Abstract
Patients with metastatic or relapsed/refractory osteosarcoma (OS) have a 5-year survival rate of <30%. This has remained unchanged over several decades. One of the factors contributing to lack of improvement in survival is the development of chemoresistance. Hence, elucidating and targeting the mechanisms that promote survival against chemotherapy and lead to chemoresistance is pivotal to improving outcomes for these patients. We identified that endoplasmic reticulum (ER) stress-activated transcription factor, ATF6α, is essential for the survival of OS cells against chemotherapy induced cell death. ATF6α cleavage and activity were enhanced in OS cells compared to normal osteoblasts and knockdown of ATF6α expression enhanced sensitivity of OS cells against chemotherapy induced cell death. This was in part due to increased Bax activation. Pharmacologic inhibition or knock-down of downstream targets of ATF6α, protein disulfide isomerases (PDI) and ERO1β, a thiol oxidase that is involved in the re-oxidation of PDIs also independently induced pronounced killing of OS cells following chemotherapy. Analysis of primary tumors from OS patients reveals that patients with high levels of nuclear ATF6α: (1) also had increased expression of its downstream targets the chaperone BiP and enzyme PDI, (2) had a significant likelihood of developing metastasis at diagnosis, (3) had significantly poorer overall and progression free survival, and (4) had poorer response to chemotherapy. These findings suggest that targeting survival signaling by the ATF6α pathway in OS cells may favor eradication of refractory OS tumor cells and ATF6α could be a useful predictor for chemo-responsiveness and prognosis.
Introduction
Osteosarcoma is the most common and aggressive primary bone cancer in children and adolescents, with 400 new cases per year [1]. Although less common than brain tumors or acute lymphoblastic leukemia, OS accounts for a disproportionate number of the cancer mortality observed in children. The standard treatment strategy for patients with newly diagnosed OS consists of surgery in combination with multi-agent chemotherapy consisting of doxorubicin, cisplatin, methotrexate, and ifosfamide, which have remained unchanged over the past 30 years [1], [2]. Although this therapy helps tumor cytoreduction and remission rate, the long-term survival has plateaued and remains at 60–70% [2], [3]. Additionally, prognosis for patients who have progressive or recurrent disease is less than 20% [3], [4]. OS has a complex karyotype and sequencing of tumors has revealed significant tumor-to-tumor variability through diverse and numerous structural variations with the exception of dysfunctional p53 in virtually all clinical cases with frequent translocations in intron 1 of the TP53 gene [5]. As a result, identifying a consistent therapeutic target that can improve outcome for these patients has proven to be elusive. Since tumors that do not respond to initial therapy or recur have mechanisms that are integral to pathogenesis and survival/resistance against therapy, delineating such mechanisms will yield not only a greater knowledge of the tumor biology of OS but will also be indicative of methods of circumventing the mechanisms of resistance.
The ER is the primary organelle where the folding of secretory proteins occurs [6]. Several physiological and pathological conditions such as cancer, perturb the cellular microenvironment causing protein misfolding and accumulation of unfolded proteins referred to as ER stress and activation of the unfolded protein response (UPR). UPR is an adaptive signaling pathway that results in the coordinated activation of three ER transmembrane proteins, protein kinase-like endoplasmic reticulum kinase (PERK), inositol-requiring 1α (IRE1α) and activating transcription factor 6α (ATF6α), which allows for protein folding in the ER by up-regulating chaperones such as BiP/GRP78 [6]. Activation of PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α) that attenuates protein synthesis. Activation of IRE1α leads to the non-canonical splicing and activation of the transcription factor X-box-binding protein-1 (XBP-1) as well as mRNA expression levels through regulated IRE1-dependent mRNA decay (RIDD) and controls the activation of the c-jun N-terminal kinase (JNK) pathway [7]. The third arm of the UPR, ATF6α, is a type II trans-membrane protein that contains a cytosolic cAMP-responsive element-binding protein (CREB)/ATF basic leucine zipper (bZIP) domain. Under non-stressed conditions, ATF6α is retained in the ER through interaction with BIP [8]. During ER stress ATF6α is released from BiP and translocates to the Golgi apparatus via COPII mediated vesicular transport [9], where it is activated via regulated intermembrane proteolysis by Site-1 and Site-2 proteases (S1P and S2P). The cleaved N-terminal cytoplasmic domain of ATF6α [pATF6α(N)], which has the bZIP DNA-binding domain and a transcriptional activation domain, translocates into the nucleus and activates the transcription of its target genes by binding to a cis-acting element, known as the ER stress response element (ERSE) and UPR element (UPRE) [10], [11]. Although PERK, IRE-1α and ATF6α activated target genes such as BiP, are widely associated with the survival and therapy resistance of several different cancers, few studies have looked at the direct impact of ATF6α and its underlying mechanisms in promoting tumor cell survival and therapy resistance. ATF6α expression and activity were shown to be elevated during carcinogenesis and cisplatin resistance of HCC; however, the functional relevance of this increase in ATF6α to these processes was not examined [12]. Recently, a few studies have demonstrated a functional role for ATF6α in in tumor cell survival and therapy resistance. In head and neck tumors AFT6α promoted survival of dormant tumor cells by regulating RHEB expression and mTOR signaling [13]. Higa et al showed that activation of ATF6α In leukemia cells was essential for resistance to imatinib treatment [14]. While in glioblastoma ATF6α was also implicated in promoting survival against radiation therapy via regulation of NOTCH expression [15]. Although UPR has been shown to be activated in osteosarcoma [16], whether it has a role in conferring resistance to chemotherapy has not yet been established.
Here we report that ATF6α activation promoted survival against cisplatin and irinotecan induced apoptosis in OS cells. We further demonstrate that this survival is mediated in part by ATF6α regulated targets, BiP, PDI and ERO1β. We also provide the first evidence that the levels of active nuclear ATF6α is an independent prognostic indicator irrespective of metastatic status and histologic response to treatment for overall and progression-free survival in patients with OS.
Materials and Methods
Reagents and Antibodies
The following antibodies were obtained from Cell Signaling (Danvers, MA); rabbit anti-p-eIF2α (Ser51), total eIF2α, anti-cleaved caspase-3, GM130 and rabbit anti-calreticulin, and PDI. Mouse monoclonal Anti-ATF6α antibody that recognizes both full-length and cleaved nuclear forms of ATF6α was from Novus Biologicals (for IF and IHC analysis) and Bioacademia (for WB). Anti- GAPDH was from Life Technologies. Anti-BiP and active Bax antibodies were from BD Biosciences. HRP conjugated anti-mouse and anti-rabbit IgG Abs were from Vector Laboratories (Burlingame, CA). Fluorescence-conjugated secondary antibodies were from Molecular Probes (Thermo Fisher). Cisplatin, dithiothreitol (DTT), tunicamycin™, and PDI inhibitor 16F16 were from Sigma (St. Louis, MO). GSK2606414 was from Tocris, STF083010 and irinotecan were from Calbiochem and Thapsigargin (Tg) was from Fisher Scientific.
Cell Lines
U2OS and 143b cells were a kind gift from Dr. Richard Gorlick's laboratory.
and hFOB cells were obtained from ATCC. The cells were maintained in DMEM supplemented with 10% fetal bovine serum under standard conditions, hFOB cells were maintained in DMEM-Ham’s F-12 medium according to ATCC's instructions. All cell lines were authenticated by our experimental mouse shared resource core facility (www.emss.azcc.arizona.edu) to match cells to their previously published identities and to detect any cross-contamination.
Cytotoxicity Assay
The OS cell lines and osteoblasts were plated at 4x103 cells/well into 96-well white-walled plates and allowed to adhere for 24 hours before drug treatments. The cells were then exposed to serial dilutions of cisplatin or irinotecan and were incubated at 37oC for an additional 24 hours. Cell viability was then determined using the CellTiter-Glo Luminescent Cell Viability assay (Promega).
Immunoblotting
After treatments, cells were placed on ice, washed twice with PBS, and lysed by boiling for 5-10mins in SDS sample buffer without reducing agent (50 mM Tris [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol) supplemented with Complete mini protease and phosphatase inhibitor cocktail tablets (Roche Diagnostics, and Pierce), 20 mM β-glycerophosphate, and 10 mM NaF for Western analysis. Lysates were centrifuged at 13,000 rpm for 15min at 4°C. Protein concentrations of each sample were determined using BCA protein assay kit as recommended by the manufacturer (Pierce, Hercules, CA). Proteins were resolved on sodium dodecyl sulfate–8% polyacrylamide gels and transferred onto Immun-Blot® PVDF Membrane with a wet transfer system (Bio-Rad). Membranes were blocked in blocking buffer (PBS containing 0.1% Tween 20 and 5% skim milk). Primary antibodies were used at indicated dilutions (Table S2). The blots were incubated in blocking buffer for 1h at room temperature in blocking buffer (for ATF6α) or overnight at 40C followed by incubations with secondary antibodies at room temperature for 1h. The proteins were then analyzed by immunoblotting and detected using LumiGLO chemiluminescent substrate system (Kirkegaard & Perry Laboratories) and visualized by exposure to autorad blue X-ray film (Research Products International).
Luciferase Assays
The cells were transiently transfected with either pGEM3-BiP-Luc (a gift from Dr. Linda Hendershot, St Jude’s Children’s Hospital) or 5xATF6-GL3 reporter construct (a gift from Dr. Ron Prywes, Columbia University, New York). Transfection and luciferase assays were performed using Viafect and Dual Luciferase Assay kit (Promega), respectively, following manufacturer's instructions.
Small Interfering RNA Transfections
siRNAs to ATF6α were obtained from Santa Cruz Biotechnology (ATF6α#1 cat. #sc-37699), and Thermor Fisher Scientific (ID# s223543). siRNAs to ERO1β and control siRNA were also from Thermo Fisher Scientific (ID# s32182 and cat.#AM4611). siRNAs (30 nM) were delivered into OS cells using siPORT NeoFx (Thermo Fisher) following manufacturer's instructions. The down-regulation of ATF6α and ERO1β mRNA and protein levels were analyzed by qPCR using the 2-ΔΔCt method, western blotting and immunofluorescence, respectively.
Immunohistochemistry (IHC)
IRB approval from Phoenix Children’s Hospital (PCH) was received prior to accessing patient data and tumor samples. Patients with a diagnosis of OS who were treated at PCH and had follow-up data available were included in this study. Formalin-fixed paraffin embedded (FFPE) archived primary tissue samples collected at the time of diagnosis as well as some available metastatic samples were retrieved from the institutional tumor bank and de-identified. Normal age matched liver and lung tissue were used as the control specimens. Immunohistochemical analysis was performed using anti-ATF6α mouse monoclonal antibody (Novus Biologics). Briefly, following deparaffinization the antigens were retrieved using antigen retrieval buffer (10mM Sodium Citrate, pH 6.0) by heating the samples to 90°C in a microwave oven for 12 min; the samples were then cooled and washed three times with PBS. The samples were then pretreated with 6% hydrogen peroxide solution in methanol for 30 min and then blocked for one hour at room temperature in blocking buffer (5% NGS in PBS-0.4% Triton X-100). The sections were then incubated with the primary antibodies at the indicated dilutions (Table S2) (1% NGS in PBS-0.4% Triton X-100) overnight at 4°C and washed with washing buffer (PBS-0.4% Triton X-100). The sections were then exposed to biotin-labeled goat secondary antibodies for 1h and immunoreactions were developed with an avidin-biotin complex (Vectastain ABC elite kit, Vector Laboratories, CA, USA) and the sites of peroxidase binding were demonstrated with diaminobenzidine. After counterstaining with hematoxylin, slides were dehydrated in ascending concentrations of ethanol and xylene and mounted, cover-slipped and labeled. IHC stained slides from each subject were reviewed by the institutional sarcoma pathologist. Based on the percentage of tumor cells positive for nuclear ATF6α (ATF6α(N)), tumors were graded as high (≥50% of tumor cells positive for ATF6α(N)) or low (<50% of tumor cells positive for ATF6α(N)) expressers. The percent of ATF6α(N) positive cells was assessed using two independent methods: grading by the institutional pathologist, and manual quantification by an independent observer both of whom were blinded to the clinical data. We had pathologist verification on all of the 40 samples, but manual quantification was only done on a 35 of 40 samples. The manual quantification of percent of tumor cells positive for AFT6α(N) was done by counting cells in two fields, double stained for ATF6α(N) and hemotoxylin over the total number of cells. The agreement between ATF6α(N) levels as observed by manual counting method and reading by a single pathologist was assessed using weighted kappa statistic. The κ coefficient was 0.44 (95% confidence interval (95%CI) 0.21-0.65) indicating moderate agreement. For all models and Kaplan-Meier analysis only the pathologist’s score was used as the pathological determinate.
