Abstract
Colorectal cancer (CRC) is the third most common malignance. Although great efforts have been made to understand the pathogenesis of CRC, the underlying mechanisms are still unclear. It is now clear that more than 90% of the total genome is actively transcribed, but lack of protein‐coding potential. The massive amount of RNA can be classified as housekeeping RNAs (such as ribosomal RNAs, transfer RNAs) and regulatory RNAs (such as microRNAs [miRNAs], PIWI‐interacting RNA [piRNAs], tRNA‐derived stress‐induced RNA, tRNA‐derived small RNA [tRFs] and long non‐coding RNAs [lncRNAs]). Small non‐coding RNAs are a group of ncRNAs with the length no more than 200 nt and they have been found to exert important regulatory functions under many pathological conditions. In this review, we summarize the biogenesis and functions of regulatory sncRNAs, such as miRNAs, piRNA and tRFs, and highlight their involvements in cancers, particularly in CRC.
Keywords: colorectal cancer, miRNA, piRNA, small non‐coding RNA, tRF
1. INTRODUCTION
Colorectal cancer (CRC) is the third most common malignance and is one leading cause of cancer‐related deaths in the world.1 Despite great efforts that have been made to understand the pathogenesis of CRC, the underlying mechanisms are still largely unknown. Increasing evidence has demonstrated that CRC is a heterogeneous disease, and its pathogenesis is involved with the activation of oncogenes and inactivation of tumour‐suppressive genes, which are mostly resulting from genetic mutations and epigenetic alterations, the latter including DNA methylation, histone modification and non‐coding RNAs (ncRNAs).2, 3, 4, 5, 6, 7
Large‐scale genome sequencing has indicated that the human genome encodes approximately 20 000 protein‐coding transcripts, which account for only around 2% of the genome, while more than 90% of the total genome is actively transcribed, but lack of protein‐coding potential, referred to as ncRNAs.8, 9 For several decades, ncRNAs were considered as ‘evolutionary junk’. However, more and more evidence has shown that a part of ncRNAs are functional RNA molecules.10 These functional RNA transcripts are composed by housekeeping ncRNAs, including highly abundant ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), as well as regulatory ncRNAs, such as microRNAs (miRNAs), PIWI‐interacting RNA (piRNAs), tRNA‐derived small RNA (tRFs), small nucleolar RNA (snoRNAs), siRNAs, long non‐coding RNAs (lncRNAs) and circular RNAs (circRNAs). In addition, according to the length of ncRNAs, they are divided into two subclasses: small or short non‐coding RNAs (sncRNAs, 18‐200 nt) and lncRNAs (>200 nt).
Long non‐coding RNAs are a subgroup of non‐coding RNAs, with more than 200 nucleotides in length and no protein coding potential. Long non‐coding RNAs have tissue‐specific expression and exert regulatory functions in many biological and pathological processes. They can function as both tumour suppressors and promoters in CRC development.11 For example, H19 is an imprinted oncofoetal ncRNA, but it is hypomethylated and thus up‐regulated in CRC. H19 can promote the development of CRC via generating miRNA or by serving as ceRNA.12 Down‐regulation of lncRNA MEG3 can promote colorectal adenocarcinoma cell proliferation and inhibit the apoptosis by up‐regulating TGF‐β1 and its downstream sphingosine kinase 1.13 Koduru et al identified differentially expressed lncRNAs in CRC samples. It revealed 18 lncRNAs in tumour vs benign, 89 in metastasis vs benign, and 15 in metastasis vs tumour groups being significantly expressed.14
Circular RNAs are another type of RNA with their 3′‐ and 5′‐ends joined together to form a covalently closed loop. They are widely expressed in human cells and have essential roles in the progression of CRC. Circular RNAs can function as a sponge for miRNAs to modulate gene expression by eliminating the inhibitory effect of miRNAs on their target genes. For instance, circRNA hsa_circ_0000523 can regulate the proliferation and apoptosis of CRC cells as miRNA sponge.15 Circ‐ZNF609 promotes migration of CRC by inhibiting Gli1 expression via microRNA‐150.16
Recent studies have revealed that regulatory sncRNAs (miRNAs, piRNAs, tRFs and snoRNAs) can also function as important regulators in gene expression, and play crucial roles in many physiological and pathological processes. And the abnormal expression of these sncRNAs is involved in many human diseases, including cancers.17 In this review, we provide an overview of the representative classes of sncRNAs, including miRNAs, piRNAs and tRFs, and summarize their involvements in CRC, focusing on their roles in the initiation and development of CRC, as well as their biomarker potential (Table 1).
Table 1.
Dysregulated small non‐coding RNAs in CRC
| Up‐regulated | Reference | Down‐regulated | Reference |
|---|---|---|---|
| MicroRNAs | |||
| let‐7b/g | 38 | let‐7a | 56 |
| miR‐7 | 37 | miR‐7 | 57 |
| miR‐9 | 39 | miR‐9 | 37 |
| miR‐17 | 40 | miR‐18a* | 59 |
| miR‐20 | 41 | miR‐26b | 60 |
| miR‐21 | 39, 42 | miR‐27b | 61 |
| miR‐23a | 35 | miR‐29b | 37 |
| miR‐31 | 40, 42 | miR‐34a | 62 |
| miR‐92a | 43 | miR‐101 | 63 |
| miR‐96 | 44 | miR‐125 | 64 |
| miR‐106 | 84 | miR‐138 | 65 |
| miR‐135 | 45, 75 | miR‐143 | 66, 67 |
| miR‐141 | 46 | miR‐144 | 68 |
| miR‐155 | 47 | miR‐145 | 67, 69 |
| miR‐193a‐3p | 35 | miR‐194 | 70 |
| miR‐205 | 40 | miR‐195 | 71 |
| miR‐214 | 48 | miR‐320a | 72 |
| miR‐224 | 49 | miR‐365 | 73 |
| miR‐338‐5p | 35 | miR‐491 | 74 |
| miR‐372 | 50 | ||
| miR‐708 | 51 | ||
| piRNAs | |||
| piR‐651 | 91 | piR‐015551 | 101 |
| piR‐823 | 100 | ||
| piR‐54878 | 91 | ||
| piR59056 | 91 | ||
| piR‐62701 | 91 | ||
| tRNA‐derived fragments | |||
| tRF‐3LeuCAG | 116 | tRF/miR‐1280 | 117 |
piRNA, PIWI‐interacting RNA, tRNA, transfer RNA.
