Abstract
Chronic and acute agonism as well as acute antagonism of CB1 receptors reveal modulation of learning and memory during stable performance of a delayed-nonmatch-to-sample (DNMS) memory task. However, it remains unclear how chronic blockade of the CB1 receptor alters acquisition of the behavioral task. We examined the effects of chronic rimonabant exposure during DNMS task acquisition to determine if blockade of the CB1 receptor with the antagonist rimonabant enhanced acquisition of operant task. Long-Evans rats, trained in the DNMS task before imposition of the trial delay, were surgically implanted with osmotic mini pumps to administer rimonabant (1.0 mg/kg/day) or vehicle (dimethyl sulfoxide/Tween-80/Saline). Following surgical recovery, DNMS training was resumed with the imposition of gradually longer delays (1–30 sec). The number of days required to achieve stable performance with either increasing length of delay or reversal of task contingency was compared between vehicle and rimonabant-treated rats. Following the completion of DNMS training, animals were euthanized, and both hippocampi were harvested for gene expression assay analysis. Rimonabant treatment animals required more time to achieve stable DNMS performance than vehicle-treated controls. Quantitative real-time polymerase chain reaction analysis revealed that the expressions of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit, brain-derived neurotrophic factor, and synapsin 1 (Syn1) were significantly increased. These results are consistent with rimonabant increasing mRNAs for proteins associated with hippocampal synapse remodeling, but that those alterations did not necessarily accelerate the acquisition of an operant behavioral task that required learning new contingencies.
Keywords: brain-derived neurotrophic factor, delayed-nonmatch-to-sample, gene expression, hippocampus, memory, rimonabant
Introduction
Delta 9-tetrahydrocannabinol (Δ9-THC) administration, in addition to other cannabinoid agonists, disrupts learning and memory1 through interactions with the endogenous cannabinoid system that plays a crucial role in the functional foundation of learning and memory.2 These disruptions consistently impair working memory while sparing long-term or reference memory.3
The delayed-nonmatch-to-sample (DNMS) task is a behavioral paradigm for studying hippocampal function in working memory.4–6 We have previously shown that hippocampal lesions produce delay-dependent impairment in the DNMS task.7 A similar decrement was observed in animals acutely administered Δ9-THC.8 Moreover, the administration of CB1 receptor antagonist rimonabant blocked these delay-dependent behavioral deficits.8 Rimonabant was also shown to improve performance in an olfactory recognition task9 and reduce errors in the eight-arm radial maze.10,11 However, no improvement in memory was observed in a two-component instrumental discrimination task.12 The ability of rimonabant to enhance memories that persist for minutes to hours and are not subject to rapid forgetting has led to the hypothesis that rimonabant administration could enhance learning and memory.13
Cellular mechanisms underlying synaptic plasticity (such as gene expression) may underscore the time-dependent ability of the endocannabinoid system to modulate learning and memory. Chronic cannabinoid receptor activation differentially alters the expression of genes required for synaptic transmission and trophic response in the rat hippocampus, including synaptotagmin, neuromodulin, cAMP response element binding protein 1, and the tyrosine receptor kinase B (TrkB) receptor for brain-derived neurotrophic factor (BDNF).14,15 The acquisition of a hippocampal-dependent learning task, as well as the expression of genes required for task acquisition during continuous CB1 receptor antagonism, has not been characterized.
This study demonstrates that continuous systemic rimonabant administration increases the expression of genes that influence synaptic remodeling, neuronal survival, and the expression of long-term potentiation (LTP). Moreover, the rimonabant treatment significantly increased the number of days to acquire stable performance at the longest delay increment of the DNMS task. These results show that systemic blockade of the CB1 receptor during the acquisition of a learning task produces a phenotype that is pharmacologically distinct from the genetic knockout of CB1 receptors.
Materials and Methods
All animal care and experimental procedures, including water deprivation and surgery, conformed to the National Institutes of Health (NIH) and Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) regulations. All surgical procedures conformed to NIH and AAALAC guidelines and were performed in a rodent surgical facility approved by the Wake Forest University IACUC.
Drug preparation and administration
Rimonabant (provided by RTI International, RTP, NC, USA, via the NIDA Drug Distribution program) was prepared for administration in a suspension of 10% dimethyl sulfoxide (DMSO) and 0.1% Tween-80 in sterile saline (% v/v). The suspension was loaded into an Alzet osmotic mini-pump (Model 2ML2; DURECT Corporation, Cupertino, CA) that delivered a continuous flow rate of 1.0 mg/kg/day for 28 days at ∼2.5 μL/h. Control solution consisted of DMSO/Tween-80/saline vehicle. In a second set of acute experiments, rimonabant (5.0 mg/kg) was prepared in a suspension of pluronic F68/saline and delivered by intraperitoneal injection. Control solution consisted of pluronic F68/saline vehicle.
Subjects
Male Long–Evans rats (n=26) were deprived of water for 15–20 h daily to facilitate DNMS/delayed-match-to-sample (DMS) task performance for water reward but were allowed free access to food. Total water intake was adjusted daily to maintain the rats at 85% of normal body weight. Animals were divided into vehicle (n=13) and rimonabant treatment groups (n=13) for both experimental procedures. Within these two groups the animals were further divided into a DNMS acquisition group (n=10) and a DNMS/DMS reversal group (n=16).
Surgery
All animals were trained to perform the DNMS task (described in DNMS cognitive task) at the “0” sec delay criterion level before the surgical procedure was implemented. Animals in the DNMS acquisition group were anesthetized with ketamine (100 mg/kg, i.p.) and xylazine (10 mg/kg, i.p.), and an ALZET osmotic mini-pump (Model 2ML2) loaded with DMSO/Tween-80/saline (vehicle) or vehicle plus rimonabant (1.0 mg/kg/day) was implanted subcutaneously on the dorsal area near the spinal cord. Behavioral testing resumed following a 2-day recovery period. In three of the rimonabant treatment animals a second osmotic mini-pump was implanted because DNMS training was not successfully completed within the functional lifespan on the initial osmotic mini-pump.
