrate specificities. Moreover, the hydrolase activity of AT1G52760 is supported by the occurrence of the Gly–Xaa–Ser–Xaa–Gly lipase motif conserved among the 16 AtMAGL members and the His–Xaa4–Asp acyltransferase motif conserved in some other AtMAGLs (20, 41). Besides the in vitro enzyme assays, we have previously demonstrated the physiological role for AtLYSOPL2 in phospholipid repair following lipid peroxidation under metal-induced stress in planta using Arabidopsis mutants and overexpressors (19), in agreement with the observation that AT1G52760 is a membrane-associated protein by confocal laser-scanning microscopy (19, 20).
In site-directed mutagenesis experiments on Rattus norvegicus MGL and Mus musculus lysophospholipase (lysoPLA I), the catalytic triad (Ser–His–Asp) was shown to be essential for lipase activities (37, 38, 43). Like the mRNA encoding MGL that is ubiquitously expressed in all tissues (43), AtLYSOPL2 mRNA was detected in roots, stems, and flowers (19), particularly higher in roots and stems than other organs (20). Its protein was observed to accumulate in Arabidopsis roots (Fig. S6). Similar to murine lysoPLA I, which has been proposed to function in the removal of LPLs produced by murine phospholipase A1 and A2 (37), AtLYSOPL2 contains an α/β hydrolase fold and may share similar roles in lysoPC removal by phospholipase hydrolysis.
Besides Ser-147, which is located in the conserved GXSXG motif, the other two conserved catalytic residues (Asp-268 and His-298) of AtLYSOPL2 were also proven essential for lysophospholipase activity (Fig. 4). LysoPC was not efficiently cleaved by the AtLYSOPL2 mutant proteins S147A, D268A, and H298A in GC-MS assays (Fig. 4). In this respect, AtLYSOPL2 resembles R. norvegicus MGL of which substitutions in the corresponding residues rendered it ineffective (21, 38, 43).
Thermodynamic analysis by ITC was carried out to study the relationship between lysoPC and its target protein complex (AtACBP270–354–AtLYSOPL2) and demonstrated that AtACBP270–354 binds to AtLYSOPL2 and lysoPC with similar KD values, both in the micromolar concentration range (Fig. S7). Although the crystal structures of the lysoPC-bound states have not been solved with either AtACBP2 or AtLYSOPL2, a common feature was observed based on thermodynamic analysis on AtACBP270–354 binding to AtLYSOPL2 and lysoPC. Their interactions were endothermic and were driven solely by a large change in entropy (−TΔS), which usually suggests that conformational change and hydrophobicity are the main driving forces in such interactions. A large amount of ordered water molecules localized on the hydrophobic interfaces of the binding molecules that are disseminated into the bulk solvent drive the hydrophobic interactions, and conformational changes of binding molecules occur accompanied by release of water molecules (44). Our results suggest that the interactions of AtACBP270–354 to lysoPC and AtACBP270–354 to AtLYSOPL2 led to conformational changes to form hydrophobic binding surfaces, which associate with the dissipation of water molecules. Interestingly, the binding affinity of AtACBP270–354 premixed with lysoPC to AtLYSOPL2 mutants S147A, D268A, or H298A (KD = 1.69, 1.38, and 0.89 μm, respectively) were 12-, 21-, and 25-fold higher than the binding affinity of unliganded AtACBP270–354 to AtLYSOPL2 mutants S147A, D268A, or H298A (KD = 20.7, 12.39, and 25.64 μm, respectively) (Table 3). These results argue that the conformational change caused by AtACBP270–354 binding to lysoPC enhanced the hydrophobic interaction of AtACBP270–354 with AtLYSOPL2 mutants (S147A, D268A, or H298A) (Fig. 6A and Table 3). Hence, it appears that the AtACBP270–354–lysoPC interaction could strengthen the hydrophobic interaction between AtACBP270–354and AtLYSOPL2. Similarly, the type 2C protein phosphatases (PP2Cs) are considered to be co-receptors in the abscisic acid (ABA)-signaling transduction pathway because their presence improved ABA perception by the ABA receptor protein family, PYRABACTIN RESISTANCE1(PYR1)/PYR1-like(PYL)/regulatory components of ABA receptor (RCAR) (PYR/PYL/RCAR) (45, 46). The availability of protein surfaces arising from conformational changes upon the interaction of PYR/PYL/RCAR to ABA promoted the binding of ABA to PP2Cs (45, 46).
In contrast, the binding of AtLYSOPL2 mutants S147A, D268A, or H298A to lysoPC was exothermic with weak binding affinities of KD values of 101.72, 99.67, and 137.37 μm, respectively (Fig. 6B and Table 4). The lower binding affinity of lysoPC to AtLYSOPL2 mutants (S147A, D268A, or H298A), which compared with the AtACBP270–354–lysoPC complex to S147A, D268A, or H298A, suggests that AtLYSOPL2 interaction with AtACBP270–354 benefits from the pre-binding of AtACBP270–354 to lysoPC. In other words, AtACBP270–354 likely plays a role in facilitating AtLYSOPL2 and lysoPC interactions (Fig. S7). Consistently, it has been recently demonstrated that site-directed mutagenesis of AtACBP1 (Y171A), a homologue of AtACBP2, weakened its interaction with STEROL C4-METHYL OXIDASE1 in vivo (47, 48). Thus, it appears to be a general observation that the liganded state of AtACBP1 (47, 48) and AtACBP2 (this study) promotes their association with protein partners, accounting for the essence of both the ankyrin repeats and the acyl-CoA–binding domain for protein–protein interactions (Fig. S4 and Table 3).
Binding of acyl-CoA thioesters to AtACBP270–354 was favored by both entropy and enthalpy, indicating that hydrogen bonding and hydrophobic forces supported the interaction. The thermodynamic analysis obtained here is consistent with data from analyses on the co-crystal structure of human cytosolic liver ACBP with myristoyl-CoA (49). Conserved positively-charged residues (Tyr-29, Lys-33, and Lys-55) in hL-ACBP form hydrogen bonds with the 3′-phosphate moiety in myristoyl-CoA, whereas a hydrophobic groove in hL-ACBP interacts with the hydrophobic acyl moiety (49). Meanwhile, lysoPC is composed of a hydrophilic head (glycerol 3-phosphocholine) and a hydrophobic fatty acid tail that should be crucial in binding AtACBP2, as the hydrophobic force mainly contributes to interaction of AtACBP2 with lysoPC from ITC analysis. However, the interaction of AtLYSOPL2 and lysoPC was favored by both entropy (−TΔS) and enthalpy (ΔH). Thus, both the hydrophilic head and hydrophobic fatty acid tail of lysoPC are important for the weak interaction of lysoPC with AtLYSOPL2.
AtACBP2 contains a transmembrane domain, whereas AtLYSOPL2 possesses a lysoPC interactive hydrophobic region that is potentially involved in membrane binding, suggesting that AtACBP2 and AtLYSOPL2 perform their biological functions at cellular membranes (19). An Arabidopsis patatin-related phospholipase, pPLAIIIβ, was reported to be associated with the plasma membrane (50), resembling the co-localization of AtACBP2 and AtLYSOPL2 at the plasma membrane (19). pPLAIIIβ also accumulated in Arabidopsis roots resembling AtLYSOPL2 and is thus considered to act upstream of AtLYSOPL2 and AtACBP2 (50). LysoPC is a major signaling molecule in cellular membranes produced by phospholipases (3), in response to environmental stresses (6, 7). Taken together, the in vitro thermodynamic data suggest that AtACBP2 binds lysoPC to facilitate AtLYSOPL2 action following environmental stimuli (Fig. 7).
Figure 7.
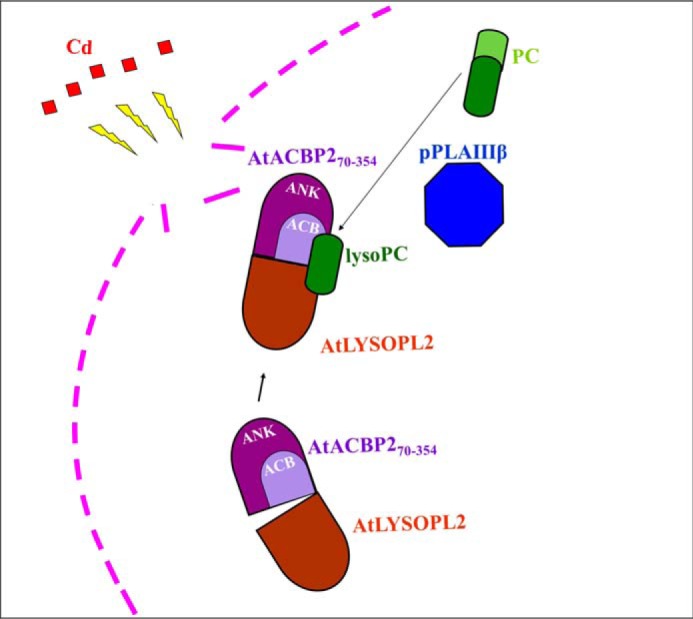
Proposed working model for AtLYSOPL2, AtACBP2, and lysoPC interaction. LysoPC, generated by pPLAIIIβ action on phosphatidylcholine (PC), binds to AtACBP2 and promotes the formation of an AtLYSOPL2–AtACBP2 complex. This complex could improve efficiency in membrane repair following Cd-induced stress. Cd, red; plasma membrane, pink; lysoPC, dark green; PC, light green; ankyrin-repeat (ANK) domain, purple; acyl-CoA-binding (ACB) domain, light purple; PLAIII β, blue; AtLYSOPL2, brown.
Experimental procedures
Generation of protein expression constructs
All primers are listed in Table S1. The AtLYSOPL2 and AtACBP2 cDNA fragments were amplified from plasmids pAT426 (19) and pAT377 (19), respectively, by PCR using Q5® high-fidelity polymerase (New England Biolabs) with denaturation at 95 °C for 5 min, followed by 31 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min 30 s, and an extension at 72 °C for 10 min. Site-directed mutagenesis of AtLYSOPL2 cDNA was performed according to Ho et al. (51). Overlapping forward and reverse oligonucleotide primers were designed at the target point mutation sites (Table S1). The PstI/NcoI-digested fragments encoding WT, D268A, H298A, and S147A AtLYSOPL2 (corresponding to aa 1–332) were inserted into the pRSETAHisSUMO expression vector (52) to generate plasmids pAT670, pAT780, pAT781, and pAT798, respectively. The AgeI/HindIII fragment encoding AtACBP270–354 (corresponding to aa 70–354, inclusive of the acyl-CoA–binding domain and the ankyrin repeats) was inserted into the pRSETAHisSUMO vector (52) to generate plasmid pAT672. The 435-bp cDNA fragment encoding AtACBP270–214 (corresponding to aa 70–214, inclusive of the acyl-CoA–binding domain) and the 421-bp fragment encoding AtACBP2215–354 (corresponding to aa 215–354 inclusive of the ankyrin repeats) were amplified from plasmid pAT672 and cloned into the pGEM®-T Easy vector (Promega) to generate plasmids pAT939 and pAT940, respectively. The BamHI/HindIII fragment and EcoRI/HindIII fragment were excised from pAT939 and pAT940 and inserted into the pRSETAHisSUMO vector (52) to generate plasmids pAT941 and pAT942, respectively. All plasmids were used for E. coli DH5α transformation. The transformants harboring the AtLYSOPL2 and AtACBP2 constructs were screened by colony-PCR using the T7/ML2041 and T7/ML2039 primer pairs, respectively. All constructs were verified by DNA sequencing across the cloning sites to ensure in-frame protein expression with the polyhistidine tag.
Protein expression
E. coli BL21 (DE3) pLysS was transformed with plasmids pAT670, pAT672, pAT780, pAT781, pAT798, pAT941, and pAT942. Transformants were selected on LB agar plates supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Ten single colonies per transformation were screened for the highest protein expression. A single colony of the high-expression clone was used to inoculate 50 ml of LB medium containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol and incubated with shaking overnight at 220 rpm at 37 °C. The pre-culture was used to inoculate 4 liters of LB medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 1:100 dilution. When the culture reached an OD600 value of 0.6, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.8 mm. The culture was grown for 16 h at 220 rpm at 16 °C for recombinant AtLYSOPL2 and 24 °C for recombinant AtACBP2. Cells were harvested by centrifugation at 9,000 rpm at 4 °C for 5 min. The cell pellet was used immediately or stored in −80 °C.
Protein purification
The cell pellet was resuspended in buffer A (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 50 mm imidazole). After sonication, the cell lysate was centrifuged at 18,000 × g at 4 °C for 50 min to spin down the cell debris. The cell-free extract was applied onto a buffer A-equilibrated nickel (Ni)-charged resin column. After washing with 10 column volumes of buffer A, buffer B (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 300 mm imidazole) was used to elute recombinant AtLYSOPL2 WT protein, AtLYSOPL2 mutant proteins (S147A, D268A, and H298A), and AtACBP270–354 protein. Buffer B eluate was collected. Dialysis was conducted using dialysis buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl) to remove imidazole. SUMO protease was applied at the same time to cleave the HisSUMO tag from the target protein. The dialyzed solution was applied onto a Ni-charged resin column again to remove the cleaved HisSUMO tag and SUMO protease. AtLYSOPL2 WT protein, AtLYSOPL2 mutant proteins (S147A, D268A, and H298A), and AtACBP270–354 protein were collected from the flow-through fraction.
