Abstract
The National Institutes of Health-sponsored Epilepsy Connectome Project aims to characterize connectivity changes in temporal lobe epilepsy (TLE) patients. The magnetic resonance imaging protocol follows that used in the Human Connectome Project, and includes 20 min of resting-state functional magnetic resonance imaging acquired at 3T using 8-band multiband imaging. Glasser parcellation atlas was combined with the FreeSurfer subcortical regions to generate resting-state functional connectivity (RSFC), amplitude of low-frequency fluctuations (ALFFs), and fractional ALFF measures. Seven different frequency ranges such as Slow-5 (0.01–0.027 Hz) and Slow-4 (0.027–0.073 Hz) were selected to compute these measures. The goal was to train machine learning classification models to discriminate TLE patients from healthy controls, and to determine which combination of the resting state measure and frequency range produced the best classification model. The samples included age- and gender-matched groups of 60 TLE patients and 59 healthy controls. Three traditional machine learning models were trained: support vector machine, linear discriminant analysis, and naive Bayes classifier. The highest classification accuracy was obtained using RSFC measures in the Slow-4 + 5 band (0.01–0.073 Hz) as features. Leave-one-out cross-validation accuracies were ∼83%, with receiver operating characteristic area-under-the-curve reaching close to 90%. Increased connectivity from right area posterior 9-46v in TLE patients contributed to the high accuracies. With increased sample sizes in the near future, better machine learning models will be trained not only to aid the diagnosis of TLE, but also as a tool to understand this brain disorder.
Keywords: ALFF, connectome, functional connectivity, machine learning, resting-state fMRI, temporal lobe epilepsy
Introduction
Epilepsy, a brain disorder characterized by recurring seizures, affects an estimated 1.2% of the United States population (3.4 million persons) and is associated with a high risk of cognitive and psychosocial dysfunction, and enormous health care costs (Zack and Kobau, 2017). Powerful imaging tools are now available for quantitatively characterizing the structural and functional connections between brain regions that make up epileptic networks (Holmes and Tucker, 2013), providing a promising new approach for understanding, predicting, and treating refractory epilepsy.
Temporal lobe epilepsy (TLE) is the most common form of epilepsy in adults, and the largest group among those with medically refractory seizures (Tellez-Zenteno and Hernandez-Ronquillo, 2012). Finding reliable biomarkers is crucial in prevention therapy and drug development, but has so far been only modestly successful (Engel et al., 2013). The National Institutes of Health-sponsored Epilepsy Connectome Project (ECP) is a longitudinal study with the specific aim of characterizing brain connectivity abnormalities in people with TLE (Cook et al., 2018). ECP magnetic resonance imaging (MRI) protocols follow those used in the Human Connectome Project (HCP) (Van Essen et al., 2013) and include a substantial resting-state functional MRI (rs-fMRI) acquisition collected using simultaneous multislice (SMS) magnetic resonance sequences (Moeller et al., 2010) with a high temporal resolution implemented specifically for Connectome studies on 3T GE 750 scanners.
The human brain is a complex dynamic system characterized by spontaneous oscillations in multiple frequency bands (Buzsaki and Draguhn, 2004). Traditionally, analysis of rs-fMRI data includes application of a bandpass filter in the low-frequency oscillation (LFO) range (0.01–0.1 Hz, although exact cutoffs vary slightly), because this frequency region is less contaminated by low-/high-frequency noise and captures relevant resting-state information (Biswal et al., 1995). Some investigators have tested narrower frequency bands within and around the LFO, labeled Slow-5 (0.01–0.027 Hz), Slow-4 (0.027–0.073 Hz), Slow-3 (0.073–0.198 Hz), and Slow-2 (0.198–0.50 Hz) by Buzsaki and Draguhn (2004). Zuo and associates (2010) suggested that the Slow-4 and Slow-5 bands reflect signal changes from the gray matter, and Slow-2 and Slow-3 from the white matter. Recent study by Gohel and Biswal (2015) revealed that functional integration between brain regions at rest occurs in multiple frequency bands.
Based on these findings, our hypothesis was that seizure activity in TLE patients, which generally occurs at much higher frequencies than these slow bands, produces alterations in gray matter connectivity that can be detected at lower frequencies with fMRI. Since the raw voxel-based signal data are four-dimensional (4D), highly complicated, and very large in size, three summary measures were calculated: resting-state functional connectivity (RSFC) (Biswal et al., 1995), amplitude of low-frequency fluctuations (ALFFs) (Biswal et al., 1995; Zang et al., 2007), and fractional ALFFs (fALFFs) (Zou et al., 2008). RSFC measures correlations between blood-oxygen-level-dependent (BOLD) time series of two brain regions, whereas ALFFs and fALFFs capture intensity-based measures of BOLD change at a single region of interest. The goal was to reveal which combinations of a resting-state measure and a frequency band capture the most valuable information to discriminate between TLE patients and healthy controls.
Previous studies that investigated these measures in TLE patients reported abnormalities in different regions of the resting brain. These abnormalities include decreased RSFC within the epileptic temporal lobe, between hippocampi, and between the hippocampus and the orbitofrontal region (Centeno and Carmichael, 2014), and increased RSFC in the lateral portions of the nonepileptic hemisphere (Kucukboyaci et al., 2013). Zhang and associates (2010) reported that TLE patients with medial temporal sclerosis (a common structural abnormality in TLE) show increased ALFFs in the medial temporal lobe and thalamus and decreased ALFFs in the default-mode network. A difference in fALFFs was noted between left and right TLE patients in the thalamus (Yang et al., 2015).
The Glasser parcellation atlas (Glasser et al., 2016) was used for this study. This parcellation is a recent development from the HCP consortium for surface-based morphometry. It consists of 180 cortical parcels per hemisphere. These parcels were delineated using a multimodal approach and the authors reported that the parcellation is highly reproducible (Glasser et al., 2016). One limiting factor is that this parcellation only contains cortical brain regions. Therefore, 19 subcortical regions from the FreeSurfer subcortical segmentation (Dale et al., 1999; Fischl et al., 2002) were added for the current analysis.
