Abstract
Alzheimer's disease (AD) has a long neuropathological accumulation phase before the onset of dementia. Such AD neuropathological deposition between neurons impairs the synaptic communication, resulting in networks disorganization. Our study aimed to explore the evolution patterns of gray matter structural covariance networks (SCNs) along AD continuum. Based on the AT(N) (i.e., Amyloid/Tau/Neurodegeneration) pathological classification system, we classified subjects into four groups using cerebrospinal fluid amyloid-beta1–42 (A) and phosphorylated tau protein181 (T). We identified 101 subjects with normal AD biomarkers (A-T-), 40 subjects with Alzheimer's pathologic change (A + T−), 101 subjects with biological AD (A + T+) and 91 AD with dementia (demented subjects with A + T+). We used four regions of interest to anchor default mode network (DMN, medial temporal subsystem and midline core subsystem), salience network (SN) and executive control network (ECN). Finally, we used a multi-regression model-based linear-interaction analysis to assess the SCN changes. Along the disease progression, DMN and SN showed increased structural association at the early stage while decreased structural association at the late stage. Moreover, ECN showed progressively increased structural association as AD neuropathological profiles progress. In conclusion, this study found the dynamic trajectory of SCNs changes along the AD continuum and support the network disconnection hypothesis underlying AD neuropathological progression. Further, SCN may potentially serve as an effective AD biomarker.
Keywords: Alzheimer's disease continuum, Gray matter structural covariance, AT(N) classification system, CSF, Tau protein, Amyloid
Highlights
-
•
To explore the AD continuum accurately by using the latest ATN classification system (based on neuropathological biomarkers).
-
•
Using SCNs analysis to reflect the brain network changes, which may further lead to cognition alternations in AD.
-
•
Results supported network disconnection hypothesis and showed a dynamic trajectory of SCNs changes along the AD continuum.
1. Introduction
Alzheimer's disease (AD) is the most common form of dementia, which was characterized clinically by progressive memory and other cognitive abilities decline. At the neuropathological level, AD neuropathological accumulation begins years before the onset of dementia (Villemagne et al., 2013) and develops in a specific temporal-ordered manner (Sperling et al., 2014), with extracellular amyloid-β plaques deposit appears the earliest and followed by intracellular phosphorylated tau deposition and downstream neurodegeneration events (Jack Jr. et al., 2013). Hence, combining the pathological stages with the symptomatic diagnosis can help to assess AD continuum more accurately (Montal et al., 2018).
Spatially, AD neuropathological deposition around neurons impairs synaptic communication, resulting in neuronal networks disorganization (Myers et al., 2014). As such, AD is regarded as a disconnection disease featuring intrinsic network disorganization (Delbeuck et al., 2003). The most commonly affected brain regions in AD studies are the default mode network (DMN), salience network (SN) and executive control network (ECN), which cooperate to maintain cognitive abilities including memory, and executive function (Zhu et al., 2016; Seeley et al., 2007). Notably, recent studies also reported that the DMN consists of multiple, spatially dissociated but interactive subsystems, including the midline core subsystem (self-relevant decision-making) and medial temporal subsystem (episodic memory) (Andrews-Hanna et al., 2010; Uddin et al., 2009). Moreover, these network disruptions have been associated with cognitive dysfunction in AD. Therefore, brain network analysis could be an effective method to study AD neuropathology and explore further cognitive changes.
Brain networks can be constructed based on similarity in GM structure between brain areas, which was named as the gray matter (GM) structural covariance network (SCN) (Mechelli et al., 2005; Alexander-Bloch et al., 2013a; Tijms et al., 2012). Based on prior studies, the biological meaning of SCN may link to coordinated GM growth during development (Alexander-Bloch et al., 2013a), functional co-activation (Alexander-Bloch et al., 2013b), axonal connectivity (Essen, 1997; Gong et al., 2012a) and genetic factors (Chen et al., 2013; Schmitt et al., 2009; Alexander et al., 2012). Brain areas that are highly correlated in size are always part of systems that subserving specific behavioral or cognitive functions (Alexander-Bloch et al., 2013a; Zielinski et al., 2010). For example, areas involved in memory showed GM volume covariance (Bohbot et al., 2007). In AD, SCN showed disorganization which correlates with cognitive dysfunction (Dicks et al., 2018; Tijms BM Kate et al., 2016). Specifically, Spreng et al. demonstrated decreased SCN in DMN in AD patients (Spreng and Turner, 2013). Another SCN study showed that early AD patients had decreased structural association within the DMN while increased structural association in the SN and ECN (Montembeault et al., 2016). These results indicate the trend of decreased structural association in DMN while increased structural association in ECN and SN in AD patients. However, few studies considered the AD neuropathological stages. This may lead to inaccurate results considering the temporal-ordered neuropathological propagation along the network.
To address this gap, the current study aimed to explore the trajectory of SCN changes along the pathophysiological continuum of AD. Combining the neuropathological hallmarks and symptomatic status, we classified the subjects into four groups to reflect the disease progression along the AD continuum. According to the evidence that AD network highly linked to its neuropathological state (Seeley et al., 2009; Knobloch and Mansuy, 2008), we hypothesized that DMN tends to show a decreased structural association while the ECN and SN tend to show an increased structural association as AD progresses.
2. Methods and materials
2.1. Alzheimer's disease neuroimaging and initiative
Data used in the preparation of this article were obtained from the Alzheimer's disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). The ADNI was initially launched in 2004 (ADNI-1), and additional recruitment was made through ADNI-GO in 2009, ADNI-2 in 2010 and ADNI-3 in 2016. The primary goal of ADNI has been to identify serial MRI, PET, biomarkers and genetic characteristics that would support the early detection and tracking of the AD, and improved clinical trial design. For up-to-date information, see http://www.adni-info.org.
2.2. Study participants
This study was approved by the Institutional Review Boards of all participating institutions and informed written consent was obtained from all participants at each site. Similar to the previous study (Palmqvist et al., 2017), we identified 242 non-demented subjects (characterized as either cognitively unimpaired or mild cognitive impairment, MCI) and 91 demented subjects from ADNI database (Table 1, the flowchart in Supplementary Material 1). We included MCI subjects since the cognitive impairment may be not caused by an underlying AD process and, given that about a third of MCI were incorrectly diagnosed (Bangen KJ Clark et al., 2016; Edmonds EC Delano-Wood L, Galasko DR, Salmon DP, Bondi MW1; Alzheimer's Disease Neuroimaging Initiative, 2014). Furthermore, based on the pathological system (Jack et al., 2018), we identified demented subjects in the study since the Alzheimer's disease with dementia features the highest risk of clinical progression and can present the most classic AD-related changes.
Table 1.
Demographic and neuropsychological data in subjects along the AD continuum.