Indirect Immunofluorescence
U2OS and 143b cells were plated on coverslips in 24-well plates at a density of 3 to 4×104 cells/well. Cells were then transfected with EGFP-ATF6α plasmid (Addgene). Forty-eight hours post-transfection, cells were treated with DTT for the indicated time points and fixed in 4% formaldehyde for 20 min on ice and then blocked for 1h at room temperature with 5% normal goat serum in PBS containing 0.1% Tween 20. Cells were then incubated with primary anti-GM130, and secondary anti- bodies for 1 h at room temperature in blocking buffer. For analysis of ATF6α levels cells were fixed and process as described above, 48h post siRNA transfections. The cells were then stained with anti-ATF6α and anti-calreticulin antibody at the concentrations indicated in Table S2. For analysis of cleaved caspase 3 or Bax activation, 24 hrs. post siRNA transfections, cells were treated with the indicated drugs for 18-20h. Cells were then fixed and processed as previously described above. Dilutions of primary antibody were as indicated in Table S2. Coverslips were then mounted on microscope slides using Prolong-Gold antifade mounting medium with DAPI (Thermo Fisher) to stain for DNA and observed using a Zeiss confocal microscope for fluorescence detection. For quantification of ATF6α and calreticulin the MFI±S.E of 50 individual cells were measured using Zen software.
qPCR
Total RNA was extracted from cells at 48 h after siRNA transfection using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA synthesis was carried out using qScript first-strand synthesis system (Quantas). qPCR analysis was done using SYBR green (Life Technologies) in a 7500 FAST system (Applied Biosystems). The primer sequences used here are listed in Table S1 in the supplemental material. Relative mRNA levels were calculated using the 2-ΔΔCt method.
Drug Combination Studies
To evaluate whether the cytotoxic effects of dual drug combinations of 16F16 with cisplatin or irinotecan were synergistic, additive or antagonistic, drug combinations at several non-constant ratios were initially evaluated. For the median effect analysis, the compounds were combined at ratio of 50:1(cisplatin/irinotecan:16F16) in part based on CI values observed in the non-constant ratio experiments. The CI method of Chou and Talalay [18] was used to analyze the nature of the interaction between cisplatin or irinotecan and 16F16 using Compusyn software (Combosyn, Paramus, NJ). In summary, the interaction of the two drugs was quantified by determining a CI at various levels of cytotoxicity or fraction affected. CI values of less than or greater than 1 indicate synergism or antagonism, respectively, whereas a value of 1 indicates additivity. Each data point represented is the mean ± SE of at least three independent experiments, each of which was performed in triplicate. Furthermore, we evaluated the drug dose in a synergistic combination. This was designated as the dose reduction index (DRI): (DRI)1 = (Dx)1/(D)1 and (DRI)2 = (Dx)2/(D)2 where DRI >1, which showed that combinations could result in reduced drug doses compared with the doses for each drug alone. Two-way ANOVA and post hoc Tukey’s multiple comparison tests were used to determine whether the DRIs were significant.
Statistical Methods
Two clinical outcomes, progression-free survival (PFS) and overall survival, were analyzed. Time to overall survival was calculated from the date of diagnosis to the date of death (death from all causes). PFS was calculated from the date of diagnosis to the date of first documented recurrence (for recurrence-free survival). Kaplan-Meier plots were used to estimate the probabilities of overall survival and recurrence/progression-free survival for this cohort. Log-rank tests (overall and stratified) were used to compare the differences in survival or recurrence in subgroups. All P values reported in the analyses are 2-sided and values .05 or less were considered significant. The multivariate models used forward selection of factors that indicated at least a minimal univariate association (0.10). Backward selection was used to verify the variables that remained in the strongest model (assessed as accounting for the most variability). SAS version 9.4 was used for the data analysis. For our in vitro studies, data are presented as mean of 3-5 independent experiments ± standard errors of the means. All statistical analyses were performed using GraphPad Prism statistical software (GraphPad Software, San Diego, CA). The level of significance was set at P < .05. and results were compared using unpaired-t-test followed by Mann Whitney testing unless otherwise mentioned in the figure legend.
Results
Osteosarcoma Cell Lines Activate Markers of UPR
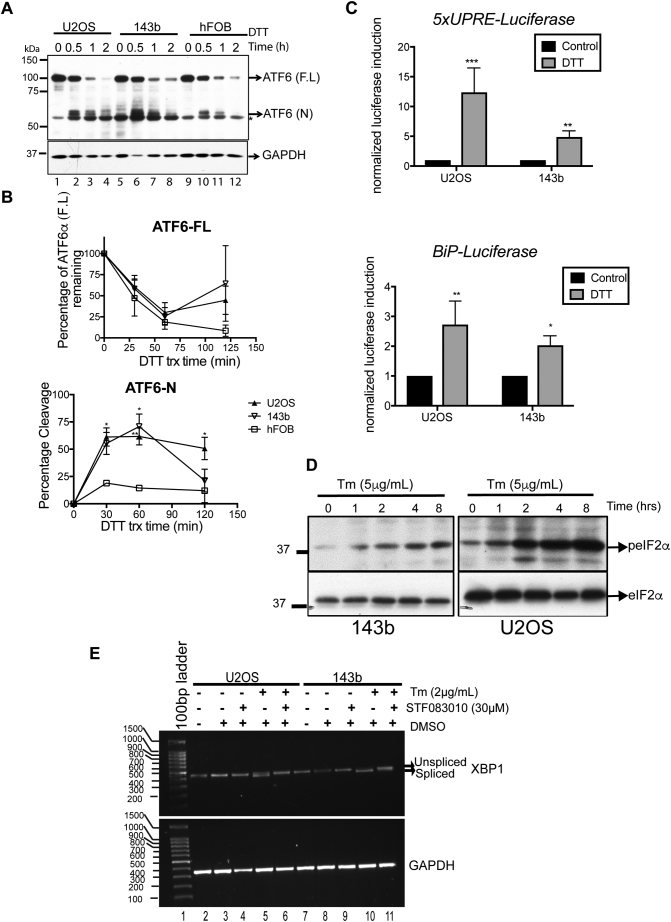
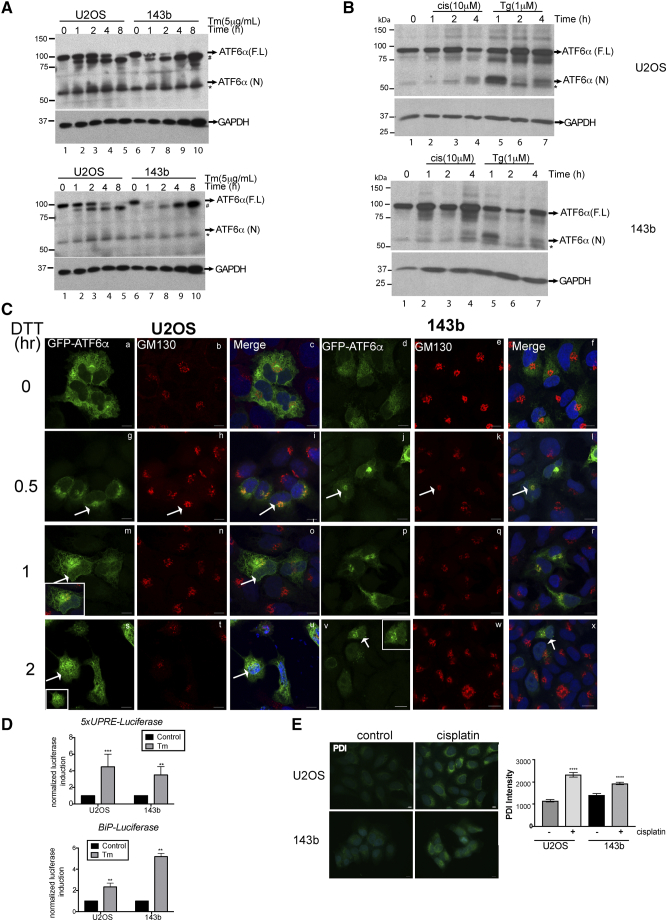
The UPR is known to promote survival against stressors such as chemotherapy in several solid tumors [19]. We therefore examined the activation of the UPR sensors ATF6α, PERK, and IRE-1α in OS cell lines following treatment with ER stress inducers DTT (a reducing agent that reduces disulfide bonds), Tm (an inhibitor of protein N-glycosylation) or Tg (an inhibitor of ER Ca2+ ATPase). To examine ATF6α activation we measured the cleavage of endogenous ATF6α, an established marker for ATF6α activation following treatment with ER stress inducers. We found that while the basal levels of full-length ATF6α were similar in both the OS cell lines and the osteoblasts, within 30 minutes of exposure to DTT, both OS cell lines showed robust increases in the cleavage of ATF6α (ATF6α(N)) when compared to the osteoblasts (Figure 1, A and B). 50-75% of ATF6α was cleaved after 1 h of treatment in the OS cells compared to osteoblasts (Figure 1A lanes 2-3,6-7 and 10-11 and 1B ). Previous studies have shown that the extent of ER stress-induced cleavage of ATF6α varied depending on inducers added, with cleavage being much more extensive in cells treated with DTT than in those treated with Tm or Tg [20], [21]. In agreement with these findings we also found that while all three ER stress inducers, DTT, Tm and Tg were able to induce cleavage of ATF6α, the extent of cleavage was more intense and rapid with DTT than Tg or Tm (Figures 1A vs S1, A and B). Next to confirm cleavage of ATF6α, we transiently expressed GFP-ATF6α fusion protein in OS cells and examined its localization following treatment with ER stress inducers. Although ATF6α is an ER localized protein, as previously reported GFP-ATF6α is expressed at much higher levels and showed a more diffused ER and golgi localization (Figure S1C, a-c), which was due to the partial degradation of GFP-ATF6α that could result in stronger GFP signal and diffuse staining [22]. However, within 30 mins of treatment with DTT, GFP-ATF6α was exported from the ER to the Golgi apparatus as the GFP fluorescence co-localized with the staining of GM130 a Golgi marker in both OS cell lines. (Figure S1C,g-l). Within 2h of treatment with DTT, we were able to detect a moderate increase in GFP signal in the nucleus as it co-localized with the nuclear stain DAPI (Figure S1C, s-x). This was more pronounced in U2OS cells than 143b cells (Figure S1C, s vs v). Tg and Tm also induced translocation of GFP-ATF6α although to a much lesser extent and with slower kinetics (data not shown).To demonstrate that cleavage leads to ATF6α activation, we monitored the activity of reporter constructs containing UPRE or ERSE [23]. Following treatment with DTT, similar to other reports [20], [21], we found an increase in transcriptional activity from both elements in OS cell lines, U2OS and 143b (Figure 1C). Treatment with Tm also resulted in an increase in transcriptional activity (Figure S1D).
Figure 1.
