Abstract
Dehydrins are intrinsically disordered proteins, generally expressed in plants as a response to embryogenesis and water-related stress. Their suggested functions are in membrane stabilization and cell protection. All dehydrins contain at least one copy of the highly conserved K-segment, proposed to be a membrane-binding motif. The dehydrin Lti30 (Arabidopsis thaliana) is up-regulated during cold and drought stress conditions and comprises six K-segments, each with two adjacent histidines. Lti30 interacts with the membrane electrostatically via pH-dependent protonation of the histidines. In this work, we seek a molecular understanding of the membrane interaction mechanism of Lti30 by determining the diffusion and molecular organization of Lti30 on model membrane systems by imaging total internal reflection– fluorescence correlation spectroscopy (ITIR-FCS) and molecular dynamics (MD) simulations. The dependence of the diffusion coefficient explored by ITIR-FCS together with MD simulations yields insights into Lti30 binding, domain partitioning, and aggregation. The effect of Lti30 on membrane lipid diffusion was studied on fluorescently labeled supported lipid bilayers of different lipid compositions at mechanistically important pH conditions. In parallel, we compared the mode of diffusion for short individual K-segment peptides. The results indicate that Lti30 binds the lipid bilayer via electrostatics, which restricts the mobility of lipids and bound protein molecules. At low pH, Lti30 binding induced lipid microdomain formation as well as protein aggregation, which could be correlated with one another. Moreover, at physiological pH, Lti30 forms nanoscale aggregates when proximal to the membrane suggesting that Lti30 may protect the cell by “cross-linking” the membrane lipids.
Keywords: membrane lipid, intrinsically disordered protein, histidine, lipid bilayer, fluorescence correlation spectroscopy (FCS), molecular dynamics, Diffusion, disorder, K-segments, Lti30, supported lipid bilayers
Introduction
Dehydrins are intrinsically disordered stress-related plant proteins that are up-regulated in cold and drought conditions (1, 2). Most of the dehydrins are composed of highly conserved sequences (S-, K-, and Y-segments) implicated in the function of these proteins (3–5). Dehydrins are extremely hydrophilic and abundant in charged residues. Although there are several experimental reports on the involvement of K-segment–membrane interactions (5–7), the molecular mechanism of how dehydrins perform cell-protective functions with their highly conserved sequences is still elusive. It has been reported for other intrinsically disordered proteins, e.g. α-synuclein, that in crowded environments they tend to gain structure or form aggregates (8, 9). However, in the case of dehydrins, it was observed that even in the presence of osmolytes or at higher concentrations of macromolecules, which might occur in the case of cellular desiccation, they do not undergo structural collapse and tend to maintain their disordered state (10). These proteins need to interact with their specific binding targets to fold and gain their biological function. It has been proposed that the function of dehydrins primarily depends on the interaction of these proteins with their specific biological targets, for example certain proteins (11, 12), metals (13), DNA/RNA (14), and membranes (15, 16). Membranes are specifically prone to be affected by environmental changes such as temperature and water content (17, 18). Some dehydrins are found localized in the vicinity of the membranes (16), and their interaction changes membrane properties such as the phase-transition temperature (6). It has also been reported that they interact with the charged lipids in membranes via electrostatic interactions (6, 19, 20). Lti30 is one such dehydrin found in Arabidopsis thaliana that shows very high expression at cold temperatures and during desiccation (21). It is composed of six conserved K-segments flanked with pairs of histidines that have been found to be responsible for membrane binding (6). Surface plasmon resonance studies have shown that Lti30 actively interacts with negatively charged lipids, thereby inducing vesicle aggregation, whereas it only weakly interacts with zwitterionic lipids (6). It has also been demonstrated by in vitro studies that Lti30 lowers the phase-transition temperature of the interacting lipids, which could be important for cold tolerance (6). A similar effect of the dehydrin K2 on the transition temperature of lipids has been shown (5). Literature suggests that the factors that influence the membrane binding of Lti30 include the protonation state of histidines flanking the K-segments, phosphorylation of Lti30 by protein kinase C on three sites within the K-segments and six sites in between the K-segments, and cleavage of Lti30 by proteases (6). Eriksson et al. (7) reported in an NMR-based study that K-segments with flanking histidines undergo conformational changes from an unstructured protein to an amphipathic helix upon membrane binding and then float on the membrane outer leaflet. It was also hypothesized that Lti30 can form oligomers or can bind other biological targets that might be important for its function. There are still questions regarding the impact of Lti30 upon binding to the membrane and whether the Lti30 conformation depends on the membrane composition.
In this study, we have used single-molecule methods to gain further insights into the membrane-interaction mechanism of Lti30. We used imaging total internal reflection–fluorescence correlation spectroscopy (ITIR-FCS)4 in combination with atomic-scale explicitly solvated MD simulations to study the effect of Lti30 on membrane lipid diffusion and membrane organization. We have also explored the conformation of individual K-segments with and without histidines within the lipid membrane environment. ITIR-FCS is a multiplexed camera-based imaging FCS modality that provides spatially resolved diffusion coefficient maps (23). This method employs total internal reflection illumination, which specifically excites molecules close to the surface, and therefore allows measurements primarily at the membrane and avoids background signal contributions by the bulk solution. However, this method is diffraction-limited, so it is unable to provide information about structures smaller than 240 nm. For this reason, ITIR-FCS is combined with the FCS Diffusion Laws to probe organization of fluorescently labeled particles below the diffraction limit. MD simulation is a theoretical approach that enables the study of the conformational dynamics of single molecules or complexes thereof in atomic resolution, which therefore provides information complementary to single molecule fluorescence experiments (24).
Our aim is to understand the molecular basis of Lti30 organization on, and its interaction with, membranes composed of different lipid types. Our results show that the membrane lipid diffusion is affected upon interaction with Lti30 in a histidine-protonation–dependent manner. Also, at low pH and upon Lti30 binding, lipid microdomains are detected. Both the presence of charged lipids and lipid fatty acid saturation alter the diffusion and binding. Notably, Lti30 is able to sense bilayer packing defects and bind to DOPC:DPPC bilayers by restricting the diffusion of membrane lipids, whereas no effect was found with DOPC bilayers. Moreover, the experimental results show that dehydrins are able to bind both zwitterionic and anionic lipids. As reported earlier, Lti30 changes its conformation in the presence of membranes from an unstructured to helical conformation. MD simulations confirm those findings and correlate K-segment helicity with the membrane composition and pH. In addition to the conformational changes observed upon membrane binding, ITIR-FCS experiments indicate that protein–protein interactions exist, which is subsequently validated using thioflavin T fluorescence experiments. Thioflavin T–positive results could imply fibril formation in Lti30. These findings indicate that Lti30 and other membrane-binding dehydrins can maintain the cell integrity during cold temperatures and drought by modulating the membrane structure and possibly via lipid “cross-linking.”
Results
Effect of Lti30 on membrane lipid diffusion is pH- and lipid composition–dependent
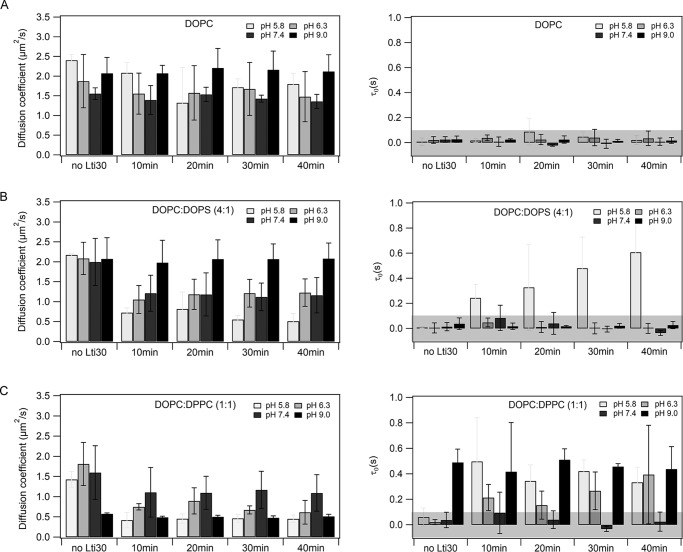
ITIR-FCS provides two important results on a sample: the spatial diffusion coefficient (D) map, and a measure of the diffusive mode of the fluorescently labeled particle provided by the diffusion law intercept (τ0, see “Experimental procedures”). In brief, values of τ0 in the range of 0 ± 0.1 indicate free Brownian diffusion; values larger 0.1 indicate diffusion with intermittent domain trapping (25). To test the effect of Lti30 on membrane lipid diffusion, ITIR-FCS measurements were performed for over 40 min on three rhodamine-PE–labeled supported lipid bilayers varying in packing and charge distribution, namely DOPC, DOPC:DOPS (4:1), and DOPC:DPPC (1:1).
The phase-transition temperature (Tm) of the constituent lipids determines the phase behavior of the membrane. DOPC bilayers are composed of zwitterionic lipids (Tm = −11 °C) and exhibit a liquid disordered phase at room temperature. DOPC:DOPS (4:1) bilayers comprise 20% negatively charged lipids and 80% zwitterionic lipids (Tm = −11 °C for DOPC and −17 °C for DOPS lipids). They also exist in a liquid-disordered phase. DOPC:DPPC (1:1) bilayers contain two types of structurally different zwitterionic lipids (Tm = −11 °C for DOPC and 41 °C for DPPC lipids) exhibiting a solid ordered–liquid disordered phase at room temperature. It is important to note that there was no visible phase separation for this bilayer composition on glass. In a previous study from our group, it was shown that D for DOPC:DPPC and DLPC:DPPC could be correlated to the line tension at the domain boundaries suggesting that Rho-PE partitions in both phases (26). However, we cannot eliminate the influence of differential Rho-PE partitioning in DOPC:DPPC SLBs on membrane dynamics readouts.
For each, measurements were conducted at four different pH conditions (5.8, 6.3, 7.4, and 9.0), as the intrinsic pKa value of histidine side chains is around 6.5, and the protonation state of the histidines is important for Lti30 binding and was shown to induce aggregation of negatively charged lipid vesicles (6).