| Demographic characteristics | Group 0 N = 101 |
Group 1 N = 40 |
Group 2 N = 101 |
Group 3 N = 91 |
P-value |
|---|---|---|---|---|---|
| Age,y, mean (SD) | 71.40 ± 6.29 | 72.70 ± 7.01 | 72.68 ± 5.70 | 73.34 ± 7.76 | G1-G0 = 0.29 |
| G2-G0 = 0.13 | |||||
| G3-G0 = 0.06 | |||||
| Female, n(%) | 51(50.50%) | 14(35.00%) | 45(44.55%) | 41(45.05%) | G1-G0 = 0.07 |
| G2-G0 = 0.24 | |||||
| G3-G0 = 0.27 | |||||
| Education,y, mean (SD) | 16.42 ± 2.66 | 16.38 ± 2.61 | 16.36 ± 2.40 | 15.80 ± 2.45 | G1-G0 = 0.93 |
| G2-G0 = 0.87 | |||||
| G3-G0 = 0.10 | |||||
| APOE ε4 status, n(%) | 16(15.84%) | 15(37.50%) | 61(60.39%) | 67(73.63%) | G1-G0 = 0.01⁎ |
| G2-G0 < 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| GDS | 1.30 ± 1.42 | 1.43 ± 1.55 | 1.50 ± 1.59 | 1.64 ± 1.38 | G1-G0 = 0.64 |
| G2-G0 = 0.35 | |||||
| G3-G0 = 0.09 | |||||
| General mental status | |||||
| MMSE | 28.77 ± 1.38 | 28.80 ± 1.27 | 28.17 ± 1.73 | 23.00 ± 2.30 | G1-G0 = 0.91 |
| G2-G0 = 0.01⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| CDR global | 0.30 ± 0.25 | 0.26 ± 0.25 | 0.36 ± 0.22 | 0.80 ± 0.28 | G1-G0 = 0.40 |
| G2-G0 = 0.08 | |||||
| G3-G0 < 0.001⁎ | |||||
| CDR sum | 0.79 ± 0.82 | 0.73 ± 0.91 | 1.18 ± 1.11 | 4.63 ± 1.80 | G1-G0 = 0.71 |
| G2-G0 = 0.01⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| Memory function | |||||
| WMS-LM immediate | 12.76 ± 3.20 | 13.18 ± 3.37 | 11.04 ± 4.21 | 3.93 ± 2.49 | G1-G0 = 0.51 |
| G2-G0 = 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| WMS-LM delay | 10.87 ± 3.44 | 11.65 ± 3.76 | 9.04 ± 4.64 | 1.47 ± 1.70 | G1-G0 = 0.24 |
| G2-G0 = 0.002⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| AVLT sum of trials 1–5 | 44.71 ± 11.64 | 41.75 ± 11.08 | 37.28 ± 10.63 | 21.96 ± 7.42 | G1-G0 = 0.169 |
| G2-G0 < 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| AVLT 30 min | 7.19 ± 4.04 | 6.60 ± 3.80 | 4.33 ± 3.92 | 0.51 ± 1.07 | G1-G0 = 0.43 |
| G2-G0 < 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| Attention | |||||
| Log-transformed TMT-A | 1.51 ± 0.14 | 1.54 ± 0.16 | 1.55 ± 0.14 | 1.71 ± 0.21 | G1-G0 = 0.34 |
| G2-G0 = 0.12 | |||||
| G3-G0 < 0.001⁎ | |||||
| Decision-making function | |||||
| Log-transformed TMT-B | 1.90 ± 0.18 | 1.94 ± 0.19 | 1.95 ± 0.20 | 2.23 ± 0.21 | G1-G0 = 0.27 |
| G2-G0 = 0.08 | |||||
| G3-G0 < 0.001⁎ | |||||
| Language | |||||
| BNT total | 27.71 ± 2.62 | 27.68 ± 2.26 | 27.73 ± 2.96 | 21.67 ± 6.00 | G1-G0 = 0.93 |
| G2-G0 = 0.96 | |||||
| G3-G0 < 0.001⁎ | |||||
| Category fluency | 19.59 ± 5.10 | 18.35 ± 5.83 | 19.26 ± 5.36 | 11.70 ± 4.89 | G1-G0 = 0.22 |
| G2-G0 = 0.65 | |||||
| G3-G0 < 0.001⁎ | |||||
| Visuospatial processing | |||||
| CDT | 4.59 ± 0.67 | 4.43 ± 0.71 | 4.57 ± 0.35 | 3.49 ± 1.45 | G1-G0 = 0.19 |
| G2-G0 = 0.84 | |||||
| G3-G0 < 0.001⁎ | |||||
| CSF | |||||
| Aβ1–42 (pg/ml) | 228.93 ± 23.66 | 159.92 ± 26.80 | 136.75 ± 25.24 | 126.71 ± 20.74 | G1-G0 < 0.001⁎ |
| G2-G0 < 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| T-Tau(pg/ml) | 43.45 ± 13.64 | 41.64 ± 13.43 | 96.30 ± 41.18 | 143.90 ± 67.02 | G1-G0 = 0.47 |
| G2-G0 < 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
| P-Tau181(pg/ml) | 17.40 ± 3.78 | 17.54 ± 3.81 | 49.77 ± 15.11 | 65.29 ± 34.24 | G1-G0 = 0.84 |
| G2-G0 < 0.001⁎ | |||||
| G3-G0 < 0.001⁎ | |||||
Data are presented as means ± standard deviations;
Abbreviation: Group 0: G0, subjects with normal AD biomarkers, non-demented individuals without abnormal CSF; Group 1: G1, subjects with Alzheimer's pathologic change, non-demented individuals with abnormal Aβ1–42 but normal P-tau181; Group 2: G2, subjects with biological Alzheimer's disease, non-demented individuals with abnormal Aβ1–42 and P-tau181; Group 3: G3, Alzheimer's disease with dementia, demented individuals with abnormal Aβ1–42 and P-tau181; MMSE, Mini-Mental State Examination; CDR: Clinical Dementia Rating; WMS-LM, Wechsler memory scale-logical memory; AVLT, Auditory Verbal Learning Test; TMT, Trail-Making Test; BNT, Boston naming test; CDT, Clock Drawing Test.
p < 0.05, significant difference between the two Groups.
Cognitively unimpaired subject was defined as having a Clinical Dementia Rating scale (CDR) score of 0, an Mini-Mental State Examination (MMSE) between 24 and 30 (inclusive), a normal Wechsler Memory Scale Logical Memory, WMS-LM, delay recall performance (in detail: ≥ 9 for subjects with 16 or more years of education; ≥ 5 for subjects with 8–15 years of education; and ≥ 3 for 0–7 years of education); absence of clinical depression (geriatric depression scale-15, GDS-15 score < 6) (Sheikh and Yesavage, 1986) and absence of dementia. MCI (Bondi et al., 2014) was defined as having preserved activities of daily living, the absence of dementia, and objective cognitive impairment as shown on the delayed recall test of the WMS-LM as well as a CDR score of 0.5. Demented individuals were defined as having an MMSE score of ≤26, and a clinical dementia rating (CDR) of ≥0.5 as well as satisfying the NINCDS/ADRDA criteria for probable AD (McKhann et al., 1984). We excluded subjects with the following manifestations: (a) significant medical, neurological, and psychiatric illness; (b) obvious head trauma history; (c) use of non-AD-related medication known to influence cerebral function; (d) clinical depression; (e) alcohol or drug abuse. Only subjects with a simultaneous structural scan, lumbar puncture, and comprehensive neuropsychological assessments were included.
2.3. Neuropsychological assessment and CSF data acquisition
All subjects underwent comprehensive neuropsychological tests (Table 1, description about demographics in Supplementary Material 2), including assessment of general mental status (Mini-Mental State Examination, MMSE; Clinical Dementia Rating, CDR) and other cognitive domains, involving memory function (Auditory Verbal Learning Test, AVLT; WMS-LM, immediate and delayed memory), attention (Trail-Making Test part A, TMT-A), visuospatial function (Clock-Drawing Test, CDT), decision-making function (Trail-Making Test part B, TMT-B), and language ability (Boston Naming Test, BNT; Category fluency).
CSF biomarkers comprise amyloid-beta1–42 (Aβ1–42), total tau (T-tau), and phosphorylated tau at position181 (P-tau181), measured by the multiplex xMAP Luminex platform as previously described (Shaw et al., 2009) (Table 1).
2.4. Group classifications
We combined the latest pathological classification model (Jack et al., 2018) with the symptomatic diagnosis to reflect the disease progression along the AD continuum. Specifically, this model uses three major neuropathological biomarkers: amyloid deposition (A), pathologic tau (T), and neurodegeneration (N) [AT(N)] to stage the disease pathology across entire AD continuum in vivo (Supplementary Material 3). As previous studies described, we set the CSF cutoff point at 192 pg/ml for Aβ1–42 and 23 pg/ml for P-tau181 (Shaw et al., 2009; Meyer et al., 2010). Subsequently, based on the pathological AT(N) system, we classified all subjects into four groups according to their CSF Aβ1–42 and P-tau181 level: (a) Group 0: subjects with normal AD biomarkers, consisting of non-demented individuals with normal Aβ1–42 and P-tau181 (A − T−); (b) Group 1: subjects with Alzheimer's pathologic change, consisting of non-demented individuals with abnormal Aβ1–42 and normal P-tau181 (A + T−); (c) Group 2: subjects with biological Alzheimer's disease, consisting of non-demented individuals with abnormal Aβ1–42 and P-tau181 (A + T+); (d) Group 3: Alzheimer's disease with dementia, consisting of demented subjects with abnormal Aβ1–42 and P-tau181 (A + T+). Notably, we excluded subjects with normal Aβ1–42 and abnormal P-tau181 (A-T+) since it was considered as a non-AD related pathology (Jack Jr. et al., 2016).
2.5. MRI acquisition and pre-processing
MRI data were acquired in 3 T scanners. Briefly, the ADNI protocol includes T1-weighted acquisition based on a sagittal volumetric magnetization-prepared rapid gradient echo sequence collected from a variety of MR systems with protocols optimized for each type of scanner. Representative imaging parameters were as follows: repetition time (TR) =2300 ms; echo time (TE) =3 ms; within plane FOV = 256 × 256 mm2; voxel size = 1.1 × 1.1 × 1.2 mm3; flip angle = 9° or 11°.
