Abstract
Methods to measure cellular target engagement are increasingly being used in early drug discovery. The Cellular Thermal Shift Assay (CETSA) is one such method. CETSA can investigate target engagement by measuring changes in protein thermal stability upon compound binding within the intracellular environment. It can be performed in high-throughput, microplate-based formats to enable broader application to early drug discovery campaigns, though high-throughput forms of CETSA have only been reported for a limited number of targets. CETSA offers the advantage of investigating the target of interest in its physiological environment and native state, but it is not clear yet how well this technology correlates to more established and conventional cellular and biochemical approaches widely used in drug discovery. We report two novel high-throughput CETSA (CETSA HT) assays for B-Raf and PARP1, demonstrating the application of this technology to additional targets. By performing comparative analyses with other assays, we show that CETSA HT correlates well with other screening technologies and can be applied throughout various stages of hit identification and lead optimization. Our results support the use of CETSA HT as a broadly applicable and valuable methodology to help drive drug discovery campaigns to molecules that engage the intended target in cells.
Keywords: CETSA, target engagement, B-Raf, PARP1
Introduction
Achieving target engagement at the desired site of action is critical for interpreting the efficacy of a drug and understanding the clinical validity of a target for treating disease.1 A thorough understanding of preclinical compound mode of action, including clear evidence of target engagement, may contribute to improved success rates for clinical candidates.2 While compound binding to the target can be measured using purified protein with a range of biochemical or biophysical assay technologies, this does not always translate into target engagement in a cellular environment.3 As such, methodologies for measuring target engagement in cells have become increasingly used in preclinical drug development. The routine use of cellular target engagement technologies throughout hit finding, lead identification, and lead optimization may expedite the identification of compounds directly engaging the target of interest, thus enabling preclinical and clinical studies to more effectively and safely bring new medicines to patients.
The Cellular Thermal Shift Assay (CETSA) has emerged as a powerful technique to monitor intracellular target engagement in a physiologically relevant and label-free manner. While early studies demonstrated the application of CETSA to monitor target engagement in a range of cellular systems, including from ex vivo samples,4 subsequent reports have demonstrated the adaption of this technology into microplate format to enable high-throughput assays with potential application in hit finding, lead identification, and lead optimization.5,6 While such assays can be constructed using tagged proteins overexpressed in cells,7–9 assays that measure label-free target engagement with endogenous protein by high-throughput CETSA (CETSA HT) have only been reported for a handful of targets to date.5,10–12 These studies have demonstrated that this technology might be used for compound screening to identify hits that are able to access and engage the target of interest in live cells, and to determine a measure of apparent potency of intracellular binding to rank compounds and inform on structure–activity relationships (SARs).5,9,13 Variations in the assay setup have also been shown to differentiate compounds based on the kinetics of intracellular target engagement.5,12 In addition, we have recently shown that this technology can be used to determine intracellular binding affinities for antagonists of the androgen receptor (AR).12
Despite the clear advantages of CETSA described in the literature, little is known about how reliably this technology can be used in screening cascades and how well it correlates to more conventional screening technologies. Here, we first report the generation of robust and novel CETSA HT assays directed against two clinically validated oncology targets: B-Raf and poly(ADP-ribose) polymerase 1 (PARP1). By screening large and diverse compound libraries, we demonstrate that CETSA HT can efficiently identify small-molecule inhibitors for both targets, and can also be used to rank cellular target engagement between compounds. In addition, by comparing CETSA HT for PARP1 with other screening approaches, we find a good correlation between these assays, indicating that CETSA HT can be used in early drug discovery to reliably identify small-molecule binders to targets of interest. These studies should serve as a useful starting point to understand both the advantages and limitations of the CETSA technology, with a particular focus on high-throughput applications for the hit identification and lead optimization stages of early drug discovery.
Materials and Methods
Cell Culture
A375 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Waltham, MA) supplemented with 1% l-glutamine (Sigma, St. Louis, MO) and 10% fetal bovine serum (FBS). MDA-MB-436 cells were cultured in RPMI1640 medium with l-glutamine, 10% FBS, and 10 µg/mL insulin (Sigma, 19278). A549 cells were cultured in DMEM F12 Hams (Sigma) supplemented with Glutamax and 10% FBS. Cells were cultured in a humidified incubator at 37 °C, 5% CO2. All cell lines were confirmed as mycoplasma free.
Identification and Optimization of AlphaScreen Antibody Pairs
Antibody pairs to quantify thermostable target protein by AlphaScreen were identified as previously described.5,12,13 Briefly, combinations of various mouse- and rabbit-derived antibodies to the target were added to cell lysates in the presence of Anti-Mouse IgG Alpha Donor Beads (PerkinElmer, Waltham, MA; AS104D) and Anti-Rabbit IgG (Fc specific) AlphaLISA Acceptor Beads (PerkinElmer, AL104C). AlphaScreen signal was analyzed using an EnVision plate reader (PerkinElmer) following overnight incubation. Signal was optimized by variation of antibody concentrations and cell density (Suppl. Figs. S1 and S5).
B-Raf CETSA HT Melt Curve
A375 cells were harvested to a density of 2 × 107 cells/mL in Hanks’ balance salt solution (HBSS) and treated with 10 µM dabrafenib or 0.1% DMSO. Cells were incubated for 1 h under tissue culture conditions with gentle continuous rotation. Cell suspensions were aliquoted into PCR strips (30 µL/tube) and each strip subjected to a 3 min heatshock at the indicated temperature using a Veriti thermal cycler (Thermo Scientific, Waltham, MA). Samples were stored on ice and lysed by three repetitive freeze–thaws in liquid nitrogen. Five microliters of lysate was transferred to a 96-well plate and anti-B-Raf antibodies Abnova (Taipei, Taiwan) H00000673-M02 and SantaCruz (Dallas, TX) sc-9002, prepared in ImmunoAssay Buffer (PerkinElmer, AL000F), were added to a final concentration of 0.3 nM in 25 µL volume. Following 1 h incubation, AlphaScreen signal was developed by addition of Anti-Mouse IgG Alpha Donor Beads to a final concentration of 80 µg/mL, and Anti-Rabbit IgG AlphaLISA Acceptor Beads to a final concentration of 20 µg/mL. After 16 h, plates were analyzed using an EnSpire Alpha Plate Reader (PerkinElmer).
PARP1 CETSA HT Melt Curve
MDA-MB-436 cells were harvested to a density of 1 × 107 cells/mL in complete media. Cells were treated with one of DMSO (0.1%), olaparib (10 µM), rucaparib (10 µM), or NMS-P118 (10 µM) and dispensed into MicroAmp 0.2 mL eight-tube PCR strips (Thermo Fisher, N8010580), 20 µL/tube. Cells were incubated 2 h under cell culture conditions. Samples were heatshocked at indicated temperatures for 3 min and then 25 °C for 1 min using a Veriti SimpliAmp PCR machine (Thermo Fisher). Heatshocked cells were lysed by the addition of 20 µL/tube 2× SureFire Lysis Buffer (PerkinElmer) and mixed, and 3 µL of lysate was transferred to a ProxiPlate384 Plus plate (PerkinElmer, 6008280). The AlphaScreen signal was developed as described below.