OS cells activate the UPR pathways (A) OS cells were treated with 1 mM DTT for the indicated times and analyzed by WB for the extent of cleavage of endogenous ATF6α. Full-length ATF6α (F.L) and cleaved nuclear fragment (N) are indicated along with a background band (*). Anti-GAPDH was used as loading control (B) Quantification of ATF6α (F.L) and ATF6α (N) over time upon treatment with the indicated ER stressor. Graph represents mean ± S.E of 6 independent experiments. Two-way ANOVA followed by Tukey’s multi-comparison tests. The level of significance was set at a P value of <.05. *P < .05, **P < .01. (C). OS cell lines were co-transfected with 5XUPRE (upper panel)- or BiP promoter-dependent luciferase reporters (lower panel) and a constitutive Renilla luciferase reporter. Forty-eight hours post transfection cells were treated with DTT (1mM) for 6 h and lysates were analyzed for luciferase expression, normalized against Renilla luciferase. Columns represent mean±S.E. of four determinations; *P < .05,**P < .01, ***P < .001. (D) OS cells were treated with Tm for the indicated times and analyzed by WB for phosph-Ser-51- eIF2α. Total eIF2α levels served as loading control.(E) RNA was extracted from OS cells following treatment with Tm in the presence or absence of STF083010. XBP1 splicing was determined by PCR, using primers that amplify both unspliced and spliced mRNA species.
Supplementary Figure 1.
(A) and (B) Effect of Tm cisplatin and Tg treatment on ATF6α cleavage. OS cells treated with Tm (A), cisplatin or Tg (B) for the indicated times were lysed and analyzed by WB for ATF6α. Full-length ATF6α (F.L) and cleaved nuclear fragment (N) are indicated along with a background band (*) # denotes an unglycosylated form of ATF6α. A, upper and lower panel represents long and short exposures of the same blot, in order to visualize the ATF6α (N). Anti-GAPDH was used as loading control. (C) OS cells were transfected with EGFP-ATF6α for 24 h and treated with 1 mM DTT for the indicated times. Cells were then immunostained with anti-GM130 (for the Golgi apparatus) and mounted with mounting media containing DAPI to stain for the nucleus. Cells were analyzed by confocal microscopy. Bars, 10 μm. The white arrows indicate the colocalization of EGFP-ATF6α with GM-130.
(D). OS cell lines were co-transfected with 5XUPRE (upper panel)- or BiP promoter-dependent luciferase reporters (lower panel) and a constitutive Renilla luciferase reporter. Twenty-four hours post transfection cells were treated with Tm (2 μg/mL) for 12 h and lysates were analyzed for luciferase expression, normalized against Renilla luciferase. Columns represent mean±S.E of four determinations; **P < .01, ***P < .001. (E) Left panel, Immunofluorescence analysis of PDI expression in U2OS and 143b cells following treatment with 10μM cisplatin and quantification of PDI fluorescence intensity as measured using Zen software (right panel). Approximately 500 cells were scored for PDI intensity, ***P < .001
We next examined if PERK and IRE-1 were also activated in OS cells. Following treatment with Tm both phosphorylation of eIF2α, a marker of PERK activation, and splicing of XBP-1, a marker of IRE-1 activation, were induced in both OS cell lines (Figure 1, D and E compare lanes 5 and 10). Together these results suggest that both U2OS and 143b cell lines robustly activate all three UPR sensors in response to ER stressors.
Down-regulation of ATF6α But Not Inhibition of PERK or IRE-1 Renders OS Cells Susceptible to Chemotherapy-Induced Apoptosis
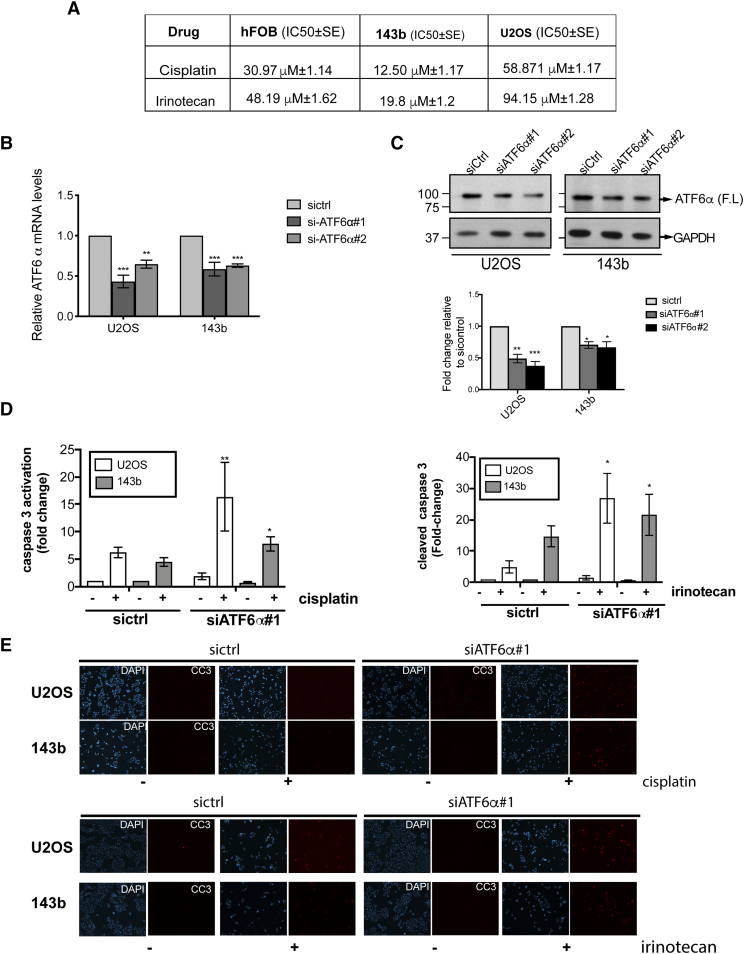
While OS is considered sensitive to triple chemotherapy with cisplatin, doxorubicin and methotrexate, resistance to this regimen is not uncommon as evidenced by variable degree of histologic necrosis noted in tumor samples after neo-adjuvant chemotherapy with about 20-30% of tumors having a poor response to upfront therapy. In addition, very few other effective chemotherapeutic agents exist at relapse. We examined if one or more of the UPR sensors could promote survival against chemotherapy. We chose cisplatin and irinotecan as our two agents as cisplatin is a known active agent and irinotecan is a known inactive agent [24] against OS and therefore were tested if we could show increased chemosensitivity following modulation of UPR sensors. Cytotoxicity assays of OS cells showed that IC50 of U2OS and 143b cells were significantly different when compared to the osteoblasts for both cisplatin and irinotecan treatment (Figures 2A and S2B). While U2OS cells had higher IC50s for both drugs than that of hFOB cells, 143b cells were more sensitive than osteoblasts to both cisplatin and irinotecan treatments (Figures 2A and S2B). The higher tolerance of osteoblasts to chemotherapy drugs compared to OS cells such as 143b is not new and has previously been demonstrated [25].
Figure 2.
Effect of ATF6α silencing on sensitivity to cisplatin treatment. (A) OS and hFOB cells were treated with increasing concentrations of cisplatin or irinotecan. Twenty-four hours post-treatment, cell viability was measured as described in methods. Data were normalized to an untreated control well and graphed, and half maximal inhibitory concentration (IC50) values were calculated from the dose-response curve as the concentration of the drug that produced a 50% decrease in the mean luminescence relative to untreated control wells. Data are presented as means ± standard errors of the means from three to 4 separate experiments. Curves were fit by non-linear regression and the significance of differences between IC50 were calculated using one-way ANOVA and post hoc Tukey’s multiple comparison tests The significance scores of all treatments versus hFOB groups are indicated; IC50 cisplatin, hFOB vs 143b, ****P < .0001, hFOB vs U2OS, ****P < .0001and IC50 irinotecan hFOB vs 143b, ****P < .0001, hFOB vs U2OS, ****P < .0001 (B) Total RNA from siControl and siATF6α expressing OS cells was extracted and expression of ATF6α levels was quantitated by qPCR, normalizing against 18s rRNA expression. Data represent mean ± SE of 3 independent experiments. **P < .01; ***P < .001. (C) Protein lysates from OS cells transfected with sicontrol and siATF6α siRNAs were analyzed by WB for the expression of ATF6α as described in methods (upper panel). GAPDH was used as a loading control and quantified using ImageJ (lower panel). Columns represent mean± SE of three independent experiments. **P < .01, *P < .05 siATF6α versus siControl (D) siControl and siATF6α expressing OS cell lines were treated with cisplatin (12.5 μM)(left panel) or irinotecan (20 μM-143b, 50 μM for U2OS) (right panel) overnight, and then fixed and stained with anti-cleaved caspase-3 antibody and analyzed for caspase-3 activation by immunofluorescence. The percent of cleaved caspase-3 positive cells was analyzed using Zen software and expressed as a fold-change after normalization to untreated siControl cells. Approximately one thousand DAPI positive cells were scored for caspase-3 activation. Columns represent mean± SE of five independent experiments performed in triplicate. **P < .01, *P < .05 siATF6α+drug versus siControl + drug. (E) Representative immunofluorescence images of cleaved caspase-3 positive cells in siControl and siATF6α expressing OS cells following treatment with cisplatin (upper panel) or irinotecan (lower panel).
Supplementary Figure 2.