In the case of DOPC bilayers, Lti30 binding reduced D of membrane lipids from 2.40 ± 0.15 to 1.74 ± 0.27 μm2/s (i.e. 30%) at pH 5.8, and at pH >5.8, the variation of D was within the experimental error limits (Fig. 1A and Table 1). The τ0 values remain within 0 ± 0.1 s showing no influence of Lti30 on the DOPC membrane organization. In the case of DOPC:DOPS (4:1) bilayers, D of membrane lipids undergoes a drop of 50–75% upon interaction with Lti30 for pH ≤7.4 (Fig. 1B and Table 1). An increase of τ0 is observed at pH 5.8 only, whereas at pH >5.8 the τ0 remains within the regime of free diffusion. The maximum influence of Lti30 binding is observed at pH 5.8 as D is reduced from 2.17 ± 0.3 to 0.51 ± 0.19 μm2/s (i.e. ∼75%), and there is an increase in τ0 from 0.006 ± 0.005 to 0.60 ± 0.30 s, which indicates the formation of some microdomains in the lipid bilayer (Fig. 1B and Table 1). At pH 9, there are no changes detected (Fig. 1B). In the case of DOPC:DPPC (1:1), D of membrane lipids decreases for pH ≤7.4 (Fig. 1C and Table 1). There is a 60% drop of D at pH 5.8 and 6.3, and a 30% drop at pH 7.4. In addition, there is a rise in τ0 from 0 to 0.4 s at pH 5.8 and 6.3, i.e. there is microdomain formation in the membrane. At pH 9, pure DOPC:DPPC (1:1) bilayers exhibit a positive τ0 of 0.5 s, i.e. there are microdomains on the membrane, as expected for this composition (27), and addition of Lti30 does not influence the bilayer. These results verify earlier reports that Lti30 binding is protonation state-dependent and show that Lti30 can induce lipid microdomain formation upon binding, at least in the case of DOPC:DPPC and DOPC:DOPS membranes at acidic pH.
Figure 1.
ITIR-FCS data of Lti30 binding with supported lipid bilayers. The effect of Lti30 on lipid mobility in supported lipid bilayers composed of different compositions and at pH 5.8, 6.3, 7.4, and 9.0 was measured. The measurements were performed at a region of 21 × 21 pixels in the form of 50,000 frames at a time exposure of 1 ms. The measurements were done up to 40 min after the addition of Lti30 over the membrane. Membrane lipid diffusion is quantified using two parameters diffusion coefficient (D) and diffusion law intercept (τ0). The protein–to–lipid ratio used is 1:130. A, DOPC; B, DOPC:DOPS (4:1); C, DOPC:DPPC (1:1). Error bars are given as standard deviations (S.D.). The experiments have been repeated three times to ensure the reproducibility. The gray-shaded area in τ0 graphs represents the range of margin of error in our setup (−0.1 s < 0 < 0.1 s) in which the particle is considered to undergo free diffusion. For representative raw data see Figs. S1–S3.
Table 1.
Lateral diffusion (D) of membrane lipids and FCS diffusion law intercept τ0 obtained from ITIR-FCS measurements on rhodamine-PE–labeled supported lipid bilayers DOPC, DOPC:DOPS (4:1), and DOPC:DPPC (1:1) interacting with Lti30
Error bars are given as S.D. calculated from three independent experiments.
| Lipid composition | pH | Dno Lti30 | D40 min | τ0, no Lti30 | τ0, 40 min |
|---|---|---|---|---|---|
| μm2/s | μm2/s | s | s | ||
| DOPC | 5.8 | 2.40 ± 0.15 | 1.79 ± 0.27 | 0.01 ± 0.02 | 0.02 ± 0.03 |
| 6.3 | 1.87 ± 0.67 | 1.47 ± 0.63 | 0.02 ± 0.03 | 0.03 ± 0.05 | |
| 7.4 | 1.55 ± 0.14 | 1.35 ± 0.17 | 0.02 ± 0.02 | 0.01 ± 0.02 | |
| 9.0 | 2.07 ± 0.40 | 2.11 ± 0.42 | 0.02 ± 0.02 | 0.02 ± 0.02 | |
| DOPC:DOPS (4:1) | 5.8 | 2.17 ± 0.03 | 0.51 ± 0.19 | 0.01 ± 0.01 | 0.60 ± 0.30 |
| 6.3 | 2.08 ± 0.40 | 1.22 ± 0.34 | 0.00 ± 0.04 | 0.00 ± 0.04 | |
| 7.4 | 1.99 ± 0.59 | 1.16 ± 0.44 | 0.01 ± 0.03 | −0.04 ± 0.02 | |
| 9.0 | 2.07 ± 0.53 | 2.08 ± 0.39 | 0.04 ± 0.05 | 0.03 ± 0.03 | |
| DOPC:DPPC (1:1) | 5.8 | 1.42 ± 0.19 | 0.45 ± 0.09 | 0.06 ± 0.07 | 0.33 ± 0.12 |
| 6.3 | 1.81 ± 0.53 | 0.61 ± 0.29 | 0.02 ± 0.02 | 0.39 ± 0.38 | |
| 7.4 | 1.59 ± 0.66 | 1.09 ± 0.45 | 0.03 ± 0.06 | 0.02 ± 0.07 | |
| 9.0 | 0.57 ± 0.02 | 0.51 ± 0.05 | 0.49 ± 0.10 | 0.43 ± 0.17 |
Flanking histidines are important for the membrane binding conformation of Lti30
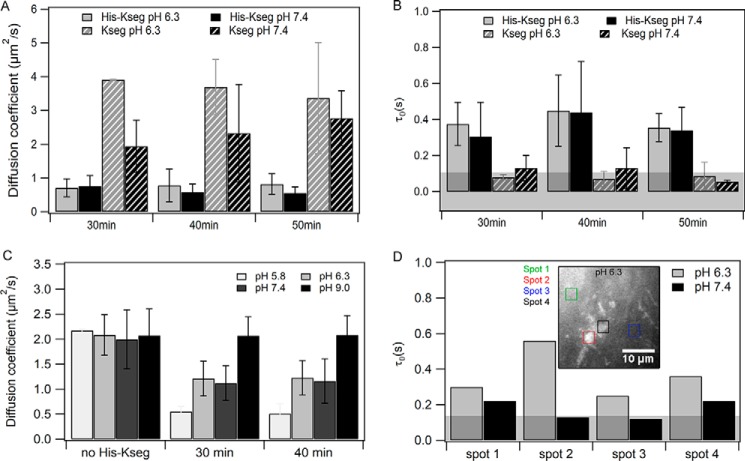
Next, we investigated which elements of Lti30 are responsible for its influence on bilayers. For this purpose, we measured the diffusion of rhodamine-labeled His–K-segments and K-segments upon interaction with unlabeled DOPC:DOPS (4:1) bilayers at pH 6.3 and 7.4, when the histidines are partially protonated.
Interestingly, D values of His–K-segments are almost equal to that of membrane lipids in the peptide-bound state of the bilayers (Fig. 2C), which indicates that these peptides are localized in the membrane environment (Fig. 2A). In addition, τ0 is positive, which indicates diffusion with intermittent domain trapping (Fig. 2B). Consistent with these observations, the membrane is not uniformly labeled but exhibits areas of different fluorescence intensity with some spots exhibiting higher rhodamine intensity than others, possibly indicating peptide aggregation as a reason for the domain-entrapped diffusion. We therefore performed measurements at various spots on the same membrane. Measurements at brighter spots yielded a higher τ0 value than what was obtained at less bright spots (Fig. 2D). This indicates that at brighter spots there is a higher degree of aggregation/oligomerization than at less bright spots.
Figure 2.
Comparison of the diffusion of K-segment and His–K-segment on DOPC:DOPS (4:1) bilayer at pH 6.3 and 7.4. The protein–to–lipid ratio used was 1:130. A, diffusion coefficient (D) of fluorescently labeled K-segment/HH on DOPC:DOPS (4:1) bilayer. B, diffusion law intercept (τ0) of fluorescently labeled K-segment/HH on the membrane. C, diffusion coefficient of membrane lipids bound to the peptide. Error bars for A–C are given as S.D. calculated from three independent experiments. D, diffusion law intercept (τ0) of fluorescently labeled His–K-segment on the membrane at different spots (one representative set). Inset is the image of the DOPC:DOPS (4:1) membrane bound with rhodamine-labeled with His–K-segment at pH 6.3. Those spots are marked for which τ0 values are reported. For representative raw data (images, ACF, and diffusion law plots) see Fig. S4.
In the case of K-segments, D is ∼3-fold higher at pH 7.4 and ∼4-fold higher at pH 6.3 (Fig. 2A) as compared with membrane lipids in a peptide-bound state. In this case, τ0 is within a range of 0 ± 0.1 s, which indicates that peptides are freely diffusing on the membrane but are not integrated into the bilayer (Fig. 2B). The autocorrelation curves recorded at pH 5.8 were excessively noisy, because of a higher degree of aggregation on the membrane, so the resulting diffusion parameters have higher errors.
Protonation driven aggregation of His–K-segment in solution
Based on structural predictions and experiments, it was earlier suggested that Lti30 has a very low aggregation propensity (3). This was attributed to its sequence, which comprises charged and hydrophilic residues. For dehydrins specifically, it was suggested that they are not prone to nonspecific structural collapse and tend to stay in their disordered form rather than attaining any tertiary structure in solution (3). The signatures of His-K–segment aggregation on the membrane compelled us to study the diffusion of these labeled peptides in solution at different pH conditions. The diffusion of individual peptides was measured using a confocal-FCS setup.
