Abstract
The RNA‐binding protein ALYREF plays key roles in nuclear export and also 3′‐end processing of polyadenylated mRNAs, but whether such regulation also extends to non‐polyadenylated RNAs is unknown. Replication‐dependent (RD)‐histone mRNAs are not polyadenylated, but instead end in a stem‐loop (SL) structure. Here, we demonstrate that ALYREF prevalently binds a region next to the SL on RD‐histone mRNAs. SL‐binding protein (SLBP) directly interacts with ALYREF and promotes its recruitment. ALYREF promotes histone pre‐mRNA 3′‐end processing by facilitating U7‐snRNP recruitment through physical interaction with the U7‐snRNP‐specific component Lsm11. Furthermore, ALYREF, together with other components of the TREX complex, enhances histone mRNA export. Moreover, we show that 3′‐end processing promotes ALYREF recruitment and histone mRNA export. Together, our results point to an important role of ALYREF in coordinating 3′‐end processing and nuclear export of non‐polyadenylated mRNAs.
Keywords: 3′‐end processing, ALYREF, mRNA export, RD‐histone mRNA, SLBP
Subject Categories: RNA Biology
Introduction
In eukaryotes, most pre‐mRNAs undergo 3′ processing in a coupled cleavage/polyadenylation (CPA) reaction, which consists of an endonucleolytic cleavage of the nascent RNAs followed by synthesis of the polyA tail (reviewed in Shi & Manley, 2015). In contrast, replication‐dependent (RD)‐histone pre‐mRNAs, which are synthesized during S‐phase of the cell cycle, undergo one‐step U7‐snRNP‐dependent 3′‐end cleavage and end in a stem‐loop (SL) structure (Mowry & Steitz, 1987; Cotten et al, 1988; Marzluff, 2005; Dominski & Marzluff, 2007; Marzluff et al, 2008). The SL structure is recognized by SL‐binding protein (SLBP), which stabilizes the interaction of U7‐snRNP with the histone downstream element (HDE) of the pre‐mRNA (Schaufele et al, 1986; Williams & Marzluff, 1995; Dominski et al, 1999; Battle & Doudna, 2001; Skrajna et al, 2017). Together, SLBP and U7‐snRNP direct RNA cleavage 4–5 nt downstream from the SL. It remains unclear how SLBP facilitates U7‐snRNP recruitment, as no physical interaction was identified between these critical processing factors.
mRNAs are transported from the nucleus to the cytoplasm through a pathway that utilizes NXF1 as an export receptor. NXF1 is recruited to polyadenylated mRNAs mostly via the highly conserved TREX complex (TREX; Strasser & Hurt, 2000; Hautbergue et al, 2008; Katahira et al, 2009; Hung et al, 2010; Viphakone et al, 2012). The core of TREX mainly comprises three parts: the RNA‐binding protein ALYREF, the RNA helicase UAP56/URH49, and the multi‐subunit THO sub‐complex (THO1/2/3/5/6/7; Strasser et al, 2002; Masuda et al, 2005; Chi et al, 2013). Among these, ALYREF and THO mediate the interaction between the mRNA and NXF1 (Strasser & Hurt, 2000; Hautbergue et al, 2008; Katahira et al, 2009; Hung et al, 2010; Viphakone et al, 2012). Different from polyadenylated mRNAs, RD‐histone mRNAs are known to recruit NXF1 via SR proteins 9G8 and SRp20, which bind to specific cis‐elements in the coding regions (Huang & Steitz, 2001). In addition to SR proteins, SLBP also plays a critical role in RD‐histone mRNA export, but the underlying mechanism remains unknown (Sullivan et al, 2009).
The physical and functional coupling between 3′‐end processing and nuclear export has been well established for polyadenylated mRNAs. In yeast, the ALYREF homologue, Yra1, is recruited via the 3′‐end processing factor Pcf11 and regulates alternative polyadenylation (APA; Johnson et al, 2009, 2011). In mammalian cells, the THO subunits associate with the 3′‐end processing machinery and influence APA (Katahira et al, 2013; Tran et al, 2014). Further, ALYREF was recently found to bind to a 3′ region of polyadenylated mRNAs in a polyA‐binding protein (PABPN1)‐dependent manner (Shi et al, 2017). Thus, the coupling between 3′‐end processing and mRNA export seems to ensure efficient mRNA export and modulate APA. Currently, the roles of TREX in regulating the metabolism of non‐polyadenylated mRNAs remain unknown, and whether nuclear export is linked to 3′‐end processing for these mRNAs is still controversial. Initially, 3′‐end processing was shown to promote histone mRNA export (Eckner et al, 1991). However, later studies reported that the RNA length, but not 3′‐end processing, is a major determinant of histone mRNA export efficiency (Erkmann et al, 2005).
In this study, a deeper analysis of ALYREF iCLIP‐seq (individual‐nucleotide‐resolution UV crosslinking and immunoprecipitation and sequencing) data led us to the finding that ALYREF universally binds to a region next to the SL on RD‐histone mRNAs (for simplicity, histone mRNAs in this study). This binding is ensured by its direct interaction with SLBP and has two functional consequences: ALYREF stimulates proper histone mRNA 3′‐end formation by facilitating efficient U7‐snRNP recruitment and promotes histone mRNA export by enhancing NXF1 recruitment. Importantly, we demonstrate that 3′‐end processing promotes ALYREF recruitment and histone mRNA export. Thus, ALYREF is shared by polyadenylated and non‐polyadenylated mRNA metabolism pathways, coordinating processing and nuclear export of both types of mRNAs.
Results
ALYREF binds to a region 5′ of the SL on histone mRNAs
Our recent iCLIP study revealed that ALYREF binds to a region near the 3′ end of polyadenylated mRNAs in a nuclear polyA‐binding protein (PABPN1)‐dependent manner (Shi et al, 2017). To investigate whether ALYREF also binds to non‐polyadenylated RNAs, we further analyzed ALYREF iCLIP data. To increase resolution of the data, we identified clustered binding sites (Konig et al, 2010; Rossbach et al, 2014). Significantly, of the total 55 histone mRNAs that were easily detected in HeLa nuclei (Fan et al, 2017), clustered ALYREF binding sites were apparently and reproducibly detected on 52 (Fig 1A). This binding was not due to the high abundance of histone mRNAs, as compared to those in nuclear total RNA‐seq, histone reads were significantly more enriched in ALYREF iCLIP‐seq (Fig 1B). Note that such enrichment was not observed with short or long ncRNAs (Fig EV1A). ALYREF binding on histone mRNAs was confirmed by RNA immunoprecipitation (RIP) and RT–qPCRs (Fig 1C). In addition to ALYREF, TREX components UAP56 and THO were also easily detected on histone mRNAs (Fig EV1B). Note that the THOC2 antibody co‐precipitates other THO subunits (Masuda et al, 2005; Chi et al, 2013).
Figure 1. ALYREF binds to a region next to the SL on histone mRNAs.
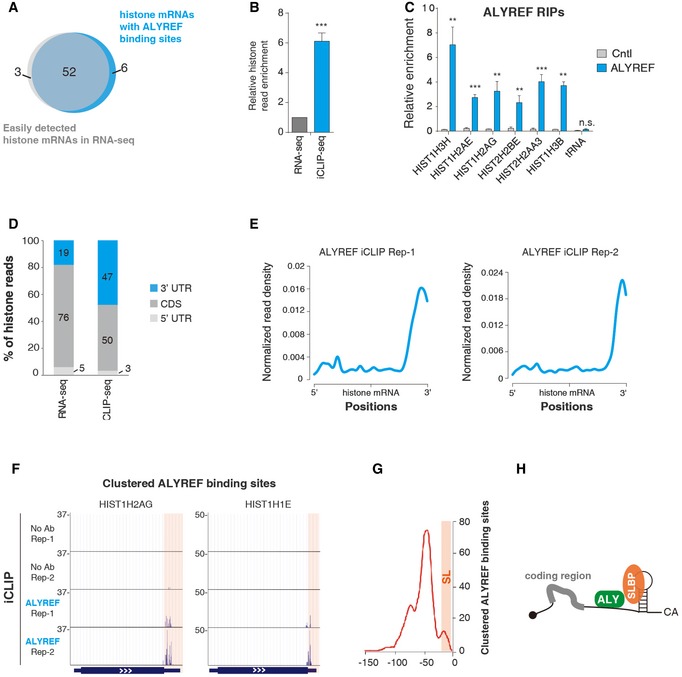
- The Venn diagram depicts the overlapping of histone mRNAs detected in a previously performed rRNA‐depleted nuclear RNA‐seq (RPM > 1) and in ALYREF iCLIP‐seq (with clustered binding sites in at least two out of three biological replicates).
- The ratio of unique histone iCLIP‐seq read population to unique histone RNA‐seq read population was calculated and is shown, with the unique histone RNA‐seq read population set as “1”. Error bars represent standard deviations from three replicates of ALYREF iCLIP data. Statistical analysis was performed using Student's t‐test. ***P < 0.001.
- ALYREF RIP–qPCRs to examine ALYREF binding on histone mRNAs. Relative RIP efficiencies are shown. A tRNA was used as a negative control for ALYREF binding. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student's t‐test. **P < 0.01, ***P < 0.001, n.s.: not significant.
- The distribution of histone reads in 5′ UTR, CDS, and 3′ UTR in rRNA‐depleted nuclear RNA‐seq and ALYREF iCLIP‐seq.
- Metagene analysis of normalized ALYREF signals on histone mRNA based on iCLIP‐seq data. iCLIP‐seq signal at each position of an mRNA is divided by the sum of signal of the mRNA to normalize each mRNA's contribution to the plot.
- Screenshots of two histone mRNAs showing clustered ALYREF binding sites. The 3′ UTR regions are highlighted.
- Distribution profile of clustered ALYREF binding sites on histone mRNA 3′ UTRs relative to the cleavage site (CS). The CS is set as “0” in the x‐axis, and the relative position to the CS is marked. The y‐axis shows the total occurrence of binding sites in three repeats at each nucleotide.