DNMS cognitive task
Complete details of apparatus design and behavioral training in the DNMS task have been reported elsewhere.4,16 Briefly, the behavioral testing chamber contained two retractable levers mounted on one wall, positioned on either side of a water trough, and a nose-poke device with a cue light positioned on the opposite wall. The animals acquired the DNMS task in three parts: sample presentation, a placeholder for delay interval, and nonmatch selection. A DNMS “trial” sequence initiated with the random presentation of either the left or right lever that when pressed (sample response [SmR]) immediately retracted, initiating the delay phase. During the delay phase, animals “nose-poked” into a photobeam under the cue light until the delay interval timed out. A final nose-poke following completion of the delay interval turned off the cue light and caused both levers to extend, initiating the nonmatch phase. A response on the lever opposite the SmR constituted the correct “nonmatch response” (NR) and was rewarded with a drop of water. An incorrect response on the same lever as the SmR (i.e., a “match response”) caused the houselights to turn off for 5 sec, and both levers retracted to delineate the error with no reward. A 10-sec intertrial interval was employed.
Animals were trained to perform the DNMS “trial” sequence, described above, with a “0” sec delay at the nose-poke. Each DNMS testing session required the animal to complete 100 trials. Successful performance during a DNMS testing session required a minimum of 80% correct trials responding within the testing session. This criterion was applied to all trials on initial training, and to trials of delay 1–10 sec when the trained delay in the task exceeded 10 sec. Once the performance criterion was achieved at the “0” sec delay, animals underwent the surgical procedure described above. Following recovery from surgery, the animals were retrained to criterion performance (at least 80% correct responding, average of one training session) at “0” sec delay before beginning the experiment and further training.
The experimental protocol required the animals to perform DNMS trials during a session with delays that were incrementally advanced to 1 and 10 sec, 1 and 20 sec, and then 1 and 30 sec (within individual trial delays randomly selected from within the specified intervals). Criterion performance was required before an animal could begin testing sessions with longer delay intervals, that is, 80% correct responding during sessions with delays between 1 and 10 sec was required before initiating sessions with delays between 1 and 20 sec.
For task contingency reversal, the animals were fully trained to 1–30 sec delays for at least 8 weeks before altering the contingency from DNMS to a DMS task. All elements of this task remained identical to the DNMS task with one exception. The rats were required to “match” instead of “nonmatch” the sample (i.e., learn the opposite rule) to obtain the water reward. Thus, nonmatching the sample was identified as an “error” response in this case. All animals were required to run 100 trials within 60 min following the treatment of either acute rimonabant or pluronic F68/saline vehicle ∼15–20 min before testing on the DMS (i.e., reversal) task for 12 successive days. Each session was terminated depending on whichever trial count (100 trials) or time limit (60 min) was reached first.
Gene expression
After completion of DNMS training (average of 14 and 28 days following mini-pump implantation, vehicle and rimonabant, respectively), rats were euthanized by isoflurane overdose followed by guillotine decapitation. The hippocampi were harvested via free-hand dissection, rapidly frozen in liquid nitrogen, and stored at −80°C until total RNA was extracted using the Trizol reagent method,17 and the RNeasy MinElute Cleanup Kit (Qiagen) was used to further purify samples. Isolated RNA concentration was assessed via the spectrophotometric quantitation of nucleic acids method18 using an Eppendorf BioPhotometer 6131, and purity was assured by an A260/A280 ratio range between 1.9 and 2.0. A portion of the total RNA (1 μg) was reverse-transcribed into cDNA using a High-Capacity cDNA Archive Kit (Applied Biosystems).
Real-time quantitative polymerase chain reaction (qPCR) was performed in triplicate for each sample using a Mastercycler EP Realplex real-time PCR system (Eppendorf) using TaqMan Gene® Expression Assays (Applied Biosystems) specific for 18S ribosomal RNA (18S), Enolase 2 (Eno2), BDNF, TrkB, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor 1 (Gria1), kainate receptor 1 (Grik1), N-methyl D-aspartate (NMDA) receptor 1 (Grin1), NMDA receptor 2A (Grin2a), Cyclic AMP-Responsive Element-Binding Protein 1 (Creb1), CRE-modulating protein (Crem), cannabinoid receptor 1 (Cnr1), cannabinoid receptor interacting protein (Cnrip1), gamma-aminobutyric acid (GABA) A receptor 1 (Gabra1), GABA B receptor 1 (Gabbr1), glutamic acid decarboxylase (Gad1), Syn1, synapsin 2 (Syn2), synapsin 3 (Syn3), and growth-associated protein-43 (Gap43).
Gene expression for each sample was normalized to 18S content, and relative quantification of gene expression levels was performed using the comparative CT method (2−ΔΔCT method) using Eno2 as the comparative control.19 Statistical differences between groups were determined using the Relative Expression Software Tool (REST©) nonparametric analysis developed for qPCR data.20
Statistical analysis for behavioral studies
Differences in the number of training days required to reach criterion performance in the DNMS acquisition study were analyzed using a two-way analysis of variance (ANOVA) (with Sidak's corrections) in GraphPad InStat statistical software (GraphPad Software, Inc., San Diego, CA). The mean percentage of correct responses and mean rate of correct responses were measured during the reversal (DMS) task across all test days. The data for each parameter were analyzed using a two-way ANOVA with Student's t-tests to compare group differences on each test day. A p-value <0.05 was considered significant.