Using the three summary measures and the surface-based morphometry analysis helps to reduce the size of the data, but the resulting data are still highly complicated (Garcia-Ramos et al., 2016; Song et al., 2015). Machine learning has proved effective at finding patterns in complex data sets in many fields and applications (Kotsiantis et al., 2006). Therefore, machine learning techniques were applied to determine which set of features would discriminate between the TLE patients and healthy controls with the highest accuracy.
Clinical application of machine learning is currently most limited by the lack of data compared with the number of potential training features (Beleites et al., 2013). To create a reliable machine learning model, one needs to select an informative set of features for training, then narrow this set down to key components (Hua et al., 2005; Vergun et al., 2016). Therefore, knowing what information is useful is essential, but typically difficult to determine a priori. In this study, 20 different combinations of resting fMRI measures and frequency bands were considered for the machine learning training. Note that it is not necessary to consider the “All” band with fALFFs, because fALFF is defined as ALFF of a specific band over that of the All band. A feature selection method using Lasso (Least Absolute Shrinkage and Selection Operator) (Tibshirani, 1996) was employed to remove uninformative features (Meier et al., 2012; Tang et al., 2013).
Given a large enough sample size, machine learning models based on artificial neural networks such as deep learning will generally outperform traditional models (Ng, 2016). In many medical imaging applications, however, there are not enough samples to train such models. In these cases, what may be tried instead is to train simpler machine learning models to get an approximate assessment of potential usefulness. This can also reveal what underlying features are able to produce good models. As more data are accumulated in the future, performance will improve and become stable with the use of deeper learning models. Any results obtained now with traditional models are setting the lower bound for the future.
Materials and Methods
Participants
The ECP (NINDS U01 NS093650) is a multisite prospective research project of the Medical College of Wisconsin (MCW) and University of Wisconsin-Madison (UW-Madison) (Cook et al., 2018). The MCW and Froedtert Hospital Institutional Review Board approved the use of human participants for this study. All participants provided written informed consent before their participation.
Epilepsy patients enrolled in the ECP are between ages of 18 and 60 (inclusive), have a full-scale IQ of 70 or above, speak English fluently, and have no medical contraindications to whole-body 3T MRI or magnetoencephalography (MEG). They must have a diagnosis of TLE supported by two or more of the following: (1) described or observed clinical semiology consistent with seizures of temporal lobe origin, (2) electroencephalography (EEG) evidence of either temporal intermittent rhythmic delta activity or temporal epileptiform discharges, (3) temporal onset of seizures captured on EEG, or (4) MRI evidence of medial temporal sclerosis or hippocampal atrophy. Patients are excluded who have any of the following: (1) presence of any lesions other than medial temporal sclerosis and nonspecific white matter abnormalities on 3T MRI with a dedicated epilepsy protocol that includes high-resolution axial and coronal fluid attenuated inversion recovery sequences, (2) an active infectious etiology of seizures, or (3) suspected or confirmed evidence of active autoimmune or inflammatory process in the central nervous system. Patients who underwent epilepsy surgery previously were also excluded.
The controls are healthy adults in the 18 to 60 years age range. Exclusion criteria for the healthy controls include: Edinburgh Laterality (Handedness) Quotient less than +50; primary language other than English; history of any learning disability, brain injury or illness, substance abuse, or major psychiatric illness (major depression, bipolar disorder, or schizophrenia); current use of vasoactive medications; and any medical contraindications to MRI or MEG.
Data from 60 consecutive TLE patients (mean age = 39.5 ± 12.0 years, 34 women, 5 left-handed, epilepsy duration = 18.7 ± 14.4 years, 38 drug-resistant TLE) and 59 healthy controls (mean age = 36.0 ± 14.4 years, 32 women) were analyzed. Supplementary Table S1 summarizes the demographics and the clinical information of the TLE patients. The two groups did not differ in the mean age (p = 0.16, two-tailed t-test), and gender ratio (p = 0.79, Chi-squared test). The patient group consisted of 29 left, 15 right, and 4 bilateral TLE patients, and 12 TLE patients of uncertain seizure laterality. To closely match the TLE and control samples, 12 of the healthy control data sets were taken from the Alzheimer Disease Connectome Project (ADCP; 1UF1AG051216-01A1) (Hwang et al., 2018), which uses the same set of MRI scanners at MCW and UW-Madison and the same imaging protocols for structural and rs-fMRI scans as the ECP. The MCW Institutional Review Board has approved the use of human participants for ADCP and the sharing of deidentified data sets from this study.
Data acquisition
MRI was performed on 3T GE 750 scanners at both institutions. Resting fMRI images were acquired using whole-brain SMS imaging (8 bands, 72 slices, repetition time (TR)/echo time (TE) = 802 ms/33.5 ms, flip angle = 50°, matrix = 104 × 104, field of view (FOV) = 20.8 cm, voxel size 2 mm isotropic) and a 32-channel receive coil. The participants were asked to fixate on a white cross at the center of a black screen during the scans. Time series from four 5-min resting-state scans acquired in a single session were concatenated. T1w structural images were acquired using a magnetization prepared gradient echo sequence (TR/TE = 604 ms/2.516 ms, inversion time [TI] = 1060.0 ms, flip angle = 8°, FOV = 25.6 cm, 0.8 mm isotropic). Cube T2w structural images were also acquired (TR/TE = 2500 ms/94.641 ms, flip angle = 90°, FOV = 25.6 cm, 0.8 mm isotropic).
Data processing
Data were preprocessed using the HCP minimal processing pipelines version 3.4.0 (Glasser et al., 2013), which is primarily based on FreeSurfer (Dale et al., 1999) and FMRIB Software Library (Jenkinson et al., 2012). In brief, the function of this pipeline is to align the T1w and T2w images, register them to the Montreal Neurological Institute space, segment the volume into predefined structures, reconstruct white and pial cortical surfaces, and perform FreeSurfer's standard folding-based surface registration to a surface atlas (the “fsaverage” template). The functional portion of the pipelines removes spatial distortions using spin echo unwarping maps, realigns volumes to compensate for subject motion, registers the fMRI data to the structural images, reduces the bias field, normalizes the 4D image to a global mean, masks the data with the final brain mask, and maps the voxels within the cortical gray matter ribbon onto the native cortical surface space. More details on the HCP processing pipelines can be found in Glasser and associates (2013).