- Graphic displays that ALYREF binds next to SLBP on histone mRNAs.
Figure EV1. TREX associates with histone mRNAs.
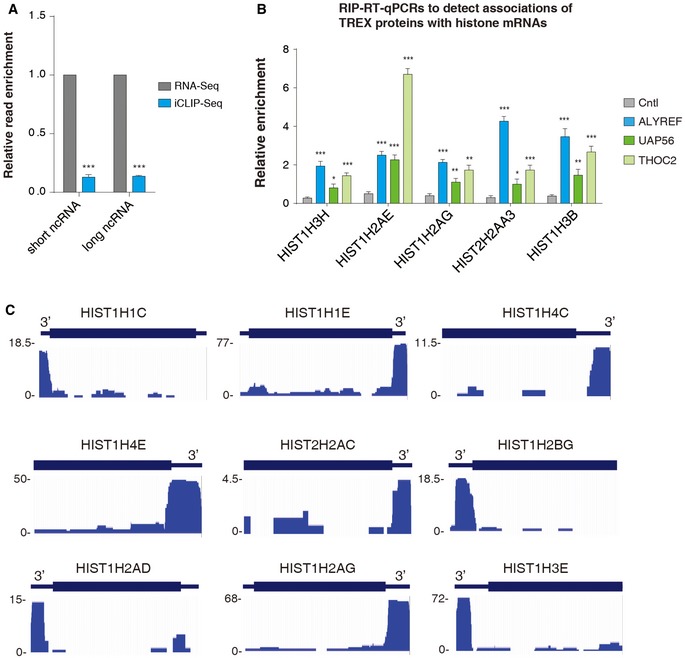
- The ratios of unique histone iCLIP‐seq read population to unique short or long ncRNA RNA‐seq read population were calculated and are shown, with the unique histone RNA‐seq read population set as “1”. Statistical analysis was performed based on three replicates of ALYREF iCLIP data using Student's t‐test. ***P < 0.001.
- ALYREF, UAP56, and THO RIP–qPCRs to examine their binding on multiple histone mRNAs. Relative RIP efficiencies are shown.
- Screenshots of several histone mRNAs with ALYREF iCLIP reads.
Relative to histone read distribution in RNA‐seq, ALYREF iCLIP reads are preferentially enriched in the 3′ UTR (Fig 1D). Interestingly, metagene analysis of iCLIP reads showed an apparent enrichment of ALYREF binding at the 3′ region of histone mRNAs (Fig 1E). Clustered ALYREF binding sites on two exemplified histone mRNAs, HIST1H2AG and HIST1H1E, are shown in Fig 1F, and ALYREF iCLIP read distribution on more histone mRNAs is shown in Fig EV1C. To understand the ALYREF binding principle at the 3′ region of histone mRNAs, we aligned clustered binding sites relative to the cleavage site (CS). Strikingly, ALYREF binds at a region spanning ~100 nt upstream of the CS, with the peak at −50 nt (Fig 1G). Considering that SLBP binds at the SL that is located at the −6~−21 region, our data demonstrate that ALYREF and SLBP bind adjacent to each other at the SL region of histone mRNAs (Fig 1H). In support of this view, a previous study identified an SLBP‐independent protected region 5′ of the SL (Brooks et al, 2015).
ALYREF interacts with SLBP in vivo and in vitro
Since histone mRNA 3′ UTR sequences are not conserved and ALYREF is a nonspecific RNA‐binding protein, it is possible that SLBP is a primary determinant of ALYREF binding neighboring to the SL. To test this possibility, we first determined whether ALYREF associates with SLBP by carrying out immunoprecipitations (IPs) from RNase A‐treated S‐phase HeLa cell lysate. Significantly, ALYREF was co‐precipitated by the SLBP antibody, but not the control IgG (Fig 2A). Conversely, when the ALYREF antibody or IgG was used for IPs, SLBP was specifically detected in the ALYREF immunoprecipitate (Fig 2B). These results indicate that ALYREF and SLBP associate with each other through protein–protein interaction. To confirm this association and examine whether UAP56 and THO associate with SLBP, we next carried out Flag IPs from Flag‐SLBP or Flag‐Cntl expression cells in the presence of RNase A. ALYREF and UAP56 and THOC2 were all co‐precipitated with Flag‐SLBP, but not Flag‐Cntl (Fig EV2A). These results indicate that ALYREF interacts with SLBP in the context of TREX.
Figure 2. SLBP interacts with ALYREF and facilitates its binding on histone mRNAs.
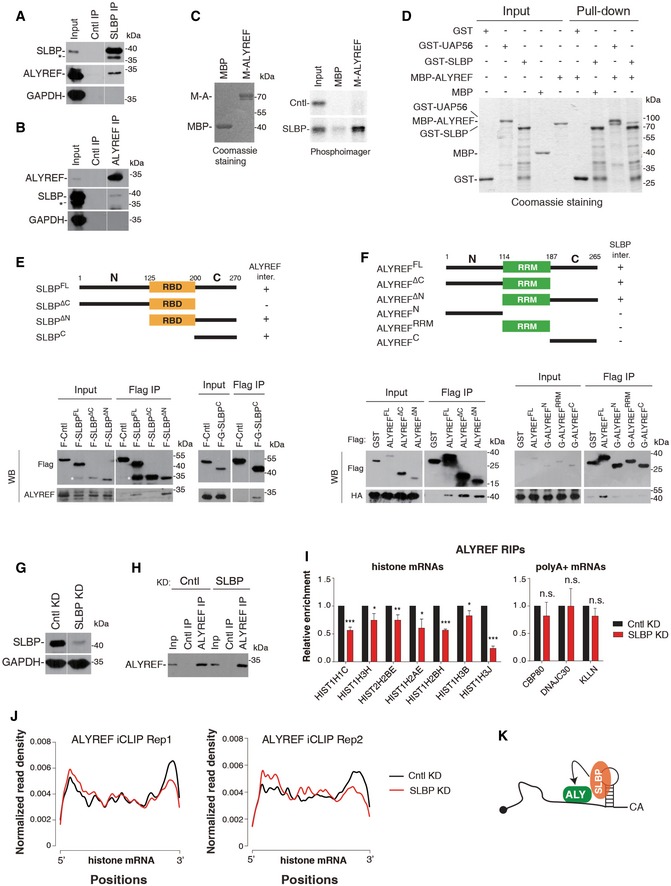
-
A, BIPs from RNase A‐treated S‐phase HeLa cell lysate using the SLBP antibody or IgG (A), or the ALYREF antibody or IgG (B). Western blotting was performed with the indicated antibodies. 2% of input was loaded. * indicates a nonspecific band. The white line delineates the boundary where irrelevant lanes have been removed from the same blots.
-
CPull‐downs of in vitro translated 35S‐labeled SLBP and luciferase (Cntl) using MBP or MBP‐ALYREF in the presence of RNase A. The proteins pulled down were visualized by Coomassie staining (left) or PhosphorImager (right). 3% of input was loaded.
-
DGST‐SLBP, GST‐UAP56, and GST were used for pull‐down of purified MBP‐ALYREF or MBP in the presence of RNase A. Proteins pulled down were separated by SDS–PAGE, followed by Coomassie staining. 37.5% of input proteins were loaded.
-
E(Top) Domain schematic representation of SLBP. (Bottom) Flag IPs from RNase A‐treated HeLa cell lysate individually expressing the indicated Flag‐tagged proteins, followed by Western blotting using Flag and ALYREF antibodies. 3% of input was loaded. * indicates a band that probably resulted from degradation of Flag‐SLBP. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
-
FSame as (E), except that Flag IPs were carried out from HA‐SLBP stable expression cells transfected with plasmids expressing ALYREF fragments.
-
GWestern blotting to examine the KD efficiency of SLBP. GAPDH was used as a loading control. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
-
H, ICntl‐ or SLBP siRNA‐treated HeLa cells were used for IPs with IgG or the ALYREF antibody. The immunoprecipitates were subjected to Western blot analysis (H) and RT–qPCRs (I). Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student's t‐test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s.: not significant.
-
JMetagene analysis of normalized ALYREF signals on histone mRNAs in Cntl and SLBP KD cells based on iCLIP‐seq data. iCLIP‐seq signal at each position of an mRNA is divided by the sum of signal of the mRNA to normalize each mRNA's contribution to the plot.
-
KGraphic displays that SLBP functions in recruiting ALYREF to histone mRNAs.
Source data are available online for this figure.
Figure EV2. TREX associates with SLBP.
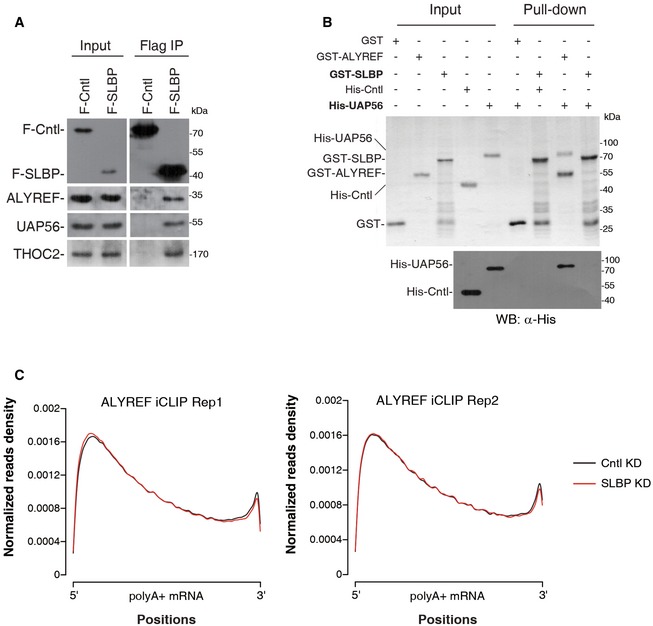
- Flag IPs from RNase A‐treated HeLa cell lysate expressing Flag‐Cntl (DDX3) or Flag‐SLBP, followed by Western blotting using the indicated antibodies. 3% of input was loaded.