Results
DNMS acquisition
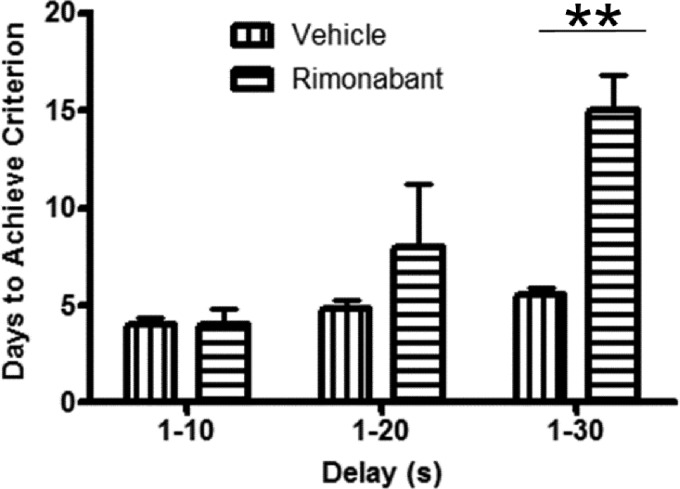
The functional relevance of continuous CB1 receptor blockade was assessed by measuring the duration to acquire the delay component of the DNMS task (Fig. 1). A significant main effect of treatment was observed (F1,9=10.8; p<0.001) as well as a significant difference in the number of days required for rimonabant-treated group to achieve criterion performance in the DNMS task (F1,12=5.13; p<0.05). However, post hoc analysis revealed no significant differences between the treatment groups at each successive training interval. This was because several animals required implantation of a second osmotic mini-pump before completion of DNMS training. An analysis of these animals' performance in the five sessions prior and following the second mini-pump implantation was conducted. This analysis did not reveal a significant difference in these animals' overall percentage of correct response rate (F1, 10=4.066; p=0.0714) during those sessions.
FIG. 1.
Effect of continuous rimonabant administration on DNMS delay acquisition. Comparison of vehicle treatment (n=5) and rimonabant treatment (n=5) on successive discrete acquisition of the delay component of the DNMS task. Vehicle was dimethyl sulfoxide/Tween-80/saline. Means±SEM; **p<0.01. DNMS, delayed-nonmatch-to-sample; SEM, standard error of the mean.
DNMS-to-DMS reversal
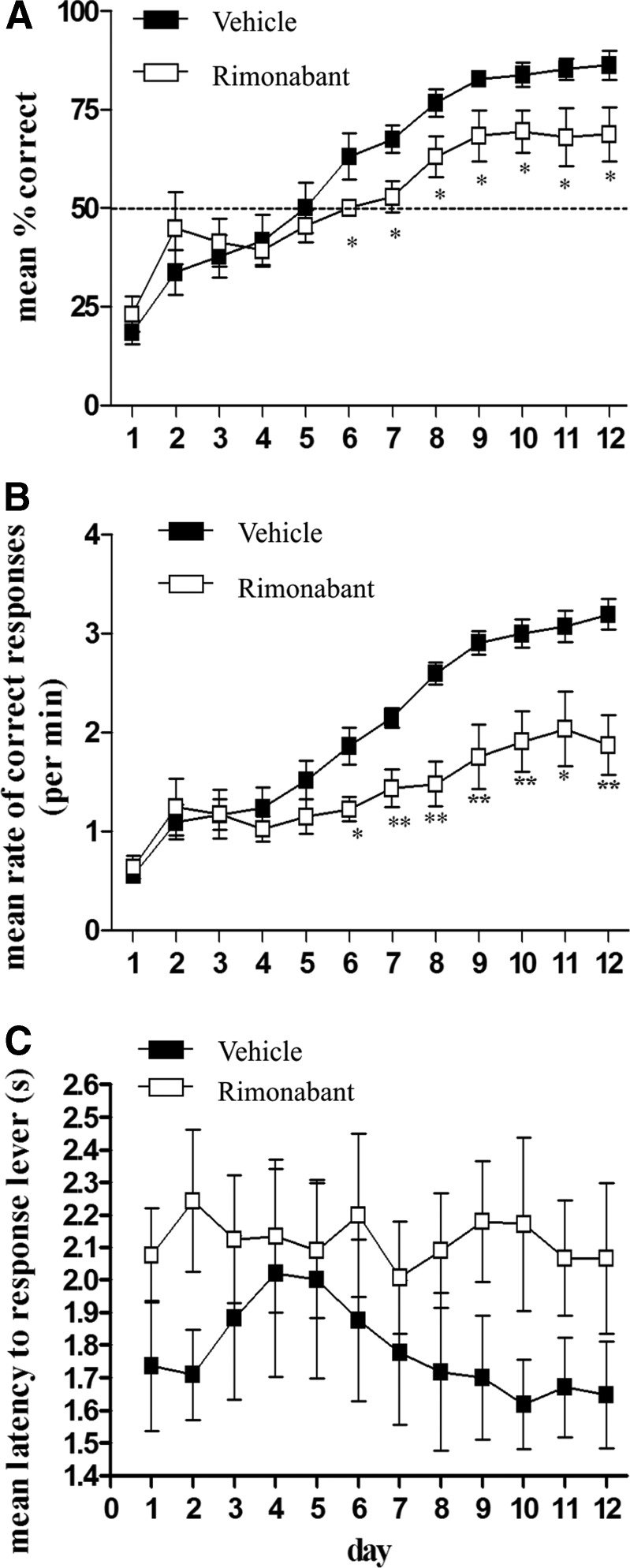
The overall mean percentage of correct responses was significantly affected by day (F11,154=43.39; p<0.001). Furthermore, a significant drug×day interaction (F11,154=2.87; p<0.01) was observed (Fig. 2A). Planned comparisons (paired t-tests) between the two groups showed no difference in learning the reversal (DMS) task on days 1–5 (all t's<1.03, all p's>0.32), but rimonabant-treated animals exhibited significant deficits in learning on days 6 (t=2.19; p<0.05), 7 (t=2.76; p<0.05), 8 (t=2.17; p<0.05), 9 (t=2.16; p<0.05), 10 (t=2.31; p<0.05), 11 (t=2.19; p<0.05), and 12 (t=2.26; p<0.05), suggesting that this rimonabant-induced deficit in reversal learning manifested with a late onset of action.
FIG. 2.
Effect of acute rimonabant administration on DNMS to DMS task contingency reversal. Comparison of vehicle treatment (n=8) and rimonabant treatment (n=8) on mean percentage of correct responses (A), mean rate of correct responses (B), and mean latency to move from the nose-poke to response lever (C) during reversal (DMS) task performance across all test days. Vehicle was pluronic F68/saline. Means±SEM; *p<0.01, **p<0.001. DMS, delayed-match-to-sample.