2. MICRORNAS
MicroRNAs are a class of sncRNAs containing approximately 18‐25 nucleotides, highly conserved and present in eukaryotic cells. In general, the majority of miRNA‐coding genes are located in intergenic or intragenic regions. The generation of mature miRNAs is a multi‐step process that starts in the nucleus and ends in the cytoplasm. Canonically, most miRNAs are transcribed as large mono‐ or polycistronic primary miRNA precursors (pri‐miRNAs) in the nucleus by RNA polymerase II (Pol. II), while other pri‐miRNAs are transcribed by RNA Pol. III. All the pri‐miRNAs contain a 5′ cap and a poly A tail at the 3′ untranslated region (UTR). Then, pri‐miRNAs are processed to generate miRNA precursors (pre‐miRNAs) by RNase III Drosha in complex with DGCR8, a RNA‐binding protein functioning as a ruler to measure cleavage point. The pre‐miRNAs are subsequently recognized and exported to the cytoplasm by Exportin‐5 (Exp5). In the cytosol, the pre‐miRNAs are cleaved by the RNase III endonuclease Dicer to yield a miRNA duplex, which contains a leading strand or miR and a passenger strand or miR*. The functional strand is loaded into the RNA induced silencing complex (RISC), while another one is destroyed following the attachment to argon‐binding proteins (AGO). miRNAs can negatively regulate gene expression in a post‐translational manner. Generally, after being loaded into RISC, the functional miRNAs recognize and basepair with specific seed sequences within the 3′ UTR of target mRNAs, resulting in direct mRNA degradation or translational silencing. When miRNAs are almost completely complementary with their mRNA targets, the targeted mRNA can be directly cleaved and degraded. However, the overwhelming majority of miRNAs and their target mRNAs are only partially complementary. In this case, the non‐perfect pairing manner, which usually involves a seed pairing of just six to seven nucleotides in length, often leads to the transcription inhibition (Figure 1).18 Additionally, recent studies have shown that miRNAs may also positively regulate gene expression. miR‐10a binds to the 5′ UTR of ribosomal protein mRNAs and promotes their translation.19 miR‐21 directly targets mitochondrial cytochrome b (mt‐Cytb) and enhances mt‐Cytb translation.20
Figure 1.
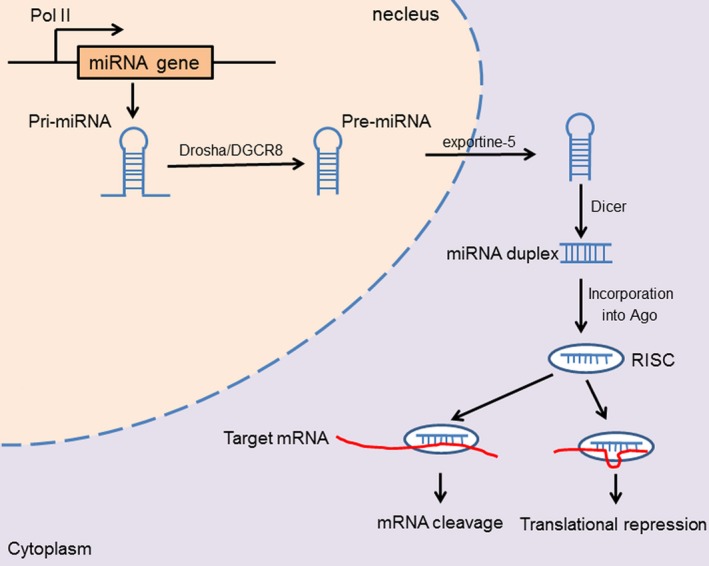
miRNA biogenesis and their functions. A miRNA gene is transcribed by RNA polymerase II (Pol II) to form a primary transcript (pri‐miRNA) with a hairpin loop. The hairpin in the pri‐miRNA is identified by the Drosha‐DGCR8 microprocessor complex, and the pri‐miRNA is converted into the pre‐miRNA. The pre‐miRNA is exported from the nucleus into the cytoplasm by Exportin‐5. Then the pre‐miRNA is processed by the RNase III endonuclease Dicer, yielding a miRNA‐double duplex, which is loaded into Argonaute (Ago) protein to generate the RNA‐induced silencing complex (RISC), then one strand of the duplex is degraded while the other guides the RISC to the target mRNA, thus leading to direct mRNA cleavage or translational repressing
As novel regulators of gene expression, miRNAs have a profound impact on the initiation and development of many cancers.17, 21, 22 A series of studies have revealed that miRNAs can act as oncogenes called ‘Oncomir’ or tumour suppressor, thus exerting an important role in many biological behaviours of cancer cells, involved with cell proliferation, apoptosis, metastasis and drug resistance. Furthermore, aberrant miRNAs can also be used as a potential diagnostic and prognostic biomarker for cancers.17, 23, 24, 25, 26, 27, 28 Recently, the involvement of miRNAs in the pathogenesis of CRC has been relatively clarified.7, 29, 30, 31, 32
By microarray‐based miRNA profiling platforms and next‐generation sequencing (NGS) approaches, a number of differentially expressed miRNAs have been identified in CRC. In 2003, Michael et al firstly reported some altered expressed miRNAs, among which, miR‐143 and miR‐145 were found to be reduced in CRC tissue compared to healthy tissue.33 In another study, Schetter et al screened 37 differentially expressed miRNAs by miRNAs microarray in 84 CRC tissues.34 In addition, a panel of dysregulated miRNAs has been detected by Real‐time PCR in CRC. Yong et al identified some differentially expressed miRNAs in both CRC tissues and blood derived from CRC patients, of which, miR‐23a, miR‐193a‐3p and miR‐338‐5p were significantly up‐regulated in CRC tissues, and the levels of these miRNAs are positively related between CRC tissue and blood.35 Meanwhile, an exosomal miRNA profile was used to highlight the most abundant miRNAs in the blood of 88 patients with CRC.36 As well as circulating miRNAs, some stool‐based miRNAs were also observed to be dysregulated and they also can be used as a biomarker for CRC. For example, miR‐7, miR‐17, miR‐20a and miR‐21 are increased in the stool of CRC patients, while miR‐9, miR‐29b, miR‐138 and miR‐143 are decreased.37