Additional preprocessing was performed using Analysis of Functional NeuroImages (Cox, 1996), which included motion regression using 12 motion parameters, and regression-based removal of signal changes in the white matter, cerebrospinal fluid, and the global signal. Bandpass filtering was applied to select frequency bands of interest: Slow-2 (0.198–0.50 Hz), Slow-3 (0.073–0.198 Hz), Slow-4 (0.027–0.073 Hz), Slow-5 (0.01–0.027 Hz), Slow-4 + 5 (covering both Slow-4 and Slow-5; 0.01–0.073 Hz), LFO (0.01–0.1 Hz), and All (no bandpass filtering; ∼0.00–0.62 Hz). For the brain atlas, a combination of 360 cortical regions defined by the HCP's Glasser parcellation and 19 subcortical regions from the FreeSurfer subcortical segmentation was used. BOLD time series from these 379 regions were extracted per subject to generate machine learning training features.
Motion outliers
A soft motion outlier criterion was implemented. It was not desired to build a classifier model based on highly selected data, because an ideal model should be able to classify participants despite moderate levels of motion in the scanner. To achieve this, the machine learning model needs to be exposed to a sufficient number of data points contaminated by motion. One needs to be cautious, however, not to train a model that classifies based on high versus low motion, instead of TLE patients versus healthy controls.
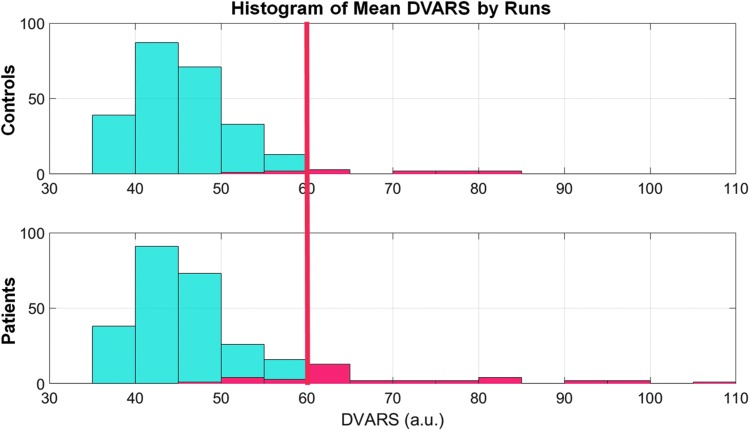
Therefore, instead of performing a rigorous motion scrubbing, only a weak threshold was selected based on the global motion. A metric called derivative of variance root mean squared (DVARS) was used, which measures the root-mean-square intensity difference between two consecutive volumes (Power et al., 2012). The mean DVARS across time was calculated for every 5-min rs-fMRI scan and, therefore, each participant received four mean DVARS scores. Figure 1 shows the histograms of mean DVARS by runs for both groups.
FIG. 1.
Histograms showing distributions of mean DVARS by runs for the two groups. Participants who had at least one run with mean DVARS >60 were excluded from the analyses. The red bars indicate the excluded participants: nine TLE patients and three healthy controls. DVARS, derivative of variance root mean squared; TLE, temporal lobe epilepsy.
Overall, the two groups had visually similar mean DVARS distributions, except for a small number of outliers who were more frequent in the TLE group. Participants who had at least one run with mean DVARS >60 were excluded from the analyses. Using this method, 9 TLE patients and 3 healthy controls were excluded as outliers, and the remaining samples consisting of 60 TLE patients (DVARS = 45.4 ± 5.7) and 59 healthy controls (DVARS = 45.7 ± 5.2) were matched statistically on mean DVARS (p = 0.48).
Machine learning training features
For RSFC measures, Pearson's correlations were computed. A total of 71,631 unique pairwise correlations were used as training features in the machine learning. For ALFFs, the filtered BOLD time series were Fourier transformed to the frequency domain, and the mean of the square root values within the frequency range of interest was calculated (Zang et al., 2007). fALFF was calculated as the ALFF of the frequency range over the ALFF of the All range (Zou et al., 2008). For ALFFs and fALFFs, the number of available features in the training was 379 for each.
Statistical analysis
Two-tailed t-tests between the groups were computed to assess group differences for each feature from each of the three measures. The Benjamini–Hochberg false discovery rate (FDR) method was used to correct for multiple comparisons (Benjamini and Hochberg, 1995).
Machine learning models
All machine learning analyses were done in MATLAB R2016a with the Statistics and Machine Learning Toolbox (MathWorks, 2017). Three different binary classifiers were examined: support vector machine (Cortes and Vapnik, 1995), linear discriminant analysis (Izenman, 2008), and naive Bayes (Friedman et al., 1997) classifiers. These three traditional classifiers were trained instead of one to get a general sense of the expected machine learning classification performance.
Leave-one-out-cross-validation (LOOCV) was used to estimate model performance (Evgeniou and Pontil, 2004). In each LOOCV loop, one participant was taken out and the machine learning model was trained with N − 1 participants. Then the left out participant was used as a testing sample for the trained model. This procedure was repeated until every participant had been left out once. The classification performances were averaged to give the LOOCV accuracy. This method is known to give the most unbiased estimate of the test error and is a good method for small sample cases (Evgeniou and Pontil, 2004; Vergun et al., 2016). Receiver operating characteristic area-under-the-curve (AUC) was also computed by adjusting the misclassification cost function during training. A random classifier would give 50% LOOCV accuracy with AUC = 0.5.
Feature selection
To reduce feature dimensionality, Lasso regression analysis was performed on the training set in each cross-validation loop, with the regularization coefficient (lambda) at 0.1 (Tang et al., 2013; Tibshirani, 1996). Only features with nonzero Lasso coefficients were used in the training of the machine learning models. Features that received nonzero coefficients in all 119 cross-validation loops were saved for further analysis. Recursive feature elimination (RFE) (Guyon et al., 2002) was employed within each loop based on the Lasso coefficients to further reduce the dimensionality.