A similar set of analyses performed on mean rates of correct responses produced a significant effect of drug (F1,154=8.88; p<0.01), day (F11,154=37.77; p<0.001), and drug×day interaction (F11,154=6.22; p<0.001) (Fig. 2B). Planned comparisons between the two treatment groups revealed no significant differences on days 1–5 (all t's<1.42, all p's>0.18) although the rimonabant-treated animals exhibited significantly reduced rates of correct responding on days 6 (t=2.88; p<0.05), 7 (t=3.30; p<0.01), 8 (t=4.42; p<0.001), 9 (t=3.32; p<0.01), 10 (t=3.22; p<0.01), 11 (t=2.53; p<0.05), and 12 (t=3.86; p<0.01). There was no significant overall effect of drug (F1,154 = 1.61; p<0.42), day (F11,154 = 1.04; p<0.22), or drug×day interaction (F11,154 = 1.03, p<0.42), on the average latency to get to the response lever during the choice or decision phase of the trial (Fig. 2C). These results rule out the possibility that the observed deficits in reversal learning with rimonabant were produced due to impairments in locomotor activity or ability to perform the task.
Gene expression
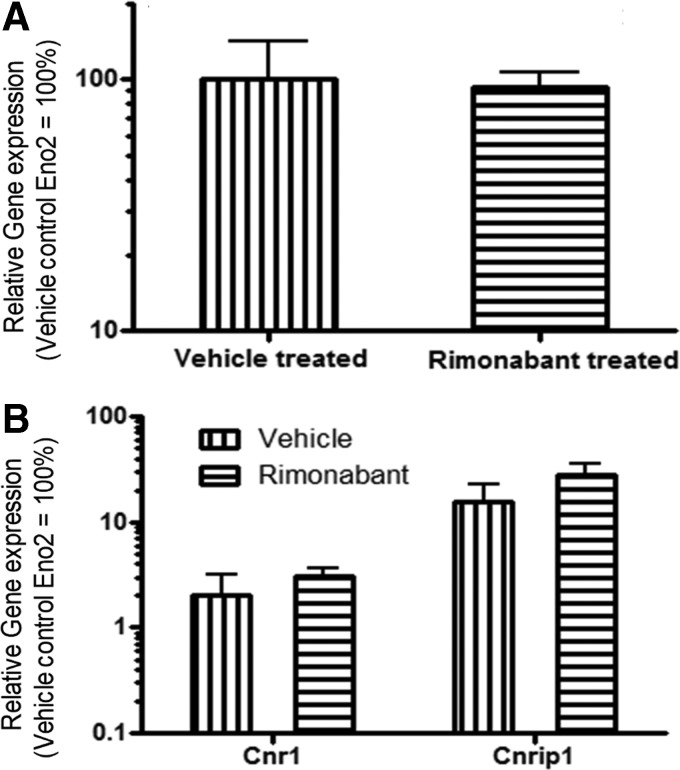
qPCR analysis revealed that continuous rimonabant treatment did not significantly alter the expression of neuron-specific Eno2 [t(7)=0.1592; p>0.05] (Fig. 3A); therefore, results were expressed relative to Eno2 values. Because previous studies indicated that CB1 receptors contribute a neuroprotective effect in mice,21,22 we questioned if continuous pharmacological blockade of CB1 receptors contributed to a measurable loss of neuronal populations in the hippocampus. We determined that continuous CB1 receptor antagonism did not alter the expression of Cnr1 or Cnrip1 (Fig. 3B).
FIG. 3.
Effect of continuous rimonabant treatment on the expression of neuronal marker and CB1 receptor. (A) Eno2 mRNA expression levels comparing rimonabant-treated rats (n=5) to vehicle-treated rats (n=5). (B) Cnr1 and Cnrip1 mRNA expression levels (n=5 per group). Values represent relative expression normalized to Eno2 mRNA expression levels. Means±SEM. Eno2, enolase 2; Cnr1, cannabinoid receptor 1; Cnrip1, cannabinoid receptor interacting protein.
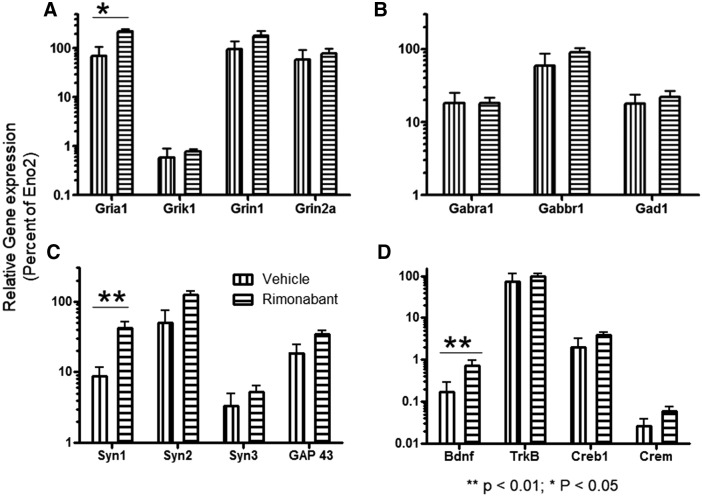
Next, we determined the effect of continuous rimonabant treatment on the expression of glutamate (Fig. 4A) and GABA (Fig. 4B) receptor subtypes and glutamate decarboxylase. The treatment failed to increase the expression of Grik1, Grin1, or Grin2a receptors. However, the expression of Gria1 was significantly increased (p<0.05). Rimonabant treatment did not significantly alter the expression of Gabra1, Gabbr1, or Gad1. Syn1, Syn2, Syn3, and Gap43 were assayed to assess changes in synaptic and neurite morphology (Fig. 4C) during rimonabant treatment. Only Syn1 expression levels were significantly increased (p<0.01). Lastly, we assessed the effects of continuous blockade of the CB1 receptor on the expression of pro-neuroprotective factors BDNF and its receptor TrkB, and transcription factors Creb1 and Crem in the hippocampus (Fig. 4D). qPCR analysis revealed that rimonabant treatment significantly increased the expression of BDNF (p<0.01).