CETSA HT Screening Assays
Test compounds were dispensed into 384-well PCR plates (4titude, Dorking, UK; 4ti-0382) using either a Tecan HP dispenser or an Echo 555 acoustic dispenser. DMSO was backfilled to standard volumes. For B-Raf CETSA HT, A375 cells were harvested to 1.5 × 107 cells/mL in complete media. For PARP1 CETSA HT, MDA-MB-436 cells were harvested to 4 × 106 cells/mL in complete media. Ten microliters per well was seeded using a Multidrop Combi dispenser. Plates were briefly centrifuged, 300g, and incubated under tissue culture conditions for 1–2 h. Plates were heatshocked at 49 °C for 3 min and then 20 °C for 1 min, using a 384-well LightCycler 480 II PCR machine (Roche, Basel, Switzerland). Twenty microliters per well of 2× SureFire Lysis Buffer was added by Multidrop Combi and incubated 10 min at room temperature (RT). A Bravo liquid handler (Agilent, Santa Clara, CA) was used to mix the lysate (10 repetitive cycles of aspirating and dispensing 7 µL) and perform a plate-to-plate transfer of 3 µL of lysate to a 384-well ProxiPlate. For B-Raf CETSA HT, the AlphaScreen signal was developed by addition of 6 µL/well of 1× ImmunoAssay Buffer (PerkinElmer, AL000F) containing 1:2888 Mouse Monoclonal Anti-B-Raf (Abnova, H00000673-M02), 1:1444 Rabbit Anti-B-Raf Antibody (SantaCruz, sc-9002), 120 µg/mL Anti-Mouse IgG Alpha Donor Beads, and 30 µg/mL Anti-Rabbit IgG AlphaLISA Acceptor Beads. For PARP1 CETSA HT, the AlphaScreen signal was developed by addition of 6 µL/well of 1× ImmunoAssay Buffer containing 1:2000 Mouse Monoclonal Anti-PARP1 Clone 3G4 (Sigma, WH0000142M1), 1:5000 Rabbit Anti-PARP Antibody (Cell Signaling Technology, Danvers, MA; 9542), 240 µg/mL Anti-Mouse IgG Alpha Donor Beads, and 60 µg/mL Anti-Rabbit IgG AlphaLISA Acceptor Beads. Following 16 h under subdued light, analysis was performed using an EnVision plate reader. EC50 was determined using GraphPad (San Diego, CA) Prism or Genedata (Basel, Switzerland) Screener.
PARP1 Biochemical Fluorescent Polarization Assay
Test compounds were dispensed into 384-well plates using an Echo 555. Recombinant PARP1 protein was prepared to 6.8 µM in 50 mM KH2PO4 (pH 7.8), 150 mM KCl, 5 mM 2-mercaptoethanol, and 10% glycerol and stored at –80 °C. Protein was diluted to 6 nM with 50 mM Tris (pH 8), 0.001% Triton X100, 10 mM MgCl2, and 150 mM NaCl and 6 µL added per well using a BioRaptr dispenser. Six microliters per well of fluorescent probe was added using a BioRaptr to a final concentration of 1 nM (1× KD). Following 4 h incubation, plates were analyzed using a BMG Pherastar FS and the IC50 determined using GraphPad Prism or Genedata Screener.
PARP Cellular PARylation Assay
A549 cells were harvested to a density of 1.125 × 105 cells/mL in complete media and 40 µL/well seeded into 384-well plates (Greiner, Kremsmunster, Austria; 781090) using a Multidrop Combi. Following incubation under tissue culture conditions overnight, test compounds were added using an Echo 555. Following 1 h compound incubation, DNA damage was induced by the addition of 5 µL/well 9 mM H2O2 for 10 min under tissue culture conditions. Media was removed and cells were fixed in 20 µL of ice-cold methanol for 15 min at 4 °C. Blocking solution (3% bovine serum albumin [BSA] in phosphate-buffered saline with Tween 20 [PBST]) was added (1 h, RT). Twenty microliters per well of rabbit anti-PAR antibody (Trevigen, Gaithersburg, MA; 4436-BPC-100) at 1:1000 was added overnight (4 °C). Following three washes in PBST, 20 µL/well AlexaFluor 488 goat anti-rabbit (Invitrogen, 1:500) and Hoescht stain (Invitrogen, 1:5000) were added (1 h, RT). Following three washes in PBST, cells were imaged in 30 µL/well PBS using a CellInsight (Thermo Fisher) fitted with a 10× objective.
Results
CETSA HT to Measure Intracellular Target Engagement with B-Raf
The RAF family of kinases plays a critical role in signaling through the RAS-RAF-MEK-ERK pathway to drive cell survival and proliferation. Components of this pathway are frequently mutated in cancer, with the prevalence of B-Raf mutations around 8% across all cancers and accounting for significant proportions of melanomas, leukemias, hepatocellular carcinomas, and thyroid cancers.14 B-Raf is a clinically validated oncology target, with second-generation inhibitors vemurafenib and dabrafenib showing impressive efficacy and approved for the treatment of BRAF-V600E melanoma alone or in combination with MEK inhibitors. Despite this, all current B-Raf inhibitors develop resistance often linked to a paradoxical induction of B-Raf dimerization, which acts to drive pathway signaling rather than inhibit it.15 Based on more than a decade of research to understand the causes of resistance to B-Raf inhibitors, numerous third-generation inhibitors are currently being progressed,14 with hopes that these molecules may prolong the efficacy of B-Raf inhibition. Understanding the effects of these compounds in cellular models is often complicated by the difficulties in achieving kinase selectivity,14 and disconnects between kinase selectivity measured against purified proteins and in cells.16 Clear quantification of B-Raf target engagement in the cell has the potential to better rationalize the therapeutic potential of these compounds.
We looked to assess B-Raf target engagement in the human melanoma cell line A375 containing the clinically relevant V600E mutation in B-Raf. In a similar manner to previous reports,5,12 we identified and optimized mouse anti-B-Raf and rabbit anti-B-Raf antibodies capable of binding to B-Raf in a manner conducive to singlet oxygen transfer to produce an AlphaScreen signal (Suppl. Fig. S1). This approach provides a high-throughput-compatible endpoint with which to quantify thermostable B-Raf.