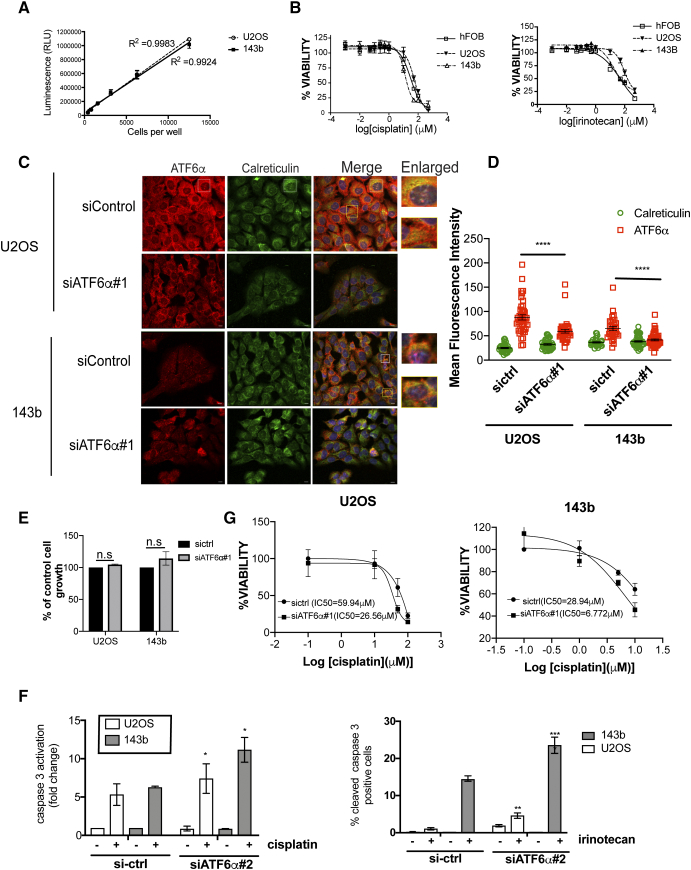
(A) Cell number correlates with luminescent output. Serial two-fold dilutions of U2OS and 143b cells were made in 96 well plates and assay were performed as described in methods. Values represent mean ± S.E of four replicates for each cell number. There is a linear relationship (r2 = 0.9924) between the luminescent signal and the number of cells from 0 to 12,500 cells/well (B) OS and hFOB cells were treated with increasing concentrations of cisplatin (upper panel) or irinotecan (lower panel). Twenty-four hours post-treatment, cell viability was measured as described in methods. Data were normalized to an untreated control well and graphed, and half maximal inhibitory concentration (IC50) values were calculated from the dose-response curve as the concentration of the drug that produced a 50% decrease in the mean luminescence relative to untreated control wells. (C). OS cell lines were transfected with a control or ATF6α siRNA as described in methods. Forty-eight hours post-transfection cells were fixed and ATF6α protein levels were measured by IF using anti-ATF6α and anti-calreticulin (for the ER) antibodies respectively, scale bar 10 μm. Confocal microscopy images of OS cells that have been stained with ATF6α (red) and the ER protein calreticulin (green) and the nucleus stained by DAPI is blue. Z-stack images were acquired to visualize the depth of the cell and confirm co-localization of ATF6α with calreticulin. (D) Scatter plot representation and statistical analysis (one-way analysis of variance [ANOVA]) of MFIs of calreticulin (green) and ATF6α (red) in more than 50 individual cells per experimental condition as shown in C. (E) Cell proliferation in OS cells expressing sictrl and siATF6α, determined by SRB assay, relative to sictrl expressing cells and its standard error. The results represent the mean and standard error of three independent experiments. (F) Effect of siATF6α#1 on the half maximal inhibitory concentration (IC50) values of OS cells following treatment with increasing concentrations of cisplatin. Data are presented as means ± standard errors of the from three separate experiments (G) Effect of ATF6αsi#2 on cisplatin or irinotecan induced apoptosis. OS cells expressing control or ATF6α#2 siRNA were treated overnight with cisplatin (left panel) irinotecan (20 μM-143b, 50 μM for U2OS) (right panel), fixed and stained for cleaved caspase-3 and analyzed by IF. Quantification represents percent of cleaved caspase-3 positive cells. Approximately one thousand DAPI positive cells were scored for caspase-3 activation. Columns represent mean ± S.E of three experiments run in triplicate; *P < .05, **P < .01, ***P < .001, siATF6α+drug versus sicontrol + drug
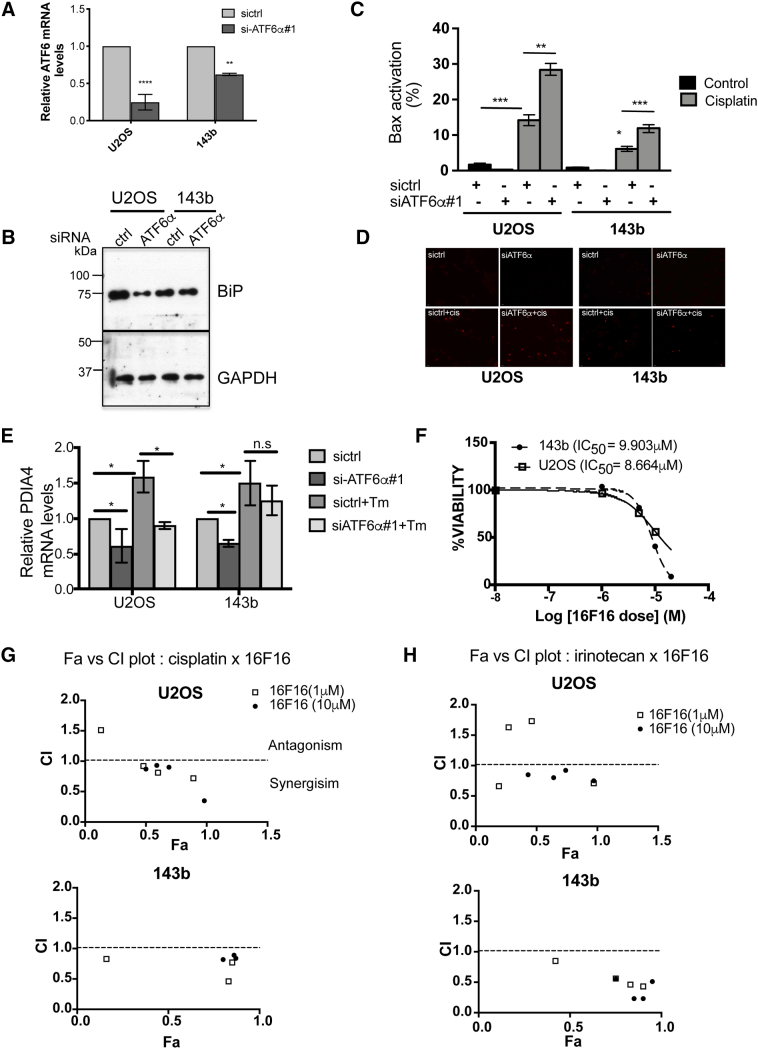
Because of the previously characterized role of ATF6α in protecting leukemia and glioblastoma cells from chemotherapy and radiation induced apoptosis [14], [15], we investigated whether or not ATF6α can similarly protect OS cells from chemotherapy induced apoptosis. We found that cisplatin treatment by itself was able to induce the cleavage of ATF6α in both OS cell lines (Figure S1B). Although the extent of cleavage was less compared to other ER stressors, it was sufficient to induce expression of PDI, a key downstream target of ATF6α activation (Figure S1E). This suggests that chemotherapeutic stress can also activate ATF6α signaling. Next, using two different siRNAs to ATF6α (siATF6α#1 and siATF6α#2) we were able to significantly down-regulate the ATF6α gene transcript and protein levels (40-50% decrease in mRNA and protein levels) (Figure 2, B and C), in both U2OS and 143b cells. Immunofluorescence analysis also showed a decrease in the intensity of ATF6α signal that co-localized with the ER marker calreticulin (Figure S2, C and D). We found that while down-regulation of endogenous ATF6α did not affect basal survival or proliferation of the OS cells in vitro, (Figure 2D and S2E) both siRNAs resulted in a significant increase in the sensitivity of U2OS and 143b cells to cisplatin induced cell death (Figures 2, D and E and S2F) as measured by an increase in caspase 3 activation using immunofluorescence as described in methods. We found that cisplatin induced a 4-6-fold increase in caspase 3 activation in OS cells expressing control siRNA. This activation was further enhanced by 2-3-fold following ATF6α down-regulation. Similarly, irinotecan induced caspase 3 activation was also significantly increased following ATF6α down-regulation by both siRNAs (Figures 2D, right panel and S2F right panel). Furthermore, ATF6α down-regulation also resulted in a 2-4-fold decrease in the IC50 for cisplatin in both OS cell lines (Figure S2G). Due to the low transfection efficiency of osteoblasts (<2%) we were unable to assess the role of ATF6α knockdown on sensitivity to cisplatin or irinotecan in these cells. The above results suggest that activation of ATF6α following chemotherapeutic stress could protect OS cells from therapy induced apoptosis.
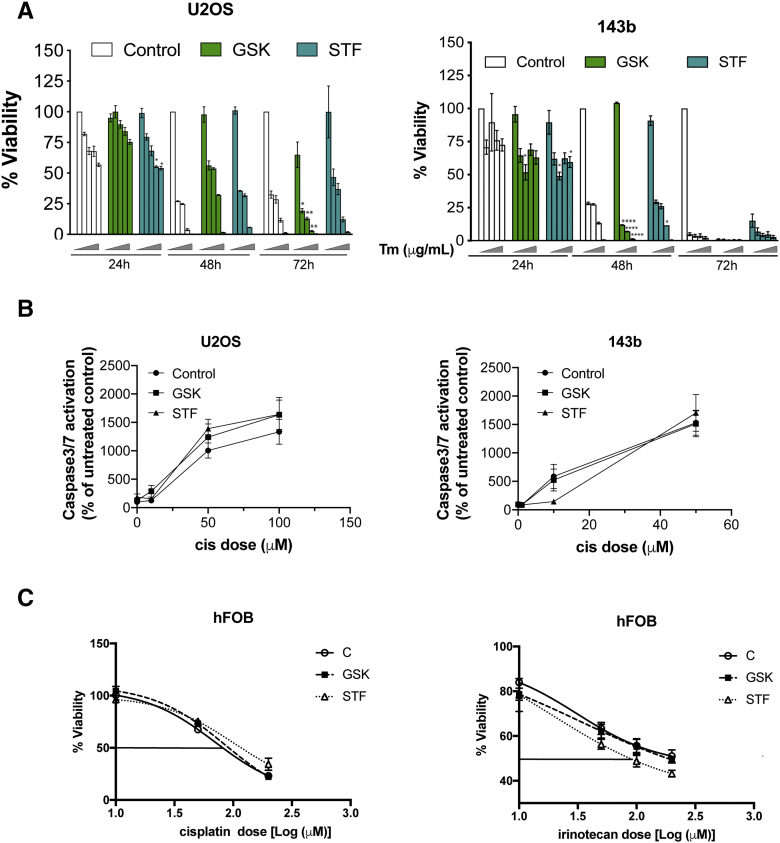
Next we examined whether PERK or IRE-1α signaling, which are well-known inducers of chemoresistance and metastasis in several cancers [26], [27], [28] also have pro-survival functions in OS cells. Utilizing the compounds GSK2606414 (GSK) and STF083010 (STF) that have been shown to effectively inhibit PERK and IRE-1 signaling respectively [27], [28], we were able to significantly reduce both basal and Tm induced GADD153 mRNA levels as well as XBP-1 splicing (Figures 3A and 1E). Both inhibitors were effective in sensitizing OS cells to ER stress mediated apoptosis (Figure S3A). Furthermore, treatment with increasing concentrations of these inhibitors did not affect OS cell viability (Figure 3B). However, co-treatment with these inhibitors did not enhance sensitivity either to cisplatin or irinotecan induced cell death or enhance caspase3/7 activation (Figure 3C, S3B and Tables S3 and S4). We also analyzed the effect of PERK and IRE1α inhibition on the ability of hFOB cells to survive cisplatin or irinotecan treatment. While PERK inhibition did not affect hFOB sensitivity to cisplatin or irinotecan, IRE-1 inhibition did affect sensitivity of hFOB cells to irinotecan induced cell death (SFigure 3C and Tables S3 and S4). These findings suggest that while IRE-1 signaling may affect tolerance of hFOB cells to chemotherapy drugs, unlike ATF6α neither PERK nor IRE-1 signaling is essential for survival against chemotherapy in OS cells.
Figure 3.
Effects of PERK and IRE1α inhibition in OS cells on cisplatin treatment. (A) Total RNA extracted from OS cells following treatment with Tm (2 μg/mL) for 12 h in the presence or absence of PERK or IRE1α inhibitors GSK2606414 (GSK) and STF083010 (STF) were quantitated for GADD153 mRNA levels by qPCR, normalizing against 18S rRNA expression. Data represent mean ± S.E of four independent experiments. *P < 0/05, **P < .01, ***P < .001. (B) Cytotoxicity assay of OS cells following treatment with increasing concentrations of GSK or STF for 24 h. Data represent mean ± SE of 3 independent experiments. (C) Cytotoxicity assay of OS cells following treatment with increasing concentrations of cisplatin in the presence or absence of GSK (100 nM) or STF (30 μM) for 24 h and cell viability was measured as described in methods. Data represent mean ±SE of three independent experiments.
Supplementary Figure 3.
(A) Inhibition of PERK and IRE1α signaling enhance sensitivity to ER stress induced cell death. U2OS and 143b cells were treated with increasing concentration of Tm (0,1,2,5 and 10μM) for the indicated times and percent cell viability was measured using Cell-Titer Glo as described in methods. (B) Measurement of cisplatin mediated caspase 3/7 activation using Caspase-Glo 3/7 assay system (Promega Inc) in OS cells following inhibition of PERK and IRE1α signaling. (C) hFOB cells were treated with or without cisplatin or irinotecan and GSK or STF. Twenty-four hours post-treatment, cell viability was measured as described in methods. In all experiments data were normalized to an untreated control cells and graphed. Data represents mean ± S.E of three experiments.
ATF6α Targets BiP, PDI and ERO1β are Required for Enhanced Survival Against Chemotherapy
The mechanisms by which ATF6α may be linked to anti-apoptotic signaling are largely mediated by its regulation of BiP/GRP78, which is well-known to suppress pro-apoptotic signals and caspase-7 activity [29] and potentiate survival signaling through Akt [30].