Results
As noted in the Materials and Methods section, the TLE patients and control participants did not differ on age, gender, or mean DVARS.
The highest LOOCV classification accuracies using RSFC were in the low to mid-80%, with the AUC close to 90%. The highest accuracies were only in the mid-70% using ALFF and fALFF measures. These results are summarized in Table 1.
Table 1.
Summary of Machine Learning Results
| SVM | LDA | NB | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Measure | Frequency band | Accuracy | AUC | Features | Accuracy | AUC | Features | Accuracy | AUC | Features |
| RSFC | Slow-2 | 57.14 | 0.52 | 57 | 63.87 | 0.62 | 3 | 61.34 | 0.60 | 3 |
| Slow-3 | 65.55 | 0.67 | 14 | 63.03 | 0.62 | 12 | 63.03 | 0.61 | 21 | |
| Slow-4 | 52.10 | 0.44 | 3 | 53.78 | 0.43 | 3 | 52.94 | 0.42 | 3 | |
| Slow-5 | 75.63 | 0.80 | 10 | 75.63 | 0.80 | 8 | 76.47 | 0.79 | 11 | |
| Slow-4 + 5 | 84.87 | 0.86 | 31 | 81.51 | 0.86 | 36 | 83.19 | 0.88 | 29 | |
| LFO | 72.27 | 0.72 | 5 | 69.75 | 0.71 | 5 | 73.95 | 0.79 | 60 | |
| All | 72.27 | 0.72 | 37 | 69.75 | 0.73 | 27 | 68.07 | 0.69 | 26 | |
| ALFFs | Slow-2 | 52.94 | 0.43 | 25 | 53.78 | 0.49 | 34 | 57.98 | 0.56 | 33 |
| Slow-3 | 63.03 | 0.59 | 4 | 67.23 | 0.69 | 1 | 66.39 | 0.67 | 1 | |
| Slow-4 | 69.75 | 0.71 | 17 | 68.91 | 0.72 | 17 | 69.75 | 0.73 | 17 | |
| Slow-5 | 78.99 | 0.81 | 11 | 77.31 | 0.81 | 13 | 73.11 | 0.76 | 12 | |
| Slow-4 + 5 | 64.71 | 0.65 | 3 | 67.23 | 0.68 | 3 | 64.71 | 0.67 | 3 | |
| LFO | 73.95 | 0.72 | 14 | 78.15 | 0.81 | 14 | 69.75 | 0.72 | 15 | |
| All | 62.18 | 0.56 | 6 | 61.34 | 0.61 | 8 | 63.87 | 0.64 | 10 | |
| fALFFs | Slow-2 | 53.78 | 0.46 | 22 | 59.66 | 0.58 | 2 | 60.50 | 0.57 | 2 |
| Slow-3 | 54.62 | 0.42 | 12 | 73.11 | 0.78 | 6 | 72.27 | 0.75 | 6 | |
| Slow-4 | 64.71 | 0.55 | 2 | 63.87 | 0.64 | 2 | 65.55 | 0.65 | 2 | |
| Slow-5 | 70.59 | 0.70 | 6 | 68.07 | 0.70 | 6 | 64.71 | 0.69 | 2 | |
| Slow-4 + 5 | 56.30 | 0.46 | 6 | 55.46 | 0.53 | 25 | 56.30 | 0.53 | 24 | |
| LFO | 58.82 | 0.50 | 16 | 55.46 | 0.54 | 3 | 63.03 | 0.57 | 20 | |
Bold values represent the best results per category.
The three resting-state measures and seven frequency bands tested are organized in the leftmost columns. The three traditional machine learning models trained are organized in the top row. The accuracies are the LOOCV accuracies. “Features” column indicates the number of features selected from the RFE feature selection. Best LOOCV accuracies were achieved with Slow-4 + 5 RSFC features.
ALFFs, amplitude of low-frequency fluctuations; AUC, area-under-the-curve; fALFFs, fractional ALFFs; LDA, linear discriminant analysis; LFO, low-frequency oscillation; LOOCV, leave-one-out cross-validation; NB, naive Bayes; RFE, recursive feature elimination; RSFC, resting-state functional connectivity; SVM, support vector machine.
Using RSFC, the Slow-4 + 5 band produced the best overall model performance in classifying the TLE patients and healthy controls, with ∼83% LOOCV accuracy. The accuracies from the three machine learning models were also consistent: 83.19% ± 1.37%. Using ALFFs and fALFFs, LOOCV accuracies were not as consistent as using RSFC.
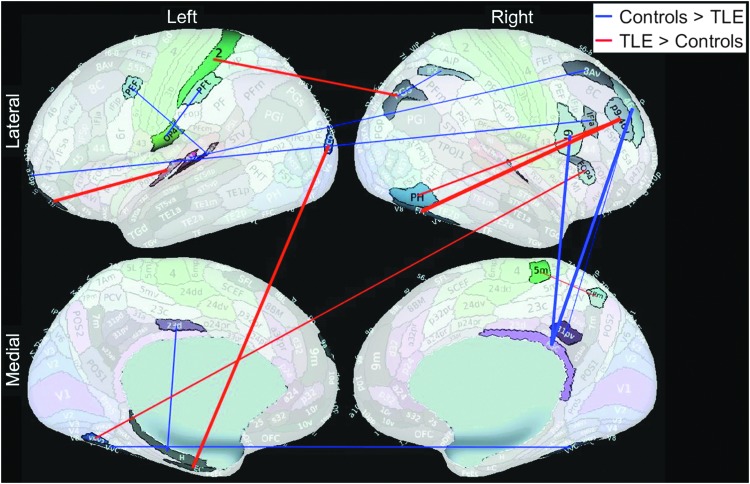
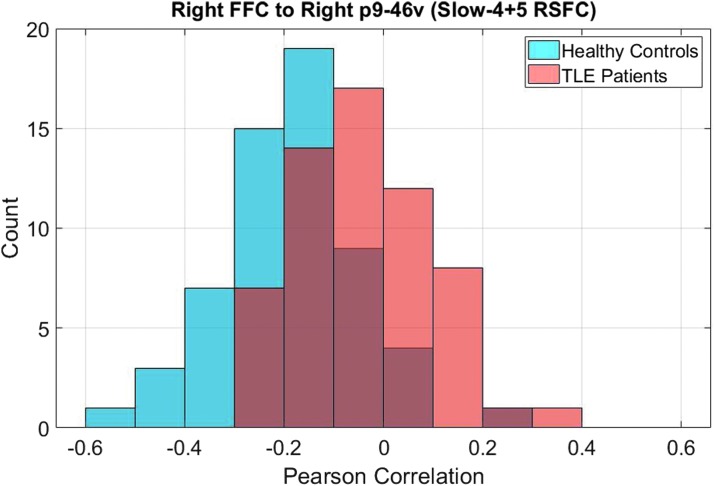
A total of 19 Slow-4 + 5 RSFC features were selected by the Lasso feature selection every time in all 119 cross-validation loops, and these are summarized in Table 2, as well as shown in Figure 2. Only five of these features also received significant p values (pfdr < 0.05) from the group t-test. Connection between right fusiform face complex (R_FFC) and right area posterior 9-46v (R_p9-46v, a part of Brodmann area 46) was the most significant feature based on both Lasso and t-test (pfdr < 0.001) analyses, and was stronger in TLE patients than in the healthy group (Fig. 3).