FIG. 4.
Effect of continuous rimonabant treatment on the expression of genes that influence learning and memory. (A) AMPA receptor 1 (Gria1), kainate receptor 1 (Grik1), NMDA receptor 1 (Grin1), and NMDA receptor 2a subunit (Grin2a) mRNA expression levels (n=5 per treatment group). (B) GABA-A receptor (Gabra1), GABA-B receptor (Gabbr1), and Gad1 mRNA expression levels (n=5 per treatment group). (C) Synapsin1 (Syn1), Synapsin2 (Syn2), Synapsin3 (Syn3), Growth Associated Protein-43 (Gap43) mRNA expression level (n = 5 per treatment group). (D) BDNF, TrkB, Creb1, and Crem1 mRNA expression levels (n=5 per treatment group). Values represent relative expression normalized to Eno2 mRNA expression levels. Means±SEM; **p<0.01; *p<0.05. TrkB, tyrosine receptor kinase B; Creb1, CRE-binding protein; Crem, CRE-modulating protein; Gad1, glutamic acid decarboxylase; BDNF, brain-derived neurotrophic factor; NMDA, N-methyl D-aspartate; GABA, gamma-aminobutyric acid.
Discussion
The current study investigated the effect of continuous osmotic mini-pump infusion of the CB1 receptor antagonist rimonabant on the acquisition of a spatial learning task as well as gene expression levels in hippocampal neurons. Our results indicate that continuous pharmacological blockade of the CB1 receptor increased the transcription of genes required for the survival of existing neurons (BDNF), the expression of late-phase LTP (Gria1), and the regulation of axogenesis and synaptogenesis (Syn1). Moreover, the infusion protocol increased the number of days required for animals to achieve criterion performance in the DNMS task. Collectively, these results suggest that continuous pharmacological blockade of the CB1 receptor enhanced the expression of genes associated with synaptic plasticity.
The current study extends the importance of evaluating changes in gene expression levels, and associated cellular processes, in brain areas relevant for learning and memory when the endocannabinoid system is pharmacologically inactivated. Several genes have previously been shown to undergo a dynamic response to acute versus chronic cannabinoid agonist administration in in vivo and in vitro preparations.15,23–31 Interestingly, the modulated genes were broadly correlated with membrane repair, synapse formation, and CB1 receptor-mediated intracellular signaling cascades. We report a similar change in the genetic profile of membrane repair and synapse formation genes, but in the current report the upregulated genes are downstream mediators of BDNF.
Before implantation of the osmotic mini-pump, CB1 receptor-mediated activation of the ERK pathway may have modulated BDNF expression levels.32 Increased BDNF signaling, through its receptor TrkB, may have activated alternate intracellular signaling pathways—that is, ras/phospholipase C γ phosphatidylinositol-3-kinase/Akt and Src pathways33—to further increase BDNF production.34 BDNF is an important mediator of neuroprotective function during reduced endocannabinoid system signaling.21,22,35 It is also critical for various CNS functions, including neuronal development and differentiation, synaptic transmission and plasticity, as well as learning and memory.34,36–38
In cultured hippocampal neurons TrkB receptor activation rapidly upregulated NMDA receptor protein levels by increasing transcription activity.33 TrkB receptor activation of the phosphatidylinositol-3-kinase signal transduction pathway induced glutamate AMPA receptor (GluR1) subunit expression.39,40 We demonstrate a similar increase in GluR1 subunit gene expression (Fig. 4A) coinciding with increased BDNF expression (Fig. 4D). GluR1 is an important subunit for the manifestation of late-phase LTP. Genetically modified mice lacking the GluR1 subunit exhibit normal acquisition of the water maze hidden platform task and spatial reference memory performance. These mice also exhibit a lack of hippocampal LTP at CA3–CA1 synapses and are impaired in hippocampus-dependent short-term working memory tasks.41–44
The other major signaling cascade activated downstream of TrkB receptors, mitogen-activated kinase, modulates synapsin-dependent vesicular dynamics.45–47 The synapsins regulate neurotransmitter release by controlling the number of available vesicles at the nerve terminus.48 Increased expression levels of synapsin I and synapsin II parallel the establishment of synaptic contacts within the hippocampus.49 The increase in synapsin I gene expression levels is consistent with the premise that CB1 receptor blockade augments the storage of memory traces through synaptic remodeling and presumably increased synaptic connections.50
To address the implications of the CB1 receptor blockade genetic profile, we examined its effect on the acquisition of a hippocampus-dependent working memory task. During the acquisition phase of a hippocampus-dependent task, BDNF mRNA expression levels peaked and subsequently declined during continued training.51,52 Elevated levels of BDNF were associated with improved performance in the water maze spatial memory task, but further enhancement of BDNF levels resulted in learning impairments.53 This suggests that the increased number of days required for rimonabant-treated rats to achieve criterion performance may have resulted from BDNF overexpression during DNMS acquisition at longer delay intervals.
Differences in drug administration protocols may explain the discrepancy with previous studies that showed that acute blockade of CB1 receptors with rimonabant enhances memory in rats.9–11 Rimonabant administered, i.p. or i.m., before or immediately following the acquisition phase improved memory in the social recognition task9 and the radial arm maze,10 but not the nonmatch-to-position12 or repeated acquisition13 operant tasks. An enhancement may not have been observed due to an inability of the animals to perform the task. We previously showed that DNMS task performance remains intact when rimonabant (1 mg/kg; i.p.) was given acutely.54 Moreover, no performance deficits were observed in the current study using the continuous osmotic mini-pump infusion protocol.