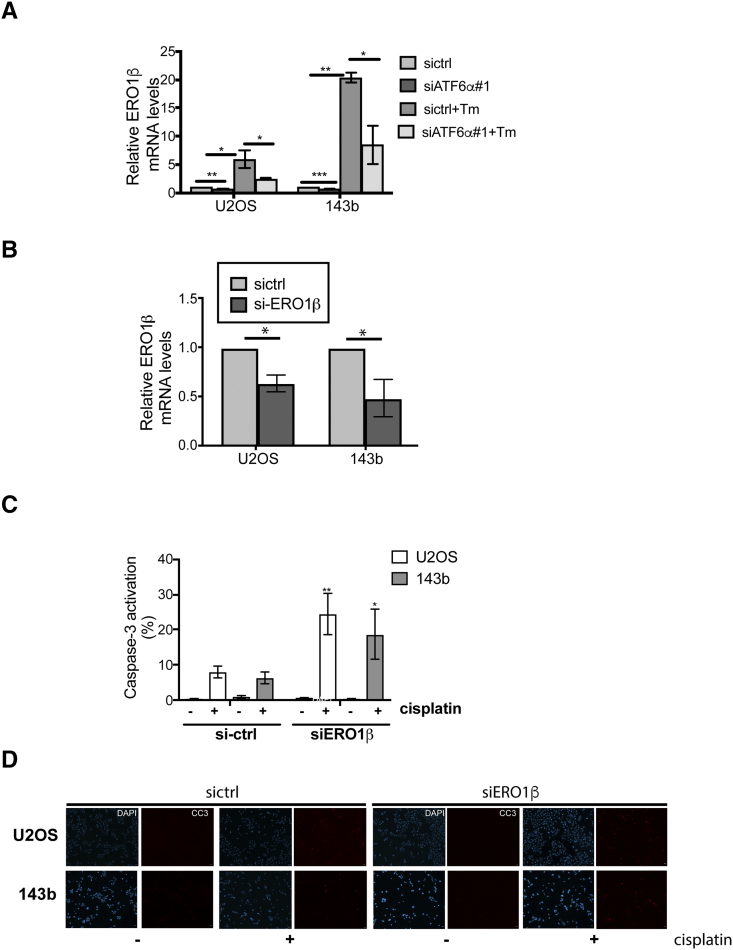
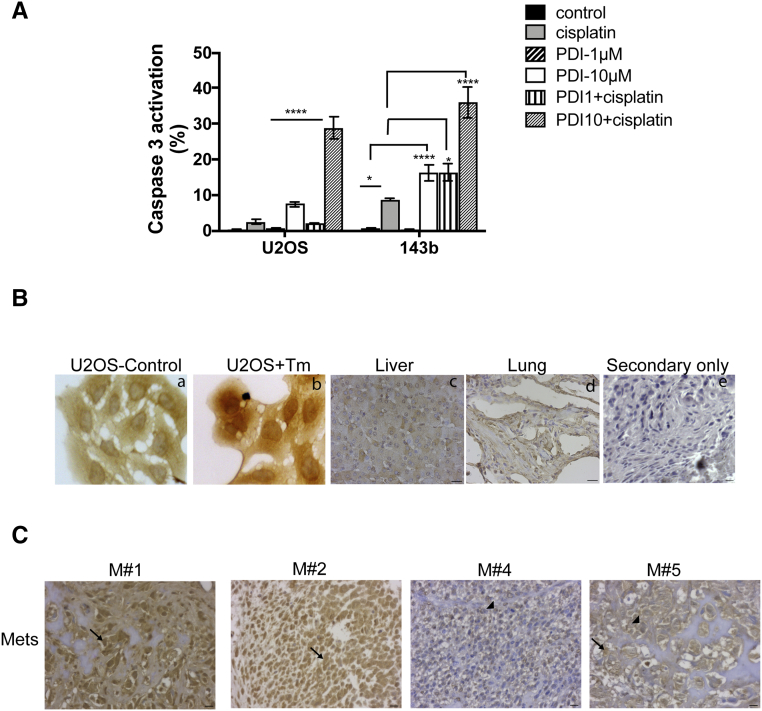
We have previously demonstrated that chemoresistance in dormant head and neck tumor cells was in part mediated by BiP-dependent inhibition of Bax activation [17]. As down-regulation of ATF6α resulted in decreased BiP expression (Figure 4, A and B), we questioned whether this would also lead to an increase in Bax activation. Bax activation was measured using an anti-human Bax 6A7 antibody, which binds to an epitope in the NH2-terminal region (amino acids 13-19) that is exposed only upon activation. As Bax is activated prior to caspase-3 activation, acute treatment with cisplatin (50μM for 8h) resulted in a significant increase in Bax activation in OS cell lines. We found that down-regulation of ATF6α further increased Bax activation in both OS cell lines by 2-fold. (Figure 4, C and D). Like BiP, other ATF6α target genes are also cytoprotective, functioning to increase the folding capacity of the ER to homeostasis [23]. qPCR analysis of some of these targets showed that both basal and Tm induced mRNA levels of genes involved in protein folding such as PDI (PDIA4) and ERO1β (Figures 4E and 5A), as well as genes involved in protein degradation such as DERL3 and HERPUD1 (data not shown), were significantly down-regulated following ATF6α knockdown. Since members of the PDI family, PDIA5, PDIA4 and PDIA6 have been shown to regulate chemosensitivity [31], we tested the inhibitory activity of 16F16, a PDI inhibitor and the cisplatin/16F16 combination on OS cell proliferation, using Cell Titer Glo assay. As shown in Figure 4F,16F16 alone inhibited the growth of U2OS, and 143b cells with IC50s of 8.66 and 9.90μM respectively. To examine whether inhibition of PDI synergizes with cisplatin or irinotecan, OS cells were co-treated for 24h with cisplatin and 16F16 at 6 different combination ratios of cisplatin:16F16 (1:1 to 500:1) ratios and the fraction affected (FA) and combination index (CI) values were calculated using Compusyn and the Chou and Talalay method [18] as described in methods. For most combinations tested synergistic interaction was noted at all effect levels (CI range 0.3 to 0.9) except in U2OS cells where some degree of antagonism was seen with 1μM 16F16 (Figure 4G and Table 1). In agreement with these findings, while the addition of both 1μM and 10μM doses of 16F16 lead to a 2 and 4-fold increase in cisplatin induced apoptosis (caspase 3 activation) in 143b cells only a combined treatment of cisplatin with 10μM 16F16 lead to a 12-fold increase in apoptosis in U2OS cells (Figure S4A). A similar combinatorial treatment with irinotecan and 16F16 also displayed synergism with combination with 16F16 at 10μM showing moderate to strong synergy in U2OS and 143b cells respectively (Figure 4G and Table 1). These results suggest that combinational use of inhibitors of PDI with chemotherapy drugs may have synergistic effect against OS. Because our aim was to achieve maximal effect of the drugs tested on OS cells, combinations that resulted in Fa <0.5 was therefore considered irrelevant. Based on these, treatment of both OS cell lines with a constant ratio combination of cisplatin or irinotecan:16F16 at 50:1 showed a significant favorable reduction in dose for both cisplatin (U2OS, ~1.1-fold; 143b, ~1.6-fold) and irinotecan (U2OS, ~1.9-fold; 143b, ~5.6-fold) for all data points that yielded a Fa >0.5 (Table 2). These data suggest that inhibition of PDIs can significantly potentiate the cytotoxic effects of both cisplatin and irinotecan on OS cells.
Figure 4.
Functional role of BiP and PDI on OS cell survival against chemotherapy. (A) qPCR analysis of ATF6α mRNA levels in OS cells and (B) corresponding decrease in BiP protein levels as measured by WB (lower panel). GAPDH served as loading control. (C) siRNA transfected OS cells were treated with or without cisplatin (50 μM) for 8 h, were fixed and stained with α-Bax 6A7 antibody and analyzed for Bax activation by immunofluorescence. Percent Bax positive cells was then quantitated. Column represents mean ± S.E of three independent experiments performed in triplicate (D) Representative immunofluorescence images of Bax activation in sictrl and siATF6α expressing cells with and without cisplatin treatment., (E) Total RNAs extracted from siRNA expressing OS cells with and without Tm treatment were quantitated for expression levels of PDIA4 mRNA levels by qPCR analysis after normalizing to 18s rRNA. (F) OS cells were treated with increasing concentrations of 16F16 for 24 h. Cell viability was measured as described in methods and IC50 was calculated as described in Figure 2. (G-H) Combination index (CI) vs fraction affected (Fa) plots obtained from the median-effect analysis program (Combosyn,). (G) cisplatin (H) irinotecan; Symbols represents actual combination data points. CI <1, =1 and >1 indicates synergism, additive effect and antagonism, respectively. *P < .05, **P < .01, ***P < .001.
Figure 5.
Functional role of ERO1β on OS cell survival against chemotherapy.(A) and (B) Total RNAs extracted from siRNA expressing OS cells with and without Tm treatment were quantitated for expression levels of ERO1β mRNA levels by qPCR analysis after normalizing to 18s rRNA Bar mean ± SE of 3 independent experiments. (C) Quantitation of caspase 3 activation in OS cells following ERO1β down-regulation in the presence or absence of cisplatin. Graph represents mean ± SE of 3 independent experiments and compared to their respective si-ctrl cisplatin treated samples for statistical analysis *P < .05, **P < .01. (D) Representative immunofluorescence images of cleaved caspase-3 positive cells in siControl and siERO1β expressing OS cells following treatment with cisplatin.
Table 1.
Combination Screening for Drug Synergy in Osteosarcoma
| U2OS |
143b |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fa (%) Cisplatin or Irinotecan Alone | 16F16 |
Fa (%) Cisplatin or Irinotecan Alone | 16F16 |
||||||||
| 1 μM |
10 μM |
1 μM |
10 μM |
||||||||
| Fa (%) | CI | Fa (%) | CI | Fa (%) | CI | Fa (%) | CI | ||||
| Cisplatin (μM) | 10 | 9.6± 10.2 | 6.9±1.6 | 1.51 | 50.1±18.8 | 0.87 | 14.2±8.1 | 15.8±7.2 | 0.83 | 79.8±4.0 | 0.82 |
| 50 | 47.2±6.7 | 47.9±11.2 | 0.92 | 59.2±13.3 | 0.93 | 79.1±6.7 | 83.2±8.3 | 0.46 | 86.0±9.3 | 0.89 | |
| 100 | 57.1±8.5 | 59.6±9.7 | 0.81 | 68.6±7.6 | 0.90 | 79.7±6.9 | 85.4±6.8 | 0.77 | 87.310.2 | 0.84 | |
| 500 | 89.3±13.6 | 89.5±14.2 | 0.72 | 98.1±1.1 | 0.35 | ||||||
| Irinotecan (μM) | 10 | 9.2±3.1 | 18.6±5.8 | 0.66 | 42.5±5.1 | 0.85 | 33.45±13.4 | 51.3±14.7 | 0.85 | 75.2±15.1 | 0.55 |
| 50 | 30.3±1.6 | 26.9±7.4 | 1.63 | 63.7±4.2 | 0.80 | 63.7±21.0 | 74.6±29.6 | 0.56 | 85.3±16.3 | 0.23 | |
| 100 | 48.8±8.2 | 45.9±5.1 | 1.73 | 74.3±4.0 | 0.92 | 72.275±11.5 | 82.6±16.5 | 0.46 | 89.7±10.9 | 0.23 | |
| 500 | 97.2±0.3 | 97.2±0.6 | 0.71 | 97.1±0.4 | 0.75 | 87.45±0.4 | 90.4±5.0 | 0.43 | 94.7±2.9 | 0.51 | |
Table with select Fa values and CI of different combinations cisplatin or irinotecan with 16F16. synergy calculations were done using the Chou and Talalay combination index (CI), based on the median-effect and mass-action principles.
Supplementary Figure 4.
(A) OS cells were treated with or without cisplatin and 16F16 for 20 h and the percent of cells expressing activated caspase-3 was quantitated by IF using α-cleaved caspase 3 antibody. Bar represents mean  ± SE of three independent experiments. Two-way Anova using Tukey’s multiple comparison test ATF6α staining by immunohistochemistry-IHC. (B) Representative images of U2OS cells, treated with or without ER stress inducer Tm, normal liver and lung tissue stained with anti-ATF6a antibody (a-d) or secondary alone (e). (C) Representative images of OS metastasis (M) from patients with high ATF6α (N) (mets from patients #s 1and 2), vs. low ATF6α(N) (mets from patients # 4 ad 5) expression. Scale Bar = 10µm.
Table 2.