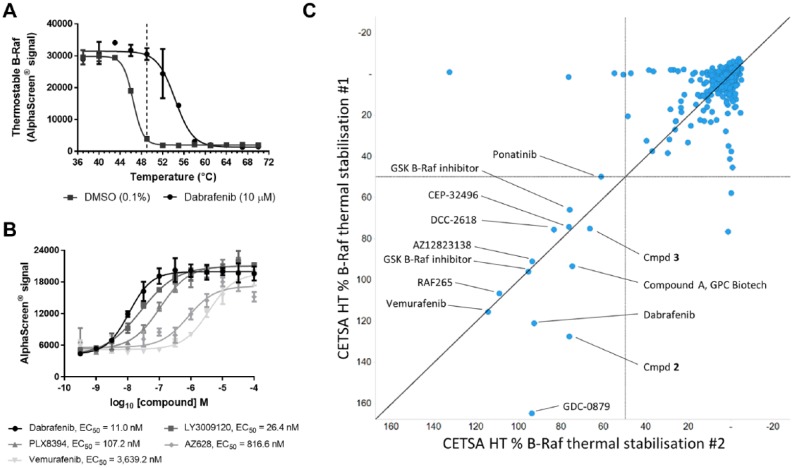
To assess the impact of compound target engagement on B-Raf thermal stability in live cells, A375 cells were incubated with DMSO control or 10 µM dabrafenib before being subjected to heatshocks across a range of temperatures from 37 to 70 °C. In untreated cells, we observed the loss of thermostable B-Raf with a Tagg of 46.4 °C, while the presence of dabrafenib caused a thermal stabilization of B-Raf (Tagg 54.3 °C; Fig. 1A ). A heatshock temperature of 49 °C was selected to perform isothermal dose–response fingerprint experiments (ITDRFCETSA)13 whereby a concentration–response series of various B-Raf inhibitors was tested to derive an apparent measure of intracellular B-Raf target engagement by determining the 50% effective concentration (EC50). A heatshock of 49 °C did not affect the integrity of A375 cell membranes (Suppl. Fig. S2). Following a 2 h incubation with A375 cells, thermal stabilization of B-Raf was evident with different degrees of potency for the second-generation B-Raf inhibitors dabrafenib and vemurafenib, as well as third-generation inhibitors LY3009120,17 PLX8394,18 and AZ62819 ( Fig. 1B ).
Figure 1.
A CETSA HT assay to screen for intracellular B-Raf target engagement. (A) A375 cells were treated for 1 h with DMSO control or dabrafenib (10 µM) before applying indicated heatshock, lysis, and quantification of thermostable B-Raf by AlphaScreen. Treatment of live cells with dabrafenib caused a thermal stabilization of B-Raf. Data are the mean ± span of n = 2. (B) ITDRFCETSA experiments to rank intracellular B-Raf target engagement. A375 cells were treated with a concentration response of B-Raf inhibitors for 2 h prior to a 49 °C heatshock, lysis, and quantification of thermostable B-Raf by AlphaScreen. Data are the mean ± standard deviation of n = 4. (C) CETSA HT screening of a library of kinase inhibitors to identify in-cell B-Raf binders. Compounds were screened at a test concentration of 30 µM for thermal stabilization of B-Raf relative to the dabrafenib control following 2 h incubation with A375 cells across two technical repeats.
To test whether B-Raf CETSA HT could successfully identify B-Raf inhibitors in a screening setup, the assay was applied to screen a focused library of 896 kinase inhibitors. Two technical repeats were performed wherein live A375 cells were incubated with compound for 2 h at a screening concentration of 30 µM prior to a 49 °C heatshock and quantification of thermostable B-Raf ( Fig. 1C ). Thirteen compounds (1.5%) reproducibly showed thermal stabilization of B-Raf. B-Raf inhibitors vemurafenib and dabrafenib were both identified as active from the screening. Other compounds identified as hits included reported B-Raf inhibitors GDC-0879,20 RAF265,21 and CEP-3249622 and two related B-Raf inhibitors described by GSK.23 A pan-kinase inhibitor, ponatinib, which has been reported to show inhibition of B-Raf,24 showed activity in the CETSA screen. Activity was also observed for a protein tyrosine kinase inhibitor reported by GPC Biotech, compound A,25 and the tyrosine kinase inhibitor DCC-2618.26 Both compound A and DCC-2618 showed thermal stabilization of purified B-Raf protein at the same concentration by differential scanning fluorimetry (data not shown). A known B-Raf inhibitor from the AstraZeneca collection, AZ12823138, and structurally related compounds 2 and 3 (Suppl. Fig. S3) were also identified as active ( Fig. 1C ). Despite a library of diverse kinase inhibitors being screened, 8 of the 13 hits identified by CETSA HT have been reported as B-Raf inhibitors, 3 of the 13 hits were internal compounds with evidence of binding and inhibiting B-Raf, and 2 compounds have been described as broad inhibitors of protein tyrosine kinases and were found to bind B-Raf. This demonstrates that the B-Raf CETSA HT assay can be applied to screening and is able to identify binders of B-Raf.
We next looked to demonstrate that B-Raf CETSA HT could reproducibly rank compounds based on their intracellular target engagement. Eighteen compounds were profiled by ITDRFCETSA to determine pEC50 across at least three technical repeats. As shown in Table 1 , the reproducible values for ITDRFCETSA pEC50 obtained allowed compounds to be successfully ranked by cellular target engagement in A375 cells. Well-described B-Raf inhibitors dabrafenib, LY3009120,17 AZ628,19 vemurafenib, PLX4720,18 and an alternative analog of vemurafenib showed clear evidence of target engagement, as did the previously described B-Raf inhibitor AZ12823138 and a structurally related compound 4 (Table 1 and Suppl. Fig. S4). The EC50 of B-Raf target engagement varied from 16 nM to 25 µM, with dabrafenib the most potent compound as measured by CETSA. Inhibitors of other components of the RAS-RAF-MEK-ERK pathway, as well as some unrelated kinases, were also profiled. Reported C-Raf-selective compounds PLX556827 and compound 10d28 were inactive in the assay, as were MEK inhibitors selumetinib (AZD6244, ARRY-142886),29 PD325901,30 and GDC-0623;31 the ERK inhibitor SCH772984;32 and an alternative ERK inhibitor, compound 3533 (Suppl. Fig. S4). The MEK inhibitor CH-5126766 showed weak activity in the assay (Table 1 and Suppl. Fig. S4) and has been reported to induce MEK-Raf complexes.34 Whether the formation of such complexes with B-Raf alters B-Raf thermal stability or the compound is directly binding to B-Raf at higher concentrations requires further investigation. Imatinib and crizotinib, inhibitors of kinases outside of the RAS-RAF-MEK-ERK pathway, were also inactive ( Table 1 ). In agreement with screening of the kinase library, the B-Raf ITDRFCETSA assay reproducibly and selectively identified B-Raf inhibitors. While weak activity was observed with CH-5126766, several inhibitors of other proteins within the RAS-RAF-MEK-ERK pathway did not show activity in CETSA HT. Hence, CETSA HT activity appeared specific to compounds binding B-Raf.
Table 1.