We pre-processed all T1-weighted images using the Computational Anatomy Toolbox (CAT12, http://dbm.neuro.uni-jena.de/cat12/) toolbox segment data pipeline implemented within SPM12 in Matlab (R2012b). First, the T1 image was spatially registered to the tissue probability maps (TPM) and then segmented into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF). We performed the affine registration to the stereotactic MNI space using ICBM152 space. Second, we performed high-dimensional DARTEL normalization and nonlinear modulation using the Jacobian determinants derived from the normalization process. To remove the MRI inhomogeneities and noise, we corrected the bias, noise and normalized the intensities. Subsequently, we smoothed the GM image with a Gaussian kernel of 8 × 8 × 8 mm3 to reduce potential inaccuracies during the normalization step. Moreover, we assessed processed image quality by visual inspection and weighted average image quality index (using the quality assurance (QA) framework in CAT 12, http://www.neuro.uni-jena.de/cat/). Here, we only include the subjects with QA better than C+. Finally, 64 subjects did not pass the image quality control.
2.6. Structural covariance gray matter network
To construct the SCNs, we chose four regions of interest (ROIs) (Montembeault et al., 2016): right entorhinal cortex (EC, MNI coordinates: 25, −9, −28) (Bernhardt et al., 2008; Eickhoff et al., 2005), left posterior cingulate cortex (PCC, MNI coordinates: −2, −36, 35) (Spreng and Turner, 2013; Zielinski et al., 2012), right frontoinsular cortex (FIC, MNI coordinates: 38, 26, −10), and right dorsolateral prefrontal cortex (DLPFC, MNI coordinates: 44, 36, 20) (Zielinski et al., 2010; Montembeault et al., 2016). These regions anchor the default mode network (DMN, medial temporal subsystem), DMN (midline core subsystem), salience network (SN) and executive control network (ECN) respectively.
To achieve SCN t-maps, we performed the general linear analysis on modulated GM images. Specifically, we extracted the GM volume from a 4-mm radius sphere (Montembeault et al., 2016; Chang et al., 2018; Chang et al., 2017) around those coordinates on modulated images. Then, we performed four separate correlation analysis by entering the GM volumes of each ROI as a regressor and the total intracranial volume, gender, age, and education as covariates. Before statistical correction, we performed Fisher's Z transformation. For each group, we performed specific contrasts to identify voxels expressing a positive correlation with each ROI. We set the threshold at P ≤ 0.01 corrected for false discovery rate (FDR). Considering the physiological meanings of the SCN clusters, we reported clusters showing cluster size >100 voxels (Chang et al., 2018; Chang et al., 2017). To achieve qualitative group-wise comparisons, we displayed the results on a standard brain template (ICBM152).
Then, we used multi-regression model-based linear interaction analysis (Bernhardt et al., 2008) to assess how the disease stage interferes with SCN, voxels showing significant differences in the regression slopes in each ROI were compared. For this study, we refer to these differences in slopes as the differences in “structural association”. We set the threshold at p < 0.05 at voxel level, p < 0.05 at cluster level, with Gaussian random field correction (GRF) corrected.
To explore the clinical significance of the identified peak voxel volume, we performed the correlation analysis (Chang et al., 2018; Chang et al., 2017). To be specific, for the peak clusters showing significant between-group differences, we extracted the GM volume using a 4-mm radius sphere placing on the peak voxel. Then, we conducted Pearson correlation analysis between the peak voxel volume and neuropsychological data within each group which showed the difference in the structural association. The threshold was set at p < 0.05, Bonferroni corrected.
3. Results
3.1. Patterns of structural association within each group
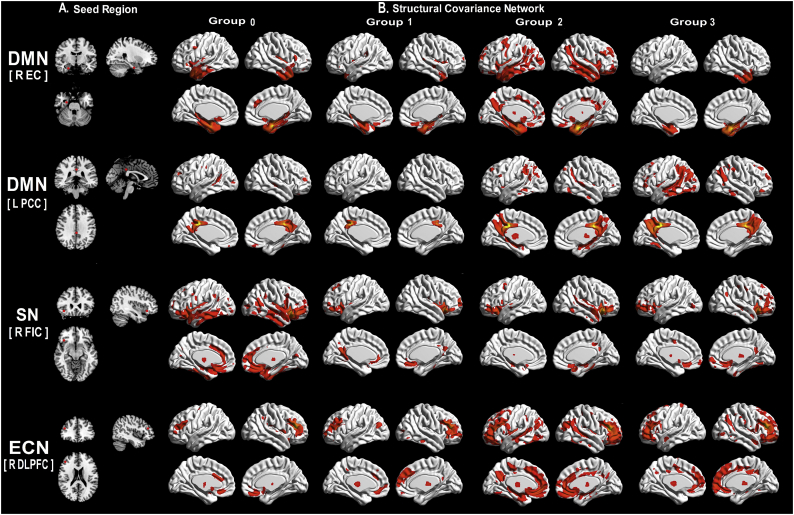
To qualitatively compare the patterns of positive correlations in subjects at all groups, we presented the statistical maps in Fig. 1. Regions presenting a structural association with the seed regions of each network in each group are listed in supplementary material 4.
Fig. 1.
Patterns of structural association within Group.
(A) the target seeds; (B) structural covariance networks (Z-statistic maps [p < 0.01, corrected with a false discovery rate with extended cluster voxels >100]). All the results were projected on a standard brain template.
Abbreviations: R: Right; L: left; DMN: default mode network; SN: salience network; ECN: executive control network; EC: entorhinal cortex; PCC: posterior cingulate cortex; FIC: frontoinsular cortex; DLPFC: dorsolateral prefrontal cortex; Group 0: subjects with normal AD biomarkers, non-demented individuals without abnormal CSF; Group 1: subjects with Alzheimer's pathologic change, non-demented individuals with abnormal Aβ1–42 but normal P-tau181; Group 2: subjects with biological Alzheimer's disease, non-demented individuals with abnormal Aβ1–42 and P-tau181; Group 3: Alzheimer's disease with dementia, demented individuals with abnormal Aβ1–42 and P-tau181.
In both DMN midline core subsystem and ECN, the subjects in Group 3 presents a greater extent of structural covariance than Group 0; In both DMN medial temporal subsystem and SN, the subjects in Group 3 presents a decreased extent of structural covariance than Group 0.
3.2. VThe difference of structural covariance gray matter network
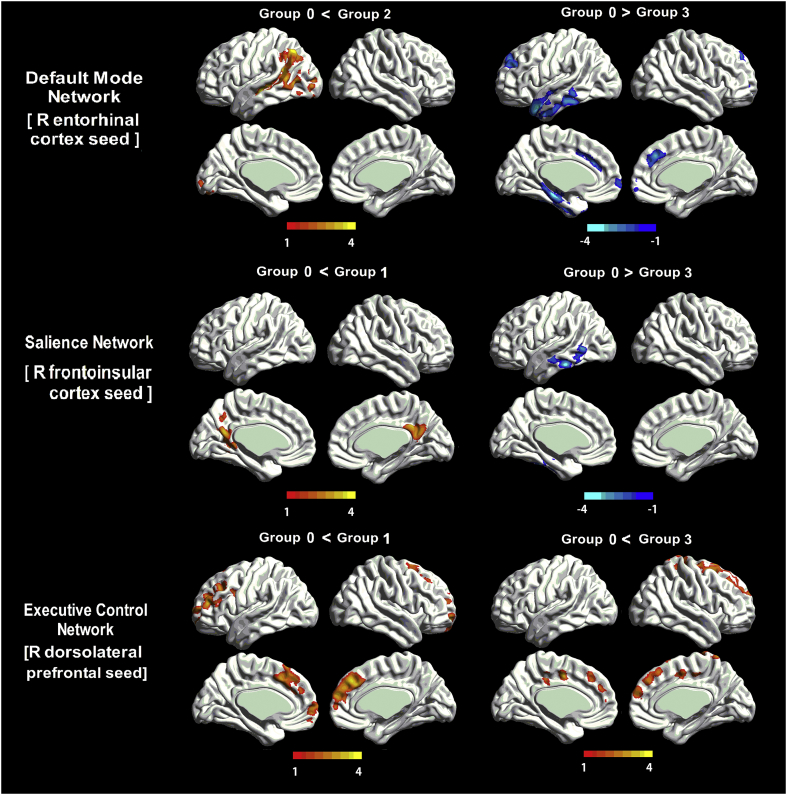
Regarding DMN medial temporal subsystem, subjects in Group 2 showed an increased structural association between the EC and middle temporal gyrus (MTG); subjects in Group 3 showed a decreased structural association between the EC and MTG as well as superior frontal gyrus (SFG) compared to subjects in Group 0. Regarding the DMN midline core subsystem, no significant difference existed in Group 1, 2, or 3 compared to Group 0. Regarding SN, subjects in Group 1 showed an increased structural association between FIC and precuneus; as pathological accumulation progressed, subjects in Group 3 showed the decreased structural association between FIC and the inferior temporal gyrus. Regarding ECN, subjects in Group 1 and Group 3 showed an increased structural association between DLPFC and SFG (Fig. 2 and Table 2). Moreover, to qualitatively depict the SCNs pattern, we also performed the group difference analysis between Group 1, 2, and 3 (Supplementary Material 8).