Combination Index and Dose Reduction Value for Cisplatin and Irinotecan at Constant Ratio of 50:1 with 16F16
| % Inhibition | U2OS |
143b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CI | Conc (μM) |
Dose Reduction | P Value | CI | Conc (μM) |
Dose Reduction | P-Value | ||||
| Alone | Combined | Alone | Combined | ||||||||
| cisplatin (uM) | 50 | 1.01 | 46.82 | 44.91 | 0.99 | 0.71 | 14.25 | 10.78 | 1.33 | n.s | |
| 75 | 0.97 | 87.18 | 81.37 | 1.10 | 0.66 | 42.20 | 27.97 | 1.51 | n.s | ||
| 90 | 0.94 | 162.35 | 147.45 | 1.12 | .0053 | 0.58 | 124.96 | 72.63 | 1.72 | <.0001 | |
| 95 | 0.92 | 247.81 | 220.92 | 1.13 | <.0001 | 0.53 | 261.46 | 139.05 | 1.88 | <.0001 | |
| irinotecan (uM) | 50 | 0.89 | 81.33 | 70.92 | 1.15 | 0.87 | 14.86 | 12.88 | 1.16 | ||
| 75 | 0.68 | 216.77 | 147.36 | 1.48 | 0.40 | 80.68 | 32.30 | 2.50 | .002 | ||
| 90 | 0.53 | 577.73 | 306.79 | 1.91 | 0.18 | 437.87 | 80.98 | 5.41 | <.0001 | ||
| 95 | 0.45 | 1125.32 | 505.69 | 2.28 | .0051 | 0.11 | 1383.50 | 151.32 | 9.15 | <.0001 | |
U2OS and 143b cells were treated with cisplatin or 16F16, alone or in combination, for 24h. CI, combination index. A CI of >1, 1, and <1 indicates antagonism, additive effect, and synergism, respectively. Dose reduction (fold) = the IC50 value of an inhibitor tested alone/the IC50 value of the same inhibitor tested in combination with another inhibitor.
We next tested whether ERO1β, a thiol oxidase that is involved in the re-oxidation of PDIs, also had a role in enhancing tumor cell survival against chemotherapy. We found that siRNA to ERO1β resulted in a significant decrease in ERO1β mRNA levels (Figure 5B) and led to a 3-6-fold increase in sensitivity to cisplatin induced caspase-3 activation in 143b and U2OS cells when compared to siRNA control cells (Figure 5, C and D). Taken together, our in vitro data support that in addition to ATF6α, inhibition of its downstream targets independently, also served as critical mediators of survival against chemotherapy in OS cells.
High nuclear ATF6α levels serve as an independent prognosticator in OS patients
To determine if there is a correlation between ATF6α signaling and response to therapy and/or outcome, we performed retrospective analysis of banked OS patient samples for the expression of ATF6α. Forty patients with OS diagnosis were eligible for this study. The median age of the patients was 13 years (range: 3-18 years). Of the 40 patients, 21 were male and 19 were female. The treatment was standardized across all patients consisting of cisplatin and doxorubicin with high dose methotrexate. All other patient characteristics such as, site of the primary tumor, presence of metastases at diagnosis, relapse, degree of histological necrosis, overall and progression-free survival outcome are tabulated in Table 3.
Table 3.
Association Between ATF6α and Characteristics of Osteosarcoma Patients (n = 40)
| Tumor Site | Overall n = 40 |
Living n = 27 |
Died n = 13 |
P Value for Survival Differences† |
|---|---|---|---|---|
| Femur | 21 | 15 | 6 | n.s. |
| Humerus | 7 | 5 | 2 | |
| Tibia | 6 | 5 | 1 | |
| Other limb | 3 | 1 | 2 | |
| Axial | 3 | 1 | 2 | |
| Overall Metastases | 19 | 7 | 12 | .0001 |
| Metastases @ diagnoses | 10 | 4 | 6 | .04 |
| % necrosis⁎ (Mean±S.D.) |
67.8+ 37.7 | 76.7 + 31.0 | 43.7 + 45.0 | .02 |
| Overall survival (months, Median ±S.D.) | 52.7 + 28.4 | 62.8 + 28.1 | 31.8 + 14.6 | <.0001 |
| Time to relapse (months) | 49.6 + 27.5 | 58.6 + 27.9 | 30.8 + 14.7 | .0002 |
| ATF6α nuclear | ||||
| Low | 30 | 24 | 6 | .003 |
| High | 10 | 3 | 7 |
Wilcoxon rank sum test.
n = 37, alive = 27, died = 10.
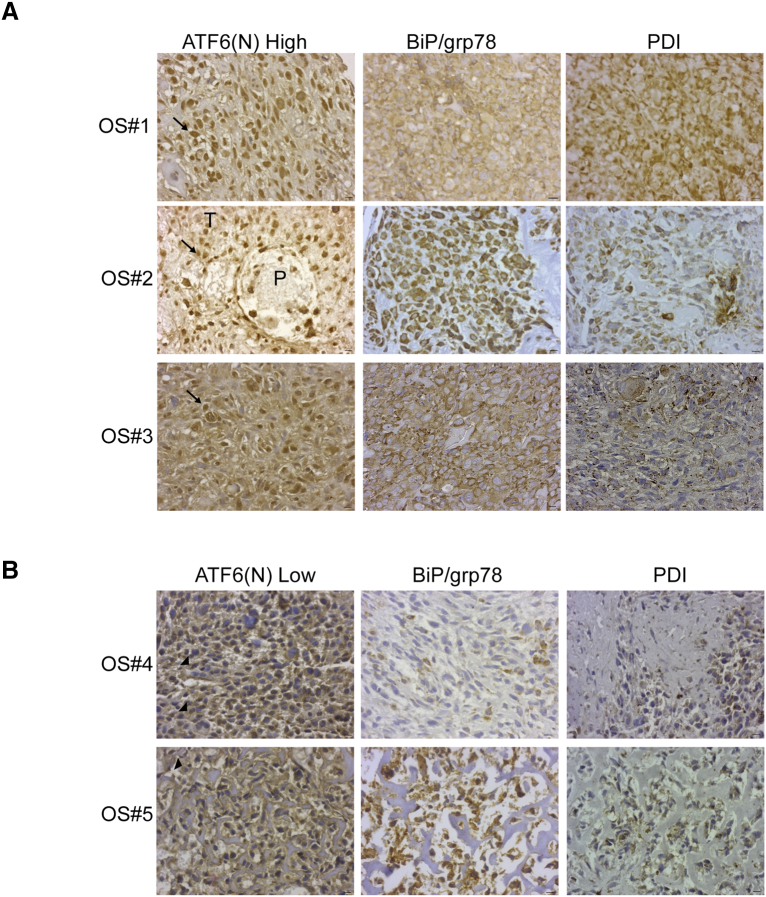
We measured the levels of ATF6α in the nucleus, which represents the cleaved and transcriptionally active form of ATF6α. The antibody used recognizes both full-length as well as the cleaved active forms of the protein (ATF6α(N) (Figure S4B). The specificity of the antibody against ATF6α was confirmed using IF (Figure 2D) and immunohistochemical staining of both OS cells treated with ER stress inducer Tm and normal tissues from age-matched patients (Figure S4B). The staining of the full-length uncleaved form of ATF6α was in the peri-nuclear/cytoplasmic region (Figure S4B). However, the staining of the cleaved transcriptionally active form of ATF6α, ATF6α(N), was predominantly nuclear. Among the 40 patients, 10 (25%) were ATF6α(N) high expressers (Figure 6A) and 30 (75%) were ATF6α(N) low expressers (Figure 6B). The ATF6α(N) expression also correlated with BiP and PDI expression levels (Figure 6, A and B), as ATF6α(N) high tumors were associated with high BiP and PDI expression when compared to ATF6α(N) low tumors. We also examined if there was any association between ATF6α(N) levels in the primary tumor and the presence of metastasis at diagnosis and overall metastases. We found a significant association between ATF6α(N) levels and the presence of metastasis at diagnosis with 5 of 10 (50%) of patients with ATF6α(N) high tumors having metastasis at diagnosis as compared to only 5 of 30 (16%) of the ATF6α(N) low tumors (P = .04). Furthermore, an additional 4 of 10 of patients who had high ATF6α (N) levels in their primary tumor at the time of diagnosis developed metastases later on during the course of the disease. Overall, 9 of 10 (90%) of patients in the ATF6α(N) high group had metastasis as compared to 10 of 30 (33%) in the ATF6α(N) low group (P = .002). These findings suggest that subgroups of patients who present with ATF6α(N) high tumors at diagnosis may be associated with high risk of developing metastatic disease overall. Analysis of ATF6α(N) levels in matched metastatic and primary lesions from the same patient showed that a large proportion of the ATF6α(N) high patients were also ATF6α(N) high in their metastasis (Figure S4C). These findings suggest that patients with ATF6α(N) high primary tumors had a higher likelihood of developing metastasis that were also ATF6α(N) high.
Figure 6.
Immunohistochemical analysis of expression of human ATF6α, BiP and PDI in OS primary tumors. (A) Representative images of diagnostic OS tumors from three patients showing high ATF6α (N) (arrow), BiP and PDI expression in the tumor cells. (B) Representative images of OS tumors with low ATF6α (N), BiP and PDI expression. Note the perinuclear staining of ATF6α indicating endoplasmic reticulum localization in these OS tumor cells (arrowheads). P, parenchyma; T, tumor. Scale = 10 μm.
Since our in vitro studies revealed that ATF6α activation promoted survival against chemotherapy, we tested whether there was an association between ATF6α(N) levels in the primary tumor and the response to chemotherapy. As the chemotherapy treatment protocol given to these patients before surgery was uniform, chemotherapy resistance was defined based on the current standard definitions of poor (<90% histologic necrosis) versus good (>90% histologic necrosis) response as seen in the primary tumor upon definitive resection after 2 cycles of chemotherapy. Using these criteria our preliminary analysis showed that patients with ATF6α(N) high tumors had a trend towards poorer histologic response when compared to the patients with ATF6α(N) low tumors (58.7 + 44.1% vs 70.7 + 35.8, respectively), however this trend did not reach statistical significance (P = .41) (Table 4).
Table 4.
Summary of Osteosarcoma Patient Characteristics by High or Low Levels of ATF6α (n = 40)
| ATF6α(N) Low n = 30 |
ATF6α(N) High n = 10 |
P value for Survival Differences† | |
|---|---|---|---|
| Proximal | 28 | 9 | n.s. |
| Axial | 2 | 1 | |
| Overall metastases | 10 | 9 | 0.002 |
| Metastases @ diagnoses | 5 | 5 | 0.04 |
| % necrosis⁎ (Mean±S.D.) |
70.7 + 35.8 | 58.7 + 44.1 | 0.41 |
| Survival months (Median±S.D.) |
58.0 + 28.9 | 36.8 + 20.7 | 0.04 |
| Months to relapse (Median+S.D.) |
55.8 + 28.0 | 30.8 + 15.5 | 0.01 |
Wilcoxon rank sum test
n = 37, ATF6α Low = 28, ATF6α High = 9.