ITDRFCETSA to Rank Intracellular B-Raf Target Engagement.
| Compound | Molecular Target | B-Raf ITDRFCETSA pEC50 |
|---|---|---|
| Dabrafenib | Pan Raf | 7.8 ± 0.1 |
| LY3009120 | Pan Raf | 7.6 ± 0.2 |
| AZ628 | Pan Raf | 6.4 ± 0.1 |
| Vemurafenib analog | B-Raf V600E | 6.2 ± 0.1 |
| PLX4720 | B-Raf V600E | 5.8 ± 0.1 |
| Vemurafenib | B-Raf V600E | 5.8 ± 0.5 |
| AZ12823138 | Pan Raf | 5.6 ± 0.1 |
| Compound 4 | Pan Raf | 4.6 ± 0.0 |
| PLX5568 | C-Raf | <4.5 |
| Compound 10d | C-Raf | <4.5 |
| CH-5126766 | Raf/MEK | 5.0 ± 0.4 |
| Selumetinib | MEK | <4.5 |
| PD325901 | MEK | <4.5 |
| GDC-0623 | MEK | <4.5 |
| SCH772984 | ERK | <4.5 |
| Compound 35 | ERK | <4.5 |
| Imatinib | Abl, c-kit, PDGFR | <4.5 |
| Crizotinib | ALK, MET | <4.5 |
pEC50 was determined following 2 h incubation with live A375 cells for a range of Pan Raf and B-Raf inhibitors as well as inhibitors of alternative targets in the RAS-RAF-MEK-ERK signaling pathway and unrelated kinase targets. Data are the mean ± standard deviation of ⩾3 technical repeats. Example raw data are reported in Supplemental Figure S4.
Taken together, these data demonstrate the potential for CETSA HT to be applied to both primary screening for hit identification and SAR profiling to support lead optimization. In the case of B-Raf, CETSA HT identified confirmed B-Raf inhibitors from a library of diverse kinase inhibitors when applied in a screening setup and was able to reproducibly rank inhibitors by the apparent potency of their in-cell B-Raf target engagement in ITDRFCETSA experiments.
CETSA HT to Measure Intracellular Target Engagement with PARP1
PARPs are a family of enzymes that catalyze a posttranslational modification critical for the repair of damaged DNA. PARPs transfer ADP-ribose from NAD+ to DNA-associated proteins such as histones to build chains of poly(ADP-ribose) (PAR), which act to signal DNA damage and recruit a variety of DNA damage proteins.35 PARP inhibitors prevent binding of NAD+, blocking the PARylation event required to initiate DNA repair, and further stall the process of DNA repair by inhibiting auto-PARylation, leading to “trapping” of PARP on the DNA.36
There are a range of PARP proteins performing several functions in cells, but the primary target of PARP inhibitors is thought to be PARP1, the most ubiquitously expressed of the PARP enzymes and the main driver of PARylation upon DNA damage.35 Inhibition of PARP1 prevents PARP-mediated DNA repair.36 However, healthy cells can utilize a number of other DNA repair pathways that allow for redundancy within the system. Where tumors have deficiencies in such pathways, such as mutations in BRCA leading to defective homologous recombination repair, cells exhibit a greater dependency on alternative DNA repair pathways. Consequently, the combination of deleterious mutations in BRCA and inhibition of PARP leads to accumulation of insurmountable DNA damage, driving BRCA-mutant cancer cell death with an exquisite selectivity.37 PARP1 has been shown to be a key driver in several cancers of unmet medical need, with drugs such as Lynparza (olaparib) approved for the treatment of BRCA-mutant or platinum-sensitive ovarian cancer and BRCA-mutant breast cancer, and a range of PARP inhibitors under evaluation in numerous disease settings. One such molecule, iniparib, did not show meaningful activity in clinical trials and subsequently failed to demonstrate conclusive evidence of on-target activity against PARP,38 further highlighting the need for assessing on-target effects and target engagement early in the drug discovery process.
To develop a disease-relevant CETSA HT assay to measure target engagement with PARP1, we utilized a triple-negative breast cancer cell line with homozygous deleterious mutations in BRCA1, MDA-MB-436.39 Panels of mouse- and rabbit-derived anti-PARP1 antibodies were screened to enable quantification of thermostable PARP1 by AlphaScreen. Compatible antibody pairs were identified and the AlphaScreen signal was optimized by varying antibody concentration and cell density (Suppl. Fig. S5).
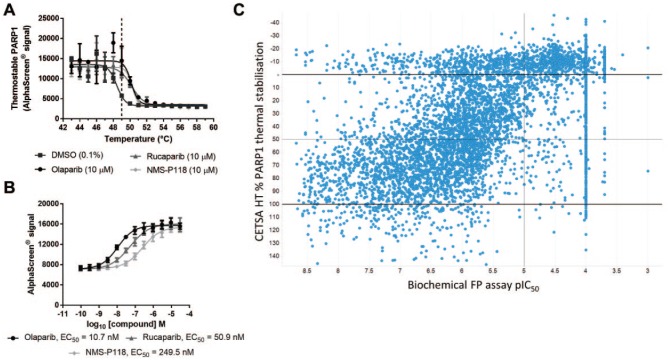
An AlphaScreen endpoint was then applied to determine the intracellular thermal aggregation behavior of PARP1 within live MDA-MB-436 cells, and the effect of the addition of PARP inhibitors on PARP1 thermal aggregation. Loss of thermostable PARP1 was observed in a temperature-dependent manner, and the treatment of live MDA-MB-436 cells with the PARP inhibitors olaparib, rucaparib, or NMS-P118 led to a thermal stabilization of roughly 2 °C, evidence of target engagement with cellular PARP1 ( Fig. 2A ). The apparent ITDRFCETSA of PARP1 target engagement could be determined and ranked by experiments performed with a 49 °C heatshock, well below temperatures that disrupt MDA-MB-436 cell membranes (Suppl. Fig. S2). This is consistent with previous reports in which the authors performed ITDRFCETSA experiments at 50 °C for CETSA Classics (Western blot) experiments on PARP1 from lysates of an alternative cell line.4 Olaparib showed intracellular PARP1 target engagement in MDA-MB-436 cells with an EC50 of 10.7 nM, while rucaparib and NMS-P118 showed intracellular PARP1 binding with reduced apparent potency of 50.9 and 249.5 nM, respectively ( Fig. 2B ).
Figure 2.
A CETSA HT assay to screen for intracellular PARP1 target engagement. (A) MDA-MB-436 cells were treated for 1 h with DMSO, olaparib (10 µM), rucaparib (10 µM), or NMS-P118 (10 µM) before applying indicated heatshock, lysis, and quantification of thermostable PARP1 by AlphaScreen. Treatment of live cells with PARP inhibitors led to a thermal stabilization of PARP1. Data are the mean ± span of n = 2. (B) ITDRFCETSA experiments to rank intracellular PARP1 target engagement. MDA-MB-436 cells were treated with a concentration response of PARP inhibitors for 1 h prior to a 49 °C heatshock, lysis, and quantification of thermostable PARP1 by AlphaScreen. Thermal stabilization as a measure of target engagement allowed compounds to be ranked by apparent potency. Data are the mean ± standard deviation of n = 10. (C) Single-concentration CETSA HT screening of a library of PARP1 binders. The affinity (pIC50) of 6288 compounds to PARP1 protein was determined by concentration–response experiments using a biochemical FP assay (x axis). The same compounds were screened for CETSA HT thermal stabilization at 10 µM, plotted as percent PARP1 thermal stabilization relative to 100% olaparib stabilized (y axis).