It has been proposed that rimonabant treatment strengthens memory consolidation processes rather than altering attentional or retrieval processes.10,11 Rimonabant administered before the test (or retention) phase failed to improve performance or reduce response errors. We previously showed that the hippocampus is imperative for proper performance of the DNMS task,7 and that activation of the endogenous cannabinoid system facilitates previously learned behaviors and associations.55,56 Moreover, the blockade of the endogenous cannabinoid system enhances memory processes in a consolidation interval-dependent manner.9,10,12,13
Our continuous osmotic mini-pump infusion protocol during task acquisition affected both new learning and reference memory in the DNMS task. However, rimonabant was supplied following the acquisition of prior task contingencies, that is, criterion performance at the 0 sec delay interval for task acquisition or following extended performance of the DNMS contingency prereversal. It is notable that the DNMS/DMS tasks involve operant conditioning, where prior contingencies must be “unlearned” to progress to the final behavioral state. CB1 receptor activation may impair the animal's ability to accommodate and successfully acquire a change in task contingency represented by an increase in delay interval or task reversal. Rimonabant reduces trial-to-trial retention of sequential memory and thus may alter the information necessary for learning new task contingencies.56 The increased time required to achieve criterion performance supports the hypothesis that rimonabant-treated rats are impaired in their ability to replace prior-learned behaviors with new contextually relevant behaviors.57–59
This agrees with reports8,12,13,57 that showed no memory-enhancing effect when a short delay (<10 sec) between successive trials as well as a small interval (<30 sec) between SmR and NR was used. Additionally, the infusion protocol reflects the net effect of CB1 receptor antagonism in all brain structures where CB1 receptors are localized, not just memory-relevant structures, that is, hippocampus. Directly targeting CB1 receptors with intra-hippocampal infusions of rimonabant may produce a memory enhancement not observed with the systemic infusion protocol.60–62
Lastly, the increased levels of BDNF, Gria1, and Syn1 may indicate that synapses previously strengthened during the acquisition of a smaller delay interval were not able to be dismantled and restructured at alternative synapses while the animal was learning to adjust to increased delay interval.
In conclusion, the significant increase in BDNF expression associated with long-term rimonabant treatment, which, as research literature suggests, can exert a beneficial effect on synaptic development, failed to correlate with an enhanced performance of a short-term working memory task. We propose that any synaptic plasticity changes represented by the gene profile may not be an advantage when acquiring a task that requires the alteration of previously learned task contingencies.63
Acknowledgments
The authors acknowledge the late Sarah Bough (Department of Biology, University of Bath) for her contribution to the collection of DNMS data. Funding sources: R01-DA08549, R01-DA03690.
Abbreviations Used
- AAALAC
Association for the Assessment and Accreditation of Laboratory Animal Care
- ANOVA
analysis of variance
- BDNF
brain-derived neurotrophic factor
- Cnr1
cannabinoid receptor 1
- Cnrip1
cannabinoid receptor interacting protein
- Creb1
CRE-binding protein
- Crem
CRE-modulating protein
- DMS
delayed-match-to-sample
- DMSO
dimethyl sulfoxide
- DNMS
delayed-nonmatch-to-sample
- Eno2
Enolase 2
- GABA
gamma-aminobutyric acid
- Gad1
glutamic acid decarboxylase
- Gap43
growth-associated protein-43
- LTP
long-term potentiation
- NIH
National Institutes of Health
- NMDA
N-methyl D-aspartate
- NR
nonmatch response
- qPCR
real-time quantitative polymerase chain reaction
- SmR
sample response
- Syn1
synapsin 1
- Syn2
synapsin 2
- Syn3
synapsin 3
- TrkB
tyrosine receptor kinase B
Author Disclosure Statement
No competing financial interests exist.
Cite this article as: Horton K-KA, Goonawardena AV, Sesay J, Howlett AC, Hampson RE (2019) Systemic blockade of the CB1 receptor augments hippocampal gene expression involved in synaptic plasticity but perturbs hippocampus-dependent learning task, Cannabis and Cannabinoid Research 4:1, 33–41, DOI: 10.1089/can.2018.0061.
References
- 1. Sullivan JM. Cellular and molecular mechanisms underlying learning and memory impairments produced by cannabinoids. Learn Mem. 2000;7:132–139 [DOI] [PubMed] [Google Scholar]
- 2. Lichtman AH, Martin BR. Marijuana withdrawal syndrome in the animal model. J Clin Pharmacol. 2002;42:20S–27S [DOI] [PubMed] [Google Scholar]
- 3. Riedel G, Davies SN. Cannabinoid function in learning, memory and plasticity. Handb Exp Pharmacol. 2005:445–477 [DOI] [PubMed] [Google Scholar]
- 4. Deadwyler SA, Bunn T, Hampson RE. Hippocampal ensemble activity during spatial delayed-nonmatch-to-sample performance in rats. J Neurosci. 1996;16:354–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rawlins JN, Maxwell TJ, Sinden JD. The effects of fornix section on win-stay/lose-shift and win-shift/lose-stay performance in the rat. Behav Brain Res. 1988;31:17–28 [DOI] [PubMed] [Google Scholar]
- 6. Squire LR. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol Rev. 1992;99:195–231 [DOI] [PubMed] [Google Scholar]