Table 2.
Nineteen Features That Were Repeatedly Selected by the Lasso Feature Selection in All 119 Cross-Validation Loops
| Lasso features | ||||||
|---|---|---|---|---|---|---|
| No | From | To | Lasso weight | t-Test p_FDR | ||
| 1 | “R_Fusiform Face Complex” | “R_FFC” | “R_Area posterior 9-46v” | “R_p9-46v” | 0.713 | <0.001* |
| 2 | “R_RetroSplenial Complex” | “R_RSC” | “R_Area 46” | “R_46” | −0.622 | 0.003* |
| 3 | “L_Entorhinal Cortex” | “L_EC” | “L_Area V3CD” | “L_V3CD” | 0.584 | >0.1 |
| 4 | “L_Area 11l” | “L_11l” | “L_Area 52” | “L_52” | 0.575 | >0.1 |
| 5 | “R_RetroSplenial Complex” | “R_RSC” | “R_Rostral Area 6” | “R_6r” | −0.552 | >0.1 |
| 6 | “L_Area 2” | “L_2” | “R_Area PGs” | “R_PGs” | 0.451 | 0.087 |
| 7 | “R_Area posterior 9-46v” | “R_p9-46v” | “R_Area PH” | “R_PH” | 0.377 | 0.005* |
| 8 | “L_Area 9 anterior” | “L_9a” | “L_Amygdala” | “L_Amygdala” | −0.376 | 0.087 |
| 9 | “L_VentroMedial Visual Area 3” | “L_VMV3” | “R_Frontal Opercular Area 4” | “R_FOP4” | 0.336 | 0.063 |
| 10 | “L_Area 23d” | “L_23d” | “L_Hippocampus” | “L_H” | −0.297 | 0.095 |
| 11 | “L_Area OP4/PV” | “L_OP4” | “L_Area PFt” | “L_PFt” | −0.292 | 0.039* |
| 12 | “L_Ventral Visual Complex” | “L_VVC” | “R_Ventral Visual Complex” | “R_VVC” | −0.290 | 0.002* |
| 13 | “R_Fusiform Face Complex” | “R_FFC” | “R_Medial Belt Complex” | “R_MBelt” | 0.274 | >0.1 |
| 14 | “L_Area anterior 10p” | “L_a10p” | “R_Area IFJa” | “R_IFJa” | −0.260 | >0.1 |
| 15 | “L_Premotor Eye Field” | “L_PEF” | “L_ParaBelt Complex” | “L_PBelt” | −0.231 | >0.1 |
| 16 | “L_Primary Auditory Cortex” | “L_A1” | “R_Area 8Av” | “R_8Av” | −0.225 | >0.1 |
| 17 | “R_Area IntraParietal 2” | “R_IP2” | “R_Area PGs” | “R_PGs” | −0.214 | >0.1 |
| 18 | “R_Medial Area 7P” | “R_7Pm” | “R_Area 5m” | “R_5m” | 0.201 | >0.1 |
| 19 | “R_Area 31p ventral” | “R_31pv” | “R_Area 46” | “R_46” | −0.191 | 0.057 |
Represents statistical significance (p < 0.05).
Features highlighted in light gray were stronger in TLE patients, and vice versa for the rest. Only five out of 19 features showed significant group differences based on t-test.
FIG. 2.
Eighteen significant Slow-4 + 5 RSFC cortical features based on Lasso feature selection are shown. Connection between left area 9 anterior (L_9a) to left amygdala is the only one not shown in the figure from Table 2. This figure suggests that the changes in the TLE brains are throughout the whole brain, not only in the temporal lobes. The background brain images were generated with the Connectome Workbench and from Glasser et al. (2016). RSFC, resting-state functional connectivity.
FIG. 3.
This is a histogram showing the distributions of Pearson correlation between signals from R_FFC and R_p9-46v. An increased correlation, or decreased negative correlation, was found in the TLE group. This feature was the most significant feature based on both Lasso and t-test analyses. R_FFC, right fusiform face complex; R_p9-46v, right area posterior 9-46v.