Subsequent optimization of this plate-based CETSA HT assay allowed for high-throughput measurement of intracellular PARP1 target engagement, although the assay achieved only moderately acceptable assay parameters. The assay robustness as measured by robust Z′ factor (RZ′) varied between ~0.25 and 0.55, driven principally by the low signal-to-noise ratio of the assay (<2) (Suppl. Fig. S6). However, as observed with the B-Raf CETSA HT assay, good reproducibility of EC50 determined from ITDRFCETSA experiments was observed with the PARP1 assay (Suppl. Fig. S6). We therefore sought to benchmark CETSA technology using PARP1 CETSA HT, with a focus on comparison with other assay formats.
We first tested PARP1 CETSA HT for successful application in a screening setup. We also looked to explore how this cellular target engagement assay (CETSA HT) compared with a biochemical binding assay. We took advantage of historical high-throughput screening (HTS) data available internally for PARP1 binders identified using a biochemical fluorescent polarization (FP) assay applied to purified recombinant PARP1. A library of 6288 compounds was prepared containing known PARP1 binders identified from historical screening and concentration–response profiling in the FP assay. This library constitutes a collection of compounds where the majority are expected to bind PARP1 with varying degrees of affinity. The library of PARP1 binders was screened by CETSA HT in an assay re-formatted for single-concentration screening at a test concentration of 10 µM.
The results of the CETSA HT screening are visualized in Figure 2C . CETSA HT percent activity was calculated based on maximal thermal stabilization induced by the olaparib control on each plate and is plotted on the y axis, against the pIC50 determined from concentration–response experiments in the original biochemical FP assay on the x axis. The majority of compounds showed similar trends between percent activity in CETSA HT and the biochemical pIC50, albeit with an apparent reduction in compound potency in CETSA. In general, less potent compounds in the biochemical FP assay showed less thermal stabilization by CETSA HT, whereas the majority of compounds with greater potency in the FP assay showed >50% thermal stabilization of PARP1 by CETSA HT ( Fig. 2C ).
Some compounds that did not show similar responses between the two assays were observed. Conclusions drawn from these comparisons should take into account that activity in CETSA HT requires the test compound to be cell permeable, which could account for compounds that show potent activity in the biochemical FP assay but are inactive by CETSA. One hundred seventy-five compounds (2.78% of all compounds) showed potent affinity for PARP1 protein (FP assay pIC50 >7) yet failed to show thermal stabilization (>50%) by CETSA HT, though only 108 (1.72%) showed <30% thermal stabilization. Additionally, 124 compounds (1.97%) failed to show any activity (pIC50 ⩽4) as a concentration response in the biochemical FP assay, despite initially being identified as a hit from single-concentration screening in the same FP assay and showing >50% PARP1 thermal stabilization by CETSA HT. These compounds most likely represent true PARP1 binders and reflect the false-negative rate of the biochemical FP assay.
Direct comparisons of expected hit rates between the assays are complicated by the reduction in observed compound potency when measured in the cellular target engagement assay (CETSA HT) compared with the biochemical FP assay. Nonetheless, using PARP1 CETSA HT as an example, the dataset suggests that some potent PARP1 binders would not have been active in a cellular target engagement assay. In addition, the reduced compound potency in the cellular target engagement assay relative to that observed in the biochemical binding assay would likely manifest as the detection of fewer hits with weak affinity from screening. However, of the 922 potent PARP1 binders in the collection (biochemical FP pIC50 >7), 747 (81%) were active (>50%) in CETSA HT. Screening by CETSA HT therefore identified the majority of potent PARP1 binders, with the advantage of immediately confirming that the compounds are cell permeable and engage the target in cells.
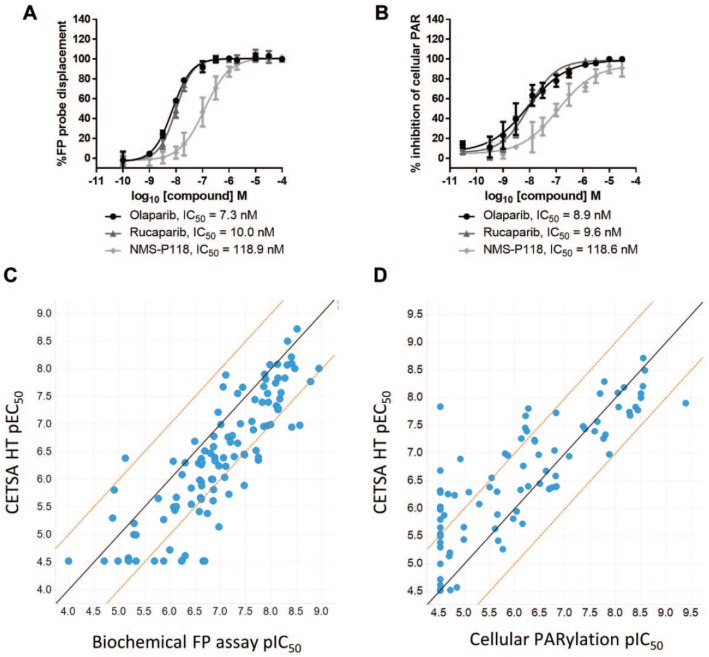
We next explored how CETSA HT might compare to other assay formats as hits were progressed into lead optimization and assessed for potency as part of SAR. The PARP1 CETSA HT assay was applied to determine the apparent potency of intracellular target engagement for 112 PARP inhibitors comprising numerous chemical series. The EC50 determined in ITDRFCETSA experiments was compared with the 50% inhibitory concentration (IC50) determined in two independent assays. First, we further compared intracellular target engagement as determined by ITDRFCETSA with the biochemical FP assay measuring binding to purified PARP1 protein ( Fig. 3A ). As shown in Figure 3C , we observed a good correlation between the CETSA HT pEC50 and the FP assay pIC50. In agreement with the dataset reported in Figure 2C , compounds were on average ninefold less potent in CETSA HT ( Fig. 3C ). The same compounds were also profiled in an alternative cellular assay measuring a functional endpoint of PARP activity. Cellular PARylation was quantified by imaging in A549 cells following 1 h compound treatment ( Fig. 3B ). A correlation was again observed between the CETSA HT pEC50 and the cellular PARylation pIC50, with more comparable potencies observed between these two cellular systems ( Fig. 3D ). A correlation was also observed between the FP assay and the cellular PARylation assay for the same compounds (Suppl. Fig. S7). A difference in compound potencies was observed between the FP assay performed with purified protein and the PARylation assay performed in cells, indicating that the reduced potency observed between CETSA HT and the FP assay (Figs. 2C and 3C) is not specific to CETSA and reflects the differences in potency measured between biochemical assays and cellular assays.
Figure 3.