The average diffusion coefficient of the His–K-segment in solution is 173 ± 9 μm2/s, which is ∼36 μm2/s lower as compared with K-segments, i.e. 209 ± 14 μm2/s. This difference in diffusion coefficient could be an indication of some conformational difference between the two peptides. An interesting observation obtained from these measurements was that the autocorrelation function (ACF) for His–K-segments at pH 5.8 is better fitted with a two-particle model with the second component having a slower diffusion coefficient of ∼0.02 ± 0.01 μm2/s. However, the ACFs for the His–K-segment at pH 6.3, 7.4, and 9.0 could be fitted to a one-particle model (Table 2). The slower diffusing component indicates peptide aggregation at pH 5.8. For the K-segment, the ACFs for all pH conditions could be fitted with a one-particle model. This is an indication that, even in solution, the protonation of histidines drives the aggregation of K-segments.
Table 2.
Diffusion coefficient of rhodamine-labeled K–segment and His-K–segment peptides in solution
Error bars are given as S.D. calculated from five independent experiments. For representative raw data, see Fig. S5. The experiments were done on a confocal-FCS setup.
| Dfast | Dslow | |
|---|---|---|
| μm2/s | μm2/s | |
| His–K-segment, pH 5.8 | 181.26 ± 3.34 | 0.02 ± 0.01 |
| His–K-segment, pH 6.3 | 167.16 ± 10.19 | |
| His–K-segment, pH 7.4 | 169.12 ± 5.98 | |
| His–K-segment, pH 9.0 | 174.34 ± 11.39 | |
| K-segment, pH 5.8 | 204.50 ± 6.75 | |
| K-segment, pH 6.3 | 204.01 ± 10.97 | |
| K-segment, pH 7.4 | 198.42 ± 6.97 | |
| K-segment, pH 9.0 | 206.58 ± 31.56 |
Protonation driven aggregation of Lti30 on the membrane
Previous reports show that His–K-segments adopt a helical conformation upon binding the membrane and float over the membrane surface, with no visible oligomerization or aggregation (7). Here, we detected domain-entrapped diffusion for pH ≤7.4 but with large aggregates visible (Fig. 3) at pH 5.8. Next, we investigated whether aggregation observed in the case of individual His–K-segments is also seen in the case of full-length Lti30 and whether it shows pH dependence. To validate the occurrence of aggregation in full-length Lti30 on the membrane we performed a ThT assay using TIRF microscopy. We use ThT as a fluorescent marker, which shows a significant increase in the intensity when in contact with protein aggregates (28). ThT was added on the supported lipid bilayer interacting with Lti30 at different pH conditions. The images were recorded 60 min after the addition of Lti30.
Figure 3.
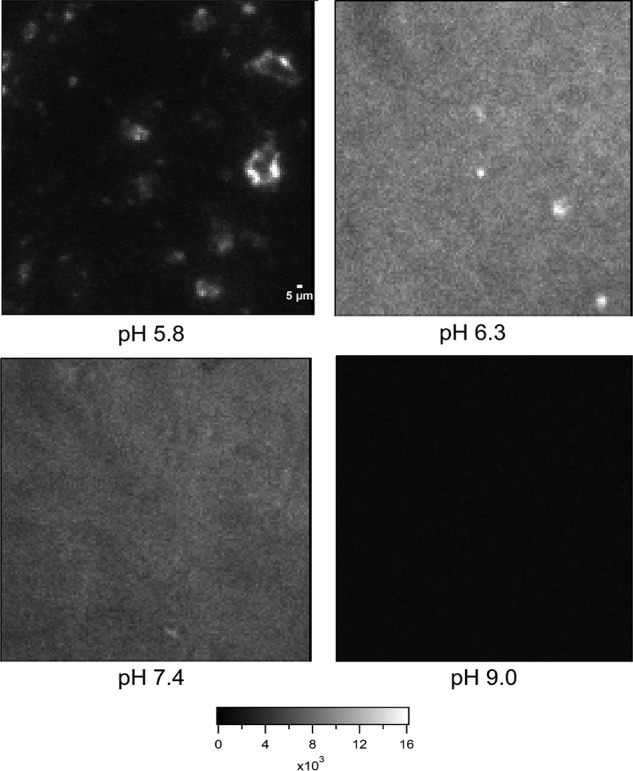
Visualization of Lti30 aggregates on DOPC:DOPS (4:1) membrane at pH 5.8, 6.3, 7.4, and 9.0 using thioflavin T dye on a TIRF microscope. First, Lti30 was incubated with the supported lipid bilayer for 60 min. Then, 5 μm of ThT dye was added, and images were recorded. Scale bar, 5 μm.
As shown in the images (Fig. 3), at pH 5.8, discrete micrometer-sized aggregates of ThT fluorescence can be observed. At pH 6.3 and 7.4, the surface looks homogeneous with few large bright spots. This indicates that there are small aggregates homogeneously spread over the whole membrane. At pH 9.0, the ThT signal is very low, consistent with the absence of Lti30 aggregation and binding.
These experiments confirm that Lti30 shows protonation-dependent aggregation on the membrane. Lti30 forms large aggregates when the histidines are fully protonated, and they form microdomains on the membrane along with a drop in D of membrane lipids. In the partially protonated state, there are smaller (typically smaller than 240 nm) Lti30 aggregates that are spread all over the membrane surface, which leads to decreased D but no permanent microdomain formation.
MD simulations highlight the electrostatic interactions of K-segment and His–K-segment binding to membranes
To gain in-depth atomic-resolution insights into the FCS experimental data, we employed MD simulations to probe peptide–membrane interactions. Because the sequence of Lti30 mainly comprises K-segments each with flanking histidines, we initially performed MD simulations on individual K-segments and His–K-segments, under conditions of extreme pH (pH values of approximately ∼4.0 and ∼10.0), to reduce computational cost and ensure efficient conformational sampling. Simulations were performed for representative lipid membrane compositions as used in equivalent fluorescence experiments, including: (i) DOPC; (ii) DOPC:DOPS (4:1); and (iii) DOPC:DPPC (1:1) (see under “Experimental procedures” for further details). This resulted in 12 peptide–membrane assembly equilibrium simulations (Table 3). All simulations were run with an ∼1:130 peptide:lipid ratio to match experimental ratios.
Table 3.
Summary of peptide–membrane MD simulations performed under extreme pH conditions in this study
| System | Sequence | pH | Overall peptide charge | Lipid bilayer | Simulation time |
|---|---|---|---|---|---|
| μs | |||||
| K-segment | EKKGMTEKVMEQLPG | 4 | +3 | DOPC | 1 |
| DOPC:DOPS (4:1) | 1 | ||||
| DOPC:DPPC (1:1) | 1 | ||||
| 10 | −3 | DOPC | 1 | ||
| DOPC:DOPS (4:1) | 1 | ||||
| DOPC:DPPC (1:1) | 1 | ||||
| His–K-segment | VHEKKGMTEKVMEQLPGHHG | 4 | +6 | DOPC | 1 |
| DOPC:DOPS (4:1) | 1 | ||||
| DOPC:DPPC (1:1) | 1 | ||||
| 1010 | −3 | DOPC | 1 | ||
| DOPC:DOPS (4:1) | 1 | ||||
| DOPC:DPPC (1:1) | 1 |
In the case of pH 4 with DOPC membranes, it was observed that the K-segment (overall peptide charge of +3) localized mostly to the region around the outer leaflet, near the phosphate groups. This is indicated by the two-dimensional partial mass density of the peptide averaged over the last 0.2 μs and the final simulation snapshots (Fig. 4A). At the same pH in the case of DOPC:DOPS (4:1), the K-segment was located slightly deeper, which indicates an increased electrostatic interaction with the membrane containing anionic lipids. In the case of DOPC:DPPC (1:1), the K-segment peptide was mostly found just above the lipid headgroups suggesting a lowered interaction with this membrane. In the case of the His–K-segments at acidic pH (overall peptide charge of +6), it appeared that the peptide also favors the lipid headgroup–water interface for pure DOPC membranes (Fig. 4B). The His–K-segment was found at a similar depth as the K-segment interacting with DOPC membranes (Fig. 4, A and B). In the case of DOPC:DOPS (4:1) and DOPC:DPPC (1:1), the averaged position density for the His–K-segment was slightly lower, below the phosphate region, suggesting a stronger interaction with these membranes. In contrast, at pH 10, it was observed that the K-segment (overall peptide charge of −3) favors the water environment and does not bind any of the membrane systems (Fig. 5A). At the same pH, the His–K-segment (similarly, overall peptide charge of −3) was observed just above the lipid phosphates in the case of DOPC and DOPC:DPPC (1:1) (Fig. 5B). In the case of DOPC:DOPS (4:1) the peptide did not bind the membrane, remaining in the solvent environment.
Figure 4.
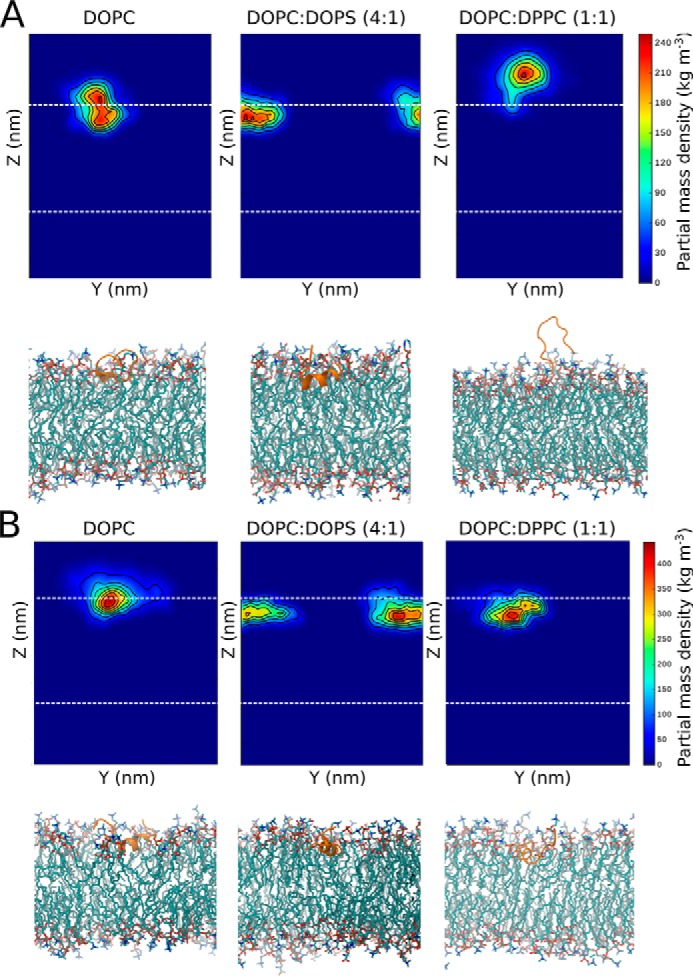
Peptide–membrane interactions during simulations at pH 4. A, K-segment, and B, His–K-segment. Each panel corresponds to a particular membrane system: DOPC, DOPC:DOPS (4:1), or DOPC:DPPC (1:1). At the top of each panel, the final 0.2-μs averaged partial mass density of the peptide is shown (with respect to Z-normal and y axis), with the outer leaflet phosphates indicated by a dashed line. The final snapshot of the simulation is shown at the bottom of each panel (peptide shown in cartoon representation in orange, lipids shown in licorice representation, with carbons colored cyan, oxygens red, and nitrogens blue).
Figure 5.
Peptide–membrane interactions during simulations at pH 10. A, K-segment, and B, His–K-segment. Each panel corresponds to a particular membrane system: DOPC, DOPC:DOPS (4:1), or DOPC:DPPC (1:1). At the top of each panel, the final 0.2-μs averaged partial mass density of the peptide (with respect to Z-normal and y axis), with the outer leaflet phosphates indicated by a dashed line. The final snapshot of the simulation is shown at the bottom of each panel (peptide shown in cartoon representation in orange, lipids shown in licorice representation, with carbons colored cyan, oxygens red, and nitrogens blue).
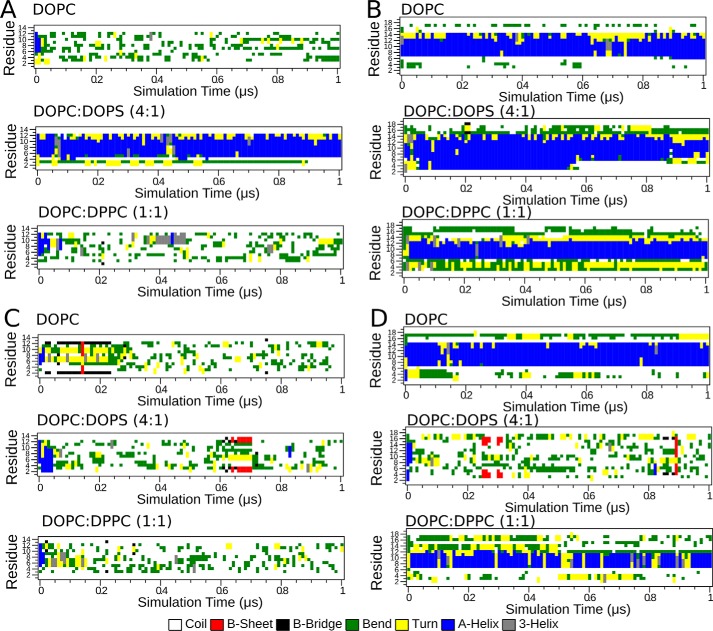
We observed that the propensity to bind membranes resulted in the formation of α-helical conformation, as illustrated in Fig. 6. Thus, irrespective of system, the average propensity for adoption of helical structure where the peptide remained in solvent, unbound from the membrane, corresponded to 4.6 ± 1.9%. In comparison, this value increased to 81 ± 25% during portions of simulation trajectories in which the peptide was bound to the lipid headgroups. This supports previous experimental findings, where membrane binding was associated with formation of helical conformation of the K-segment (7).
Figure 6.
Per-residue peptide secondary structure propensity over the simulation time in lipid membranes. A, K-segment at acidic pH; B, His–K-segment at acidic pH; C, K-segment at basic pH; D, His–K-segment at basic pH.
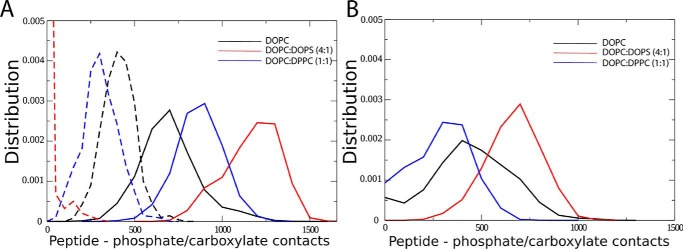
Next, we estimated the number of contacts between peptide and lipid phosphate or anionic lipid carboxylate groups. For His–K-segments, the propensity to bind membranes is reflected by the number of contacts of the peptide with phosphate and carboxylate lipid groups (Fig. 7, A and B), with the maximum number of contacts formed with DOPC:DOPS (4:1), followed by DOPC:DPPC (1:1), and the least number for DOPC membranes. Therefore, the stronger the interactions with the membrane, the more contacts with the anionic lipid groups. For K-segments, the overall number of peptide contacts with phosphate and carboxylate lipid groups is lower as compared with the His–K-segment and is consistent with the estimated partial mass density around the membranes. The length of helix formed in each peptide (Fig. 6) correlates with the depth of binding (Figs. 4 and 5) and number of interactions with the membrane lipids (Fig. 7), supporting the notion that lipid binding favors formation of helical peptide structures.
Figure 7.
Interactions of individual K-segment and His–K-segment with lipid bilayers. A, distribution of the number of contacts between peptide and lipid phosphate or anionic lipid carboxylate groups (0.6 nm cutoff) are shown for the His–K-segment at acidic pH (data shown in solid format) and basic pH (data shown in dashed format). B, distribution of the number of contacts between peptide and lipid phosphate or anionic lipid carboxylate groups (0.6 nm cutoff) are shown for the K-segment at acidic pH.
In summary, the above observations emphasize the electrostatic nature of interactions between Lti30 and negatively charged lipid headgroups. The lower the pH, the stronger the binding affinity of Lti30. The binding ability was also found to be influenced by the presence of histidines as well as the presence of anionic lipids in the membranes, consistent with the electrostatically dominant interactions. These observations are also supported by additional MD simulations performed under “less extreme” effective pH conditions equivalent to pH 5.8 and 9.0 (Table S1). The differences between acidic and basic systems were less pronounced in this case, particularly for K-segments that lack titratable histidines and therefore do not experience a change in net charge under the classical MD simulation framework (Fig. S6). Nevertheless, electrostatic interactions were similarly observed to be dominant, with a greater tendency for lipid headgroup binding upon the addition of anionic lipids or protonation of histidines in His–K-segments (Fig. S7), and a corresponding propensity for peptide helix formation following adsorption to the membrane surface (Fig. S8).
Discussion
An intrinsic consequence of drought stress is the deformation and rupture of cellular membranes. The response of the plant under such conditions involves the expression of dedicated stress proteins such as dehydrins, which seem to mitigate the water loss by safeguarding membrane integrity. In this action, the dehydrins can either act globally by interacting with and stabilizing the normal membrane arrangement or locally by selectively targeting perturbed membrane areas. To fine-tune the response, the dehydrin–membrane interaction needs also to be regulated by cellular cues beyond expression levels. Finally, it is possible that the dehydrin action involves additional components like dehydrin–dehydrin quaternary interaction to allow concerted effects over larger distances, i.e. protein scaffolding. In this study we find that, indeed, all these factors seem to be in play: (i) Lti30 recognition and binding show a response to both the specific lipid composition and membrane-packing density; (ii) the strength of Lti30–membrane interaction is regulated by pH through titration of the protein's His moieties in the conserved K-segments; and (iii) Lti30 assembles into microaggregates that uniformly decorate the membrane surface at physiological pH and ionic strength.
Role of lipid composition
It has previously been observed that some dehydrins bind membranes by electrostatic interactions between the conserved K-segment's positively charged side chains and the negatively charged lipids (5, 6). Such binding is confirmed here by ITIR-FCS experiments. Our results show that full-length Lti30 interacts most strongly with anionic DOPC:DOPS (4:1) and zwitterionic DOPC:DPPC (1:1), but not with the pure zwitterionic DOPC bilayers (Fig. 1, B and C). MD simulations were also used to analyze K-segment and His–K-segment membrane binding under varying effective acidic and basic pH conditions. Electrostatics were found to be dominant in determining peptide binding and subsequent helix formation at the membrane surface. The resulting contacts of peptide with lipid headgroups for the interaction between the protonated His–K-segments and membranes (Fig. 7), and the corresponding binding depths (Fig. 4), following spontaneous binding were observed to be greatest for anionic bilayers compared with zwitterionic ones (Fig. 7).
Role of membrane-packing density
An explanation to why Lti30 bind stronger to mixed DOPC:DPPC membranes than to pure DOPC membranes is provided by the lipid-packing geometries. Despite being zwitterionic, the ζ potential of phosphatidylcholine lipids does not always cancel to zero but varies with the specific orientations of the headgroups (30). Of particular interest here is that the orientation of phosphates and carboxylates in pure DPPC membranes, which adapt dense gel phases, renders the effective surface charge slightly negative (31, 32). These properties of DPPC lipid transpires also in mixed DOPC:DPPC membranes that gain more negative ζ potential than pure DOPC, matching their protein-binding capacity. In other words, the binding of Lti30 is sensitive to the specific packing of zwitterionic membranes through its coupling to the negative ζ potential. A conjecture of this finding is that dehydrin binding responds not only to lipid identity but also to their specific orientation and the way the membrane is packed. Because all these lipid characteristics are bound to undergo changes in membranes under stress, such manifold binding recognition can be of physiological importance. DOPC is a fluid-phase unsaturated lipid, whereas DPPC is a saturated gel-phase lipid. The difference in the structural and chemical properties of these lipids causes variation in the bilayer packing. Consistently, our FCS data and simulation results show that both the positively charged full-length Lti30 and the His–K-segments interact more favorably with the mixed DOPC:DPPC membranes.
The hydration state directly influences the surface properties of the membrane by modulating the orientations of lipid headgroups and lipid acyl chains in response to altered water–lipid contacts (33, 34). In plants, in response to dehydration and freeze stress, the fraction of phospholipids in plasma membranes is found to uniformly increase from 16 to 20 mol %. Accompanying this increase is an alteration in fatty acid saturation, for example the proportion of di-unsaturated species of PC is observed to change (35). Moreover, the plasma membrane of Arabidopsis is reported to form disruptive lipid HII phases as a response to acute freeze-induced dehydration stress. This transition between inter-bilayer–lamellar phase to HII–hexagonal phase occurs where the plasma membrane is close to other endomembranes, resulting in curvature stress (35, 36). Membranes in acclimated Arabidopsis plants, however, have a lower propensity to form the harmful HII phase, partly due to an increase in the proportion of di-unsaturated spices of PC (35). Although it is yet not established how additional factors like dehydrins affect the lamellar- to HII–phase transition, our present data show that Lti30 binds better to membranes that partly consist of saturated species of PC, as typically found under normal conditions in nonacclimated membranes, or to lipid surfaces with packing defects. The indicated preference of Lti30 is thus to bind to the nonacclimated membrane. A tentative function is to safeguard the synthesis of new lipids in the acclimation process, which is expected to be slower than the dehydrin response. Consistently, some of the dehydrins are indeed found on the same timescale as of the increase of PA (phosphatidic acid) in plant membranes (35), a lipid that not only gives negative ζ potential but also affects membrane curvature by being cone-shaped. Because PA lacks a headgroup, penetration of Lti30 into the membrane is facilitated. To this end, Arabidopsis is found to respond to freeze-induced dehydration by forming small vesicles, so-called exocytotic extrusions, which are located outside the plasma membrane. These extrusions not only increase locally the membrane–curvature tension but also increases the total membrane surface area. When the stress is then released, these vesicles re-merge with the plasma membrane (35). It is conceivable that the changes in lipid-packing and headgroup exposure accompanying these reversible membrane alterations can transiently trigger Lti30 binding. Our findings are also in agreement with other peripheral membrane-binding proteins, such as α-synuclein and CTP:phosphocholine cytidylyltransferase (CCT), that are able to sense lipid-packing defects that are normally enriched in curved surfaces (37, 38). Like the dehydrins, the disordered α-synuclein and CCT employ induced amphipathic helices that recognize the physical properties of the target membrane surfaces. The main factors driving these helical interactions are electrostatics and hydrophobicity, with hydrophobicity being the major factor if electrostatics are weak. Interestingly, the corresponding binding of Lti30 in this study seems dominated by the electrostatics in the form of a negative ζ potential. This is a new and mechanistically intriguingly finding as it highlights that Lti30 and other dehydrins can interact with any lipid membrane because of the exposed lipid headgroups irrespective of the overall zero net charge of the lipid. This, of course, widens the range of putative Lti30-interacting lipid surfaces.
pH dependence
Consistent with previous observations, experiments by ITIR-FCS suggest that Lti30-membrane binding is strongly pH-dependent. The pH switch is determined by the pKa of the histidine residues flanking the K-segments. It has been observed that when Lti30 is fully protonated, it tends to form microdomains in the lipid bilayer and makes the bilayer more rigid. In its partially protonated state, it does influence the diffusion of bilayer lipids by reducing D but does not necessarily form microdomains in the membrane (Fig. 1, B and C). Finally, at pH 9.0 when Lti30 is fully deprotonated, there is no effect of Lti30 on the diffusion of the lipid bilayer. It is important to note that the membrane properties are also modulated at various pH values, which might also contribute to the membrane interaction mechanism. For instance, in the case of DOPC:DPPC (1:1) at pH 9.0, τ0 is positive, indicating microdomains in the membrane that are not Lti30-induced, and D is also much lower (Fig. 1C) than that at other tested pH values consistent with the existing literature (27). Hence, the extent of the effect of Lti30 membrane diffusion is strongly dependent on the protonation state of the histidines. Also, results show that histidine residues significantly increase the affinity of Lti30/His–K-segment toward the membranes with negative surface charge. Notably, flanking histidines significantly alter the conformation of the K-segments so that they allow penetration of K-segments into the membranes as shown by MD simulations and FCS measurements. Therefore, histidines are important for the conformational change from a disordered Lti30 in solution to a structurally ordered assembly on the membrane. Moreover, FCS being a single molecule-sensitive method provides access to the different populations of the His–K-segment based on the difference in their diffusion profiles. These results provide an explanation for the importance of flanking histidines in the membrane interaction mechanism of Lti30.
Membrane stabilization by surface scaffolding; the quaternary component
In addition to the effect of binding on the membrane, Lti30 aggregation on the membrane is also found to be pH-dependent (Figs. 2 and 3). The degree of Lti30 aggregation can be correlated with the effect of binding on the membrane as shown by FCS experiments. Simulations have shown that the peptide helicity depends on the binding affinity (Fig. 6), and in cases of higher affinity, the peptides tend to penetrate the membrane (Figs. 4 and 5). Possibly, this membrane penetration tendency is higher with larger protein aggregates that alter the membrane structure by forming microdomains. It is now well recognized that the microdomains in the plasma membrane are critical for facilitating protein function by modulating membrane geometry (39). In this study, all the experiments have been performed at a 5 μm Lti30 concentration (1:130 protein:lipid ratio), but in the literature, even higher concentrations of Lti30 and other dehydrins are used (5–7). At higher Lti30 concentrations, even larger Lti30 aggregates can form that can lead to excessive curvature strain on the membrane. This can be essential for the biological function of Lti30, but it requires further investigation. Moreover, we have also observed that there is protonation-driven aggregation in Lti30 and His–K-segment in solution. FCS measurements show that there is a fraction of His–K-segment that forms aggregates in solution when histidines are fully protonated.
The question is what drives the aggregation of the His–K-segment and Lti30 and why histidines are crucial for the higher-order molecular interactions. To answer that, it is important to note that histidine residues can be involved in several types of possible molecular interactions because of their unique structural features (40). They have an imidazole side chain with the pKa around pH 6.5. They thus can form complexes with metallic cations, and they can be involved in hydrogen bonding. These structural features and different protonation states of histidines at various pH values can be responsible for their interaction with other residues within the protein or their interaction with the other nearby proteins. There are multiple types of possible interactions that histidine can be involved in such as the following. 1) For cation–π interactions, the side chain of histidine is an imidazole ring that serves as an organic cation in its protonated state, and it can interact with the nearby aromatic residues (41, 42) (e.g. Tyr, Trp, and Phe). Note that Lti30 itself lacks Trp and Phe residues but has three Tyr and K-segments have none. 2) For π–π interactions, the imidazole side chain of the histidine can act as a conjugative π plane and be involved in π–π interactions with the aromatic rings of the other aromatic residues (41, 42). 3) For hydrogen–π interactions, depending on the orientation of surrounding aromatic amino acids, they can interact with the histidine polar hydrogen atom (43). 4) For coordinate bond interaction (44), histidines can interact with the metallic cations (e.g. Zn2+) as a ligand coordinate using its lone electron pairs. Interaction of Lti30 with the metals has been reported in the past (45). 5) For hydrogen bond interaction, this is possible between the polar hydrogen in the imidazole ring and the basic nitrogen atom (46, 47). We speculate that these possible modes of interaction may be responsible for the wide range of molecular interactions in Lti30. This requires further experimental validation.
Eriksson et al. (6) proposed that there is a possibility of membrane cross-linking by Lti30. The observation that at physiological pH the Lti30 aggregates are bound on the whole surface of the membrane and the membrane fluidity is decreased indicates that Lti30 indeed cross-links the membrane. N-terminally acetylated α-synuclein was found to have shortened aggregation kinetics and promoted fibril formation with gel-phase vesicles as compared with fluid membranes (37). Here, we find that Lti30 gives a ThT signal mainly at low pH with fluid membranes, indicative of fibril formation. Whether Lti30 or other dehydrins are able to form fibrils under other conditions, such as altered lipid composition, has yet to be determined. Membrane cross-linking can influence the membrane permeability. During cold temperatures and dehydration, plant cell membranes get injured, as discussed above, resulting in the leakage of the cell contents (48). Regulation of membrane permeability to water and solutes is crucial for the cell survival. It is possible that during cold and drought stress, Lti30 protects the cell membrane by preserving the membrane permeability by cross-linking.
Conclusion
Dehydrins are intrinsically disordered plant stress proteins containing conserved sequence motifs. Lti30 is a dehydrin embracing six conserved K-segments with a high percentage of positively charged amino acids. Here, using fluorescence correlation spectroscopy in combination with MD simulations, we find the following points:
(i) Lti30 recognition and binding show response to both the specific lipid composition and membrane-packing density. Lti30 binds to membranes where anionic lipid headgroups are exposed and are sensitive to the structure of the lipids in the membrane and lipid composition.
(ii) The strength of Lti30–membrane interaction is regulated by pH through titration of the protein's His moieties in the conserved K-segments. Moreover, the effect of Lti30 binding on membrane diffusion depends on the protonation states of histidines, which are modulated by pH. Binding of Lti30 reduces the mobility of membrane lipids, and there is microdomain formation at acidic pH. Flanking histidines in the Lti30 sequence significantly increase the affinity of Lti30 to the membrane. Protonation of histidines causes higher order molecular interactions in Lti30 as observed by ITIR-FCS and ThT fluorescence experiments. The role of histidines can be further studied by understanding the role of π–ion interactions leading to the possibility of multiple interaction modes.
(iii) Lti30 assembles into microaggregates that decorate the whole membrane surface at physiological pH and ionic strength. The propensity of microdomain formation is related to the size of Lti30 aggregates. These observations suggest that Lti30 protects the membrane by cross-linking it. Membrane cross-linking can be important for preventing cell leakage, which generally occurs in plant cells during cold temperatures and drought.
This means that Lti30 is a peripheral membrane-binding protein that becomes functional when it interacts with its biological targets. Additional interaction of Lti30 with other biological targets (DNA/RNA, metals, etc.) can lead to several different ways it protects the plant during stress, but this needs to be further investigated.
Experimental procedures
Protein/peptide production
Lti30 expression and purification of the recombinant A. thaliana dehydrin Lti30 were performed as described by in a previous work (45), with minor changes. Glycerol stocks of the Escherichia coli strain were made, and 150 μl was spread on Luria agar plates with 150 μg of ampicillin and grown 37 °C overnight. The cells were suspended and added to 2 liters of Luria-Bertani medium containing 50 μg/ml carbenicillin and kept at 37 °C. Expression was induced at an A600 of 0.6–0.7 by adding 1 mm isopropyl β-d-thiogalactopyranoside and kept at 23 °C overnight. Cells were harvested by centrifugation at 6000 rpm for 15 min. The pellet from 1 liter of culture was suspended in 25 ml of 20 mm Na2HPO4, pH 7.2, and 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 1 tablet of Complete (Roche Applied Science). Cells were sonicated for five 1-min periods on ice followed by centrifugation at 18,000 rpm for 30 min. The supernatant was placed in an 80 °C water bath for 30 min to precipitate heat-denatured proteins and then centrifuged at 18,000 rpm for 30 min. Lti30 was purified by metal ion-affinity chromatography. The supernatant from heat precipitations were diluted 1:2 with 20 mm Na2HPO4, pH 7.2, 1.88 m NaCl, and 1 mm phenylmethylsulfonyl fluoride. The samples were loaded onto a 5-ml HiTrap IDA-Sepharose column (GE Healthcare) charged with 7 ml of 3 mg/ml CuSO4. The column was equilibrated with 5 volumes of 20 mm Na2HPO4, pH 7.2, and 1.0 m NaCl. The same buffer (40 volumes) was used to wash off unbound sample from the column followed by 2 m NH4Cl in 20 mm Na2HPO4, pH 7.2, and 1.0 m NaCl. Fractions of 5 ml were collected for analysis during the whole run. The column was then equilibrated with 10 volumes of 20 mm Na2HPO4, pH 7.2, followed by elution of Lti30 with 10 mm EDTA in 20 mm Na2HPO4, pH 7.2. Precipitation of protein was performed with 80% (NH4)2SO4 overnight, and protein was collected by centrifugation at 18,000 rpm for 45 min. Protein pellets were suspended with 2.5 ml of 5 mm MES buffer, pH 6.3. The protein was desalted on two PD-10 columns (GE Healthcare). Purity was analyzed by Ready Gel® SDS-PAGE system (Bio-Rad). Protein quantification was measured with the bicinchoninic acid (BCA) assay (Sigma). The final protein was freeze-dried and stored at −80 °C. Rhodamine-labeled K-segment peptides were purchased from PHTD Peptides, Zhengzhou City, China. Lti30 protein and K-segments with and without histidines (dry powder) were solubilized in the required amount of 5 mm MES set to the desired pH for 5.8 and 6.3, 10 mm Tris for pH 7.4, and 50 mm glycine for pH 9. Buffers were prepared in deionized water. All the buffers were filtered by a 0.45-μm Teflon filter. The protein and peptide are highly hydrophilic and thus readily dissolve in all the mentioned buffers. The concentration of protein/peptide solution was estimated by standard Coomassie assay with BSA as standard.
Lipids and dyes
DOPC, DPPC, and DOPS lipids were used in this study. The head-labeled rhodamine dye (RhoPE) and DiO are used as the fluorescent probe for supported lipid bilayers. All the lipids except DiO were purchased from Avanti Polar Lipids (Alabaster, AL). DiO was purchased from ThermoFisher Scientific.
Preparation of supported lipid bilayers of different lipid compositions at various pH conditions
The calculated amounts of the lipids and dye (the final concentration of total lipid and dye was 650 μm and 100 nm, respectively) were taken in a cleaned round bottom flask. The components were mixed thoroughly and then evaporated in a rotary evaporator (Rotavap R-210, Buchi, Switzerland) for 3–4 h to create a thin lipid film. The lipid film was resuspended in buffer (10 mm HEPES and 150 mm NaCl, pH 7.4) resulting in a turbid solution of multilamellar vesicles. This suspension was sonicated using a bath sonicator (model FB15051, ThermoFisher Scientific, Singapore) until clarity to form large unilamellar vesicles.
The buffer was mixed with vesicles obtained by the above-described procedure in a 1:1 ratio and was deposited onto a freshly cleaned glass cover slide (24 × 50–1, Fisher Brand Microscope cover glass, ThermoFisher Scientific, Singapore). It was incubated at 65 °C for 60–80 min followed by cooling at room temperature for 20 min. Then the unfused vesicles were removed by washing with buffer at the desired pH about 20 times. SLBs equilibrated at room temperature were used for ITIR-FCS measurements.
Total internal reflection imaging and imaging fluorescence correlation spectroscopy (ImFCS) set up, data acquisition, and data analysis
The experiments were done on labeled/unlabeled supported lipid bilayers with labeled/unlabeled Lti30, K-segment, and His–K-segment. The protein–to–lipid ratio for all the experiments was 1:130.
ITIR-FCS
Diffusion measurements were performed using an objective type TIRF microscope (IX-71, Olympus, Singapore) with a high NA oil immersion objective (PlanApo, ×100, NA 1.45, Olympus, Singapore). The excitation light from a 532-nm laser (Cobolt Samba, Sweden) was coupled into the microscope by a combination of two tilting mirrors. The light was focused on the back focal plane of the objective after being reflected by a dichroic mirror (Z488/532RPC, Semrock) and was totally internally reflected at the glass–water interface by controlling the incident angle of the excitation beam by the same combination of tilting mirrors. The laser power used was 0.8 to 1 milliwatts. The immersion medium of the objective was mineral oil (Olympus, refractive index 1.516). The fluorescence from the bilayers was reflected through the same objective followed by transmission through the same dichroic mirror. Finally, it was imaged on the CCD chip of a cooled (−80 °C) back-illuminated EMCCD camera (Andor iXON 860, 128 × 128 pixels, Andor Technology) after being filtered by an emission filter (Z488/532M, Semrock). The software Andor Solis for imaging (version 4.18.30004.0 and 4.24.30004.0) was used for data acquisition. The pixel side length of the CCD chip in the device is 24 μm corresponding to a pixel side length of 240 nm in the sample plane. The camera was operated in the kinetic mode, and baseline clamp was used to minimize the baseline fluctuations. The readout speed was 10 MHz with 4.7× maximum analog-to-digital gain and 25-μs vertical shift speed. An EM gain of 200–300 was used for most imaging experiments.
The signal was simultaneously recorded from a 21 × 21-pixel region of interest (ROI) in the form of a stack of 50,000 frames with 1-ms time resolution. The data were saved as a 16-bit Tiff file. The temporal intensity trace from each pixel was autocorrelated using multi-τ correlation scheme using a FIJI plug-in ImFCS 1.49 to generate ACF (49). The data were bleach-corrected using a 4th order polynomial function. The ACF for each pixel was individually fitted with the following one-particle model (Equation 1) for diffusion using the same software.
| (Eq. 1) |
Here, G(τ) is the ACF as a function of correlation time (τ), and N, a, D, and σ are the number of particles per pixel, pixel side length, diffusion coefficient, and standard deviation of the Gaussian approximation of the microscope point spread function, respectively. G∞ is the convergence value of the ACF at long correlation times.
Fitting of ACFs with theoretical models provides D and N. Because it is an imaging-based FCS modality, diffusion coefficient, and the number of particle maps are obtained for the whole ROI (49). In ITIR-FCS, the data are represented as mean ± S.D. The S.D. is obtained from the measurements over 441 pixels per experiment. The S.D. of an ITIR-FCS measurement not only contains contributions from the measurement variability but also from the heterogeneity of the peptide/bilayer system. Measurements were performed at room temperature.
For experiments with labeled SLBs, the concentration of unlabeled protein/peptides was 5 μm. The concentration of peptide in the experiments with labeled peptide was also 5 μm where 1 nm of labeled peptide was mixed with the unlabeled peptide.
Total internal reflection microscopy
Imaging experiments were conducted on an objective-type TIRF microscope (Olympus IX-83, Singapore) with a high NA oil immersion objective (ApoN, ×100, NA 1.49, Olympus, Singapore). The excitation laser used to excite ThT is 405 nm (LAS/405/100, Olympus). The penetration depth of the evanescent field was adjusted as 110 nm. The rest of the instrumentation is the same as explained above. The emission filter used is zet405/488/561/647m (Semrock). The exposure time of excitation was 10 ms.
FCS diffusion laws implemented in ITIR-FCS
For the study of organization of peptide–membrane systems below the diffraction limit, FCS diffusion laws are used in combination with ITIR-FCS. With the help of these laws one can probe whether a particle is undergoing free diffusion or is hindered by the trapping sites such as domains (50). This is achieved by analyzing the spatial dependence of the diffusion time of the labeled molecules on the observation area size. For a freely diffusing particle, the time a particle takes to diffuse through an area is directly proportional to the area (Aeff). Hence, these plots are fitted to a straight line which is mathematically expressed as shown in Equation 2,
| (Eq. 2) |
where τ0 is the FCS diffusion law intercept. Most of the microscopy methods are diffraction-limited, but molecular heterogeneities in biological samples are often in nanoscale size regime. To get sub-resolution information, the diffusion law plot is extrapolated to zero and the y-intercept (τ0) is used as a readout of membrane organization. For a freely diffusing particle, the diffusion time has a linear relationship with the observation area, and the y-intercept is zero. In case of a domain partitioning, the relationship between the diffusion time of the molecules and the observation area deviates from linearity, and the y-intercept is positive. To perform FCS Diffusion Law analysis, the same set of data that is obtained in an ITIR-FCS experiment is used. Variable observation areas (Aeff) are obtained by post-acquisition pixel binning (1 × 1 to 5 × 5) followed by convolution with the point spread function of the microscope system. The Aeff/D is plotted against Aeff, and the plot is fitted to a line with the S.E.-weighted Equation 2 to obtain the Y-intercept τ0. The typical margin of error on SLBs is ±0.1, and thus intercepts in that range are indistinguishable for free diffusion. Only intercepts larger than 0.1 can be attributed to domain trapping in our setup (25).
Confocal FCS setup, data acquisition, and data analysis
Confocal FCS experiments were performed using an Olympus FV1200 confocal laser-scanning microscope (Olympus, Tokyo, Japan) equipped with an FCS upgrade kit (Picoquant, Berlin, Germany). The green laser beam from 543-nm helium-neon laser (GLG 7000, Showa Optronics, Tokyo, Japan) was focused on the sample by a water immersion objective (×60, NA 1.2; Olympus, Tokyo, Japan) after being reflected by a dichroic mirror (560 DCLP, Omega, Burlington, VT) and a scanning unit. The laser power after the objective was measured to be 13 microwatts. The fluorescence signal from the sample was collected by the same objective, through a 120-μm pinhole in the image plane to eliminate out–of–focus light. The fluorescence emission is passed through a 405/488/543/635 major dichroic mirror (Chroma Technology, Bellows Falls, VT) and through a 600/50 (Chroma Technology) bandpass emission filter, which is then detected by avalanche photodiodes (SPCM-AQR-14; PerkinElmer Life Sciences). The photon counts from the detectors were registered by a TimeHarp 260 time-correlated single photon counting board (PicoQuant) and processed by the SymPhoTime 64 software (PicoQuant, Berlin, Germany); the same software was also used to calculate the correlation functions. The experimentally obtained ACFs were fitted using the fitting model (Equation 3) given below:
| (Eq. 3) |
where κ is the structure factor. The value of κ was kept between 5 and 7 for all the measurements. τDiff represents diffusion time of ith species; ρ denotes the contribution of ith diffusing species, T represents the fraction of triplet population, and τtrip denotes the lifetime of the dark state.
The diffusion coefficient (D) was determined from Equation 4, where ω0 was obtained from a calibration measurement with rhodamine 6G dye in solution assuming its diffusion coefficient to be 400 μm2/s.
| (Eq. 4) |
All-atom MD simulations
Lti30 is a partially and intrinsically disordered protein of which conserved sequence motifs K-segment EKKGMTEKVMEQLPG and His–K-segment VHEKKGMTEKVMEQLPGHHG sequences were initially modeled as α-helices in line with data from previous experiments (7). Peptides were modeled under effective pH conditions of approximately pH 4.0, 5.8, 9.0, or 10.0. At pH 4.0, the side chains of all ionizable residues present in the peptides, i.e. histidine, lysine, and glutamic acid, were treated in their protonated form. At pH 5.8 or 9.0, histidines were treated in their protonated or deprotonated forms, respectively, whereas lysine and glutamic acid side chains remained in their default charged states. At pH 10.0, the side chains of all the ionizable residues were treated in their deprotonated forms. Each peptide terminus was in its charged form. This led to a variable overall peptide charge, as summarized in Table 3 and Table S1.
Lipid membrane systems were built using the CHARMM-GUI membrane builder (51), consisting of 128 lipids for the pure DOPC system, or 102 DOPC and 26 DOPS lipids for the DOPC:DOPS (4:1) system, and 64 of DOPC and 64 of DPPC for DOPC:DPPC (1:1) system. Each membrane was placed in a box of dimensions 6.7 × 6.7 × 12 nm3, with the K-segment or His–K-segment ∼3 nm away from the membrane surface. Subsequently, the box was filled with ∼17,000 TIP3P water molecules. The overall charge of each system was neutralized by addition of the minimum required number of chloride or sodium ions. Energy minimization using S.D. was performed for 5000 steps with a 0.1-nm step size, using flexible TIP3P (52) water. The equilibrations in the NVT and NPT ensemble were each performed for 0.03 μs with position restraints on peptide heavy atoms. All production simulations of systems at pH 4.0 and 10.0 were run in the NPT ensemble without any restraints for 1 μs using GROMACS 5.1.4. Because of the slower convergence of production runs of systems mimicking pH 5.8 and 9.0, these were run for 1.4 μs. Equations of motion were integrated through the Verlet leapfrog algorithm with a 2-fs time step, and bonds connected to hydrogens were constrained with the LINCS algorithm. The cutoff distance was 1.2 nm for the short-range neighbor list and van der Waals interactions with a smooth switching function from 1.0 nm. The Particle Mesh Ewald method was applied for long-range electrostatic interactions with a 1.2-nm real space cutoff (53). The Nose-Hoover thermostat (54, 55) and Parinello-Rahman barostat (56) were used to maintain the temperature and pressure at 310 K and 1 bar, respectively. All peptide–membrane assembly simulations were performed on the following: (i) an in-house Linux cluster composed of seven nodes containing two GPUs (Nvidia K20) and 20 CPUs (Intel® Xeon® CPU E5-2680 version 2 @ 2.8 GHz) each; and (ii) the Singapore National Supercomputing Centre, where each simulation employed four nodes consisting of 1 GPU (Nvidia Tesla K40t) and 24 CPUs (Intel® Xeon® CPU E5-2690 version 3 @ 2.6 GHz).
Simulation analysis
The per-residue secondary structure propensity was analyzed using the DSSP (22) algorithm in GROMACS 5.1.4. The partial peptide densities were calculated using g_mydensity tool for last 0.2 μs of each run (29). The number of peptide–phosphate/carboxylate contacts was calculated based on a 0.6-nm cutoff distance over the entire trajectory per system.
Author contributions
A. G., P. H., and T. W. conceptualization; A. G., J. K. M., and D. J. data curation; A. G., J. K. M., D. J., and P. J. B. formal analysis; A. G., J. K. M., and P. H. validation; A. G., P. J. B., P. H., and T. W. investigation; A. G., J. K. M., and D. J. visualization; A. G. and J. K. M. methodology; A. G., J. K. M., P. J. B., P. H., and T. W. writing-original draft; A. G., J. K. M., P. J. B., P. H., and T. W. writing-review and editing; J. K. M., P. J. B., and T. W. software; P. J. B., P. H., and T. W. supervision; P. J. B., P. H., and T. W. funding acquisition; P. H. and T. W. resources; P. H. and T. W. project administration.
Supplementary Material
Acknowledgment
We thank the National Supercomputing Centre Singapore for computational resources.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Table S1.
- ITIR-FCS
- imaging total internal spectroscopy–fluorescence correlation spectroscopy
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine
- DPPC
- 1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- PC
- phosphatidylcholine
- SLB
- supported lipid bilayer
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- MD
- molecular dynamics
- ACF
- autocorrelation function
- CCT
- CTP:phosphocholine cytidylyltransferase
- Tm
- transition melting temperature
- ThT
- thioflavin T
- PA
- phosphatidic acid
- ROI
- region of interest
- DiO
- 3,3′-dioctadecyloxacarbocyanine perchlorate
- Rho-PE
- 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt)
- ImFCS
- imaging fluorescence correlation spectroscopy.