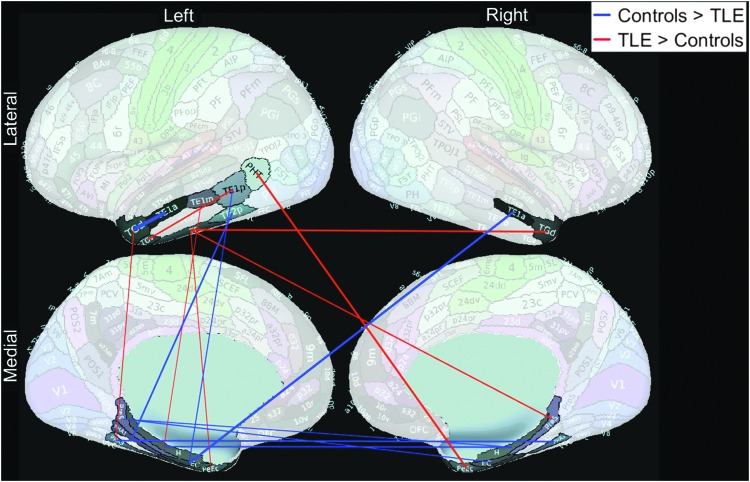
The 19 significant features did not include any exclusively temporal lobe connections. We reran the analysis using Slow-4 + 5 RSFC, but only within the temporal lobe (24 regions, 276 connections). The maximum LOOCV accuracy was only 68.91%. The connection between left area TG dorsal (L_TGd, a part of Brodmann area 38) and left area TE1 anterior (L_TE1a) was found to be the most significant temporal lobe feature based on both Lasso and t-test (pfdr = 0.006) analyses, and was stronger in the healthy group. Figure 4 shows 15 temporal lobe features that were selected by the Lasso feature selection every time in all 119 cross-validation loops.
FIG. 4.
Fifteen significant Slow-4 + 5 RSFC temporal lobe features based on Lasso feature selection are shown. Only temporal lobe features (276 out of 71,631) entered the feature selection. None of these features were selected when whole brain connectivity was used to separate between TLE patients and healthy controls.
Discussion
TLE is associated with chronic structural brain changes, particularly in medial temporal structures, but also in the thalamus, insula, and various cortical regions, which are thought to be a consequence of repeated seizure activity (Caciagli et al., 2017). Our hypothesis was that these brain alterations in TLE would also have significant effects on resting brain activity and inter-regional connectivity, regardless of their seizure focus. Therefore, we studied a heterogeneous group of TLE patients.
Seven frequency ranges with three different measures of the resting functional brain signals were tested to train three different traditional machine learning models. This extensive search for good training features was an attempt to cover all possible measures using the resting fMRI images. In brief, the results suggest that functional brain alterations in the TLE patients are indeed detectable and are captured best by RSFC using the Slow-4 + 5 range. The machine learning models can use this information to separate TLE patients from age- and gender-matched healthy controls with ∼83% accuracy. Also notably, the features separating between the TLE patients and healthy controls were located throughout the entire brain, and not just within the temporal lobe, which is consistent with previous findings (Liao et al., 2010; Morgan et al., 2015).
There have been many articles in the recent literature describing the development of reliable machine learning models to make more accurate decisions from complex clinical data sets. For example, there are reports on using machine learning to predict postsurgical outcome of TLE patients using nonimaging data (Armananzas et al., 2013), structural MRI data (Feis et al., 2013; Munsell et al., 2015), or intracranial EEG (Memarian et al., 2015). Machine learning has also been applied in the lateralization of TLE based on rs-fMRI (Yang et al., 2015) or positron emission tomography (PET) (Kerr et al., 2013), and in separating TLE patients and healthy controls using structural imaging data (Bernhardt et al., 2015), diffusion imaging data (Del Gaizo et al., 2017), or both (Focke et al., 2012). It was also applied in separating epilepsy patients overall and healthy controls using RSFC (Rajpoot et al., 2015; Zhang et al., 2012). No studies have intensively studied the classification between TLE patients and healthy controls using rs-fMRI features. The current results may guide future research in this area.
As an extensive high-quality imaging data set, the ECP will provide an unprecedented opportunity for the development of more reliable machine learning models. By the end of the project, comprehensive data from close to 200 TLE patients will be acquired, which will include demographics, cognitive test scores, structural and fMRI, MEG, and genetics information. All of this information can be regarded as potential machine learning features. In this ongoing research, it was decided to first start with the RSFC because it has shown success with machine learning as already explained.
Deep learning methods generally outperform traditional machine learning models given a sufficient sample size (Ng, 2016). However, this is often an unrealistic sample size in clinical studies with limited data. More immediate use of machine learning, therefore, may be to use traditional machine learning techniques for multidimensional analyses of given features. Without machine learning, or a similar automated method, we are limited in our ability to comprehend high-dimensional data, especially when the patterns are complex. Also, the true nature of a clinical question may be significantly more nonlinear than one may assume at the outset. Instead of trying to sort out useful information from a complex set of features, one can consult machine learning models to discover patterns within them.
The biggest limitation of traditional machine learning models is the need to select input features for the model being trained. In most cases, we do not know a priori what combination of features would contain the most useful information for those models. In this study, a Lasso-based feature selection method along with RFE was employed. At present, identifying the best set of features and the correct nonlinearity of the models (or kernels) remains a trial-and-error process. In theory, deep learning methods can perform this feature selection much efficiently given a sufficient amount of data. Given limited data, we believe that it is advisable to think broadly, considering a wide range of potential features available, while actively narrowing the set down so that the models are not clogged with noisy information.
In this study, three traditional machine learning models were trained, to get a general sense of the expected machine learning classification performance using traditional techniques. The best overall accuracy was achieved with the Slow-4 + 5 RSFC features and it was very comparable between the three classifiers. These traditional models are more straightforward and understandable than highly nonlinear models such as deep learning. Therefore, they allow us to more easily analyze what underlying features are contributing the most to the models. The set of Slow-4 + 5 RSFC features that contributed most to the models, which are visualized in Figure 2, suggest widespread functional connectivity alterations in TLE patients. This list included no exclusively temporal lobe connections, which is perhaps due to the heterogeneity of our TLE patient group. It is notable that connections outside of the temporal lobe result in better classification than temporal lobe connections. The increased connection between R_FFC and R_p9-46v is consistent with the findings of Riley and associates (2015), who reported altered functional connectivity of the cortical face processing networks in TLE patients. Abnormal structure and function of the retrosplenial cortex (RSC) have also been reported (Addis et al., 2007; Mueller et al., 2009).
These results are promising for future applications in diagnosing and understanding the basic pathophysiology of TLE. Currently TLE diagnosis is made with a combination of EEG, MEG, structural MRI, and/or single-photon emission computed tomography/PET in conjunction with clinical phenomenology, but in many cases, convincing evidence is difficult to capture noninvasively, and more invasive approaches such as subdural grids or depth electrodes are used (Pizarro et al., 2016). Diagnosis of TLE with an rs-fMRI scan could provide valuable adjunctive evidence and might eventually replace some current clinical tests with a more cost-effective method.