Comparison of PARP inhibitor potency in CETSA HT with alternative assay formats. (A) A biochemical FP assay was employed to measure binding to purified PARP1 protein via competition of an FP probe. Data for PARP inhibitors, comparable to CETSA data in Figure 2B, are shown. Data are the mean ± standard deviation of ⩾4 technical replicates. (B) A cellular PARylation assay was employed to measure cellular PARP function. A549 cells were treated with PARP inhibitors for 1 h before addition of a DNA-damaging agent and imaging and quantifying PARylation. Data are the mean ± standard deviation of four technical replicates. (C) pEC50 determined by ITDRFCETSA plotted against pIC50 determined in the biochemical FP assay for 112 PARP inhibitors. The solid line shows a 1:1 correlation, and dashed lines represent a 1 log10 shift in potency. (D) pEC50 determined by ITDRFCETSA plotted against pIC50 determined in the cellular PARylation assay for 99 of the same PARP inhibitors. The solid line shows a 1:1 correlation, and dashed lines represent a 1 log10 shift in potency.
The majority of compounds showed good correlation between the potency and rank order of intracellular target engagement as determined by CETSA HT, binding to purified PARP1 protein as determined by the biochemical FP assay, and functional cellular effects on PARP activity as determined by cellular PARylation. However, a number of compound responses stood out.
Thirteen compounds (12%) showed binding to purified PARP1 with different degrees of potency but were inactive in CETSA HT ( Fig. 3C ). The possible explanations for such an observation could include lack of compound cellular permeability, instability within the cell media or intracellular environment, rapid cellular efflux, or intracellular binding without inducing a thermal stabilization, the latter constituting a CETSA false negative. However, analysis of the same compounds within the cellular PARylation assay revealed that all 13 were inactive in this alternative cell-based assay (Suppl. Fig. S8). Therefore, while these compounds bind to purified PARP1, they neither induce PARP1 thermal stabilization in CETSA HT nor inhibit PARP-mediated PARylation in cells, strongly highlighting the possibility that these compounds are not cell permeable. In support of this, the majority of these compounds did cause thermal stabilization of purified PARP1 protein by differential scanning fluorimetry (Suppl. Fig. S9).
Within the cellular PARylation assay, 19 compounds (17%) were inactive despite different degrees of apparent potency for intracellular binding by CETSA HT ( Fig. 3D ). All 19 of these compounds showed a similar potency of binding to purified PARP1 in the biochemical FP assay (Suppl. Fig. S10). Therefore, two independent assays report binding of these compounds to PARP1, one with purified PARP1 and one measuring intracellular target engagement by CETSA HT. Yet they failed to elicit functional inhibition of cellular PARylation. This observation could be explained in a few ways: (1) these compounds bind PARP1 in a manner that does not cause functional inhibition, or (2) 1 h may not be sufficient to observe functional inhibition of PARylation with these compounds.
The data reported herein demonstrate the potential advantages of applying CETSA HT technology in combination with additional assay formats to build a correlative understanding of binding to purified protein, target engagement in cells, and translation to functional cellular effects. For the majority of compounds tested, we observed good concordance of intracellular target engagement by CETSA HT with alternative assays for binding the target and causing functional cellular effects. Overall, while different compound characteristics could be observed between the three assays, all compounds correlated with at least one other assay, and there were no examples of compounds that were considered a false positive or false negative from profiling in CETSA HT. In agreement with the previous conclusions drawn from the B-Raf CETSA HT assay, the data obtained for PARP1 suggest that CETSA HT can be reliably used to rank compound potency and drive SAR and lead optimization in drug discovery screening cascades. This dataset also indicates that additional valuable information can be obtained by evaluating compounds using several assays that include CETSA HT.
Discussion
Methods to measure intracellular target engagement have been reported using chemical modifications or probe compounds (e.g., PET imaging or chemoproteomic probes1), or approaches that require probe compounds as well as labeling the target of interest (e.g., NanoBRET16). Together with CETSA, these technologies offer a range of options with which to better inform on cellular target engagement. As early drug discovery strives to become more efficient at delivering successful clinical candidates, cell-based assays increasingly aim to better recapitulate disease. This will likely involve the use of more advanced cellular models that are often less amenable to modifications required by some technologies. The absence of labels on either a probe compound or the target of interest yields a system more representative of disease physiology. As a label-free technique, CETSA may prove valuable for assessing cellular target engagement in more complex cellular models.
To date there have been only a few reported endogenous CETSA HT assays limited to the following drug targets: p38α,13 thymidylate synthase,5 and the AR.12 Recent work has demonstrated the potential to target further protein classes with alternative high-throughput-compatible forms of CETSA. High-throughput dose–response CETSA (HTDR-CETSA) uses transient expression of a tagged target to allow thermostable protein to be quantified using enzyme complementation technology, with demonstrated application to the histone methyltransferase SMYD3 and the immune checkpoint molecule indoleamine 2,3-dioxygenase (IDO1).9 A similar approach using a split luciferase reporter has also recently been described for several targets to reveal how changes in protein thermal stability in cells can be reliably applied in screening formats.8 Such approaches remove the requirement for high-quality antibodies against the target of interest, potentially enabling application of the technology to a greater number of proteins, and are a valuable additional approach to measure target engagement in cells. Herein we have focused on a high-throughput form of CETSA that retains endogenous and label-free expression of the target, though we believe the conclusions from this study and similar comparative observations from studies using exogenous tagged proteins8 should be applicable to most systems that monitor changes in protein thermal stability within the context of live cells.
Data presented herein expand the reported CETSA HT assays to include two additional and important drug targets, B-Raf and PARP1. Target engagement of inhibitors could be monitored for both of these targets by assessing changes in the proteins’ thermal stability in the context of live cells. While CETSA HT may not be universally applicable to all targets, it is clear that it can be applied to a varied range of intracellular proteins and should be considered a broadly applicable technology for drug discovery.
Several CETSA HT assays have now been described that are sufficiently robust for screening, although the small assay window observed with PARP1 CETSA HT may highlight an area for future improvement. The key driver for this small assay window was the low AlphaScreen signal from quantification of cellular PARP1, which could be caused by inefficient singlet oxygen transfer between the two primary PARP1 antibodies used, by the nature of measuring low levels of endogenous target protein, or by a combination of the two. This may be an important consideration for future applications of CETSA technology where assays should ideally retain the endogenous nature of the target protein. CETSA HT would benefit greatly from endpoints allowing greater amplification of signal when quantifying low levels of endogenous thermostable protein, allowing a larger assay window and reducing requirements for cell numbers. Recent reports of identification of individual antibodies capable of selectively quantifying thermostable protein for p38α10 and Chk111 have enabled application of CETSA with an imaging endpoint for these targets. This endpoint both allows single-cell resolution of target engagement and reduces requirements for cell numbers, though the successful application of this approach to measure binding to targets with low expression remains to be seen.
To build confidence in the application of CETSA HT to early drug discovery, we performed comparative analyses with more traditional drug discovery assay formats. For hit identification, application of both B-Raf CETSA HT and PARP1 CETSA HT for compound screening was able to identify genuine binders of the target. Comparison of a cellular target engagement assay (CETSA HT) with a biochemical binding assay using purified protein highlighted differences between the systems that should be considered before applying cellular target engagement for hit identification. First, we observed a reduction in apparent compound potency in CETSA when compared with an assay using purified protein, which would likely reduce the hit rate of screening and fail to identify binders with weaker affinity ( Fig. 2C ). For PARP1 this effect appeared relevant to all cell-based assays (Suppl. Fig. S7) rather than specific to CETSA, and comparisons of CETSA with an alternative cellular assay showed similar compound potencies ( Fig. 3D ). Second, a small number of compounds (~2%) showed potent binding to purified protein but were inactive in the cellular target engagement assay. It is possible that these compounds lack sufficient cellular permeability to exert activity in cell assays. Testing such compounds in cell lysate CETSA may help resolve such effects for other targets. Unfortunately, due to the instability of PARP1 in cell lysates, we were unable to generate a lysate form of the CETSA assay for this target. Third, CETSA screening would have identified the majority of potent PARP1 binders that were active in the biochemical binding assay. One consideration might be to preferentially apply cellular target engagement assays for primary screening during hit finding campaigns where the objective is to rapidly identify a potent, cell-permeable compound, and there is a reasonable expectation that such chemistry exists within the collection. Alternatively, as demonstrated in Figure 2C , a cellular target engagement assay can be employed immediately post-HTS to rapidly annotate which hits are binding the target in cells. Our data suggest that CETSA HT would be a robust and reliable technology to apply in these cases.
We also found CETSA HT to be suitable for lead optimization and ranking of compound potency as part of SAR. In addition to confirming good correlations between ITDRFCETSA and potency determined from other assays ( Fig. 3 ), we also explored the potential for false positives or false negatives from CETSA HT. Using the described PARP1 assays, we did not observe any ITDRFCETSA responses that did not correlate with at least one other assay, indicating that for this target at least CETSA HT can be considered highly reliable for profiling compounds. Nonetheless, as with other thermal shift assays, the possibility of compound binding without affecting the thermal stability of the protein has to be considered, an artifact that conceivably could be more pronounced in the complex environment of the cell where a protein’s thermal stability may already be influenced by interactions with other proteins or metabolites.40 Our own experience suggests that such situations are more likely to result in a failure to construct a CETSA HT assay in the first place, rather than false negatives being missed while screening, but whether CETSA HT responds differently to alternative modes of action, for example, allosteric binders, requires further investigation.
In conclusion, using B-Raf and PARP1 as two example targets, we show that CETSA HT is a highly robust and reliable approach that can be applied across numerous stages of early drug discovery. Both B-Raf CETSA HT and PARP1 CETSA HT were able to identify intracellular binders from screening, and rank binders to drive SAR. For PARP inhibitors, we observed good correlations between CETSA HT and more traditional assay formats. We propose that the inclusion of CETSA HT in screening cascades offers two advantages: (1) confirmation that compound-mediated functional cellular effects are a consequence of binding the intended target in cells, and (2) understanding of disconnects when binding to purified protein does not translate into desired functional cellular effects. As a technology broadly applicable to a large number of protein targets, CETSA HT may be a valuable addition to screening cascades in drug discovery projects. By bridging the gap between target engagement and the desired functional effect, CETSA HT could help expedite the identification and progression of novel small-molecule drugs.
Supplemental Material
Supplemental material, Shaw_et_al_SLAS_Discovery_Supplementary_Information for Positioning High-Throughput CETSA in Early Drug Discovery through Screening against B-Raf and PARP1 by Joseph Shaw, Ian Dale, Paul Hemsley, Lindsey Leach, Nancy Dekki, Jonathan P. Orme, Verity Talbot, Ana J. Narvaez, Michal Bista, Daniel Martinez Molina, Michael Dabrowski, Martin J. Main and Davide Gianni in SLAS Discovery
Acknowledgments
We thank Jeffrey Johannes and Andy Thomas (AstraZeneca) for help with generating suitable compound libraries. We also acknowledge Thomas Lundbäck and Paola Castaldi (AstraZeneca) for advice and comments on the manuscript.
Footnotes
Declaration of Conflicting Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: J.S., I.D., P.H., L.L., J.P.O., V.T., A.J.N., M.B., M.J.M., and D.G. are employees of AstraZeneca and potentially own stock and/or stock options in the company. N.D., D.M.M., and M.D. are employees of Pelago Bioscience AB and own stock in the company.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Supplemental material is available online with this article.
References
- 1. Simon G. M., Niphakis M. J., Cravatt B. F. Determining Target Engagement in Living Systems. Nat. Chem. Biol. 2013, 9, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morgan P., Brown D. G., Lennard S., et al. Impact of a Five-Dimensional Framework on R&D Productivity at AstraZeneca. Nat. Rev. Drug Discov. 2018, 17, 167–181. [DOI] [PubMed] [Google Scholar]
- 3. Smyth L. A., Collins I. Measuring and Interpreting the Selectivity of Protein Kinase Inhibitors. J. Chem. Biol. 2009, 2, 131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martinez Molina D., Jafari R., Ignatushchenko M., et al. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science 2013, 341, 84–87. [DOI] [PubMed] [Google Scholar]
- 5. Almqvist H., Axelsson H., Jafari R., et al. CETSA Screening Identifies Known and Novel Thymidylate Synthase Inhibitors and Slow Intracellular Activation of 5-Fluorouracil. Nat. Commun. 2016, 7, 11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seashore-Ludlow B., Lundback T. Early Perspective. J. Biomol. Screen. 2016, 21, 1019–1033. [DOI] [PubMed] [Google Scholar]
