Abstract
Nucleosomes represent the fundamental repeating unit of eukaryotic DNA, and comprise eight core histones around which DNA is wrapped in nearly two superhelical turns. Histones do not have the intrinsic ability to form nucleosomes; rather, they require an extensive repertoire of interacting proteins collectively known as ‘histone chaperones’. At a fundamental level, it is believed that histone chaperones guide the assembly of nucleosomes through preventing non-productive charge-based aggregates between the basic histones and acidic cellular components. At a broader level, histone chaperones influence almost all aspects of chromatin biology, regulating histone supply and demand, governing histone variant deposition, maintaining functional chromatin domains and being co-factors for histone post-translational modifications, to name a few. In this essay we review recent structural insights into histone-chaperone interactions, explore evidence for the existence of a histone chaperoning ‘pathway’ and reconcile how such histone-chaperone interactions may function thermodynamically to assemble nucleosomes and maintain chromatin homeostasis.
Keywords: Chaperone, chromatin, HIstone chaperone, histones, Nucleosome
Introduction
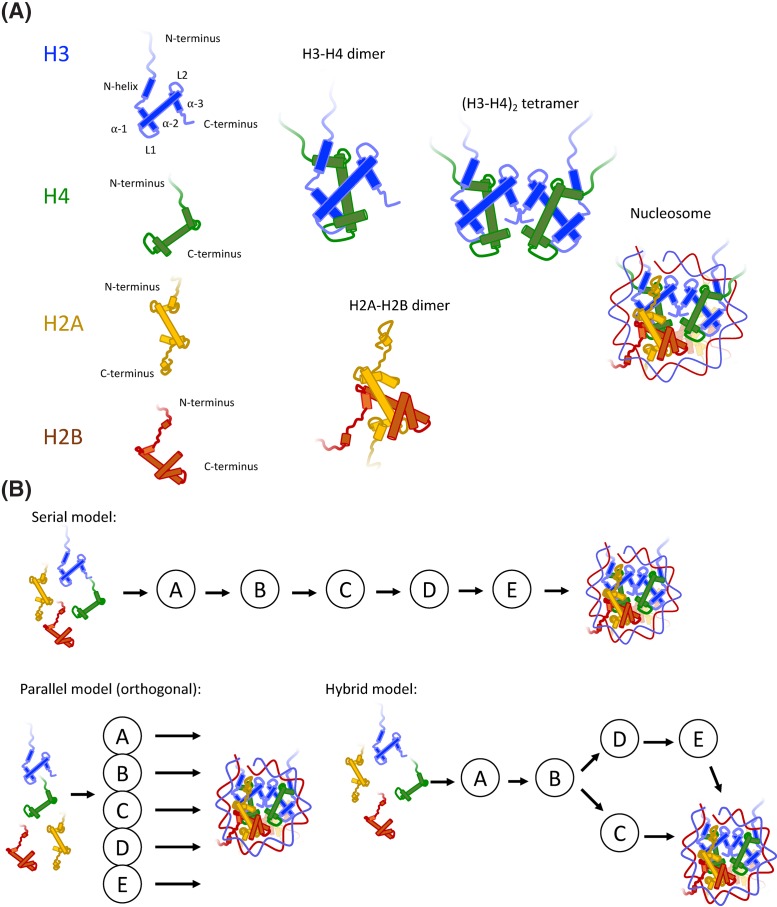
Eukaryotes package their genome within the confines of the cell nucleus into a structure broadly defined as chromatin. Chromatin is composed of an array of proteins, RNA and genomic DNA, whose integrity is crucial for genomic regulation, stability, replication and repair [1,2]. The most abundant proteins in chromatin are histones. These form an octamer around which genomic DNA wraps. Two copies of Histone H2A, H2B, H3 and H4 are encircled by a strand of DNA that is wrapped 1.65 times, corresponding to ∼147 base pairs of DNA [3]. These core histones, together with their associated DNA form the basic repeating unit of chromatin – the nucleosome (Figure 1A). Core histones contain a globular region that is characterized by a conserved histone-fold domain. Histone-fold domains comprise approximately 65 amino acids that form three α-helices in the presence of a dimerization partner [4]. On the N-terminus, histones present an unstructured tail that protrudes from the nucleosome when fully assembled, and can also contain C-terminal extensions [5]. Dimerization of histones occurs by the antiparallel association of histone monomers in what is often referred to as a ‘handshake’ motif [6] (Figure 1A).
Figure 1. Components of the nucleosome and considerations for multichaperone networks.
(A) Nucleosomes comprise of four core histones – H2A, H2B, H3 and H4. The central histone fold comprises three helices, α1, α2 and α3, which can be flanked by extension on the N and C termini. N-terminal extensions are in the form of basic tails, and in the case of H3, an additional α-helix (αN-helix). H3 and H4 fold to form an H3–H4 dimer, two of which can form an (H3–H4)2-heterotetramer. The tetramer adopts the central location in the nucleosome, known as a ‘tetrasome’ when bound to DNA on its own, and is capped by two H2A–H2B dimers to form the nucleosome core particle, which wraps 145–147 base pairs of DNA in almost two superhelical turns. (B) Histones associate with a large repertoire of chaperoning complexes, represented as A, B, C, D and E. Assuming mutual exclusivity in binding, histones may transition through multiple chaperoning complexes before their incorporation in chromatin (a serial pathway), or each chaperoning complex may represent an orthologous route to deposition (a parallel pathway), or indeed, a mixture of the two (a hybrid model). In reality, a hybrid model which bifurcates depending of histone isotype best explains the H3–H4 deposition pathway (see Figure 2).
Histones are positively charged at physiological pH, enabling them to interact with the negatively charged DNA backbone. However, this also means histones are prone to promiscuous interactions with acidic cellular components, which may result in spurious protein aggregates. As such, regulating histone interactions from their synthesis in the cytoplasm and subsequent integration into nucleosomes is paramount. This function is performed by proteins that interact with soluble (i.e. non-chromatinized) histones, broadly defined as ‘histone chaperones’.
Histone chaperones originate from a wide range of protein families, spanning diverse protein folds [7]. Yet, they share the key common feature of binding histones and protecting them from undesirable interactions, be that during storage, transport, or nucleosome assembly [8]. Here we present an overview regarding the life of a histone, from synthesis and ribosomal exit, through import and nuclear processing, and finally deposition onto DNA at sites of DNA replication or histone turnover.
The challenges in observing transitions between histone chaperones
Core histones stably interact with a large repertoire of structurally distinct histone chaperones [9]. As histones are small proteins (a histone fold dimer is approximately 25 kDa) it is unlikely that all of these proteins can bind at once, and indeed, more detailed biochemical investigations have demonstrated, for H3 and H4 at least, both compatibility and mutual exclusivity in histone binding [10–15]. These findings as a whole have been corroborated through high-resolution structures demonstrating both competition and compatibility. The question thus arises of whether mutual exclusivity in histone chaperone binding manifests as multiple parallel pathways for histone deposition, or whether a serial pathway prevails, in which histones transition between chaperoning components, or whether a mixture of the two exists in the cell (Figure 1B).
Experimentally, gaining kinetic information on the protein-protein interactions that histones make from the point of synthesis to the point of deposition has been difficult. Early pulse-chase analysis using radiolabelled amino acids and cultured mammalian cells revealed that newly synthesised histones are incorporated into chromatin in less than a minute after their synthesis [16]. The challenging nature of performing a pulse-chase experiment, that also relays protein-protein interaction information on such short time scales means the majority of our understanding has been derived through structural and biochemical observations, often of individual components in isolation. Taking histones H3 and H4 as examples, this has resulted in a number of common themes: (1) Histones are imported through interaction with importin-β proteins, namely IPO4, (2) a multi-chaperoning complex involving ASF1a/b, NASP, HAT1 and RbAp46 forms the major soluble pool of H3 and H4, (3) ASF1 is a central hub for the histone chaperoning network and is regulated by a number of co-chaperoning factors including the Tousled-like kinases 1 and 2 (TLK1 and TLK2), and Codanin-1 (CDAN1) in humans, and (4) H3–H4 are transferred to variant specific chaperones, often tethered to chromatin, for incorporation at sites of DNA replication or histone turnover (Figures 2 and 3). Thus, it appears that histones H3 and H4 pass through a common soluble complex, regardless of their variant type (excluding the centromeric variant CENPA), before they bifurcate depending on isoform and/or genomic context into a number of different deposition complexes. Much less is known about potential hand-off between H2A–H2B chaperoning complexes, therefore, in this article we will focus predominantly on the chaperoning pathways concerned with H3 and H4.
Figure 2. Histone synthesis and nuclear import.
After synthesis histones H3 and H4 interact in the cytosol with common folding chaperones HSC70 and HSP90, with H3 being mono-methylated on K9 co-translationally by SetDB1. Two pathways have been proposed for the import of H3 and H4: folding of an H3–H4 dimer in the cytosol and subsequent binding to ASF1 before import by IPO4, or import of monomeric histones bound directly to importins, with H3–H4 dimerisation occurring in the nucleus. ASF1’s interaction with H3–H4 in the cytosol can be regulated by the protein CDAN1, which competes with histone for ASF1 binding. H2A and H2B are imported as dimers bound to NAP1 and IPO9. In the nucleus, NASP interacts with H3 downstream of ASF1 through a high affinity interaction with the H3 α3 helix, and may thereby function as a receptor of H3 from the import machinery. Whether a monomeric H4 chaperone exists is not known. RbAp46 and HAT1 associate with H4, acetylating K5 and K12, and together with NASP and ASF1 form the major soluble H3–H4 complex. The S-phase-specific TLK1 and TLK2 can increase the affinity between histones and ASF1 through ASF1-specific phosphorylation marks.
Figure 3. Histone chaperoning in the nucleus.
Histone deposition can be split into replication dependent, and replication independent mechanisms. The variants H3.1/H3.2–H4 are incorporated into chromatin predominantly at the replication fork during replication dependent deposition, in which a number of chaperoning components have been implicated, including FACT (SpT16-SSRP1), CAF1 (p150, p60, RbAp48), and the replication proteins POLE3–4, RPA, MCM2 (N-terminal tail) and TONSL. TONSL is also involved in the DNA damage response, a role that may be related to its chromatin assembly function. ASF1 may act as a bridging factor between soluble pool and deposition factors. In addition to H3–H4, the FACT complex also binds to H2A–H2B through its U-turn motif and C-terminal acidic stretch, and may be responsible for governing H2A–H2B deposition post replication, or indeed, the chaperones NAP1 and Nucleoplasmin may play a role. Replication independent deposition occurs throughout the cell cycle, utilising the H3.3 variant. At sites of transcription both the HIRA complex (HIRA, UBN1 and CABIN1), which can bind naked DNA, and FACT complex, which associates with elongating RNA pol II, are involved in H3–H4 deposition, with the FACT complex also extending its role to H2A–H2B incorporation. At other sites in the genome, including heterochromatic repetitive elements, the chaperone DAXX and chromatin remodeller ATRX govern the incorporation of H3.3. Histone chaperones also play a central role during DNA repair, omitted here for brevity. For a detailed description we refer the reader to some recent excellent reviews [131–133].
Histone synthesis and nuclear import
Shortly after translation, histones interact with canonical folding chaperones. H4 has been shown to interact with HSC70 and HSP90 [11], whereas H3 interacts solely with HSC70 [17]. Attempts have been made to separate out pre- and post-import machineries through biochemical fractionation of the cytosolic fraction. However, a number of factors identified to be cytosolic in these studies have been found to be nuclear when probed in intact cells; namely NASP [15,18], ASF1 [19–21], HAT1 [22,23] and RbAp46 [15,24,25]. The reason for this discrepancy could lie in the soluble nature of these proteins, rapidly leaking into the cytosolic fraction upon common sub-cellular fractionation methods, with insoluble proteins, such as lamins and tubulin, being poor controls for reporting on such mixings [15,26].
Biochemical data showing a soluble pool of importin-ASF1 bound to the H3–H4 dimer suggested a fast mechanism for histone H3 and H4 co-folding and import [10,11,17]. Recently, evidence combining cell microscopy and a novel pulse-chase system to deploy fluorophore-labelled histones, suggested that human histones H3 and H4 can be translocated by importin proteins as monomers to the nucleus [15]. It should be highlighted, however, that the approach required a synthetic tethering system for observation, and therefore its relevance to endogenous histone import had to be implied. Promisingly, however, it was also found that H3 exists in a stoichiometric excess of H4 when bound to the ubiquitous nuclear chaperone NASP, suggesting an endogenous pool of NASP-H3 monomer may exist (Figure 2) [15]. This was in support of previous biochemical analysis of NASP showing that it could form both a tetrameric complex with H3–H4 and ASF1, and a stable dimeric complex with H3 on its own, albeit using purified components in vitro [27]. Further investigation is needed to discriminate the utilisation of these two potential pathways for histone import.
NASP is the human homolog of the first H3–H4-specific chaperone to be identified, N1/N2 [28,29], and contains four conserved TPR repeat motifs. Stacking of TPR repeats forms a superhelical groove that often serves as a peptide binding pocket in mediating protein-protein interactions [30]. Consistently, it was found that NASP binds to a short motif found at the very C-terminus of histone H3 through a canonical TPR-peptide interaction [31], but with much greater affinity than is typical of TPR–peptide interactions. Interestingly, the crystal structure of ASF1–H3–H4 shows that the residues of H3 involved in NASP interaction are the same residues necessary for binding to ASF1 [32–34]. NASP and ASF1 can compete for this binding site, with NASP out-competing ASF1 when an H3 monomer is used as a substrate [27]. Surprisingly, however, NASP can also bind an H3–H4 dimer in a conformation that is compatible with ASF1 [27]. This disparity was resolved with the discovery of a second H3–H4 specific interaction site on NASP involving its unstructured acidic domain [27]. Interestingly, the ability to chaperone both H3 monomers and H3–H4 dimers implies that NASP may play a role in the folding of H3 with H4. Its ability, in concert with ASF1, to produce folded H3–H4 dimers in vitro from monomeric substrates lends support to this idea [27]. Governing the transition from monomer to dimer may also aid in NASP’s ability to protect a soluble pool of H3–H4 in vivo [35]. Thus, NASP may function as a receptor for incoming monomeric H3 in the nucleus, accepting H3 from importins, and guiding its folding with H4. In addition to NASP, the histone chaperone ASF1 has the ability to destabilize the importin-histone H3 interaction in vitro [36], suggesting a possible role for importin-histone dissociation upon nuclear localization.
HAT1 and RbAp46 form the dimeric HAT1 complex, which can interact with H3–H4 together with ASF1 and NASP [13–15,37]. This is likely due to the binding sites of HAT1 and RbAp46 residing in the N-terminal tail [38] and first α-helix of H4 [23,39,40], respectively, being distinct from the binding sites of NASP and ASF1. A recent co-crystal structure of the yeast HAT1 complex demonstrated how binding of the α1 helix by HAT2 (the yeast homolog of RbAp46) positions the H4 tail for acetylation on K12 by the enzyme HAT1 [41]. Interestingly, the binding site of HAT2 (RbAp46), the α1 helix of H4, is positioned against the H3–H4 histone fold dimer in all known crystal structures. Thus, in order to interact with RbAp46 the helix must rotate outwards, suggesting that the H3–H4 dimer may be in a destabilised form when associated with the HAT1 complex.
Diacetylation of newly synthesised H4 is highly conserved throughout evolution, however, its mechanistic function is still unclear, with the modification being removed soon after deposition. In addition to diacetylation of H4 [42], recent biochemical analysis of H3 has proposed methylation of K9 by SETD1B in the cytosol, occurring co-translationally [43]. Such methylation on newly synthesised H3 has been proposed to potentiate their final epigenetic fate [44–46]. Similarly, the enzymes PRDM3 and PRDM16 have been shown to mono-methylate H3K9 in the cytosol of mouse cells, a necessary precursor of tri-methylation in the nucleus [47]. HAT2 was shown to interact with the N-terminus of H3 through interacting with unmethylated R2 via the pocket formed at the centre of its WD40 repeats [41]. As newly synthesised H3 molecules are devoid of R2 methylation, it was suggested that this could be a mechanism to discriminate between new and old histones. In conclusion, the major soluble complex of H3–H4 in human cells is non-variant specific in relation to H3, and contains the chaperoning components ASF1, NASP, RbAp46 and HAT1 (Figure 2).
Incorporation of newly synthesized histones
Numerous H3–H4 specific chaperones, in addition to established replication components with novel chaperoning ability, are stable constituents of the replication fork, including CAF1, FACT, MCM2, ASF1, RPA, POLE3–4 and TONSL [48–56]. During replication-dependent deposition both parental and newly synthesised histones must be used as substrates to assemble the newly replicated DNA into chromatin.
H3–H4 dimers can be provided by de novo nucleosome assembly through ASF1. Interestingly, ASF1, a component of the major soluble complex of H3–H4 (Figure 3), can interact with a number of deposition chaperones including CAF1, MCM2 and HIRA. This suggests a role for ASF1 in bridging the soluble and deposition phases of the histone chaperoning pathway. ASF1 in humans is present in two non-allelic isoforms, ASF1a and ASF1b. These paralogs share 70% sequence identity, and appear to be predominantly redundant in their roles as histone chaperones, although ASF1b is associated with cellular proliferation, whereas ASF1a is also expressed in cells that have become quiescent [57].
ASF1 can interact with an H3–H4 dimer at the same time as the unstructured N-terminal region of MCM2 and the ankyrin repeat domain (ARD) of TONSL [52,58]. In turn, MCM2 has been shown to bind to both the (H3–H4)2 histone tetramer and to ASF1 bound to an H3–H4 dimer [59,60], the latter suggesting a mechanism for the recycling of (H3–H4)2 histone tetramers across the replication fork [59,61]. TONSL interacts with an H4 tail peptide through its ARD domain, which can occur concomitantly with MCM and ASF1 binding [58]. Interestingly, the MMS22L–TONSL complex remains bound to H4 after its incorporation into chromatin, marking post-replicative chromatin through its ability to discriminate newly synthesized, non-methylated H4K20 [58]. TONSL has an additional, well documented, role in stimulating replication fork recovery after collapse, a function that may be linked to its chromatin assembly ability [52,53]. As TONSL interacts selectively with newly synthesised H4, and is found in complex with MCM2, it follows that MCM2 may also interact with newly synthesized histones in addition to parental histones.
ASF1 also interacts with yeast CAF1 subunit Cac2 [62–65] and its human [21] and fly counterparts [19] through a weak interaction distinct from its H3–H4 binding interface [66,67], providing the H3–H4 dimer for the concerted H3–H4 tetramerization on DNA by a CAF1 dimer. The small subunit of CAF1, RbAp48, interacts with the α1 helix of H4 [23] and likely represents a secondary hand-off event from its paralog RbAp46 in the HAT1 complex [39,68]. The CAF1 complex is recruited to active replication foci [69], where it binds to the DNA polymerase complex through its PCNA interacting peptide (PIPs) motifs [49,70–73]. In vivo [74] and in vitro [75] studies have shown that CAF1 is necessary and sufficient for nucleosome deposition after DNA replication. In yeast, Cac1, the largest subunit, acts as a scaffolding for the Cac2 and Cac3 subunits as well as for the H3–H4 dimer [76]. The H3–H4 dimer binds extensively to the Cac1’s C-terminus, including an acidic domain that releases intramolecular interactions with the Winged-helix DNA Binding domain, or WDB [77]. In the proposed model, the freed WDB domain can then bind to DNA, stabilising the complex at the replication fork [78]. The Cac1 subunit undertakes most of the chaperoning activity, enabling CAF1 complex dimerization, and promoting the deposition of H3–H4 onto DNA as a tetramer (H3–H4)2 [76,77].
Histone recycling during replication
Early experiments established the ‘tetramer conservation model’, where (H3–H4)2 tetramers are formed either with parental H3–H4 or de novo synthesized histones, but not randomly mixed [79,80]. Parental and new tetramers are found evenly partitioned across the leading and lagging strands, forming full nucleosomes in a second step with available H2A–H2B dimers [80–82]. However, the tetramer conservation model is not completely universal, but rather restricted to replication, since highly translated genes tend to present a certain degree of tetramer mixing [83], contrary to lowly expressed loci [84]. In agreement with this, tetramers with histone variant H3.3, associated with the replication-independent pathway, are more likely to mix than tetramers with variant H3.1 deposited during DNA replication [85].
Nucleosome recycling constitutes a potential mechanism to enable the perpetuation of the chromatin landscape with its implications in epigenetic inheritance. Experiments recreating replication in vitro suggest that parental nucleosomes tend to remain at nearby loci when a rich extract containing all the replication-fork interacting chaperones (such as the aforementioned FACT, CAF1, or MCM2) is used [86]. On top of this, nascent chromatin sequencing paired with amino acid radio-labelling suggests that PTM are kept on parental histones with chromatin landscape reconstruction within one generation [87]. Due to the speed with which replication occurs, it has been difficult to gain direct information about the order of binding events that must follow to process both newly synthesised and parental histones, and the mechanisms that may keep them partitioned. However, significant advances have been made from recent structural, biochemical and novel next-generation sequencing.
Replication protein A complex (RPA, a complex that binds and protects single strand DNA), can also function as a binding partner for an H3–H4 dimer [55]. Possibly related to these findings, recent studies have speculated that histones may stably interact with single-stranded DNA [88,89]. Whether this is mediated through their interaction with RPA, and what role this would have in vivo (perhaps protecting the lagging strand or conserving the original nucleosome position), is yet to be solidified. Interestingly, DNA polymerase ε (POLE) small subunits POLE3 and POLE4 have recently been reported to act as H3–H4 histone chaperones at the replication fork during histone recycling [56] (Figure 3). Work done on yeast homologs Dpb3 and Dpb4 suggests that these subunits of DNA polε favour parental histone loading on the leading strand to counter a lagging strand bias [90]. Conversely, MCM2 has been implicated in the even distribution of parental (H3–H4)2 tetramers across both DNA strands, showing a preference for parental histone recycling to the lagging strand in mice to counter a leading strand bias [91,92]. Future work is necessary to clarify in vivo deposition partners and their handover mechanisms. For instance, do POLD3 and POLD4 (equivalent subunits to POLE3 and 4 in polymerase δ) have similar roles in the lagging strand?
Nucleosome disassembly, re-assembly and H2A–H2B histone chaperones
Another highly conserved chaperoning complex at the replication fork is FACT. FACT comprises three subunits in yeast, Spt16, Pob3 and Nhp6, but only two, SPT16 and SSRP1, in metazoans [93]. Originally identified in its role as FACilitating Transcription (FACT) through its interaction with RNA polymerase II [94–96], FACT has an essential role in the removal [97] and deposition of nucleosomes [98], and localizes to the replication fork predominantly through interactions with DNA polymerase α [50,99], but also through MCM2 [54] and RPA [100]. FACT can bind to H2A–H2B dimers through both the U-turn motif of the Spt16 M-domain, and through hydrophobic residues within the C-terminal acidic domains of both Spt16 and Pob3 [99,101]. In addition, the Spt16 subunit can bind to the (H3–H4)2 tetramer through the M-domain, contacting the same surface used to mediate histone–DNA interactions [102]. SSRP1 can bind to both H3–H4 and H2A–H2B [103]. FACT binds simultaneously to the (H3–H4)2 tetramer bound to DNA and to H2A–H2B [104], possibly interacting with the H2A–H2B dimer in concert with DNA polymerase α [105]. The structure of Spt16 bound to the (H3–H4)2 tetramer is in contrast with that of MCM2 in that it contacts both H3–H4 dimers across the dyad interface, rather than a single H3–H4 dimer. This may have implications in the re-deposition of the parental tetramer across cell generations as mentioned above [80,84,85].
Functionally, in vitro nucleosome reconstitution and electrophoretic mobility assays showed that FACT can bind transiently to partially dissociated nucleosomes [101,104]. Structural modelling suggested that FACT acts as a wedge binding to the (H3–H4)2-tetramer whilst still associated with DNA to favour H2A–H2B dissociation [102]. Furthermore, micrococcal nuclease treatment of FACT-bound nucleosomes and nucleosome fragments evidence that FACT binds preferentially to hexasomes or partially digested nucleosomal DNA rather than to whole nucleosomes [102,104]. These observations, together with the ability to reconstruct nucleosomes in vitro, where DNA ultimately outcompetes FACT [104], suggests that FACT is capable of disassembling nucleosomes only when the process is energetically favourable (potentially fuelled through the CMG helicase, a processive polymerase or an ATP-dependent chromatin remodelling factor).
In addition to FACT, NAP1 and Nucleoplasmin, as discussed in the next section, are H2A–H2B binding chaperones that could provide H2A–H2B for nucleosome assembly behind the replication fork. Although they have been isolated as constituent parts of the replisome, they could be in high enough concentration in the nucleus that free diffusion is sufficiently rapid to allow the final step of nucleosome assembly to occur within a sufficient time frame.
Histone chaperoning during transcription
Outside of S-phase, histones are deposited at regions of high nucleosome turnover. This occurs as a consequence of processes requiring access to DNA, such as transcription, DNA repair, recombination and also variant replacement under specific genomic contexts [106]. HIRA is a histone chaperone complex, named after its principle subunit, dedicated to nucleosome formation in a replication-independent manner [13,107]. HIRA functions by chaperoning H3.3 variant to exposed DNA and transcriptionally active regions [108–110]. The HIRA complex is made of three HIRA subunits, two CABIN1 subunits and Ubinuclein-1 or UBN1 [111]. ASF1 serves as histone H3.3–H4 dimer donor through the B-domain of HIRA, analogous to that of the recruitment to CAF1 [112,113]. The specificity of binding towards H3.3 is conferred to the HIRA complex through the UBN1 subunit, directly interacting with ASF1–H3.3–H4 [114]. HIRA complex can be tethered to actively transcribed DNA through RPA [115], highlighting the universal nature of RPA tethering chromatin formation to single stranded DNA. Indeed, a gap-filling mechanism has been proposed for HIRA in which it senses naked DNA through its intrinsic DNA-binding ability and directs deposition of an H3.3 containing nucleosome [109].
In addition to being a component of the replisome, FACT is also an essential chaperone that associates with elongating RNA polymerase II, as its name suggests (FACilitating Transcription) [94,116], as well as with RNA polymerase I [117], and III [95]. One can assume that the underlying biochemical mechanism for nucleosome reorganisation by FACT is the same for both its functions at the replication fork and at sites of transcription, and again suggests that FACT is only capable of outcompeting DNA for its substrate when fuelled by a processive enzyme such as RNA polymerase. Kinetic analysis suggests that human FACT decreases the non-productive time of RNA polymerase II [118] while ChIP-seq analysis showed a marked correlation between FACT associated with gene transcription start sites and gene transcription [117]. Taken together, these data suggest a role for FACT in helping RNA polymerase II processivity and ensuring the prompt reassembly of the nucleosome (reviewed in [119]).
Structurally similar to FACT, ANP32E is a histone chaperone dedicated to the removal of histone variant H2A.Z–H2B dimers, located to actively transcribed genes and regulatory elements such as enhancers and insulators [120]. Another H2A–2B chaperone that has been implicated in transcription is nucleosome assembly protein 1 (Nap1). Nap1 envelops the H2A–H2B dimer similarly to DNA [121], preventing non-productive associations and thereby driving correct nucleosome assembly [122]. ∆Nap1 assays in yeast showed an increase in the presence of hexasomes in the genome, suggesting a preferential role in depositing the second of the two H2A–H2B dimers [121]. Nucleoplasmin (or NPM2 in humans) is a pentameric (or decameric) complex [123,124] that can reconstitute nucleosomes in vitro [125]. The potential interactions and dynamics between H2A–H2B-interacting chaperones, such as FACT, Nap1 and nucleoplasmin remain to be clarified.
Histone replacement at repetitive elements
Interestingly, the H3.3 variant accumulates not only on actively transcribed genes, but also over largely silenced regions such as telomeres and pericentromeric regions [103]. This observation led to the discovery of the histone-binding protein DAXX and chromatin remodeler ATRX, that form a dimeric complex involved in H3.3 deposition and remodelling [23,29]. DAXX envelops the H3.3–H4 dimer in a way incompatible with ASF1 interaction, and correspondingly ASF1 and DAXX are not found in complex with each other in vivo. Whether H3.3–H4 must first pass through ASF1 and the major soluble H3–H4 complex, or whether DAXX represents an orthologous pathway to H3.3 deposition has yet to be determined. The specificity of DAXX to histone H3.3 over H3.1 relies on the presence of a glycine residue at position 90 of histone H3.3 (one of the five amino acids that vary between H3.1 and H3.3) in such a way that any substitution abrogates the complex formation [126]. ATRX recruits DAXX to regions enriched in heterochromatic marks such as H3K9me3 [127], the presence of heterochromatin protein 1 HP1 [128] or through the mediation of long non-coding RNAs [129]. In turn, ATRX/DAXX complex plays a role in maintaining and propagating this repressive chromatin state [130], working as a positive feedback loop.
Conclusions and future outlook
A gargantuan effort has been made regarding the discovery and characterisation of histone chaperoning proteins in the last 40 years since the discovery of Nucleoplasmin in 1978. This has culminated in the characterisation of a number of histone chaperones at atomic resolution in complex with their histone cargo, the spatio-temporal ordering of histone hand-off events (Figure 4A) and the realisation of the role of histone chaperones in development and disease. We expect that this will continue apace in the coming decade. The recent revolution in structure determination by cryo-EM, coupled to the biological importance of many large chaperoning complexes, such as CAF1, HIRA and FACT, means that it may not be long before we have a detailed understanding of how even the largest histone chaperoning complexes mediate interactions with their histone cargo. One key question still to be address is how histone hand-off between chaperoning components relates to the thermodynamic assembly of nucleosome in the context of the cellular environment (Figure 4B). Ongoing advances in live-cell imaging and super-resolution microscopy, combined with novel pulse-chase strategies and biochemical methods will hopefully result in new tools to probe the rapid kinetics of histone transfer between chaperones in living cells.
Figure 4. Transitioning through the thermodynamic landscape of nucleosome assembly.
Histones must transition through a number of protein complexes in order to fold with DNA into nucleosomes without the input of energy from ATP hydrolysis. Immediately after synthesis histones contain a high free energy. This energy may be captured by histone chaperones and utilised in a way which drives correct folding and oligomerisation of histone subunits. This is mostly likely achieved through extensive and specific interactions covering all transitions states of histone intermediates. In such a scenario, accepting potential ATP-driven input from canonical protein folding chaperones in the initial stages, the histone chaperoning pathway may represent an efficient way to assemble a highly abundant cellular complex.
Summary
Core histones have a set of dedicated molecular chaperones that associate tightly with them from synthesis to chromatin incorporation.
Under certain ionic buffering conditions, histones and DNA form nucleosomes in vitro. Histone chaperones, with their disparate domain structures, facilitate this process in vivo.
Structural biology is spreading ever more light on the molecular details of histone–histone–chaperone interactions. To date this has been predominantly limited to individual domains, yet with the revolution in cryo-electron microscopy we anticipate greater understanding at the larger complex level in the near future.
Transitions between chaperoning complexes are poorly understood, but fundamental to gaining an understanding of the thermodynamic rules governing nucleosome assembly.
The fast kinetics of histone deposition makes it challenging to study in living cells, and thus new techniques are required to be able to test histone chaperoning models directly.
Abbreviations
- ARD
ankyrin repeat domain
- CDAN1
Codanin-1
- FACT
FACilitating Transcription
- Nap1
nucleosome assembly protein 1
- PCNA
proliferating cell nuclear antigen
- PIP
PCNA interacting peptide
- RPA
replication protein A
- TLK1 and TLK2
Tousled-like kinases 1 and 2
- UBN1
Ubinuclein-1
Funding
This work was supported by the Wellcome Trust Sir Henry Dale Fellowship to A.J.B. [grant number 208801/Z/17/Z (to A.J.B., A.J.P.)]; and the MRC Doctoral Training Partnership [grant number MR/N014294/1 (to F.F.-D.)].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Hübner M.R., Eckersley-Maslin M.A. and Spector D.L. (2013) Chromatin organization and transcriptional regulation. Curr. Opin. Genet. Dev. 23, 89–95 10.1016/j.gde.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mondal T., Rasmussen M., Pandey G.K., Isaksson A. and Kanduri C. (2010) Characterization of the RNA content of chromatin. Genome Res. 20, 899–907 10.1101/gr.103473.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luger K., Mäder A.W., Richmond R.K., Sargent D.F. and Richmond T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 4.Arents G. and Moudrianakis E.N. (1995) The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. U. S. A. 92, 11170–11174 10.1073/pnas.92.24.11170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey C.A., Sargent D.F., Luger K., Maeder A.W. and Richmond T.J. (2002) Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution. J. Mol. Biol. 319, 1097–1113 10.1016/S0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- 6.Mariño-Ramírez L., Hsu B., Baxevanis A.D. and Landsman D. (2006) The Histone Database: a comprehensive resource for histones and histone fold-containing proteins. Proteins 62, 838–842 10.1002/prot.20814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsässer S.J. and D’Arcy S. (2013) Towards a mechanism for histone chaperones. Biochim. Biophys. Acta 1819, 211–221 10.1016/j.bbagrm.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren C. and Shechter D. (2017) Fly fishing for histones: catch and release by histone chaperone intrinsically disordered regions and acidic stretches. J. Mol. Biol. 429, 2401–2426 10.1016/j.jmb.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond C.M., Strømme C.B., Huang H., Patel D.J. and Groth A. (2017) Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 10.1038/nrm.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos E.I., Smits A.H., Kang Y.-H., Landry S., Escobar T.M., Nayak S.. et al. (2015) Analysis of the histone H3.1 interactome: a suitable chaperone for the right event. Mol. Cell 60, 697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campos E.I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W.-H.W.. et al. (2010) The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 10.1038/nsmb.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drané P., Ouararhni K., Depaux A., Shuaib M. and Hamiche A. (2010) The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tagami H., Ray-Gallet D., Almouzni G. and Nakatani Y. (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 10.1016/S0092-8674(03)01064-X [DOI] [PubMed] [Google Scholar]
- 14.Ask K., Jasencakova Z., Menard P., Feng Y., Almouzni G. and Groth A. (2012) Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBO J. 31, 2013–2023 10.1038/emboj.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apta-Smith M.J., Hernandez-Fernaud J.R. and Bowman A.J. (2018) Evidence for the nuclear import of histones H3.1 and H4 as monomers. EMBO J. 37, 10.15252/embj.201798714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonner W.M., Wu R.S., Panusz H.T. and Muneses C. (1988) Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry 27, 6542–6550 10.1021/bi00417a052 [DOI] [PubMed] [Google Scholar]
- 17.Alvarez F., Muñoz F., Schilcher P., Imhof A., Almouzni G. and Loyola A. (2011) Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286, 17714–17721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dannah N.S., Nabeel-Shah S., Kurat C.F., Sabatinos S.A. and Fillingham J. (2018) Functional analysis of Hif1 histone chaperone in Saccharomyces cerevisiae. G3 (Bethesda)., Genetics Society of America 8, 1993–2006 10.1534/g3.118.200229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyler J.K., Collins K.A., Prasad-Sinha J., Amiott E., Bulger M., Harte P.J.. et al. (2001) Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21, 6574–6584 10.1128/MCB.21.19.6574-6584.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galvani A., Courbeyrette R., Agez M., Ochsenbein F., Mann C. and Thuret J.-Y. (2008) In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells. Mol. Cell. Biol. 28, 3672–3685 10.1128/MCB.00510-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mello J.A., Silljé H. H.W., Roche D. M.J., Kirschner D.B., Nigg E.A. and Almouzni G. (2002) Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3, 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barman H.K., Takami Y., Ono T., Nishijima H., Sanematsu F., Shibahara K.. et al. (2006) Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem. Biophys. Res. Commun. 345, 1547–1557 10.1016/j.bbrc.2006.05.079 [DOI] [PubMed] [Google Scholar]
- 23.Verreault A., Kaufman P.D., Kobayashi R. and Stillman B. (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8, 96–108 10.1016/S0960-9822(98)70040-5 [DOI] [PubMed] [Google Scholar]
- 24.Giri R., Yeh H.-H., Wu C.-H. and Liu H.-S. (2008) SUMO-1 overexpression increases RbAp46 protein stability and suppresses cell growth. Anticancer Res. 28, 3749–3756 [PubMed] [Google Scholar]
- 25.Creekmore A.L., Walt K.A., Schultz-Norton J.R., Ziegler Y.S., McLeod I.X., Yates J.R.. et al. (2008) The role of retinoblastoma-associated proteins 46 and 48 in estrogen receptor alpha mediated gene expression. Mol. Cell. Endocrinol. 291, 79–86 10.1016/j.mce.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paine P.L., Austerberry C.F., Desjarlais L.J. and Horowitz S.B. (1983) Protein loss during nuclear isolation. J. Cell Biol. 97, 1240–1242 10.1083/jcb.97.4.1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowman A., Koide A., Goodman J.S., Colling M.E., Zinne D., Koide S.. et al. (2017) sNASP and ASF1A function through both competitive and compatible modes of histone binding. Nucleic Acids Res. 45, 643–656 10.1093/nar/gkw892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinschmidt J.A., Fortkamp E., Krohne G., Zentgraf H. and Franke W.W. (1985) Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J. Biol. Chem. 260, 1166–1176 [PubMed] [Google Scholar]
- 29.Kleinschmidt J.A. and Franke W.W. (1982) Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell 29, 799–809 10.1016/0092-8674(82)90442-1 [DOI] [PubMed] [Google Scholar]
- 30.Zeytuni N. and Zarivach R. (2012) Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405 10.1016/j.str.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 31.Bowman A., Lercher L., Singh H.R., Zinne D., Timinszky G., Carlomagno T.. et al. (2016) The histone chaperone sNASP binds a conserved peptide motif within the globular core of histone H3 through its TPR repeats. Nucleic Acids Res. 44, 3105 10.1093/nar/gkv1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antczak A.J., Tsubota T., Kaufman P.D. and Berger J.M. (2006) Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct. Biol. 6, 26 10.1186/1472-6807-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.English C.M., Adkins M.W., Carson J.J., Churchill M.E.A. and Tyler J.K. (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M. and Senda T. (2007) Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 10.1038/nature05613 [DOI] [PubMed] [Google Scholar]
- 35.Cook A.J.L., Gurard-Levin Z.A., Vassias I. and Almouzni G. (2011) A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol. Cell 44, 918–927 10.1016/j.molcel.2011.11.021 [DOI] [PubMed] [Google Scholar]
- 36.Yoon J., Kim S.J., An S., Cho S., Leitner A., Jung T.. et al. (2018) Integrative structural investigation on the architecture of human Importin4_Histone H3/H4_Asf1a complex and its histone H3 tail binding. J. Mol. Biol. 430, 822–841 10.1016/j.jmb.2018.01.015 [DOI] [PubMed] [Google Scholar]
- 37.Groth A., Corpet A., Cook A.J.L., Roche D., Bartek J., Lukas J.. et al. (2007) Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 10.1126/science.1148992 [DOI] [PubMed] [Google Scholar]
- 38.Wu H., Moshkina N., Min J., Zeng H., Joshua J., Zhou M.-M.. et al. (2012) Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc. Natl. Acad. Sci. 110, 8925–8930 10.1073/pnas.1114117109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song J.-J., Garlick J.D. and Kingston R.E. (2008) Structural basis of histone H4 recognition by p55. Genes Dev. 22, 1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murzina N.V., Pei X.Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J.V.. et al. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 10.1016/j.str.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Zhang L., Liu T., Chai C., Fang Q., Wu H.. et al. (2014) Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 28, 1217–1227 10.1101/gad.240531.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makowski A.M., Dutnall R.N. and Annunziato A.T. (2001) Effects of acetylation of histone H4 at lysines 8 and 16 on activity of the Hat1 histone acetyltransferase. J. Biol. Chem. 276, 43499–43502 10.1074/jbc.C100549200 [DOI] [PubMed] [Google Scholar]
- 43.Rivera C., Saavedra F., Alvarez F., Díaz-Celis C., Ugalde V., Li J.. et al. (2015) Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res. 43, 9097–9106 10.1093/nar/gkv929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loyola A., Bonaldi T., Roche D., Imhof A. and Almouzni G. (2006) PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24, 309–316 10.1016/j.molcel.2006.08.019 [DOI] [PubMed] [Google Scholar]
- 45.Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J.P., Imhof A.. et al. (2009) The HP1α-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10, 769–775 10.1038/embor.2009.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivera C., Gurard-Levin Z.A., Almouzni G. and Loyola A. (2014) Histone lysine methylation and chromatin replication. Biochim. Biophys. Acta 1839, 1443–1439 10.1016/j.bbagrm.2014.03.009 [DOI] [PubMed] [Google Scholar]
- 47.Pinheiro I., Margueron R., Shukeir N., Eisold M., Fritzsch C., Richter F.M.. et al. (2012) Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150, 948–960 10.1016/j.cell.2012.06.048 [DOI] [PubMed] [Google Scholar]
- 48.Stillman B. (1986) Chromatin assembly during SV40 DNA replication in vitro. Cell 45, 555–565 10.1016/0092-8674(86)90287-4 [DOI] [PubMed] [Google Scholar]
- 49.Kaufman P.D., Kobayashi R., Kessler N. and Stillman B. (1995) The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81, 1105–1114 10.1016/S0092-8674(05)80015-7 [DOI] [PubMed] [Google Scholar]
- 50.Wittmeyer J. and Formosa T. (1997) The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17, 4178–4190 10.1128/MCB.17.7.4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittmeyer J., Joss L. and Formosa T. (1999) Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry 38, 8961–8971 10.1021/bi982851d [DOI] [PubMed] [Google Scholar]
- 52.Duro E., Lundin C., Ask K., Sanchez-Pulido L., MacArtney T.J., Toth R.. et al. (2010) Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol. Cell 40, 632–644 10.1016/j.molcel.2010.10.023 [DOI] [PubMed] [Google Scholar]
- 53.O’Donnell L., Panier S., Wildenhain J., Tkach J.M., Al-Hakim A., Landry M.C.. et al. (2010) The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol. Cell 40, 619–631 10.1016/j.molcel.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foltman M., Evrin C., De Piccoli G., Jones R.C., Edmondson R.D., Katou Y.. et al. (2013) Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 3, 892–904 10.1016/j.celrep.2013.02.028 [DOI] [PubMed] [Google Scholar]
- 55.Liu S., Xu Z., Leng H., Zheng P., Yang J., Chen K.. et al. (2017) RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science 355, 415–420 10.1126/science.aah4712 [DOI] [PubMed] [Google Scholar]
- 56.Bellelli R., Belan O., Pye V.E., Clement C., Maslen S.L., Skehel J.M.. et al. (2018) POLE3-POLE4 is a histone H3-H4 chaperone that maintains chromatin integrity during DNA replication. Mol. Cell 72, 112–126 10.1016/j.molcel.2018.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corpet A., De Koning L., Toedling J., Savignoni A., Berger F., Charlé L.. et al. (2011) Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 30, 480–493 10.1038/emboj.2010.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saredi G., Huang H., Hammond C.M., Alabert C., Bekker-Jensen S., Forne I.. et al. (2016) H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature 534, 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang H., Strømme C.B., Saredi G., Hödl M., Strandsby A., González-Aguilera C.. et al. (2015) A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 22, 618–626 10.1038/nsmb.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., Wang M., Yang N. and Xu R.-M. (2015) Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell 6, 693–697 10.1007/s13238-015-0190-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richet N., Liu D., Legrand P., Velours C., Corpet A., Gaubert A.. et al. (2015) Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 43, 1905–1917 10.1093/nar/gkv021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malay A.D., Umehara T., Matsubara-Malay K., Padmanabhan B. and Yokoyama S. (2008) Crystal structures of fission yeast histone chaperone Asf1 complexed with the Hip1 B-domain or the Cac2 C Terminus. J. Biol. Chem. 283, 14022–14031 10.1074/jbc.M800594200 [DOI] [PubMed] [Google Scholar]
- 63.Liu W.H., Roemer S.C., Port A.M. and Churchill M.E.A. (2012) CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 40, 11229–11239 10.1093/nar/gks906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D., Setiaputra D., Jung T., Chung J., Leitner A., Yoon J.. et al. (2016) Molecular architecture of yeast chromatin assembly factor 1. Sci. Rep. 6, 26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tyler J.K., Adams C.R., Chen S.-R., Kobayashi R., Kamakaka R.T. and Kadonaga J.T. (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 10.1038/990147 [DOI] [PubMed] [Google Scholar]
- 66.Donham D.C., Scorgie J.K., Churchill M.E.A. and Churchill M.E.A. (2011) The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 39, 5449–5458 10.1093/nar/gkr097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sauer P.V., Timm J., Liu D., Sitbon D., Boeri-Erba E., Velours C.. et al. (2017) Insights into the molecular architecture and histone H3-H4 deposition mechanism of yeast Chromatin assembly factor 1. Elife 6, 10.7554/eLife.23474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kadyrova L.Y., Rodriges Blanko E. and Kadyrov F.A. (2013) Human CAF-1-dependent nucleosome assembly in a defined system. Cell Cycle 12, 3286–3297 10.4161/cc.26310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krude T. (1995) Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res. 220, 304–311 10.1006/excr.1995.1320 [DOI] [PubMed] [Google Scholar]
- 70.Shibahara K. and Stillman B. (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 10.1016/S0092-8674(00)80661-3 [DOI] [PubMed] [Google Scholar]
- 71.Zhang Z., Shibahara K. and Stillman B. (2000) PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221–225 10.1038/35041601 [DOI] [PubMed] [Google Scholar]
- 72.Gérard A., Koundrioukoff S., Ramillon V., Sergère J.-C., Mailand N., Quivy J.-P.. et al. (2006) The replication kinase Cdc7-Dbf4 promotes the interaction of the p150 subunit of chromatin assembly factor 1 with proliferating cell nuclear antigen. EMBO Rep. 7, 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rolef Ben-Shahar T., Castillo A.G., Osborne M.J., Borden K.L.B., Kornblatt J. and Verreault A. (2009) Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol. Cell. Biol. 29, 6353–6365 10.1128/MCB.01051-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoek M. and Stillman B. (2003) Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. U. S. A. 100, 12183–12188 10.1073/pnas.1635158100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith S. and Stillman B. (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25 10.1016/0092-8674(89)90398-X [DOI] [PubMed] [Google Scholar]
- 76.Liu W.H., Roemer S.C., Zhou Y., Shen Z.-J., Dennehey B.K., Balsbaugh J.L.. et al. (2016) The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones. Elife 5, 10.7554/eLife.18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mattiroli F., Gu Y., Yadav T., Balsbaugh J.L., Harris M.R., Findlay E.S.. et al. (2017) DNA-mediated association of two histone-bound complexes of yeast Chromatin Assembly Factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. Elife 6, 10.7554/eLife.22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang K., Gao Y., Li J., Burgess R., Han J., Liang H.. et al. (2016) A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks. Nucleic Acids Res. 44, 5083–5094 10.1093/nar/gkw106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jackson V. (1988) Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27, 2109–2120 10.1021/bi00406a044 [DOI] [PubMed] [Google Scholar]
- 80.Prior C.P., Cantor C.R., Johnson E.M. and Allfrey V.G. (1980) Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell 20, 597–608 10.1016/0092-8674(80)90306-2 [DOI] [PubMed] [Google Scholar]
- 81.Almouzni G., Clark D.J., Méchali M. and Wolffe A.P. (1990) Chromatin assembly on replicating DNA in vitro. Nucleic Acids Res. 18, 5767–5774 10.1093/nar/18.19.5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith S. and Stillman B. (1991) Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 10.1002/j.1460-2075.1991.tb08031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katan-Khaykovich Y. and Struhl K. (2011) Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc. Natl. Acad. Sci. U. S. A. 108, 1296–1301 10.1073/pnas.1018308108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Radman-Livaja M., Verzijlbergen K.F., Weiner A., van Welsem T., Friedman N., Rando O.J.. et al. (2011) Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 9, e1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu M., Long C., Chen X., Huang C., Chen S. and Zhu B. (2010) Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 10.1126/science.1178994 [DOI] [PubMed] [Google Scholar]
- 86.Madamba E.V., Berthet E.B. and Francis N.J. (2017) Inheritance of histones H3 and H4 during DNA replication in vitro. Cell Rep. 21, 1361–1374 10.1016/j.celrep.2017.10.033 [DOI] [PubMed] [Google Scholar]
- 87.Alabert C., Barth T.K., Reverón-Gómez N., Sidoli S., Schmidt A., Jensen O.N.. et al. (2015) Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y., van Merwyk L., Tönsing K., Walhorn V., Anselmetti D. and Fernàndez-Busquets X. (2017) Biophysical characterization of the association of histones with single-stranded DNA. Biochim. Biophys. Acta 1861, 2739–2749 10.1016/j.bbagen.2017.07.018 [DOI] [PubMed] [Google Scholar]
- 89.Huang T.-H., Fowler F., Chen C.-C., Shen Z.-J., Sleckman B. and Tyler J.K. (2018) The histone chaperones ASF1 and CAF-1 promote MMS22L-TONSL-mediated Rad51 loading onto ssDNA during homologous recombination in human cells. Mol. Cell 69, 879.e5–892.e5 10.1016/j.molcel.2018.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu C., Gan H., Serra-Cardona A., Zhang L., Gan S., Sharma S.. et al. (2018) A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 10.1126/science.aat8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gan H., Serra-Cardona A., Hua X., Zhou H., Labib K., Yu C.. et al. (2018) The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell 72, 140.e3–151.e3 10.1016/j.molcel.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petryk N., Dalby M., Wenger A., Stromme C.B., Strandsby A., Andersson R.. et al. (2018) MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 10.1126/science.aau0294 [DOI] [PubMed] [Google Scholar]
- 93.McCullough L.L., Connell Z., Xin H., Studitsky V.M., Feofanov A.V., Valieva M.E.. et al. (2018) Functional roles of the DNA-binding HMGB domain in the histone chaperone FACT in nucleosome reorganization. J. Biol. Chem. 293, 6121–6133 10.1074/jbc.RA117.000199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D.. et al. (2003) Tracking FACT and the RNA polymerase II elongation Complex Through Chromatin in Vivo. Science 301, 1094–1096 10.1126/science.1085712 [DOI] [PubMed] [Google Scholar]
- 95.Birch J.L., Tan B.C.-M., Panov K.I., Panova T.B., Andersen J.S., Owen-Hughes T.A.. et al. (2009) FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 28, 854–865 10.1038/emboj.2009.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tessarz P., Santos-Rosa H., Robson S.C., Sylvestersen K.B., Nelson C.J., Nielsen M.L.. et al. (2014) Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kurat C.F., Yeeles J. T.P., Patel H., Early A. and Diffley J. F.X. (2017) Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65, 117–130 10.1016/j.molcel.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morillo-Huesca M., Maya D., Muñoz-Centeno M.C., Singh R.K., Oreal V., Reddy G.U.. et al. (2010) FACT prevents the accumulation of free histones evicted from transcribed chromatin and a subsequent cell cycle delay in G1. PLoS Genet. 6, e1000964 10.1371/journal.pgen.1000964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., Zhang E.T.. et al. (2013) Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 499, 111–114 10.1038/nature12242 [DOI] [PubMed] [Google Scholar]
- 100.VanDemark A.P., Blanksma M., Ferris E., Heroux A., Hill C.P. and Formosa T. (2006) The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22, 363–374 10.1016/j.molcel.2006.03.025 [DOI] [PubMed] [Google Scholar]
- 101.Kemble D.J., McCullough L.L., Whitby F.G., Formosa T. and Hill C.P. (2015) FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsunaka Y., Fujiwara Y., Oyama T., Hirose S. and Morikawa K. (2016) Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Winkler D.D., Muthurajan U.M., Hieb A.R. and Luger K. (2011) Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang T., Liu Y., Edwards G., Krzizike D., Scherman H. and Luger K. (2018) The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci. Alliance 1, e201800107 10.26508/lsa.201800107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Evrin C., Maman J.D., Diamante A., Pellegrini L. and Labib K. (2018) Histone H2A‐H2B binding by Pol α in the eukaryotic replisome contributes to the maintenance of repressive chromatin. EMBO J. 37, e99021 10.15252/embj.201899021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Talbert P.B. and Henikoff S. (2017) Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126 10.1038/nrm.2016.148 [DOI] [PubMed] [Google Scholar]
- 107.Ray-Gallet D., Quivy J.-P., Scamps C., Martini E.M.-D., Lipinski M. and Almouzni G. (2002) HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 10.1016/S1097-2765(02)00526-9 [DOI] [PubMed] [Google Scholar]
- 108.Goldberg A.D., Banaszynski L.A., Noh K.-M., Lewis P.W., Elsaesser S.J., Stadler S.. et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A.. et al. (2011) Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 110.Schneiderman J.I., Orsi G.A., Hughes K.T., Loppin B. and Ahmad K. (2012) Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc. Natl. Acad. Sci. U. S. A. 109, 19721–19726 10.1073/pnas.1206629109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ray-Gallet D., Ricketts M.D., Sato Y., Gupta K., Boyarchuk E., Senda T.. et al. (2018) Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat. Commun. 9, 3103 10.1038/s41467-018-05581-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Green E.M., Antczak A.J., Bailey A.O., Franco A.A., Wu K.J., Yates J.R.. et al. (2005) Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15, 2044–2049 10.1016/j.cub.2005.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tang Y., Poustovoitov M.V, Zhao K., Garfinkel M., Canutescu A., Dunbrack R.. et al. (2006) Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13, 921–929 10.1038/nsmb1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ricketts M.D., Frederick B., Hoff H., Tang Y., Schultz D.C., Singh Rai T.. et al. (2015) Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6, 7711 10.1038/ncomms8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang H., Gan H., Wang Z., Lee J.-H., Zhou H., Ordog T.. et al. (2017) RPA interacts with HIRA and regulates H3.3 deposition at gene regulatory elements in mammalian cells. Mol. Cell 65, 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Orphanides G., LeRoy G., Chang C.H., Luse D.S. and Reinberg D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 10.1016/S0092-8674(00)80903-4 [DOI] [PubMed] [Google Scholar]
- 117.Mylonas C. and Tessarz P. (2018) Transcriptional repression by FACT is linked to regulation of chromatin accessibility at the promoter of ES cells. Life Sci. Alliance 1, e201800085 10.26508/lsa.201800085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hsieh F.-K., Kulaeva O.I., Patel S.S., Dyer P.N., Luger K., Reinberg D.. et al. (2013) Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 110, 7654–7659 10.1073/pnas.1222198110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Formosa T. (2012) The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255 10.1016/j.bbagrm.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Obri A., Ouararhni K., Papin C., Diebold M.-L., Padmanabhan K., Marek M.. et al. (2014) ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 10.1038/nature12922 [DOI] [PubMed] [Google Scholar]
- 121.Aguilar-Gurrieri C., Larabi A., Vinayachandran V., Patel N.A., Yen K., Reja R.. et al. (2016) Structural evidence for Nap1-dependent H2A-H2B deposition and nucleosome assembly. EMBO J. 35, 1465–1482 10.15252/embj.201694105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Andrews A.J., Chen X., Zevin A., Stargell L.A. and Luger K. (2010) The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dutta S., Akey I.V, Dingwall C., Hartman K.L., Laue T., Nolte R.T.. et al. (2001) The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8, 841–853 10.1016/S1097-2765(01)00354-9 [DOI] [PubMed] [Google Scholar]
- 124.Platonova O., Akey I.V, Head J.F. and Akey C.W. (2011) Crystal structure and function of human nucleoplasmin (npm2): a histone chaperone in oocytes and embryos. Biochemistry 50, 8078–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Earnshaw W.C., Honda B.M., Laskey R.A. and Thomas J.O. (1980) Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell 21, 373–383 10.1016/0092-8674(80)90474-2 [DOI] [PubMed] [Google Scholar]
- 126.Elsässer S.J., Huang H., Lewis P.W., Chin J.W., Allis C.D. and Patel D.J. (2012) DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature, NIH Public Access 491, 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iwase S., Xiang B., Ghosh S., Ren T., Lewis P.W., Cochrane J.C.. et al. (2011) ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 18, 769–776 10.1038/nsmb.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lechner M.S., Schultz D.C., Negorev D., Maul G.G. and Rauscher F.J. (2005) The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 331, 929–937 10.1016/j.bbrc.2005.04.016 [DOI] [PubMed] [Google Scholar]
- 129.Park J., Lee H., Han N., Kwak S., Lee H.-T., Kim J.-H.. et al. (2018) Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization. Nucleic Acids Res. 46, 11759–11775 10.1093/nar/gky923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Voon H. P.J., Hughes J.R., Rode C., DeLaRosa-Velázquez I.A., Jenuwein T., Feil R.. et al. (2015) ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 11, 405–418 10.1016/j.celrep.2015.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dabin J. and Polo S.E. (2017) Choreography of parental histones in damaged chromatin. Nucleus 8, 255–260 10.1080/19491034.2017.1292192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Polo S.E. and Almouzni G. (2015) Chromatin dynamics after DNA damage: the legacy of the access-repair-restore model. DNA Repair (Amst) 36, 114–121 10.1016/j.dnarep.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hauer M.H. and Gasser S.M. (2017) Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 52, 295–319 10.1101/gad.307702.117 [DOI] [PMC free article] [PubMed] [Google Scholar]