Fig. 2.
The difference of Gray Matter Structural Covariance network between subjects in AD continuum and Group 0.
(A) As for DMN medial temporal subsystem, subjects in Group 2 showed an increased structural association between the EC and MTG; subjects in Group 3 showed a decreased structural association between the EC and MTG as well as SFG when compared to subjects in Group 0. (B) As for SN, subjects in Group 1 showed an increased structural association between FIC and precuneus; subjects in Group 3 showed decreased association between FIC and the inferior temporal gyrus. (C) As for ECN, subjects in Group 1 in Group 3 showed an increased structural association in the SFG.
Abbreviations: DMN: default mode network; SN: salience network; ECN: executive control network; EC: entorhinal cortex; FIC: frontoinsular cortex; DLPFC: dorsolateral prefrontal cortex; MTG: middle temporal gyrus; SFG: superior frontal gyrus; Group 0: subjects with normal AD biomarkers, non-demented individuals without abnormal CSF; Group 1: subjects with Alzheimer's pathologic change non-demented individuals with abnormal Aβ1–42 but normal P-tau181; Group 2: subjects with biological Alzheimer's disease, non-demented individuals with abnormal Aβ1–42 and P-tau181; Group 3: Alzheimer's disease with dementia, demented individuals with abnormal Aβ1–42 and P-tau181.
Table 2.
The difference of gray matter structural covariance network between subjects in AD continuum and Group 0.
| Network | Group | Peak region | MNI coordinates |
Extent | Peak intensity | ||
|---|---|---|---|---|---|---|---|
| X | Y | Z | |||||
| Default mode (medial temporal lobe subsystem) (R EC) | Group 2 VS. Group 0 | L Middle Temporal Gyrus | −49.5 | −19.5 | −4.5 | 8811 | 4.81 |
| Group 3 VS. Group 0 | L Middle Temporal Gyrus | −45 | 4.5 | −31.5 | 9380 | −4.25 | |
| L Superior Frontal Gyrus | 12 | 36 | 36 | 6031 | −3.80 | ||
| Salience (R FIC) | Group 1 VS. Group 0 | L Precuneus | −12 | −58.5 | 36 | 3992 | 3.80 |
| Group 3 VS. Group 0 | L Inferior Temporal Gyrus | −55.5 | −33 | −22.5 | 4167 | −4.24 | |
| Executive Control (R DLPFC) | Group 1 VS. Group 0 | R Superior Frontal Gyrus | 12 | 27 | 36 | 3208 | 3.85 |
| Group 3 VS. Group 0 | R Superior Frontal Gyrus | 18 | 6 | 55.5 | 9977 | 4.50 | |
Abbreviation: R: right; EC: entorhinal cortex; FIC: frontoinsular cortex; DLPFC: dorsolateral prefrontal cortex; Group 0: subjects with normal AD biomarkers, non-demented individuals without abnormal CSF; Group 1: subjects with Alzheimer's pathologic change, non-demented individuals with abnormal Aβ1–42 but normal P-tau181; Group 2: subjects with biological Alzheimer's disease, non-demented individuals with abnormal Aβ1–42 and P-tau181; Group 3: Alzheimer's disease with dementia, demented individuals with abnormal Aβ1–42 and P-tau181.
Additionally, we performed an analysis using the contralateral ROI seeds by changing the sign on each seed's x coordinate (Zielinski et al., 2010). DMN showed initially increased and then decreased structural association between EC and frontal region along AD continuum. SN showed the increased structural association between FIC and inferior parietal gyrus at the early neuropathological stage (Supplementary Material 5).
To test the reliability, we adopted different statistical thresholds. Results remained mostly unchanged (Supplementary Material 6). Moreover, we repeated the analysis based on ADNI1 database (acquired from 1.5 T MRI scanner). Similarly, DMN showed a trend of the decreased structural association while ECN showed a trend of increased structural association (Supplementary Material 7).
3.3. Correlations of peak cluster volumes with neuropsychological scores
We then performed the Pearson correlation analysis between the peak cluster volumes and neuropsychological scores. The correlation mainly located at the EC anchored DMN medial temporal subsystem, concerning memory and language (Table 3). To be specific, in Group 3 subjects, the peak volume of MTG significantly correlated with memory (WMS-LM immediate and delay (r = 0.34, p < 0.005, r = 0.30, p < 0.005, respectively); AVLT sum of trials 1–5 (r = 0.34, p < 0.005)), language (category fluency (r = 0.42, p < 0.001), BNT total (r = 0.39, p < 0.001)), and executive function (log-transformed TMT-B (r = −0.33, p < 0.005)). Moreover, regarding DLPFC anchored ECN, in Group 3 subjects, the peak volume of SFG negatively correlated with log-transformed TMT-A (r = −0.35, p < 0.005) and log-transformed TMT-B (r = −0.48, p < 0.001).
Table 3.
Correlation coefficients between peak cluster volume and neuropsychological data.
| Seed region |
EC (25, −9, −28) |
DLPFC (44, 36, 20) |
FIC (38, 26, −10) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak Cluster | MTG (G0 < G2) | MTG (G0 > G3) | SFG (G0 > G3) | SFG (G0 < G1) | SFG (G0 < G3) | precuneus (G0 < G1) | ITG (G0 > G3) | |||||||
| Group | G0 | G2 | G0 | G3 | G0 | G3 | G0 | G1 | G0 | G3 | G0 | G1 | G0 | G3 |
| General cognitive state | ||||||||||||||
| MMSE | 0.06 | 0.30a | 0.07 | 0.21 | 0.11 | 0.03 | 0.08 | 0.38 | −0.36 | 0.14 | 0.19 | 0.22 | 0.04 | 0.26 |
| CDR global | 0.16 | −0.22 | −0.04 | −0.23 | 0.08 | −0.02 | 0.09 | −0.07 | 0.04 | −0.04 | 0.18 | −0.13 | 0.14 | −0.21 |
| CDR sum | 0.10 | −0.30a | −0.06 | −0.23 | −0.06 | −0.10 | 0.02 | −0.01 | 0.02 | 0.06 | 0.10 | −0.17 | 0.04 | −0.19 |
| Memory | ||||||||||||||
| WMS-LM immediate | −0.03 | 0.19 | 0.22 | 0.34b | −0.02 | 0.21 | −0.05 | 0.03 | 0.05 | 0.25 | −0.14 | 0.00 | 0.10 | 0.22 |
| WMS-LM delay | −0.01 | 0.24 | 0.21 | 0.30a | 0.05 | 0.01 | 0.01 | 0.16 | 0.01 | 0.07 | −0.17 | 0.11 | 0.03 | 0.21 |
| AVLT sum of trials 1–5 | −0.07 | 0.21 | 0.00 | 0.34b | 0.05 | 0.12 | 0.17 | 0.27 | −0.21 | 0.12 | 0.03 | 0.16 | −0.14 | 0.16 |
| AVLT 30 min | −0.03 | 0.17 | 0.06 | 0.33b | 0.13 | 0.02 | 0.16 | 0.24 | −0.16 | 0.09 | −0.05 | 0.03 | −0.12 | 0.22 |
| Language | ||||||||||||||
| BNT total | 0.10 | 0.29a | 0.38b | 0.39b | 0.21 | 0.04 | 0.19 | 0.05 | 0.07 | 0.14 | 0.23 | −0.16 | 0.36b | 0.21 |
| Category fluency | 0.20 | 0.35b | 0.29a | 0.42b | 0.30a | 0.09 | 0.36b | 0.41 | 0.13 | 0.11 | 0.20 | 0.32 | 0.22 | 0.35b |
| Attention | ||||||||||||||
| Log-transformed TMT-A | −0.18 | −0.12 | −0.26 | −0.17 | −0.19 | −0.13 | −0.10 | −0.19 | −0.01 | −0.35b | −0.12 | −0.02 | −0.11 | −0.10 |
| Decision-making function | ||||||||||||||
| Log-transformed TMT-B | −0.09 | −0.30a | −0.19 | −0.33a | −0.07 | −0.22 | −0.11 | −0.19 | 0.02 | −0.48b | −0.14 | −0.07 | −0.13 | −0.33a |
| Visual-spacial processing | ||||||||||||||
| CDT | −0.12 | 0.15 | −0.02 | 0.27 | −0.09 | 0.10 | 0.03 | 0.38 | 0.004 | 0.16 | −0.12 | 0.16 | −0.03 | 0.25 |
Numbers indicate Pearson correlation coefficients, a: p < .005, b: p < .01.