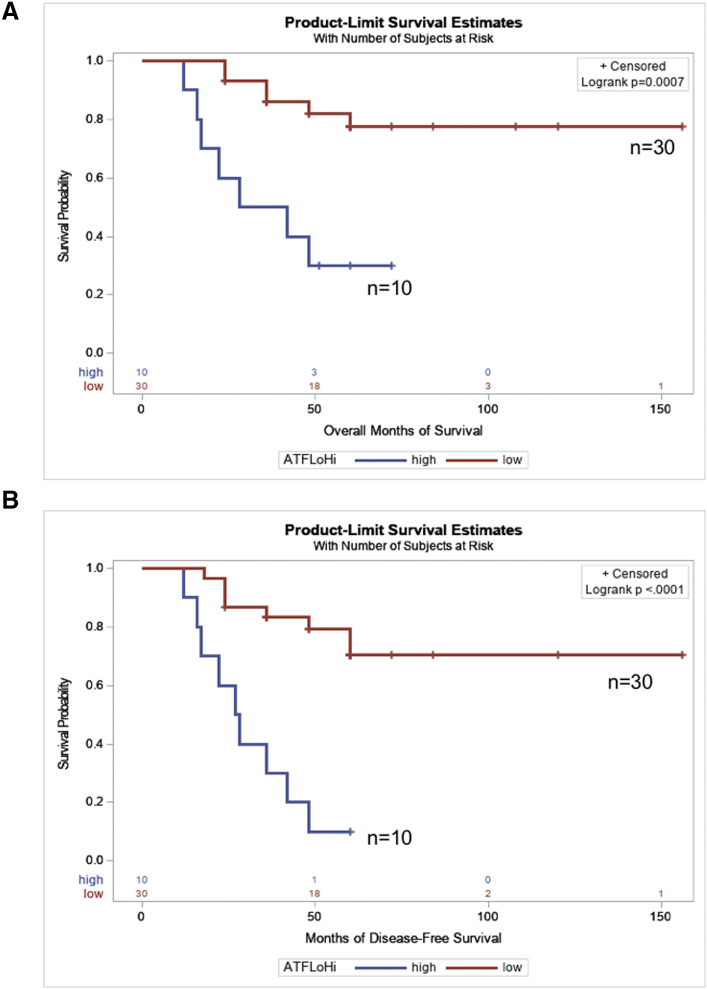
Despite the small number of patients with ATF6α(N) high tumors, 70% (7/10) of these patients died from the disease compared to 20% (6/30) of patients whose tumors had low ATF6α(N) levels (median overall survival = 36.8 months vs >58.0 ± 28.9 months, P = .04). Moreover, patients with ATF6α(N) high tumors also had a significantly shorter time to relapse as compared to patients with ATF6α(N) low tumors (30.8 ± 15.5 vs. 55.8± 28.0, P = .01, Table 3). Analyses of overall (Table 5) and PFS (Table 6) in patients who had ATF6α(N) high vs. low tumors, showed that patients who had ATF6α(N) high tumors had significantly worse overall survival (univariate-HR, 5.446; 95% CI,1.814- 16.344; P = .003) as well as PFS (univariate-HR, 6.69; 95% CI, 2.502- 17.888; P = .0002) than those with ATF6α(N) low tumors (Figure 7, A and B). In a multivariate analysis the magnitude of the effect of ATF6α(N) high tumors on overall survival (HR, 5.105; 95% CI, 1.151- 22.648; P = .0001) and PFS (HR, 5.76; 95% CI,1.654-20.111; P < .0001) was more significant even after adjusting for the presence of metastasis at the time of diagnosis and histologic necrosis. These results suggest that ATF6α levels could be an independent prognostic indicator for patients with OS.
Table 5.
Association between ATF6α (N) levels and models of overall survival among forty osteosarcoma patients
| Characteristics | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| ATF6α(N) Low (0) or High (1) | 5.446 (1.814, 16.344) | .003 | 4.045 (1.248, 13.109) | |
| Metastasis at diagnosis | 4.207 (1.395, 12.689) | .011 | 2.670 (0.821, 8.689) | .004 |
| ATF6α(N) Low (0) or High (1) | 8.608 (2.248, 32.965) | |||
| Percentage of necrosis* | 0.982 (0.967, 0.997) | .017 | 0.978 (0.962, 0.994) | .0005 |
| ATF6α(N) Low (0) or High (1) | 5.105 (1.151, 22.648) | |||
| Metastasis at diagnosis | 7.196 (1.247, 41.506) | |||
| Percentage of necrosis⁎ | 0.969 (0.948, 0.990) | .0001 | ||
Including ATF6α in the analysis leads to a 4.5- fold improvement in the model of overall survival of osteosarcoma.
results and models that include the percentage of necrosis are based on the data of n = 37.
Table 6.
Association Between ATF6α (N) Levels and Models of Progression-Free Survival Among Osteosarcoma Patients (n = 40)
| Characteristics | Univariate Analysis |
Multivariate Analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| ATF6α Low (0) or High (1) | 6.690 (2.502, 17.888) | .0002 | 4.912 (1.735, 13.904) | |
| Metastasis at diagnosis | 5.029 (1.910, 13.239) | .001 | 3.283 (1.180, 9.138) | .0001 |
| ATF6α Low (0) or High (1) | 9.182 (2.926, 28.814) | |||
| Percentage of necrosis⁎ | 0.988 (0.975, 1.001) | .071 | 0.985 (0.972, 0.999) | .0002 |
| ATF6α Low (0) or High (1) | 5.767 (1.654, 20.111) | |||
| Metastasis at diagnosis | 5.316 (1.484, 19.044) | |||
| Percentage of necrosis⁎ | 0.980 (0.943, 0.996) | <.0001 | ||
Including ATF6α in the analysis leads to a 7.6-fold improvement in the model of PFS.
Results and models that include the percentage of necrosis are based on the data of n = 37.
Figure 7.
Probability of overall and progression free-survival according to ATF6α(N) expression in patients with OS. A, Overall Survival. B, Progression-free survival.
Discussion
The present study demonstrates that activation of UPR transcription factor ATF6α in OS cells protects them from chemotherapy induced apoptosis via BiP, PDI and ERO1β dependent pro-survival mechanisms. Furthermore, we show that ATF6α activation is an indicator of poor prognosis in osteosarcoma irrespective of the metastatic status or the histological response to treatment, two well characterized indicators of OS prognosis.
While the role of UPR transducers in chemoresistance and tumor progression have been extensively studied [19] there is limited evidence for the role of ATF6α in promoting survival and chemoresistance of tumor cells. Here we demonstrate that all three arms of the UPR, PERK, IRE-1 and ATF6α are activated in OS cells upon exposure to inducers of ER stress. In agreement with these findings preliminary in silico analysis of gene expression datasets from patients with primary OS indicated that the expression of UPR genes such as those examined in our manuscript, ATF6α, XBP, PDI and ERO1β were induce 1.5-6-fold. Furthermore, analysis of pathway enrichment of these datasets using Enrichr, indicated that Protein processing in ER pathway was among the top five pathways enriched in tumors from osteosarcoma patients (P = 2×10-5), which is a strong indicator of UPR activation. Furthermore, vast majority of the genes enriched in the ER protein folding machinery pathway (such as PDIA4, PDIA6, ERO1β, grp78, DNAJC1 etc., which were induced 4-10-fold) were also key players of the UPR (unpublished observations).
ATF6α was robustly activated in both the OS cell lines tested when compared to human osteoblasts. While the levels of full-length ATF6α were similar between the normal osteoblasts and the OS cell lines, the extent of cleavage of ATF6α was more robust in the OS cells compared to the osteoblasts. These data suggest that the increased cleavage of ATF6α could be due to increased ER➔ Golgi trafficking of ATF6α. Our preliminary bioinformatics analysis of gene expression profiles from osteosarcoma patients compared to normal tissue shows an enrichment in expression of genes involved in ER➔ Golgi protein trafficking (unpublished observations). Our findings here suggest that increased trafficking of ATF6α could account for the increased ATF6α activation in OS cells.
Although the ATF6α pathway functionally overlaps to varying degrees with both the PERK and IRE-1 pathways [32], animals that lack one or both copies of ATF6α fail to survive persistent stress insults to the ER even in the presence of functional IRE-1α and PERK pathways [23]. This suggests that ATF6α could play a critical role in the chronic adaptive response, by regulating cell survival and could be a beneficial response for tumor cells when facing cytotoxic stress induced by strong chemotherapeutic drugs. In agreement with this, we found that while OS cells have an active and intact UPR, down-regulation of ATF6α was sufficient to enhance sensitivity to chemotherapy mediated cell death by activating pro-apoptotic mechanisms. While PERK and IRE-1 are also active in OS cells, inhibition of these pathways did not enhance chemosensitivity to cisplatin or irinotecan. We find that similar to BiP knockdown ATF6α knockdown also led to increased Bax activation. While this could be due to ATF6α regulation of BiP, whether ATF6α can also regulate Bax directly needs to be elucidated. Studies in Arabidopsis, have shown that the Arabidopsis homolog of Bax inhibitor -1 (AtBI1) contains ER stress response elements (ERSEs) in its promoter region that are conserved in other ATF6α regulated targets such as BiP, PDI, calnexin etc. [33]. Whether such ERSEs are present in the human homolog is unknown, but the possibility that BI-1 could be a direct transcriptional target of ATF6α is interesting and warrants further investigation.
We further elaborated on the pro-survival function of ATF6α by examining the role of another key transcriptional targets PDI and ERO1β in this process. ERO1s reoxidize PDIs during protein folding. The increased sensitivity of OS cells to cisplatin following PDI inhibition, as well as ERO1β knockdown, suggest that these enzymes could also have pro-survival functions in OS. Since PDIA5 catalyzed disulfide bond formation of ATF6α was shown to regulate ATF6α trafficking and activation [14], the enhanced sensitivity of OS cells to chemotherapy following PDI inhibition could be in part due to its regulation of ATF6α and therefore needs to be further examined. But whether these enzymes can have anti-tumor effects independent of ATF6α is not known.
Our findings here suggest that PDI inhibitors could be a potential drug candidate that can be used in combination with chemotherapy drugs to potentiate cytotoxic killing of OS cells. Since PDI physiology where cysteine oxidation, reduction, and isomerization of disulfide bonds is accomplished in short sequence and is influenced by additional enzymes, like ERO1-α and β that restore oxidative potential of PDI, the extent to which these drugs can impair PDI activity or how profoundly restoration of enzymatic activity by ERO1 is affected in vivo cannot be elucidated as to date there are no reliable method for measuring PDI activity in cells. However, given the synergism of PDI inhibitors with chemotherapeutic drugs, the effects of these inhibitors as a combinatorial treatment for OS patients is worth further examination.
In the case of ERO1s, studies have shown that ERO1α promoted tumor cell survival and therapy resistance by regulating the oxidative protein folding of molecules such as VEGF, MHC-Class-I molecules, PD-L1 and cytokines [34], [35], [36]. Although a role for ERO1β in cancer progression and chemoresistance is yet to be delineated, ERO1β could function similar to ERO1α in mediating oxidative protein folding. Recently, Tien et al. demonstrated that oxidative folding of NOTCH protein, a critical mediator of OS pathogenesis [37], is severely impaired in drosophila larvae that lack the ERO1 gene, leading to accumulation of improperly folded NOTCH in the ER compartment, induction of UPR, and developmental impairment [38]. Our preliminary analysis revealed that cisplatin induced NOTCH signaling was attenuated in OS cells following down-regulation of ERO1β as well as ATF6α (unpublished observations). While the ATF6α regulation of NOTCH could be transcriptional as previously suggested [15], it is possible that regulation of oxidative protein folding of NOTCH via the ERO1β-PDI axis could also have a role and contribute to decreased survival against cisplatin induced cell death and subsequent chemoresistance.
The cytoprotective role of ATF6α activation extends beyond tumor cells and chemoresistance. For instance, cardiac tissue from ATF6α knockout mice displayed increased damage and decreased function following myocardial ischemia/reperfusion injury, compared to wildtype littermates [39]. Also, ATF6α signaling has also been shown to have a cytoprotective/neuroprotective role in certain glomerular diseases and nervous tissue disorders such as Huntington’s disease [40], [41]. Furthermore, conditional activation of ATF6α in the forebrain neurons was also protective as it reduced infarct volume and improved functional recovery in a mouse model of stroke [42]. Additionally, ATF6α activation also plays a critical role during normal development and tissue homeostasis. Most notably during vertebrate embryogenesis, bone morphogenetic protein stimulates osteoblast differentiation and mineralization through Runx2 induced ATF6α activation [43]. But ATF6α activation can also contribute to enhanced apoptosis, during mouse embryonic muscle development via caspase 12 activation [44]. These finding suggest that ATF6α serves as an important homeostatic regulator operating in a cell and tissue specific manner, however significant gaps in knowledge still remain to filled.
Chemoresistance is a significant problem for patients with OS. The histologic response to treatment at the time of resection is a known prognostic factor for OS patients. Attempts to improve outcome of poor responders by escalating or intensifying the doses or using postoperative chemotherapy regimens that are different from the preoperative treatment failed although their histologic response predicted improved prognosis [45]. These studies highlighted the need for not only new therapeutic agents, but also the need for a prognostic marker other than histologic response that can accurately predict at diagnosis both the prognosis as well as the sensitivity to chemotherapy. Our in vitro findings suggest ATF6α signaling could predict chemoresponse. Although the association between ATF6α(N) levels and percent necrosis in patient samples did not achieve statistical significance, these pilot studies are promising and warrants further examination.