References
- 1. Nylander M., Svensson J., Palva E. T., and Welin B. V. (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 45, 263–279 10.1023/A:1006469128280 [DOI] [PubMed] [Google Scholar]
- 2. Chakrabortee S., Boschetti C., Walton L. J., Sarkar S., Rubinsztein D. C., and Tunnacliffe A. (2007) Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl. Acad. Sci. U.S.A. 104, 18073–18078 10.1073/pnas.0706964104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mouillon J. M., Gustafsson P., and Harryson P. (2006) Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol. 141, 638–650 10.1104/pp.106.079848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Graether S. P., and Boddington K. F. (2014) Disorder and function: a review of the dehydrin protein family. Front. Plant Sci. 5, 576 DOI not found. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clarke M. W., Boddington K. F., Warnica J. M., Atkinson J., McKenna S., Madge J., Barker C. H., and Graether S. P. (2015) Structural and functional insights into the cryoprotection of membranes by the intrinsically disordered dehydrins. J. Biol. Chem. 290, 26900–26913 10.1074/jbc.M115.678219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eriksson S. K., Kutzer M., Procek J., Gröbner G., and Harryson P. (2011) Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell. 23, 2391–2404 10.1105/tpc.111.085183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eriksson S., Eremina N., Barth A., Danielsson J., and Harryson P. (2016) Membrane-induced folding of the plant-stress protein Lti30. Plant Physiol. 171, 932–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goers J., Uversky V. N., and Fink A. L. (2003) Polycation-induced oligomerization and accelerated fibrillation of human α-synuclein in vitro. Protein Sci. 12, 702–707 10.1110/ps.0230903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uversky V. N., M Cooper E., Bower K. S., Li J., and Fink A. L. (2002) Accelerated α-synuclein fibrillation in crowded milieu. FEBS Lett. 515, 99–103 10.1016/S0014-5793(02)02446-8 [DOI] [PubMed] [Google Scholar]
- 10. Mouillon J.-M., Eriksson S. K., and Harryson P. (2008) Mimicking the plant cell interior under water stress by macromolecular crowding: disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 148, 1925–1937 10.1104/pp.108.124099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kovacs D., Kalmar E., Torok Z., and Tompa P. (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147, 381–390 10.1104/pp.108.118208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hara M., Terashima S., and Kuboi T. (2001) Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol. 158, 1333–1339 10.1078/0176-1617-00600 [DOI] [Google Scholar]
- 13. Nachbar M., Mozafari M., Krull F., Krull F., Maul K. J., Preu L., Hara M., and Wätzig H. (2017) Metal ion–dehydrin interactions investigated by affinity capillary electrophoresis and computer models. J. Plant Physiol. 216, 219–228 10.1016/j.jplph.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 14. Hara M., Shinoda Y., Tanaka Y., and Kuboi T. (2009) DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ. 32, 532–541 10.1111/j.1365-3040.2009.01947.x [DOI] [PubMed] [Google Scholar]
- 15. Koag M. C., Fenton R. D., Wilkens S., and Close T. J. (2003) The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 131, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danyluk J., Perron A., Houde M., Limin A., Fowler B., Benhamou N., and Sarhan F. (1998) Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 10, 623–638 10.2307/3870737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steponkus P. L., and Lynch D. V. (1989) Freeze thaw-induced destabilization of the plasma membrane and the effects of cold acclimation. J. Bioenerg. Biomembr. 21, 21–41 10.1007/BF00762210 [DOI] [PubMed] [Google Scholar]
- 18. Crowe L. M., and Crowe J. H. (1992) Anhydrobiosis: a strategy for survival. Adv. Space Res. 12, 239–247 10.1016/0273-1177(92)90178-Z [DOI] [PubMed] [Google Scholar]
- 19. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- 20. Koag M.-C., Wilkens S., Fenton R. D., Resnik J., Vo E., and Close T. J. (2009) The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 150, 1503–1514 10.1104/pp.109.136697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Puhakainen T., Hess M. W., Mäkelä P., Svensson J., Heino P., and Palva E. T. (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 54, 743–753 10.1023/B:PLAN.0000040903.66496.a4 [DOI] [PubMed] [Google Scholar]
- 22. Joosten R. P., te Beek T. A., Krieger E., Hekkelman M. L., Hooft R. W., Schneider R., Sander C., and Vriend G. (2011) A series of PDB related databases for everyday needs. Nucleic Acids Res. 39, D411–D419 10.1093/nar/gkq1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bag N., Sankaran J., Paul A., Kraut R. S., and Wohland T. (2012) Calibration and limits of camera-based fluorescence correlation spectroscopy: a supported lipid bilayer study. Chemphyschem 13, 2784–2794 10.1002/cphc.201200032 [DOI] [PubMed] [Google Scholar]
- 24. Sharma K. K., Marzinek J. K., Tantirimudalige S. N., Bond P. J., and Wohland T. (2018) Single-molecule studies of flavivirus envelope dynamics: experiment and computation. Prog. Biophys. Mol. Biol. 2018, S0079–6107(18)30181–0 10.1016/j.pbiomolbio.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 25. Sankaran J., Bag N., Kraut R. S., and Wohland T. (2013) Accuracy and precision in camera-based fluorescence correlation spectroscopy measurements. Anal. Chem. 85, 3948–3954 10.1021/ac303485t [DOI] [PubMed] [Google Scholar]
- 26. Bag N., Yap D. H., and Wohland T. (2014) Temperature dependence of diffusion in model and live cell membranes characterized by imaging fluorescence correlation spectroscopy. Biochim. Biophys. Acta 1838, 802–813 10.1016/j.bbamem.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 27. Lucero A., Rodríguez Niño M. R., Gunning A. P., Morris V. J., Wilde P. J., and Rodríguez Patino J. M. (2008) Effect of hydrocarbon chain and pH on structural and topographical characteristics of phospholipid monolayers. J. Phys. Chem. B 112, 7651–7661 10.1021/jp8013157 [DOI] [PubMed] [Google Scholar]
- 28. Ban T., Hamada D., Hasegawa K., Naiki H., and Goto Y. (2003) Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J. Biol. Chem. 278, 16462–16465 10.1074/jbc.C300049200 [DOI] [PubMed] [Google Scholar]
- 29. Castillo N., Monticelli L., Barnoud J., and Tieleman D. P. (2013) Free energy of WALP23 dimer association in DMPC, DPPC, and DOPC bilayers. Chem. Phys. Lipids 169, 95–105 10.1016/j.chemphyslip.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 30. Makino K., Yamada T., Kimura M., Oka T., Ohshima H., and Kondo T. (1991) Temperature- and ionic strength-induced conformational changes in the lipid headgroup region of liposomes as suggested by ζ potential data. Biophys. Chem. 41, 175–183 10.1016/0301-4622(91)80017-L [DOI] [PubMed] [Google Scholar]
- 31. Chibowski E., and Szcześ A. (2016) ζ potential and surface charge of DPPC and DOPC liposomes in the presence of PLC enzyme. Adsorption 22, 755–765 10.1007/s10450-016-9767-z [DOI] [Google Scholar]
- 32. Sierra M. B., Pedroni V. I., Buffo F. E., Disalvo E. A., and Morini M. A. (2016) The use of ζ potential as a tool to study phase transitions in binary phosphatidylcholines mixtures. Colloids Surf. B Biointerfaces 142, 199–206 10.1016/j.colsurfb.2016.02.061 [DOI] [PubMed] [Google Scholar]
- 33. Disalvo E. A., Lairion F., Martini F., Tymczyszyn E., Frías M., Almaleck H., and Gordillo G. J. (2008) Structural and functional properties of hydration and confined water in membrane interfaces. Biochim. Biophys. Acta 1778, 2655–2670 10.1016/j.bbamem.2008.08.025 [DOI] [PubMed] [Google Scholar]
- 34. Abusharkh S. E., Erkut C., Oertel J., Kurzchalia T. V., and Fahmy K. (2014) The role of phospholipid headgroup composition and trehalose in the desiccation tolerance of Caenorhabditis elegans. Langmuir 30, 12897–12906 10.1021/la502654j [DOI] [PubMed] [Google Scholar]
- 35. Uemura M., Joseph R. A., and Steponkus P. L. (1995) Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol. 109, 15–30 10.1104/pp.109.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sugawara Y. S. (1990) Effect of cold acclimation and modification of the plasma membrane lipid composition on lamellar-to-hexagonal II phase transitions in rye protoplasts. Cryobiology 27, 667 [Google Scholar]
- 37. O'Leary E. I., Jiang Z., Strub M. P., and Lee J. C. (2018) Effects of phosphatidylcholine membrane fluidity on the conformation and aggregation of N-terminally acetylated α-synuclein. J. Biol. Chem. 293, 11195–11205 10.1074/jbc.RA118.002780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hou Q., Ufer G., and Bartels D. (2016) Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 39, 1029–1048 10.1111/pce.12666 [DOI] [PubMed] [Google Scholar]
- 39. Koldsø H., Shorthouse D., Hélie J., and Sansom M. S. (2014) Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput. Biol. 10, e1003911 10.1371/journal.pcbi.1003911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liao S. M., Du Q. S., Meng J. Z., Pang Z. W., and Huang R. B. (2013) The multiple roles of histidine in protein interactions. Chem. Cent J. 7, 44 10.1186/1752-153X-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Engerer L. K., and Hanusa T. P. (2011) Geometric effects in olefinic cation-π interactions with alkali metals: a computational study. J. Org. Chem. 76, 42–49 10.1021/jo101307z [DOI] [PubMed] [Google Scholar]
- 42. Priyakumar U. D., Punnagai M., Mohan G. P., and Sastry G. N. (2004) A computational study of cation-π interactions in polycyclic systems: exploring the dependence on the curvature and electronic factors. Tetrahedron 60, 3037–3043 10.1016/j.tet.2004.01.086 [DOI] [Google Scholar]
- 43. Meyer E. A., Castellano R. K., and Diederich F. (2003) Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 42, 1210–1250 10.1002/anie.200390319 [DOI] [PubMed] [Google Scholar]
- 44. Remko M., and Rode B. M. (2006) Effect of metal ions (Li+, Na+, K+, Mg2+, Ca2+, Ni2+, Cu2+, and Zn2+) and water coordination on the structure of glycine and zwitterionic glycine. J. Phys. Chem. A 110, 1960–1967 10.1021/jp054119b [DOI] [PubMed] [Google Scholar]
- 45. Svensson J., Palva E. T., and Welin B. (2000) Purification of recombinant Arabidopsis thaliana dehydrins by metal ion affinity chromatography. Protein Expr. Purif. 20, 169–178 10.1006/prep.2000.1297 [DOI] [PubMed] [Google Scholar]
- 46. Du Q. S., Li D. P., Liu P. J., and Huang R. B. (2008) Molecular potential energies in dodecahedron cell of methane hydrate and dispersion correction for DFT. J. Mol. Graph. Model. 27, 140–146 10.1016/j.jmgm.2008.03.008 [DOI] [PubMed] [Google Scholar]
- 47. Sirois S., Proynov E. I., Truchon J. F., Tsoukas C. M., and Salahub D. R. (2003) A density functional study of the hydrogen-bond network within the HIV-1 protease catalytic site cleft. J. Comput. Chem. 24, 1110–1119 10.1002/jcc.10176 [DOI] [PubMed] [Google Scholar]
- 48. Simon E. W. (1974) Phospholipids and plant membrane permeability. New Phytol. 73, 377–420 10.1111/j.1469-8137.1974.tb02118.x [DOI] [Google Scholar]
- 49. Sankaran J., Shi X., Ho L. Y., Stelzer E. H., and Wohland T. (2010) ImFCS: a software for imaging FCS data analysis and visualization. Opt. Express 18, 25468–25481 10.1364/OE.18.025468 [DOI] [PubMed] [Google Scholar]
- 50. Wawrezinieck L., Rigneault H., Marguet D., and Lenne P. F. (2005) Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89, 4029–4042 10.1529/biophysj.105.067959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jo S., Kim T., and Iyer V. G., and Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- 52. Berendsen H. J., Postma J. P., van Gunsteren W. F., and Hermans J. (1981) Interaction models for water in relation to protein hydration. Intermol. Forces 31, 331–342 [Google Scholar]
- 53. Essmann U., Perera L., Berkowitz M. L., Darden T., and Lee H. P. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 10.1063/1.470117 [DOI] [Google Scholar]
- 54. Nose S, and Nosé S. N. S. (1984) A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511 10.1063/1.447334 [DOI] [Google Scholar]
- 55. Hoover W. G. (1985) Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A 31, 1695–1697 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]
- 56. Parrinello M. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 10.1063/1.328693 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.