Abbreviation: EC: entorhinal cortex; DLPFC: dorsolateral prefrontal cortex; FIC: frontoinsular cortex; MTG: middle temporal gyrus; SFG: superior frontal gyrus; ITG: inferior temporal gyrus; Group 0: G0, subjects with normal AD biomarkers, non-demented individuals without abnormal CSF; Group 1: G1, subjects with Alzheimer's pathologic change, non-demented individuals with abnormal Aβ1–42 but normal P-tau181; Group 2: G2, subjects with biological Alzheimer's disease, non-demented individuals with abnormal Aβ1–42 and P-tau181; Group 3: G3, Alzheimer's disease with dementia, demented individuals with abnormal Aβ1–42 and P-tau181. MMSE, Mini-Mental State Examination; CDR: Clinical Dementia Rating; WMS-LM, Wechsler memory scale-logical memory; AVLT, Auditory Verbal Learning Test; TMT, Trail-Making Test; BNT, Boston naming test; CDT, Clock Drawing Test.
4. Discussion
Our study explored the SCN changes along the AD continuum based on the neuropathological classification system. Regarding the DMN and SN, subjects at the early and late stage of AD successively showed increased and decreased structural association. As for ECN, subjects with abnormal CSF showed progressively increased structural association as AD neuropathological profiles progress. Our results suggest the dynamic trajectory of AD progression based on GM SCN technique and provide support for the disconnection hypothesis underlying AD neuropathological progression.
These results are partially in line with previous studies which reported the DMN, SN (Supekar et al., 2008; Zhou et al., 2010) and the ECN changes (Weiler et al., 2014; Agosta et al., 2012) in AD by using the functional (Zhou et al., 2010; Brier et al., 2012; Luo X Qiu et al., 2016; Luo et al., 2016) or white matter connectivity analysis (Garces et al., 2014). The possible mechanism is the high metabolic load in DMN (Buckner et al., 2005) and pathogenic molecules spread via synaptic connections (Zhou et al., 2012; Raj et al., 2012; Koch et al., 2015; Klunk et al., 2004; Molinuevo et al., 2014; Racine et al., 2014). Although SCN could not be considered as a direct measure of connectivity, convergent studies reported the spatial overlap between intrinsic connectivity and structural covariance, thus demonstrating that these patterns mirror each other (Mechelli et al., 2005; Seeley et al., 2009; Segall et al., 2012; Zhang et al., 2011; Lerch et al., 2006). This could be explained by the fact that synchronous neuronal firing promotes network based synaptogenesis (Katz and Shatz, 1996; Bi and Poo, 1999). Accordingly, GM SCN could indicate the connectivity to some extent and provide additional insight into the network topographical organization (Alexander-Bloch et al., 2013a; Bassett and Bullmore, 2009; Gong et al., 2012b). Here, decreased correlations between brain regions may be suggestive of disconnectivity or localization degeneration while increased correlations may indicate overconnectivity or correlated gray matter loss in regions targeted by the same neurodegenerative process.
Regarding the DMN, subjects at the early stage of AD showed increased structural association while decreased association in subjects at the late stage. This result is in line with previous studies which reported the early involvement of DMN in AD (Seeley et al., 2009; He et al., 2007; Luo et al., 2018a; Luo et al., 2018b; Gili et al., 2011), and the progressive SCN DMN score decline through the AD progression (Spreng and Turner, 2013). The mechanism may be the activity-dependent or metabolism-dependent pathology hypothesis: the DMN's continuous activity may lead to the early formation and diffusion of the AD neuropathological hallmarks (Palmqvist et al., 2017; Buckner et al., 2005; Buckner et al., 2009). To be specific, we found an increased structural association between the EC and MTG in Group 2 and decreased structural association between EC and MTG as well as SFG in Group 3. Although no significant difference in Group 1, these subjects show the trend of an increased structural association involving the temporal and frontal regions. These changes could be described as a biphasic trajectory which was resulted by neuropathological progression. Moreover, such DMN changes always correlate with cognitive status and CSF level (Spreng and Turner, 2013; Allen et al., 2007; Zhang et al., 2010). One SCN study found a decreased association between EC and prefrontal region in early AD patients and proposed it as the underlying reason for cognitive deficits (Montembeault et al., 2016). Similarly, we also found that entorhinal-anchored peak clusters linked to the clinical scores in the subjects along the AD continuum. These findings highlighted the primary role of the entorhinal-anchored network (DMN medial temporal subsystem) in cognition maintenance. Conclusively, we proposed that DMN showed a structural hyperconnectivity at the early stage and then hypoconnectivity as AD progresses; furthermore, structural connectivity changes may indicate the cognition decline.
SN is in charge of diverse homeostatically relevant internal and external stimuli, and always shows alternated function in AD patients. Here, we observed a dynamic structural association in this intrinsic network: increased structural association between FIC and precuneus in Group 1 subjects, while decreased structural association in ITG in Group 3 subjects. These results are in line with one functional study, which reported firstly increased (between CDR 0 and CDR 0.5) and then decreased correlation (at CDR 1) in SN in subjects along AD continuum (Brier et al., 2012). Similarly, the work of Schultz AP also reported hyperconnectivity in SN in subjects with amyloid positive and then hypoconnectivity when neocortical tau levels are high (Schultz et al., 2017). Conclusively, these findings suggested the dynamic changes in SN along AD neuropathological progression.
Regarding the ECN, we observed a continuously increased association in Group 1 and Group 3 subjects. Based on the previous study, we speculated it as a compensatory network associated with cognitive reservation in AD patients (Grady et al., 2003; Stern, 2006). Similar results can be found in another study which observed extensive SCN in ECN in AD patients with positive Aβ status (Montembeault et al., 2016). Our data extended this work by demonstrating a gradually increased structural association between DLPFC and frontal regions along the AD continuum. Here, stronger covariance strength between the seed and peak clusters indicated more intra-network connections to maintain cognition (Lin et al., 2016). Our further correlation analysis showed that DLPFC-anchored peak cluster volumes correlated to the cognitive performance, suggesting a compensatory clinical meaning of the DLPFC-anchored ECN in AD continuum. By coincidence, SFG presents a significant decreased structural association with the EC in AD patients. This may partially support the hypothesis that AD features opposing connectivity in the DMN and ECN (Zhou et al., 2010; Agosta et al., 2012).
The DMN, SN, and ECN showed interactive and dynamic changes with neuropathological progression. Regarding the increased structural association in DMN and SN at the early stage, possible explanations may be the compensatory processes in response to toxic effects of Aβ (Supekar et al., 2008; Zhou et al., 2010; Matura et al., 2014) or the amyloid-induced inflammatory response (Montal et al., 2018), especially in Group 1. Such inflammation would trigger changes in cell volume (neuronal and glia swelling) as well as number (glia recruitment and activation) at the early stages of AD pathology and lead to the GM volume changes (Fortea et al., 2010; Rodriguez-Vieitez et al., 2016). As the neuropathological progression, the synergy between amyloid and tau pathologies starts (Schultz et al., 2017; Pascoal et al., 2017), such compensation mechanism become gradually invalid, ending with the whole structural connectivity in chaos. This could be proved the progressively decreased cognition in Group 2 and Group 3, reflecting the progressive decompensation process.
Conclusively, our study suggested a dynamic SCN pattern changes along the temporal pathological accumulation. As the AD progresses, the DMN and SN showed phases of hyperconnectivity and then hypoconnectivity while ECN showed the potentially compensatory role of the ECN in AD patients.
5. Limitation
There were some limitations to this study. Firstly, our study is cross-sectional research. Although we try to use the subjects with different disease stages to depict the AD continuum, a further longitudinal study should be done. Moreover, our SCN analysis was constructed in a group level which may not allow the assessment of the relationship between single-subject gray matter network measures with cognition. Further SCN analysis based on single-subject should be done.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance by the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Written informed consent was obtained from all participants and authorized representatives, and the study partners before any protocol-specific procedures were carried out in the ADNI study. More details in http://www.adni-info.org.
Consent for publication
Not applicable.
Availability of data and material
The datasets generated and analyzed during the current study are available in the ADNI study. More details in www.adni-info.org.
Acknowledgments
KL designed the study and wrote the first draft of the manuscript. XL analyzed the MRI data and wrote the protocol. QZ and JX collected clinical and MRI data. JZ, CW, XX, PH, ZS and MZ assisted with study design and interpretation of findings. All authors have contributed to and approved the final manuscript. All authors read and approved the final manuscript. The authors declare no conflict of interest.
This study was funded by National Key Research and Development Program of China (Grant No. 2016YFC1306600), Zhejiang Provincial Natural Science Foundation of China (Grant No LQ19H180006), Young Research Talents Fund, Chinese Medicine Science, and Technology Project of Zhejiang Province (Grant No. 2018ZQ035), the Fundamental Research Funds for the Central Universities (No.2017XZZX001-01), Zhejiang Medicine and Health Science and Technology Program (2018KY418 and 2016KYA099). Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.;Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; ElanPharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & D development t, LLC.; J Johnson & Johnson PharmaceuticalResearch & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; MesoScaleDiagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis PharmaceuticalsCorporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; andTransition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego.ADNI data are disseminated by the Laboratory for NeuroImaging at the University ofSouthern California.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2019.101828.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- Agosta F., Pievani M., Geroldi C., Copetti M., Frisoni G.B., Filippi M. Resting state fMRI in Alzheimer's disease: beyond the default mode network. Neurobiol. Aging. 2012;33:1564–1578. doi: 10.1016/j.neurobiolaging.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Alexander G.E., Bergfield K.L., Chen K., Reiman E.M., Hanson K.D., Lin L., Bandy D., Caselli R.J., Moeller J.R. Gray matter network associated with risk for Alzheimer's disease in young to middle-aged adults. Neurobiol. Aging. 2012;33:2723–2732. doi: 10.1016/j.neurobiolaging.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A., Giedd J.N., Bullmore E. Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci. 2013;14:322e336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A., Raznahan A., Bullmore E., Giedd J. The convergence of maturational change and structural covariance in human cortical networks. J. Neurosci. 2013;33:2889–2899. doi: 10.1523/JNEUROSCI.3554-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G., Barnard H., McColl R., Hester A.L., Fields J.A., Weiner M.F., Ringe W.K., Lipton A.M., Brooker M., McDonald E., Rubin C.D., Cullum C.M. Reduced hippocampal functional connectivity in Alzheimer disease. Arch. Neurol. 2007;64:1482–1487. doi: 10.1001/archneur.64.10.1482. [DOI] [PubMed] [Google Scholar]
- Andrews-Hanna J.R., Reidler J.S., Sepulcre J., Poulin R., Buckner R.L. Functional-anatomic fractionation of the brain's default network. Neuron. 2010;65:550–562. doi: 10.1016/j.neuron.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangen KJ Clark A.L., Werhane M., Edmonds E.C., Nation D.A., Evangelista N., Libon D.J., Bondi M.W., Delano-Wood L., Alzheimer's Disease Neuroimaging Initiative Cortical amyloid burden differences across empirically-derived mild cognitive impairment subtypes and interaction with APOE ɛ4 genotype. J. Alzheimers Dis. 2016;52:849–861. doi: 10.3233/JAD-150900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D.S., Bullmore E.T. Human brain networks in health and disease. Curr. Opin. Neurol. 2009;22:340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt B.C., Worsley K.J., Besson P., Concha L., Lerch J.P., Evans A.C., Bernasconi N. Mapping limbic network organization in temporal lobe epilepsy using morphometric correlations: insights on the relation between mesiotemporal connectivity and cortical atrophy. Neuroimage. 2008;42:515–524. doi: 10.1016/j.neuroimage.2008.04.261. [DOI] [PubMed] [Google Scholar]
- Bi G., Poo M. Distributed synaptic modification in neural networks induced by patterned stimulation. Nature. 1999;401:792–796. doi: 10.1038/44573. [DOI] [PubMed] [Google Scholar]
- Bohbot V.D., Lerch J., Thorndycraft B., Iaria G., Zijdenbos A.P. Gray matter differences correlate with spontaneous strategies in a human virtual navigation task. J. Neurosci. 2007;27:10078–10083. doi: 10.1523/JNEUROSCI.1763-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondi M.W., Edmonds E.C., Jak A.J., Clark L.R., DelanoWood L., McDonald C.R., Nation D.A., Libon D.J., Au R., Galasko D., Salmon D.P. Neuropsychological criteria for mild cognitive impairment improves diagnostic precision, biomarker associations, and progression rates. J. Alzheimers Dis. 2014;42:275–289. doi: 10.3233/JAD-140276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier M.R., Thomas J.B., Snyder A.Z., Benzinger T.L., Zhang D., Raichle M.E., Holtzman D.M., Morris J.C., Ances B.M. Loss of intranetwork and internetwork resting state functional connections with Alzheimer's disease progression. J. Neurosci. 2012;32:8890–8899. doi: 10.1523/JNEUROSCI.5698-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R.L., Snyder A.Z., Shannon B.J., LaRossa G., Sachs R., Fotenos A.F., Sheline Y.I., Klunk W.E., Mathis C.A., Morris J.C., Mintun M.A. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J. Neurosci. 2005;25:7709e7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R.L., Sepulcre J., Talukdar T., Krienen F.M., Liu H., Hedden T., Andrews-Hanna J.R., Sperling R.A., Johnson K.A. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J. Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.T., Hsu S.W., Tsai S.J., Chang Y.T., Huang C.W., Liu M.E., Chen N.C., Chang W.N., Hsu J.L., Lee C.C., Chang C.C. Genetic effect of MTHFR C677T polymorphism on the structural covariance network and white-matter integrity in Alzheimer's disease. Hum. Brain Mapp. 2017;38:3039–3051. doi: 10.1002/hbm.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C., Chang Y.T., Huang C.W., Tsai S.J., Hsu S.W., Huang S.H., Lee C.C., Chang W.N., Lui C.C., Lien C.Y. Associations of Bcl-2 rs956572 genotype groups in the structural covariance network in early-stage Alzheimer's disease. Alzheimers Res. Ther. 2018;10:17. doi: 10.1186/s13195-018-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.H., Fiecas M., Gutiérrez E.D., Panizzon M.S., Eyler L.T., Vuoksimaa E., Thompson W.K., Fennema-Notestine C., Hagler D.J., Jr., Jernigan T.L., Neale M.C., Franz C.E., Lyons M.J., Fischl B., Tsuang M.T., Dale A.M., Kremen W.S. Genetic topography of brain morphology. Proc. Natl. Acad. Sci. U. S. A. 2013;15 doi: 10.1073/pnas.1308091110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbeuck X., Van der Linden M., Collette F. Alzheimer's disease as a disconnection syndrome? Neuropsychol. Rev. 2003;13:79–92. doi: 10.1023/a:1023832305702. [DOI] [PubMed] [Google Scholar]
- Dicks E., Tijms B.M., Ten Kate M., Gouw A.A., Benedictus M.R., Teunissen C.E., Barkhof F., Scheltens P., van der Flier W.M. Gray matter network measures are associated with cognitive decline in mild cognitive impairment. Neurobiol. Aging. 2018;61:198–206. doi: 10.1016/j.neurobiolaging.2017.09.029. [DOI] [PubMed] [Google Scholar]
- Edmonds E.C., Delano-Wood L., Galasko D.R., Salmon D.P., Bondi M.W., Alzheimer's Disease Neuroimaging Initiative Subjective cognitive complaints contribute to misdiagnosis of mild cognitive impairment. J. Int. Neuropsychol. Soc. 2014;20:836–847. doi: 10.1017/S135561771400068X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S.B., Stephan K.E., Mohlberg H., Grefkes C., Fink G.R., Amunts K., Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Van Essen D.C. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Fortea J., Sala-Llonch R., Bartrés-Faz D., Bosch B., Lladó A., Bargalló N., Molinuevo J.L., Sánchez-Valle R. Increased cortical thickness and caudate volume precede atrophy in PSEN1 mutation carriers. J. Alzheimers Dis. 2010;22:909–922. doi: 10.3233/JAD-2010-100678. [DOI] [PubMed] [Google Scholar]
- Garces P., Angel Pineda-Pardo J., Canuet L., Aurtenetxe S., Lopez M.E., Marcos A., Yus M., Llanero-Luque M., Del-Pozo F., Sancho M., Maestu F. The default mode network is functionally and structurally disrupted in amnestic mild cognitive impairment - a bimodal MEG-DTI study. Neuroimage Clin. 2014;6:214–221. doi: 10.1016/j.nicl.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gili T., Cercignani M., Serra L., Perri R., Giove F., Maraviglia B., Caltagirone C., Bozzali M. Regional brain atrophy and functional disconnection across Alzheimer's disease evolution. J. Neurol. Neurosurg. Psychiatry. 2011;82:58–66. doi: 10.1136/jnnp.2009.199935. [DOI] [PubMed] [Google Scholar]
- Gong G., He Y., Chen Z.J., Evans A.C. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. Neuroimage. 2012;59:1239–1248. doi: 10.1016/j.neuroimage.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Gong G., He Y., Chen Z., Evans A. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. Neuroimage. 2012;59:1239–1248. doi: 10.1016/j.neuroimage.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Grady C.L., McIntosh A.R., Beig S., Keightley M.L., Burian H., Black S.E. Evidence from functional neuroimaging of a compensatory prefrontal network in Alzheimer's disease. J. Neurosci. 2003;23:986–993. doi: 10.1523/JNEUROSCI.23-03-00986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wang L., Zang Y., Tian L., Zhang X., Li K., Jiang T. Regional coherence changes in the early stages of Alzheimer's disease: a combined structural and resting-state functional MRI study. Neuroimage. 2007;35:488–500. doi: 10.1016/j.neuroimage.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., Shaw L.M., Vemuri P., Wiste H.J., Weigand S.D., Lesnick T.G., Pankratz V.S., Donohue M.C., Trojanowski J.Q. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Chetelat G., Dickson D., Fagan A.M., Frisoni G.B., Jagust W., Mormino E.C., Petersen R.C., Sperling R.A., van der Flier W.M., Villemagne V.L., Visser P.J., Vos S.J. Suspected non-Alzheimer disease pathophysiology--concept and controversy. Nat. Rev. Neurol. 2016;12:117–124. doi: 10.1038/nrneurol.2015.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J., Liu E., Molinuevo J.L., Montine T., Phelps C., Rankin K.P., Rowe C.C., Scheltens P., Siemers E., Snyder H.M., Sperling R. NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L.C., Shatz C.J. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Klunk W.E., Engler H., Nordberg A., Wang Y., Blomqvist G., Holt D.P., Bergstrom M., Savitcheva I., Huang G.F., Estrada S., Ausen B., Debnath M.L., Barletta J., Price J.C., Sandell J., Lopresti B.J., Wall A., Koivisto P., Antoni G., Mathis C.A., Langstrom B. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann. Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- Knobloch M., Mansuy I.M. Dendritic spine loss and synaptic alterations in Alzheimer's disease. Mol. Neurobiol. 2008;37:73–82. doi: 10.1007/s12035-008-8018-z. [DOI] [PubMed] [Google Scholar]
- Koch K., Myers N.E., Gottler J., Pasquini L., Grimmer T., Forster S., Manoliu A., Neitzel J., Kurz A., Forstl H., Riedl V., Wohlschlager A.M., Drzezga A., Sorg C. Disrupted intrinsic networks link amyloid-beta pathology and impaired cognition in prodromal Alzheimer's disease. Cereb. Cortex. 2015;25:4678–4688. doi: 10.1093/cercor/bhu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch J.P., Worsley K., Shaw W.P., Greenstein D.K., Lenroot R.K., Giedd J., Evans A.C. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Lin P.H., Tsai S.J., Huang C.W., Mu-En L., Hsu S.W., Lee C.C., Chen N.C., Chang Y.T., Lan M.Y., Chang C.C. Dose-dependent genotype effects of BDNF Val66Met polymorphism on default mode network in early stage Alzheimer's disease. Oncotarget. 2016;7:54200–54214. doi: 10.18632/oncotarget.11027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X Qiu T., Xu X., Huang P., Gu Q., Shen Z., Yu X., Jia Y., Guan X., Song R., Zhang M., Alzheimer's Disease Neuroimaging Initiative Decreased inter-hemispheric functional connectivity in cognitively intact elderly APOE ɛ4 carriers: a preliminary study. J. Alzheimers Dis. 2016;50:1137–1148. doi: 10.3233/JAD-150989. [DOI] [PubMed] [Google Scholar]
- Luo X., Qiu T., Jia Y., Huang P., Xu X., Yu X., Shen Z., Jiaerken Y., Guan X., Zhou J., Zhang M., ADNI Intrinsic functional connectivity alterations in cognitively intact elderly APOE ε4 carriers measured by eigenvector centrality mapping are related to cognition and CSF biomarkers: a preliminary study. Brain Imaging Behav. 2016;11:1290–1301. doi: 10.1007/s11682-016-9600-z. [DOI] [PubMed] [Google Scholar]
- Luo X., Li K., Jia Y.L., Zeng Q., Jiaerken Y., Qiu T., Huang P., Xu X., Shen Z., Guan X., Zhou J., Wang C., Xu J.J., Zhang M., Alzheimer's Disease Neuroimaging Initiative Altered effective connectivity anchored in the posterior cingulate cortex and the medial prefrontal cortex in cognitively intact elderly APOE epsilon4 carriers: a preliminary study. Brain Imaging Behav. 2018;13:270–282. doi: 10.1007/s11682-018-9857-5. [DOI] [PubMed] [Google Scholar]
- Luo X., Li K., Zeng Q., Huang P., Jiaerken Y., Qiu T., Xu X., Zhou J., Xu J., Zhang M. Decreased bilateral FDG-PET uptake and inter-hemispheric connectivity in multi-domain amnestic mild cognitive impairment patients: a preliminary study. Front. Aging Neurosci. 2018;10:161. doi: 10.3389/fnagi.2018.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matura S., Prvulovic D., Butz M., Hartmann D., Sepanski B., Linnemann K., Oertel-Knochel V., Karakaya T., Fusser F., Pantel J., van de Ven V. Recognition memory is associated with altered resting-state functional connectivity in people at genetic risk for Alzheimer's disease. Eur. J. Neurosci. 2014;40:3128–3135. doi: 10.1111/ejn.12659. [DOI] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Mechelli A., Friston K.J., Frackowiak R.S., Price C.J. Structural covariance in the human cortex. J. Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer G., Shapiro F., Vanderstichele H., Vanmechelen E., Engelborghs S., De Deyn P. P., Coart E., Hansson O., Minthon L., Zetterberg H., Blennow K., Shaw L., Trojanowski J. Q., Alzheimer's Disease Neuroimaging Initiative (2010) Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch. Neurol. 67, 949–956. [DOI] [PMC free article] [PubMed]
- Molinuevo J.L., Ripolles P., Simo M., Llado A., Olives J., Balasa M., Antonell A., Rodriguez-Fornells A., Rami L. White matter changes in preclinical Alzheimer's disease: a magnetic resonance imaging-diffusion tensor imaging study on cognitively normal older people with positive amyloid beta protein 42 levels. Neurobiol. Aging. 2014;35:2671–2680. doi: 10.1016/j.neurobiolaging.2014.05.027. [DOI] [PubMed] [Google Scholar]
- Montal V., Vilaplana E., Alcolea D., Pegueroles J., Pasternak O., González-Ortiz S., Clarimón J., Carmona-Iragui M., Illán-Gala I., Morenas-Rodríguez E., Ribosa-Nogué R., Sala I., Sánchez-Saudinós M.B., García-Sebastian M., Villanúa J., Izagirre A., Estanga A., Ecay-Torres M., Iriondo A., Clerigue M., Tainta M., Pozueta A., González A., Martínez-Heras E., Llufriu S., Blesa R., Sanchez-Juan P., Martínez-Lage P., A1 Lleó, Fortea J. Cortical microstructural changes along the Alzheimer's disease continuum. Alzheimers Dement. 2018;14:340–351. doi: 10.1016/j.jalz.2017.09.013. [DOI] [PubMed] [Google Scholar]