- 7. Dart M. L., Machleidt T., Jost E., et al. Homogeneous Assay for Target Engagement Utilizing Bioluminescent Thermal Shift. ACS Med. Chem. Lett. 2018, 9, 546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez N. J., Asawa R. R., Cyr M. G., et al. A Widely-Applicable High-Throughput Cellular Thermal Shift Assay (CETSA) Using Split Nano Luciferase. Sci. Rep. 2018, 8, 9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McNulty D. E., Bonnette W. G., Qi H., et al. A High-Throughput Dose-Response Cellular Thermal Shift Assay for Rapid Screening of Drug Target Engagement in Living Cells, Exemplified Using SMYD3 and IDO1. SLAS Discov. 2018, 23, 34–46. [DOI] [PubMed] [Google Scholar]
- 10. Axelsson H., Almqvist H., Otrocka M., et al. In Situ Target Engagement Studies in Adherent Cells. ACS Chem. Biol. 2018, 13, 942–950. [DOI] [PubMed] [Google Scholar]
- 11. Massey A. J. A High Content, High Throughput Cellular Thermal Stability Assay for Measuring Drug-Target Engagement in Living Cells. PLoS One 2018, 13, e0195050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shaw J., Leveridge M., Norling C., et al. Determining Direct Binders of the Androgen Receptor Using a High-Throughput Cellular Thermal Shift Assay. Sci. Rep. 2018, 8, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jafari R., Almqvist H., Axelsson H., et al. The Cellular Thermal Shift Assay for Evaluating Drug Target Interactions in Cells. Nat. Protoc. 2014, 9, 2100–2122. [DOI] [PubMed] [Google Scholar]
- 14. Karoulia Z., Gavathiotis E., Poulikakos P. I. New Perspectives for Targeting RAF Kinase in Human Cancer. Nat. Rev. Cancer 2017, 17, 676–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lito P., Rosen N., Solit D. B. Tumor Adaptation and Resistance to RAF Inhibitors. Nat. Med. 2013, 19, 1401–1409. [DOI] [PubMed] [Google Scholar]
- 16. Vasta J. D., Corona C. R., Wilkinson J., et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem. Biol. 2018, 25, 206–214.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng S. B., Henry J. R., Kaufman M. D., et al. Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-Tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell 2015, 28, 384–398. [DOI] [PubMed] [Google Scholar]
- 18. Zhang C., Spevak W., Zhang Y., et al. RAF Inhibitors That Evade Paradoxical MAPK Pathway Activation. Nature 2015, 526, 583–586. [DOI] [PubMed] [Google Scholar]
- 19. Hatzivassiliou G., Song K., Yen I., et al. RAF Inhibitors Prime Wild-Type RAF to Activate the MAPK Pathway and Enhance Growth. Nature 2010, 464, 431–435. [DOI] [PubMed] [Google Scholar]
- 20. Hoeflich K. P., Herter S., Tien J., et al. Antitumor Efficacy of the Novel RAF Inhibitor GDC-0879 Is Predicted by BRAFV600E Mutational Status and Sustained Extracellular Signal-Regulated Kinase/Mitogen-Activated Protein Kinase Pathway Suppression. Cancer Res. 2009, 69, 3042–3051. [DOI] [PubMed] [Google Scholar]
- 21. Williams T. E., Subramanian S., Verhagen J., et al. Discovery of RAF265: A Potent mut-B-RAF Inhibitor for the Treatment of Metastatic Melanoma. ACS Med. Chem. Lett. 2015, 6, 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. James J., Ruggeri B., Armstrong R. C., et al. CEP-32496: A Novel Orally Active BRAF(V600E) Inhibitor with Selective Cellular and In Vivo Antitumor Activity. Mol. Cancer Ther. 2012, 11, 930–941. [DOI] [PubMed] [Google Scholar]
- 23. Hammond M., Kallander L. S., Lawhorn B. L., et al. Compounds and Methods. WO2011149827, December 1, 2011. [Google Scholar]
- 24. Mian A. A., Rafiei A., Haberbosch I., et al. PF-114, a Potent and Selective Inhibitor of Native and Mutated BCR/ABL Is Active against Philadelphia Chromosome-Positive (Ph+) Leukemias Harboring the T315I Mutation. Leukemia 2015, 29, 1104–1114. [DOI] [PubMed] [Google Scholar]
- 25. Daub H., Wissing J., Missio A., et al. Pyridopyrimidines for Treating Inflammatory and Other Diseases. WO2005105097, November 10, 2005. [Google Scholar]
- 26. BLU-285, DCC-2618 Show Activity against GIST. Cancer Discov. 2017, 7, 121–122. [DOI] [PubMed] [Google Scholar]
- 27. Buchholz B., Klanke B., Schley G., et al. The Raf Kinase Inhibitor PLX5568 Slows Cyst Proliferation in Rat Polycystic Kidney Disease but Promotes Renal and Hepatic Fibrosis. Nephrol. Dial. Transplant. 2011, 26, 3458–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aman W., Lee J., Kim M., et al. Discovery of Highly Selective CRAF Inhibitors, 3-Carboxamido-2H-Indazole-6-Arylamide: In Silico FBLD Design, Synthesis and Evaluation. Bioorg Med. Chem. Lett. 2016, 26, 1188–1192. [DOI] [PubMed] [Google Scholar]
- 29. Yeh T. C., Marsh V., Bernat B. A., et al. Biological Characterization of ARRY-142886 (AZD6244), a Potent, Highly Selective Mitogen-Activated Protein Kinase Kinase 1/2 Inhibitor. Clin. Cancer Res. 2007, 13, 1576–1583. [DOI] [PubMed] [Google Scholar]
- 30. Sebolt-Leopold J. S., Herrera R. Targeting the Mitogen-Activated Protein Kinase Cascade to Treat Cancer. Nat. Rev. Cancer 2004, 4, 937–947. [DOI] [PubMed] [Google Scholar]
- 31. Robarge K. D., Lee W., Eigenbrot C., et al. Structure Based Design of Novel 6,5 Heterobicyclic Mitogen-Activated Protein Kinase Kinase (MEK) Inhibitors Leading to the Discovery of Imidazo[1,5-a] Pyrazine G-479. Bioorg. Med. Chem. Lett. 2014, 24, 4714–4723. [DOI] [PubMed] [Google Scholar]
- 32. Morris E. J., Jha S., Restaino C. R., et al. Discovery of a Novel ERK Inhibitor with Activity in Models of Acquired Resistance to BRAF and MEK Inhibitors. Cancer Discov. 2013, 3, 742–750. [DOI] [PubMed] [Google Scholar]
- 33. Ward R. A., Bethel P., Cook C., et al. Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point. J. Med. Chem. 2017, 60, 3438–3450. [DOI] [PubMed] [Google Scholar]
- 34. Lito P., Saborowski A., Yue J., et al. Disruption of CRAF-Mediated MEK Activation is Required for Effective MEK Inhibition in KRAS Mutant Tumors. Cancer Cell 2014, 25, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davar D., Beumer J. H., Hamieh L., et al. Role of PARP Inhibitors in Cancer Biology and Therapy. Curr. Med. Chem. 2012, 19, 3907–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pommier Y., O’Connor M. J., de Bono J. Laying a Trap to Kill Cancer Cells: PARP Inhibitors and Their Mechanisms of Action. Sci. Transl. Med. 2016, 8, 362ps17. [DOI] [PubMed] [Google Scholar]
- 37. Farmer H., McCabe N., Lord C. J., et al. Targeting the DNA Repair Defect in BRCA Mutant Cells as a Therapeutic Strategy. Nature 2005, 434, 917–921. [DOI] [PubMed] [Google Scholar]
- 38. Mateo J., Ong M., Tan D. S., et al. Appraising Iniparib, the PARP Inhibitor That Never Was—What Must We Learn? Nat. Rev. Clin. Oncol. 2013, 10, 688–696. [DOI] [PubMed] [Google Scholar]
- 39. Chavez K. J., Garimella S. V., Lipkowitz S. Triple Negative Breast Cancer Cell Lines: One Tool in the Search for Better Treatment of Triple Negative Breast Cancer. Breast Dis. 2010, 32, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dai L., Zhao T., Bisteau X., et al. Modulation of Protein-Interaction States through the Cell Cycle. Cell 2018, 173, 1481–1494.e13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Shaw_et_al_SLAS_Discovery_Supplementary_Information for Positioning High-Throughput CETSA in Early Drug Discovery through Screening against B-Raf and PARP1 by Joseph Shaw, Ian Dale, Paul Hemsley, Lindsey Leach, Nancy Dekki, Jonathan P. Orme, Verity Talbot, Ana J. Narvaez, Michal Bista, Daniel Martinez Molina, Michael Dabrowski, Martin J. Main and Davide Gianni in SLAS Discovery