Future models can build on our current model by increasing the sample size by incorporating additional data sets of TLE patients, other type of epilepsies, and healthy controls, so a machine learning classifier with high accuracy, sensitivty, and specificity can be constructed. It should also be noted that the current results are from studying TLE patients regardless of their patient subtypes. With increasing sample sizes, it will allow one to subclassify the TLE patient group based on clinical subgroupings such as left versus right seizure foci, those with versus without medial temporal sclerosis, and those with well-controlled versus uncontrolled seizures, which will provide more comprehensive information regarding functional changes associated with this brain disorder.
Other future direction for this line of research is to test a wider variety of training features. From this study, it was learned that RSFC measures contained more useful information than ALFFs or fALFFs for the traditional machine learning models tested. Potential training features from other modalities (e.g., MEG, diffusion tensor imaging) in the ECP data set will be tested in future analyses. It is also interesting to combine different feature sets to potentially produce better models, because the goal of ECP is to use all the information available to make comprehensive statements about the TLE population. Once we settle on the best set of training features that can discriminate TLE patients from the healthy controls, they can then be used to predict clinical outcomes often found in this patient population by training regression type models, which may guide clinical decisions in the future.
Conclusion
The current results suggest that oscillations in the low-frequency range may provide valuable information about altered brain connectivity patterns in TLE patients. Using only RSFC features generated from the Slow-4 + 5 range, between 0.01 and 0.073 Hz, traditional machine learning models were able to separate a heterogeneous sample of TLE patients from healthy controls with 83% accuracy. Increased connectivity from R_p9-46v and decreased connectivity from RSC in the TLE group contributed most to these classification models. ALFF and fALFF measures produced less accurate and more unstable models than RSFC. More effective and stable classification models are expected in the future with additional data and with higher machine learning model complexity. The ECP protocol with its sensitive state-of-the-art methods allows one to capture with higher spatial and temporal resolution abnormal network function in TLE than existing literature.
Supplementary Material
Acknowledgments
The authors thank all the participants and their families. In addition, the authors thank Taylor McMillan for recruitment and participant acquisition, MRI technologists for their assistance in scanning, and other support staff. Funding for healthy control subjects' data acquisition was provided in part by the Department of Radiology at the University of Wisconsin-Madison. This study was supported by grant numbers U01NS093650 and 1UF1AG051216-01A1 from the National Health Institute.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- Addis DR, Moscovitch M, McAndrews MP. 2007. Consequences of hippocampal damage across the autobiographical memory network in left temporal lobe epilepsy. Brain 130:2327–2342 [DOI] [PubMed] [Google Scholar]
- Armananzas R, Alonso-Nanclares L, DeFelipe-Oroquieta J, Kastanauskaite A, de Sola RG, DeFelipe J, et al. 2013. Machine learning approach for the outcome prediction of temporal lobe epilepsy surgery. PLos One 8:e62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleites C, Neugebauer U, Bocklitz T, Krafft C, Popp J. 2013. Sample size planning for classification models. Anal Chim Acta 760:25–33 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B Methodol 57:289–300 [Google Scholar]
- Bernhardt BC, Hong SJ, Bernasconi A, Bernasconi N. 2015. Magnetic resonance imaging pattern learning in temporal lobe epilepsy: classification and prognostics. Ann Neurol 77:436–446 [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34:537–541 [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Draguhn A. 2004. Neuronal oscillations in cortical networks. Science 304:1926–1929 [DOI] [PubMed] [Google Scholar]
- Caciagli L, Bernasconi A, Wiebe S, Koepp MJ, Bernasconi N, Bernhardt BC. 2017. A meta-analysis on progressive atrophy in intractable temporal lobe epilepsy: time is brain? Neurology 89:506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno M, Carmichael DW. 2014. Network connectivity in epilepsy: resting state fMRI and EEG-fMRI contributions. Front Neurol 5:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CJ, Hwang G, Mathis J, Nair VA, Conant L, Allen L, et al. 2018. Effective connectivity within the default mode network in left temporal lobe epilepsy: findings from the Epilepsy Connectome Project. Brain Connect. DOI: 10.1089/brain.2018.0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. Mach Learn 20:273–297 [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173 [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9:179–194 [DOI] [PubMed] [Google Scholar]
- Del Gaizo J, Mofrad N, Jensen JH, Clark D, Glenn R, Helpern J, Bonilha L. 2017. Using machine learning to classify temporal lobe epilepsy based on diffusion MRI. Brain Behav 7:e00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J, Jr., Pitkanen A, Loeb JA, Dudek FE, Bertram EH, 3rd, Cole AJ, et al. 2013. Epilepsy biomarkers. Epilepsia 54 Suppl 4:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgeniou T, Pontil M. 2004. Leave one out error, stability, and generalization of voting combinations of classifiers. Mach Learn 55:71–97 [Google Scholar]
- Feis DL, Schoene-Bake JC, Elger C, Wagner J, Tittgemeyer M, Weber B. 2013. Prediction of post-surgical seizure outcome in left mesial temporal lobe epilepsy. Neuroimage Clin 2:903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355 [DOI] [PubMed] [Google Scholar]
- Focke NK, Yogarajah M, Symms MR, Gruber O, Paulus W, Duncan JS. 2012. Automated MR image classification in temporal lobe epilepsy. Neuroimage 59:356–362 [DOI] [PubMed] [Google Scholar]
- Friedman N, Geiger D, Goldszmidt M. 1997. Bayesian network classifiers. Mach Learn 29:131–163 [Google Scholar]
- Garcia-Ramos C, Song J, Hermann BP, Prabhakaran V. 2016. Low functional robustness in mesial temporal lobe epilepsy. Epilepsy Res 123:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, et al. 2016. A multi-modal parcellation of human cerebral cortex. Nature 536:171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, et al. 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80:105–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohel SR, Biswal BB. 2015. Functional integration between brain regions at rest occurs in multiple-frequency bands. Brain Connect 5:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. 2002. Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422 [Google Scholar]