- Montembeault M., Rouleau I., Provost J.S., Brambati S.M., Alzheimer's Disease Neuroimaging Initiative Altered gray matter structural covariance networks in early stages of Alzheimer's disease. Cereb. Cortex. 2016;26:2650–2662. doi: 10.1093/cercor/bhv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N., Pasquini L., Gottler J., Grimmer T., Koch K., Ortner M., Neitzel J., Muhlau M., Forster S., Kurz A., Forstl H., Zimmer C., Wohlschlager A.M., Riedl V., Drzezga A., Sorg C. Within-patient correspondence of amyloid-beta and intrinsic network connectivity in Alzheimer's disease. Brain. 2014;137:2052–2064. doi: 10.1093/brain/awu103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist Sebastian, Schöll Michael, Strandberg Olof, Mattsson Niklas, Stomrud Erik, Zetterberg Henrik, Blennow Kaj, Landau Susan, Jagust William, Hansson Oskar. Earliest accumulation of β-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat. Commun. 2017;8:1214. doi: 10.1038/s41467-017-01150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal T.A., Mathotaarachchi S., Mohades S., Benedet A.L., Chung C.O., Shin M., Wang S., Beaudry T., Kang M.S., Soucy J.P., Labbe A., Gauthier S., Rosa-Neto P. Amyloid-β and hyperphosphorylated tau synergy drives metabolic decline in preclinical Alzheimer's disease. Mol. Psychiatry. 2017;22:306–311. doi: 10.1038/mp.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine A.M., Adluru N., Alexander A.L., Christian B.T., Okonkwo O.C., Oh J., Cleary C.A., Birdsill A., Hillmer A.T., Murali D., Barnhart T.E., Gallagher C.L., Carlsson C.M., Rowley H.A., Dowling N.M., Asthana S., Sager M.A., Bendlin B.B., Johnson S.C. Associations between white matter microstructure and amyloid burden in preclinical Alzheimer's disease: a multimodal imaging investigation. Neuroimage Clin. 2014;4:604–614. doi: 10.1016/j.nicl.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Kuceyeski A., Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73:1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Vieitez E., Saint-Aubert L., Carter S.F., Almkvist O., Farid K., Schöll M., Chiotis K., Thordardottir S., Graff C., Wall A., Långström B., Nordberg A. Diverging longitudinal changes in astrocytosis and amyloid PET in autosomal dominant Alzheimer's disease. Brain. 2016;139:922–936. doi: 10.1093/brain/awv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J.E., Lenroot R.K., Ordaz S.E., Wallace G.L., Lerch J.P., Evans A.C., Prom E.C., Kendler K.S., Neale M.C., Giedd J.N. Variance decomposition of MRIbased covariance maps using genetically informative samples and structural equation modeling. Neuroimage. 2009;47:56–64. doi: 10.1016/j.neuroimage.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A.P., Chhatwal J.P., Hedden T., Mormino E.C., Hanseeuw B.J., Sepulcre J., Huijbers W., LaPoint M., Buckley R.F., Johnson K.A., Sperling R.A. Phases of Hyperconnectivity and Hypoconnectivity in the default mode and salience networks track with amyloid and tau in clinically Normal individuals. J. Neurosci. 2017;37:4323–4331. doi: 10.1523/JNEUROSCI.3263-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley W.W., Menon V., Schatzberg A.F., Keller J., Glover G.H., Kenna H., Reiss A.L., Greicius M.D. Dissociable intrinsic connectivity networks for salience processing and executive control. J. Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley W.W., Crawford R.K., Zhou J., Miller B.L., Greicius M.D. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J.M., Allen E.A., Jung R.E., Erhardt E.B., Arja S.K., Kiehl K., Calhoun V.D. Correspondence between structure and function in the human brain at rest. Front Neuroinform. 2012;6:10. doi: 10.3389/fninf.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L.M., Vanderstichele H., Knapik-Czajka M., Clark C.M., Aisen P.S., Petersen R.C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V.M., Trojanowski J.Q., Alzheimer's Disease Neuroimaging Initiative Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J.I., Yesavage J.A. Geriatric depression scale (GDS): Recent evidence and development of a shorter version. In: Brink T.L., editor. Clinical Gerontology: A Guide to Assessment and Intervention. The Haworth Press; New York, NY: 1986. pp. 165–173. [Google Scholar]
- Sperling R., Mormino E., Johnson K. The evolution of preclinical Alzheimer's disease: implications for prevention trials. Neuron. 2014;84:608–622. doi: 10.1016/j.neuron.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng R.N., Turner G.R. Structural covariance of the default network in healthy and pathological aging. J. Neurosci. 2013;33:15226–15234. doi: 10.1523/JNEUROSCI.2261-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve and Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2006;20:S69–S74. doi: 10.1097/00002093-200607001-00010. [DOI] [PubMed] [Google Scholar]
- Supekar K., Menon V., Rubin D., Musen M., Greicius M.D. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms BM Kate M.T., Wink A.M., Visser P.J., Ecay M., Clerigue M., Estanga A., Garcia Sebastian M., Izagirre A., Villanua J., Martinez Lage P., van der Flier W.M., Scheltens P., Sanz Arigita E., Barkhof F. Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiol. Aging. 2016:154–160. doi: 10.1016/j.neurobiolaging.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Tijms B.M., Series P., Willshaw D.J., Lawrie S.M. Similarity-based extraction of individual networks from gray matter MRI scans. Cereb. Cortex. 2012;22:1530–1541. doi: 10.1093/cercor/bhr221. [DOI] [PubMed] [Google Scholar]
- Uddin L.Q., Kelly A.M., Biswal B.B., Castellanos F.X., Milham M.P. Functional connectivity of default mode network components: correlation, anticorrelation, and causality. Hum. Brain Mapp. 2009;30:625–637. doi: 10.1002/hbm.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O., Szoeke C., Macaulay S.L., Martins R., Maruff P., Ames D., Rowe C.C., Masters C.L., Australian Imaging Biomarkers, Lifestyle Research Group Amyloid beta deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- Weiler M., Fukuda A., Massabki L.H., Lopes T.M., Franco A.R., Damasceno B.P., Cendes F., Balthazar M.L. Default mode, executive function, and language functional connectivity networks are compromised in mild Alzheimer's disease. Curr. Alzheimer Res. 2014;11:274–282. doi: 10.2174/1567205011666140131114716. [DOI] [PubMed] [Google Scholar]
- Zhang H.Y., Wang S.J., Liu B., Ma Z.L., Yang M., Zhang Z.J., Teng G.J. Resting brain connectivity: changes during the progress of Alzheimer disease. Radiology. 2010;256:598–606. doi: 10.1148/radiol.10091701. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liao W., Zuo X.N., Wang Z., Yuan C., Jiao Q., Chen H., Biswal B.B., Lu G., Liu Y. Resting-state brain organization revealed by functional covariance networks. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Greicius M.D., Gennatas E.D., Growdon M.E., Jang J.Y., Rabinovici G.D., Kramer J.H., Weiner M., Miller B.L., Seeley W.W. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer's disease. Brain. 2010;133:1352–1367. doi: 10.1093/brain/awq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gennatas E.D., Kramer J.H., Miller B.L., Seeley W.W. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhou P., Alcauter S., Chen Y., Cao H., Tian M., Ming D., Qi H., Wang X., Zhao X., He F., Ni H., Gao W. Changes of intranetwork and internetwork functional connectivity in Alzheimer's disease and mild cognitive impairment. J. Neural Eng. 2016;13 doi: 10.1088/1741-2560/13/4/046008. [DOI] [PubMed] [Google Scholar]
- Zielinski B.A., Gennatas E.D., Zhou J., Seeley W.W. Network-level structural covariance in the developing brain. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18191–18196. doi: 10.1073/pnas.1003109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski B.A., Anderson J.S., Froehlich A.L., Prigge M.B., Nielsen J.A., Cooperrider J.R., Cariello A.N., Fletcher P.T., Alexander A.L., Lange N., Bigler E.D., Lainhart J.E. scMRI reveals large-scale brain network abnormalities in autism. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Data Availability Statement
The datasets generated and analyzed during the current study are available in the ADNI study. More details in www.adni-info.org.