Dysregulated miRNAs have significant effects on the biology of cancer cells, and they can act as oncogenes or tumour suppressor genes, therefore contributing to the development of CRC. Typically, oncogenic miRNAs are up‐regulated in CRC and often target endogenous tumour suppressors. Some of these miRNAs are let‐7b and 7 g,38 miR‐9,39 miR‐17,40 miR‐20,41 miR‐21,39, 42 miR‐31,40, 42 miR‐92a,43 miR‐96,44 miR‐135,45 miR‐141,46 miR‐155,47 miR‐205,40 miR‐214,48 miR‐224,49 miR‐37250 and miR‐708.51 For example, miR‐21, which has been proven as an oncogenic miRNA in several cancers, can promote the proliferation and metastasis of CRC cells by targeting some key tumour suppressive genes, including PDCD4, TIAM1, SPRTY, PTEN, TGFBR2 and CDC25A.52, 53, 54, 55, 56 miR‐92a has been shown to promote the epithelial‐mesenchymal transition (EMT) process through the suppression of E‐cadherin in CRC.43 miR‐96 is up‐regulated in CRC and has been shown to target p53 inducible nuclear protein 1 (TP53INP1), exerting suppressive effects on p53 activity.44 miR‐224 has also recently been found to enhance the metastasis of CRC through its modulation of SMAD4.49 On the other hand, tumour‐suppressive miRNAs are commonly down‐regulated in CRC tissues, and play a crucial role in down‐regulating oncogenes in CRC. These miRNAs are let‐7a,57 miR‐7,58 miR‐18a*,59 miR‐26b,60 miR‐27b,61 miR‐34a,62 miR‐101,63 miR‐125,64 miR‐138,65 miR‐143,66, 67 miR‐144,68 miR‐145,67, 69 miR‐194,70 miR‐195,71, miR‐320a,72 miR‐36573 and miR‐491.74 miRNAs let 7a, miR‐143, miR‐18a* and miR‐145 suppress the development of CRC by down‐regulating RAS.57, 59, 66, 69 In addition, miR‐143 and miR‐145 also exert anti‐tumour roles through their down‐regulation of insulin‐like growth factor 1 receptor (IGF1R).67 miR‐195 and miR‐491 have been found to promote apoptosis in CRC cells through the targeting of B‐Cell CLL/Lymphoma 2 (BCL2) and BCL2‐Like 1 (BCLXL), respectively.71, 74
The onset and development of CRC are involved with the activation of many oncogenic signalling pathways, such as Wnt, Ras, TGF‐β and inflammatory signalling pathways. And these signalling pathways can be regulated by miRNAs.7 Wnt signalling is one of the most frequently changed pathways in CRC. Previous studies have revealed that miRNAs play an important role in regulating the Wnt signalling pathway through targeting the key molecules of the Wnt pathway. miR‐135a/b is highly expressed in CRC and is able to directly target APC, resulting in the reduction in APC expression and in the up‐regulation of Wnt signalling. miR‐135a/b is also predicted to target secreted frizzled‐related protein 4 (SFRP4), which is a Wnt/β‐catenin inhibitor. Meanwhile, recent studies have showed that miR‐135b can be transcriptionally activated by β‐catenin/TCF4, which leads to the up‐regulation of miR‐135b in CRC.45, 75 Unlike miR‐135, miR‐21 can increase β‐catenin nuclear translocation and promote tumorigenesis in CRC.76 miR‐155 is also a Wnt/β‐catenin stimulator by targeting HMGB1 and indirectly increasing Wnt/β‐catenin expression.77, 78 The RAS gene family, such as NRAS, HRAS and KRAS, and KRAS has been frequently mutated in CRCs. The dysregulation of KRAS family has been shown to be well‐associated with poorer outcomes, in terms of shorter survival times, and being more aggressive and drug‐resistant. The RAS genes have several miRNA let‐7 binding sites at their 3′ UTR. The reduction in let‐7 in cancer tissues is correlated with higher KRAS mRNA expression; and let‐7 miRNA suppressed colon cancer growth and proliferation.79 miR‐143 has been also shown down‐regulated in CRC tissues, and reduced expression of miR‐143 led to cell proliferation in vitro, which is linked to the increased expression of KRAS.66 In contrast, miR‐31 has been shown to negatively regulate KRAS inhibitor RASA1; thus, miR‐31 could be a potent enhancer of KRAS in CRC. miRNAs also play important roles in regulating TGF‐β signalling.80, 81 The miR‐17 family has a crosstalk with the TGF‐β signalling pathway. The miR‐17 family has eight miRNAs, including miR‐17, miR‐18a/b, miR‐20a/b, miR‐93 and miR‐106a/b. miR‐17 targets and inhibits PTEN and RHOE (RND3).42, 82 miR‐20a promotes cancer progression by facilitating CRC cell line migration and invasion and up‐regulating the expression of EMT markers.83 miR‐106a/b seems also to enhance EMT and metastasis by targeting TGF‐β receptor TGFBR2.84
3. PIWI‐INTERACTING RNAS
P‐element induced wimpy testis (PIWI)‐interacting RNAs are a class of small ncRNAs of 26‐31 nucleotides, which are defined because of their interaction with PIWI subfamily of Argonaute proteins.85, 86, 87 PIWI‐interacting RNA shows a very diverse set of nucleotide sequences compared with other known cellular RNA family members. According to their origin, piRNAs are classified into three groups: transposon‐derived piRNAs, mRNA‐derived piRNAs and lncRNAs‐derived piRNAs.88, 89, 90 PIWI‐interacting RNA are mostly transcribed as large single‐stranded transcripts, which are then exported to the cytoplasm and processed into mature piRNAs independently from Dicer. It is currently known that two mechanisms are involved in the generation of mature piRNAs: the primary synthesis mechanism and the ‘ping‐pong’ amplification mechanism. First, the primary transcript is cleaved by the riboendonuclease Zucchini, and the 3′ fragment of the product is incorporated into PIWI proteins and trimmed by a 3′ to 5′ exonuclease. Then, the 2′ hydroxy group at the 3′ end is methylated by the enzyme Hen1. The piRNA/PIWI complex migrates back to the nucleus, which reaches their target transcripts and mobilizes the silencing machinery to block the transcription of transposable elements, maintaining genome integrity. In addition, the ‘ping‐pong’ amplification mechanism is involved in the process of piRNAs' accumulation. As well as migrating into the nucleus, piRNAs can also bind with AGO3 or AUB proteins to form piRNA/Ago or piRNA/Aub complexes in the cytoplasm, which contain complementary sequences and provide substrate to each other, therefore producing new antisense piRNAs. The ping‐pong framework has been identified in zebrafish, D. melanogaster and other various species. However, previous data have revealed that piRNAs biogenesis during adult spermatogenesis in mice is independent of the ping‐pong mechanism.88, 89, 90 Therefore, the biogenesis of piRNAs is still not clear in mammals, which needs further investigation. Actually, the functions of piRNAs are still not perfectly clear. Up to date, piRNAs have been shown to be involved in transposon silencing, epigenetic regulation, gene and protein regulation, genome rearrangement, spermatogenesis and germ stem‐cell maintenance.88, 89, 90 As reviewed above, piRNAs can bind with PIWI proteins and direct them to their transposon targets, therefore contributing to genetic diversity and genetic instability. Additionally, PIWI can serve as an epigenetic activator and so, as a partner of PIWI proteins, piRNAs are likely to be implicated in transcriptional silencing or activation. PIWI‐interacting RNA is also found to regulate gene expression and protein stability. For example, piRNAs can regulate the ubiqutination level of mouse PIWI protein MIWI by enhancing MIWI interaction with an APC/C substrate‐binding subunit. Recently, some piRNAs have been found to be dysregulated in tumour tissues, and altered piRNAs play an important role in cancer cell proliferation, apoptosis and metastasis, and they may become potential prognostic and diagnostic biomarkers during cancer development.17, 91 Previous studies have shown that piR‐34377, piR‐34736, piR‐36249, piR‐35407 and piR‐36318 are significantly down‐regulated, while piR‐651, piR‐932, piR‐4987, piR‐20365, piR‐20485, piR‐20582, piR‐36743, piR‐36026, piR‐31106 and piR‐021285 are up‐regulated in breast cancer.92, 93 piR‐651, piR‐32105, piR‐58099 and piR‐59056 are increased in gastric cancer, whereas piR‐823 is decreased.94, 95 piR‐55490 is down‐regulated in lung cancer, while piR‐651 is up‐regulated96, 97; piR‐55490 inhibits lung cancer cell proliferation and the mechanism is associated with its binding to 3′ UTR of mTOR messenger RNA (mRNA).97 piR‐32051, piR‐39894 and piR‐43607 were found up‐regulated in kidney cancer tissues.98 PIWI‐interacting RNA DQ594040 is one down‐regulated piRNA in bladder cancer, and its overexpression can inhibit bladder cancer cell proliferation, colony formation and promote cell apoptosis through up‐regulating TNFSF4 protein.99 As for CRC, piR‐651 was found to be up‐regulated in tumour tissues, which is associated with metastasis state. piR59056, piR‐54878 and piR‐62701 are also highly expressed in CRC, and are associated with recurrence‐free survival.91 piR‐823 contributes to colorectal tumorgenesis by enhancing the transcriptional activity of HSF1.100 Chu et al observed that an lncRNA, LNC00964‐3, which includes the piR‐015551 sequence, is significantly lower in CRC tissues. Meanwhile, the expression of piR‐015551 is positively correlated with LNC00964‐3, indicating that piR‐015551 might be generated from LNC00964‐3, and may be implicated in the development of CRC.101
4. TRNA‐DERIVED FRAGMENTS
Transfer RNAs are a group of ncRNAs that function as a fundamental component of the translation machinery, which can help deliver amino acids to the ribosome. Traditionally, tRNAs have been considered as key actors in protein synthesis.102, 103 However, recent studies have revealed that a mass of non‐coding small RNAs are derived from tRNAs, namely which serve as novel regulators in some pathophysiologic processes.104 On basis of the length and cleavage sites of tRNAs, small non‐coding RNA derived from tRNAs can be classified into two major groups: tRNA‐derived stress‐induced RNA (tiRNAs), also named tRNA‐halves, produced by specific cleavage of the anticodon loops of mature tRNAs, with the length of 28‐36 nt105, 106; tRNA‐derived fragment (tRFs), about 14‐30 nt length, derived from the mature or primary tRNAs (Figure 2).107 Herein, we will focus on the roles of tRFs in the development of CRC.
Figure 2.
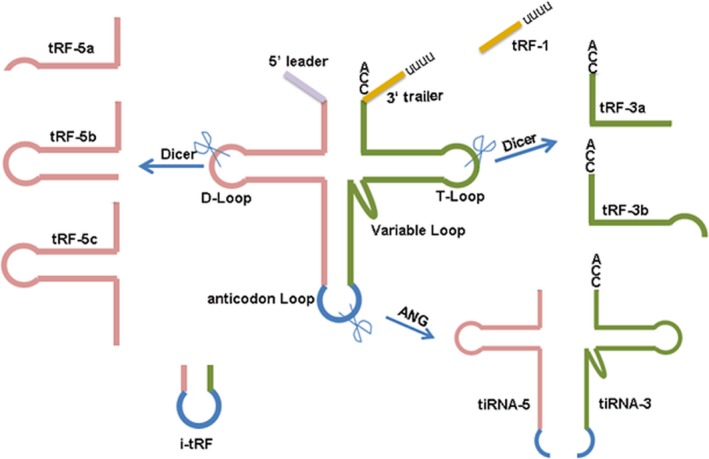
The types of tsRNAs are classified by size and sequence location in the tRNA structure. Based on the length and cleavage sites of tRNAs, small non‐coding RNA derived from tRNAs can be classified into two major groups: tRNA‐derived stress‐induced RNA (tiRNAs), with the length of 28‐36 nt, and tRNA‐derived fragment (tRFs), about 14‐30 nt length
tRNA‐derived fragments have three subtypes: tRF‐5s, tRF‐3s and inter tRFs.107, 108 The tRF‐5s are usually formed by a cleavage in the D‐loop or arm region between D‐loop and anticodon loop of a mature tRNA. The tRF‐3s are generated from a cleavage in the T‐loop, with an end trinucleotides ‘CCA’. The inter tRFs, also termed as tRF‐1s, originate from internal region of mature tRNA, which includes anticodon loop and part of D‐loop and T‐loop.109 Based on the cleavage site and sequence length, these fragments are further sub‐classified: tRFs‐5a (14‐16 nt), tRFs‐5b (22‐24 nt), tRFs‐5c (28‐30 nt), tRFs‐3a (18 nt) and tRFs‐3b (22 nt) and most tRF‐1s identified in human cells are 20 and 36 nt in length.109 The production of tRF‐5s and tRF‐3s has reported to be dependent on Dicer.109 However, the specific process of generating tRF‐5s and tRF‐3s is still not clear. Meanwhile, the biogenesis mechanism of tRF‐1s is also unknown. Recently, tRFs have been found to play an important role in many biological and pathological processes, and they are associated with many diseases, such as virus infection, metabolic disorder, neurodegenerative diseases and cancers.17, 109, 110 To date, the regulatory mechanisms of tRFs mainly involve several strategies as follows. tRNA‐derived fragments have also been reported to be involved in global translational inhibition in human cells. A 19‐nt long tRF‐5 derived from tRNAGln down‐regulates the expression of a reporter gene, but which does not have a reverse complementary target sequence, suggesting that the translational repression mediated by this tRF‐5 is non‐specific. The translational inhibition of tRFs required a conserved ‘GG’ dinucleotide for their activity, and it may exert the function through interacting with the ribosome.111 In addition to global translational inhibition, some tRFs mediate gene silencing similarly to miRNAs.112 For instance, upon infection of respiratory syncytial virus (RSV), tRF‐5GluCTC is drastically induced and exhibits a gene‐silencing role by targeting mRNA.113
Recently, a series of tRFs have been shown to be altered in cancer tissues, and they are implicated in the development of cancers. tRF‐1001, one of tRF‐1s, is highly expressed in prostate cancer cells and other several cancer cell lines. Knockdown of tRF‐1001 can repress the proliferation of prostate cancer cells, which is associated with DNA synthesis suppression and G2 stage arrest.107 CU1276, a tRF‐3 derived from tRNA(Gly), was firstly identified in human mature B cells. Overexpression of CU1276 in lymphoma cells can suppress cell proliferation. Meanwhile, lymphomas cells with CU1276 knockdown are sensitive to DNA damage, resulting in an accumulation of mutations and genomic aberrations during tumour development.17 By small RNA‐sequencing of breast cancer cells, it was found that inter tRFs derived from mature tRNA(Glu), tRNA(Asp), tRNA(Gly) and tRNA(Tyr) are induced under hypoxic stress and these inter tRFs are also found to repress cell growth under serum‐starvation, cancer cell invasion and metastasis in breast cancer cells.114 The tRF‐3ThrTGT is significantly down‐regulated in primary breast cancer and metastatic tumours; overexpression of tRF‐3ThrTGT in breast cancer cells remarkably inhibits cell invasion and migration.115 tRF‐3LeuCAG, identified in HeLa and HCT‐16 cells, can promote cell viability.116 In CRC, tRF/miR‐1280 was found to be decreased in tumour tissues. The overexpression of tRF/miR‐1280 can reduce cell proliferation and colony formation, whereas knockdown of it would reverse these effects. tRF/miR‐1280 reduces tumour formation and metastasis by directly targeting the Notch ligand JAG2. Interestingly, Notch signalling pathways are essential for cancer stem‐like cells (CSC) in CRC progression. Therefore, tRF/miR‐1280 suppresses CRC growth and metastasis by repressing Notch signalling pathways that support CSC phenotypes.117
5. CONCLUSION
Although great achievements have been made in understanding the pathogenesis of CRC, the underlying mechanisms are still needed for further investigations. For decades, many studies have provided a lot of evidence that sncRNAs are important regulatory molecules and play a crucial role in cancer development and progression. So, it is believed that sncRNAs are involved in the pathogenesis of CRC. Notably, many miRNAs have been demonstrated to be dysregulated in CRC, and they may exert important functions in the development of CRC. Furthermore, they have been found to act as potential diagnostic biomarkers for CRC. In addition, some piRNAs and tRFs have also been proven to be implicated in the development of CRC. However, studies on them are still at a very early stage, and many questions still need to be addressed. In conclusion, considering sncRNAs as important regulators in CRC, we need to shed light on many aspects of their biogenesis and functions in CRC.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
This work is supported by the fundament of People's Hospital of Taizhou.
Chen H, Xu Z, Liu D. Small non‐coding RNA and colorectal cancer. J Cell Mol Med. 2019;23:3050–3057. 10.1111/jcmm.14209
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. El Bairi K, Tariq K, Himri I, et al. Decoding colorectal cancer epigenomics. Cancer Genet. 2018;220:49‐76. [DOI] [PubMed] [Google Scholar]
- 3. Porcellini E, Laprovitera N, Riefolo M, Ravaioli M, Garajova I, Ferracin M. Epigenetic and epitranscriptomic changes in colorectal cancer: diagnostic, prognostic, and treatment implications. Cancer Lett. 2018;419:84‐95. [DOI] [PubMed] [Google Scholar]
- 4. Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8(12):686‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pellegrini ML, Argibay P, Gomez DE. Genetics and epigenetics of colorectal cancer. Acta Gastroenterol Latinoam. 2011;41(3):247‐261. [PubMed] [Google Scholar]
- 6. Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo Y, Bao Y, Yang W. Regulatory miRNAs in colorectal carcinogenesis and metastasis. Int J Mol Sci. 2017;18(4):E890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. International Human Genome Sequencing Consortium . Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931‐945. [DOI] [PubMed] [Google Scholar]
- 9. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337(6099):1159, 1161. [DOI] [PubMed] [Google Scholar]
- 11. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253‐1261. [DOI] [PubMed] [Google Scholar]
- 12. Li CF, Li YC, Wang Y, Sun LB. The effect of lncRNA H19/miR‐194‐5p axis on the epithelial‐mesenchymal transition of colorectal adenocarcinoma. Cell Physiol Biochem. 2018;50(1):196‐213. [DOI] [PubMed] [Google Scholar]
- 13. Dong X, Wang J, Li T, Xu YP, Li SY. Down regulation of lncRNA MEG3 promotes colorectal adenocarcinoma cell proliferation and inhibits the apoptosis by up‐regulating TGF‐beta1 and its downstream sphingosine kinase 1. Eur Rev Med Pharmacol Sci. 2018;22(23):8265‐8272. [DOI] [PubMed] [Google Scholar]
- 14. Koduru SV, Tiwari AK, Hazard SW, Mahajan M, Ravnic DJ. Exploration of small RNA‐seq data for small non‐coding RNAs in Human Colorectal Cancer. J Genomics. 2017;5:16‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51(12):e7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu L, Xia J, Yang J, et al. Circ‐ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA‐150. J BUON. 2018;23(5):1343‐1349. [PubMed] [Google Scholar]
- 17. Romano G, Veneziano D, Acunzo M, Croce CM. Small non‐coding RNA and cancer. Carcinogenesis. 2017;38(5):485‐491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281‐297. [DOI] [PubMed] [Google Scholar]
- 19. Orom UA, Nielsen FC, Lund AH. MicroRNA‐10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460‐471. [DOI] [PubMed] [Google Scholar]
- 20. Li H, Zhang X, Wang F, et al. MicroRNA‐21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation. 2016;134(10):734‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang Y, Fang D, Hu J. MicroRNA and its roles in esophageal cancer. Med Sci Monit. 2012;18(3):RA22‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chu Y, Zhu H, Lv L, Zhou Y, Huo J. MiRNA s in oesophageal squamous cancer. Neth J Med. 2013;71(2):69‐75. [PubMed] [Google Scholar]
- 23. Catela Ivkovic T, Voss G, Cornella H, Ceder Y. microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. 2017;407:113‐122. [DOI] [PubMed] [Google Scholar]
- 24. Orellana EA, Kasinski AL. MicroRNAs in cancer: a historical perspective on the path from discovery to therapy. Cancers (Basel). 2015;7(3):1388‐1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113(4):569‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anfossi S, Fu X, Nagvekar R, Calin GA. MicroRNAs, regulatory messengers inside and outside cancer cells. Adv Exp Med Biol. 2018;1056:87‐108. [DOI] [PubMed] [Google Scholar]
- 27. Oom AL, Humphries BA, Yang C. MicroRNAs: novel players in cancer diagnosis and therapies. Biomed Res Int. 2014;2014:959461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blandino G, Fazi F, Donzelli S, et al. Tumor suppressor microRNAs: a novel non‐coding alliance against cancer. FEBS Lett. 2014;588(16):2639‐2652. [DOI] [PubMed] [Google Scholar]
- 29. Orang AV, Barzegari A. MicroRNAs in colorectal cancer: from diagnosis to targeted therapy. Asian Pac J Cancer Prev. 2014;15(17):6989‐6999. [DOI] [PubMed] [Google Scholar]
- 30. Muhammad S, Kaur K, Huang R, et al. MicroRNAs in colorectal cancer: role in metastasis and clinical perspectives. World J Gastroenterol. 2014;20(45):17011‐17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705‐713. [DOI] [PubMed] [Google Scholar]
- 32. Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J. 2012;18(3):244‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882‐891. [PubMed] [Google Scholar]
- 34. Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier: miR‐193a‐3p, miR‐23a and miR‐338‐5p for early detection of colorectal cancer. BMC Cancer. 2013;13:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogata‐Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE. 2014;9(4):e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahmed FE, Ahmed NC, Vos PW, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10(3):93‐113. [PubMed] [Google Scholar]
- 38. Agostini M, Pucciarelli S, Calore F, Bedin C, Enzo M, Nitti D. miRNAs in colon and rectal cancer: a consensus for their true clinical value. Clin Chim Acta. 2010;411(17‐18):1181‐1186. [DOI] [PubMed] [Google Scholar]
- 39. Bandres E, Cubedo E, Agirre X, et al. Identification by Real‐time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non‐tumoral tissues. Mol Cancer. 2006;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687‐3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Earle JS, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12(4):433‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR‐21, miR‐31, miR‐143 and miR‐145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5‐6):397‐402. [DOI] [PubMed] [Google Scholar]
- 43. Zhang G, Zhou H, Xiao H, Liu Z, Tian H, Zhou T. MicroRNA‐92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59(1):98‐107. [DOI] [PubMed] [Google Scholar]
- 44. Gao F, Wang W. MicroRNA‐96 promotes the proliferation of colorectal cancer cells and targets tumor protein p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1) and FOXO3a. Mol Med Rep. 2015;11(2):1200‐1206. [DOI] [PubMed] [Google Scholar]
- 45. Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR‐135 family in colorectal cancer. Cancer Res. 2008;68(14):5795‐5802. [DOI] [PubMed] [Google Scholar]
- 46. Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5‐fluorouracil in vitro. Pharmacol Res. 2007;56(3):248‐253. [DOI] [PubMed] [Google Scholar]
- 47. Valeri N, Gasparini P, Fabbri M, et al. Modulation of mismatch repair and genomic stability by miR‐155. Proc Natl Acad Sci U S A. 2010;107(15):6982‐6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Polytarchou C, Hommes DW, Palumbo T, et al. MicroRNA214 Is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis‐associated cancer in mice. Gastroenterology. 2015;149(4):981‐992.e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ling H, Pickard K, Ivan C, et al. The clinical and biological significance of MIR‐224 expression in colorectal cancer metastasis. Gut. 2016;65(6):977‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu J, Jin L, Jiang L, et al. Serum miR‐372 is a diagnostic and prognostic biomarker in patients with early colorectal cancer. Anticancer Agents Med Chem. 2016;16(4):424‐431. [DOI] [PubMed] [Google Scholar]
- 51. Lei SL, Zhao H, Yao HL, et al. Regulatory roles of microRNA‐708 and microRNA‐31 in proliferation, apoptosis and invasion of colorectal cancer cells. Oncol Lett. 2014;8(4):1768‐1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA‐21 (miR‐21) post‐transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128‐2136. [DOI] [PubMed] [Google Scholar]
- 53. Yu Y, Kanwar SS, Patel BB, et al. MicroRNA‐21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis. 2012;33(1):68‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sayed D, Rane S, Lypowy J, et al. MicroRNA‐21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19(8):3272‐3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cottonham CL, Kaneko S, Xu L. miR‐21 and miR‐31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285(46):35293‐35302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xiong B, Cheng Y, Ma L, Zhang C. MiR‐21 regulates biological behavior through the PTEN/PI‐3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42(1):219‐228. [DOI] [PubMed] [Google Scholar]
- 57. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let‐7 microRNA family. Cell. 2005;120(5):635‐647. [DOI] [PubMed] [Google Scholar]
- 58. Suto T, Yokobori T, Yajima R, et al. MicroRNA‐7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36(3):338‐345. [DOI] [PubMed] [Google Scholar]
- 59. Tsang WP, Kwok TT. The miR‐18a* microRNA functions as a potential tumor suppressor by targeting on K‐Ras. Carcinogenesis. 2009;30(6):953‐959. [DOI] [PubMed] [Google Scholar]
- 60. Ma YL, Zhang P, Wang F, et al. Human embryonic stem cells and metastatic colorectal cancer cells shared the common endogenous human microRNA‐26b. J Cell Mol Med. 2011;15(9):1941‐1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ye J, Wu X, Wu D, et al. miRNA‐27b targets vascular endothelial growth factor C to inhibit tumor progression and angiogenesis in colorectal cancer. PLoS ONE. 2013;8(4):e60687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamakuchi M, Ferlito M, Lowenstein CJ. miR‐34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105(36):13421‐13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen MB, Yang L, Lu PH, et al. MicroRNA‐101 down‐regulates sphingosine kinase 1 in colorectal cancer cells. Biochem Biophys Res Commun. 2015;463(4):954‐960. [DOI] [PubMed] [Google Scholar]
- 64. Chen H, Xu Z. Hypermethylation‐associated silencing of miR‐125a and miR‐125b: a potential marker in colorectal cancer. Dis Markers. 2015;2015:345080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Long L, Huang G, Zhu H, Guo Y, Liu Y, Huo J. Down‐regulation of miR‐138 promotes colorectal cancer metastasis via directly targeting TWIST2. J Transl Med. 2013;11:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen X, Guo X, Zhang H, et al. Role of miR‐143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385‐1392. [DOI] [PubMed] [Google Scholar]
- 67. Su J, Liang H, Yao W, et al. MiR‐143 and MiR‐145 regulate IGF1R to suppress cell proliferation in colorectal cancer. PLoS ONE. 2014;9(12):e114420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Iwaya T, Yokobori T, Nishida N, et al. Downregulation of miR‐144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391‐2397. [DOI] [PubMed] [Google Scholar]
- 69. Yin Y, Yan ZP, Lu NN, et al. Downregulation of miR‐145 associated with cancer progression and VEGF transcriptional activation by targeting N‐RAS and IRS1. Biochim Biophys Acta. 2013;1829(2):239‐247. [DOI] [PubMed] [Google Scholar]
- 70. Zhao HJ, Ren LL, Wang ZH, et al. MiR‐194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathway. Theranostics. 2014;4(12):1193‐1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu L, Chen L, Xu Y, Li R, Du X. microRNA‐195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400(2):236‐240. [DOI] [PubMed] [Google Scholar]
- 72. Sun JY, Huang Y, Li JP, et al. MicroRNA‐320a suppresses human colon cancer cell proliferation by directly targeting beta‐catenin. Biochem Biophys Res Commun. 2012;420(4):787‐792. [DOI] [PubMed] [Google Scholar]
- 73. Nie J, Liu L, Zheng W, et al. microRNA‐365, down‐regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl‐2. Carcinogenesis. 2012;33(1):220‐225. [DOI] [PubMed] [Google Scholar]
- 74. Nakano H, Miyazawa T, Kinoshita K, Yamada Y, Yoshida T. Functional screening identifies a microRNA, miR‐491 that induces apoptosis by targeting Bcl‐X(L) in colorectal cancer cells. Int J Cancer. 2010;127(5):1072‐1080. [DOI] [PubMed] [Google Scholar]
- 75. Valeri N, Braconi C, Gasparini P, et al. MicroRNA‐135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin PL, Wu DW, Huang CC, et al. MicroRNA‐21 promotes tumour malignancy via increased nuclear translocation of beta‐catenin and predicts poor outcome in APC‐mutated but not in APC‐wild‐type colorectal cancer. Carcinogenesis. 2014;35(10):2175‐2182. [DOI] [PubMed] [Google Scholar]
- 77. Wan J, Xia L, Xu W, et al. Expression and function of miR‐155 in diseases of the gastrointestinal tract. Int J Mol Sci. 2016;17(5):E709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Itou J, Taniguchi N, Oishi I, Kawakami H, Lotz M, Kawakami Y. HMGB factors are required for posterior digit development through integrating signaling pathway activities. Dev Dyn. 2011;240(5):1151‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Akao Y, Nakagawa Y, Naoe T. let‐7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29(5):903‐906. [DOI] [PubMed] [Google Scholar]
- 80. Kent OA, Mendell JT, Rottapel R. Transcriptional regulation of miR‐31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res. 2016;14(3):267‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sun D, Yu F, Ma Y, et al. MicroRNA‐31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol Chem. 2013;288(13):9508‐9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fang L, Li H, Wang L, et al. MicroRNA‐17‐5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974‐2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sokolova V, Fiorino A, Zoni E, et al. The effects of miR‐20a on p21: two mechanisms blocking growth arrest in TGF‐beta‐responsive colon carcinoma. J Cell Physiol. 2015;230(12):3105‐3114. [DOI] [PubMed] [Google Scholar]
- 84. Petrocca F, Vecchione A, Croce CM. Emerging role of miR‐106b‐25/miR‐17‐92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68(20):8191‐8194. [DOI] [PubMed] [Google Scholar]
- 85. Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203‐207. [DOI] [PubMed] [Google Scholar]
- 86. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline‐specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199‐202. [DOI] [PubMed] [Google Scholar]
- 87. Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363‐367. [DOI] [PubMed] [Google Scholar]
- 88. Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141(18):3458‐3471. [DOI] [PubMed] [Google Scholar]
- 89. Fu Q, Wang PJ. Mammalian piRNAs: biogenesis, function, and mysteries. Spermatogenesis. 2014;4:e27889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Han YN, Li Y, Xia SQ, Zhang Y‐Y, Zheng J‐H, Li W. PIWI proteins and PIWI‐interacting RNA: emerging roles in cancer. Cell Physiol Biochem. 2017;44(1):3050‐20. [DOI] [PubMed] [Google Scholar]
- 92. Hashim A, Rizzo F, Marchese G, et al. RNA sequencing identifies specific PIWI‐interacting small non‐coding RNA expression patterns in breast cancer. Oncotarget. 2014;5(20):9901‐9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15(7):563‐568. [DOI] [PubMed] [Google Scholar]
- 94. Cui L, Lou Y, Zhang X, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44(13):1050‐1057. [DOI] [PubMed] [Google Scholar]
- 95. Cheng J, Deng H, Xiao B, et al. piR‐823, a novel non‐coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315(1):12‐17. [DOI] [PubMed] [Google Scholar]
- 96. Li D, Luo Y, Gao Y, et al. piR‐651 promotes tumor formation in non‐small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38(3):927‐936. [DOI] [PubMed] [Google Scholar]
- 97. Peng L, Song L, Liu C, et al. piR‐55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016;37(2):2749‐2756. [DOI] [PubMed] [Google Scholar]
- 98. Li Y, Wu X, Gao H, et al. Piwi‐interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer‐specific survival. Mol Med. 2015;21:381‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356(2 Pt B):561–567. [DOI] [PubMed] [Google Scholar]
- 100. Yin J, Jiang XY, Qi W, et al. piR‐823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017;108(9):1746‐1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chu H, Xia L, Qiu X, et al. Genetic variants in noncoding PIWI‐interacting RNA and colorectal cancer risk. Cancer. 2015;121(12):2044‐2052. [DOI] [PubMed] [Google Scholar]
- 102. Banerjee R, Chen S, Dare K, et al. tRNAs: cellular barcodes for amino acids. FEBS Lett. 2010;584(2):387‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16(2):98‐112. [DOI] [PubMed] [Google Scholar]
- 104. Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2018;19(1):45‐58. [DOI] [PubMed] [Google Scholar]
- 105. Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583(2):437‐442. [DOI] [PubMed] [Google Scholar]
- 106. Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14(10):2095‐2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA‐derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639‐2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kumar P, Mudunuri SB, Anaya J, et al. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43(Database issue):D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhu L, Liu X, Pu W, Peng Y. tRNA‐derived small non‐coding RNAs in human disease. Cancer Lett. 2018;419:3050‐7. [DOI] [PubMed] [Google Scholar]
- 110. Li S, Xu Z, Sheng J. tRNA‐derived small RNA: a novel regulatory small non‐coding RNA. Genes (Basel). 2018;9(5):E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sobala A, Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10(4):553‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA‐derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21(2):368‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA‐derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li LZ, Zhang CZ, Liu LL, et al. miR‐720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35(2):469‐478. [DOI] [PubMed] [Google Scholar]
- 116. Kim HK, Fuchs G, Wang S, et al. A transfer‐RNA‐derived small RNA regulates ribosome biogenesis. Nature. 2017;552(7683):57‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Huang B, Yang H, Cheng X, et al. tRF/miR‐1280 suppresses stem cell‐like cells and metastasis in colorectal cancer. Cancer Res. 2017;77(12):3194‐3206. [DOI] [PubMed] [Google Scholar]