- GST‐SLBP, GST‐ALYREF, and GST were used for pull‐down of purified His‐UAP56 or His‐Cntl (PGK1) in the presence of RNase A. Proteins pulled down were separated by SDS–PAGE, followed by Coomassie staining and Western blotting. 18.75% of input was loaded.
- Metagene analysis of normalized ALYREF signals on polyadenylated mRNAs in Cntl and SLBP KD cells based on two replicates of iCLIP‐seq data. iCLIP‐seq signal at each position of an mRNA is divided by the sum of signal of the mRNA to normalize each mRNA's contribution to the plot.
To further examine the ALYREF‐SLBP interaction, we in vitro translated SLBP and luciferase (Cntl) and carried out pull‐downs using MBP‐ALYREF or MBP. Significantly, SLBP, but not Cntl, was pulled down by MBP‐ALYREF, whereas neither of these in vitro translated proteins interacted with MBP (Fig 2C). This result provides additional evidence for the ALYREF‐SLBP interaction and suggests that this interaction might be direct. Indeed, GST‐SLBP, but not GST, pulled down purified MBP‐ALYREF (Fig 2D). In contrast, His‐UAP56 did not interact with GST‐SLBP, although it was efficiently pulled down by GST‐ALYREF (Fig EV2B). Together, these data demonstrate that ALYREF interacts with SLBP in vivo and in vitro.
SLBP interacts with ALYREF via its C‐terminal region
To decipher how SLBP interacts with ALYREF, we constructed two Flag‐SLBP mutants, with either the N‐ or C‐terminal region truncated (ΔN or ΔC; Fig 2E). These constructs, as well as Flag‐SLBPFL (full‐length SLBP) and Flag‐Cntl, were separately expressed in HeLa cells, followed by Flag IPs. Western blot analysis showed that ALYREF was efficiently co‐precipitated with Flag‐SLBPFL and Flag‐SLBPΔN, but not Flag‐SLBPΔC or Flag‐Cntl, indicating that the C‐terminal region (SLBPC) is required for interacting with ALYREF (Fig 2E, left lower panel). Further, using Flag‐GST‐SLBPC, we found that SLBPC is sufficient for this interaction (Fig 2E, right lower panel). Thus, SLBP interacts with ALYREF via its C‐terminal region. Notably, this region is important for facilitating U7‐snRNP recruitment (Skrajna et al, 2017).
We also mapped the interaction of ALYREF with SLBP. Neither N‐ nor C‐terminal region of ALYREF is required for the SLBP interaction (Fig 2F, left lower panel), raising the possibility that the RRM domain might be important. However, when ALYREF was separated into N‐terminal, RRM, and C‐terminal fragments, none of them was associated with SLBP (Fig 2F, right lower panel). It is possible that both the RRM and the surrounding residues are important for ALYREF interacting with SLBP.
SLBP is required for ALYREF binding at the SL region on histone mRNAs
To examine whether SLBP is required for ALYREF binding on histone mRNAs, we carried out ALYREF RIP in Cntl‐ and SLBP‐knockdown (KD) cells. SLBP was efficiently knocked down, and ALYREF was equally immunoprecipitated from these cells (Fig 2G and H). Significantly, RT–qPCR data showed that ALYREF association with histone mRNAs was reproducibly reduced in SLBP KD cells, as compared to that in Cntl cells (Fig 2I). In contrast, its binding with the three randomly picked polyadenylated mRNAs, including CBP80, DNAJC30, and KLLN, was not apparently affected (Fig 2I). To examine how SLBP KD impacts ALYREF distribution along the histone mRNA, we next carried out ALYREF iCLIP in Cntl and SLBP KD cells. Low RNase I digestion efficiency could possibly result in biased iCLIP read enrichment at the SL region. We have thus optimized the experiment and ensured the digestion efficiency of the 3′ region containing the SL is not generally lower than that of a 5′ region of histone mRNAs (Appendix Fig S1; See Materials and Methods). Under this condition, in Cntl cells, ALYREF binding was still mostly enriched at the 3′ region, while it was also partially detected at other regions (Fig 2J). Significantly, in both biological replicates, SLBP KD led to a preferential reduction in ALYREF binding at the 3′ region (Fig 2J). In contrast, ALYREF distribution along the polyadenylated mRNA was not apparently affected (Fig EV2C). Taken together, the data suggest that SLBP plays a determinant role in ALYREF binding at the SL region of histone mRNAs (Fig 2K).
ALYREF facilitates 3′‐end processing of histone pre‐mRNAs
Considering the critical roles of SLBP in histone mRNA 3′‐end formation, we reasoned that ALYREF might also be involved in this process. RD‐histone genes have a canonical polyadenylation signal downstream of the CS. Defects in histone mRNA 3′‐end processing usually cause the usage of this signal, resulting in upregulation in polyA+ forms and concomitant downregulation in polyA− forms (Narita et al, 2007; Yang et al, 2009; Romeo et al, 2014; Brodersen et al, 2016). We thus isolated polyA+ and polyA− RNAs from Cntl and ALYREF KD cells and carried out RNA‐seq separately (Figs 3A and EV3A). In the polyA− dataset, 27 histone genes showed apparent downregulation (< 0.7‐fold) in ALYREF KD (Dataset EV1). Among these, 12 were significantly upregulated (> 1.5‐fold) in the polyA+ dataset (Fig 3B), indicating that 3′‐mRNA processing of these genes was disrupted by ALYREF KD. Two exemplified histone mRNAs, HIST2H2BE and HIST1H1C, with 3′‐end processing defects are shown in Fig 3C.
Figure 3. ALYREF is required for proper 3′‐end processing of histone pre‐mRNAs.
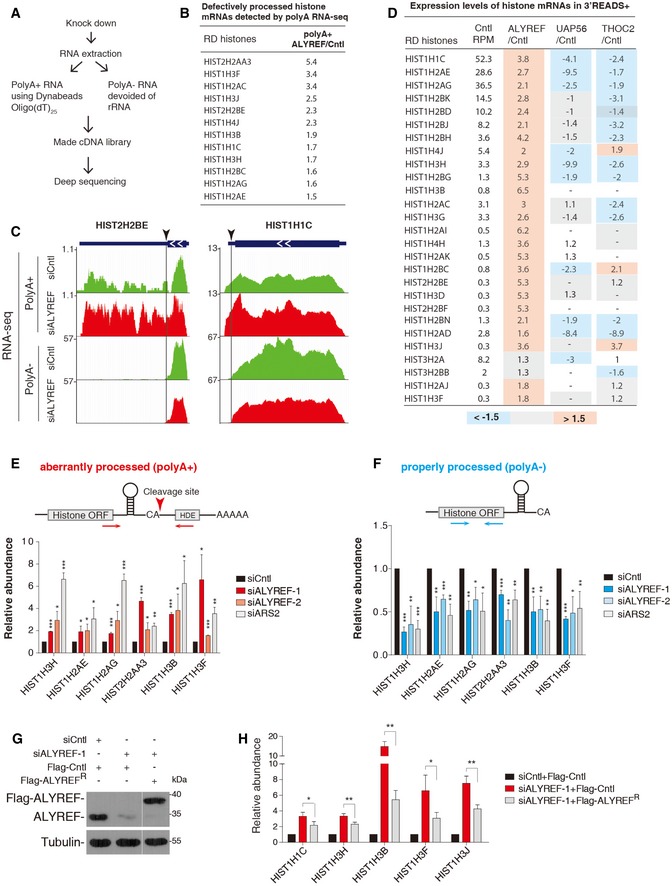
-
AA diagram of the RNA‐seq experimental approach.
-
BList of histone genes with defective 3′‐end processing detected by polyA+ and polyA− RNA‐seq. PolyA+ ALYREF/Cntl shows polyA+ RNA‐seq RPM ratio of each histone gene in ALYREF KD versus Cntl.
-
CScreenshots of two exemplified histone mRNAs with 3′‐end processing defects are shown. Transcription direction is indicated. Black arrowheads and the corresponding lines mark the position of CS.
-
DRPM of each histone mRNA in Cntl cells and RPM ratio of each histone mRNA in KDs to Cntl detected in 3′READS+.
-
E, FRT–qPCRs to detect histone mRNA levels in the polyA+ (E) or the polyA− fraction (F) in cells treated with Cntl, ALYREF, or ARS2 siRNA. The arrowhead and arrows indicate the cleavage site and primer location, respectively. The bars show the relative abundance of polyA+ forms to the actin mRNA (E) and that of polyA− forms relative to the 18S rRNA (F).
-
G, HWestern blotting to examine ALYREF protein level (G) and RT–qPCRs to detect polyA+ histone mRNA levels (H) in Flag‐Cntl (eIF4A3) or siRNA‐resistant Flag‐ALYREF stable expression cells treated with Cntl or ALYREF siRNA. The bars show the relative abundance of histone mRNAs to the GAPDH mRNA. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
Figure EV3. Depletion of UAP56 and THOC2 did not result in enhanced level of polyA+ histone transcripts.