Another prognostic indicator is the presence of metastasis. While approximately 20% to 25% of newly diagnosed patients present with overt metastasis, a similar percentage of patients with localized disease also relapse within 5 years and develop pulmonary metastasis. Recently, expression of ATF6α regulated chaperone BiP/grp78 was shown to be enhanced and necessary for lung metastatic progression of OS cells [46]. In agreement with these findings we also found that the levels of grp78 were up-regulated in lung metastasis (unpublished observations). Our findings here suggest that measuring the levels of grp78 activator ATF6α(N) levels could be a good predictor for the occurrence of metastasis at diagnosis and later during disease progression. While the late recurrence could be due to the development of resistance, it also suggests that these OS patients are harboring dormant disease at the time of diagnosis [47]. Studies have shown the existence of disseminated OS tumor cells in the bone marrow at the time of diagnosis in patients who do not present with detectable metastasis and this correlated with relapses in these patients [48]. Therefore, identification of factor(s) that can stratify patients with localized disease for the presence or absence of latent micrometastatic disease will potentially be useful for tailoring therapies. Given the role of ATF6α in the survival of dormant disseminated tumor cells [13], we hypothesize that high ATF6α(N) levels could also predict the existence of latent disseminated disease at the time of diagnosis in subgroups of OS patients that can have late recurrences.
Our study demonstrates that ATF6α levels could be a good prognostic indicator. However, it is limited by the fact that it is a small sample number, which curtails the power of this study. Also, it is a retrospective analysis of an existing patient cohort from one institution. Therefore, confirmation in a subsequent larger study will be critical to determine if the associations identified here are indeed causal. Despite these limitations, the results obtained here are promising. Knowing the nuclear levels of ATF6α in OS tumors at the time of diagnosis can be useful tool to help make better assessments of the prognosis and treatment response for subgroups of patients and tailor therapies accordingly, if validated in a larger study. Moreover, identifying and targeting ATF6α modulated survival mechanisms could be a more suitable way to re-establish sensitivity to existing treatments and improve therapeutic efficacy.
In summary our data establish an important role for UPR activated transcription factor ATF6α in OS response to chemotherapy and prognosis. The results of this study show that active ATF6α (ATF6α(N)) level is up-regulated in OS tumors and is associated with decreased overall and progression free survival, poor response to chemotherapy and increase incidence of metastasis. In agreement with these findings genetic or pharmacologic inhibition of ATF6α or its downstream effectors such as ERO1β and PDI increased the sensitivity of OS cells to cisplatin and irinotecan treatment. Taken together our data suggest that ATF6α activation may serve as a potential target to limit OS metastasis and decrease therapeutic resistance.
The following are the supplementary data related to this article.
Supplementary tables
Acknowledgements
We would like to thank Dr Kurt Gustin and Carol Haussler for critical reading and editing of the manuscript and helpful suggestions. This work was supported by Department of Child Health and Basic Medical Sciences (DoCH-BMS) grant and Valley Research Partnership VRP23-P2 grant to A.R.S and P.H.
Footnotes
This work was supported by Department of Child Health and Basic Medical Sciences (DoCH-BMS) grant and Valley Research Partnership VRP23-P2 grant to A.R.S and P.H.
The authors declare no potential conflicts of interest.
Contributor Information
Pooja Hingorani, Email: phingorani@phoenixchildrens.com.
Aparna R Sertil, Email: arsertil@email.arizona.edu.
References
- 1.Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs. 2008;10:315–327. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 2.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9:422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 3.Meyers PA, Heller G, Healey JH, Huvos A, Applewhite A, Sun M. Osteogenic sarcoma with clinically detectable metastasis at initial presentation. J Clin Oncol. 1993;11:449–453. doi: 10.1200/JCO.1993.11.3.449. [DOI] [PubMed] [Google Scholar]
- 4.Kager L, Zoubek A, Pötschger U, Kastner U, Flege S, Kempf-Bielack B. Vol. 21. American Society of Clinical Oncology; 2003. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols; pp. 2011–2018. (Journal of Clinical Oncology). [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 7.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Vol. 186. Rockefeller University Press; 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells; pp. 323–331. (J Cell Biol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 9.Schindler AJ, Schekman R. In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc Natl Acad Sci U S A. 2009;106:17775–17780. doi: 10.1073/pnas.0910342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T. Vol. 16. Cold Spring Harbor Lab; 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response; pp. 452–466. (Genes Dev). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 12.Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, Mori K. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–614. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 13.Schewe DM, Aguirre-Ghiso JA. ATF6 alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Natl Acad Sci. 2008;105:10519–10524. doi: 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higa A, Taouji S, Lhomond S, Jensen D, Fernandez-Zapico ME, Simpson JC. Endoplasmic reticulum stress-activated transcription factor ATF6 requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2014;34:1839–1849. doi: 10.1128/MCB.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dadey DYA, Kapoor V, Khudanyan A, Urano F, Kim AH, Thotala D. The ATF6 pathway of the ER stress response contributes to enhanced viability in glioblastoma. Oncotarget. 2016;7:2080–2092. doi: 10.18632/oncotarget.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji G-R, Yu N-C, Xue X, Li Z-G. PERK-mediated Autophagy in Osteosarcoma Cells Resists ER Stress-induced Cell Apoptosis. Int J Biol Sci. 2015;11:803–812. doi: 10.7150/ijbs.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006;66:1702–1711. doi: 10.1158/0008-5472.CAN-05-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzym Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045–13052. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- 21.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadanaka S, Yoshida H, Kano F, Murata M, Mori K. Vol. 15. American Society for Cell Biology; 2004. Activation of mammalian unfolded protein response is compatible with the quality control system operating in the endoplasmic reticulum; pp. 2537–2548. (Mol Biol Cell). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Crews KR, Stewart CF, Liu T, Rodriguez-Galindo C, Santana VM, Daw NC. Effect of fractionated ifosfamide on the pharmacokinetics of irinotecan in pediatric patients with osteosarcoma. J Pediatr Hematol Oncol. 2004;26:764–767. doi: 10.1097/00043426-200411000-00016. [DOI] [PubMed] [Google Scholar]
- 25.Evdokiou A, Bouralexis S, Atkins GJ, Chai F, Hay S, Clayer M. Chemotherapeutic agents sensitize osteogenic sarcoma cells, but not normal human bone cells, to Apo2L/TRAIL-induced apoptosis. Int J Cancer. 2002;99:491–504. doi: 10.1002/ijc.10376. [DOI] [PubMed] [Google Scholar]
- 26.Kusio-Kobialka M, Podszywalow-Bartnicka P, Peidis P, Glodkowska-Mrowka E, Wolanin K, Leszak G. The PERK-eIF2α phosphorylation arm is a pro-survival pathway of BCR-ABL signaling and confers resistance to imatinib treatment in chronic myeloid leukemia cells. Cell Cycle (Georgetown, Tex) 2012;11:4069–4078. doi: 10.4161/cc.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkins C, Liu Q, Minthorn E, Zhang S-Y, Figueroa DJ, Moss K. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 29.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 30.Wey S, Luo B, Tseng C-C, Ni M, Zhou H, Fu Y. Inducible knockout of GRP78/BiP in the hematopoietic system suppresses Pten-null leukemogenesis and AKT oncogenic signaling. Blood. American Society of. Hematology. 2012;119:817–825. doi: 10.1182/blood-2011-06-357384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tufo G, Jones AWE, Wang Z, Hamelin J, Tajeddine N, Esposti DD. Vol. 21. Nature Publishing Group; 2014. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma; pp. 685–695. (Cell Death Differ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe N, Lam E. Vol. 283. American Society for Biochemistry and Molecular Biology; 2008. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis; pp. 3200–3210. (J Biol Chem). [DOI] [PubMed] [Google Scholar]
- 34.Kajiwara T, Tanaka T, Kukita K, Kutomi G, Saito K, Okuya K. Hypoxia augments MHC class I antigen presentation via facilitation of ERO1-α-mediated oxidative folding in murine tumor cells. Eur J Immunol. 2016;46:2842–2851. doi: 10.1002/eji.201646525. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka T, Kajiwara T, Torigoe T, Okamoto Y, Sato N, Tamura Y. Cancer-associated oxidoreductase ERO1-α drives the production of tumor-promoting myeloid-derived suppressor cells via oxidative protein folding. J Immunol. 2015;194:2004–2010. doi: 10.4049/jimmunol.1402538. [DOI] [PubMed] [Google Scholar]
- 36.May D, Itin A, Gal O, Kalinski H, Feinstein E, Keshet E. Ero1-L alpha plays a key role in a HIF-1-mediated pathway to improve disulfide bond formation and VEGF secretion under hypoxia: implication for cancer. Oncogene. 2005;24:1011–1020. doi: 10.1038/sj.onc.1208325. [DOI] [PubMed] [Google Scholar]
- 37.Tao J, Jiang MM, Jiang L, Salvo JS, Zeng H-C, Dawson B. Notch activation as a driver of osteogenic sarcoma. Cancer Cell. 2014;26:390–401. doi: 10.1016/j.ccr.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tien A-C, Rajan A, Schulze KL, Ryoo HD, Acar M, Steller H. Ero1L, a thiol oxidase, is required for Notch signaling through cysteine bridge formation of the Lin12-Notch repeats in Drosophila melanogaster. J Cell Biol. 2008;182:1113–1125. doi: 10.1083/jcb.200805001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin J-K, Blackwood EA, Azizi K, Thuerauf DJ, Fahem AG, Hofmann C. Vol. 120. Lippincott Williams & Wilkins Hagerstown; MD: 2017. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart; pp. 862–875. (Circ Res). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elimam H, Papillon J, Takano T, Cybulsky AV. Vol. 290. American Society for Biochemistry and Molecular Biology; 2015. Calcium-independent phospholipase A2γ enhances activation of the ATF6 transcription factor during endoplasmic reticulum stress; pp. 3009–3020. (J Biol Chem). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naranjo JR, Zhang H, Villar D, González P, Dopazo XM, Morón-Oset J. Vol. 126. American Society for Clinical Investigation; 2016. Activating transcription factor 6 derepression mediates neuroprotection in Huntington disease; pp. 627–638. (J Clin Invest). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Z, Sheng H, Liu S, Zhao S, Glembotski CC, Warner DS. Activation of the ATF6 branch of the unfolded protein response in neurons improves stroke outcome. J Cereb Blood Flow Metab. 2017;37:1069–1079. doi: 10.1177/0271678X16650218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang W.-G, Kim E.-J, Kim D.-K, Ryoo H.-M, Lee K.-B, Kim S.-HÂ. BMP2 protein regulates osteocalcin expression via Runx2-mediated ATF6 gene transcription. J Biol Chem. 2012;287:905–915. doi: 10.1074/jbc.M111.253187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakanishi K, Sudo T, Morishima N. Vol. 169. Rockefeller University Press; 2005. Endoplasmic reticulum stress signaling transmitted by ATF6 mediates apoptosis during muscle development; pp. 555–560. (J Cell Biol). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whelan JS, Bielack SS, Marina N, Smeland S, Jovic G, Hook JM. EURAMOS-1, an international randomised study for osteosarcoma: results from pre-randomisation treatment. Ann Oncol. 2015;26:407–414. doi: 10.1093/annonc/mdu526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lizardo MM, Morrow JJ, Miller TE, Hong ES, Ren L, Mendoza A. Upregulation of glucose-regulated protein 78 in metastatic cancer cells is necessary for lung metastasis progression. Neoplasia. 2016;18:699–710. doi: 10.1016/j.neo.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorlick R, Khanna C. Osteosarcoma. J Bone Miner Res. 2010;25:683–691. doi: 10.1002/jbmr.77. [DOI] [PubMed] [Google Scholar]
- 48.Bruland OS, Høifødt H, Sæter G, Smeland S, Fodstad O. Vol. 11. American Association for Cancer Research; 2005. Hematogenous micrometastases in osteosarcoma patients; pp. 4666–4673. (Clin Cancer Res). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables