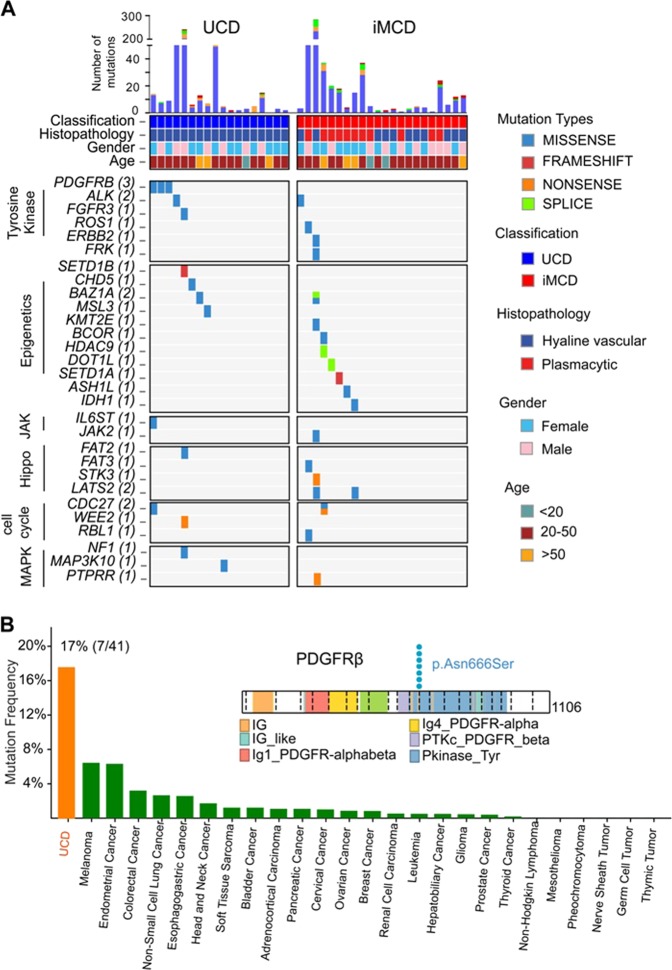
Fig. 1.
Whole-exome sequencing in 40 cases of CD. a Heat map of mutated genes in CD cases. These cases are separated into UCD set (n = 18) and iMCD set (n = 22). Each row represents a mutated gene in CD. Each column represents a patient sample. Blocks are color-coded by functional type of mutation. Top panel shows the number and type of nonsilent somatic mutations in each case. The mutations most likely to be related to CD pathogenesis are classified into the categories (indicated by different colors on the left): tyrosine kinase, epigenetic modifiers, Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway, Hippo pathway, cell cycle, and mitogen-activated protein kinase (MAPK) pathway. b Frequencies of PDGFRB mutations across cancer entities. Mutation frequencies obtained from http://www.cbioportal.org/public-portal/. UCD show the highest PDGFRB mutations frequency in cancer. Right panel shows the mapping of PDFGRB mutation sites in all of the UCD cases. Functional domains of the altered proteins are based on the UniProt database