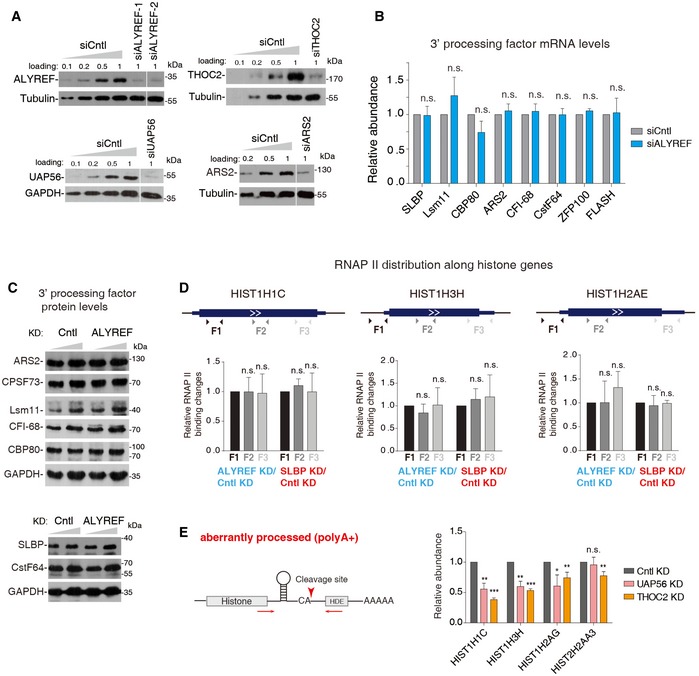
- Western blotting to examine the KD efficiencies of ALYREF, UAP56, THOC2, and ARS2. Tubulin or GAPDH was used as a loading control. Different amounts of Cntl KD samples were loaded to estimate the KD efficiencies. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
- RT–qPCRs to examine mRNA levels of histone 3′‐end processing factors in Cntl and ALYREF KD cells.
- Western blot analysis to examine protein levels of histone 3′‐end processing factors in Cntl and ALYREF KD cells. GAPDH was used as a loading control.
- (Top) Illustration of the locations of primer sets. (Bottom) ChIP–PCR to examine RNAP II distribution change along several histone genes in cells treated with Cntl, ALYREF, or SLBP siRNAs. RNAP II binding change (KD/Cntl) at F1 was set as “1”, and the relative ratios of F2/F1 and F3/F1 were calculated and are shown.
- RT–qPCRs to detect polyA+ histone mRNAs in cells depleted of Cntl, UAP56/URH49, or THOC2. The arrowhead and arrows indicate the correct cleavage site and the location of primers, respectively. The bars show the relative abundance of polyA+ forms to the actin mRNA.
The abundance of polyA+ histone mRNAs is usually very low. 3′READS+ (an updated version of 3′ Region Extraction And Deep Sequencing) is a sensitive and effective method that selectively sequences the 3′‐end region of polyA+ RNAs (Zheng et al, 2016). To further study the role of ALYREF in histone mRNA processing and to examine whether other TREX components are also required for this process, we analyzed 3′READS+ data obtained from Cntl‐, ALYREF‐, UAP56‐, or THOC2‐depleted HeLa cells (Fig EV3A). We note that THOC2 depletion results in co‐depletion of other THO subunits (Chi et al, 2013). As shown in Fig 3D, of the 27 histone mRNAs detected by 3′READS+, 25 were accumulated more than 1.5‐fold upon ALYREF KD (Dataset EV2). In contrast, none was apparently accumulated in UAP56 KD and only three were accumulated more than 1.5‐fold in THO KD (Fig 3D), indicating that they are not required for histone mRNA 3′‐end processing. It was possible that ALYREF KD impedes histone mRNA 3′‐end processing by reducing the levels of processing factors. However, this possibility was not supported by the unchanged mRNA and protein levels of all 3′‐end processing factors we examined in ALYREF KD versus Cntl cells (Fig EV3B and C). Recently, it was reported that slow RNAP II transcription elongation disrupts histone mRNA 3′‐end processing (Saldi et al, 2018). To examine whether ALYREF KD impairs 3′‐end processing by altering transcription elongation, we carried out RNAP II ChIP–qPCRs using primers spanning several histone genes exhibiting significant 3′‐end processing defects. No apparent RNAP II distribution change was observed in ALYREF KD. Similar results were also obtained with SLBP KD that is not known to impact RNAP II elongation (Fig EV3D). These data suggest that histone mRNA processing defects in ALYREF KD cannot be mainly ascribed to altered RNAP II transcription elongation.
To confirm the genome‐wide data, we selected several histone genes, including four upregulated in both polyA+ RNA‐seq and 3′READS+ and two upregulated only in 3′READS+, for validation. To distinguish aberrantly processed from properly processed histone mRNAs, we separated polyA+ and polyA− RNAs. All six genes showed elevated polyA+, and concomitantly reduced polyA−, transcripts in cells treated with siARS2 (a positive control; Gruber et al, 2012) or siALYREF‐1, which was used in the genome‐wide studies (Fig 3E and F). Similar effects were also observed with another ALYREF siRNA (siALYREF‐2; Figs 3E and F, and EV3A). Importantly, polyA+ transcript accumulation of all histone genes we examined was rescued to some extent by the expression of siRNA‐resistant ALYREF, further excluding the possible siRNA off‐target effect (Fig 3G and H). Consistent with the 3′READS+ data, no polyA+ transcript upregulation was detected in UAP56 and THO KD (Fig EV3E). Together, we conclude that ALYREF, but not other TREX components, facilitates 3′‐end processing of histone mRNAs.
ALYREF directly interacts with Lsm11 and facilitates U7‐snRNP recruitment
To decipher the molecular mechanism for ALYREF promoting histone mRNA processing, we examined whether it associates with U7‐snRNP. To this end, we carried out co‐IPs using antibodies to ALYREF and Lsm11, which is a specific U7‐snRNP component (Pillai et al, 2003; Godfrey et al, 2009), in RNase A‐treated S‐phase HeLa cell lysate. The Lsm11 antibody, but not IgG, co‐immunoprecipitated ALYREF (Fig 4A). Conversely, Lsm11 was specifically detected in the immunoprecipitate of ALYREF (Fig 4B), indicating that ALYREF indeed associates with U7‐snRNP. We further validated this association using Flag‐Lsm11. In addition to ALYREF, UAP56 and THOC2 were also co‐precipitated with Flag‐Lsm11 (Fig EV4A and B). Further, RIP‐RT–qPCR data showed that ALYREF, UAP56, and THO all associated with histone pre‐mRNAs (Fig EV4C). These data together suggest that the whole TREX complex associates with U7‐snRNP on histone pre‐mRNAs.
Figure 4. ALYREF directly interacts with Lsm11 and facilitates U7‐snRNP recruitment.
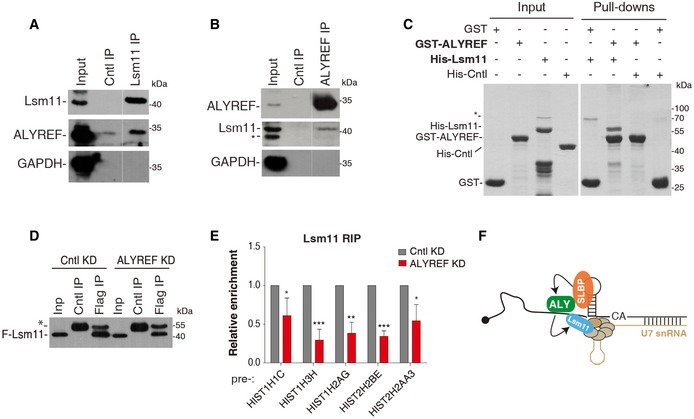
-
A, BIPs were carried out from RNase A‐treated S‐phase HeLa cell lysate using the Lsm11 antibody and IgG (A), or the ALYREF antibody and IgG (B), followed by Western blotting with the indicated antibodies. 2% of input was loaded. * indicates a nonspecific band. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
-
CPull‐down of purified His‐Cntl (PGK1) or His‐Lsm11 using equal amount of GST and GST‐ALYREF proteins in the presence of RNase A. Proteins pulled down were separated by SDS–PAGE followed by Coomassie staining. * indicates a nonspecific band.
-
D, ECntl‐ or ALYREF siRNA‐treated Flag‐Lsm11 stable expression cells were used for IPs with IgG or the Flag antibody. The immunoprecipitates were subjected to RT–qPCRs (E) and Western blot analysis (D). * indicates the antibody heavy chain. The bars show the relative IP efficiencies of histone pre‐mRNAs to U7 snRNA. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student's t‐test. *P < 0.05, **P < 0.01, ***P < 0.001.
-
FGraphic displays that ALYREF interacts with Lsm11 and facilitates U7‐snRNP recruitment.
Source data are available online for this figure.
Figure EV4. TREX associates with Lsm11 and histone pre‐mRNAs.
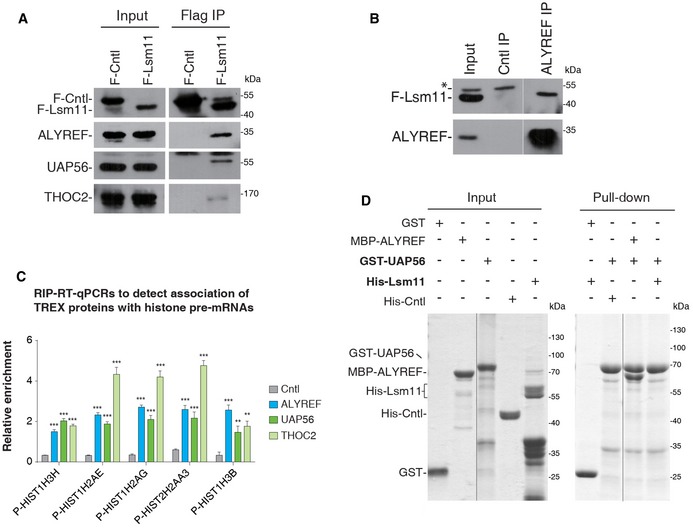
- Flag IPs from RNase A‐treated HeLa cell lysate expressing Flag‐Cntl (eIF4A3) or Flag‐Lsm11, followed by Western blotting with the indicated antibodies. 2.5% of input was loaded.
- IPs with IgG or the ALYREF antibody from RNase A treated Flag‐Lsm11 stable expression cells, followed by Western blotting using the Flag and ALYREF antibodies. 3% of input was loaded. * indicates a nonspecific band. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
- ALYREF, UAP56, and THO RIP–qPCRs to examine their binding on multiple histone pre‐mRNAs. Relative RIP efficiencies are shown. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student's t‐test. **P < 0.01, ***P < 0.001.
- GST‐UAP56 and GST were used for pull‐down of purified His‐Lsm11, His‐Cntl (PGK1), or MBP‐ALYREF in the presence of RNase A. Proteins pulled down were separated by SDS–PAGE, followed by Coomassie staining. 37.5% of input was loaded. The black line delineates the boundary where irrelevant lanes have been removed from the same gel.
Source data are available online for this figure.
Given the association of all TREX components with U7‐snRNP, why is only ALYREF required for 3′‐end processing? It is possible that ALYREF, but not other TREX components, directly interacts with U7‐snRNP. To test this possibility, we carried out pull‐downs of purified His‐Lsm11 using GST‐ALYREF, GST‐UAP56, and GST. Coomassie staining showed that GST‐ALYREF efficiently pulled down His‐Lsm11, but not His‐Cntl (Fig 4C). In contrast, GST‐UAP56 did not interact with His‐Lsm11, although it efficiently pulled down ALYREF (Fig EV4D). These data suggest that ALYREF, but not UAP56, makes direct contact with U7‐snRNP, thus providing an explanation for the specific role of ALYREF in histone mRNA processing.
We next examined whether ALYREF facilitates U7‐snRNP recruitment by carrying out Flag RIPs from Flag‐Lsm11 stable expression cells depleted of Cntl or ALYREF. Both mRNA and protein levels of Flag‐Lsm11 were reproducibly decreased in ALYREF KD for some unknown reason (Appendix Fig S2), resulting in reduced Flag‐Lsm11 immunoprecipitated in these cells (Fig 4D). We thus used co‐immunoprecipitated U7 snRNA as an internal control. Significantly, relative to that of U7 snRNA, the IP efficiencies of all histone pre‐mRNAs we examined were apparently reduced in ALYREF KD (Fig 4E). Taken together, these results indicate that ALYREF promotes histone mRNA 3′‐end processing by directly interacting with U7‐snRNP and facilitating its recruitment (Fig 4F).
The TREX complex promotes histone mRNA export
Considering the key role of ALYREF in polyadenylated mRNA export, we speculated that after 3′‐end processing, ALYREF might promote mature histone mRNA export together with other TREX components. To study this, we constructed an HIST2H2AA3 reporter construct (Fig 5A). When transfected into HeLa cells, the HIST2H2AA3 mRNA was easily detected by a 5′ probe, but not by a 3′ probe to the CS downstream sequence (Appendix Fig S3A and B), indicative of efficient 3′‐end processing. When this construct was microinjected, the HIST2H2AA3 mRNA was mostly accumulated in the cytoplasm of Cntl cells (Fig 5A). In contrast, it was apparently detected in the nuclei of cells depleted of ALYREF, THO, and UAP56 (Fig 5A). It has been reported that improperly processed transcripts are mostly accumulated in the nucleus (Romeo et al, 2014). Although it is hard to separate the role of ALYREF in histone mRNA processing and export, the export defects observed in UAP56 and THO KD, which do not exhibit processing defects, suggest that TREX plays a direct role in histone mRNA export. This conclusion was further validated with two endogenous histone mRNAs, HIST2H2AA3 and HIST1H3H. KD of ALYREF, UAP56, and THO all resulted in elevated nuclear mRNA signals and concomitantly reduced cytoplasmic signals (Figs 5B and EV5A). Further, cell fractionation data revealed that the nuclear/cytoplasmic (N/C) ratios of all histone mRNAs we examined significantly increased in UAP56 KD, as compared to Cntl cells (Fig 5C and D). Together, these results indicate that TREX components function in histone mRNA export as an integrated complex (Fig 5E). This view was further supported by the reduced THO association with histone mRNAs in ALYREF KD (Fig EV5B and C). Notably, ALYREF overexpression did not apparently affect HIST2H2AA3 mRNA N/C distribution (Fig EV5D and E), suggesting that it is not a limiting factor for histone mRNA export.
Figure 5. The TREX complex promotes histone mRNA export.
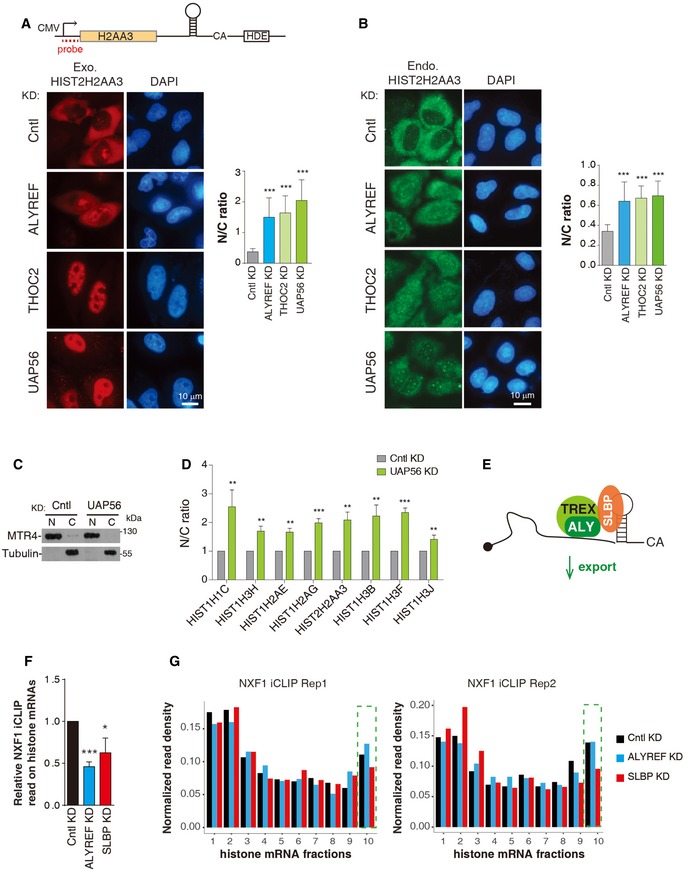
-
A(Top) Illustration of the HIST2H2AA3 reporter construct. (Bottom) FISH to detect the distribution of the HIST2H2AA3 mRNA in HeLa cells depleted of Cntl, ALYREF, THOC2 (THO), and UAP56. DAPI staining served as a nuclear marker. N and C indicate nuclear and cytoplasmic FISH signals, respectively. N/C ratios were determined for 20 cells in each experiment.
-
BDistribution of the endogenous HIST2H2AA3 mRNA was detected with the transcript‐specific probe in HeLa cells depleted of Cntl, ALYREF, THOC2 (THO), and UAP56. N and C indicate nuclear and cytoplasmic FISH signals, respectively. N/C ratios were calculated the same as (A).
-
C, DWestern blot analysis to examine the purities of nuclear and cytoplasmic fractions (C), and RT–qPCRs to examine N/C distribution of each histone mRNA (D) in Cntl and UAP56 KD cells. MTR4 and tubulin were used as the nuclear and cytoplasmic marker, respectively.
-
EGraphic displays that TREX promotes histone mRNA export.
-
FRelative NXF1 iCLIP read populations on histone mRNAs in Cntl, ALYREF, and SLBP KD cells.
-
GNXF1 iCLIP signals along the histone mRNA. The histone mRNA is divided into 10 fractions based on their mRNA size. Each fraction represents 10% of each histone mRNA. The tenth fraction that contains the SL is boxed by green dashed lines.
Figure EV5. TREX functions in histone mRNA export.
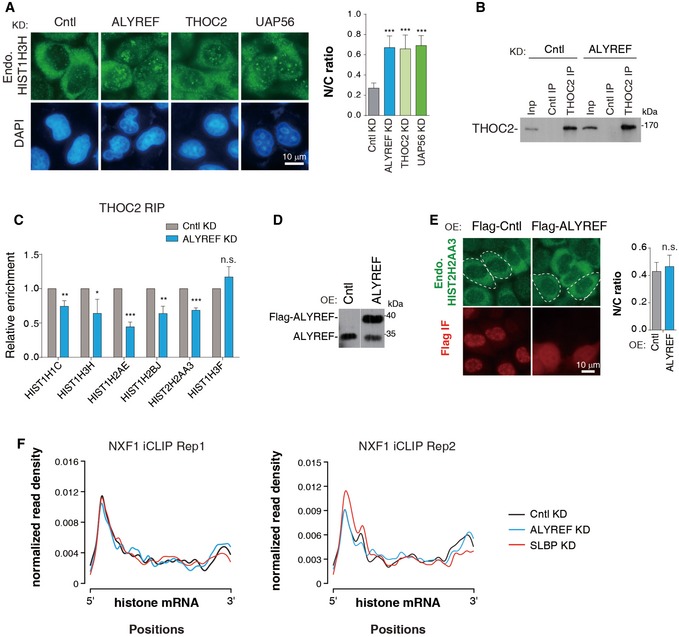
-
ADistribution of the endogenous HIST1H3H mRNA was detected with the transcript‐specific probe in HeLa cells depleted of Cntl, ALYREF, THOC2 (THO), and UAP56. DAPI staining served as a nuclear marker. N and C indicate nuclear and cytoplasmic FISH signals, respectively. N/C ratios were determined for 20 cells in each experiment.
-
B, CCntl‐ or ALYREF siRNA‐treated HEK293 cells were used for IPs with IgG or the THOC2 antibody. The immunoprecipitates were subjected to Western blot analysis (B) and RT–qPCRs (C).
-
DWestern blot analysis to examine the overexpression of Flag‐ALYREF. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
-
EDistribution of endogenous HIST2H2AA3 mRNA was detected with the transcript‐specific probe in Cntl (eIF4A3) and ALYREF overexpression cells. N/C ratios were quantified the same as (A).
-
FMetagene analysis of normalized NXF1 signals on histone mRNAs in Cntl, ALYREF, and SLBP KD cells based on iCLIP‐seq data. iCLIP‐seq signal at each position of an mRNA is divided by the sum of signal of the mRNA to normalize each mRNA's contribution to the plot.
We next wanted to ask whether ALYREF promotes histone mRNA export by enhancing NXF1 binding. To this end, we knocked down ALYREF in cells stably expressing Flag‐NXF1 at the physiological level (Appendix Fig S4) and carried out iCLIP with the Flag antibody. Considering the important role of SLBP in ALYREF recruitment, we also included SLBP KD. Significantly, KD of both ALYREF and SLBP reproducibly reduced NXF1 iCLIP read population on histone mRNAs, indicative of reduced NXF1 binding (Fig 5F). While SLBP KD preferentially reduced NXF1 binding at the 3′ region, ALYREF KD did not show such a trend (Figs 5G and EV5F). This result suggests that SLBP plays a prominent role in NXF1 binding at the 3′ region, whereas ALYREF probably promotes its binding throughout the full length of the histone mRNA. Taken together, our data indicate that TREX promotes histone mRNA export by recruiting NXF1.
3′‐end processing significantly enhances histone mRNA export
Previous studies on whether 3′‐end processing promotes histone mRNA export have reached contradictory conclusions (Eckner et al, 1991; Erkmann et al, 2005). The roles of ALYREF in both 3′‐end processing and nuclear export of histone mRNAs promoted us to revisit this question. We first compared the export efficiency of the HIST2H2AA3 mRNA transcribed from the mature sequence (H2AA3‐SL) with that processed from the precursor (H2AA3‐SLD, Stem‐Loop and Histone Downstream Element; Fig 6A). Significantly, when microinjected into HeLa nuclei, the H2AA3‐SL mRNA was mostly nuclear, whereas the H2AA3‐SLD mRNA was largely cytoplasmic (Fig 6A). We next constructed a pair of HIST1H1C reporters, containing either the mature (H1C‐SL‐Rb) or the precursor (H1C‐SLD‐Rb) sequence, followed by the hepatitis D viral ribozyme (Fig 6B). Efficient ribozyme cleavage of H1C‐SL‐Rb and U7‐snRNP processing of H1C‐SLD‐Rb mRNAs were confirmed (Appendix Fig S5A). When transfected into the cells, the processed H1C mRNA (pH1C), but not the ribozyme cleaved one (cH1C), was efficiently accumulated in the cytoplasm (Fig 6B). These data together indicate that 3′‐end processing in general promotes histone mRNA export. This promotion is not dependent on the coding sequence, as replacement of the coding sequence in the HIST2H2AA3 reporters with that of β‐globin (cG) did not affect processing‐dependent mRNA export (Fig 6C).
Figure 6. 3′‐end processing promotes TREX recruitment and histone mRNA export.
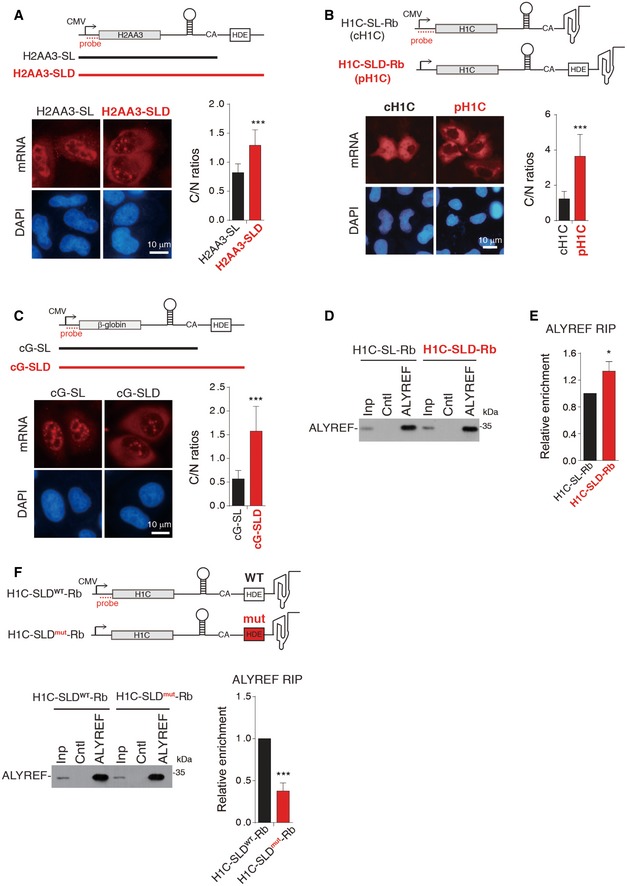
-
A(Top) Illustration of the reporter constructs. The black and red lines indicate H2AA3‐SL and H2AA3‐SLD PCR products, respectively. The dashed line indicates the FISH probe. (Bottom) FISH to detect the distribution of the corresponding mRNA at 2 h after injection of H2AA3‐SL and H2AA3‐SLD PCR products. C and N indicate cytoplasmic and nuclear FISH signals, respectively. C/N ratios were determined for 20 cells in each experiment.
-
B(Top) Illustration of the reporter constructs. The dashed line indicates the FISH probe. FISH to detect mRNA distribution at 24 h after transfection of the H1C‐SL‐Rb (cH1C) and H1C‐SLD‐Rb (pH1C) constructs. C/N ratios were quantified the same as (A).
-
CSame as (A), except that the PCR products of cG‐SL and cG‐SLD were used instead. cG, β‐globin cDNA.
-
D, EThe H1C‐SL‐Rb or H1C‐SLD‐Rb construct, together with VSV M, was co‐transfected into HeLa cells, followed by IPs with IgG or the ALYREF antibody at 24 h post‐transfection. The immunoprecipitates were subjected to Western blot analysis to detect ALYREF (D) and to RT–qPCRs to detect RNAs (E). The bars in the graph show the relative ALYREF IP efficiency, with that for the H1C‐SL‐Rb mRNA set as “1”.
-
FThe H1C‐SLD‐Rb construct with wild‐type (H1C‐SLDWT‐Rb) or mutant (H1C‐SLDmut‐Rb) U7‐binding sequence, together with VSV M, was co‐transfected into HeLa cells, followed by IPs with IgG or the ALYREF antibody at 24 h post‐transfection. The immunoprecipitates were subjected to Western blot analysis (left panel) and RT–qPCRs (right panel). The bars in the right graph show the relative ALYREF IP efficiency, with that for the H1C‐SLDWT‐Rb mRNA set as “1”.
3′‐end processing promotes ALYREF recruitment to histone mRNAs via U7‐snRNP
To examine whether 3′‐end processing enhances ALYREF recruitment, we carried out ALYREF RIP to compare its association with the pH1C versus the cH1C mRNA. Note that for a fair comparison, both mRNAs were blocked in the nucleus by overexpression of vesicular stomatitis virus (VSV) M protein, which impedes mRNA export by targeting Nup98 and Rae1 (von Kobbe et al, 2000; Faria et al, 2005). Importantly, RT–qPCR data showed that compared to the cH1C, the pH1C mRNA was reproducibly more enriched with ALYREF, supporting the notion that 3′‐end processing promotes ALYREF recruitment (Fig 6D and E).
How could 3′‐end processing enhance ALYREF recruitment? One possibility is that U7‐snRNP plays a role in stabilizing ALYREF binding on histone mRNAs. To examine this, we mutated U7‐binding sequence in the H1C‐SLD‐Rb construct. As expected, this mutation impedes both 3′‐end processing and nuclear export of the H1C mRNA (Appendix Fig S5B). When the wild‐type (WT) and the mutant H1C‐SLD‐Rb constructs were separately co‐transfected with VSV M into the cells, the mutant mRNA apparently associates with less ALYREF, as compare to the WT (Fig 6F). Together, our data suggest that 3′‐end processing promotes histone mRNA export at least partially through U7‐snRNP‐dependent ALYREF binding.
ALYREF ensures histone protein levels
Finally, we determined how histone protein levels are impacted by ALYREF KD. We examined the levels of histone proteins H2B and H4 in HeLa cells treated with siCntl, siALYREF‐1, and siALYREF‐2. ARS2 KD cells were used as a positive control (Gruber et al, 2012). As observed in Fig 7A, akin to ARS2 KD, ALYREF KD resulted in moderately but reproducibly reduced protein levels of both H2B and H4. Consistent with their inability to produce sufficient histone proteins to support cell growth, we repeatedly observed proliferation defect in cells treated with ALYREF siRNAs (Fig 7B). Similar to ARS2 KD cells, ALYREF KD cells did not accumulate in the S‐phase of the cell cycle (Fig 7C). Instead, these cells arrested throughout the cell cycle.
Figure 7. ALYREF ensures histone protein production.
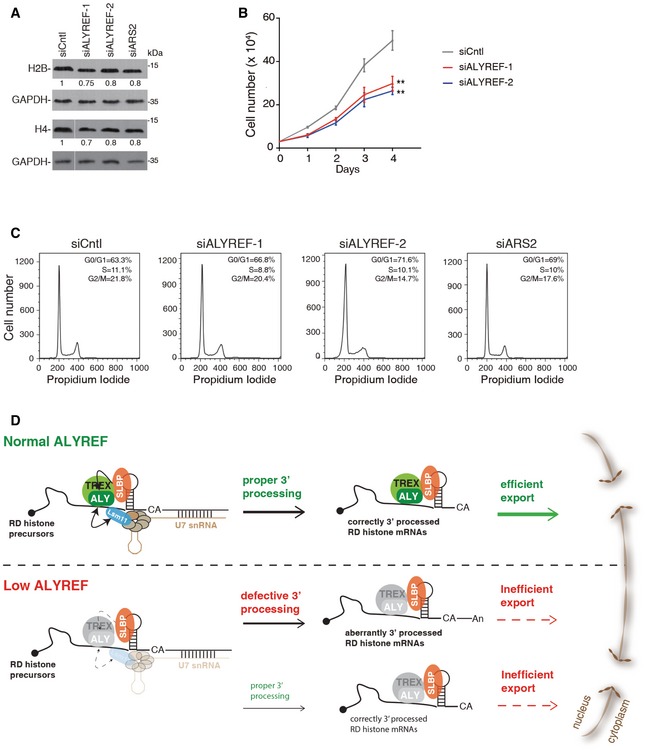
- Western blotting to examine the expression levels of H2B and H4 in Cntl, ALYREF, and ARS2 KD cells. GAPDH was used as a loading control. The white line delineates the boundary where irrelevant lanes have been removed from the same blot.
- Growth curve of Cntl and ALYREF KD cells. Error bars represent standard deviations from biological repeats (n = 3). Statistical analysis was performed using Student's t‐test. **P < 0.01.
- Cell cycle analysis of Cntl, ALYREF, and ARS2 KD cells. Quantifications of cells in G0/G1 (1N)‐, G2 (2N)‐, and S‐phase (intermediate) of the cell cycle are shown in the upper right corner of each histogram.
- Model for ALYREF promoting histone mRNA 3′‐end processing and nuclear export. See text for details.
Source data are available online for this figure.
Discussion
The highly conserved TREX complex has been implicated in coordinating 3′‐end processing and nuclear export of polyadenylated mRNAs. In this study, we show that TREX component ALYREF is important for two aspects of non‐ployadenylated histone mRNA metabolism, namely 3′‐end processing and nuclear export. Further, our data demonstrate that a parallel coordination mechanism exists for histone mRNAs and ALYREF makes important contribution to this mechanism.
A model for ALYREF promoting histone mRNA processing and export
Based on our data, we propose a model for ALYREF orchestrating 3′‐end processing and nuclear export of histone mRNAs (Fig 7D). During 3′‐end processing, SLBP recruits ALYREF as well as other TREX components to a region near SL on histone pre‐mRNAs through its direct interaction with ALYREF. This recruitment has two functional consequences. On histone pre‐mRNAs, ALYREF ensures efficient 3′‐end processing by facilitating U7‐snRNP recruitment via its direct interaction with Lsm11. After processing, ALYREF, together with other TREX components, facilitates histone mRNA export through recruiting NXF1. When ALYREF is limited, 3′‐end processing cannot proficiently occur due to the impaired U7‐snRNP recruitment, resulting in the usage of downstream polyA site and the production of polyadenylated histone mRNAs that are mostly retained in the nucleus (Romeo et al, 2014). Without ALYREF and other TREX proteins, histone mRNAs cannot be efficiently exported to the cytoplasm.
Previous studies found specific SR‐binding motifs present in the coding region of histone mRNAs also assist histone mRNA export (Huang & Carmichael, 1997; Huang & Steitz, 2001). Except for the coding region, SR proteins are also recruited to the SL region (Anko et al, 2012), raising the possibility that they also contribute to NXF1 binding to this region. Consistent with this possibility, ALYREF KD did not lead to a preferential reduction of NXF1 binding at 3′ region of histone mRNAs.
ALYREF might contribute to SLBP‐mediated histone mRNA processing and export
It has been ~20 years since the initial discovery of SLBP as a factor promoting histone mRNA 3′‐end processing by facilitating U7‐snRNP recruitment (Dominski et al, 1999). However, it is still unclear how SLBP is linked to U7‐snRNP recruitment. FLASH, which interacts with U7‐snRNP and facilitates 3′‐end processing, has been thought to be the linker (Yang et al, 2009; Burch et al, 2011). However, so far the interaction between SLBP and FLASH has not been found. Another candidate SLBP‐U7 linker is ZFP100 that interacts with both SLBP and Lsm11 (Dominski et al, 2002; Azzouz et al, 2005; Wagner & Marzluff, 2006). However, mutations in Lsm11 that inhibit histone mRNA 3′‐end processing did not influence its interaction with ZFP100 (Azzouz et al, 2005). Our findings here that ALYREF directly interacts with both SLBP and U7‐snRNP and facilitates U7‐snRNP recruitment suggest that ALYREF contributes to linking SLBP to U7‐snRNP. In agreement with this possibility, depletion of ALYREF results in widespread histone mRNA processing defects. Further, ALYREF interacts with the C‐terminal region of SLBP that is important for U7‐snRNP recruitment (Skrajna et al, 2017). However, ALYREF has not been identified in the purified histone mRNA processing complex. Further studies are required to determine whether ALYREF serves as a key linker for SLBP and U7‐snRNP.
Our data demonstrating that SLBP directly interacts with ALYREF and facilitates ALYREF recruitment suggest that SLBP promotes histone mRNA export by recruiting ALYREF. In support of this view, TREX is required for efficient histone mRNA export. Post‐translational modifications occur to both ALYREF and SLBP (Borchers et al, 2006; Okada et al, 2008; Hung et al, 2010; Brodersen et al, 2016). It is possible that these modifications could regulate their interaction and modulate histone mRNA export.
Physical and functional coupling of histone mRNA processing and export
Our work demonstrates that in mammalian cells, histone mRNA processing is physically and functionally linked to nuclear export, and suggests that ALYREF contributes to this linkage. We also provide evidence that U7‐snRNP is important for 3′‐end processing‐dependent ALYREF recruitment. In addition to SLBP and Lsm11, ALYREF was found to directly interact with ARS2 and CstF64 (Fan et al, 2017; Shi et al, 2017), which have been implicated in histone mRNA processing as well (Gruber et al, 2012; Romeo et al, 2014). It is possible that these processing factors also contribute to 3′‐end processing‐dependent ALYREF recruitment.
It has been believed that nuclear export of polyadenylated and non‐polyadenylated mRNAs occurs via TREX and SR proteins, respectively. Our study suggests that nuclear export machinery is extensively shared by the polyadenylated and the non‐polyadenylated mRNAs. In this vein, SR proteins were recently found to promote polyadenylated mRNA export (Muller‐McNicoll et al, 2016). Although distinct in the 3′ end, polyadenylated and non‐polyadenylated mRNAs evolve resembling mechanisms for recruiting ALYREF to the 3′ region: polyA/PABPN1‐dependent ALYREF recruitment to polyadenylated mRNAs (Shi et al, 2017) and SL/SLBP‐dependent ALYREF recruitment to non‐polyadenylated histone mRNAs. mRNA export factors have been implicated in the regulation of 3′‐end processing of polyadenylated mRNAs (Johnson et al, 2011; Katahira et al, 2013; Tran et al, 2014). Thus, our work suggests that 3′‐end processing regulation by nuclear export factors is likely a common mechanism for both polyadenylated and non‐polyadenylated mRNAs. This mechanism could be important to integrate processes in gene expression to obtain precise regulation.
Materials and Methods
RNA extraction and RT–qPCRs
RNA extracted using TRIzol (Invitrogen) was treated with RNase‐free RQ1 DNase I (Promega) to remove genomic DNA. Oligo dT (for polyA+ RNA only) or the random primer was used for reverse transcription by M‐MLV reverse transcriptase (Promega). Quantitative PCR was carried out using GoTaq Master Mix (Promega) according to the manufacturer's instruction. The primers used for PCR are listed in Appendix Table S1.
PolyA+ and PolyA− RNA preparation
For polyA+ RNA selection, 7.5 μg total RNA was heated to 65°C for 2 min and then placed on ice. The RNA was rotated with oligo (dT)25 Dynabeads in 10 μl of binding buffer (20 mM Tris at pH 7.5, 1 M LiCl, 2 mM EDTA) for 5 min at room temperature. After two washes with 20 μl of washing buffer (10 mM Tris at pH 7.5, 0.15 M LiCl, 1 mM EDTA), polyA+ RNA was eluted from the beads using 10 μl of elution buffer (10 mM Tris at pH 7.5). The supernatant after the initial binding was further cleaned with two incubations with the beads and kept as the polyA− RNA.
Protein immunoprecipitations
For each assay, 106 cells were suspended in the lysis buffer (20 mM Tris–HCl at pH 7.4, 150 mM NaCl, 2 mM EDTA at pH 8.0, 0.1% Triton, 1 mM DTT, 1 mM PMSF). After sonication and centrifugation, the lysate was treated with RNase A for 20 min at 30°C, followed by incubation with antibody‐crosslinked beads at 4°C overnight. The beads were washed four times with the lysis buffer, and proteins were eluted with sodium dodecyl sulfate (SDS) loading buffer.
RNA immunoprecipitations
RNA immunoprecipitations were performed as previously described (Fan et al, 2017). Briefly, cells grown on 10‐cm dishes were transfected with the corresponding siRNAs. Seventy‐two hours later, cells were re‐suspended in 1 ml of NET‐2 buffer (50 mM Tris–HCl at pH 7.4, 150 mM NaCl, 0.1% NP‐40, 0.2 mM PMSF), followed by sonication and centrifugation. The lysates were incubated with antibodies for 2 h, followed by rotation with nProtein A Sepharose (GE) for another 2 h at 4°C. The immunoprecipitates were washed three times with the NET‐2 buffer, followed by Western blot analysis or RNA extraction.
Protein pull‐downs
For each pull‐down, 8 μg of purified GST‐ or MBP‐tagged proteins bound to 20 μl of Glutathione Sepharose 4B or Amylose resins was used. For pull‐downs of purified proteins, 8 μg of each protein was mixed together with protein‐bound beads in pull‐down buffer (1 × PBS with 0.1% Triton, 0.2 mM PMSF, 50 ng/ml RNase A and protease inhibitor). The mixtures were rotated overnight at 4°C, and beads were washed five times. Proteins were eluted with SDS loading buffer and separated by SDS–PAGE, followed by Coomassie staining and Western blot analyses. For pull‐downs of in vitro translated proteins, 35S methionine‐labeled proteins were produced using the TNT T7 Quick Coupled Transcription/Translation Kit (Promega). The translation mix was then incubated with RNase A to a final concentration of 0.35 ng/ml at 30°C for 20 min. 10 μl of this reaction mixture was incubated with protein‐bound beads. The rest of the experiment was the same as pull‐down of purified proteins. Proteins pulled down were separated by SDS–PAGE and visualized by Coomassie staining and autoradiography.
RNA‐seq
For polyA+ RNA sequencing, 5 μg of total RNA was used for polyA+ RNA selection. After selection, the rest of the RNA was depleted of rRNA and was treated as polyA− RNA. Stranded cDNA libraries were generated for both polyA+ and polyA− RNA with TruSeq Stranded Total RNA Sample Prep Kit (Illumina) according to the manufacturer's instruction. The libraries were then sequenced on an Illumina HiSeq 2000 using a single‐read protocol of 100 cycles with v3 chemistry at CAS‐MPG Partner Institute for Computational Biology Omics Core, Shanghai, China.
iCLIP‐seq
The iCLIP assay was carried out as previously described, with modifications (Shi et al, 2017). Note that NXF1 iCLIP was carried out in Flag‐NXF1 stable expression cells. Briefly, 1 × 104 cells were irradiated with UV light at 200 mJ/cm2 or 400 mJ/cm2 for ALYREF or NXF1 iCLIP, respectively. After cell lysis, RNAs were partially fragmented using RNase I (2.5 U/ml). IPs with the ALYREF antibody immobilized on protein A Dynabeads (Life Technologies) or the Flag antibody immobilized on protein G Dynabeads (Life Technologies). After extensive washing, immunoprecipitated RNAs were ligated at the 3′ ends to an RNA adapter and radioactively labeled by T4 polynucleotide kinase. The protein–RNA complexes were then transferred to a nitrocellulose membrane. For iCLIP cDNA library preparation, fragmented RNAs were purified and reverse‐transcribed with a primer containing a barcode. The resulting cDNAs were purified by PAGE, circularized by single‐stranded DNA ligase (Epicentre), linearized by restriction enzyme cleavage, and amplified by PCRs. High‐throughput sequencing of iCLIP cDNA libraries from two biological replicates was performed on an Illumina HiSeq X flow cell with a 150 nt run length.
Further methods can be found in the Appendix Supplementary Methods. SiRNAs used in this study are listed in Appendix Table S2 and probes used in this study in Appendix Table S3.
Author contributions
JF and HC conceived the study. JF and HC designed experiments. JF, KW, JW, SC, YW, MS, LZ, DZ, CW, and LW performed experiments. XD, XW, GL, BT, and YZ analyzed the data. JF and HC wrote the paper. HC supervised the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Eric Wagner for advices on obtaining S‐phase cells, HongLi Hu for providing the His‐PGK1 protein, and Dangsheng Li and Cheng Lab members for useful discussion. This work was supported by grants from the National Key R&D Program of China (2017YFA0504400), National Natural Science Foundation of China (31570822, 31770880 and 31800686), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), and China Postdoctoral Science Foundation (2018M630481, BX20180333).
The EMBO Journal (2019) 38: e99910
Data availability
The data associated with the manuscript are available under accession numbers GSE117701 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117701) and GSE125005 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125005).
References
- Anko ML, Muller‐McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM (2012) The RNA‐binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz TN, Gruber A, Schumperli D (2005) U7 snRNP‐specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100‐independent function in histone RNA 3′ end processing. Nucleic Acids Res 33: 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, Doudna JA (2001) The stem‐loop‐binding protein forms a highly stable and specific complex with the 3′ stem‐loop of histone mRNAs. RNA 7: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers CH, Thapar R, Petrotchenko EV, Torres MP, Speir JP, Easterling M, Dominski Z, Marzluff WF (2006) Combined top‐down and bottom‐up identifies a phosphorylation proteins that contributes to proteomics site in stem‐loop‐binding high‐affinity RNA binding. Proc Natl Acad Sci USA 103: 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen MM, Lampert F, Barnes CA, Soste M, Piwko W, Peter M (2016) CRL4(WDR23)‐mediated SLBP ubiquitylation ensures histone supply during DNA replication. Mol Cell 62: 627–635 [DOI] [PubMed] [Google Scholar]
- Brooks L, Lyons SM, Mahoney JM, Welch JD, Liu ZL, Marzluff WF, Whitfield ML (2015) A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA 21: 1943–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z (2011) Interaction between FLASH and Lsm11 is essential for histone pre‐mRNA processing in vivo in Drosophila . RNA 17: 1132–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi BK, Wang QL, Wu GF, Tan M, Wang LT, Shi M, Chang XY, Cheng H (2013) Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res 41: 1294–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel ML (1988) Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the invitro 3′ RNA processing reaction. EMBO J 7: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Zheng LX, Sanchez R, Marzluff WF (1999) Stem‐loop binding protein facilitates 3′‐end formation by stabilizing U7 snRNP binding to histone pre‐mRNA. Mol Cell Biol 19: 3561–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Erkmann JA, Yang XC, Sanchez R, Marzluff WF (2002) A novel zinc finger protein is associated with U7 snRNP an interacts with the stem‐loop binding protein in the histone pre‐mRNP to stimulate 3′‐end processing. Genes Dev 16: 58–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF (2007) Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Ellmeier W, Birnstiel ML (1991) Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J 10: 3513–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann JA, Sanchez R, Treichel N, Marzluff WF, Kutay U (2005) Nuclear export of metazoan replication‐dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA 11: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Kuai B, Wu GF, Wu XD, Chi BK, Wang LT, Wang K, Shi ZB, Zhang H, Chen S, He ZS, Wang SY, Zhou ZC, Li GH, Cheng H (2017) Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J 36: 2870–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM (2005) VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17: 93–102 [DOI] [PubMed] [Google Scholar]
- Godfrey AC, White AE, Tatomer DC, Marzluff WF, Duronio RJ (2009) The Drosophila U7 snRNP proteins Lsm10 and Lsm11 are required for histone pre‐mRNA processing and play an essential role in development. RNA 15: 1661–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB (2012) Ars2 promotes proper replication‐dependent histone mRNA 3′ end formation. Mol Cell 45: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA (2008) Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci USA 105: 5154–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Carmichael GG (1997) The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV‐1‐related mRNAs. Proc Natl Acad Sci USA 94: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Steitz JA (2001) Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell 7: 899–905 [DOI] [PubMed] [Google Scholar]
- Hung ML, Hautbergue GM, Snijders APL, Dickman MJ, Wilson SA (2010) Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res 38: 3351–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL (2009) Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Kim H, Erickson B, Bentley DL (2011) The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol 18: 1164–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y (2009) Adaptor Aly and co‐adaptor Thoc5 function in the Tap‐p15‐mediated nuclear export of HSP70 mRNA. EMBO J 28: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Okuzaki D, Inoue H, Yoneda Y, Maehara K, Ohkawa Y (2013) Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res 41: 7060–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo‐Fonseca M, Izaurralde E (2000) Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell 6: 1243–1252 [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–U166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF (2005) Metazoan replication‐dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol 17: 274–280 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R (2005) Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA (1987) Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNAs. Science 238: 1682–1687 [DOI] [PubMed] [Google Scholar]
- Muller‐McNicoll M, Botti V, Domingues AMD, Brandl H, Schwich OD, Steiner MC, Curk T, Poser I, Zarnack K, Neugebauer KM (2016) SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev 30: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H (2007) NELF interacts with CBC and participates in 3′ end processing of replication‐dependent histone mRNAs. Mol Cell 26: 349–365 [DOI] [PubMed] [Google Scholar]
- Okada M, Jang SW, Ye K (2008) Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc Natl Acad Sci USA 105: 8649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D (2003) Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev 17: 2321–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo V, Griesbach E, Schumperli D (2014) CstF64: cell cycle regulation and functional role in 3′ end processing of replication‐dependent histone mRNAs. Mol Cell Biol 34: 4272–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Khrameeva E, Schreiner S, Konig J, Curk T, Zupan B, Ule J, Gelfand MS, Bindereif A (2014) Crosslinking‐immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol 11: 146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T, Fong N, Bentley DL (2018) Transcription elongation rate affects nascent histone pre‐mRNA folding and 3′ end processing. Genes Dev 32: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML (1986) Compensatory mutations suggest that base‐pairing with a small nuclear‐RNA is required to form the 3′ end Of H‐3 messenger‐RNA. Nature 323: 777–781 [DOI] [PubMed] [Google Scholar]
- Shi Y, Manley JL (2015) The end of the message: multiple protein‐RNA interactions define the mRNA polyadenylation site. Genes Dev 29: 889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Zhang H, Wu XD, He ZS, Wang LT, Yin SY, Tian B, Li GH, Cheng H (2017) ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo . Nucleic Acids Res 45: 9640–9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrajna A, Yang XC, Bucholc K, Zhang J, Hall TMT, Dadlez M, Marzluff WF, Dominski Z (2017) U7 snRNP is recruited to histone pre‐mRNA in a FLASH‐dependent manner by two separate regions of the stem‐loop binding protein. RNA 23: 938–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E (2000) Yra1p, a conserved nuclear RNA‐binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez‐Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ (2009) Knockdown of SLBP results in nuclear retention of histone mRNA. RNA 15: 459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DDH, Saran S, Williamson AJK, Pierce A, Dittrich‐Breiholz O, Wiehlmann L, Koch A, Whetton AD, Tamura T (2014) THOC5 controls 3′ end‐processing of immediate early genes via interaction with polyadenylation specific factor 100 (CPSF100). Nucleic Acids Res 42: 12249–12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA (2012) TREX exposes the RNA‐binding domain of Nxf1 to enable mRNA export. Nat Commun 3: 1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Marzluff WF (2006) ZFP100, a component of the active U7 snRNP limiting for histone pre‐mRNA processing, is required for entry into S phase. Mol Cell Biol 26: 6702–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AS, Marzluff WF (1995) The sequence of the stem and flanking sequences at the 3′ end of histone messenger‐RNA are critical determinants for the binding of the stem‐loop binding‐protein. Nucleic Acids Res 23: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z (2009) FLASH, a proapoptotic protein involved in activation of caspase‐8, is essential for 3′ end processing of histone pre‐mRNAs. Mol Cell 36: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Liu X, Tian B (2016) 3′READS+, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA 22: 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
The data associated with the manuscript are available under accession numbers GSE117701 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117701) and GSE125005 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125005).