- 7. Hampson RE, Jarrard LE, Deadwyler SA. Effects of ibotenate hippocampal and extrahippocampal destruction on delayed-match and -nonmatch-to-sample behavior in rats. J Neurosci. 1999;19:1492–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hampson RE, Deadwyler SA. Cannabinoids reveal the necessity of hippocampal neural encoding for short-term memory in rats. J Neurosci. 2000;20:8932–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Terranova JP, Storme JJ, Lafon N, et al. Improvement of memory in rodents by the selective CB1 cannabinoid receptor antagonist, SR 141716. Psychopharmacology (Berl). 1996;126:165–172 [DOI] [PubMed] [Google Scholar]
- 10. Lichtman AH. SR 141716A enhances spatial memory as assessed in a radial-arm maze task in rats. Eur J Pharmacol. 2000;404:175–179 [DOI] [PubMed] [Google Scholar]
- 11. Wolff MC, Leander JD. SR141716A, a cannabinoid CB1 receptor antagonist, improves memory in a delayed radial maze task. Eur J Pharmacol. 2003;477:213–217 [DOI] [PubMed] [Google Scholar]
- 12. Mallet PE, Beninger RJ. The cannabinoid CB 1 receptor antagonist SR141716A attenuates the memory impairment produced by? 9 -tetrahydrocannabinol or anandamide. Psychopharmacology. 1998;140:11–19 [DOI] [PubMed] [Google Scholar]
- 13. Brodkin J, Moerschbaecher JM. SR141716A antagonizes the disruptive effects of cannabinoid ligands on learning in rats. J Pharmacol Exp Ther. 1997;282:1526–1532 [PubMed] [Google Scholar]
- 14. Butovsky E, Juknat A, Goncharov I, et al. In vivo up-regulation of brain-derived neurotrophic factor in specific brain areas by chronic exposure to Delta-tetrahydrocannabinol. J Neurochem. 2005;93:802–811 [DOI] [PubMed] [Google Scholar]
- 15. Grigorenko E, Kittler J, Clayton C, et al. Assessment of cannabinoid induced gene changes: tolerance and neuroprotection. Chem Phys Lipids. 2002;121:257–266 [DOI] [PubMed] [Google Scholar]
- 16. Deadwyler SA, Hampson RE. Differential but complementary mnemonic functions of the hippocampus and subiculum. Neuron. 2004;42:465–476 [DOI] [PubMed] [Google Scholar]
- 17. Nolan RL, Teller JK. Diethylamine extraction of proteins and peptides isolated with a mono-phasic solution of phenol and guanidine isothiocyanate. J Biochem Biophys Methods. 2006;68:127–131 [DOI] [PubMed] [Google Scholar]
- 18. Sauer T, Beraki K, Jebsen PW, et al. Numerical aberrations of chromosome 17 in interphase cell nuclei of breast carcinoma cells: lack of correlation with abnormal expression of p53, neu and nm23 protein. APMIS. 1998;106:921–927 [DOI] [PubMed] [Google Scholar]
- 19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 20. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilbert GL, Kim HJ, Waataja JJ, et al. Delta9-tetrahydrocannabinol protects hippocampal neurons from excitotoxicity. Brain Res. 2007;1128:61–69 [DOI] [PubMed] [Google Scholar]
- 22. Khaspekov LG, Brenz Verca MS, Frumkina LE, et al. Involvement of brain-derived neurotrophic factor in cannabinoid receptor-dependent protection against excitotoxicity. Eur J Neurosci. 2004;19:1691–1698 [DOI] [PubMed] [Google Scholar]
- 23. Aquino DA, Peng D, Lopez C, et al. The constitutive heat shock protein-70 is required for optimal expression of myelin basic protein during differentiation of oligodendrocytes. Neurochem Res. 1998;23:413–420 [DOI] [PubMed] [Google Scholar]
- 24. Becker CG, Artola A, Gerardy-Schahn R, et al. The polysialic acid modification of the neural cell adhesion molecule is involved in spatial learning and hippocampal long-term potentiation. J Neurosci Res. 1996;45:143–152 [DOI] [PubMed] [Google Scholar]
- 25. Duggan DJ, Bittner M, Chen Y, et al. Expression profiling using cDNA microarrays. Nat Genet. 1999;21(1 Suppl):10–14 [DOI] [PubMed] [Google Scholar]
- 26. Kittler JT, Grigorenko EV, Clayton C, et al. Large-scale analysis of gene expression changes during acute and chronic exposure to [Delta]9-THC in rats. Physiol Genomics. 2000;3:175–185 [DOI] [PubMed] [Google Scholar]
- 27. Mailleux P, Verslype M, Preud'homme X, et al. Activation of multiple transcription factor genes by tetrahydrocannabinol in rat forebrain. Neuroreport. 1994;5:1265–1268 [DOI] [PubMed] [Google Scholar]
- 28. Martin KC, Kandel ER. Cell adhesion molecules, CREB, and the formation of new synaptic connections. Neuron. 1996;17:567–570 [DOI] [PubMed] [Google Scholar]
- 29. McGregor IS, Arnold JC, Weber MF, et al. A comparison of delta 9-THC and anandamide induced c-fos expression in the rat forebrain. Brain Res. 1998;802:19–26 [DOI] [PubMed] [Google Scholar]
- 30. Southern E, Mir K, Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21(1 Suppl):5–9 [DOI] [PubMed] [Google Scholar]
- 31. Zhuang S, Kittler J, Grigorenko EV, et al. Effects of long-term exposure to delta9-THC on expression of cannabinoid receptor (CB1) mRNA in different rat brain regions. Brain Res Mol Brain Res. 1998;62:141–149 [DOI] [PubMed] [Google Scholar]
- 32. Derkinderen P, Valjent E, Toutant M, et al. Regulation of extracellular signal-regulated kinase by cannabinoids in hippocampus. J Neurosci. 2003;23:2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caldeira MV, Melo CV, Pereira DB, et al. BDNF regulates the expression and traffic of NMDA receptors in cultured hippocampal neurons. Mol Cell Neurosci. 2007;35:208–219 [DOI] [PubMed] [Google Scholar]
- 34. Xiong H, Futamura T, Jourdi H, et al. Neurotrophins induce BDNF expression through the glutamate receptor pathway in neocortical neurons. Neuropharmacology. 2002;42:903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marsicano G, Goodenough S, Monory K, et al. CB1 cannabinoid receptors and on-demand defense against excitotoxicity. Science. 2003;302:84–88 [DOI] [PubMed] [Google Scholar]
- 36. Lu Y, Christian K, Lu B. BDNF: a key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol Learn Mem. 2008;89:312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakajo Y, Miyamoto S, Nakano Y, et al. Genetic increase in brain-derived neurotrophic factor levels enhances learning and memory. Brain Res. 2008;1241:103–109 [DOI] [PubMed] [Google Scholar]