- Holmes MD, Tucker DM. 2013. Identifying the epileptic network. Front Neurol 4:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JP, Xiong ZX, Lowey J, Suh E, Dougherty ER. 2005. Optimal number of features as a function of sample size for various classification rules. Bioinformatics 21:1509–1515 [DOI] [PubMed] [Google Scholar]
- Hwang G, Cook CJ, Nair VA, Alexander A, Antuono PG, Asthana S, et al. 2018. Characterizing structural brain alterations in alzheimer's disease patients with machine learning. Alzheimers Dement 14:P135–P136 [Google Scholar]
- Izenman AJ. 2008. Linear discriminant analysis. In Modern Multivariate Statistical Techniques: Regression, Classification, and Manifold Learning. New York, NY: Springer; pp. 237–280 [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. 2012. FSL. Neuroimage 62:782–790 [DOI] [PubMed] [Google Scholar]
- Kerr WT, Nguyen ST, Cho AY, Lau EP, Silverman DH, Douglas PK, et al. 2013. Computer-aided diagnosis and localization of lateralized temporal lobe epilepsy using interictal FDG-PET. Front Neurol 4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsiantis SB, Zaharakis ID, Pintelas PE. 2006. Machine learning: a review of classification and combining techniques. Artif Intell Rev 26:159–190 [Google Scholar]
- Kucukboyaci NE, Kemmotsu N, Cheng CE, Girard HM, Tecoma ES, Iragui VJ, McDonald CR. 2013. Functional connectivity of the hippocampus in temporal lobe epilepsy: feasibility of a task-regressed seed-based approach. Brain Connect 3:464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Zhang Z, Pan Z, Mantini D, Ding J, Duan X, et al. 2010. Altered functional connectivity and small-world in mesial temporal lobe epilepsy. PLoS One 5:e8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MathWorks. 2017. Statistics and machine learning toolbox release notes. https://www.mathworks.com/help/pdf_doc/stats/rn.pdf (accessed February6, 2019)
- Meier TB, Desphande AS, Vergun S, Nair VA, Song J, Biswal BB, et al. 2012. Support vector machine classification and characterization of age-related reorganization of functional brain networks. Neuroimage 60:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarian N, Kim S, Dewar S, Engel J, Staba RJ. 2015. Multimodal data and machine learning for surgery outcome prediction in complicated cases of mesial temporal lobe epilepsy. Comput Biol Med 64:67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. 2010. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med 63:1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan VL, Abou-Khalil B, Rogers BP. 2015. Evolution of functional connectivity of brain networks and their dynamic interaction in temporal lobe epilepsy. Brain Connect 5:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Laxer KD, Barakos J, Cheong I, Garcia P, Weiner MW. 2009. Widespread neocortical abnormalities in temporal lobe epilepsy with and without mesial sclerosis. Neuroimage 46:353–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsell BC, Wee CY, Keller SS, Weber B, Elger C, da Silva LAT., et al. 2015. Evaluation of machine learning algorithms for treatment outcome prediction in patients with epilepsy based on structural connectome data. Neuroimage 118:219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A. 2016. Nuts and bolts of applying deep learning. Paper presented at the Bay Area Deep Learning School, Stanford University, CA Conference Lecture retrieved from https://www.youtube.com/watch?v=F1ka6a13S9I (accessed February6, 2019) [Google Scholar]
- Pizarro R, Nair V, Meier T, Holdsworth R, Tunnell E, Rutecki P, et al. 2016. Delineating potential epileptogenic areas utilizing resting functional magnetic resonance imaging (fMRI) in epilepsy patients. Neurocase 22:362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59:2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpoot K, Riaz A, Majeed W, Rajpoot N. 2015. Functional connectivity alterations in epilepsy from resting-state functional MRI. PLoS One 10:e0134944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JD, Fling BW, Cramer SC, Lin JJ. 2015. Altered organization of face-processing networks in temporal lobe epilepsy. Epilepsia 56:762–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Nair VA, Gaggl W, Prabhakaran V. 2015. Disrupted brain functional organization in epilepsy revealed by graph theory analysis. Brain Connect 5:276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Alelyani S, Liu H. 2014. Feature selection for classification: a review. In: Data Classification: Algorithms and Applications. CRC Press; pp. 37–64 [Google Scholar]
- Tellez-Zenteno JF, Hernandez-Ronquillo L. 2012. A review of the epidemiology of temporal lobe epilepsy. Epilepsy Res Treat 2012:630853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. 1996. Regression shrinkage and selection via the Lasso. J R Stat Soc B Methodol 58:267–288 [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, WU-Minn HCP Consortium. 2013. The WU-Minn human connectome project: an overview. Neuroimage 80:62–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergun S, Gaggl W, Nair VA, Suhonen JI, Birn RM, Ahmed AS, et al. 2016. Classification and extraction of resting state networks using healthy and epilepsy fMRI data. Front Neurosci 10:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Choupan J, Reutens D, Hocking J. 2015. Lateralization of temporal lobe epilepsy based on resting-state functional magnetic resonance imaging and machine learning. Front Neurol 6:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack MM, Kobau R. 2017. National and state estimates of the numbers of adults and children with active epilepsy—United States, 2015. MMWR Morb Mortal Wkly Rep 66:821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang YF, He Y, Zhu CZ, Cao QJ, Sui MQ, Liang M, et al. 2007. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev 29:83–91 [DOI] [PubMed] [Google Scholar]
- Zhang J, Cheng W, Wang ZG, Zhang ZQ, Lu WL, Lu GM, Feng JF. 2012. Pattern classification of large-scale functional brain networks: identification of informative neuroimaging markers for epilepsy. PLoS One 7:e36733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lu G, Zhong Y, Tan Q, Chen H, Liao W, et al. 2010. fMRI study of mesial temporal lobe epilepsy using amplitude of low-frequency fluctuation analysis. Hum Brain Mapp, 31, 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou QH, Zhu CZ, Yang Y, Zuo XN, Long XY, Cao QJ, et al. 2008. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods 172:137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Di Martino A, Kelly C, Shehzad ZE, Gee DG, Klein DF, et al. 2010. The oscillating brain: complex and reliable. Neuroimage 49:1432–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.