Western blot analysis
The protein assay kit from Bio-Rad was used to estimate total protein separated on an SDS-polyacrylamide gel before electrophoretic transfer to the Hybond-C membrane (Amersham Biosciences) by the Trans-Blot Turbo Transfer System (Bio-Rad). Twenty μg of total protein was loaded per well in an SDS-polyacrylamide gel. AtLYSOPL2-specific antibodies generated previously in rabbits using a synthetic peptide (REWIDEKVKKYGSKT) corresponding to aa 317–331 of AtLYSOPL2 (19) were used to identify the AtLYSOPL2 cross-reacting bands in Western blot analysis. The ECL Western blotting detection kit (Amersham Biosciences) was used for detection of cross-reacting bands in Western blot analysis.
Lysophospholipase activity assays
LysoPC (C16:0) from soybean (Avanti Polar Lipids Inc.) was used as a substrate for measuring lysophospholipase activities by GC-MS following previous methods with modification (53, 54). Purified AtLYSOPL2 (50 μg) was assayed in 1 ml of reaction mixture containing 50 mm potassium-phosphate buffer, pH 8.0, 100 μm substrate, and 0.2% (v/v) Triton X-100. Gum arabic (5% w/v) was used to emulsify the substrate by sonication for 30 s before addition of AtLYSOPL2. The reaction was performed at 33 °C for 1 h with 100-μl aliquots withdrawn at 10-min intervals. Free fatty acids were purified from the reaction mixtures by TLC, and fatty acid methyl esters were prepared by methanolysis at 80 °C for 3 h in 5% (v/v) HCl/methanol, extracted with hexane, and then quantified by a trace GC-polaris Q mass spectrometer (Finnigan-Spectronex, ThermoFisher Scientific), equipped with a DB-5 column (30 m × 0.25 film thickness; J&W Scientific, Folsom, CA), following a temperature program described previously (55).
ITC measurements
The experiments were conducted using a MicroCal ITC200 system (GE Healthcare). Purified recombinant proteins were dialyzed against a buffer containing 20 mm Tris (pH 7.5) and 20 mm NaCl before ITC. AtACBP270–354 (0.4–0.5 mm), AtACBP270–214 lacking ankyrin repeats (0.4–0.5 mm), AtACBP2215–354 lacking the acyl-CoA–binding domain (0.4 mm), a mixture of AtACBP270–354 with lysoPC (C16:0) (molar ratio 1:1), AtACBP270–214–lysoPC complex (molar ratio 1:1), or AtACBP2215–35-lysoPC complex (molar ratio 1:1) was titrated automatically into a solution of AtLYSOPL2 WT protein, AtLYSOPL2 mutant proteins (S147A, D268A, and H298A) (30–40 μm), a mixture of AtLYSOPL2 with lysoPC (C16:0) (molar ratio 1:1), or a mixture of each AtLYSOPL2 mutant protein with lysoPC (C16:0), respectively. The interactions of AtACBP270–354 with lysoPC (C16:0) and C16:0–CoA (Avanti Polar Lipids Inc.) were also performed by ITC. AtACBP270–354 and C16:0–CoA interaction was measured by titrating 30–40 μm AtACBP270–354 in the chamber with 0.6–0.8 mm C16:0–CoA in the syringe. For AtACBP270–354 and lysoPC interaction, 30–40 μm AtACBP270–354 in the chamber was titrated with 0.6–0.8 mm lysoPC in the syringe, and the dialysis buffer was supplemented with 5% (v/v) acetonitrile (56). The interactions of AtLYSOPL2 WT protein or AtLYSOPL2 mutant proteins (S147A, D268A, and H298A) and lysoPC (C16:0) were each measured by titrating 30–40 mm AtLYSOPL2 WT protein or AtLYSOPL2 mutant proteins (S147A, D268A, and H298A) in the chamber with 0.6–0.8 mm lysoPC in the syringe, and the dialysis buffer was also mixed with 5% (v/v) acetonitrile. Typically, each injection was performed at 25 °C with 17 injections of 2-μl aliquots of the ligand into the protein solution (250 μl in the cell) at 300-s intervals. The association constant (KA), binding stoichiometry (n), and enthalpy changes (ΔH) upon binding were derived directly, and the Gibbs energy (ΔG) and entropy (−TΔS) changes were calculated from the equation, ΔG0 = ΔH0 − TΔS0 = −RTlnKA. Data were collected and analyzed using the ORIGIN software version 7.0 (MicroCal).
Homology modeling and dock simulation
The tertiary structural model of AtLYSOPL2 was created using the software MODELLER version 9.14. The X-ray crystal structure of human MGL (Protein Data Bank code 3JW8) served as a template for homology modeling (57–59). The lysoPC-binding pocket was identified using automated ligand docking by the software AutoDock version 4.0 (http://autodock.scripps.edu/)3 (60). The ligand of lysoPC (C16:0) was generated by the software ChemDraw version 8.0. The program allowed the ligand to be flexible, whereas the side chains in the protein were fixed. The grid box covered the entire protein of AtLYSOPL2. The ligand lysoPC (C16:0) was then docked using genetic algorithm/local search hybrid simulation. The lowest energy configuration was assumed to be the best docking pose for AtLYSOPL2.
Author contributions
R. M., X. D. L., and M.-L. C. conceptualization; X. D. L. and M.-L. C. resources; R. M., S.-C. L., X. L., X. D. L., and M.-L. C. data curation; R. M. and M.-L. C. software; R. M., S.-C. L., X. L., X. D. L., and M.-L. C. formal analysis; R. M., S.-C. L., X. D. L., and M.-L. C. validation; R. M., S.-C. L., X. L., X. D. L., and M.-L. C. investigation; R. M., S.-C. L., X. D. L., and M.-L. C. visualization; R. M., S.-C. L., X. L., X. D. L., and M.-L. C. methodology; R. M., X. D. L., and M.-L. C. writing-original draft; R. M. and M.-L. C. project administration; R. M., S.-C. L., X. D. L., and M.-L. C. writing-review and editing; X. D. L. and M.-L. C. supervision; M.-L. C. funding acquisition.
Supplementary Material
Acknowledgments
We are grateful to Wing-Ki Chan for technical support and Prof. Kam-Bo Wong for gift of the pRSETAHisSUMO vector.
This work was supported by the Wilson and Amelia Wong Endowment Fund, Research Grants Council of the Hong Kong Special Administrative Region, China, Grants 17101818, AoE/M-05/12, and AoE/M-403/16, and Innovation Technology Fund of the Innovation Technology Commission (Funding Support to State Key Laboratory of Agrobiotechnology in Hong Kong). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- LPL
- lysophospholipid
- lysoPC
- lysophosphatidylcholine
- ITC
- isothermal titration calorimetry
- aa
- amino acid
- ROS
- reactive oxygen species
- MAGL
- monoacylglycerol lipase
- MGL
- monoglyceride lipase
- ACBP
- acyl-CoA–binding protein
- ABA
- abscisic acid
- RCAR
- regulatory component of ABA receptor.
References
- 1. de Kroon A. I., Rijken P. J., and De Smet C. H. (2013) Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog. Lipid Res. 52, 374–394 10.1016/j.plipres.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 2. Grechkin A. (1998) Recent developments in biochemistry of the plant lipoxygenase pathway. Prog. Lipid Res. 37, 317–352 10.1016/S0163-7827(98)00014-9 [DOI] [PubMed] [Google Scholar]
- 3. Wang X. (2004) Lipid signaling. Curr. Opin. Plant Biol. 7, 329–336 10.1016/j.pbi.2004.03.012 [DOI] [PubMed] [Google Scholar]
- 4. Ryu S. B. (2004) Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9, 229–235 10.1016/j.tplants.2004.03.004 [DOI] [PubMed] [Google Scholar]
- 5. Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H. E., Rajashekar C. B., Williams T. D., and Wang X. (2002) Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002 10.1074/jbc.M205375200 [DOI] [PubMed] [Google Scholar]
- 6. D'Arrigo P., and Servi S. (2010) Synthesis of lysophospholipids. Molecules 15, 1354–1377 10.3390/molecules15031354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Torkhovskaya T. I., Ipatova O. M., Zakharova T. S., Kochetova M. M., and Khalilov E. M. (2007) Lysophospholipids receptors in cell signaling. Biochemistry 72, 125–131 [DOI] [PubMed] [Google Scholar]
- 8. Wi S. J., Seo Sy., Cho K., Nam M. H., and Park K. Y. (2014) Lysophosphatidylcholine enhances susceptibility in signaling pathway against pathogen infection through biphasic production of reactive oxygen species and ethylene in tobacco plants. Phytochemistry 104, 48–59 10.1016/j.phytochem.2014.04.009 [DOI] [PubMed] [Google Scholar]
- 9. Moolenaar W. H. (2000) Development of our current understanding of bioactive lysophospholipids. Ann. N.Y. Acad. Sci. 905, 1–10 [DOI] [PubMed] [Google Scholar]
- 10. Asaoka Y., Oka M., Yoshida K., Sasaki Y., and Nishizuka Y. (1992) Role of lysophosphatidylcholine in T-lymphocyte activation: involvement of phospholipase A2 in signal transduction through protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 89, 6447–6451 10.1073/pnas.89.14.6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murohara T., Scalia R., and Lefer A. M. (1996) Lysophosphatidylcholine promotes P-selectin expression in platelets and endothelial cells: possible involvement of protein kinase C activation and its inhibition by nitric oxide donors. Circ. Res. 78, 780–789 10.1161/01.RES.78.5.780 [DOI] [PubMed] [Google Scholar]
- 12. Wuhanqimuge, Itakura A., Matsuki Y., Tanaka M., and Arioka M. (2013) Lysophosphatidylcholine enhances NGF-induced MAPK and Akt signals through the extracellular domain of TrkA in PC12 cells. FEBS Open Bio. 3, 243–251 10.1016/j.fob.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makide K., Uwamizu A., Shinjo Y., Ishiguro J., Okutani M., Inoue A., and Aoki J. (2014) Novel lysophospholipid receptors: their structure and function. J. Lipid Res. 55, 1986–1995 10.1194/jlr.R046920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McIntyre T. M., Pontsler A. V., Silva A. R., St Hilaire A., Xu Y., Hinshaw J. C., Zimmerman G. A., Hama K., Aoki J., Arai H., and Prestwich G. D. (2003) Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARγ agonist. Proc. Natl. Acad. Sci. U.S.A. 100, 131–136 10.1073/pnas.0135855100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasternack S. M., von Kügelgen I., Al Aboud K., Lee Y. A., Rüschendorf F., Voss K., Hillmer A. M., Molderings G. J., Franz T., Ramirez A., Nürnberg P., Nothen M. M., and Betz R. C. (2008) G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat. Genet. 40, 329–334 10.1038/ng.84 [DOI] [PubMed] [Google Scholar]
- 16. Kawaguchi M., Okabe T., Okudaira S., Hanaoka K., Fujikawa Y., Terai T., Komatsu T., Kojima H., Aoki J., and Nagano T. (2011) Fluorescence probe for lysophospholipase C/NPP6 activity and a potent NPP6 inhibitor. J. Am. Chem. Soc. 133, 12021–12030 10.1021/ja201028t [DOI] [PubMed] [Google Scholar]
- 17. Wang A., and Dennis E. A. (1999) Mammalian lysophospholipases. Biochim. Biophys. Acta 1439, 1–16 10.1016/S1388-1981(99)00063-3 [DOI] [PubMed] [Google Scholar]
- 18. de Torres Zabela M., Fernandez-Delmond I., Niittyla T., Sanchez P., and Grant M. (2002) Differential expression of genes encoding Arabidopsis phospholipases after challenge with virulent or avirulent Pseudomonas isolates. Mol. Plant Microbe Interact. 15, 808–816 10.1094/MPMI.2002.15.8.808 [DOI] [PubMed] [Google Scholar]
- 19. Gao W., Li H. Y., Xiao S., and Chye M. L. (2010) Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J. 62, 989–1003 [DOI] [PubMed] [Google Scholar]
- 20. Kim R. J., Kim H. J., Shim D., and Suh M. C. (2016) Molecular and biochemical characterizations of the monoacylglycerol lipase gene family of Arabidopsis thaliana. Plant J. 85, 758–771 10.1111/tpj.13146 [DOI] [PubMed] [Google Scholar]
- 21. Bertrand T., Augé F., Houtmann J., Rak A., Vallée F., Mikol V., Berne P. F., Michot N., Cheuret D., Hoornaert C., and Mathieu M. (2010) Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 396, 663–673 10.1016/j.jmb.2009.11.060 [DOI] [PubMed] [Google Scholar]
- 22. Vanholme R., Cesarino I., Rataj K., Xiao Y., Sundin L., Goeminne G., Kim H., Cross J., Morreel K., Araujo P., Welsh L., Haustraete J., McClellan C., Vanholme B., Ralph J., et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106 10.1126/science.1241602 [DOI] [PubMed] [Google Scholar]
- 23. Gao W., Xiao S., Li H. Y., Tsao S. W., and Chye M. L. (2009) Arabidopsis thaliana acyl-CoA–binding protein ACBP2 interacts with heavy-metal–binding farnesylated protein AtFP6. New Phytol. 181, 89–102 10.1111/j.1469-8137.2008.02631.x [DOI] [PubMed] [Google Scholar]
- 24. Clemens S. (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88, 1707–1719 10.1016/j.biochi.2006.07.003 [DOI] [PubMed] [Google Scholar]
- 25. Wong C. K. E., and Cobbett C. S. (2009) HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 181, 71–78 10.1111/j.1469-8137.2008.02638.x [DOI] [PubMed] [Google Scholar]
- 26. Clemens S., and Ma J. F. (2016) Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 67, 489–512 10.1146/annurev-arplant-043015-112301 [DOI] [PubMed] [Google Scholar]
- 27. Park J., Song W. Y., Ko D., Eom Y., Hansen T. H., Schiller M., Lee T. G., Martinoia E., and Lee Y. (2012) The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288 10.1111/j.1365-313X.2011.04789.x [DOI] [PubMed] [Google Scholar]
- 28. Khairnar N. P., Joe M. H., Misra H. S., Lim S. Y., and Kim D. H. (2013) FrnE, a cadmium-inducible protein in Deinococcus radiodurans, is characterized as a disulfide isomerase chaperone in vitro and for its role in oxidative stress tolerance in vivo. J. Bacteriol. 195, 2880–2886 10.1128/JB.01503-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DalCorso G., Farinati S., Maistri S., and Furini A. (2008) How plants cope with cadmium: staking all on metabolism and gene expression. J. Integr. Plant Biol. 50, 1268–1280 10.1111/j.1744-7909.2008.00737.x [DOI] [PubMed] [Google Scholar]
- 30. Verbruggen N., Hermans C., and Schat H. (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 12, 364–372 10.1016/j.pbi.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 31. Zhai Z., Gayomba S. R., Jung H. I., Vimalakumari N. K., Piñeros M., Craft E., Rutzke M. A., Danku J., Lahner B., Punshon T., Guerinot M. L., Salt D. E., Kochian L. V., and Vatamaniuk O. K. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26, 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Michele R., Vurro E., Rigo C., Costa A., Elviri L., Di Valentin M., Careri M., Zottini M., Sanità di Toppi L., and Lo Schiavo F. (2009) Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol. 150, 217–228 10.1104/pp.108.133397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kulik A., Anielska-Mazur A., Bucholc M., Koen E., Szymanska K., mieńko A., Krzywińska E., Wawer I., McLoughlin F., Ruszkowski D., Figlerowicz M., Testerink C., Sklodowska A., Wendehenne D., and Dobrowolska G. (2012) SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 160, 868–883 10.1104/pp.112.194472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., and Provart N. J. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pierce M. M., Raman C. S., and Nall B. T. (1999) Isothermal titration calorimetry of protein–protein interactions. Methods 19, 213–221 10.1006/meth.1999.0852 [DOI] [PubMed] [Google Scholar]
- 36. Abdullah N., Padmanarayana M., Marty N. J., and Johnson C. P. (2014) Quantitation of the calcium and membrane binding properties of the C2 domains of dysferlin. Biophys. J. 106, 382–389 10.1016/j.bpj.2013.11.4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang A., Deems R. A., and Dennis E. A. (1997) Cloning, expression, and catalytic mechanism of murine lysophospholipase I. J. Biol. Chem. 272, 12723–12729 10.1074/jbc.272.19.12723 [DOI] [PubMed] [Google Scholar]
- 38. Wang A., Loo R., Chen Z., and Dennis E. A. (1997) Regiospecificity and catalytic triad of lysophospholipase I. J. Biol. Chem. 272, 22030–22036 10.1074/jbc.272.35.22030 [DOI] [PubMed] [Google Scholar]
- 39. Meier R., Drepper T., Svensson V., Jaeger K. E., and Baumann U. (2007) A calcium-gated lid and a large β-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens. J. Biol. Chem. 282, 31477–31483 10.1074/jbc.M704942200 [DOI] [PubMed] [Google Scholar]
- 40. Gutiérrez-Fernández J., Vaquero M. E., Prieto A., Barriuso J., Martínez M. J., and Hermoso J. A. (2014) Crystal structures of Ophiostoma piceae sterol esterase: structural insights into activation mechanism and product release. J. Struct. Biol. 187, 215–222 10.1016/j.jsb.2014.07.007 [DOI] [PubMed] [Google Scholar]
- 41. Vijayaraj P., Jashal C. B., Vijayakumar A., Rani S. H., Venkata Rao D. K., and Rajasekharan R. (2012) A bifunctional enzyme that has both monoacylglycerol acyltransferase and acyl hydrolase activities. Plant Physiol. 160, 667–683 10.1104/pp.112.202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee L. C., Lee Y. L., Leu R. J., and Shaw J. F. (2006) Functional role of catalytic triad and oxyanion hole-forming residues on enzyme activity of Escherichia coli thioesterase I/protease I/phospholipase L1. Biochem. J. 397, 69–76 10.1042/BJ20051645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karlsson M., Contreras J. A., Hellman U., Tornqvist H., and Holm C. (1997) cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase: evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 272, 27218–27223 10.1074/jbc.272.43.27218 [DOI] [PubMed] [Google Scholar]
- 44. Hariharan P., Balasubramaniam D., Peterkofsky A., Kaback H. R., and Guan L. (2015) Thermodynamic mechanism for inhibition of lactose permease by the phosphotransferase protein IIAGlc. Proc. Natl. Acad. Sci. U.S.A. 112, 2407–2412 10.1073/pnas.1500891112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., and Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 46. Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S. Y., Márquez J. A., Cutler S. R., and Rodriguez P. L. (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60, 575–588 10.1111/j.1365-313X.2009.03981.x [DOI] [PubMed] [Google Scholar]
- 47. Lung S. C., Liao P., Yeung E. C., Hsiao A. S., Xue Y., and Chye M. L. (2017) Acyl-CoA-binding protein ACBP1 modulates sterol synthesis during embryogenesis. Plant Physiol. 174, 1420–1435 10.1104/pp.17.00412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lung S. C., Liao P., Yeung E. C., Hsiao A. S., Xue Y., and Chye M. L. (2018) Arabidopsis ACYL-COA–BINDING PROTEIN1 interacts with STEROL C4–METHYL OXIDASE1–2 to modulate gene expression of homeodomain-leucine zipper IV transcription factors. New Phytol. 218, 183–200 10.1111/nph.14965 [DOI] [PubMed] [Google Scholar]
- 49. Taskinen J. P., van Aalten D. M., Knudsen J., and Wierenga R. K. (2007) High resolution crystal structures of unliganded and liganded human liver ACBP reveal a new mode of binding for the acyl-CoA ligand. Proteins 66, 229–238 [DOI] [PubMed] [Google Scholar]
- 50. Li M., Bahn S. C., Guo L., Musgrave W., Berg H., Welti R., and Wang X. (2011) Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23, 1107–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 52. Fong Y. H., Wong H. C., Chuck C. P., Chen Y. W., Sun H., and Wong K. B. (2011) Assembly of preactivation complex for urease maturation in Helicobacter pylori: crystal structure of UreF–UreH protein complex. J. Biol. Chem. 286, 43241–43249 10.1074/jbc.M111.296830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hong Y., Wang T. W., Hudak K. A., Schade F., Froese C. D., and Thompson J. E. (2000) An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proc. Natl. Acad. Sci. U.S.A. 97, 8717–8722 10.1073/pnas.140213697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., and Okada K. (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hsu K. H., Wang S. Y., Chu F. H., and Shaw J. F. (2010) Characterization and heterologous expression of a novel lysophospholipase gene from Antrodia cinnamomea. J. Appl. Microbiol. 108, 1712–1722 10.1111/j.1365-2672.2009.04569.x [DOI] [PubMed] [Google Scholar]
- 56. Suzuki T., Shibuya Y., Sato T., Nishizawa S., Sato I., and Yamaguchi A. (2016) Thermodynamics of complexation between thiourea-based receptor and acetate in water/acetonitrile mixture. Anal. Sci. 32, 741–744 10.2116/analsci.32.741 [DOI] [PubMed] [Google Scholar]
- 57. Fiser A., and Šali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 10.1016/S0076-6879(03)74020-8 [DOI] [PubMed] [Google Scholar]
- 58. Sali A., Potterton L., Yuan F., van Vlijmen H., and Karplus M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23, 318–326 10.1002/prot.340230306 [DOI] [PubMed] [Google Scholar]
- 59. Sánchez R., and Šali A. (2000) Comparative protein structure modeling: introduction and practical examples with modeller. Methods Mol. Biol. 143, 97–129 [DOI] [PubMed] [Google Scholar]
- 60. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tzeng S. R., and Kalodimos C. G. (2009) Dynamic activation of an allosteric regulatory protein. Nature 462, 368–372 10.1038/nature08560 [DOI] [PubMed] [Google Scholar]
- 62. Tzeng S. R., and Kalodimos C. G. (2012) Protein activity regulation by conformational entropy. Nature 488, 236–240 [DOI] [PubMed] [Google Scholar]
- 63. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.