- 38. Yamada K, Mizuno M, Nabeshima T. Role for brain-derived neurotrophic factor in learning and memory. Life Sci. 2002;70:735–744 [DOI] [PubMed] [Google Scholar]
- 39. Mizuno M, Yamada K, Takei N, et al. Phosphatidylinositol 3-kinase: a molecule mediating BDNF-dependent spatial memory formation. Mol Psychiatry. 2003;8:217–224 [DOI] [PubMed] [Google Scholar]
- 40. Strutz-Seebohm N, Seebohm G, Mack AF, et al. Regulation of GluR1 abundance in murine hippocampal neurones by serum- and glucocorticoid-inducible kinase 3. J Physiol. 2005;565:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reisel D, Bannerman DM, Schmitt WB, et al. Spatial memory dissociations in mice lacking GluR1. Nat Neurosci. 2002;5:868–873 [DOI] [PubMed] [Google Scholar]
- 42. Sanderson DJ, Good MA, Seeburg PH, et al. Chapter 9 The role of the GluR-A (GluR1) AMPA receptor subunit in learning and memory, in Progress in Brain Research. Amsterdam, NY: Elsevier, 2008:159–178 [DOI] [PubMed] [Google Scholar]
- 43. Schmitt WB, Sprengel R, Mack V, et al. Restoration of spatial working memory by genetic rescue of GluR-A-deficient mice. Nat Neurosci. 2005;8:270–272 [DOI] [PubMed] [Google Scholar]
- 44. Zamanillo D, Sprengel R, Hvalby O, et al. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science. 1999;284:1805–1811 [DOI] [PubMed] [Google Scholar]
- 45. Jovanovic JN, Benfenati F, Siow YL, et al. Neurotrophins stimulate phosphorylation of synapsin I by MAP kinase and regulate synapsin I-actin interactions. Proc Natl Acad Sci U S A. 1996;93:3679–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jovanovic JN, Czernik AJ, Fienberg AA, et al. Synapsins as mediators of BDNF-enhanced neurotransmitter release. Nat Neurosci. 2000;3:323–329 [DOI] [PubMed] [Google Scholar]
- 47. Yamada K, Nabeshima T. Brain-derived neurotrophic factor/TrkB signaling in memory processes. J Pharmacol Sci. 2003;91:267–270 [DOI] [PubMed] [Google Scholar]
- 48. Ferreira A, Rapoport M. The synapsins: beyond the regulation of neurotransmitter release. Cell Mol Life Sci. 2002;59:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Melloni RH, Jr, DeGennaro LJ. Temporal onset of synapsin I gene expression coincides with neuronal differentiation during the development of the nervous system. J Comp Neurol. 1994;342:449–462 [DOI] [PubMed] [Google Scholar]
- 50. Bannerman DM, Rawlins JN, Good MA. The drugs don't work-or do they? Pharmacological and transgenic studies of the contribution of NMDA and GluR-A-containing AMPA receptors to hippocampal-dependent memory. Psychopharmacology (Berl). 2006;188:552–566 [DOI] [PubMed] [Google Scholar]
- 51. Kesslak JP, So V, Choi J, et al. Learning upregulates brain-derived neurotrophic factor messenger ribonucleic acid: a mechanism to facilitate encoding and circuit maintenance? Behav Neurosci. 1998;112:1012–1019 [DOI] [PubMed] [Google Scholar]
- 52. Rapanelli M, Frick LR, Zanutto BS. Differential gene expression in the rat hippocampus during learning of an operant conditioning task. Neuroscience. 2009;163:1031–1038 [DOI] [PubMed] [Google Scholar]
- 53. Pietropaolo S, Paterna JC, Bueler H, et al. Bidirectional changes in water-maze learning following recombinant adenovirus-associated viral vector (rAAV)-mediated brain-derived neurotrophic factor expression in the rat hippocampus. [Article]. Behav Pharmacol. 2007;18:533–547 [DOI] [PubMed] [Google Scholar]
- 54. Hampson RE, Deadwyler SA. Cannabinoids, hippocampal function and memory. Life Sci. 1999;65:715–723 [DOI] [PubMed] [Google Scholar]
- 55. Deadwyler SA, Goonawardena AV, Hampson RE. Short-term memory is modulated by the spontaneous release of endocannabinoids: evidence from hippocampal population codes. Behav Pharmacol. 2007;18:571–580 [DOI] [PubMed] [Google Scholar]
- 56. Deadwyler SA, Hampson RE. Endocannabinoids modulate encoding of sequential memory in the rat hippocampus. Psychopharmacology (Berl). 2008;198:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Niyuhire F, Varvel SA, Thorpe AJ, et al. The disruptive effects of the CB1 receptor antagonist rimonabant on extinction learning in mice are task-specific. Psychopharmacology (Berl). 2007;191:223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shiflett MW, Rankin AZ, Tomaszycki ML, et al. Cannabinoid inhibition improves memory in food-storing birds, but with a cost. Proc Biol Sci. 2004;271:2043–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Varvel SA, Lichtman AH. Evaluation of CB1 receptor knockout mice in the Morris water maze. J Pharmacol Exp Ther. 2002;301:915–924 [DOI] [PubMed] [Google Scholar]
- 60. Egashira N, Mishima K, Iwasaki K, et al. Intracerebral microinjections of delta 9-tetrahydrocannabinol: search for the impairment of spatial memory in the eight-arm radial maze in rats. Brain Res. 2002;952:239–245 [DOI] [PubMed] [Google Scholar]
- 61. Lichtman AH, Dimen KR, Martin BR. Systemic or intrahippocampal cannabinoid administration impairs spatial memory in rats. Psychopharmacology (Berl). 1995;119:282–290 [DOI] [PubMed] [Google Scholar]
- 62. Robinson L, Goonawardena AV, Pertwee RG, et al. The synthetic cannabinoid HU210 induces spatial memory deficits and suppresses hippocampal firing rate in rats. Br J Pharmacol. 2007;151:688–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goonawardena AV, Sesay J, Riedel G, et al. The CB1 receptor antagonist, Rimonabant, impairs reversal learning by altering task-specific firing correlates in the hippocampus, 883.14. In: Society for neuroscience. Online: Chicago, IL, 2009 [Google Scholar]